Abstract
A series of novel 4-anilinoquinazoline derivatives were designed and synthesized from benzoic acid through ring closure, chlorination or nucleophilic substitution. The structures of compounds were characterized by IR, 1H-NMR and mass spectroscopy. All synthesized derivatives were screened for their antimicrobial activities against Gram-positive (Staphylococcus aurous, Bacillus subtilis, Listeria monocitogenes) and Gram-negative (Escherichia coli, Pseudomonas aeruginosa, Salmonella entritidis) bacteria and also for antifungal activities against Candida albicans using the conventional micro dilution method. Most of the compounds have shown good antibacterial activities, especially compound 4c having highest activities against E. coli at 32 μg/mL concentration while the tested compounds did not exhibited remarkable antifungal activities. The potential DNA gyrase inhibitory activity of these compounds was investigated in silico using molecular docking simulation method. All compounds showed good results especially compound 4c which showed the lowest ΔGbind results (-8.16 Kcal/mol).
Keywords: 4-Anilinoquinazoline, Synthesis, Antimicrobial activity, Antifungal activity, Molecular docking
INTRODUCTION
The increasing rates of resistance to antimicrobial agents have caused severe health problems. Consequently, there is a rapid need of design and synthesis of newer antimicrobial agents (1). Some of the useful approaches for the discovery of new drugs are based on investigations of drug targets like enzymes or receptors (2,3). DNA gyrase is one of the attractive targets in E. coli which is involved in replication and transcription. This enzyme contains an ATPase activity which introduces negative supercoiling of circular DNA. The enzyme belongs to a superfamily of ATPases which is a known target for antibacterial agents since its blocking induces bacterial death (4,5). Quinazolines are a class of fused pyrimidine derivatives, which show a wide range of biological activities and used widely in the pharmaceutical industry, medicine and agriculture (6,7,8,9,10,11). Quinazolines act as an important backbone for a range of inhibitors of enzymes such as tyrosine kinase, thymidylate synthase and dihydrofolate reductase (12,13,14). In this heterocyclic family, 4-anilinoquinazoline derivatives have been reported as potent and selective inhibitors of protein kinases, such as epidermal growth factor receptor (EGFR). For example gefitinib and erlotinib are as dual EGFR-human epidermal growth factor receptor 2 (HER2) inhibitors, which are used for certain breast, lung and other cancers (15). Docking technique is a very important tool in the rational design of drugs which helps to predict the interactions between a ligand and a receptor molecule in order to predict the affinity and the activity of the small molecules. To the best of our knowledge, docking studies of 4-anilinoquinazoline derivatives with DNA-gyrase have not yet been studied (16).
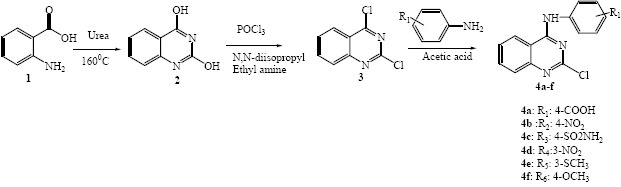
In the present work, we have focused on the effect of substitution of different aniline derivatives at the 4th position of the quinazoline ring and also on the antibacterial activities of these compounds. Compounds were prepared through the routine synthetic procedure in which anthranilic acid was cyclized with urea to yield quinazoline-2, 4-dione (17). The synthesis of 2, 4-dichloroquinazoline as the key intermediate was performed by reaction of quinazoline-2, 4-dione with phosphorus oxychloride (18). Aniline substitution occurred selectively at position 4 through nucleophilic aromatic substitution (Scheme 1). All 2-chloro-4-anilino-quinazoline derivatives were purified and structurally confirmed by mass spectrometry, infrared spectroscopy and 1H nuclear magnetic resonance (1H-NMR). Antimicrobial effects were evaluated using the serial dilution method against three Gram-positive bacteria (Staphylococcus aureu, Bacillus subtilis, and Listeria monocitogenes) and three Gram-negative bacteria (Escherichia coli, Pseudomonas aeruginosa, Salmonella entritidis) and also on the Candida albicansas, an yeast-like fungi strain. Synthesized analogues were docked into the binding pocket of DNA gyrase protein and their binding energies were calculated.
Scheme 1.
General reaction schemes for the synthesis of the target compounds 2, 3 and 4a-f.
MATERIALS AND METHODS
All commercially available reagents and solvents used in this study were purchased from Merck Co (Merck, Germany). The reactions were monitored by thin-layer chromatography (TLC) on silica gel (F245 Merck plates). Melting points of the synthesized compounds were determined in open capillaries using electrothermal 9200 melting point apparatus (England) and uncorrected. 1H-NMR spectra were obtained on a Bruker 400 MHz spectrometer (Germany) using TMS as internal reference with chemical shifts are reported in δ scale (ppm). Mass spectra were recorded on the Shimadzu Mass Spectrometer (Japan). IR spectra in KBr were recorded on a WQF-510 FT-IR spectrophoto-meter (China).
Antibacterial studies
The in vitro antimicrobial activity of the synthesized compounds were carried out by the serial dilution method against microorganism obtained from the Persian Type Culture Collection. Sabouraud dextrose agar and Mueller Hinton agar were used to culture fungal strains and bacterial strains, respectively. Standard antibacterial drug (ciprofloxacin) and antifungal drug (ketoconazole) were used for comparison.
Molecular docking studies
Molecular docking studies were performed in order to predict the interaction of synthesized compounds with the binding sites of DNA-gyrase (19,20). The crystal structure of the enzyme (PDB code 1KZN) with resolution 2.3 Å was chosen as the protein model for the current study. The structures of ligands were optimized using the HyperChem 7.0 software (http://www.hyper.com) as was explained previously (21). Auto Dock Tools were used to prepare the molecules and parameters before submitting it for docking analysis with Auto Dock (21).
Polar hydrogen atoms were added while non-polar hydrogen atoms were merged and then, Gasteiger partial atomic charges were assigned to the ligands.
All rotatable bonds of ligands, defined by default of the program, were allowed to rotate during the automated docking process and then prepared protein and ligand structures were saved in the PDBQT format suitable for calculating energy grid maps. A grid box size of 46 × 46 × 46 Å points with a grid spacing of 0.375 Å was considered.
Lamarckian genetic algorithm (LGA) program with an adaptive whole method search in the Auto Dock was chosen to calculate the different ligand conformers (21,22). After 200 independent docking runs for each ligand, a cluster analysis was done. In according to the root mean square deviation (RMSD) tolerance of 2.0 Å conformations were clustered and were ranked by energy of which the conformation with the best scored pose with the lowest binding energy was selected for these ligands (22).
Chemistry
The synthetic route for the novel compounds is shown in Scheme 1. Synthesis of novel 4-anilino quinazoline derivatives was initiated from benzoic acid in three steps ring closure, chlorination and nucleophilic substitution.
The compounds 2 and 3 were synthesized in accordance with a previously reported method (17,18). Compound 3 as the key intermediate was treated with different substituted aniline derivatives in the presence of acetic acid to form title compound 4a-f in high yield in order to obtain biologically active compounds.
Synthesis of quinazoline-2, 4-dione (2)
A mixture of 2-aminobenzoic acid (1) (68.5 g, 0.5 mol) and urea (210 g, 3.5 mol) was stirred at 160 °C for 12 h. The reaction mixture was filtered and the filtrate was washed with water to afford title compound (2) as a white solid in 90% yield (17).
Synthesis of 2, 4-dichloroquinazoline (3)
In a flask equipped with a reflux condenser, the reaction mixture containing the quinazolinone 2 (48.6 g, 0.3 mol) and excess amount of POCI3 (200 mL) was stirred at room temperature, and then N,N-Diisopropyl-ethylamine (DIPEA) (77.6 g, 0.3 mol) was added dropwise to the mixture. The reaction was monitored by TLC and after complete consumption of quinazolinone 2 the product was triturated with n-hexane and isolated by filtration to yield compound 3 as light yellow crystals in 80% yield (17,18).
General procedure for synthesis of new 4-anilino quinazoline derivatives (4a-f)
Aniline derivatives (1 mmol) and 2, 4-dichloroquinazoline (3) (1 mmol) were dissolved in acetic acid and stirred at room temperature. Through the progress of reaction an insoluble product was produced which had a different Rf in TLC plate. The precipitate was collected by filtration, recrystallized in ethanol and characterized by different techniques.
Antibacterial evaluation
Following approach was used to demonstrate the minimal inhibitory concentrations (MICs) of the synthesized compounds using Microplate Alamar Blue Assay (MABA) method. All synthesized compounds were dissolved in DMSO and diluted with water to obtain concentration of 5120 µg/mL as a stock solution. The stock solution was diluted to obtain 2560 to 320 µg/mL concentrations. Mueller Hinton broth was used as a medium for bacterial growth and 96-well microtiter plates with U-shaped wells were used in this method. Each well was inoculated with 20 µL of each concentration with the exception of those wells acting as a growing control. After adding Alamar Blue reagent, the plates were sealed with parafilm and incubated at 37 °C for 24 h for bacteria and 48 h at 25 °C for the fungus (23,24). The MIC was defined as the lowest concentration that shows no growth by visual reading, which avoids discoloration from blue to pink. MBC and MFC results were obtained from each well that show no growth by Mueller Hinton agar plates and Sabouraud Dextrose agar for bacteria and fungi, respectively (25).
RESULTS
All synthesized compounds were docked into the active site of DNA gyrase B subunit and then were analyzed for binding free energy and their interactions with the receptor. Table 1 summarizes the binding free energy in Kcal/mole and interactions between 6 synthesized compounds and DNA-gyrase. In this table, all ligands interact through hydrogen bonding with the DNA-gyrase binding site. The distance of hydrogen bonds was shorter than 3.5 Å.
Table 1.
Energy-based interactions for 6 novel 4-anilinoquinazoline derivatives docked into DNA gyrase.

The best conformations from the docking procedure with the best scored pose and the lowest binding energy (~-7 – -8 kcal/mol) were selected for these ligands (4a-f) (Figs. 1-4).
Fig. 1.
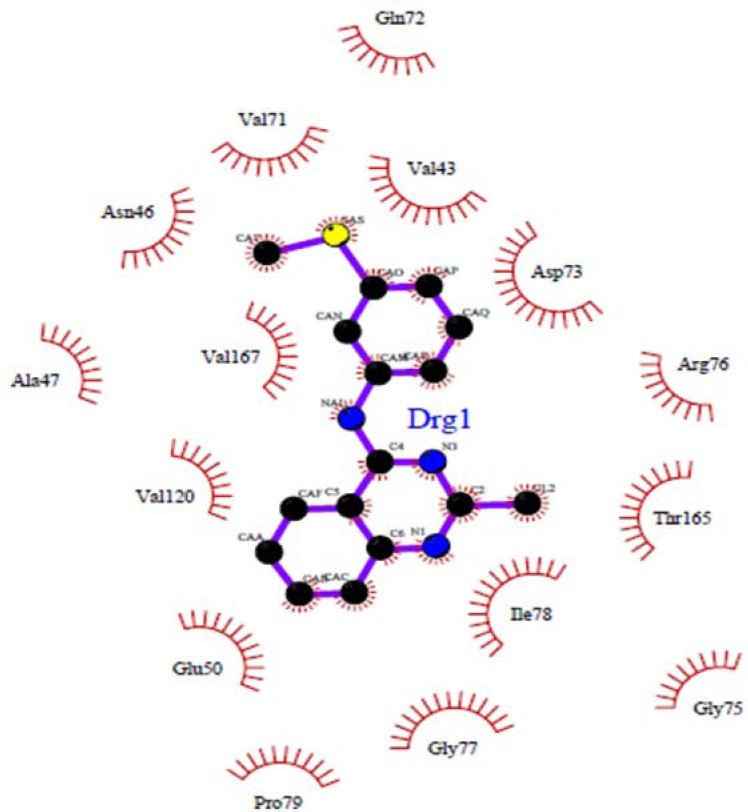
Docked conformations of ligand structure 4e in the binding site of DNA gyrase.
Fig. 4.
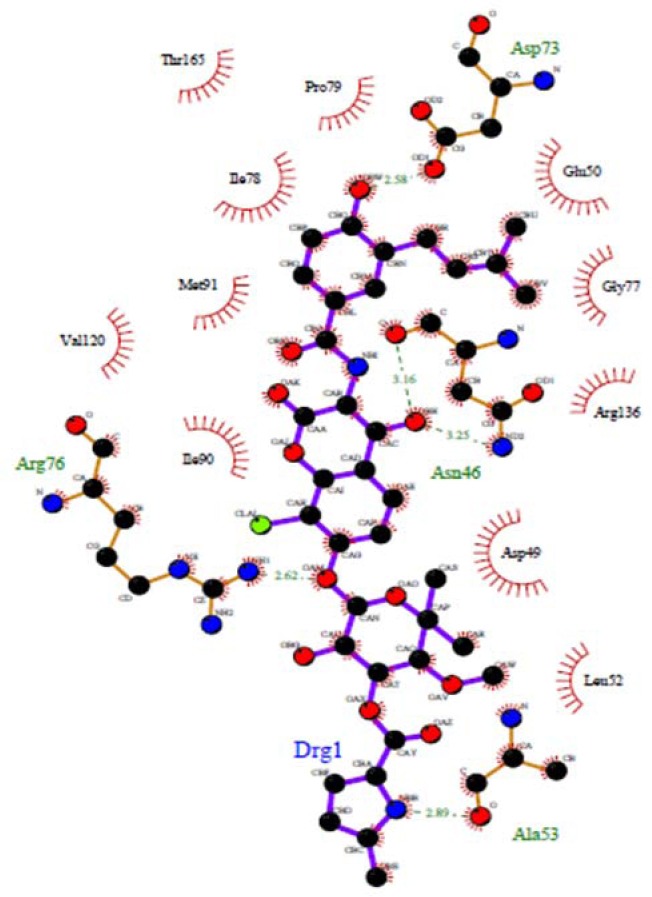
Redocking results of clorobiocin the active site of DNA gyrase. Hydrogen bonds are shown by green dashed line.
The designed compounds were synthesized through conventional synthetic procedures and characterized by different methods. Structural properties of synthesized compounds are shown below.
Synthesis of 4-(2-chloroquinazolin-4-yl amino) benzoic acid (4a)
White solid (79%). M.p: 260-261 °C, (MS: m/z (%): 298 (M, 100), 300 (M+2), C14H9N4O2Cl M.W. 299, vmax, 3423.99 (N-H), 2923.56 (CH-arom), 1685.48 (C=O), 1606.41 (C=N), 758.85 (C-Cl) cm-1; δH (400 MHz; DMSO-d6): 7.8 (1H, t, J = 8 Hz, H-Qu), 7.9 (1H, d, J = 8 Hz, H-Qu), 8.0 (1 H, t, J = 8 Hz, H-Qu), 8.2 (2 H, d, J=8 Hz, H-Ar), 8.4 (2 H, d, J = 8 Hz, H-Ar), 8.7 (1 H, d, J = 8 Hz, H-Qu), 10.6 (1 H, s, NH-Ph, exchangeable with D2O).
2-chloro-N-(4-nitrophenyl) quinazolin-4-amine (4b)
Light yellow crystals (88%). M.p: 235-236 °C, (MS: m/z (%): 300 (M+, 100), 302 (M+2), C14H9N4O2C1M.W. 300, vmax, 3397.96 (N-H), 3050.83 (CH-arom), 2923.56 (CH-aliph), 1566.88 (C=N), 759.81 (C-Cl) cm-1; δH (400 MHz; DMSO-d6): 7.0 (1 H, d, J = 6.8 Hz, H-Ar), 7.70 (1 H, d, J = 6.8 Hz, H-Ar), 7.72 (1 H, t, J = 8 Hz, H-Qu), 7.76 (1 H, d, J = 8 Hz, H-Qu), 7.9 (1 H, t, J = 8 Hz, H-Qu), 8.6 (1 H, d, J = 8 Hz, H-Qu), 10.2 (1 H, s, NH-ph, exchangeable with D2O).
4-amino-N-(2-chloroquinazolin-4-yl) benzene-sulfonamide (4c)
([A-Z])([0-9])White crystal (90%). M.p. 250-251 °C, (MS:334 m/z (%): 335 (M+, 100), 337 (M+2), C14H11N4SO2Cl, M.W. 334, vmax, 3354.57 (N-H), 3276.47 (CH-arom), 2925.48 (CH-aliph), 1571.7 (C=N), 1238.08 (C=S), 1147.44 (SO2), 766.56 (C-Cl) cm-1; δH (400 MHz; DMSO-d6): 7.4 (1 H, s, NH-ph, exchangeable with D2O), 7.7 (1 H, t, J = 7.6 Hz, H-Qu), 7.8 (1 H, d, J = 7.6 Hz, H-Qu), 7.9 (1 H, d, J = 8.8 Hz, H-Ar), 8.0 (1 H, t, J = 7.2 Hz, H-Qu), 8.06 (1 H, d, J = 6.8 Hz, H-Ar), 8.6 (1 H, d, J = 8 Hz, H-Qu), 10.5 (1 H, s, SO2NH, exchangeable with D2O).
2-chloro-N-(3-nitrophenyl) quinazolin-4-amine (4d)
Yellow crystal (90%). M.p. 195-196 °C. (MS: m/z (%): 301 (M+, 100), 303 (M+2), C14H9N4O2Cl M.W. 300). vmax, 3395.07 (N-H), 2920.66 (CH-arom), 2574.5 (CH-aliph), 1623.77 (C=N), 1530.24 (NO2) 765.60 (C-Cl) cm-1; δH (400 MHz;CDCl3): 7.6 (2 H, t, J = 8Hz, H-Qu), 7.7 (1 H, s, NH-Ph, exchangeable with D2O), 7.8 (3 H, d, J = 8 Hz, H-Qu, Ar), 8.0 (1 H, d, J = 8.0 Hz, H-Qu), 8.3 (1 H, d, J = 8 Hz, H-Ar), 8.5 (1 H, s, H-Ar).
2-chloro-N-(3-(methylthio) phenyl) quinazolin-4-amine (4e)
White solid (90%), M.p. 169-170 °C, (MS: m/z (%):300 (M+, 100), 302 (M+2), (C15H12N3Cl M.W. 300). vmax, 3322.75 (N-H), 2921.63 (CH-arom), 2493.51 (CH-aliph), 1573.63 (C=N), 766.56 (C-Cl) cm-1; δH (400 MHz; DMSO-d6): 3.4 (s, 3H, SCH3), 7.1 (1H, d, J = 8 Hz, H-Ar), 7.4 (1H, t, J = 8 Hz, H-Ar), 7.6 (1 H, d, J = 8 Hz, H-Ar), 7.75 (1 H, t, J = 8 Hz, H-Qu), 7.78 (1 H, d, J = 8 Hz, H-Qu), 7.8 (1 H, s, H-Ar), 7.9 (1 H, t, J = 8 Hz, H-Qu), 8.6 (1 H, d, J = 8 Hz, H-Qu), 10.2 (s, 1 H, NH-ph, exchangeable with D2O).
2-chloro-N-(4-methoxyphenyl) quinazolin-4-amine (4f)
Yellow solid (80%). M.p. 178-180 °C, (MS: m/z (%):286 (M+, 100), 288 (M+2), (C15H12N3OCl M.W. 285) Vmax, 3421.1 (N-H), 3252.36 (CH-arom), 2964.05 (CH-aliph), 1575.56 (C=N), 759.81 (C-Cl) cm-1; δH (400 MHz; DMSO-d6): 2.0 (s, 3H, OCH3), 7.7 (1 H, t, J = 8 Hz, H-Qu), 7.8 (1 H, d, J = 8 Hz, H-Qu), 7.9 (1 H, t, J = 8 Hz, H-Qu), 8.0 (4 H, s, H-Ar), 8.7 (1 H, d, J = 8 Hz, H-Qu).
All synthesized compounds were tested for antibacterial activity against Gram-positive (Staphylococcus aurous, Bacillus subtilis, Listeria monocitogenes), Gram-negative bacteria (Escherichia coli, Pseudomonas aeruginosa, Salmonella entritidis) and also for antifungal activities against Candida albicans. The results of biological effects are shown in Tables 2 and 3.
Table 2.
Minimum inhibitory concentration and minimum bactericidal concentration results of synthesized compounds against bacteria.
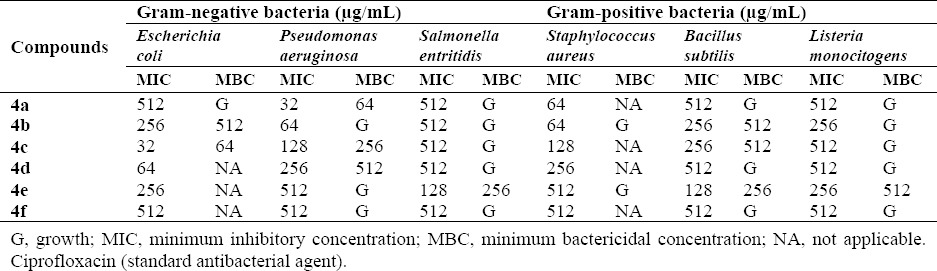
Table 3.
Minimum inhibitory concentration (μg/mL) and minimum fungicidal concentration (μg/mL) results of synthesized compounds against fungi.

DISCUSSION
In the majority of the structures, hydrophobic sites of the ligands are conserved. The residue that interacts with the acceptor/donor site of the ligand through a hydrogen bonding differs depending on the ligand. The binding patterns of different ligands are also slightly different.
The nitrogen atom in the middle ring of all compounds except 4e can form a strong hydrogen bond with Asp73 at distance 2.54 Å-3.23 Å, which is consistent with the decomposition analysis of the electrostatic interaction (Fig. 1). It is interesting that more complex stabilization might result from the hydrogen bonds between these ligands and Arg136 via electron withdrawing substitution on the phenyl ring. Although these interactions were also recognized for these derivatives, these are different from those present in 4c (Glu50, Arg 76) and 4d (Gly 77) and may be responsible for the activity changes (Figs. 2 and 3). These results are compatible with the X-ray cocrystal structure and earlier studies prove the important roles of those residues (26,27,28).
Fig. 2.
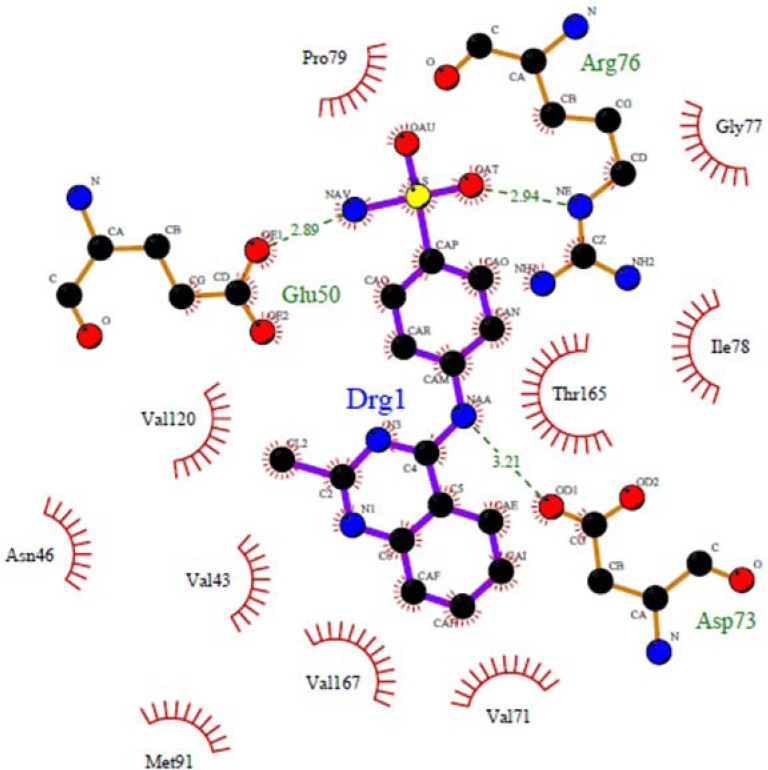
Docked conformations of ligand structure 4c in the binding site of DNA gyrase. Hydrogen bonds are shown by green dashed line.
Fig. 3.
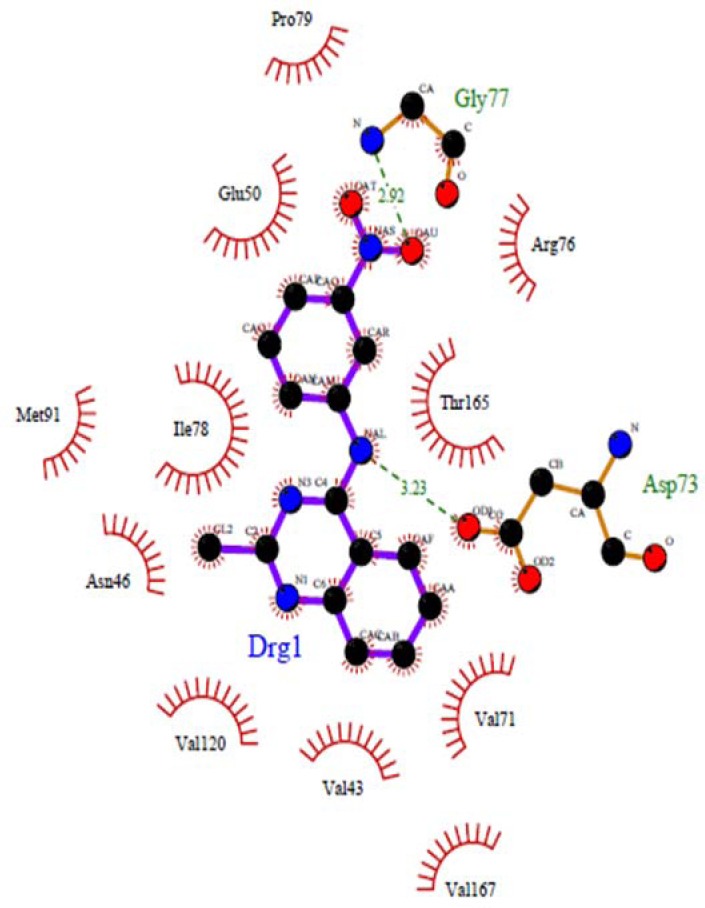
Docked conformations of ligand structure 4d in the binding site of DNA gyrase. Hydrogen bonds are shown by green dashed line.
No hydrogen bond interaction between the 4e compound and discussed residues was predicted. Because of the long distance (more than 3.5 Å) between the nitrogen atom at the middle ring of 4e and the oxygen atom of Asp73, this ligand in the best pose with the lowest binding energy could not form a hydrogen bonding interaction, but in other compounds such as 4c and 4d could establish a hydrogen bond with Glu50, Arg76 (4c) and Gly77 (4d).
These compounds also significantly preserve the DNA gyrase through hydrophobic contacts with Asn40, Val43, Val71, Arg76, Gly77, Ile78, Pro79, Met91, Val120, Thr163, Thr165 and Val167 that are important in hydrophobic interactions.
In all compounds it is clear that the hydrophobic pocket of the inhibitor binding site was occupied by 4-anilinoquinazoline or phenyl plus the groups substituted on these rings (28,29,30).
The docking procedure in this research was validated by redocking of chlorobiocin as a well-known inhibitor to the energy minimized DNA gyrase protein. The residues Asp73, Asn46 and Arg136 are important in developing hydrogen bond (Fig. 4) (30,31,32). The same is mainly true for our quinazoline derivatives, the only exception is the relatively weak hydrogen bond with Asn46. The best molecule in these series (4c) showed a high dock score of -8.16 kcal/mol in docking protocol. Rest of molecules showed an appropriate dock score ranging from –7.07 to - 8.16 kcal/mol. So, the binding mode reported here suggests that these 4-anilinoquinazoline compounds act as DNA-gyrase inhibitors and show some key structural points to be considered in future optimization. The synthetic pathways to the intermediates and final compounds (2-4f) are presented in Scheme 1.
At first, anthranilic acid 1 was condensed with urea to produce quinazoline-2, 4-dione. Amino group in anthranilic acid acts as a nucleophile and attacks to carbonyl group of urea to produce the intermediate upon elimination of ammonia group. Simultaneous nucleophilic attacks of amino group in urea resulted in the production of the product 2 upon elimination of water molecule (33,34). Chlorination reaction was performed with POCl3 to form intermediate 3. In this reaction, diisopropyl ethylamine was used as a strong base (35). Finally the addition of different aniline derivatives provided the compound 4a-f through SNAr reaction as presented in Scheme 1. The reaction was initiated by the nucleophilic attack of NH2 to the fourth position of the quinazoline ring to displace the chlorine moiety (36).
All synthesized compounds were tested for antibacterial activity against Gram-positive (Staphylococcus aurous, Bacillus subtilis, Listeria monocitogenes) and Gram-negative bacteria (Escherichia coli, Pseudomonas aeruginosa, Salmonella entritidis) and also for antifungal activities against (Candida albicans).
Most of the synthesized compounds showed good activity against both the Gram-positive and Gram-negative bacterial species. Obtained results of screened compounds against Gram-negative organisms showed that compounds 4a, 4b and 4c had the highest activities against E. coli at 32 μg/mL concentration. In case of Gram-positive bacteria, compounds 4a and 4b showed acceptable activity results against Staphylococcus aureus. Antibacterial study revealed that compounds showed better bacteriostatic activity than bactericidal activity.
Structure–activity relationship study based on the observed results indicated that in 4a-f, the type of aryl substitution plays a controlling role in developing the exhibited biological properties. It has been noticed that, substitution on the phenyl group with an electron-withdrawing group such as a COOH, NO2 or a sulfonamide group 4a, 4b, 4c and 4d (MIC:512, 256, 32 and 64 μg/mL) seems more favorable for enhancing the antibacterial activity than substitution with an electron-donating group such as methylthio and methoxy groups 4e, 4f (MIC:, 256 and 512 μg/mL). Compound with the COOH group in the para position of the phenyl ring showed high activity which could be due to the participation in hydrogen bonding interactions and improved solubility. NO2 substitution at the para position of phenyl ring has also resulted in high activity against the bacteria. Compound with a sulfonamide group has also good effect against the bacteria which might be due to the increasing of ionization of the N-H group. Interestingly, introducing of SCH3 group at the meta position, reduced antibacterial activity. This might be attributed to the donation of electrons to the benzene ring through inductive effect. Similarly, the compound containing OCH3 group in para position has the least effect (37,38,39). Results of antifungal study showed that almost all of the screened compounds have no antifungal activity against C. albicans, except compound 4e which showed moderate activity at 256 µg/mL (Table 3).
CONCLUSION
In this study, in silico design, synthesis and evaluation of antimicrobial activity of six novel 4-anilinoquinazoline derivatives were reported. All compounds showed good antibacterial activity, especially against E. coli at 32 µg/mL concentration while no remarkable antifungal activities were observed for these compounds. Careful investigation in this series gave the compound 4c as the most promising inhibitor of DNA-gyrase based on the docking score energies, hydrogen bonds distances and antimicrobial evaluation. Further developments are in progress to optimize new 4-anilinoquinazoline derivatives as potential antibiotic drug candidates in the future.
ACKNOWLEDGEMENTS
The content of this paper is extracted from the PhD thesis (No. 394158) submitted by Rezvan Rezaee Nasab which was financially supported by the Isfahan University of Medical Sciences, Isfahan, Iran.
REFERENCES
- 1.Gupta B, Singh S. Antibiotics-new tools in current and future drug therapy. Journal Of Scientific & Innovative Research. 2012;1(1):27–42. [Google Scholar]
- 2.Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl Microbiol Biotechnol. 2011;92(3):479–497. doi: 10.1007/s00253-011-3557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrov DA, Prada JAH, Corsino PE, Finton KA, Le N, Rowe TC. Discovery of novel DNA gyrase inhibitors by high-throughput virtual screening. Antimicrob Agents Chemother. 2007;51(10):3688–3698. doi: 10.1128/AAC.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heddle J, Maxwell A. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob Agents Chemother. 2002;46(6):1805–1815. doi: 10.1128/AAC.46.6.1805-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R, Madhumathi BS, Nagaraja V. Molecular basis for the differential quinolone susceptibility of mycobacterial DNA gyrase. Antimicrob Agents Chemother. 2014;58(4):2013–2020. doi: 10.1128/AAC.01958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A, Sharma P, Kumari P, Kalal BL. Exploration of antimicrobial and antioxidant potential of newly synthesized 2, 3-disubstituted quinazoline-4 (3H)-ones. Bioorg Med Chem Lett. 2011;21(14):4353–4357. doi: 10.1016/j.bmcl.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Khodarahmi G, Jafari E, Hakimelahi G, Abedi D, Khajouei MR, Hassanzadeh F. Synthesis of some new quinazolinone derivatives and evaluation of their antimicrobial activities. Iran J Pharm Res. 2012;11(3):789–797. [PMC free article] [PubMed] [Google Scholar]
- 8.Laddha SS, Wadodkar SG, Meghal SK. Studies on some biologically active substituted 4(3H)-quinazolinones. Part 1. Synthesis, characterization and anti-inflammatory#-antimicrobial activity$ of 6,8-disubstituted 2-phenyl-3-[substituted-benzothiazol-2-yl]-4(3H)-quinazolinones. ARKIVOC. 2006;Xi:1–20. [Google Scholar]
- 9.Abouzid K, Shouman S. Design, synthesis and in vitro antitumor activity of 4-aminoquinoline and 4-aminoquinazoline derivatives targeting EGFR tyrosine kinase. Bioorg. Med Chem. 2008;16(16):7543–7551. doi: 10.1016/j.bmc.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 10.Jafari E, Khodarahmi G, Hakimelahi G, Tsai F, Hassanzadeh F. Synthesis of some new tricyclic 4 (3H)-quinazolinone derivatives. Res Pharm Sci. 2011;6(2):93–100. [PMC free article] [PubMed] [Google Scholar]
- 11.Rezaee Nasab R, Karami B, Khodabakhshi S. Selective solvent-free biginelli condensation using Tungestat Sulfuric Acid as powerful and reusable catalyst. Bulletin of chemical reaction engineering& catalysis. 2014;9(2):148–154. [Google Scholar]
- 12.Gilbert IH. Inhibitors of dihydrofolate reductase in Leishmania and trypanosomes. Biochim Biophys Acta. 2002;1587(2):249–257. doi: 10.1016/s0925-4439(02)00088-1. [DOI] [PubMed] [Google Scholar]
- 13.Jackman AL, Taylor GA, Gibson W, Kimbell R, Brown M, Calvert AH, et al. ICI D1694, a quinazoline antifolate thymidylate synthase inhibitor that is a potent inhibitor of L1210 tumor cell growth in vitro and in vivo: a new agent for clinical study. Cancer Res. 1991;51(20):5579–5586. [PubMed] [Google Scholar]
- 14.Ismail RS, Ismail NS, Abuserii S, El Ella DAA. Recent advances in 4-aminoquinazoline based scaffold derivatives targeting EGFR kinases as anticancer agents. Future Journal of Pharmaceutical Sciences. 2016;2(1):9–19. [Google Scholar]
- 15.de Castro Barbosa ML, Lima LM, Tesch R, Sant'Anna CMR, Totzke F, Kubbutat MH, et al. Novel 2-chloro-4-anilino-quinazoline derivatives as EGFR and VEGFR-2 dual inhibitors. Eur J Med Chem. 2014;71:1–14. doi: 10.1016/j.ejmech.2013.10.058. [DOI] [PubMed] [Google Scholar]
- 16.Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 17.Jiang N, Zhai X, Zhao Y, Liu Y, Qi B, Tao H, et al. Synthesis and biological evaluation of novel 2-(2-arylmethylene) hydrazinyl-4-aminoquinazoline derivatives as potent antitumor agents. Eur J Med Chem. 2012;54:534–541. doi: 10.1016/j.ejmech.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 18.Pawar VG, Sos ML, Rode HB, Rabiller M, Heynck S, Van Otterlo WA, et al. Synthesis and biological evaluation of 4-anilinoquinolines as potent inhibitors of epidermal growth factor receptor. J Med Chem. 2010;53(7):2892–2901. doi: 10.1021/jm901877j. [DOI] [PubMed] [Google Scholar]
- 19.Bansal S, Kumar S, Aggarwal V, Joseph A. Design, synthesis, docking study & antibacterial evaluation of 1, 3-diarylpyrazolyl substituted indolin-2-ones. Indo Global Journal of Pharmaceutical Sciences. 2014;4(1):1–7. [Google Scholar]
- 20.Jayashree B, Thomas S, Nayak Y. Design and synthesis of 2-quinolones as antioxidants and antimicrobials: a rational approach. Med Chem Res. 2010;19(2):193–209. [Google Scholar]
- 21.Mansourian M, Fassihi A, Saghaie L, Madadkar-Sobhani A, Mahnam K, Abbasi M. QSAR and docking analysis of A2B adenosine receptor antagonists based on non-xanthine scaffold. Med Chem Res. 2015;24:394–407. [Google Scholar]
- 22.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639–1662. [Google Scholar]
- 23.Jayalakshmi B, Raveesha K, Amruthesh K. Evaluation of antibacterial and antioxidant potential of Euphorbia cotinifolia linn.leaf extracts. Chem Ind Chem Eng Q. 2014;20(1):19–28. [Google Scholar]
- 24.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3(2):163–715. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 25.Baker CN, Banerjee SN, Tenover FC. Evaluation of Alamar colorimetric MIC method for antimicrobial susceptibility testing of Gram-negative bacteria. J Clin Microbiol. 1994;32(5):1261–1267. doi: 10.1128/jcm.32.5.1261-1267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kampranis SC, Gormley NA, Tranter R, Orphanides G, Maxwell A. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry. 1999;38:1967–1976. doi: 10.1021/bi982320p. [DOI] [PubMed] [Google Scholar]
- 27.Lewis RJ, Singh OMP, Smith CV, Skarzynski T, Maxwell A, Wonacott AJ, et al. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 1996;15:1412–1420. [PMC free article] [PubMed] [Google Scholar]
- 28.Blance SJ, Williams NL, Preston ZA, Bishara J, Smyth MS, Maxwell A. Temperature-sensitive suppressor mutations of the Escherichia coli DNA gyrase B protein. Protein Sci. 2000;9(5):1035–1037. doi: 10.1110/ps.9.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyapati S, Kulandaivelu U, Sangu S, Vanga MR. Synthesis, antimicrobial evaluation, and docking studies of novel 4-substituted quinazoline derivatives as DNA-gyrase inhibitors. Archiv der Pharmazie. 2010;343(10):570–576. doi: 10.1002/ardp.201000065. [DOI] [PubMed] [Google Scholar]
- 30.Mladenovic M, Vukovic N, Sukdolak S, Solujic S. Design of novel 4-hydroxy-chromene-2-one derivatives as antimicrobial agents. Molecules. 2010;15(6):4294–4308. doi: 10.3390/molecules15064294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perumal P, Pandey VP, Parasuraman P. Docking studies on some novel piperidine analogues against DNA gyrase enzyme. Inventi Rapid Molecular Modeling. 2014;1:1–4. [Google Scholar]
- 32.Patil RB, Sawant SD. Synthesis, characterization, molecular docking and evaluation of antimicrobial activity of some 3-heteroaryl substituted chromen-2-one derivatives. Int J Pharm Tech Res 2014. 2015;7(3):471–480. [Google Scholar]
- 33.Salih NA. Synthesis of some new quinazolin-4 (3h)- one derivatives and study of their some antibacterial activity. Journal of Al-Nahrain University. 2008;11(2):16–23. [Google Scholar]
- 34.Li F, Feng Y, Meng Q, Li W, Li Z, Wang Q, et al. An efficient construction of quinazolin-4 (3H)-ones under microwave irradiation. Arkivoc. 2007;1:40–50. [Google Scholar]
- 35.Arnott EA, Chan LC, Cox BG, Meyrick B, Phillips A. POCl3 chlorination of 4-quinazolones. J Org Chem. 2011;76(6):1653–1661. doi: 10.1021/jo102262k. [DOI] [PubMed] [Google Scholar]
- 36.El-Azab AS, Al-Omar MA, Alaa A-M, Abdel-Aziz NI, Magda A-A, Aleisa AM, et al. Design, synthesis and biological evaluation of novel quinazoline derivatives as potential antitumor agents: molecular docking study. Eur J Med Chem. 2010;45(9):4188–4198. doi: 10.1016/j.ejmech.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Van Horn KS, Burda WN, Fleeman R, Shaw LN, Manetsch R. Antibacterial activity of a series of N 2, N 4-disubstituted quinazoline-2, 4-diamines. J Med Chem. 2014;57(7):3075–3093. doi: 10.1021/jm500039e. [DOI] [PubMed] [Google Scholar]
- 38.Gottasová R, Kubiková J. Antibacterial effect of some 2, 6-disubstituted 4-anilinoquinazolines. Folia Microbiologica. 1998;43(6):679–682. doi: 10.1007/BF02816389. [DOI] [PubMed] [Google Scholar]
- 39.Parhi AK, Zhang Y, Saionz KW, Pradhan P, Kaul M, Trivedi K, et al. Antibacterial activity of quinoxalines, quinazolines, and 1, 5-naphthyridines. Bioorg Med Chem Lett. 2013;23(17):4968–4974. doi: 10.1016/j.bmcl.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]


