Abstract
Gliomas are the most common primary malignant brain tumor with poor prognosis, characterized by a highly heterogeneous cell population, extensive proliferation, and migration. A lot of molecular mechanisms regulate gliomas development and invasion, including abnormal expression of oncogenes and variation of epigenetic modification. MicroRNAs could affect cell growth and functions. Several reports have demonstrated that miR-139 plays multifunctions in kinds of solid tumors through different pathways. However, the antitumor mechanisms of this miR-139 are not unveiled in detail. In this study, we not only validated the low expression level of miR-139 in glioma tissues and cell lines but also detected the effect of miR-139 on modulating gliomas proliferation and invasion both in vitro and in vivo. We identified insulin-like growth factor 1 receptor, associate of Myc 1, and peroxisome proliferator-activated receptor γ coactivator 1β as direct targets of miR-139 and the levels of them were all inversely correlated with miR-139 in gliomas. Insulin like growth factor 1 receptor promoted gliomas invasion through Akt signaling and increased proliferation in the peroxisome proliferator-activated receptor γ coactivator 1β-dependent way. Associate of Myc 1 also facilitated gliomas progression by activating c-Myc pathway. Overexpression of the target genes could retrieve the antitumor function of miR-139, respectively, in different degrees. The nude mice transplantation tumor experiment displayed that glioma cells stably expressed miR-139 growth much slower in vivo than the negative control cells. Taken together, these findings suggested miR-139 acted as a favorable factor against gliomas progression and uncovered a novel regulatory mechanism, which may provide a new evidenced prognostic marker and therapeutic target for gliomas.
Keywords: miR-139, glioma, tumor progression, IGF-1 R, AMY-1, PGC-1β
Introduction
Malignant gliomas, with an annual incidence of 5 per 100 thousand individuals, are the most common type of primary central nervous system tumor in humans. According to the classification of World Health Organization (WHO) based on the malignancy of the neoplasms, gliomas range from grade I, which assigned to low-proliferative and noninvasive tumors, up to grade IV glioblastoma multiforme (GBM) with the high level of penetration, mitotic activation, distal spread, and poorest survival rates.1 The patients with high-grade gliomas always have poor outcomes, and the median survival time of GBM is less than 1 year.2–4 The major reasons that make the gliomas to be incurable are the tendency of glioblastoma cells to infiltrate into surrounding brain tissue5,6 and the extreme molecular heterogeneity of this cancer intratumourally.7–9 Despite the fact that the current standard treatment with surgery, radiotherapy, and chemotherapy could increase the median survival time of patients with gliomas, the gliomas recurrence is inevitable. The new effective therapeutic targets and anti-glioma strategies are urgent to be investigated to combat this disease.
The recent studies have demonstrated that several microRNAs (miRNAs) are associated with the invasion process in gliomas and other tumor cells.10,11 MicroRNAs are highly conserved endogenous small noncoding RNAs, which contain 19 to 21 nucleotides and suppress gene expression posttranscriptionally by targeting the 3′ untranslated regions of messenger RNAs (mRNAs).12 MicroRNAs have gained more attention in regulating molecular pathogenesis of cancer recently, because each miRNA could suppress hundreds of targets that may play various roles in different tissues or cells. Many miRNAs act as crucial proto-oncogenes or tumor suppressors on tumor progression, especially gliomas. Recent study has demonstrated that the expression of miR-21, miR-10b, miR-155, and miR-210 was increased in gliomas and promoted the tumor progression.13–16 On the other hand, miR-181 and miR-128 were consistently downregulated in glioblastomas or glioma cell lines.17,18 The functional diversity of miRNAs makes it possible to be considered as early diagnosis biomarkers or new therapeutic targets.
It has been reported that expression of miR-139 was lower in many kinds of tumor cells and displayed a tumor suppressor function in early cancer development.19–21 Li’s research indicated that miR-139 could promote temozolomide (TMZ)-induced apoptosis of gliomas by targeting Mcl-1.22 However, previous report has not clearly determined the relationship between miR-139 expression and gliomas malignancy. Here, we demonstrated miR-139 expression was lower in malignant glioma tissues and glioma cell lines. Our study also identified new target genes of miR-139 that participated in regulating gliomas proliferation, migration, and invasion. miR-139 could act as a tumor suppressor by directly targeting insulin-like growth factor 1 receptor (IGF-1 R), associate of Myc 1 (AMY-1), and peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β) and thus inhibiting PI3K/AKT and c-Myc signaling pathways. These results suggested miR-139 could attenuate gliomas growth and may provide the novel strategies for gliomas therapy.
Materials and Methods
Human Tissue Samples
All human glioma tissues were obtained from patients undergoing surgery for glioma in the Department of Neurosurgery, Xijing Hospital, Fourth Military Medical University. The normal brain tissues were collected as negative controls from patients with cerebral trauma. The glioma samples were histologically classified by the diagnosis of clinical and pathological grading according to WHO guidelines. Written informed consent conforming to the tenets of the Declaration of Helsinki was obtained from each participant, and the study procedures were approved by the Institutional Review Board of the hospital.
Plasmid Construction, Cell Culture, and Transfection
The fragments of IGF-1 R, AMY-1, and PGC-1β 3′ untranslated regions (3′ UTRs) were amplified by polymerase chain reaction (PCR) from human complementary DNA library and inserted into pGL3-promotor vector (Promega, Madison, Wisconsin). The coding sequence region of IGF-1 R, AMY-1, and PGC-1β was also generated using PCR amplification and cloned in the expression vector pCMV-Myc (Clontech Laboratories, Inc, Mountain View, California).
Human cortical neuron cell line HCN-2 and glioma cell lines U251 and U87MG cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mmol/L glutamine (Invitrogen Life Technologies, Carlsbad, California). The passaged cells were seeded into 6-well plates and transfected on the next day using Lipofectamine 2000 (Invitrogen, Grand Island, NY) at a confluence of about 70% to 80%. Oligonucleotides were chemically synthesized and transfected into glioma cells at a final concentration of 50 nmol/L according to the manufacturer’s instructions (RiboBio, Guangzhou, China). The sequences of synthetic RNAs (siRNAs) for IGF-1 R, AMY-1, and PGC-1β are shown as follows: siIGF-1 R 1, 5′-tcttcaagggcaatttgctcattaa; siIGF-1 R 2, 5′-cagctagaagggaattactccttct; siAMY-1 1, 5′-gcatcacttaggagctgctactcca; siAMY-1 2, 5′-tcacttaggagctgctactccagaa; siPGC-1β 1, 5′-ccttgtcacaaagcgacccaacttt; and siPGC-1β 2, 5′-gaagagctttgagcagaccttgaca. After being transfected, cells were cultured in complete DMEM medium for definite periods of time and then harvested for further experiments. All cells were incubated at 37°C in an atmosphere of 5% carbon dioxide.
RNA Extraction and Quantification Assay
Total RNAs were extracted from human tissue specimens or cell lines with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Complementary DNA reverse transcribed by PrimerScript RT Reagent Kit (Takara, Dalian, China) was used to detect the expression levels of miR-139 downstream molecules and the epithelial–mesenchymal transition-related genes. Real-time reverse transcription PCR was performed using an SYBR Premix EX Taq kit (Takara) and the ABI PRISM 7500 Real-time PCR system. The primers used are as follows: IGF-1 R forward, 5′-tcgacatccgcaacgactatc-3′ and reverse, 5′-ccagggcgtagttgtagaagag-3′; AMY-1 forward, 5′-atggcccattacaaagccg-3′ and reverse, 5′-tttctggagtagcagctcctaa-3′; PGC-1β forward, 5′-gatgccagcgactttgactc-3′ and reverse, 5′-gatgccagcgactttgactc-3′; Zonula Occludens protein-1 forward, 5′-caacatacagtgacgcttcaca-3′ and reverse, 5′-cactattgacgtttccccactc-3′; E-cadherin forward, 5′-cgagagctacacgttcacgg-3′ and reverse, 5′-gggtgtcgagggaaaaatagg-3′; vimentin forward, 5′-gacgccatcaacaccgagtt-3′ and reverse, 5′-ctttgtcgttggttagctggt-3′; snail forward, 5′-tcggaagcctaactacagcga-3′ and reverse, 5′-agatgagcattggcagcgag-3′; slug forward, 5′-cgaactggacacacatacagtg-3′ and reverse, 5′-ctgaggatctctggttgtggt-3′; twist forward, 5′-gtccgcagtcttacgaggag-3′ and reverse, 5′-gcttgagggtctgaatcttgct-3′; glyceraldehyde-3-phosphate dehydrogenase primer forward, 5′-cttcaacgaccactttgt-3′ and reverse, 5′-tggtccaggggtcttact-3′. To analyze miR-139 expression levels, the Bulge-Loop miRNA quantitative real-time polymerase chain reaction (qRT-PCR) primer kits (RiboBio) were utilized according to the manufacturer’s instructions. RNA input was normalized to the level of human U6 small nuclear RNA.
Western Blotting
Cells were harvested and lysed on ice for 30 minutes in radioimmunoprecipitation assay buffer supplemented with protease inhibitors (100 mmol/L Tris-hydrochloric acid at pH 7.4, 150 mmol/L sodium chloride, 5 mmol/L EDTA, 1% Triton X-100, 1% deoxycholate acid, 0.1% sodium dodecyl sulfate, 2 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L sodium orthovanadate, 2 mmol/L dithiothreitol, 2 mmol/L leupeptin, and 2 mmol/L pepstatin). Lysates were centrifuged at 12 000 rpm for 10 minutes, and supernatants were collected as total proteins. Protein concentrations were determined by the bicinchoninic acid assay method (Beyotime, Haimen, China), and aliquots of protein lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to the polyvinylidene difluoride membrane. Membranes were blocked with 5% nonfat dried milk solution for 2 hours and then probed with primary antibodies, respectively. The antibodies used were against IGF-1 R, AMY-1, phospho-AKT, total AKT (Abcam, Cambridge, Massachusetts), PGC-1β, phospho-p38 MAPK, total p38 MAPK (Sigma-Aldrich, St Louis, Missouri), and β-actin (Boster Bio Tec, Wuhan, China). The proteins were then incubated with horseradish peroxidase-conjugated secondary antibodies and visualized with an enhanced chemiluminescence detection system. Protein expression was measured by ImageJ software.
Luciferase Reporter Assay
U251 cells were seeded in a 24-well plate 24 hours prior to the experiment and the pGL3-promoter vector bearing miR-139 recognized sequences, or the mutated sequences of the target genes were transfected by using Lipofectamine2000 reagent with miR-139 oligonucleotides and pRL-TK vector. Cells were harvested 24 hours later, and the relative luciferase activity was read out using the Dual Luciferase Reporter Assay System (Promega, Madison, Wisconsin) and the relative activity of Renilla luciferase was normalized to that of firefly luciferase. Each experiment was performed at least 3 times, and the data were analyzed with the Student’s t test.
Proliferation Assays
U251 and U87MG cells were seeded in 96-well plates with 5 × 103 cells/well and transfected with miR-139 oligonucleotides or negative control. Cell proliferation was evaluated at 24, 48, 72, and 96 hours after transfection using the methyl thiazolyl tetrazolium (MTT) reagent (5.0 mg/mL in phosphate-buffered saline [PBS]). After the incubation for 4 hours at 37°C, the supernatant was discarded, and 100 μL dimethyl sulfoxide was added in each well to dissolve the precipitate. Spectrophotometric absorbance was measured at the wavelength of 570 nm.
Cell Cycle Assay
Twenty-four hours after transfected with miR-139 or control, U251 and U87MG cells were collected in ice-cold PBS and fixed with 70% ice-cold ethanol for at least 12 hours at 4°C. The fixed cells were incubated with 50 mg/mL propidium iodide (PI; Sigma-Aldrich) and 20 g/mL RNase A (Takara) for 1 hours at 37°C in the dark. The DNA content was analyzed by flow cytometry.
Wound-Scratch Assay
U251 and U87MG cells were seeded in 6-well plates at a density of 2 × 105 cells/well and cultured overnight. Cells were transfected with miR-139 mimics or control at 80% confluence and scratched uniformly by using sterile plastic 200 μL micropipette tips 6 hours after the transfection. Then, the medium was replaced with fresh medium containing 10% FBS to wash out the exfoliated cells. The cells were photographed under a microscope (Olympus, Japan) 24 hours after the scratch. Each group was analyzed by measuring the gap area of wound healing and the migrated cells numbers in 3 different fields.
Cell Invasion and Migration Assay
U251 and U87MG cells (5 × 104) were loaded into the top chamber of a 24-wells invasion chamber assay plate in DMEM without serum. The lower chamber wells contained complete DMEM medium to induce cell invasion. After incubation for 24 hours at 37°C, cells that did not invade into the lower wells were removed from the top wells with a cotton swab, while the bottom cells were fixed with paraformaldehyde and then stained with 0.1% crystal violet. The migration and invasion cell numbers were measured by photographing at 5 random fields.
In Vivo Tumor Growth
Six week-old nude mice (Male BALB/cA-nu) were purchased from the Shanghai Experimental Animal Center (Chinese Academy of Sciences, Shanghai, China) and maintained in specific pathogen-free conditions. Ten mice were randomly divided into 2 groups. U87MG cells stably expressing miR-139 and negative control were injected subcutaneously into the right flank of each mouse with 5 × 106 cells in 2 groups. Tumor size was monitored using sliding caliper measurements every 3 days, and tumor volume was calculated according to the formula: volume = 0.51 × length × width2. The mice were killed after 20 days. The tumors were excised to weigh, and portions were frozen in liquid nitrogen for further analyses.
Statistical Analysis
Data were analyzed with the SPSS 12.0 software. Comparisons between groups were performed with an unpaired Student’s t test using Graph Pad Prism 5 software, version 5.0. Results were expressed as mean ± SD. The value of P of less than .05 was considered statistically significant.
Results
Aberrant Expression of miR-139 in Human Gliomas and Cell Lines
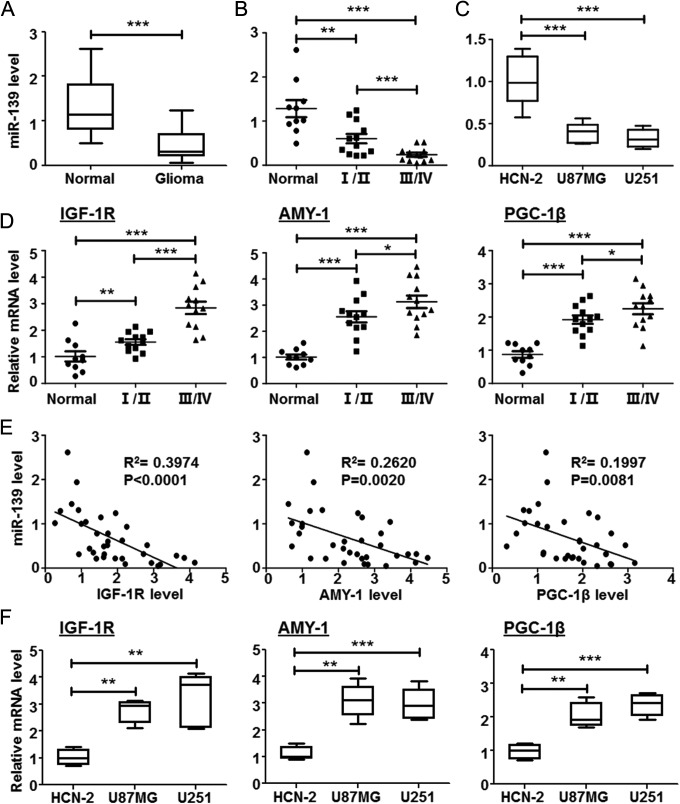
Accumulating evidence has demonstrated that miRNAs play important roles in human glioma progression. The miRNA array from several groups indicated that miR-139 decreased expression in glioma patients’ tumor tissues.23–26 To confirm the miR-139 level in gliomas, we examined its expression in frozen glioma tissues and normal brain tissues by qRT-PCR. Compared with normal brain tissues, miR-139 was significantly downregulated in glioma tissues, with a greater decrease in high-grade (WHO grades III and IV) than low-grade (WHO grades I and II) gliomas, indicating that miR-139 expression correlated with glioma malignancy (Figure 1A and B). Consistently, we further detected miR-139 in different human glioma cell lines and normal brain cells. Our results displayed that miR-139 expression was lower in U87MG and U251, compared with that in normal brain cells (Figure 1C).
Figure 1.
miR-139 expression in glioma tissues and cell lines. A, The total RNA was extracted from the normal and glioma tissues excised from patients undergoing surgery. The miR-139 expression levels were detected by reverse transcription polymerase chain reaction (RT-PCR; normal: n = 10; gliomas: n = 24). B, The gliomas were divided into 2 groups according to the World Health Organization (WHO) guidelines. miR-139 expression was compared in gliomas with different grades (grade I/II: n = 12; grade III/IV: n = 12). C, miR-139 expression was compared in brain cells and different glioma cell lines (n = 6). D, The expression of insulin-like growth factor type 1 receptor (IGF-1 R), associate of Myc-1 (AMY-1), and peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β) in normal brain tissues and different grades glioma tissues. E, The linear regression relationship between the expression of miR-139 and its target genes. F, The expression of IGF-1 R, AMY-1, and PGC-1β in brain cells and glioma cell lines. Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
To fully understand the molecular mechanisms of glioma cells progression modulating by miR-139, we identified the target genes predicted by several bioinformatic algorithms (Target Scan, PicTar, and miRDB) to increase the specificity and accuracy of prediction. All of the 3 programs predicted IGF-1 R, AMY-1, and PGC-1β as candidate targets of miR-139. Meanwhile, the mRNA levels of IGF-1 R, AMY-1, and PGC-1β were detected in human glioma tissues, and the expressions were both increased along with the malignant grade raising (Figure 1D), which were inversely correlated with the expression level of miR-139 (Figure 1E). The similar expression profile was also validated in U87MG and U251 cells (Figure 1F). Taken together, these results indicated expression of miR-139, and its predicted targets was altered as gliomas progression.
Overexpression of Decreased Proliferation in Glioma
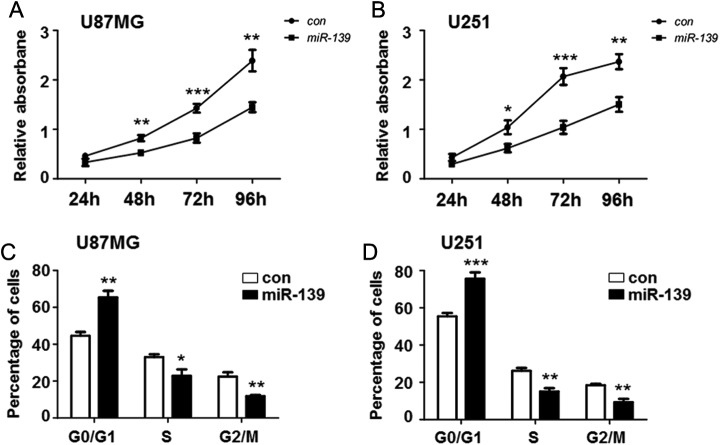
We had demonstrated the downregulation of miR-139 in glioma cells. However, the biological functions of miR-139 in glioma progression and whether it could act as an anti-oncomir remained unclear. To investigate the effect of miR-139 on glioma cells proliferation, U87MG and U251 cells were transfected with miR-139 mimics or scramble control to analyze viability and growth. Methyl thiazolyl tetrazolium assay showed that the viability of U87MG and U251 cells was significantly lower in the miR-139 overexpression group than the control group (Figure 2A and B).
Figure 2.
miR-139 repressed gliomas proliferation. A and B, U251 and U87MG cells were seeded in 96-well plates after transfecting miR-139 or control oligonucleotide. The cell proliferation was evaluated at 24, 48, 72, and 96 hours (n = 5). C and D, U251 and U87MG cells were overexpressing miR-139 as above and collected for further flow cytometry analysis to detect the cell cycle (n = 4). Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
To provide additional evidence to support this result, cell cycle analyses were performed using flow cytometry. The PI staining showed that after miR-139 overexpressing, the numbers of U87MG and U251 cells in G0/G1 phases were significantly increased, whereas those in S phase and G2/M phases were significantly reduced (Figure 2C and D). These results showed that miR-139 overexpression contributes to regulation of glioma cell proliferation in vitro.
Overexpression of miR-139 Suppressed Glioma Cells Migration and Invasion
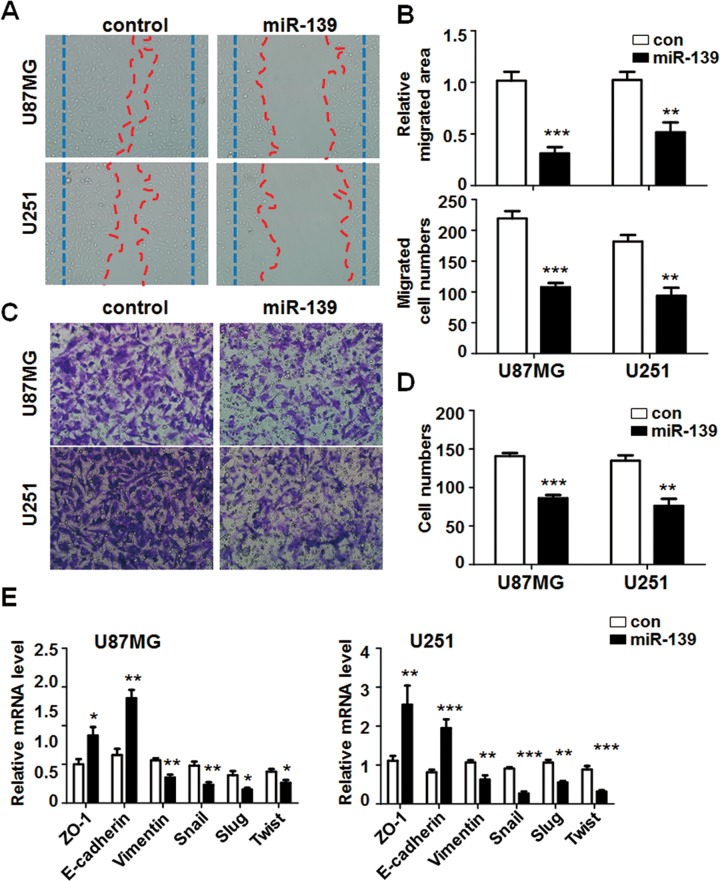
Except the sustaining proliferation, activating invasion and metastasis capability are also significant hallmarks of cancer, especially for malignant gliomas. To evaluate the effect of miR-139 expression on glioma cells migration and invasion, wound-healing assay and matrigel invasion assay were performed. We transfected miR-139 oligonucleotide or control into U87MG and U251 cells and analyzed the wound-healing areas and the migrated cell numbers. The results showed that artificial wound gap was almost completely closed in the control group; nevertheless that of miR-139 overexpressed U87MG cells remained wide open (Figure 3A, top panel). The migration areas and migrated cell numbers were both reduced in miR-139 overexpressed cells (Figure 3B). The consistent results were observed at U251 cells (Figure 3A, bottom panel and 3B). In addition, we seeded U87MG and U251 cells overexpressing miR-139 or control in the chamber to detect the invasion of glioma cells. The transwell matrix penetration assay indicated that miR-139 upregulation in U87MG and U251 cells significantly reduced cell invasive ability in vitro (Figure 3C and D).
Figure 3.
miR-139 repressed gliomas migration and invasion. A and B, U87MG and U251 cells were transfected miR-139 or control. The wound-healing assay was performed to evaluate the migration ability (n = 4). C and D, U87MG and U251 cells were treated as above, and the invasion was detected by using transwell assay (n = 4). E, Total RNA was extracted from U87MG and U251 cells transfected miR-139 or control. The expression of epithelial-mesenchymal transition (EMT markers was tested (n = 4). Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
The EMT is one of the major reasons of glioblastomas recurrence.27 Inhibiting EMT could repress the motility of gliomas. We detected the EMT markers’ expression level in miR-139-transfected glioma cells, which showed miR-139 suppressed gliomas EMT obviously (Figure 3E and F). These results indicated that miR-139 could inhibit the migration and invasion capacity of glioma cells.
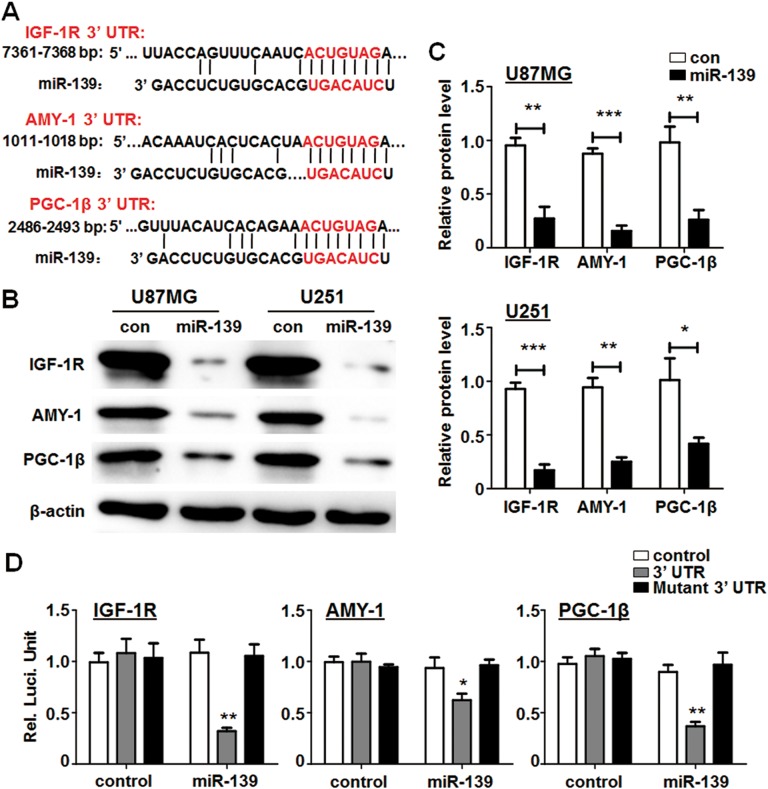
miR-139 Directly Targeted IGF-1 R, AMY-1, and PGC-1β
Figure 4A shows the 3′ UTR of IRG-1 R, AMY-1, and PGC-1β harbored conserved miR-139 binding site. Further detection suggested that the protein levels of IGF-1 R, AMY-1, and PGC-1β were significantly reduced in U87MG and U251 cells after transfected with miR-139 for 48 hours using Western blotting analysis (Figure 4B and C). Moreover, luciferase assays were performed using the reporter vector containing the 3′ UTR sequences or the mutated versions of the 3 target genes. The results displayed miR-139 could modulate IGF-1 R, AMY-1, and PGC-1β expression dependent on specific binding on the 3′ UTRs (Figure 4D). The above results demonstrated that IGF-1 R, AMY-1, and PGC-1β were primary targets of miR-139 in regulating the activity of glioma cells.
Figure 4.
miR-139 reduced the expression of its target genes. A, Sequence of the 3′ untranslated region (3′ UTR) of insulin-like growth factor type 1 receptor (IGF-1 R), associate of Myc-1 (AMY-1), and peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β) matched with the recognition site of miR-139. The seed sequence was marked in red color. B and C, Protein levels of IGF-1 R, AMY-1, and PGC-1β were regulated by miR-139 both in U87MG and U251 cells (n = 4). D, pGL3-promoter vector with the 3′ UTR of IGF-1 R, AMY-1, and PGC-1β (gray box), the mutant 3′ UTR regions (black box) or the control (white box) were co-transfected into U87MG cells with an miR-139 mimic or control mimic respectively. Twenty-four hours after transfection, luciferase activity was determined (n = 5). Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
Molecular Mechanisms of miR-139 Modulating Glioma Cells Progression Through Target Genes
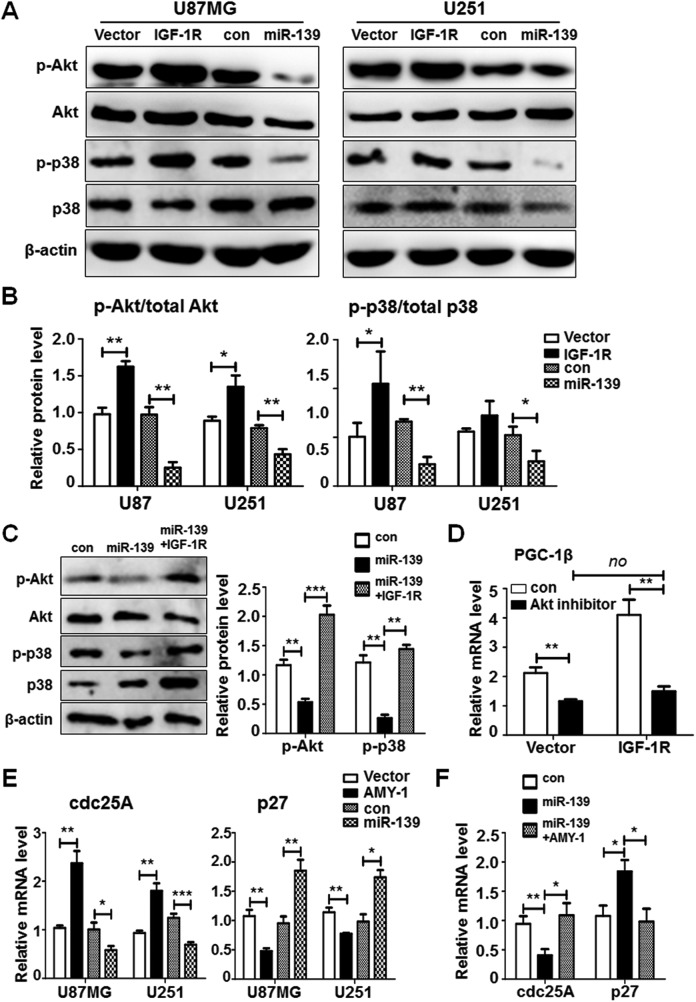
Insulin-like growth factor 1 receptor belongs to a member of the insulin receptor subsets of receptor tyrosine kinases and is critical for cells’ growth, development, and oncogenic transformation.28 Several reports have shown that the IGF-1 R is highly activated in tumor cells. Insulin-like growth factor 1 receptor-induced activation of PI3K-Akt and MAPK signaling took rise to GBM growth by preventing apoptosis and promoting cell survival.29,30 We demonstrated that the activation of Akt and MAPK signaling was precisely increased in IGF-1 R overexpressed U87MG and U251 cells and repressed in miR-139 transfected cells (Figure 5A and B). What’s more, the overexpression of IGF-1 R could rescue the level of phosphorylated Akt and p38, which indicated miR-139 prevented Akt and MAPK activation through targeting IGF-1 R (Figure 5C). Peroxisome proliferator-activated receptor γ coactivator 1β is a coactivator of peroxisome proliferator-activated receptor γ (PPARγ) and its function remains unclear. However, the PGC-1β protein family were found high expressed in all kinds of solid tumors.31,32 Our data manifested that PGC-1β expression level was also elevated when IGF-1 R was overexpressed (Figure 5D). Further detection suggested the inhibitor of Akt could suppress the PGC-1β expression even when IGF-1 R was transfected (Figure 5D), which was also validated in breast cancer.33 The increased expression of PGC-1β by activation of IGF-1R-Akt axis may be the new mechanism of IGF-1 R modulation on tumor formation.
Figure 5.
miR-139 inhibited Akt, MAPK, and c-Myc signaling through insulin-like growth factor type 1 receptor (IGF-1 R) and associate of Myc-1 (AMY-1). A and B, U87MG and U251 cells were transfected with IGF-1 R or miR-139 and empty vector or scramble as negative controls. The protein expression of downstream signaling was detected (n = 4). C, Insulin-like growth factor type 1 receptor was transfected into miR-139 overexpressing U87MG cells and the total and phosphorylation level of Akt and MAPK were detected (n = 3). D, The glioma cells were overexpressing IGF-1 R and added with Akt inhibitor at the same time. The peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β) expression was detected (n = 4). E, The glioma cells were transfected with AMY-1 or miR-139 and the expression level of c-Myc signaling molecules was evaluated (n = 4). F, AMY-1 was transfected into miR-139 overexpressing U87MG cells and the RNA levels of cell division cycle 25 homologue A (cdc25A) and p27 were detected (n = 3). Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
AMY-1 has been identified to be a coactivator of c-Myc, which is known to contribute to cell cycle progression, transformation, and apoptosis induction as an oncogene.34 It has been reported that c-Myc signaling could activate cell division cycle 25 homologue A (cdc25A) transcription while inhibiting p27 expression, which were important to cell cycles.35–37 We detected the RNA levels of c-Myc signaling downstream molecules after overexpressing AMY-1 or miR-139. Cell division cycle 25 homologue A was increased by AMY-1 and partially suppressed by miR-139, whereas the p27 expression was opposite to cdc25A (Figure 5E). When transfected AMY-1 as well as miR-139 in U87MG cells, the expression of cdc25A and p27 was retrieved, suggesting miR-139 modulates c-Myc signaling in glioma cells through AMY-1 dependent way (Figure 5F). These results suggested that miR-139 modulated IGF-1 R, AMY-1, and PGC-1β to suppress glioma progression by different molecular mechanisms.
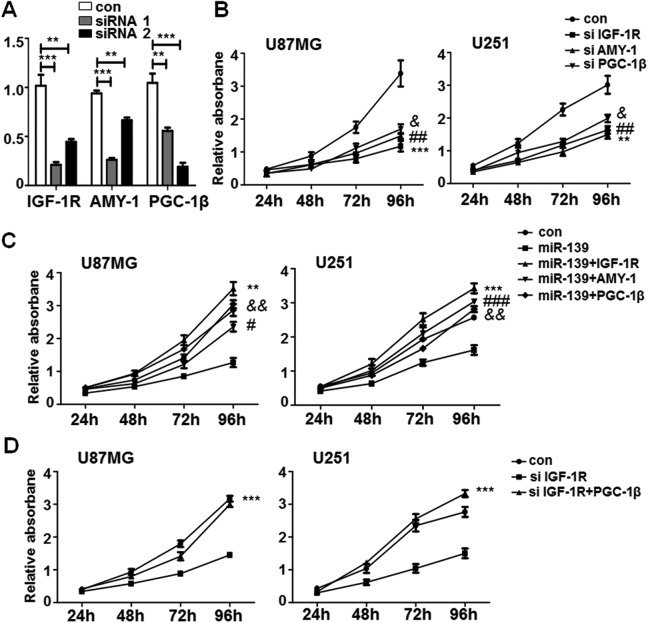
To confirm the functions of miR-139 in glioma cells were based on the regulation of its targets, the specific siRNAs for IGF-1 R, AMY-1, and PGC-1β were designed to knockdown their expression. Figure 6A showed that siRNA 1# for IGF-1 R, siRNA 1# for AMY-1, and siRNA 2# for PGC-1β were more effective. We transfected these assessed siRNAs into U87MG and U251 cells to evaluate the proliferation and invasion ability by MTT and transwell assay. The MTT results indicated that siRNAs of IGF-1 R, AMY-1, and PGC-1β could repress glioma cells proliferation (Figure 6B), and the effects of miR-139 on U87MG and U251 proliferation were restored when overexpressing IGF-1 R, AMY-1, or PGC-1β (Figure 6C). We have demonstrated IGF-1 R increased PGC-1β expression through Akt signaling pathway (Figure 5D). However, whether IGF-1 R promoting gliomas growth by regulating PGC-1β remains unclear. We increased PGC-1β expression on the condition of IGF-1 R was knocked down. The proliferation rate of both U87MG and U251 was improved obviously in the PGC-1β transfection group (Figure 6D). These results indicated that the capability of IGF-1 R on promoting gliomas growth was due to upregulating PGC-1β.
Figure 6.
miR-139 repressed glioma cells proliferation through insulin-like growth factor type 1 receptor (IGF-1 R), associate of Myc-1 (AMY-1), and peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β). A, The knock down efficiency of synthetic RNAs (siRNAs) for IGF-1 R, AMY-1, and PGC-1β (n = 4). B, The glioma cells were transfected with effective siRNAs of IGF-1 R, associate of Myc-1 (AMY-1), and PGC-1β and detected the proliferation by methyl thiazolyl tetrazolium (MTT; n = 5). C, The glioma cells were transfected IGF-1 R, associate of Myc-1 (AMY-1), and PGC-1β in the condition of miR-139 overexpressed. The MTT assay was performed to tested proliferation of glioma cells (n = 5). D, The glioma cells were transfected siRNA of IGF-1 R with PGC-1β together and tested the proliferation (n = 5). Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
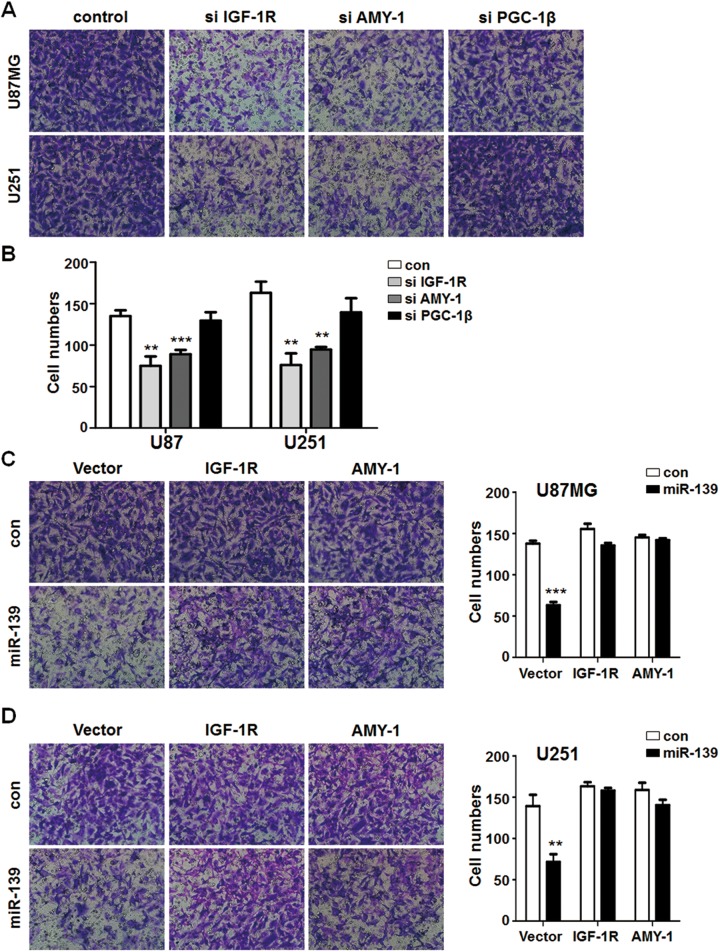
Simultaneously, siRNA of IGF-1 R and AMY-1 but not PGC-1β decreased glioma cells invasion function (Figure 7A and B). Meanwhile, overexpression of IGF-1 R and AMY-1 reversed the effects of miR-139 oligonucleotides in U87MG and U251 cells. The invasion ability of glioma cells was completely retrieved with different degrees in different groups (Figure 7C and D). The consequence above suggested miR-139 modulated glioma cells proliferation through IGF-1 R, AMY-1, and PGC-1β directly or indirectly, and modulated those migration through IGF-1 R and AMY-1.
Figure 7.
miR-139 reduced glioma cells invasion through IGF-1 R and associate of Myc-1 (AMY-1). A and B, U87MG and U251 cells were transfected the synthetic RNAs (siRNAs) for target genes of miR-139 and seeded into the transwell chambers. The cells numbers of infiltrated gliomas were counted (n = 4). C and D, The different glioma cell lines that were transfected miR-139 were overexpressed IGF-1 R and associate of Myc-1 (AMY-1) to rescue. The migrated cell numbers were calculated (n = 4). Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
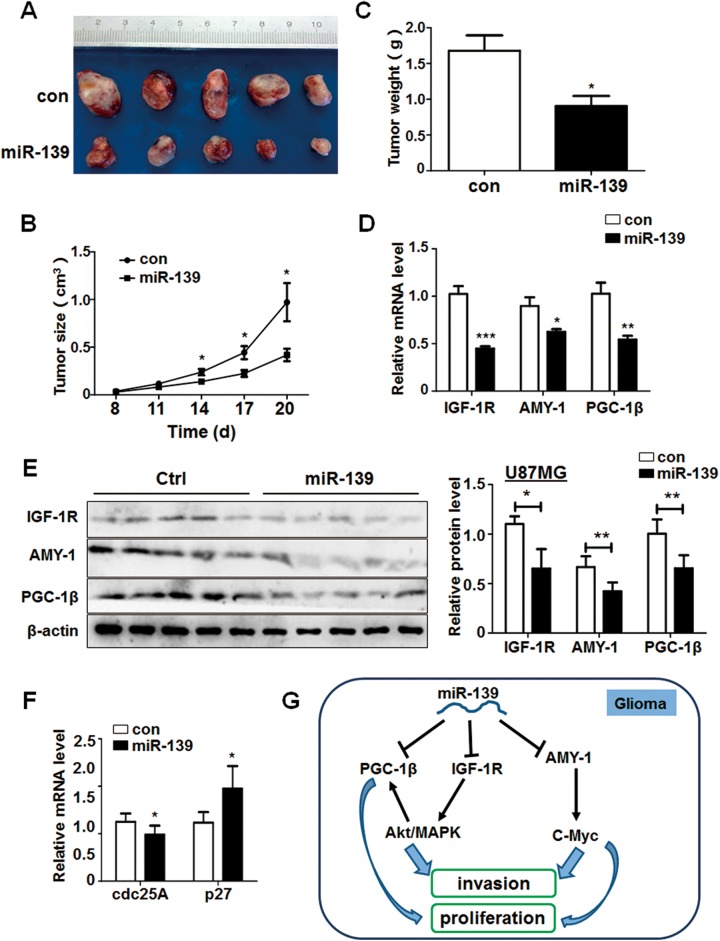
miR-139 Suppressed Glioma Growth in Vivo Through Akt Signaling
We have demonstrated the noticeable antitumor effect of miR-139 to gliomas in vitro. To further validate its function, an in vivo tumor model was employed. We constructed U87MG cells that stably expressed miR-139 or negative control and injected the modified glioma cells subcutaneously into the right flank of nude mice. The size of tumors was monitored at 10 days after injection and measured every 3 days. The tumors of miR-139 overexpressed group developed slowly than the control mice (Figure 8A and B). Meanwhile, the comparison of tumors weight between the 2 groups was consistent with the tumors volume (Figure 8C). The mRNA and protein expression levels of IGF-1 R, AMY-1, and PGC-1β in xenografts from the miR-139 group were significantly lower than those from the control group (Figure 8D and E). Furthermore, the expression of c-Myc associated molecules altered noticeably. Cell division cycle 25 homologue A was decreased, and p27 was increased simultaneously in miR-139 overexpression group (Figure 8F), confirming that miR-139 suppressed gliomas progression in vivo through target genes and regulation c-Myc signaling pathway.
Figure 8.
miR-139 plays an antitumor function and inhibit the Akt signaling in vivo. A, U87MG cells stably expressing miR-139 and negative control were injected subcutaneously in 2 groups, respectively. The phenotype of tumors was observed after 20 days (n = 5). B and C, The growth of tumors was monitored by measuring tumor size every 3 days from the 8th day, and the tumors’ weight was measured after the mice were killed. D and E, The RNA and protein were extracted from the tumors of different groups, and the expression of IGF-1 R, associate of Myc-1 (AMY-1), and peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β) were detected. F, The RNA levels of c-Myc pathway downstream genes were determined. G, The schematic diagram of miR-139 modulated gliomas progression. Bars represent means ± SD, *P < .05, **P < .01, ***P < .001.
Discussion
MicroRNAs abnormal expression is a common consequence or character of human tumors and other diseases. With the development of detecting method, we could evaluate the total miRNAs expression through microarrays and next generation sequencing. Several groups’ miRNAs profile data displayed that miR-139 was decreased in the high-grade gliomas compared with the normal brain or tumor-adjacent tissues.23–26 Recent research indicated miR-139 suppressed the proliferation of glioma cells and enhanced the proapoptosis effect of TMZ through direct posttranscriptional regulation of Mcl-1.22 However, the roles and mechanisms of miR-139 in gliomas progression are not completely known. Our study confirmed the downregulation of miR-139 in high-grade glioma tissues and glioma cell lines. Furthermore, we demonstrated IGF-1, AMY-1, and PGC-1β were target genes of miR-139 using Western blot and reporter assay. What is more, all of the 3 genes were inversely correlated to miR-139 in gliomas and regulated gliomas proliferation, migration, and invasion, respectively.
Insulin-like growth factor 1 receptor is essential for the development and progression of cancer cells. Meanwhile, it is a crucial determinant of the response to radiation and chemotherapy for patients with glioma. The dysregulation of IGF-1 R has always suggested an aggressive phenotype and poor prognostic outcome in various tumor types. A mass of evidence has displayed the activation of PI3K/Akt and p38 MAPK signaling when IGF-1 R is elevated in cancer cells. The activated Akt was considered to be associated with the grade of gliomas malignancy. In vitro, Akt could promote proliferation and invasiveness by regulating cyclin D and MMP9, respectively.38,39 Besides, the phosphorylated Akt could also suppress the inhibitory effects on cell cycle through phosphorylating p27 and p21 in turn.38 Associate of Myc 1 was also known as c-Myc-binding protein, which was firstly identified to interact with c-Myc and enhance its E-box-dependent transactivation. A large amount of experimental data clearly demonstrated that c-Myc protein regulated multiple cellular functions correlated with the malignant phenotype, including cancer cells proliferation, transformation, and tumor metastasis.37 Our study demonstrated that miR-139-induced potent suppression in glioma cell lines proliferation, survival, migration, and invasion, which is similar to the results of siRNAs for IGF-1 R and AMY-1. Simultaneously, overexpression of IGF-1 R or AMY-1 could completely rescue the phenotypes forced by miR-139.
However, we discovered that PGC-1β was also downregulated by miR-139 by direct targeting. PGC-1β belongs to the PPARγ co-activators 1 (PGC-1s) family, which has multiple functions on regulating cellular metabolism and survival. It has also been reported that PGC-1s were upregulated and played an important role in kinds of cancer. The high expression of PGC-1β always accompany with malignant proliferation and a poor prognosis in breast cancer.40,41 When PGC-1β was suppressed by specific siRNA, the MCF-7 cells resistant to tamoxifen were retrieved the sensitivity.42 Besides, McDonnell’s research showed that (IGF-1 R)-(c-Myc) regulating axis could activate the expression of PGC-1β in breast cancer, which had demonstrated to be one of the mechanisms to elevate PGC-1β.33 Our study demonstrated that PGC-1β played a broadly role on gliomas growth. Synthetic RNA of PGC-1β could inhibit the proliferation of glioma cell lines but no effect on gliomas motivation capacity; meanwhile overexpression of PGC-1β restored the proliferation deficiency phenotype induced by miR-139 or siRNAs of IGF-1 R. According to these above data, we made the conclusion that miR-139 attenuated the development and progression of glioma cells by targeting the downstream genes IGF-1 R, AMY-1, and PGC-1β. What’s more, IGF-1 R suppressed gliomas proliferation through a PGC-1β–dependent pathway. Since IGF-1 R could activate PI3K/Akt signaling simultaneously, we considered that IGF-1 R inhibited the migration of gliomas mediated by other mechanisms except PGC-1β.
In summary, our results first describe the tumor-suppressing function of miR-139 and its molecular mechanisms in gliomas (Figure 8G). The in vivo experiments revealed the fact that gliomas overexpressed miR-139 displayed a weaker tumorigenic ability, which indicated miR-139 would be beneficial for cancer treatment. Although many of the mechanisms still remain unclear for miR-139-based suppression of gliomas, our findings are encouraging and might provide a new therapeutic strategy to gliomas prevention and treatment.
Acknowledgments
This study was supported by grants from NSFC 81172095.
Abbreviations
- AMY-1
associate of Myc-1
- Cdc25A
cell division cycle 25 homologue A
- cDNA
complementary DNA
- CNS
central nervous system
- DMEM
Dulbecco’s Modified Eagle’s Medium
- EMT
epithelial-mesenchymal transition
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GBM
glioblastoma multiforme
- IGF-1 R
insulin-like growth factor-1 receptor
- Mcl-1
myeloid cell leukemia 1
- miRNA
microRNA
- MMP9
matrix metalloproteinase 9
- MTT
methyl thiazolyl tetrazolium
- PCR
polymerase chain reaction
- PGC-1β
peroxisome proliferator-activated receptor γ coactivator 1β
- PI
propidium iodide
- PPARγ
peroxisome proliferator-activated receptor γ
- qRT-PCR
quantitative real-time polymerase chain reaction
- siRNA
synthetic RNA
- TMZ
temozolomide
- UTR
untranslated region
- WHO
World Health Organization
- ZO-1
Zonula Occludens protein 1
- miR-139
MicroRNA-139-5p
- Akt
phosphatidylinositol 3 kinase /protein kinase B
- MAPK
mitogen-activated protein kinases
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 4. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 5. Butowski NA, Sneed PK, Chang SM. Diagnosis and treatment of recurrent high-grade astrocytoma. J Clin Oncol. 2006;24(8):1273–1280. [DOI] [PubMed] [Google Scholar]
- 6. Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lopez-Ochoa S, Ramirez-Garcia M, Castro-Sierra E, Arenas-Huertero F. Analysis of Chromosome 17 miRNAs and Their Importance in Medulloblastomas. BioMed Res Int. 2015;2015:717509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwak SY, Kim BY, Ahn HJ, et al. Ionizing radiation-inducible miR-30e promotes glioma cell invasion through EGFR stabilization by directly targeting CBL-B. FEBS J. 2015;282(8):1512–1525. [DOI] [PubMed] [Google Scholar]
- 12. Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. [DOI] [PubMed] [Google Scholar]
- 13. Zhang KL, Han L, Chen LY, et al. Blockage of a miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in glioblastomas. Cancer Lett. 2014;342(1):139–149. [DOI] [PubMed] [Google Scholar]
- 14. Liu Q, Zou R, Zhou R, et al. miR-155 Regulates Glioma Cells Invasion and Chemosensitivity by p38 Isforms in vitro. J Cellular Biochem. 2015;116(7):1213–1221. [DOI] [PubMed] [Google Scholar]
- 15. Guessous F, Alvarado-Velez M, Marcinkiewicz L, et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112(2):153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lai NS, Wu DG, Fang XG, et al. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer. 2015;112(suppl):1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peruzzi P, Bronisz A, Nowicki MO, et al. MicroRNA-128 coordinately targets Polycomb Repressor Complexes in glioma stem cells. Neuro Oncol. 2013;15(9):1212–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi ZM, Wang XF, Qian X, et al. MiRNA-181b suppresses IGF-1 R and functions as a tumor suppressor gene in gliomas. RNA. 2013;19(4):552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krishnan K, Steptoe AL, Martin HC, et al. miR-139-5p is a regulator of metastatic pathways in breast cancer. RNA. 2013;19(12):1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yue S, Wang L, Zhang H, et al. miR-139-5p suppresses cancer cell migration and invasion through targeting ZEB1 and ZEB2 in GBM. Tumour Biol. 2015;36(9):6741–6749. [DOI] [PubMed] [Google Scholar]
- 21. Zhang HD, Jiang LH, Sun DW, Li J, Tang JH. MiR-139-5p: promising biomarker for cancer. Tumour Biol. 2015;36(3):1355–1365. [DOI] [PubMed] [Google Scholar]
- 22. Li RY, Chen LC, Zhang HY, et al. MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neurosci Ther. 2013;19(7):477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lavon I, Zrihan D, Granit A, et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol. 2010;12(5):422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lages E, Guttin A, El Atifi M, et al. MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PloS One. 2011;6(5):e20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PloS One. 2011;6(9):e24248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. [DOI] [PubMed] [Google Scholar]
- 27. Kubelt C, Hattermann K, Sebens S, Mehdorn HM, Held-Feindt J. Epithelial-to-mesenchymal transition in paired human primary and recurrent glioblastomas. Int J Oncol. 2015;46(6):2515–2525. [DOI] [PubMed] [Google Scholar]
- 28. Girnita L, Worrall C, Takahashi S, Seregard S, Girnita A. Something old, something new and something borrowed: emerging paradigm of insulin-like growth factor type 1 receptor (IGF-1 R) signaling regulation. Cell Mol Life Sci. 2014;71(13):2403–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osuka S, Sampetrean O, Shimizu T, et al. IGF1 receptor signaling regulates adaptive radioprotection in glioma stem cells. Stem Cells. 2013;31(4):627–640. [DOI] [PubMed] [Google Scholar]
- 30. Guo T, Feng Y, Liu Q, et al. MicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1 R. Tumour Biol. 2014;35(11):11269–11275. [DOI] [PubMed] [Google Scholar]
- 31. Ellis HP, Kurian KM. Biological Rationale for the Use of PPARgamma Agonists in Glioblastoma. Front Oncol. 2014;4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones AW, Yao Z, Vicencio JM, Karkucinska-Wieckowska A, Szabadkai G. PGC-1 family coactivators and cell fate: roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion. 2012;12(1):86–99. [DOI] [PubMed] [Google Scholar]
- 33. Chang CY, Kazmin D, Jasper JS, Kunder R, Zuercher WJ, McDonnell DP. The metabolic regulator ERRalpha, a downstream target of HER2/IGF-1 R, as a therapeutic target in breast cancer. Cancer Cell. 2011;20(4):500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taira T, Maeda J, Onishi T, et al. AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC. Genes Cells. 1998;3(8):549–565. [DOI] [PubMed] [Google Scholar]
- 35. Zornig M, Evan GI. Cell cycle: on target with Myc. Current biology: CB. 1996;6(12):1553–1556. [DOI] [PubMed] [Google Scholar]
- 36. Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66(3):1289–1293. [DOI] [PubMed] [Google Scholar]
- 37. Pelengaris S, Khan M. The many faces of c-MYC. Arch Biochem Biophys. 2003;416(2):129–136. [DOI] [PubMed] [Google Scholar]
- 38. Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal. 2002;14(5):381–395. [DOI] [PubMed] [Google Scholar]
- 39. Das G, Shiras A, Shanmuganandam K, Shastry P. Rictor regulates MMP-9 activity and invasion through Raf-1-MEK-ERK signaling pathway in glioma cells. Mol Carcinog. 2011;50(6):412–423. [DOI] [PubMed] [Google Scholar]
- 40. Li Y, Wedren S, Li G, et al. Genetic variation of ESR1 and its co-activator PPARGC1B is synergistic in augmenting the risk of estrogen receptor-positive breast cancer. Breast Cancer Res. 2011;13(1):R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez-Nava GA, Burguete-Garcia AI, Lopez-Carrillo L, Hernandez-Ramirez RU, Madrid-Marina V, Cebrian ME. PPARgamma and PPARGC1B polymorphisms modify the association between phthalate metabolites and breast cancer risk. Biomarkers. 2013;18(6):493–501. [DOI] [PubMed] [Google Scholar]
- 42. Deblois G, Chahrour G, Perry MC, Sylvain-Drolet G, Muller WJ, Giguere V. Transcriptional control of the ERBB2 amplicon by ERRalpha and PGC-1beta promotes mammary gland tumorigenesis. Cancer Res. 2010;70(24):10277–10287. [DOI] [PubMed] [Google Scholar]