Abstract
Lowered activity of the insulin/IGF signalling (IIS) network can ameliorate the effects of ageing in laboratory animals and, possibly, humans. Although transcriptome remodelling in long‐lived IIS mutants has been extensively documented, the causal mechanisms contributing to extended lifespan, particularly in specific tissues, remain unclear. We have characterized the proteomes of four key insulin‐sensitive tissues in a long‐lived Drosophila IIS mutant and control, and detected 44% of the predicted proteome (6,085 proteins). Expression of ribosome‐associated proteins in the fat body was reduced in the mutant, with a corresponding, tissue‐specific reduction in translation. Expression of mitochondrial electron transport chain proteins in fat body was increased, leading to increased respiration, which was necessary for IIS‐mediated lifespan extension, and alone sufficient to mediate it. Proteasomal subunits showed altered expression in IIS mutant gut, and gut‐specific over‐expression of the RPN6 proteasomal subunit, was sufficient to increase proteasomal activity and extend lifespan, whilst inhibition of proteasome activity abolished IIS‐mediated longevity. Our study thus uncovered strikingly tissue‐specific responses of cellular processes to lowered IIS acting in concert to ameliorate ageing.
Keywords: ageing, insulin/IGF, mitochondria, proteasome, proteome
Subject Categories: Ageing; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Introduction
Reduced activity of the highly evolutionarily conserved insulin/IGF‐like signalling (IIS) network in laboratory model organisms can extend lifespan (Fontana et al, 2010; Kenyon, 2010), maintain function in older ages (Barzilai et al, 2012) and ameliorate pathology associated with multiple age‐related diseases (Niccoli & Partridge, 2012; Johnson et al, 2015). Furthermore, lowered activity of the IIS network is implicated in human survival to advanced ages (Suh et al, 2008; Tazearslan et al, 2011; Deelen et al, 2013; He et al, 2014). Understanding exactly how reducing IIS network activity ameliorates the effects of ageing could hence pave the way to prevention of ageing‐related disease in humans.
The IIS network responds to nutrient availability, growth factors and stress signals to regulate multiple processes, including development, growth, metabolism, stress resistance, reproduction and lifespan (Fontana et al, 2010; Kenyon, 2010). These highly pleiotropic phenotypes make it difficult to pinpoint the mechanisms that specifically ameliorate ageing and hence to understand whether they can be separated from other, not necessarily desirable, pleiotropic effects.
Long‐lived IIS mutants show a major and tissue‐specific rearrangement of RNA transcript expression, as a consequence of alteration of the activity of target transcription factors (TFs; Fontana et al, 2010; Kenyon, 2010). In Caenorhabditis elegans and Drosophila, the single Forkhead boxO, FoxO, transcription factor (DAF‐16 and dFOXO, respectively) is required for blunted IIS to extend lifespan (Kenyon et al, 1993; Slack et al, 2011; Yamamoto & Tatar, 2011), with other TFs also required in both organisms (Kenyon, 2010; Tepper et al, 2013; Slack et al, 2015). In Drosophila, dFOXO is required only for longevity and xenobiotic resistance, and not for other phenotypes of reduced IIS (Slack et al, 2011). Hence, in Drosophila, dfoxo dependence triages at least some of the gene expression changes potentially causal in longevity from several other phenotypes caused by IIS mutants, and facilitates identification of the mechanisms involved.
Profiling of the RNA transcriptome has identified genes and molecular mechanisms that may ameliorate ageing in IIS mutants in C. elegans (Murphy et al, 2003; Halaschek‐Wiener et al, 2005; Oh et al, 2006; McElwee et al, 2007; Ewald et al, 2015; Kaletsky et al, 2016) and Drosophila (Teleman et al, 2008; Alic et al, 2011). Increased detoxification (Amador‐Noguez et al, 2007; Selman et al, 2009; Ewald et al, 2015; Afschar et al, 2016) and reduced translation (Selman et al, 2009; Afschar et al, 2016; Kaletsky et al, 2016) are two functional signatures consistently associated with longevity across different model organisms (McElwee et al, 2007). However, in general RNA profiling in C. elegans has been carried out on whole worms, potentially leaving tissue‐specific mechanisms undetected. Furthermore, a wide range of post‐transcriptional mechanisms may modify expression of proteins (Barrett et al, 2012; Batista & Chang, 2013; Cech & Steitz, 2014; Liu et al, 2016; MacInnes, 2016), with recent reports of the correlation between expression levels in the transcriptome and the proteome ranging from 40 to 84% (Wolkow, 2000; Schwanhäusser et al, 2011; Li et al, 2014).
Tissue‐specific modulation of IIS can also extend lifespan, in the worm, Drosophila and the mouse (Wolkow, 2000; Blüher et al, 2003; Giannakou, 2004; Demontis & Perrimon, 2010; Alic et al, 2014). Conditional knockout models in mice, and studies in humans, have revealed that the responses of gene expression to reduced IIS are tissue‐specific (Nandi et al, 2004; Rask‐Madsen & Kahn, 2012). Recently, tissue‐specific, global regulation of RNA expression in IIS mutants has been undertaken in the Drosophila gut and fat body (Alic et al, 2014) and in neurons in C. elegans (Alic et al, 2014; Kaletsky et al, 2016). However, proteomic profiling of individual tissues in the key invertebrate model organisms Drosophila and C. elegans has been limited, due to their small size, and the requirement for large quantities of starting material. Initial investigations into the proteome of wild‐type Drosophila focused on protein identification, using samples of low complexity, and either subcellular or biochemical fractionation to achieve increasing depth (Veraksa et al, 2005; Brunner et al, 2007; Aradska et al, 2015). Recent advances in mass spectrometry, sample preparation techniques and data analysis now allow the quantification of near complete proteomes and proteomic expression profiling (Nagaraj et al, 2012; Mann et al, 2013; Azimifar et al, 2014; Kim et al, 2014; Deshmukh et al, 2015). These new developments have reduced the amount of starting material required for shotgun proteomics and created an opportunity to investigate how attenuated IIS affects the proteome of individual fly tissues and hence to identify candidate mechanisms that ameliorate the effects of ageing.
In this study, we profiled the changes in the tissue‐specific proteomes of IIS mutant Drosophila and their responses to removal of the key dFOXO transcription factor. We profiled four key insulin‐sensitive tissues: the brain, gut, fat body and muscle. The majority of the responses to a systemic reduction in IIS were highly tissue‐specific, and 60% of them were not detected in previous transcriptional studies. Proteins associated with the ribosome and the mitochondrial electron transport chain were differentially expressed in the fat body, and led to a reduction in translation and increased mitochondrial respiration. The gut showed a proteomic signature of proteasome–ubiquitin‐mediated catabolism, and was associated with elevated proteasomal assembly and activity. Increased respiration and proteasome activity in the fat body and gut, respectively, were required for the increased longevity of IIS mutants. Importantly, manipulation of mitochondrial biogenesis in the fat body and of proteasomal activity in the gut could each extend lifespan in wild‐type flies. Our tissue‐specific, proteomic analysis has thus revealed how individual tissues of an organism can act in concert, through diverse tissue‐specific responses, to ameliorate the ageing of the whole organism in response to reduced IIS.
Results
The tissue‐specific proteome of wild‐type flies
We first characterized the tissue‐specific proteomes of control, wild‐type flies. We dissected the brain, thorax (containing predominately muscle), intestinal tract with Malpighian tubules (referred to as gut from here on) and abdominal fat body of young (10 days), adult, female flies. We then performed single‐shot, label‐free proteomics with a Q‐Exactive benchtop quadrupole‐Orbitrap mass spectrometer (Fig 1A). We achieved 99% confidence of identification at both protein and peptide levels, and in total identified and quantified 96,404 peptides corresponding to 6,085 proteins, representing 44% coverage of the predicted, protein‐coding genes (Fig 1B, Dataset EV1).
Figure 1. Drosophila tissues contain common and tissue‐specific proteomes.
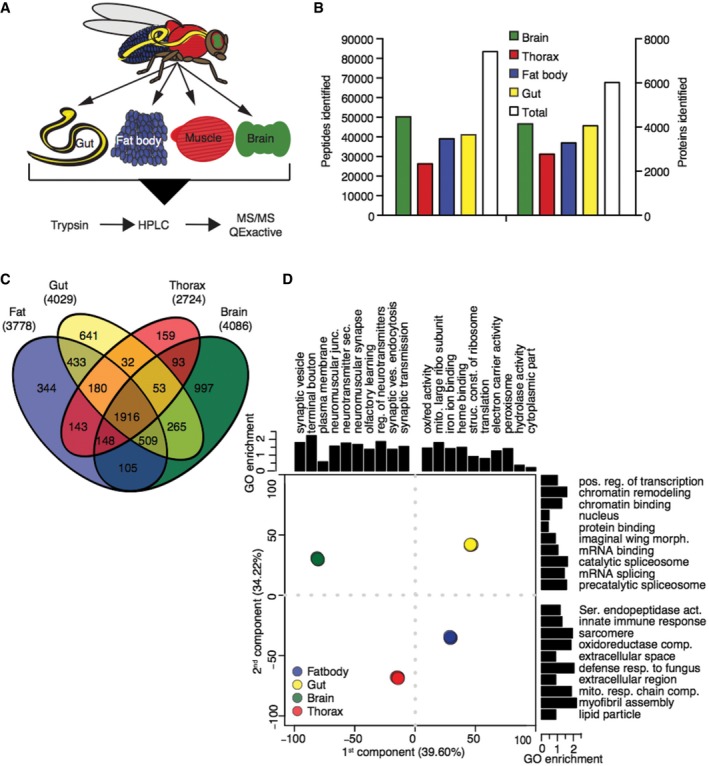
- Schematic of experimental design.
- Tissue‐specific and total number of detected peptides and proteins.
- Venn diagram of proteins detected in wild‐type fly tissues.
- Principal component analysis of the four wild‐type tissues GO terms enriched in the 5% tails of contribution along each dimension. Replicates show little variation and overlay each other.
The four tissues shared a common core of 1,916 proteins (Fig 1C). Gene ontology (GO) enrichment analysis showed that these served a variety of house‐keeping functions, and included mitochondrial and ribosomal constituents (see Dataset EV2 for GO analysis). The four tissues also showed marked differences, with 6–26% of the proteins identified as present in a tissue being unique to it (Fig 1C). To further understand the proteomic features that distinguish individual tissues, we performed principal component (PC) analysis. 74% of the variation between tissues could be captured by the first two PCs. Mapping GO terms onto the proteins contributing to PC1 revealed that proteins with neuronal functions accounted for much of the effect, separating the brain, which had the largest number of unique proteins, from the other three tissues. PC2 detected mainly differences in developmental origins, structure and metabolic activity (Fig 1D and Dataset EV3). Biological replicates from each wild‐type tissue clustered tightly together (PCA, Fig 1D), confirming reproducibility (Fig EV1).
Figure EV1. Tissue‐specific proteomic analysis in Drosophila .
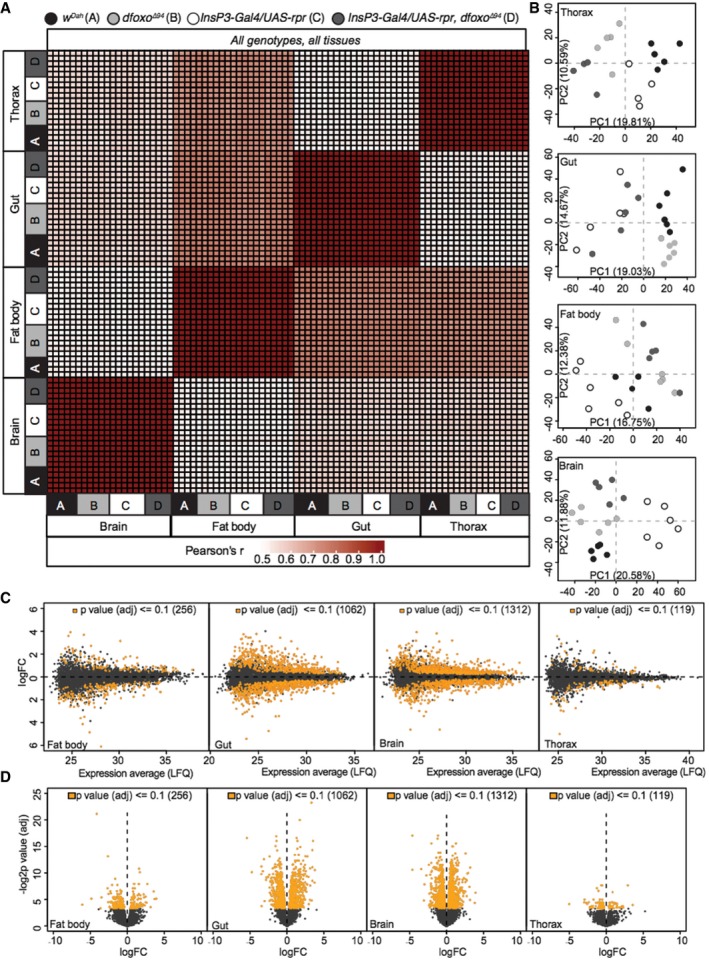
- Correlation heatmap (Pearson's correlation coefficient) across all tissues (thorax, gut, fat body and brain) and all genotypes (A: w Dah , B: dfoxo Δ94 , C: InsP3‐Gal4/UAS‐rpr and D: InsP3‐Gal4/UAS‐rpr, dfoxo Δ94).
- Tissue‐specific PCA plots showing inter‐genotype clustering of individual samples.
- Tissue‐specific MA plots of log2 fold change and average expression level (LFQ) for all detected (black) and significantly regulated (orange) proteins between InsP3‐Gal4/UAS‐rpr vs. w Dah flies (FDR = 0.1).
- Tissue‐specific volcano plots of −log2 P‐value (adjusted) and log2FC for all detected (black) and significantly regulated (orange) proteins between InsP3‐Gal4/UAS‐rpr vs. w Dah flies (FDR = 0.1).
Reduced IIS remodels tissue‐specific proteomes
We next identified the effect on the proteomes of down‐regulation of IIS. Ablation of the median neurosecretory cells (mNSCs) in the fly brain, which secrete three of the seven Drosophila insulin‐like peptides (Dilps) into circulation, causes a robust extension of lifespan (Broughton et al, 2010). We compared protein expression levels in mNSC‐ablated (InsP3‐Gal4/UAS‐rpr) flies to those of wild‐type controls (w Dah). In total, expression of 2,372 proteins was significantly altered upon reduced IIS (10% FDR, Fig 2 and Dataset EV4), 982 of which showed absolute fold changes larger than two in at least one tissue. 60% of these changes were not previously identified at the RNA transcript level in whole IIS mutant flies (McElwee et al, 2007; Alic et al, 2011). Thus substantial, new information was gained by tissue‐specific proteome analysis.
Figure 2. Proteome response to reduced IIS is highly tissue‐specific.
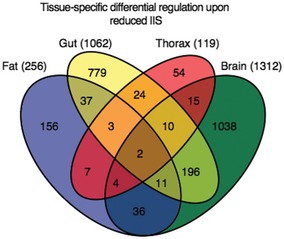
Venn diagram of differentially expressed proteins upon reduced IIS, comparing tissues from control (w Dah) and mNSC‐ablated flies. Total number of differentially expressed, tissue‐specific proteins is shown in parentheses.
The brain and gut showed the highest numbers of differentially regulated proteins (1,312 and 1,062, respectively), whilst the fat body (256) and thorax (119) showed comparatively few. Changes in expression were almost entirely tissue‐specific (85%), with only two proteins, β‐tubulin (60D) and l(1)G0230, showing changes in all four (Dataset EV4). Only 13% of the proteins that were differentially regulated in a single tissue were also detected as solely expressed in that tissue, and thus, the tissue‐specific nature of the differential regulation by IIS was mainly due to tissue‐specific responses rather than tissue‐specific expression patterns (Dataset EV4).
Identifying tissue‐specific, differential protein expression potentially causal in extension of lifespan
To identify candidate proteins that could be causal in the increased longevity of mNSC‐ablated flies, we classified them according to their requirement for dfoxo for their differential regulation. We took the set of proteins that changed expression with ablation of mNSCs in wild‐type flies and asked whether their response to mNSC ablation was abrogated in a dfoxo‐null background. Accordingly, we profiled the tissue‐specific proteomes of mNSC‐ablated flies lacking dfoxo (Insp3‐Gal4/UAS‐rpr; dfoxo Δ/Δ) and the corresponding dfoxo Δ/Δ controls. We constructed a linear model using all four genotypes and identified proteins whose expression was significantly affected by the interaction between ablation of mNSCs and dfoxo deletion. We excluded from this set any proteins whose response to reduced IIS was exaggerated, as opposed to abrogated, in the dfoxo‐null background. We thus identified 361 proteins that required dfoxo for their change in expression in IIS mutant flies (Fig EV2). To assess whether those 361 proteins were more likely to be direct or indirect targets of dfoxo, we searched for predicted dfoxo‐binding motifs within 1 kb of their transcriptional start sites using MEME (Bailey et al, 2009). 45% of the 361 proteins came from genes with dfoxo‐binding motifs (Dataset EV4) and may therefore be directly regulated by dFOXO. However, the remaining 55% of those 361 proteins came from genes lacking dfoxo‐binding motifs which suggest that although their expression was dfoxo‐dependent, they were not directly regulated transcriptionally by dFOXO.
Figure EV2. Bioinformatic scheme for determining dfoxo‐dependent and ‐independent protein expression under reduced IIS .
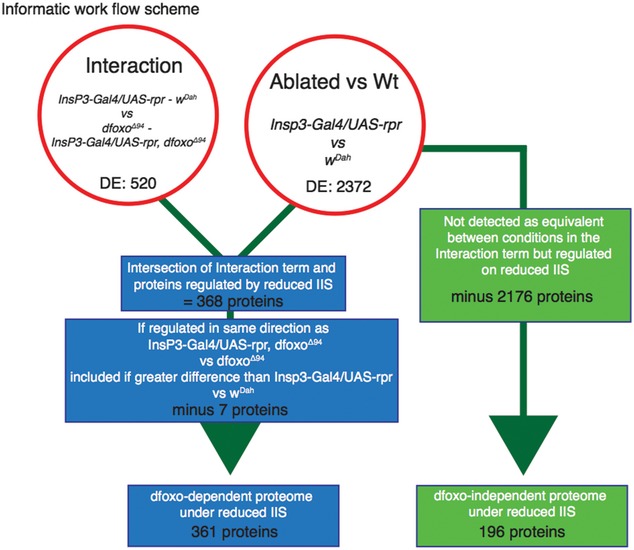
Genotypes defined as w Dah, dfoxo Δ94 mutant, InsP3‐Gal4/UAS‐rpr and InsP3‐Gal4/UAS‐rpr, dfoxo Δ94. Reduced IIS‐mediated changes in the wild‐type background (InsP3‐Gal4/UAS‐rpr vs. w Dah) and in the dfoxo Δ94 background (InsP3‐Gal4/UAS‐rpr, dfoxo Δ94 vs. dfoxo Δ94) or their interaction term (InsP3‐Gal4/UAS‐rpr ‐ w Dah) vs. InsP3‐Gal4/UAS‐rpr, dfoxo Δ94‐dfoxo Δ94. dfoxo‐dependent proteins were defined as proteins regulated between InsP3‐Gal4/UAS‐rpr and w Dah and within the interaction term (InsP3‐Gal4/UAS‐rpr ‐ w Dahvs. InsP3‐Gal4/UAS‐rpr, dfoxo Δ94‐dfoxo Δ94), but also if the expression response was the same in InsP3‐Gal4/UAS‐rpr vs. w Dah and InsP3‐Gal4/UAS‐rpr, dfoxo Δ94 vs. dfoxo Δ94 providing the response was greater in InsP3‐Gal4/UAS‐rpr vs. w Dah. Proteins were identified as dfoxo‐independent if they were regulated between InsP3‐Gal4/UAS‐rpr and w Dah, and were not subject to interaction effects (equivalence given a tissue‐specific threshold). Also included were proteins with a stronger same directional response in InsP3‐Gal4/UAS‐rpr, dfoxo Δ94 vs. dfoxo Δ94 than in InsP3‐Gal4/UAS‐rpr vs. w Dah, regardless of interaction. See Materials and Methods section for full descriptions.
The proteins whose expression showed no significant interaction between response to mNSC ablation and the presence of dfoxo could have been truly dfoxo‐independent or false negatives. To define a set of proteins that are most likely dfoxo‐independent, but ablation responsive, we identified those for which the fold difference in responsiveness to mNSC ablation in the wild‐type and dfoxo‐null background was significantly less than the minimum fold change of dfoxo‐dependent proteins in the same tissue. Additionally, we included the proteins that showed an exaggerated responsiveness to the ablation in the dfoxo‐null background. In this way, we defined a set of 196 proteins as dfoxo‐independent, with high confidence. Indeed, 52% of these showed significant differences in expression even between the dfoxo Δ/Δ and dfoxo Δ/Δ Insp3‐Gal4/UAS‐rpr (Fig EV2).
To further increase the power of our analysis of dfoxo dependency, we employed network propagation, which combines differential expression with physical interactors of individual proteins to identify altered subnetworks in each tissue (Vanunu et al, 2010). Differentially expressed dfoxo‐dependent or dfoxo‐independent proteins were mapped independently onto the Drosophila protein–protein interaction network (Murali et al, 2011), and the associated P‐value of each protein was negative logarithmic transformed and propagated to adjacent interacting proteins. We then clustered the dfoxo‐dependent and dfoxo‐independent responses to reduced IIS in each tissue and identified functional categories of these clusters with GO enrichment analysis (Dataset EV5).
We focused on clusters whose regulation was predicted as dfoxo‐dependent within the network propagation analysis, since these should include processes causally linked to longevity. dfoxo‐dependent responses in the brain were enriched for mitochondrial electron transport chain, mRNA splicing and nucleosome components (Fig 3), whilst in the gut they were enriched for proteasome and ubiquitin‐mediated protein catabolism (Fig 3). Mitochondrial electron transport chain was also enriched within dfoxo‐dependent responses in the fat body, as were ribosomal constituents and proteins involved in nucleosome assembly. Amongst the functional groups identified by network propagation, we further focused on three strong candidates for mediating the longevity of IIS mutant flies. First, we chose the ribosome, which we identified as an almost uniquely fat body‐specific response. Second, we focused on the mitochondrial electron transport chain, the only response identified as common to multiple tissues, the brain, gut and fat body. Finally, we examined proteasome–ubiquitin protein catabolism as the most enriched cluster in the gut.
Figure 3. Hierarchical clustering and GO enrichment analysis of dFOXO‐dependent and dFOXO‐independent IIS‐mediated regulation of the proteome.
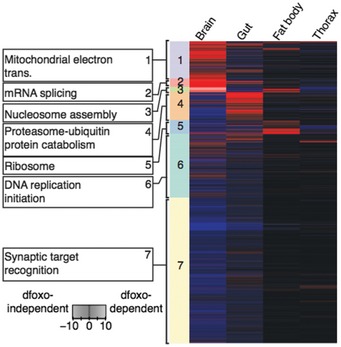
Tissue‐specific heatmap of significantly regulated dfoxo‐dependent (red) and dfoxo‐independent (blue) proteins in response to reduced IIS. Coloured side bars represent network propagated score clustering and associated most significantly enriched GO terms.
Reduced IIS alters fat body‐specific, dfoxo‐dependent protein expression to regulate translation
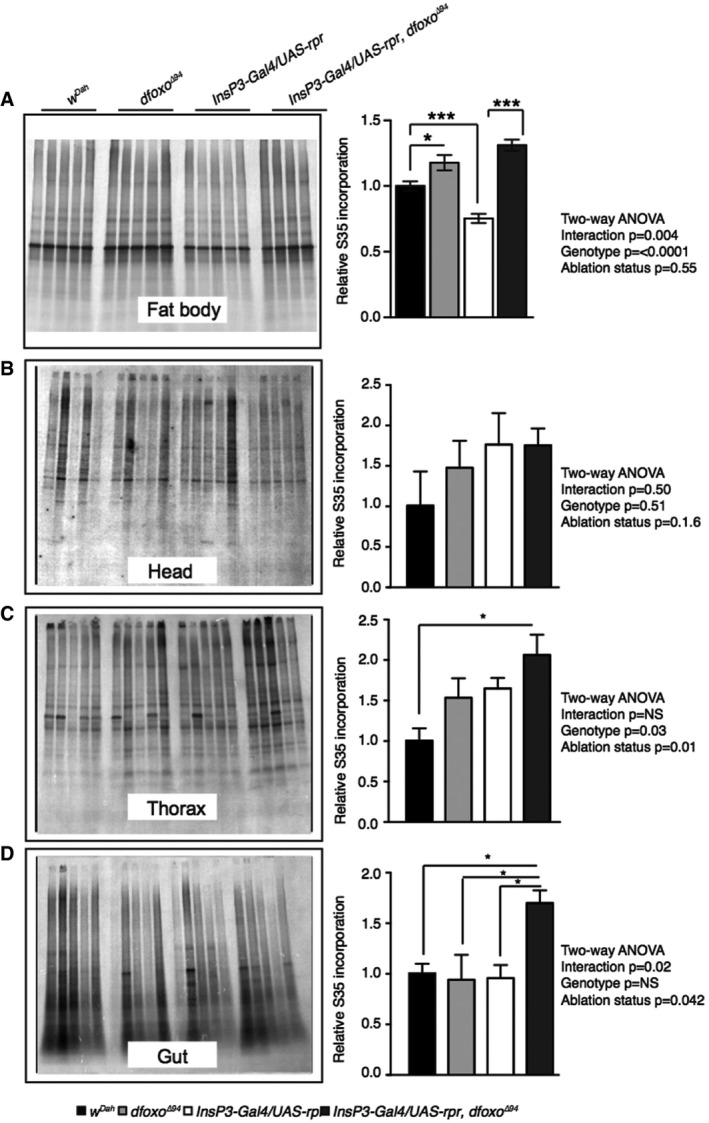
We next determined if the tissue‐specific, dfoxo‐dependent proteomic responses led to tissue‐specific functional changes. Our proteomic and bioinformatics analysis identified a fat body‐specific, dfoxo‐dependent reduction in ribosome‐associated proteins in response to reduced IIS (Fig 3). To determine whether this signature was reflected in altered physiology, we quantified tissue‐specific translation, using 35S incorporation to detect de novo protein synthesis. Individual tissues were dissected and incubated in medium containing 35S‐labelled methionine and cysteine. We separated samples by SDS–PAGE and quantified de novo labelled proteins (Fig EV3). No significant change was detected in head, thorax or gut (Fig EV3), but there was significantly less incorporation of labelled amino acids into proteins in the fat body of the IIS mutants, and that reduction was dependent on dfoxo (two‐way ANOVA P < 0.05; Fig EV3). This finding suggests that IIS‐mediated, dfoxo‐dependent reduction in translation in the fat body may contribute to longevity.
Figure EV3. Translational activity in the fat body under reduced IIS is reduced in a dfoxo‐dependent manner.
-
A–DDe novo protein synthesis as measured by incorporation of 35S into proteins from ex vivo (A) fat body, (B) head, (C) thorax and (D) gut tissue. Equal levels of protein were loaded per lane. Representative gel exposures show fat body 35S incorporation from w Dah , dfoxo Δ94, InsP3‐Gal4/UAS‐rpr and InsP3‐Gal4/UAS‐rpr, dfoxo Δ94 flies. Quantification of gel exposures normalized to total protein and corresponding two‐way ANOVA, as well as Bonferroni corrected post hoc tests are shown alongside each gel exposure. Error bars show SEM, n = 11 for w Dah and InsP3‐Gal4/UAS‐rpr fat body samples, for all other genotypes and tissues n = 5, *P < 0.05, ***P < 0.001.
Reduced IIS tissue‐specifically and dfoxo‐dependently regulates respiration
Changes in mitochondrial function have been linked to ageing in several model organisms (Bratic & Larsson, 2013). Paradoxically, however, both reduced mitochondrial respiratory chain protein levels (Copeland et al, 2009) and increased mitochondrial biogenesis (Rera et al, 2011) can extend lifespan. Our analysis highlighted a tissue‐specific regulation of mitochondrial proteins that was dfoxo‐dependent in the brain and fat body and independent of dfoxo in the gut (Fig 3). In total, we detected 60 proteins belonging to the annotation mitochondrial electron transport chain, with 6–35% of them in different tissues showing significantly altered levels upon IIS reduction (Fig 4A). They were coordinately up‐regulated in the fat body, with no clear coordinated change in other organs (Fig 4A). Using Western blotting, we directly confirmed that NDUFS3, a complex I subunit, was up‐regulated in a dfoxo‐dependent manner in the fat body (Fig 4B).
Figure 4. Reduced IIS differentially regulates tissue‐specific mitochondrial function and biogenesis.
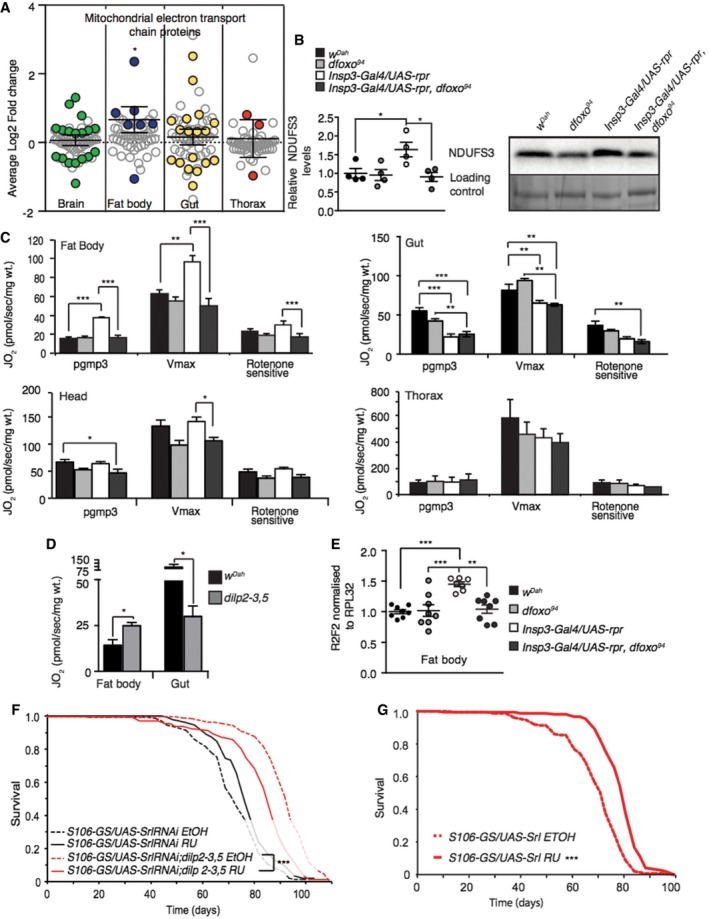
- Average log2 fold change of IIS‐regulated proteins associated with the functional term mitochondrial electron transport chain. One‐sample t‐test shows directional significance. Grey circles show all detected proteins, and coloured circles show significantly regulated proteins.
- Western blot analysis of fat body NDUFS3, and stain free loading control (n = 4), significance determined by t‐test.
- Fat body, gut, head and thorax oxygen consumption normalized to dry wt. in 10 days old control, dfoxo Δ94 mutants, mNSC ablation, mNSC‐ablated flies lacking dfoxo. Oxygen consumption was assessed by using substrates entering the level of complex I (PGMP3), complex I+II once uncoupled by CCP (Vmax) and rotenone‐sensitive complex I+II + rotenone (n = 5).
- Respiratory measurements of an independent IIS mutant (dilp2‐3,5) compared to controls (w Dah) (n = 5), significance determined by t‐test.
- Relative mtDNA levels compared to nuclear DNA in fat body (n = 8). Relative mtDNA levels in the gut, head and thorax are shown in Fig EV4A. Significance determined by one‐way ANOVA.
- Lifespan analysis of dilp2‐3,5 mutant and control flies with reduced expression of Spargel in the fat body (S106‐GS/UAS‐SrlRNAi) induced by RU or none induced (n = 150/genotype/treatment). Lifespan analysis of genetic controls shown in Fig EV4D. Statistical significance was determined by Log Rank test.
- Lifespan analysis of flies over‐expressing Spargel, in the adult fat body (n = 150/genotype/treatment). Genetic control lifespan shown in Fig EV4G. Statistical significance was determined by Log Rank test.
To determine the functional significance of the observed changes in protein levels, we provided substrates for the individual complexes of the electron transport chain to measure respiration in actual respiratory state (PGMP3), the potential maximum respiratory state (Vmax) and role of complex I (rotenone sensitive). In agreement with our proteomic and bioinformatic analysis, endogenous respiration (PGMP3) was up‐regulated by reduced IIS in the fat body, a response that was completely dependent on dfoxo (two‐way ANOVA interaction term P < 0.01; Fig 4C). This was also the case for the Vmax and rotenone‐sensitive respiratory states, suggesting that the increased respiration was due to increased capacity of the respiratory chain, at least in part through complex I (Fig 4C). In contrast, endogenous (PGMP3) and Vmax respiratory states in the gut were reduced in mNSC‐ablated flies and, as predicted by our analysis of the proteome, this occurred independently of dfoxo (Fig 4C). Respiration in the head and thorax was unchanged by reduced IIS (Fig 4C). We confirmed the increase in endogenous respiration of the fat body, and the reduced endogenous respiration of the gut, in an independent model of reduced IIS, long‐lived dilp2‐3,5 mutant flies (Gronke et al, 2010; Fig 4D).
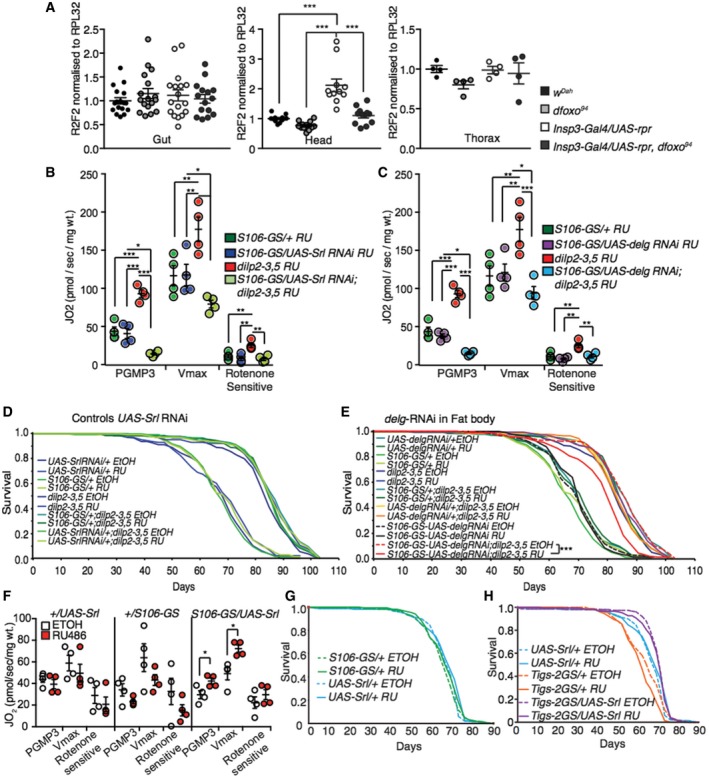
To further understand the mechanistic basis for increased respiration in the fat bodies of mNSC‐ablated flies, we examined mitochondrial biogenesis. We first examined mitochondrial DNA (mtDNA) levels in the fat body, in the presence and absence of dfoxo, and found that they were increased in the MNC‐ablated flies in a dfoxo‐dependent manner (two‐way ANOVA interaction term P = < 0.01; Figs 4E and EV4A). Mitochondrial biogenesis is driven by nuclear transcription factors, including nuclear respiratory factors 1 and 2 (nrf; Evans & Scarpulla, 1990; Virbasius et al, 1993), which are co‐activated by PGC‐1 to regulate the expression of mitochondrial proteins (Puigserver & Spiegelman, 2003; Scarpulla, 2008; Tiefenböck et al, 2010). Drosophila has single homologs of PGC‐1 and nrf‐2 (Gershman et al, 2007), named spargel and delg, respectively (Tiefenböck et al, 2010), and loss of either reduces transcription of nuclear encoded mitochondrial genes (Tiefenböck et al, 2010). Spargel and delg are both expressed at low levels in adult Drosophila fat body (Graveley et al, 2011), and we did not detect them, or their regulation in our analysis (Dataset EV1). However, as with many TFs and cofactors, the activity of Spargel and delg could be regulated on the post‐translational level (Li et al, 2007). We tested whether spargel or delg were required for the increased respiration in the fat bodies of IIS mutant flies. To achieve fat body‐specific knockdown of spargel and delg, we performed this analysis in dilp2‐3,5 mutants. We found that increased respiration in the fat body of the IIS mutant was entirely dependent on Spargel and delg expression in the fat body (two‐way ANOVA interaction term P = < 0.001; Fig EV4B and C). Furthermore, knockdown of expression of Spargel or delg in the fat body of dilp2‐3,5 mutants reduced the extent of their increased longevity, but did not affect the lifespan of wild‐type controls (Figs 4F, and EV4D and E). Experimentally increased expression of Spargel in the adult fat body, using the GeneSwitch driver S106‐GS, was sufficient to increase respiration (Fig EV4F) and lifespan (Figs 4G and EV4G), whilst over‐expression in the gut alone was not (Fig EV4H).
Figure EV4. Reduced IIS does not regulate mtDNA levels in the gut or head.
-
ARelative mtDNA levels compared to nuclear DNA in the gut (n = 17), head (n = 12) and thorax (n = 4). Fat body mtDNA levels shown in Fig 5E, significance determined by two‐way ANOVA.
-
B, CFat body oxygen consumption normalized to dry wt. of flies with fat body‐specific reduction of either (B) Spargel levels (S106GS/UAS‐Srl RNAi + RU) (n = 4) or (C) delg (S106GS/UAS‐delg RNAi + RU) in the presence and absence of dilp2‐3,5 and genetic controls (n = 4), significance determined by two‐way ANOVA. Oxygen consumption was assessed by using substrates entering the level of complex I (PGMP3), complex I+II once uncoupled by CCP (Vmax) and rotenone‐sensitive complex I+II + rotenone (n = 4), significance determined by two‐way ANOVA.
-
DLifespan analysis of genetic control flies with fat body‐specific reduction in Spargel levels (S106‐GS/UAS‐Srl RNAi + RU) (Fig 4F) (n > 150).
-
ELifespan analysis of dilp2‐3,5 mutant and control flies with reduced expression of Delg in the fat body (S106‐GS/UAS‐Delg RNAi) induced by RU or none induced (n = 150). Statistical significance was determined by Log Rank test.
-
FFat body oxygen consumption normalized to dry wt. of flies with induced (RU) or non‐induced (EtOH) fat body‐specific over‐expression of Spargel (S106GS/UAS‐Srl) and genetic controls (+UAS‐Srl and +/S106GS) (n = 4), significance determined by one‐way ANOVA.
-
GLifespan analysis of genetic control flies from over‐expression of Spargel, specifically in the adult fat body (Fig 4G) (n = 150).
-
HLifespan analysis of flies with induced (RU) or non‐induced (EtOH) over‐expression of Spargel in the adult gut (Tigs‐2GS/UAS‐Srl), along with genetic controls (n = 150).
Overall, our data reveal that lowered systemic IIS coopts different mechanisms to regulate respiration in different tissues. Reduced IIS decreased respiration in the gut independently of dfoxo, whilst simultaneously increasing respiration in the fat body in a dfoxo‐ and spargel/delg‐dependent manner. Furthermore, increased mitochondrial biogenesis, and thus respiration, in the fat body is both necessary and sufficient to extend lifespan.
Reduced IIS alters the dfoxo‐dependent gut proteome, increasing proteostasis to maintain gut health
The proteasome is a major site of protein degradation in the cell (Lecker et al, 2006), and its dysfunction is associated with ageing and disease (Vilchez et al, 2014). Our proteomic and bioinformatics analysis identified a prominent, gut‐specific, dfoxo‐dependent regulation of proteasomal proteins in response to reduced IIS (Fig 3). The 26S proteasome consists of 33 proteins, 14 of which (7‐α & 7‐β subunits) constitute the 20S proteasome core, whilst 19 constitute the 19S regulatory particle (Tomko & Hochstrasser, 2013). In total, we detected 94% of the proteasomal subunits, and 36% were differentially regulated specifically in the gut in response to reduced IIS (Fig 5A). However, we did not detect a consistent pattern or direction of regulation (4 up‐ and 8 down‐regulated, Fig 5A). Proteasomal subunit Rpt6R showed the greatest degree of regulation, increasing 2.6‐fold in the gut. We confirmed this change and its dfoxo dependency, by Western blot analysis of the guts of mNSC‐ablated flies compared to controls (Fig 5B). Rpt6, a 19S proteasomal subunit, plays a key role, along with Rpn6, in regulating the assembly and activity of the 26S proteasome holoenzyme (Park et al, 2011; Pathare et al, 2012; Sokolova et al, 2015). Hence, up‐regulation of this subunit could indicate an up‐regulation of proteasomal assembly. Indeed, the guts of mNSC‐ablated flies showed a threefold increase in assembly of the 26S proteasome compared to controls (Fig 5C), as measured using an in‐gel assay (Vernace et al, 2007). These data suggest that reduced IIS results in increased proteasomal assembly in the gut, possibly through increased levels of Rpt6.
Figure 5. IIS‐dfoxo‐dependent regulation of proteasome activity in the gut enhances proteome maintenance.
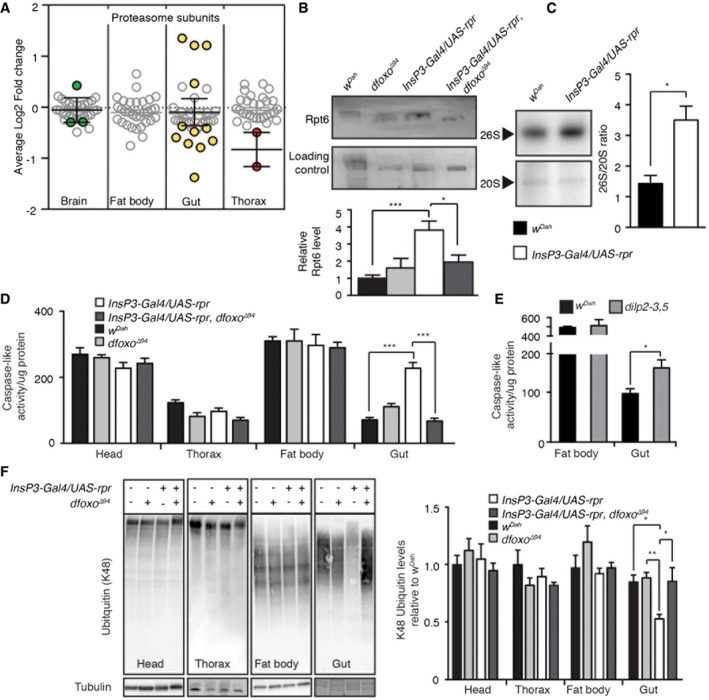
- Tissue‐specific average log2 fold change of IIS‐regulated proteins associated to the proteasome.
- Western blot and quantification analysis of Rpt6 in wild‐type (w Dah) (n = 8), dfoxo Δ94 mutants (n = 3), mNSC‐ablated flies (InsP3‐Gal4/UAS‐rpr) (n = 7) and mNSC‐ablated flies lacking dfoxo (InsP3‐Gal4/UAS‐rpr, dfoxo Δ94) (n = 4), normalized to protein loading and significance determined by one‐way ANOVA.
- Assessment of proteasome assembly by in‐gel caspase‐like (LLE) proteasome activity in mNSC‐ablated fly gut and control flies (w Dah). Quantification shows the activity ratio 26S/20S (n = 5), significance determined by t‐test.
- Tissue‐specific caspase‐like activity was assessed using fluorogenic substrate (LLE‐AMC, Enzo Life Science) in wild‐type (w Dah; n = head 14, fat body 5, gut 8, thorax 8), dfoxo Δ94 mutants (n = head 9, fat body 5, gut 7, thorax 8), mNSC‐ablated flies (InsP3‐Gal4/UAS‐rpr; n = head 14, fat body 5, gut 7, thorax 9) and mNSC‐ablated flies lacking dfoxo (InsP3‐Gal4/UAS‐rpr, dfoxo Δ94; n = head 7, fat body 5, gut 9, thorax 9), significance determined by two‐way ANOVA and post hoc pairwise tests.
- Proteasomal caspase‐like activity of an independent IIS mutant (dilp2‐3,5, grey) compared to controls (w Dah) (gut n = 9, fat body n = 7). Statistical significance was determined by t‐test.
- Representative Western blot showing tissue‐specific levels of K48‐linked poly‐ubiquitin and their quantification (head, thorax and fat body Westerns performed in quadruplicate). For gut samples wild‐type (w Dah) and dfoxo Δ94 mutants n = 11, InsP3‐Gal4/UAS‐rpr n = 12 and InsP3‐Gal4/UAS‐rpr, dfoxo Δ94 n = 10, significance determined by two‐way ANOVA and post hoc pairwise tests.
We next determined whether the increased proteasomal assembly in the gut affected proteasomal function. To measure proteasome activity, isolated tissue homogenates were incubated with a fluorogenic (AMC, 7‐Amino‐4‐methylcoumarin) proteasome substrate (Z‐Leu‐Leu‐Glu‐AMC), allowing the quantification of peptidylglutamyl‐peptide hydrolysing (caspase‐like) activity of the proteasome. As predicted by our bioinformatic analysis (Fig 3), no differences in activity were detected in the fat body, head or thorax in response to reduced IIS (Fig 5D). However, in agreement with our assembly data, reduced IIS resulted in a highly significant, dfoxo‐dependent, threefold increase in proteasomal activity in the gut (two‐way ANOVA interaction term P < 0.001; Fig 5D). To establish the generality of this finding, we measured proteasomal activity in long‐lived dilp2‐3,5 mutants (Gronke et al, 2010) and found that, again, proteasome activity was increased in gut but not fat body (Fig 5E). Thus, increased proteasome activity in the gut in response to reduced IIS is a candidate mechanism for increased longevity.
To determine the functional relevance of gut‐specific increased proteasomal activity, we quantified the level of ubiquitinated proteins. Ubiquitination serves many functions in the cell (Grabbe et al, 2011). Poly‐ubiquitination, specifically on Lysine 48 (K48), however, targets the protein for degradation by the proteasome, and accumulation of poly‐ubiquitinated proteins, often seen with increasing age, indicates loss of proteostasis (Tonoki et al, 2009). Based on our tissue‐specific proteasomal activity assay (Fig 5D), we hypothesized that K48‐ubiquitinated proteins levels would not differ in the head, thorax or fat body as a result of altered IIS activity, but would be reduced in the gut. We performed Western blot analysis against K48 poly‐ubiquitinated proteins (Fig 5F) and, in agreement with our hypothesis, we detected a gut‐specific, dfoxo‐dependent reduction in K48 poly‐ubiquitinated proteins in response to reduced IIS (Fig 5F). Therefore, increased proteasomal assembly/activity, and thus clearance of K48 poly‐ubiquitinated proteins, may underlie IIS‐mediated longevity through enhanced proteome maintenance.
Increased gut proteasomal activity maintains gut integrity and extends lifespan
We next determined the role of increased proteasomal activity in the extended lifespan of IIS mutants. We orally administered bortezomib, a potent inhibitor of the proteasome, to flies with reduced IIS and controls, for the duration of their lifespan. At a concentration of 2 μM, bortezomib did not reduce the lifespan of wild‐type flies (Fig 6A), but significantly reduced the lifespan‐extension of mNSC‐ablated flies (Fig 6A) and, independently, of dilp2‐3,5 mutants (Fig EV5A). Using Cox proportional hazards, we confirmed that treatment with bortezomib had a significantly different effect on survival in the two models of reduced IIS flies compared to their wild‐type controls (P < 0.01). Higher concentrations of bortezomib (3 μM) completely abolished the difference in lifespan between wild‐type and ablated flies, but also reduced the survival of wild‐type flies (Fig EV5B).
Figure 6. Proteasome activity is necessary for IIS‐mediated longevity, and sufficient to extend lifespan.
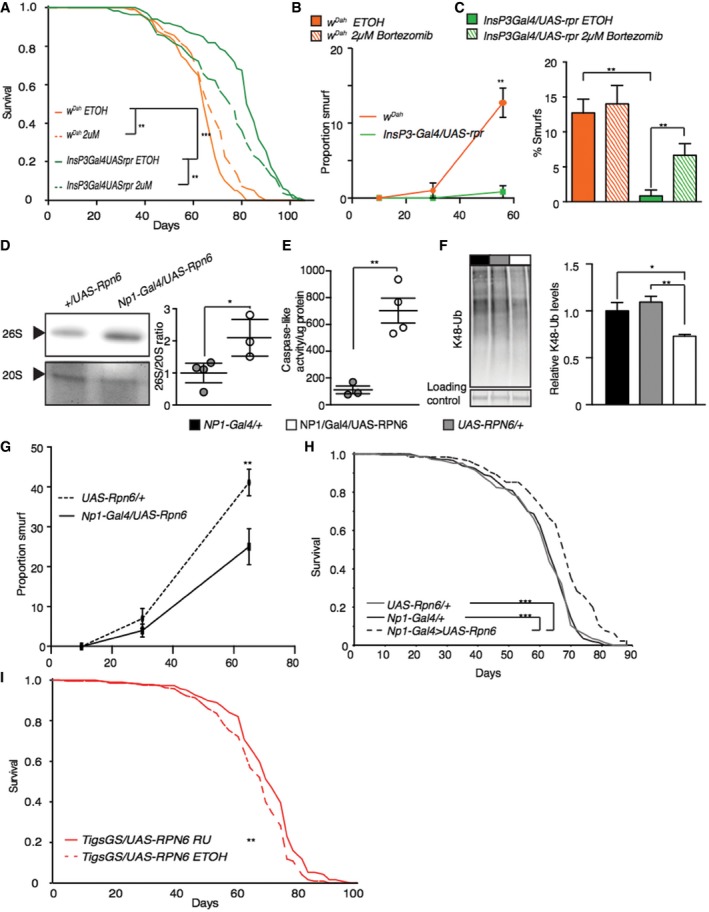
-
ALifespan analysis of wild‐type flies and mNSC‐ablated flies treated with 2 μM proteasome inhibitor (bortezomib) or vehicle (EtOH) (n = 100).
-
B, CAssessment of age‐related gut integrity through “Smurf” assay at either different ages (10, 30, 65 days, n = 100), significance determined by t‐test, or in aged (55 days, n = 90) flies in the presence of 2 μM proteasome inhibitor (bortezomib), two‐way ANOVA.
-
DAssessment of proteasome assembly by in‐gel caspase‐like (LLE) proteasome activity in NP1‐Gal4/UAS‐rPN6 (n = 3) flies compared to +/UAS‐RPN6 control flies (n = 4), significance determined by t‐test. Quantification shows the activity ratio 26S/20S (n > 4).
-
EGut proteasome activity (caspase‐like) of NP1‐Gal4/UAS‐rPN6 (n = 4) flies compared to +/UAS‐RPN6 control flies (n = 3), significance determined by t‐test.
-
FRepresentative Western blot showing gut‐specific levels of K48‐linked poly‐ubiquitin and their quantification (n = 6), significance determined by one‐way ANOVA.
-
GAssessment of age‐related gut integrity through “Smurf” assay at difference ages (10, 30, 65 days) in NP1‐Gal4/UAS‐rPN6 flies and +/UAS‐RPN6 control flies (n > 100), significance determined by t‐test.
-
H, ILifespan analysis of flies with gut‐specific over‐expression of UAS‐RPN6 using a constitutive gut‐specific driver (Np1‐Gal4) or in the adult using Tigs‐Gal4‐GS in the presence of RU486 (200 μm) or vehicle (EtOH) (n = 150).
Figure EV5. Proteasome activity is necessary for IIS‐mediated longevity, in an independent model of reduced IIS .
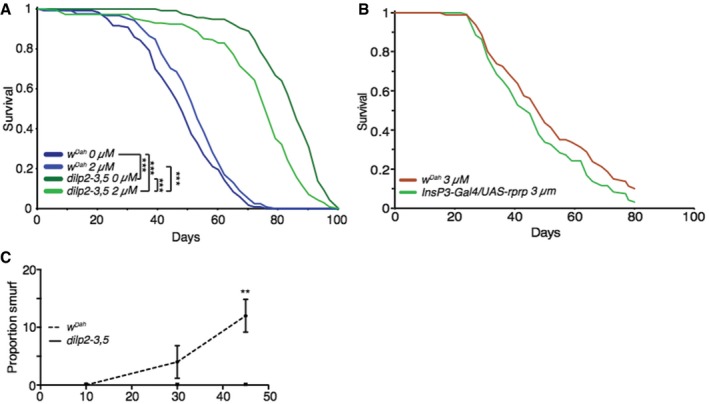
- Lifespan analysis of control flies (w Dah) and dilp2‐3,5 mutant flies treated with 2 μM proteasome inhibitor (bortezomib) or vehicle (EtOH) (n > 125). Statistical significance was determined by Log Rank test.
- Lifespan analysis of wild‐type flies (w Dah) and mNSC‐ablated flies treated with 3 μM proteasome inhibitor (bortezomib) (n > 80).
- Assessment of age‐related gut integrity through “Smurf” assay at different ages (10, 30, 45 days) in wild‐type flies (w Dah) and dilp2‐3,5 mutant flies (n = 100), significance determined by t‐test at each age.
To determine the functional consequence of increased proteasome activity in the gut, we quantified age‐associated changes in gut integrity in control and long‐lived flies. Feeding non‐absorbable blue food dye to flies allows direct assessment of gut integrity, with loss of integrity resulting in blue dye leaking into the hemolymph giving “smurf” flies (Rera et al, 2012). As previously reported (Rera et al, 2012), control flies show an age‐associated loss of gut integrity, with the proportion of “smurfs” in the population increasing with age; however, this was suppressed in mNSC‐ablated flies (Fig 6B), and independently in dilp2‐3,5 mutants (Fig EV5C). Whilst treatment with bortezomib (2 μM) did not affect aged control flies, it dramatically increased the proportion of “smurfs” in mNSC‐ablated flies (Fig 6C). These effects of the proteasomal inhibitor on lifespan and gut integrity indicate that increased proteasomal activity in the gut is required for the lifespan extension of IIS mutants because it increases gut integrity.
Together, the dfoxo dependence of the increased proteasomal activity and increased gut integrity of mNSC‐ablated flies, as well as the known role of proteostasis in ageing (Lopez‐Otin et al, 2013), prompted us to experimentally test whether increasing proteasomal assembly and activity in the adult gut was sufficient to extend lifespan. RPN6 aids in stabilizing the assembly of the 26S proteasome (Pathare et al, 2012) and can increase lifespan when over‐expressed ubiquitously in C. elegans (Vilchez et al, 2012). We therefore over‐expressed RPN6 specifically in the gut, using the constitutive, mid‐gut‐specific Gal4 driver Np1, which drives expression exclusively in enterocytes (Jiang et al, 2009; Alic et al, 2014). Constitutive, gut‐specific over‐expression of RPN6 was sufficient to significantly increase proteasome assembly in the gut (Fig 6D), increase proteasomal activity (Fig 6E) and induce a 26% reduction in K48 poly‐ubiquitinated proteins (Fig 6F), suggesting increased protein clearance and increased gut integrity (Fig 6G). Thus, increased levels of RPN6 are sufficient to increase proteasome assembly and activity, and increased clearance of K48 poly‐ubiquitinated proteins, enhancing proteome maintenance, and thus the health of the gut.
We next determined whether over‐expression of RPN6 was sufficient to extend the fly lifespan. Constitutive, gut‐specific over‐expression of RPN6 resulted in a significant 5% increase in mean lifespan, and up to a 19% increase in maximal lifespan compared to controls (Fig 6H). We independently confirmed this finding by over‐expressing RPN6 in the adult gut using the inducible, gut‐specific GeneSwitch driver TIGS‐2, which was sufficient to increase proteasome activity, increase gut integrity and induce a small, but significant, extension of lifespan (Figs 6I and EV6A–C). Hence, increased gut proteome maintenance, through increased proteasome function, can increase gut integrity and extend lifespan.
Figure EV6. Control lifespan analysis for adult gut‐specific expression of proteasomal subunit RPN6, proteasomal activity and gut integrity.
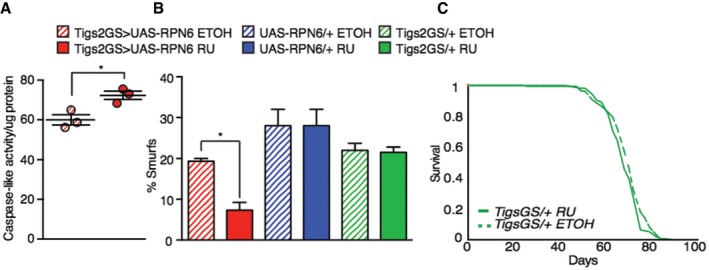
- Gut proteasome activity (caspase‐like) of Tigs2‐Geneswitch/UAS‐RPN6 flies compared to +/UAS‐RPN6 control flies (n = 3), significance determined by t‐test.
- Assessment of gut integrity though “Smurf” assay (65 days) between induced (RU treated) and uninduced (ETOH treated) Tigs2‐Geneswitch/UAS‐RPN6 (n = 200) flies and induced (RU treated) and uninduced (ETOH treated) driver controls Tigs2‐Geneswitch/+ (n = 200) and UAS (+UAS‐RPN6, n = 75) control flies.
- Gut‐specific driver (Tigs‐Gal4‐GS/+) control lifespan in the presence of RU486 (200 μm) or vehicle (EtOH) (n > 180).
Discussion
Reduced activity of the evolutionarily conserved insulin/IGF‐like signalling network extends longevity and reduces age‐associated pathologies, extending health and vitality (Fontana et al, 2010). Several studies have examined transcriptome remodelling in response to reduced IIS, to uncover the genes and molecular mechanisms underlying the longevity phenotype. However, the specific combination of expression changes and physiological processes that are required to prolong healthy lifespan remains elusive. Here, we have quantified the tissue‐specific proteome of Drosophila in response to reduced IIS, using genetic manipulations that allowed us to focus on expression profiles associated with longevity.
Establishing the tissue‐specific proteome of Drosophila
In recent years, the technology for proteomics has begun to reach the accessibility, sensitivity and reproducibility of genomic studies (Mann et al, 2013). Taking advantage of advancing technologies, we have performed an in‐depth, tissue‐specific proteomic screen in Drosophila, detecting over 6,000 proteins, ~40% of the predicted proteome. This dataset emphasizes the importance of tissue‐specific profiling and provides a unique and freely available resource for future expression studies in Drosophila (Dataset EV1).
Our proteomic analysis revealed that ~15% of the predicted fly proteome is remodelled in response to reduced IIS, with 2,372 proteins showing significant up‐ or down‐regulation. The RNA transcripts encoding 60% of these proteins have not previously been identified as differentially regulated by reduced IIS in whole fly profiles. This may be due to the highly tissue‐specific nature of the changes, with only two proteins being regulated in all tissues. Differences between patterns of expression of RNA and protein may also be important, due to post‐transcriptional regulation and changes in protein stability (Liu et al, 2016). For example, expression of GCN4 in nutritionally challenged yeast increases due to increased translation, not transcription (Ingolia et al, 2009), whilst in flies subject to dietary restriction several mitochondrial genes are regulated by translational, not transcriptional, control (Zid et al, 2009).
Confirming the quality of this dataset, proteins associated with several IIS‐mediated processes, including development, growth, sleep, lipid metabolism, translation and adult lifespan, were detected and differentially regulated. We were able to pinpoint these responses to specific tissues. For example, global reduction in translation can extend lifespan in C. elegans and Drosophila (Hansen et al, 2007; Pan et al, 2007; Wang et al, 2014) and is an evolutionarily conserved response to reduced IIS (McElwee et al, 2007; Stout et al, 2013; Essers et al, 2016), and our study showed this response to be specific to the fat body.
Our dataset also identified possible novel mediators of responses to reduced IIS. For example, our proteomic analysis suggested a gut‐specific regulation of proteasomal function in response to reduced IIS, which was confirmed by finding a corresponding gut‐specific proteasomal phenotype. We have also characterized dfoxo‐dependent changes in protein expression in IIS mutant flies, separating those changes associated with IIS‐mediated longevity from those changes associated to other IIS‐mediated phenotypes. Furthermore, we determined which IIS responsive protein‐coding genes contain predicted dFOXO‐binding motifs, identifying possible direct and indirect targets of dFOXO. Additionally, we examined transcriptional changes in several regulated candidate genes associated with mitochondria and the proteasome (Fig EV7A–C). Some candidate genes were regulated in a dfoxo‐dependent manner, consistent with a direct regulation by dFOXO; however, many did not reflect the changes seen at the protein level, suggesting indirect regulation by dFOXO, possibly though post‐transcriptional and/or post‐translational regulation (Fig EV7A–C). Importantly, we have thus identified longevity‐associated changes in the proteome of IIS mutant flies and the processes that they regulate, and we have experimentally demonstrated the role of some of these processes in extension of lifespan.
Figure EV7. qRT–PCR analysis of tissue‐specific candidate gene expression in response to reduced IIS.
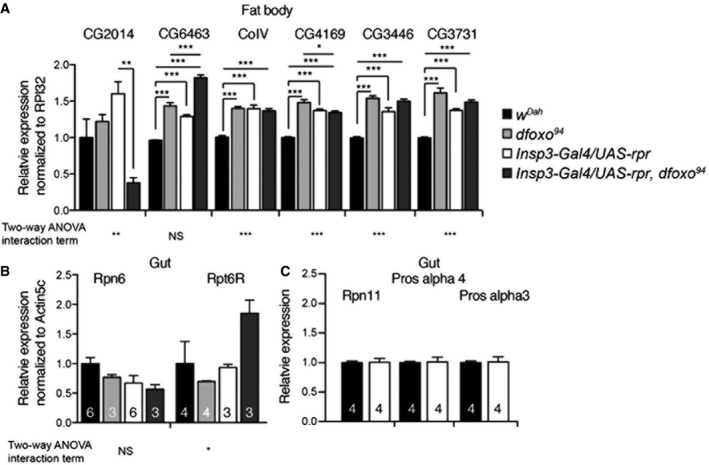
-
A–CqRT–PCR analysis shows gene expression levels of CG2014, CG6463, CoIV, CG4169, CG3731 in the fat body (A), or of Rpn6, Rpt6R, Rpn11 and Pros‐alpha3 in the gut (B, C) of control (w Dah), dfoxo mutant (dfoxo 94), mNSC‐ablated (InsP3‐Gal4/UAS‐rpr) and mNSC‐ablated flies in the absence of dfoxo (InsP3‐Gal4/UAS‐rpr, dfoxo 94). Relative expression levels are normalized to either RPL32 (A) or Actin5c (B, C). Significance established by two‐way ANOVA and post hoc pairwise tests (n = 3, or otherwise shown). Bars indicate mean SEM (*P < 0.05; **P < 0.01; ***P < 0.001).
Tissue‐specific regulation of mitochondrial number and respiration as a regulator of the IIS longevity response
Mitochondrial respiration and the production of reactive oxygen species (ROS) have been proposed as a mechanism of ageing (Bratic & Larsson, 2013). According to this theory, ROS are produced as a by‐product of mitochondrial respiration and on interaction with cellular macromolecules they induce damage (Harman, 1956). However, the link between mitochondria and ageing is not simple, and the validity of this theory has been questioned (Bratic & Larsson, 2013).
Seemingly in agreement with reduced ROS being beneficial, reducing mitochondrial respiration extends lifespan in several organisms (Dillin et al, 2002; Rea et al, 2007; Copeland et al, 2009). In contrast, many pro‐longevity interventions are associated with increased activation of mitochondrial respiration (Evans & Scarpulla, 1990; Virbasius et al, 1993; Ristow & Schmeisser, 2011), including models of dietary/caloric restriction (Mootha et al, 2003; Baker et al, 2006) and IIS, as found here and in C. elegans (Zarse et al, 2012). Declining mitochondrial number and function is associated with ageing (Yen et al, 1989; Shigenaga et al, 1994; Hebert et al, 2015). Thus, increasing or maintaining mitochondrial biogenesis with age may be beneficial.
We found increased, fat body‐specific respiration that was causal in the longevity of IIS mutants. Furthermore, we and others (Rera et al, 2011), have shown that increasing mitochondrial biogenesis, extends lifespan, although we found the effect to be confined to the fat body, whilst Rera et al (2011) found an effect in both the fat body and the gut, the discrepancy possibly due to the use of different fly stains. We also found that the increased fat body‐specific respiration and longevity of IIS mutant flies are dependent on PGC1‐α in the fat body, indicating that PGC1‐α is a downstream mediator of IIS, not only for growth (Tiefenböck et al, 2010; Mukherjee & Duttaroy, 2013), but also, as found here, longevity.
How increased respiration can, directly or indirectly, extend lifespan remains unclear. Growing evidence suggests that ROS, produced by mitochondrial respiration, is increased by pro‐longevity interventions and may act as signalling molecules to activate the expression of pro‐longevity genes (Ristow & Schmeisser, 2011). On the other hand, inhibition of target of rapamycin (TOR) in yeast increases respiration, but reduces ROS production, due to increased electron transport chain subunit expression and increased efficiency of electron transport, preventing a protracted electron transit time and ROS formation (Bonawitz et al, 2007). Furthermore, in flies, increased mitochondrial biogenesis protects against high fat diets, preventing lipid accumulation and heart dysfunction (Diop et al, 2015). To determine the exact mechanisms involved, analysis of tissue‐specific determination of ROS production and electron transport chain complex status under impaired IIS and other pro‐longevity interventions will be needed.
Interestingly, although not associated with longevity, we also found that respiration in the gut is decreased under reduced IIS, independently of dfoxo activity. Our analysis suggested that this response was associated with down‐regulation of Complex III protein subunits. When Complex III subunits are ubiquitously reduced throughout development and adulthood, flies become considerably longer lived and produce fewer offspring (Copeland et al, 2009). However, adult‐specific expression fails to extend lifespan, yet flies remain less fecund (Copeland et al, 2009). Since the down‐regulation of respiration we observed on reducing IIS in the gut was not associated with increased lifespan, and occurred independently of dfoxo, the reduction in gut respiration may underlie the reduced fecundity of IIS mutants.
Tissue‐specific proteostasis as a regulator of the IIS longevity response
Gradual loss of proteostasis is considered a hallmark of ageing (Lopez‐Otin et al, 2013). We found that, under reduced IIS, Drosophila tissue specifically regulate proteostasis, an evolutionarily conserved response that correlates with longevity. In flies, the age‐associated decline of proteostasis has been linked to declining proteasome function (Tsakiri et al, 2013), whilst maintaining proteasome function is associated with longevity (Chondrogianni et al, 2000; Pérez et al, 2009; Vilchez et al, 2012; Ungvari et al, 2013). In yeast, C. elegans, and Drosophila, ubiquitous over‐expression of proteasomal subunits can extend lifespan, possibly through increased maintenance of the cellular proteome (Chen et al, 2006; Tonoki et al, 2009; Kruegel et al, 2011; Vilchez et al, 2012). Increasing the maintenance of the cellular proteome and extending life span can also be achieved by reducing translation (Hansen et al, 2007; Pan et al, 2007; Wang et al, 2014). We found in Drosophila that increased proteasome activity and decreased translation correlated with longevity. However, these responses were highly tissue‐specific. Only the fat body of Drosophila responded to reduced IIS by decreasing translation, and only the gut responded by increasing proteasome activity. We were able to recapitulate increased proteasome activity by gut‐specific over‐expression of RPN6, which was sufficient to extend lifespan in wild‐type flies. Importantly, we could also show that increased proteasomal activity is required for the longevity of IIS mutants. Understanding how manipulation of one, or relatively few, protein subunits affects the function of a large protein complex is important for translating functional studies towards amelioration of the effects of human ageing. Whilst each subunit plays a role, only some subunits, that is the catalytic subunits, are essential for proteasome activity. However, Rpt3, Rpt6 and RPN6 play an equally vital role, those of opening the catalytic core and assembly maintaining structural integrity of the proteasome (Murata et al, 2009; Tian et al, 2011; Pathare et al, 2012; Sokolova et al, 2015). RPN6 over‐expression also influences lifespan (Vilchez et al, 2012), and we suggest through increased 26S proteasomal assembly, which recapitulates the phenotype seen in IIS mutant flies. It will therefore be important to determine whether similar regulation occurs in the tissues of long‐lived mammalian IIS models.
Tissue‐tissue communication and rebuilding the fly
Tissue‐specific reduction in IIS activity can improve function in that specific tissue, and can also act at a distance to improve the function of other tissues through systemic effects (Zhang et al, 2013; García‐Cáceres et al, 2016; Kaletsky et al, 2016). In these cases, tissue‐to‐tissue communication, involving FoxO and other key regulators, promotes longevity (Murphy et al, 2007; Bai et al, 2012; Zhang et al, 2013; Alic et al, 2014). The fly genome encodes ~300 proteins predicted to be targeted for secretion. We found 35% of these proteins to be regulated in response to reduced IIS, but very few dependent upon dfoxo, and thus associated with longevity. Future investigations to directly assay the proteomic changes in circulating hemolymph will help to determine which proteins may be involved in communication between tissues.
A system‐level approach to analyse the tissue‐specific proteome response to reduced IIS, followed by functional genetic and molecular analysis, has allowed the identification of IIS‐mediated, tissue‐specific pro‐longevity processes. Despite their highly specific responses to lowered IIS, individual tissues each contributed to the increase in lifespan in the IIS mutant flies. As well as its effect on ageing, IIS co‐ordinates tissue‐specific processes to regulate early life traits such as development, growth and reproduction in response to nutrition and other cues. The level of IIS appears to have evolved to optimize these early life fitness traits, but evidently this level is too high for optimal health at later ages not normally subject to natural selection, resulting in pro‐longevity effects of reduced IIS.
Here, we focused on two pro‐longevity effects, gut‐specific proteasome assembly/activity and fat body‐specific respiration/mitochondrial biogenesis. Increasing proteasome activity in the gut was sufficient to extend lifespan by 11%, and elevating mitochondrial respiration in the fat body can extend lifespan to the same extent (11%). However, reducing IIS extends lifespan to a greater extent. For example, mNSCs‐ablated flies are ~30% longer lived than controls. Thus, it is tempting to suggest that increasing both proteasome activity and mitochondrial respiration in the same fly could lead to an additive increase in lifespan, and that manipulation of other processes identified here, such as reduced translation in the fat body, may extend lifespan even further (Fig 7), a hypothesis that should be tested in future experiments.
Figure 7. Model of tissue‐specific responses to reduced IIS that may mediate longevity.
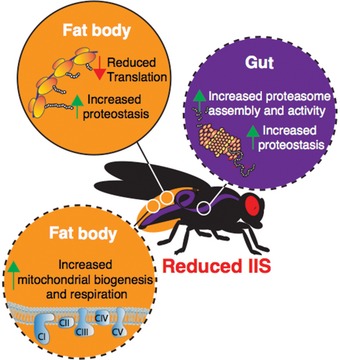
Colours denote different tissues (fat body, orange; gut, purple). Dashed lines show tissue‐specific responses that are both necessary and sufficient to extend lifespan.
In summary, tissue‐specific proteomic analysis of the responses to reduced IIS in Drosophila revealed that proteins from 15% of Drosophila's protein‐coding genes were regulated in response to reduced IIS across the four tissues examined here. Surprisingly, our study detected considerably more regulated proteins than our previous RNA transcript analyses in IIS mutants, highlighting the importance of tissue‐specific, and proteome, profiling to understand the effects of IIS. Here we focused specifically on those underlying responses to reduced IIS that are causal for longevity. Reducing translation, specifically in the fat body, is associated with longevity and not other IIS phenotypes, and tissue‐specific respiration was also increased through elevated mitochondrial biogenesis. Elevated mitochondrial biogenesis and respiration in the fat body were required for IIS‐mediated longevity, and directly increasing mitochondrial biogenesis in the fat body extended life span. Expression of proteasomal subunits was modulated in the gut of the fly by reduced IIS. These changes resulted in increased proteasome assembly and activity, increased proteome maintenance and increased gut integrity with age. Inhibition of these changes abolished IIS‐mediated longevity, and directly increasing gut‐specific proteasome function extended life span. Importantly, we showed that these responses are pro‐longevity and largely, tissue‐specific. Due to the level of conservation of these processes, our study suggests the development of tissue‐targeted pharmacological agents via homing peptides (Svensen et al, 2012) or chemical hybridization (Finan et al, 2016) to inhibit translation, activate the proteasome or increase respiration may be beneficial in prolonging longevity and health in mammals.
Materials and Methods
Fly stocks and fly husbandry
All mutants and transgenes were backcrossed into a white Dahomey (w Dah) wild‐type strain for at least eight generations. Fly stocks were kept at 25°C on a 12‐h light and 12‐h dark cycle and fed a standard sugar/yeast/agar diet (SYA; Bass et al, 2007). In all experiments, once‐mated females were reared at controlled larval densities. Adult flies were aged (10 days) in SYA food vials (10–25 flies per vial) prior to analysis. Dissections were carried out in cold phosphate‐buffered saline (PBS) and tissues either directly analysed or frozen on dry ice. Fly strains used in this study were as follows: Tigs‐Geneswitch (Tigs‐GS) (Buch et al, 2008; Poirier et al, 2008), UAS‐RPN6, UAS‐rpr, InsP3‐Gal4 (Buch et al, 2008; Slack et al, 2011), UAS‐Srl (Tiefenböck et al, 2010), dfoxo Δ94 (Slack et al, 2011).
Generation of UAS‐RPN6 transgenic flies
To generate a transgenic fly stock for conditional over‐expression of Drosophila Rpn6, the Rpn6 ORF was PCR amplified with primers SOL710 (ATGAATTCGCAAGATGGCCGGAGCAACAC) and SOL711 (ATGGTACCTTACGACAGCTTCTTAGCCTTC) and cDNA LD18931 as template (Drosophila Genomics Resource Center) and subsequently cloned into the pUAST attB vector using the EcoRI and KpnI restriction sites of the primers. Transgenic flies were generated using the φC31 and attP/attB integration system (Bischof et al, 2007), and transgenes were inserted into the attP40 landing‐site.
Peptide generation
Fly tissues (50/sample) from six biological replicates were lysed in pre‐heated (95°C) 6M guanidine hydrochloride, 10 mM TCEP, 40 mM CAA, 100 mM Tris pH 8,5 lysis buffer. Following shaking at 18,407 g (95°C), tissues were sonicated for five cycles (Bioruptor plus). Lysis buffer was then diluted 11‐fold in digestion buffer (25 mM Tris 8.5 pH, 10% acetyl nitride) and vortexed prior to a trypsin gold (Promega) overnight (37°C) digest (1 μg trypsin/50 μg protein). Samples were sonicated again for five cycles, prior to a further trypsin gold (Promega) overnight (37°C) digest (0.5 μg trypsin/50 μg protein) for 4 h (37°C) with gentle agitation. Samples were then placed in a SpeedVac (5 min, 37°C) to remove acetyl nitride. Peptides were desalted using SDB.XC Stage Tips (Rappsilber et al, 2003). Peptides were then eluted using (80% acetyl nitride, 0.1% formic acid) and placed in a SpeedVac (55 min, 29°C) to remove acetyl nitride, and quantified via Nanodrop.
MS/MS
Peptides were loaded on a 50‐cm column with 75 μm inner diameter, packed in‐house with 1.8‐μm C18 particles (Dr Maisch GmbH, Germany). Reversed phase chromatography was performed using the Thermo EASY‐nLC 1000 with a binary buffer system consisting of 0.1% formic acid (buffer A) and 80% acetonitrile in 0.1% formic acid (buffer B). The peptides were separated by a segmented gradient of 0 to 20 to 40% buffer B in 0 to 85 to 140 min for a 160‐min gradient run with a flow rate of 350 nl/min. The Q‐Exactive was operated in the data‐dependent mode with survey scans acquired at a resolution of 120,000; the resolution of the MS/MS scans was set to 15,000. Up to the 20 most abundant isotope patterns with charge ≥ 2 and < 7 from the survey scan were selected with an isolation window of 1.5 Th and fragmented by HCD (20) with normalized collision energies of 27. The maximum ion injection times for the survey scan and the MS/MS scans were 50 and 100 ms, respectively, and the AGC target value for the MS and MS/MS scan modes was set to 1E6 and 1E5, respectively. The MS AGC underfill ratio was set to 20% or higher. Repeat sequencing of peptides was kept to a minimum by dynamic exclusion of the sequenced peptides for 45 s.
Protein identification and quantification
The .raw data files were analysed. Protein identification was carried out using MaxQuant (Cox & Mann, 2008) version 1.5.0.4 using the integrated Andromeda search engine (Cox et al, 2011). The data were searched against the canonical and isoform, Swiss‐Prot and TrEMBL, Uniprot sequences corresponding to Drosophila melanogaster (20,987 entries). The database was automatically complemented with sequences of contaminating proteins by MaxQuant. For peptide identification, cysteine carbamidomethylation was set as “fixed” and methionine oxidation and protein N‐terminal acetylation as “variable” modification. The in‐silico digestion parameter of the search engine was set to “Trypsin/P”, allowing for cleavage after lysine and arginine, also when followed by proline. The minimum number of peptides and razor peptides for protein identification was 1; the minimum number of unique peptides was 0. Protein and peptide identification was performed with FDR of 0.01. The “second peptide” option was on, allowing for the identification of co‐fragmented peptides. In order to transfer identifications to non‐sequenced peptides in the separate analyses, the option “Match between runs” was turned on using a “Match time window” of 0.5 min and “Alignment time window” of 20 min. Protein and peptide identifications were performed within, not across, tissue groups. Label‐free quantification (LFQ) and normalization was done using MaxQuant (Cox et al, 2014). The default parameters for LFQ were used, except that the “LFQ min. ratio count” parameter was set to 1. Unique plus razor peptides were used for protein quantification. LFQ analysis was done separately on each tissue.
Perseus informatics analysis
The results of the LFQ analyses were loaded into the Perseus statistical framework (http://www.perseus-framework.org/) version 1.4.1.2. Protein contaminants, reverse database identifications and proteins “Only identified by site” were removed, and LFQ intensity values were log2 transformed. After categorical annotation into four categories based on genotype (dfoxo Δ94/w Dah) and ablation of mNSCs (yes/no), the data were filtered in order to contain a minimum of four valid values in at least one category. The remaining missing values were replaced, separately for each column, from normal distribution using width of 0.3 and down shift of 1.8. Based on quality control analysis, seven of the 90 samples (three fat body samples, three thorax samples and one brain sample) were excluded from the subsequent analysis due to technical failure of the chromatography. With those exceptions, the data were of high quality and reproducible between replicates. Information on Genotype and tissue‐specific average LFQ values and associated SEM values are included in Dataset EV1.
Bioinformatics
PCA analysis was carried out using the FactoMineR R package (Lê et al, 2008). The LIMMA package (Bioconductor suite, R) was used for differential expression analysis (Schwämmle et al, 2013). Multiplicity‐corrected P‐values were calculated using the Benjamini‐Hochberg procedure and significance determined using an adjusted P‐value cut‐off of 0.1.
Experimental groups were specified as follows: (A) w Dah, (B) dfoxo Δ94 mutant, (C) InsP3‐Gal4/UAS‐rpr and (D) InsP3‐Gal4/UAS‐rpr, dfoxo Δ94. Hypotheses were tested as linear combinations against the null hypothesis of no change: ablation‐induced changes in the wild‐type background (C vs. A), ablation‐induced changes in the dfoxo Δ94 background (D vs. B) and their interaction term (C–A vs. D–B). The subset of proteins differentially expressed in C vs. A was further subdivided into two categories: Proteins that changed in response to mNSC ablation in a dfoxo‐dependent manner, and those that behaved similarly regardless of genetic background were dfoxo‐independent: Foxo‐dependent: Differential in C vs. A and the interaction term. If a protein was regulated in the same direction in C vs. A and D vs. B, it was required to show a stronger response in the first contrast. Foxo‐independent: Differential in C vs. A. Protein expression levels were required to be equivalent with regard to differential expression in the interaction term. In any given tissue, a protein was identified as equivalent if the 95% CI of its log2 fold change fell within an interval ([−t; t]). The parameter t was determined as minimum log‐fold change of any significantly differentially expressed protein detected in the interaction term of each respective tissue: brain (t = 0.072), fat body (t = 0.177), gut (t = 0.155) and thorax (t = 0.216). Additionally, this subset included proteins found to be significant in the interaction term if they showed a stronger response to mNSC ablation in equal direction to that in the dfoxo Δ94 background compared to the wild‐type background.
Binding site identification
We used FIMO, MEME suite (Bailey et al, 2009) to identify genes whose transcription start site was within 1,000 bp of a dFOXO binding motif, using the default P‐value threshold (1e−4). Sequences corresponding to genes of measured proteins were extracted from the BDGP6 reference genome, whilst the dFOXO binding motif was taken from Fly Factor Survey (Zhu et al, 2011). We then mapped FIMO hits to proteins whose expression changed under reduced IIS conditions or those that were identified as dfoxo‐dependent and used a hypergeometric test to evaluate the significance of the overlap. Selection and background were limited to the set of genes corresponding to measured proteins.
Gene ontology term enrichment
topGO was used for GO term enrichment analysis and gene annotation performed using the Bioconductor annotation package org.dm.e.g.db. To identify enriched GO terms, the one‐sided elim Fisher procedure was used (α ≤ 0.05; Alexa et al, 2006). The enrichment score of a GO term is defined as log2 (#Detected significant genes/#Expected significant genes). For individual tissue‐specific contrasts, the subsets of differentially expressed proteins were tested for term enrichment against a tissue‐specific reference background. Cell plots show the most specific significantly enriched categories with a minimum of five significant associated proteins. To functionally characterize the individual tissues of wild‐type flies, GO enrichment analysis was carried out for proteins detected exclusively in the respective tissue, against the background of all detected proteins. For PCA map functional annotation (Fig 1D), the top 5% and lowest 5% quantiles of proteins according to their contributions to each dimension were tested for GO term enrichment, against the background of all detected proteins. The plot shows the 10 most significant terms with a minimum of 10 significant proteins, for each direction of the two dimensions.
Network propagation
Network propagation (Vanunu et al, 2010) was done using the D. melanogaster PPI network DroID (Murali et al, 2011) filtered for high confidence edges (40% confidence, n = 34,866). The network was converted to an adjacency matrix and normalized with the Laplacian transformation (using the graph.laplacian function in the igraph R package). Differentially regulated proteins were classified into dfoxo‐dependent and dfoxo‐independent (see Bioinformatics section above). For the analysis of dfoxo‐dependent proteins, the P‐values of proteins not belonging to this group were excluded (set to 1). Likewise, for the evaluation of the dfoxo‐independent set, P‐values of proteins not detected as such were excluded. Finally, the P‐values were −log2 transformed and mapped to the network. After mapping, the transformed P‐values on the individual nodes were diffused to their adjacent nodes using the spreading coefficient of 0.8 (corresponds to the percentage of sharing to neighbours). This spreading changed the scores of all proteins in the network, and it was iteratively repeated until protein scores do not change anymore.
We noticed that the network topology (i.e. the network structure) induces a bias: certain regions of the network tend to accumulate high scores during the propagation even if there is no actual signal. In order to correct for this bias, we computed a node‐specific topology bias in the following way: we started with equal scores on each node (e.g. each node has a score of 1). Then, we perform the propagation as described above. The resulting scores reflect the bias and are subtracted from the node scores after propagating the actual node scores. Subsequently, the corrected propagated scores were clustered based on their Euclidean distances (using the hclust function in R) allowing for the identification of enriched clusters for each tissue. For visualizing the enriched clusters from dfoxo‐dependent and dfoxo‐independent proteins on the same heatmap, the propagated profiles were subtracted (dfoxo‐dependent smoothed scores and dfoxo‐independent smoothed scores). GO enrichment analysis of individual clusters was done using the topGO R package defining all proteins that are part of the network as background.
35S‐methionine/35S‐cysteine incorporation assay
Incorporation assays were performed as previously described. Briefly, 5–8 tissues were dissected and collected in DMEM (#41965‐047, Gibco). For labelling, DMEM was replaced with methionine and cysteine free DMEM (#21‐013‐24, Gibco), supplemented with 35S‐labelled methionine and cysteine (#NEG772, Perkin‐Elmer) and incubated in uncapped Eppendorf tubes on a shaking platform at room temperature (60 min). Samples were then washed in ice cold PBS and lysed in RIPA buffer (150 mM sodium chloride, 1.0% NP‐40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0). Lysates were cleared by centrifugation at 15,871 g, 4°C for 10 min. Protein was precipitated by adding 1 volume of 20% TCA, incubating for 15 min on ice and centrifugation at 15,871 g, 4°C for 15 min. The pellet was washed twice in acetone and resuspended in 4 M guanidine HCl. Half the sample was added to 10 ml of scintillation fluid (Ultima Gold, Perkin‐Elmer) and counted for 5 min per sample in a scintillation counter (Perkin‐Elmer). The remaining sample was used to determine protein content using BCA (Pierce), following the manufacturers protocol. Scintillation counts were then normalized to total protein content prior to statistical analysis.
Respiratory rate measurements
Flies (10 days) were dissected in PBS and transferred to respiratory buffer (120 mM sucrose, 50 mM KCl, 20 mM Tris–HCl, 4 mM KH2PO4, 2 mM MgCl2, 1 mM EGTA, 0.01% digitonin, 0.05% BSA, pH 7.2). Oxygen consumption was measured using an oxygraph chamber (OROBOROS) at 25°C. Complex I‐dependent respiration was assessed using the substrates proline (10 mM), pyruvate (10 mM), malate (5 mM) and glutamate (5 mM), along with ADP (1.25 mM). The respiration was uncoupled by the addition of CCCP (0.3 μM). Maximum flux was then measured by adding complex II substrates succinate (10 mM) and glycerol‐3‐phosphate (5 mM). Rotenone‐sensitive flux was measured in the presence of rotenone (3 μM). Finally, dry weights of tissues were determined to normalize the oxygen consumption.
20s proteasome activity assay
Proteasome caspase‐like activity was measured as previously described (Vernace et al, 2007). Briefly, tissues were dissected in PBS and homogenized in 25 mM Tris (pH 7.5), and debris cleared by centrifugation. Protein content was measured, and 15 μg used to measure activity by incubating with 12.5 μM Z‐Leu‐Leu‐Glu‐AMC (Enzo Life Sciences) at a total volume of 200 μl. AMC fluorescence was measured at 360 nM excitation and 460 nM emission using a spectrofluorimeter (Tecan). Free AMC was used as a standard every 2 min for 30 min.
In‐gel proteasome assembly/activity assay
In‐gel activity/assembly assay was performed as previously described (Vernace et al, 2007). Briefly, individual tissues were dissected in PBS and homogenized in proteasome buffer (50 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 1 mM ATP, 1 mM DTT and 10% glycerol) on ice, centrifuged at 16,000 g (4°C, 10 min) and equal amounts of proteins loaded on a Bio‐Rad TGX 7.5% precast gel. Gels were incubated in proteasome buffer containing 0.4 mM Z‐Leu‐Leu‐Glu‐AMC (Enzo Life Sciences) for 15 min (37°C). Proteasome bands were visualized with UV light (360 nm) on a ChemiDoc station (Bio‐Rad). Differential activity was estimate by densitometry using ImageJ.
Western blotting
Western blots were carried out on protein extracts of individual dissected tissues. Proteins were quantified using BCA (Pierce), and equal amounts loaded on Any‐KD pre‐stained SDS–PAGE gels (Bio‐Rad) and blotted according to standard protocols. Antibody dilutions were as follows: K48‐poly Ubiquitin (Cell Signaling Technology D9D5 #8081; 1:1,000), Rpt6 (Santa Cruz biotechnology 9E3 #65752; 1:100), NDUFS3 (Abcam 17D95 #14711; 1:1,000). Appropriate secondary antibodies conjugated to horseradish peroxidase were used at a dilution of 1:10,000.
Life span analysis
Once‐mated flies, reared at standard densities, were transferred to vials (10‐25/vial). Flies were transferred to fresh vials three times a week, and deaths scored on transferal. Standard SYA food (Bass et al, 2007) was used throughout but, where needed, RU486, EtOH (vehicle control) was added to the food at a final concentration of 200 μM, or for bortezomib at 1–3 μm.
Gut integrity assay (“Smurf” assay)
Flies were grown and aged on standard SYA food until 48 h prior to the assay and then switched to SYA food containing 2.5% (w/v) Brilliant Blue FCF (AppliChem). Flies were then examined for “smurfing” and were recorded as “smurf” or not “smurf”, as previously described (Rera et al, 2012; Vizcaíno et al, 2014), except flies were kept on dyed food for 48 h.
Quantitative real‐time PCR
Total RNA was extracted using Trizol (Invitrogen Corp.) according to the manufacturer's instructions, including a DNase treatment. cDNA was prepared using SuperScript III first‐strand synthesis kit (Invitrogen Corp.). Quantitative real‐time PCR was performed in a 7900HT real‐time PCR system (Applied Biosystems). Relative expression (fold induction) was calculated using the ΔΔC T method and Rpl32 or Actin5c as a normalization control.
Statistical analysis
Statistical analysis was performed using Graphpad Prism and JMP. Individual statistical tests are mentioned in appropriate figure legends. Lifespan assays were recorded using Excel and survival was analysed using log rank test.
Data availability
The mass spectrometry proteomics data from this publication have been deposited to the ProteomeXchange Consortium (Vizcaíno et al, 2014) via the PRIDE archive database (Vizcaíno et al, 2016) https://www.ebi.ac.uk/pride/archive/ and assigned the accession PXD006225.
Author contributions
LST, LP designed the experiments. LST, CJ, NN, PE, MR, SG, and JF performed the experiments. LST, RS, MC, NN, CD, MM, NA, AB, and LP designed the data analysis. LST, RS, and MC performed the analysis. LST, NA, and LP wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Review Process File
Acknowledgements
Stocks from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We would also like to acknowledge Ilian Atanassov and the Proteomic facility at the Max Planck Institute for Biology of Ageing for help during MS analysis. We acknowledge funding from the Max Planck Society (to LT, RS, CJ, MR, SG, JF, NN, MM, LP), a Wellcome Trust Strategic Award (WT098565/Z/12/Z) (to LP), the European Union's Framework Programme for Research and Innovation Horizon (2014–2020) under the Marie Sklodowska‐Curie Grant Agreement no. 655623 (to PE), a Bundesministerium für Bildung und Forschung Grant SyBACol 0315893A‐B (to AB, CD, MC, LT, LP). We also acknowledge funding form Biotechnology and Biological Sciences Research Council (NA: BB/M029093/1) (to NA), the Medical Research Council (NA: MR/L018802/1) (to NA) and the Royal Society (NA: RG140694) (to NA). CD was supported by the Klaus Tschira Stiftung gGmbH [00.219b.2013]. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007‐2013)/ERC grant agreement number 268739 (to LP).
Mol Syst Biol. (2017) 13: 939
Contributor Information
Andreas Beyer, Email: andreas.beyer@uni-koeln.de.
Linda Partridge, Email: partridge@age.mpg.de.
References
- Afschar S, Toivonen JM, Hoffmann JM, Tain LS, Wieser D, Finlayson AJ, Driege Y, Alic N, Emran S, Stinn J, Froehlich J, Piper MD, Partridge L (2016) Nuclear hormone receptor DHR96 mediates the resistance to xenobiotics but not the increased lifespan of insulin‐mutant Drosophila . Proc Natl Acad Sci USA 113: 1321–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Alic N, Andrews TD, Giannakou ME, Papatheodorou I, Slack C, Hoddinott MP, Cocheme HM, Schuster EF, Thornton JM, Partridge L (2011) Genome‐wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7: 502–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N, Tullet JM, Niccoli T, Broughton S, Hoddinott MP, Slack C, Gems D, Partridge L (2014) Cell‐nonautonomous effects of dFOXO/DAF‐16 in aging. Cell Rep 6: 608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador‐Noguez D, Dean A, Huang W, Setchell K, Moore D, Darlington G (2007) Alterations in xenobiotic metabolism in the long‐lived Little mice. Aging Cell 6: 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aradska J, Bulat T, Sialana FJ, Birner‐Gruenberger R, Erich B, Lubec G (2015) Gel‐free mass spectrometry analysis of Drosophila melanogaster heads. Proteomics 15: 3356–3360 [DOI] [PubMed] [Google Scholar]
- Azimifar SB, Nagaraj N, Cox J, Mann M (2014) Cell‐type‐resolved quantitative proteomics of murine liver. Cell Metab 20: 1076–1087 [DOI] [PubMed] [Google Scholar]
- Bai H, Kang P, Tatar M (2012) Drosophila insulin‐like peptide‐6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin‐like peptide‐2 from the brain. Aging Cell 11: 978–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Betik AC, Krause DJ, Hepple RT (2006) No decline in skeletal muscle oxidative capacity with aging in long‐term calorically restricted rats: effects are independent of mitochondrial DNA integrity. J Gerontol A Biol Sci Med Sci 61: 675–684 [DOI] [PubMed] [Google Scholar]
- Barrett LW, Fletcher S, Wilton SD (2012) Regulation of eukaryotic gene expression by the untranslated gene regions and other non‐coding elements. Cell Mol Life Sci 69: 3613–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai N, Huffman DM, Muzumdar RH, Bartke A (2012) The critical role of metabolic pathways in aging. Diabetes 61: 1315–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass TM, Grandison RC, Wong R, Martinez P, Partridge L, Piper MDW (2007) Optimization of dietary restriction protocols in Drosophila . J Gerontol A Biol Sci Med Sci 62: 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152: 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K (2007) An optimized transgenesis system for Drosophila using germ‐line‐specific phiC31 integrases. Proc Natl Acad Sci USA 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M, Kahn BB, Kahn CR (2003) Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299: 572–574 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay‐Lapointe M, Pan Y, Shadel GS (2007) Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab 5: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic A, Larsson N‐G (2013) The role of mitochondria in aging. J Clin Invest 123: 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton SJ, Slack C, Alic N, Metaxakis A, Bass TM, Driege Y, Partridge L (2010) DILP‐producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell 9: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Ahrens CH, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch EW, Panse C, de Lichtenberg U, Rinner O, Lee H, Pedrioli PGA, Malmstrom J, Koehler K, Schrimpf S, Krijgsveld J, Kregenow F, Heck AJR, Hafen E, Schlapbach R et al (2007) A high‐quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol 25: 576–583 [DOI] [PubMed] [Google Scholar]
- Buch S, Melcher C, Bauer M, Katzenberger J, Pankratz MJ (2008) Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin‐like peptide signaling. Cell Metab 7: 321–332 [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA (2014) The noncoding RNA revolution‐trashing old rules to forge new ones. Cell 157: 77–94 [DOI] [PubMed] [Google Scholar]
- Chen Q, Thorpe J, Dohmen JR, Li F, Keller JN (2006) Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic Biol Med 40: 120–126 [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Petropoulos I, Franceschi C, Friguet B, Gonos ES (2000) Fibroblast cultures from healthy centenarians have an active proteasome. Exp Gerontol 35: 721–728 [DOI] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW (2009) A mitochondrial ATP synthase subunit interacts with TOR signaling to modulate protein homeostasis and lifespan in Drosophila . Curr Biol 19: 1591–1598 19747824 [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Uh H‐W, Monajemi R, van Heemst D, Thijssen PE, Böhringer S, van den Akker EB, de Craen AJM, Rivadeneira F, Uitterlinden AG, Westendorp RGJ, Goeman JJ, Slagboom PE, Houwing‐Duistermaat JJ, Beekman M (2013) Gene set analysis of GWAS data for human longevity highlights the relevance of the insulin/IGF‐1 signaling and telomere maintenance pathways. Age 35: 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Perrimon N (2010) FOXO/4E‐BP signaling in Drosophila muscles regulates organism‐wide proteostasis during aging. Cell 143: 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh AS, Murgia M, Nagaraj N, Treebak JT, Cox J, Mann M (2015) Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol Cell Proteomics 14: 841–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu A‐L, Arantes‐Oliveira N, Lehrer‐Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C (2002) Rates of behavior and aging specified by mitochondrial function during development. Science 298: 2398–2401 [DOI] [PubMed] [Google Scholar]
- Diop SB, Bisharat‐Kernizan J, Birse RT, Oldham S, Ocorr K, Bodmer R (2015) PGC‐1/spargel counteracts high‐fat‐diet‐induced obesity and cardiac lipotoxicity downstream of TOR and brummer ATGL lipase. Cell Rep 10: 1572–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers P, Tain LS, Nespital T, Goncalves J, Froehlich J, Partridge L (2016) Reduced insulin/insulin‐like growth factor signaling decreases translation in Drosophila and mice. Sci Rep 6: 30290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC (1990) NRF‐1: a trans‐activator of nuclear‐encoded respiratory genes in animal cells. Genes Dev 4: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Ewald CY, Landis JN, Porter Abate J, Murphy CT, Blackwell TK (2015) Dauer‐independent insulin/IGF‐1‐signalling implicates collagen remodelling in longevity. Nature 519: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan B, Clemmensen C, Zhu Z, Stemmer K, Gauthier K, Müller L, De Angelis M, Moreth K, Neff F, Perez‐Tilve D, Fischer K, Lutter D, Sánchez‐Garrido MA, Liu P, Tuckermann J, Malehmir M, Healy ME, Weber A, Heikenwalder M, Jastroch M et al (2016) Chemical hybridization of glucagon and thyroid hormone optimizes therapeutic impact for metabolic disease. Cell 167: 843–850.e14 [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD (2010) Extending healthy life span‐from yeast to humans. Science 328: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Cáceres C, Quarta C, Varela L, Gao Y, Gruber T, Legutko B, Jastroch M, Johansson P, Ninkovic J, Yi C‐X, Le Thuc O, Szigeti‐Buck K, Cai W, Meyer CW, Pfluger PT, Fernandez AM, Luquet S, Woods SC, Torres‐Alemán I, Kahn CR et al (2016) Astrocytic insulin signaling couples brain glucose uptake with nutrient availability. Cell 166: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M, Garofalo RS (2007) High‐resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol Genomics 29: 24–34 [DOI] [PubMed] [Google Scholar]
- Giannakou ME (2004) Long‐lived Drosophila with overexpressed dFOXO in adult fat body. Science 305: 361 [DOI] [PubMed] [Google Scholar]
- Grabbe C, Husnjak K, Dikic I (2011) The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 12: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, May G, Brooks AN, Carlson JW, Cherbas L, Davis CA, Duff M, Eads B, Landolin J, Sandler J, Wan KH, Andrews J, Brenner SE, Cherbas P, Gingeras TR, Hoskins R, Kaufman T, Celniker SE (2011) The developmental transcriptome of Drosophila melanogaster . Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L (2010) Molecular evolution and functional characterization of Drosophila insulin‐like peptides. PLoS Genet 6: e1000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaschek‐Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJM, Marra MA, Brooks‐Wilson AR, Riddle DL (2005) Analysis of long‐lived C. elegans daf‐2 mutants using serial analysis of gene expression. Genome Res 15: 603–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee S‐J, Kenyon C (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans . Aging Cell 6: 95–110 [DOI] [PubMed] [Google Scholar]
- Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11: 298–300 [DOI] [PubMed] [Google Scholar]
- He Y‐H, Lu X, Yang L‐Q, Xu L‐Y, Kong Q‐P (2014) Association of the insulin‐like growth factor binding protein 3 (IGFBP‐3) polymorphism with longevity in Chinese nonagenarians and centenarians. Aging (Albany NY) 6: 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SL, Marquet‐de Rougé P, Lanza IR, McCrady‐Spitzer SK, Levine JA, Middha S, Carter RE, Klaus KA, Therneau TM, Highsmith EW, Nair KS (2015) Mitochondrial aging and physical decline: insights from three generations of women. J Gerontol A Biol Sci Med Sci 70: 1409–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA (2009) Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Dong X, Vijg J, Suh Y (2015) Genetic evidence for common pathways in human age‐related diseases. Aging Cell 14: 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, Murphy CT (2016) The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 529: 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild‐type. Nature 366: 461–464 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ (2010) The genetics of ageing. Nature 464: 504–512 [DOI] [PubMed] [Google Scholar]
- Kim M‐S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal‐Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN et al (2014) A draft map of the human proteome. Nature 509: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, Sutphin G, Carr D, Tar K, Dittmar G, Kaeberlein M, Kennedy BK, Schmidt M (2011) Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae . PLoS Genet 7: e1002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25: 1–18 [Google Scholar]
- Lecker SH, Goldberg AL, Mitch WE (2006) Protein degradation by the ubiquitin‐proteasome pathway in normal and disease states. J Am Soc Nephrol 17: 1807–1819 [DOI] [PubMed] [Google Scholar]
- Li X, Monks B, Ge Q, Birnbaum MJ (2007) Akt/PKB regulates hepatic metabolism by directly inhibiting PGC‐1α transcription coactivator. Nature 447: 1012–1016 [DOI] [PubMed] [Google Scholar]
- Li JJ, Bickel PJ, Biggin MD (2014) System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2: e270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165: 535–550 [DOI] [PubMed] [Google Scholar]
- Lopez‐Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The hallmarks of aging. Cell 153: 1194–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes AW (2016) The role of the ribosome in the regulation of longevity and lifespan extension. Wiley Interdiscip Rev RNA 7: 198–212 [DOI] [PubMed] [Google Scholar]
- Mann M, Kulak NA, Nagaraj N, Cox J (2013) The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell 49: 583–590 [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D (2007) Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol 8: R132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K‐F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D et al (2003) PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Duttaroy A (2013) Spargel/dPGC‐1 is a new downstream effector in the insulin‐TOR signaling pathway in Drosophila . Genetics 195: 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali T, Pacifico S, Yu J, Guest S, Roberts GG, Finley RL (2011) DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila . Nucleic Acids Res 39: D736–D743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K (2009) Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10: 104–115 [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C (2003) Genes that act downstream of DAF‐16 to influence the lifespan of Caenorhabditis elegans . Nature 424: 277–283 [DOI] [PubMed] [Google Scholar]
- Murphy CT, Lee S‐J, Kenyon C (2007) Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans . Proc Natl Acad Sci USA 104: 19046–19050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M (2012) System‐wide perturbation analysis with nearly complete coverage of the yeast proteome by single‐shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics 11: M111.013722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A, Kitamura Y, Kahn CR, Accili D (2004) Mouse models of insulin resistance. Physiol Rev 84: 623–647 [DOI] [PubMed] [Google Scholar]
- Niccoli T, Partridge L (2012) Ageing as a risk factor for disease review. Curr Biol 22: R741–R752 [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA (2006) Identification of direct DAF‐16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: 251–257 [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P (2007) Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans . Aging Cell 6: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kim W, Tian G, Gygi SP, Finley D (2011) Structural defects in the regulatory particle‐core particle interface of the proteasome induce a novel proteasome stress response. J Biol Chem 286: 36652–36666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Körner R, Nickell S, Lasker K, Sali A, Tamura T, Nishioka T, Förster F, Baumeister W, Bracher A (2012) The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci USA 109: 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A (2009) Protein stability and resistance to oxidative stress are determinants of longevity in the longest‐living rodent, the naked mole‐rat. Proc Natl Acad Sci USA 106: 3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier L, Shane A, Zheng J, Seroude L (2008) Characterization of the Drosophila gene‐switch system in aging studies: a cautionary tale. Aging Cell 7: 758–770 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM (2003) Peroxisome proliferator‐activated receptor‐gamma coactivator 1 alpha (PGC‐1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev 24: 78–90 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Rask‐Madsen C, Kahn CR (2012) Tissue‐specific insulin signaling, metabolic syndrome, and cardiovascular disease. Arterioscler Thromb Vasc Biol 32: 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE (2007) Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans . PLoS Biol 5: e259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jones DL, Walker DW (2011) Modulation of longevity and tissue homeostasis by the Drosophila PGC‐1 homolog. Cell Metab 14: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Clark RI, Walker DW (2012) Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila . Proc Natl Acad Sci USA 109: 21528–21533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S (2011) Extending life span by increasing oxidative stress. Free Radic Biol Med 51: 327–336 [DOI] [PubMed] [Google Scholar]
- Scarpulla RC (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638 [DOI] [PubMed] [Google Scholar]
- Schwämmle V, León IR, Jensen ON (2013) Assessment and improvement of statistical tools for comparative proteomics analysis of sparse data sets with few experimental replicates. J Proteome Res 12: 3874–3883 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al‐Qassab H, Carmignac D, Ramadani F, Woods A, Robinson IC, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L et al (2009) Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326: 140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenaga MK, Hagen TM, Ames BN (1994) Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA 91: 10771–10778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C, Giannakou ME, Foley A, Goss M, Partridge L (2011) dFOXO‐independent effects of reduced insulin‐like signaling in Drosophila . Aging Cell 10: 735–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C, Alic N, Foley A, Cabecinha M, Hoddinott MP, Partridge L (2015) The Ras‐Erk‐ETS‐signaling pathway is a drug target for longevity. Cell 162: 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova V, Li F, Polovin G, Park S (2015) Proteasome activation is mediated via a functional switch of the Rpt6 C‐terminal tail following chaperone‐dependent assembly. Sci Rep 5: 14909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout GJ, Stigter ECA, Essers PB, Mulder KW, Kolkman A, Snijders DS, van den Broek NJF, Betist MC, Korswagen HC, MacInnes AW, Brenkman AB (2013) Insulin/IGF‐1‐mediated longevity is marked by reduced protein metabolism. Mol Syst Biol 9: 679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho M‐O, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P (2008) Functionally significant insulin‐like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci USA 105: 3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensen N, Walton JGA, Bradley M (2012) Peptides for cell‐selective drug delivery. Trends Pharmacol Sci 33: 186–192 [DOI] [PubMed] [Google Scholar]
- Tazearslan C, Huang J, Barzilai N, Suh Y (2011) Impaired IGF1R signaling in cells expressing longevity‐associated human IGF1R alleles. Aging Cell 10: 551–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila . Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ (2013) PQM‐1 complements DAF‐16 as a key transcriptional regulator of DAF‐2‐mediated development and longevity. Cell 154: 676–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Park S, Lee MJ, Huck B, McAllister F, Hill CP, Gygi SP, Finley D (2011) An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol 18: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiefenböck SK, Baltzer C, Egli NA, Frei C (2010) The Drosophila PGC‐1 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J 29: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Hochstrasser M (2013) Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem 82: 415–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M (2009) Genetic evidence linking age‐dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol 29: 1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri EN, Sykiotis GP, Papassideri IS, Gorgoulis VG, Bohmann D, Trougakos IP (2013) Differential regulation of proteasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress. FASEB J 27: 2407–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungvari Z, Csiszar A, Sosnowska D, Philipp EE, Campbell CM, McQuary PR, Chow TT, Coelho M, Didier ES, Gelino S, Holmbeck MA, Kim I, Levy E, Sonntag WE, Whitby PW, Austad SN, Ridgway I (2013) Testing predictions of the oxidative stress hypothesis of aging using a novel invertebrate model of longevity: the giant clam (Tridacna derasa). J Gerontol A Biol Sci Med Sci 68: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R (2010) Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol 6: e1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraksa A, Bauer A, Artavanis‐Tsakonas S (2005) Analyzing protein complexes in Drosophila with tandem affinity purification‐mass spectrometry. Dev Dyn 232: 827–834 [DOI] [PubMed] [Google Scholar]
- Vernace VA, Arnaud L, Schmidt‐Glenewinkel T, Figueiredo‐Pereira ME (2007) Aging perturbs 26S proteasome assembly in Drosophila melanogaster . FASEB J 21: 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues APC, Manning G, Dillin A (2012) RPN‐6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489: 263–268 [DOI] [PubMed] [Google Scholar]
- Vilchez D, Saez I, Dillin A (2014) The role of protein clearance mechanisms in organismal ageing and age‐related diseases. Nat Commun 5: 1–13 [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Virbasius CA, Scarpulla RC (1993) Identity of GABP with NRF‐2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev 7: 380–392 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz P‐A, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus H‐J, Albar JP, Martinez‐Bartolomé S et al (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, Del‐Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez‐Riverol Y, Reisinger F, Ternent T, Xu Q‐W, Wang R, Hermjakob H (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: 11033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cui Y, Jiang Z, Xie W (2014) Knockdown expression of eukaryotic initiation factor 5 C‐terminal domain containing protein extends lifespan in Drosophila melanogaster . Biochem Biophys Res Commun 446: 465–469 [DOI] [PubMed] [Google Scholar]
- Wolkow CA (2000) Regulation of C. elegans life‐span by insulinlike signaling in the nervous system. Science 290: 147–150 [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Tatar M (2011) Insulin receptor substrate chico acts with the transcription factor FOXO to extend Drosophila lifespan. Aging Cell 10: 729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TC, Chen YS, King KL, Yeh SH, Wei YH (1989) Liver mitochondrial respiratory functions decline with age. Biochem Biophys Res Commun 165: 944–1003 [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M (2012) Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L‐proline catabolism to induce a transient ROS signal. Cell Metab 15: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee S‐J, Kenyon C (2013) Direct and indirect gene regulation by a life‐extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab 17: 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Christensen RG, Kazemian M, Hull CJ, Enuameh MS, Basciotta MD, Brasefield JA, Zhu C, Asriyan Y, Lapointe DS, Sinha S, Wolfe SA, Brodsky MH (2011) FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one‐hybrid system. Nucleic Acids Res 39: D111–D117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P (2009) 4E‐BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila . Cell 139: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Review Process File
Data Availability Statement
The mass spectrometry proteomics data from this publication have been deposited to the ProteomeXchange Consortium (Vizcaíno et al, 2014) via the PRIDE archive database (Vizcaíno et al, 2016) https://www.ebi.ac.uk/pride/archive/ and assigned the accession PXD006225.


