Abstract
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous in the human environment. Since they are present in crude oilfractions used for the production of rubber and plastics, consumers may come into direct dermal contacts with these compounds (e.g., via tool handles) on a daily basis. Some individual PAHs are identified as genotoxic mutagens thereby prompting particular toxicological and environmental concern. Among this group, benzo[a]pyrene (BAP) constitutes a model carcinogen which is also used as reference compound for risk assessment purposes. It acts as a strong agonist of the aryl hydrocarbon receptor (AHR) and becomes metabolically activated toward mutagenic and carcinogenic intermediates by cytochrome P450-dependent monooxygenases (CYPs). While BAP has been exhaustively characterized with regard to its toxicological properties, there is much less information available for other PAHs. We treated an AHR-proficient immortal human keratinocyte cell line (i.e., HaCaT) with three selected PAHs: BAP, chrysene (CRY) and dibenzo[a,l]pyrene (DALP). Compound-mediated alterations of endogenous metabolites were investigated by an LC–MS/MS-based targeted approach. To examine AHR-dependent changes of the measured metabolites, AHR-deficient HaCaT knockdown cells (AHR-KD) were used for comparison. Our results reveal that 24 metabolites are sufficient to separate the PAH-exposed cells from untreated controls by application of a multivariate model. Alterations in the metabolomics profiles caused by each PAH show influences on the energy and lipid metabolism of the cells indicating reduced tricarboxylic acid (TCA) cycle activity and β-oxidation. Up-regulation of sphingomyelin levels after exposure to BAP and DALP point to pro-apoptotic processes caused by these two potent PAHs. Our results suggest that in vitro metabolomics can serve as tool to develop bioassays for application in hazard assessment.
Abbreviations: AAs, amino acids; ACs, acyl carnitines; AHR, aryl hydrocarbon receptor; BAs, biogenic amines; BAP, benzo[a]pyrene; CRY, chrysene; DALP, dibenzo[a,l]pyrene; LPCs, lysophosphatidylcholines; PAHs, polycyclic aromatic hydrocarbons; PCs, phosphatidylcholines; SMs, sphingomyelines
Keywords: Polycyclic aromatic hydrocarbons, Metabolomics, Aryl hydrocarbon receptor, Keratinocytes
1. Introduction
In daily life, consumers are exposed to a variety of potentially toxic compounds that may be released from consumer products [1], [2]. For instance, polycyclic aromatic hydrocarbons (PAHs) are ubiquitous in the human environment. Generated from incomplete combustion of organic matter like coal, grilled meat or cigarettes they are present in air, food and soil [3], [4], [5]. Furthermore, PAHs occur in crude oil fractions and carbon black used for the production of rubber, certain plastics and elastomers [6], [7]. Dermal contact with consumer products containing petroleum and subsequent dermal uptake of diesel oil and other lubricants were identified in human volunteer trial experiments [8]. Therefore, direct skin contact (e.g., via tool handles or lubricants and fuels) in combination with possible migration is supposed to be an important route of exposure for various PAHs and percutaneous absorption of PAHs has been shown before [9], [10], [11]. Moreover, a strong correlation between dermal exposure to PAHs and the occurrence of skin cancer has been reported in the past [12].
Carcinogenic PAHs require activation through biotransformation to form reactive intermediates and to target biological macromolecules such as DNA [5]. Cytochrome P450-dependent monooxygenases (CYPs), in particular CYP1A1, 1A2 and 1B1, are primarily involved in the metabolism of PAHs [13], [14]. Enzymatic generation of sterically hindered bay or fjord region diol-epoxides and subsequent DNA adduct generation is considered the dominant mechanism in chemical carcinogenesis resulting in mutational miscoding [15]. Many bay region PAHs such as benzo[a]pyrene (BAP) are activated through CYP1A1 [16]. By contrast, CYP1B1 seems to play a more important role in the metabolism of fjord region PAHs such as dibenzo[a,l]pyrene (DALP; IUPAC notation: dibenzo[def,p]chrysene) [17]. Besides activation through CYP enzymes, two other activation pathways are important in PAH metabolism: the formation of radical cations by one electron oxidation via peroxidases [18], and the formation of o-quinones by dihydrodiol dehydrogenases [19]. It is assumed that all three pathways—to different extents—might contribute to PAH-mediated carcinogenesis [20]. Most PAHs act as activators of the aryl hydrocarbon receptor (AHR) [21]. AHR activation then leads to gene expression of metabolizing enzymes such as CYPs, which—in turn—contribute to the formation of activated PAH metabolites and subsequent DNA damage. Besides its role in the enzymatic biotransformation of xenobiotics, the AHR may also contribute more directly to alterations in cell signaling and proliferation [22]. The complexity of PAH toxicity is further illustrated by the fact that the level of DNA adduct formation does not necessarily correlate with the extent of tumor formation in skin [23], [24].
BAP was classified as human carcinogen by IARC [25]. By contrast, chrysene (CRY) has less carcinogenic activity in comparison to BAP. It also contains a bay region that can be converted into reactive diol-epoxide moieties though [26], which then may induce cancer in mice [27], [28]. DALP is one of the strongest carcinogenic PAHs containing a fjord region [29]. The described toxicological mechanisms triggered by different PAHs might lead to interactions and non-additive effects if such compounds occur in mixtures [30]. Hence risk assessment strategies for exposures to PAH mixtures need to be developed.
The three selected PAHs applied in our study are present in the environment and may therefore come into contact with human skin. All three PAHs possess different chemical structures and carcinogenic potencies,5 but all of them are metabolized by CYP enzymes. We used an immortal human keratinocyte cell line (HaCaT) as an in vitro model to study the effects that occur upon exposure to these PAHs. HaCaT cells have been shown to be a suitable metabolically competent model system to mimic human skin cells [31]. We examined the effects of the three selected PAHs using a targeted metabolomics approach by analyzing 188 endogenous metabolites. Thereby we analyzed the end-products of cellular metabolism via comparative analysis of endogenous metabolites and examined AHR-dependent changes induced in HaCaT cells. The overall aim of our studies is to gain more molecular information on the adverse signaling pathways associated with dermal exposures to PAHs.
2. Materials and methods
2.1. Chemicals
Cell culture media and supplements were purchased from PAN Biotech (Aidenbach, Germany). All other chemicals were purchased from Sigma Aldrich with the highest purity available (Munich, Germany). Substances were routinely dissolved as stock solutions in dimethylsulfoxide (DMSO).
2.2. Cell lines
Human adult wild-type (HaCaT WT) and AHR knockdown (HaCaT AHR-KD) keratinocyte cell lines were kindly provided by Prof. Ellen Fritsche (IUF – Leibniz Research Institute for Environmental Medicine, Germany) and are described elsewhere [32]. Both cell lines were maintained in DMEM (Dulbecco's Modified Eagle Medium supplemented with 10% fetal calf serum (FCS, v/v), 2 mM l-glutamine, penicillin (100 U/mL), streptomycin (0.1 mg/mL) at 37 °C in humidified atmosphere with 5% CO2. After 24 h of initial incubation the cells were exposed to the test substances.
2.3. MTT assay
In brief, cells (2 × 105 cells/mL) were seeded in 24-well plates and exposed to various concentrations (0–20 μM) of the chemicals (BAP, CRY, DALP). The final concentration of DMSO did not exceed 0.1% (v/v). After 24 h and 48 h of exposure, MTT reagent (0.5 mg/mL) was added and incubated for 2 h at 37 °C, 5% CO2. Afterwards, formazan crystals were dissolved by adding 400 μL DMSO into each well. The absorption of the samples was measured in triplicates on a microplate TECAN reader at 595 nm.
2.4. Real-time PCR
For gene expression studies cells (2 × 105 cells/mL) were seeded in 12-well plates and incubated in DMEM. Cells were exposed to the test substances for 24 h or 48 h, respectively. Following substance treatment the total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA). RNA quantity and quality was determined using a Nanodrop-1000 (Thermo Fisher Scientific, Asheville, NC, USA). Subsequently, cDNA was synthesized of 1 μg of RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). Levels of individual transcripts were then quantified using SYBR®Green Fast (Applied Biosystems). Quantitative real time-polymerase chain reaction (qRT-PCR) was performed according to the manufacturer’s instructions on an ABI-7500 Fast Real Time PCR system (Applied Biosystems). Gene selective primers for CYP1A1 (forward 5′-CCAAGAGTCCACCCTTCCCAGCT-3′, reverse 5′-GAGGCCAGAAGAAACTCCG-TGGC-3′), CYP1B1 (forward 5′-TGGATTTGGAGAACGTACCG-3′, reverse 5′-CCACG-ACCTGATCCAATTCT-3′) and HPRT (forward 5′-GTTCTGTGGCCATCTGCTTAG-3′, reverse 5′-GCCCAAAGGGAACTGATAGTC-3′) were used. Relative gene expression was calculated using the ΔΔCt method and normalized to expression levels of HPRT. Vehicle control (0.1% DMSO) sample expressions were set to a fold change value of 1 and used as comparison for treated cells.
2.5. Western blots
Equal amounts of proteins (30 μg) obtained from RIPA buffer lysed cells were applied to SDS-PAGE, transferred onto nitrocellulose membranes and immune-blotted according to the manufacturer’s instructions. Antibodies against CYP1A1 (sc-20772) and CYP1B1 (sc-32882) were used (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Primary antibody probed blots were visualized with appropriate horseradish peroxidase-coupled secondary antibodies (Santa Cruz Biotechnology) using enhanced chemo-luminescence (34078; Thermo Scientific, Waltham, MA, USA) for detection.
2.6. Cell culture for metabolomics approach
Cells (1 × 105 cells/mL) were seeded in 10 cm cell culture dishes and incubated in DMEM. Cells were exposed to the test substances (4 replicates for each treatment) for 48 h. After substance treatment, cells were quickly washed twice with ice-cold ammonium formate buffer solution (155 mM, pH 7.4). Cells were detached from the culture dish by scraping into 1 mL of buffer solution, transferred to 1.5 mL safe-lock tubes (Eppendorf, Hamburg, Germany) and centrifuged (500 × g, 5 min, 4 °C). After centrifugation the supernatant was removed and the cell pellet weight was recorded before freezing them in liquid nitrogen. The frozen samples were stored until analysis at −80 °C A volume of 0.5 mL UHPLC grade ice-cold methanol (J.T. Baker, Deventer, Holland) was added for cell pellet extraction followed by 3 freeze and thaw cycles: cells were frozen for 5 min in liquid nitrogen and afterwards thawed for 5 min at room temperature followed by 1 min of ultrasonication in a water bath. After centrifugation (12,000 x g, 30 min, 4 °C), supernatants were immediately analyzed using the Biocrates AbsoluteIDQ p180 kit.
2.7. Targeted metabolomics approach
The AbsoluteIDQ platform (Kit p180, Biocrates, Innsbruck, Austria) was employed for targeted metabolite profiling as described by the manufacturer. This platform detects a total of 188 metabolites, including 42 amino acids (AAs) and biogenic amines (BAs), 40 acylcarnitines (Cx:y), hydroxyacylcarnitines (Cx:y-OH) and dicarboxyacylcarnitines (Cx:y-DC) (ACs), the sum of hexoses, 15 sphingomyelins (SM_Cx:y) and sphingomyelin derivatives [SM(OH)Cx:y], as well as 14 lysophosphatidylcholines (LPC_Cx:y) and 76 phosphatidylcholines (PC_Cx:y). LPCs and PCs were further differentiated with respect to the presence of ester (“a”) or ether (“e”) bonds in the glycerol moiety (“a” = acyl, “aa” = diacyl, and “ae” = acyl-alkyl). The lipid side chain composition is abbreviated with “Cx:y”; “x” denotes the number of carbons in the side chain and “y” the number of double bonds. Detailed information on the metabolites analyzed along with the protocol of metabolite quantification is presented elsewhere [33]. The cellular extracts were prepared following the manufacturer’s protocol. Briefly, samples were prepared as follows: (a) pipetting of a standard mixture labeled with stable isotopes onto the filter paper inserts of the 96-well plate of the kit and drying under a stream of nitrogen using an EVA LS evaporator (VLM, Bielefeld, Germany), (b) adding of 20 μL cell extract onto the filter inserts and drying under a stream of nitrogen, (c) derivatization of AAs using phenylisothiocyanate, (d) extraction of metabolites and internal standards with 5 mM ammonium acetate in methanol, (e) centrifugation through a membrane filter followed by dilution with solvent for mass spectrometry. One part of the eluate was diluted with 40% methanol in water (1/10 (v/v)) for LC–MS/MS analysis, the other part with Biocrates kit running solvent (1/50 (v/v)) for FIA-MS/MS. The final extracts were then analyzed using a QTRAP 5500 (AB Sciex, Darmstadt, Germany) triple quadrupole mass spectrometer coupled to an Agilent 1200 Series HPLC (Agilent, Waldbronn, Germany) using ESI MS/MS. For the mass spectrometry analysis the software package Analyst 1.6 was used (AB Sciex, Darmstadt, Germany). LC–MS/MS analysis was performed in the positive mode which enables the quantification of 42 metabolites, covering 21 AAs and 21 BAs. A Zorbax Eclipse XDB-C18 column (3 × 100 mm, 3.5 μm; Agilent, Waldbronn, Germany) was used for metabolite separation by LC. Subsequently, the FIA-MS/MS procedure was used for the quantification of 146 additional metabolites, covering 40 ACs including free carnitine, 38 PCaa’s, 38 PCae’s, 14 LPCs), 15 SMs, and the sum of hexoses in both positive and negative ion mode. Multiple reaction monitoring (MRM) was used for quantification and compound identification using the MetIDQ™ software package which is an integral part of the AbsoluteIDQ kit. Concentrations of all metabolites were calculated in micromolar units. The method is in concordance with FDA guidelines, warranting reproducibility within a given error range [34]. Metabolite concentrations were standardized to cell pellet weight. For statistical analyses, only metabolites were chosen which were present in each sample, thus restricting the profile to a total of 166 metabolites.
2.8. Statistical analysis
Processing of metabolomics data was carried out using the statistical package SPSS + (version 12.0.2G) as described elsewhere [35]. In brief, each sample was standardized to the mean of the control (set to 100%) and normalized using z-score values. Then, a MANOVA (multivariate analysis of variance) including Bonferroni correction was used for compound selection and reduction of the dataset to metabolites that are significantly changed in at least one exposure group compared to controls. Metabolites that showed a significant group difference (p ≤ 0.05) were selected for multivariate analysis. Principal component analysis (PCA) was performed to achieve dimension reduction of the datasets and the resulting factors were used for a post-hoc Fisher’s Discriminant analysis as described elsewhere [36] for group separation. The performance of the discriminant models was subsequently verified by applying the “leave-one-out” cross-validation formalism [37].
3. Results
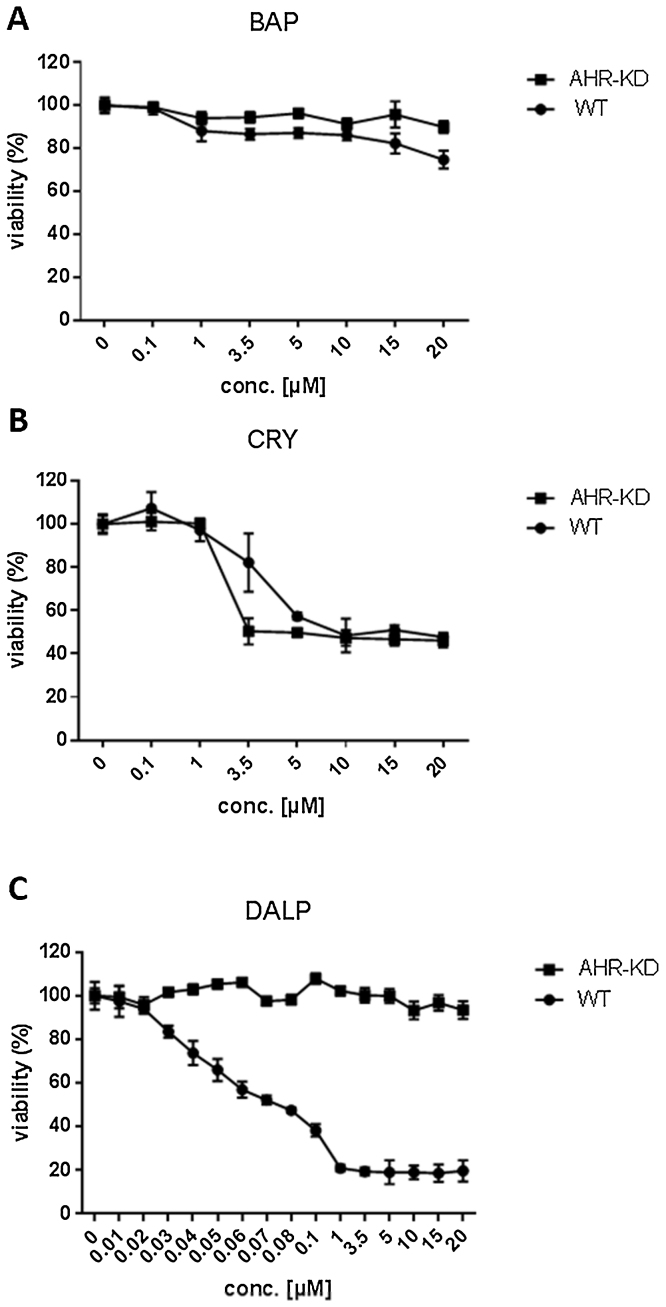
Prior to the metabolomics analysis, dose-dependent cytotoxicity was investigated in HaCaT WT and AHR-KD cells upon exposure to each of the three PAHs (Fig. 1).
Fig. 1.
Toxicity of BAP, CRY and DALP in HaCaT wild-type (WT) or AHR knockdown (AHR-KD) cells. Viability was determined via MTT assay. Cells were treated with 7 different concentrations (0.1–20 μM) of BAP (A) and CRY (B) for 48 h. For DALP (C) 8 additional concentrations (0.01–0.08 μM) were tested due to the strong cytotoxicity of this compound emerging in WT cells. Controls contained 0.1% DMSO (vehicle). Each experiment was repeated three times and data were expressed as mean ± SD. The x-axis depicts the concentrations of the respective PAH applied, the y-axis shows the viability of the treated cells in percent compared to the controls (set to 100%).
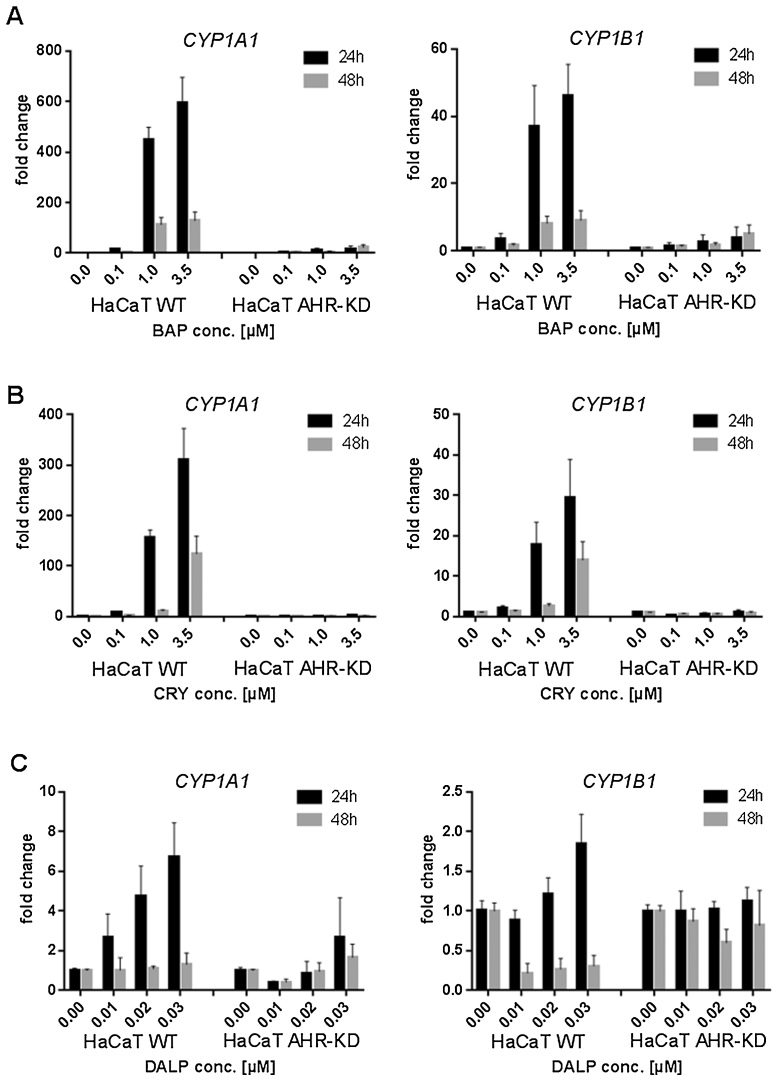
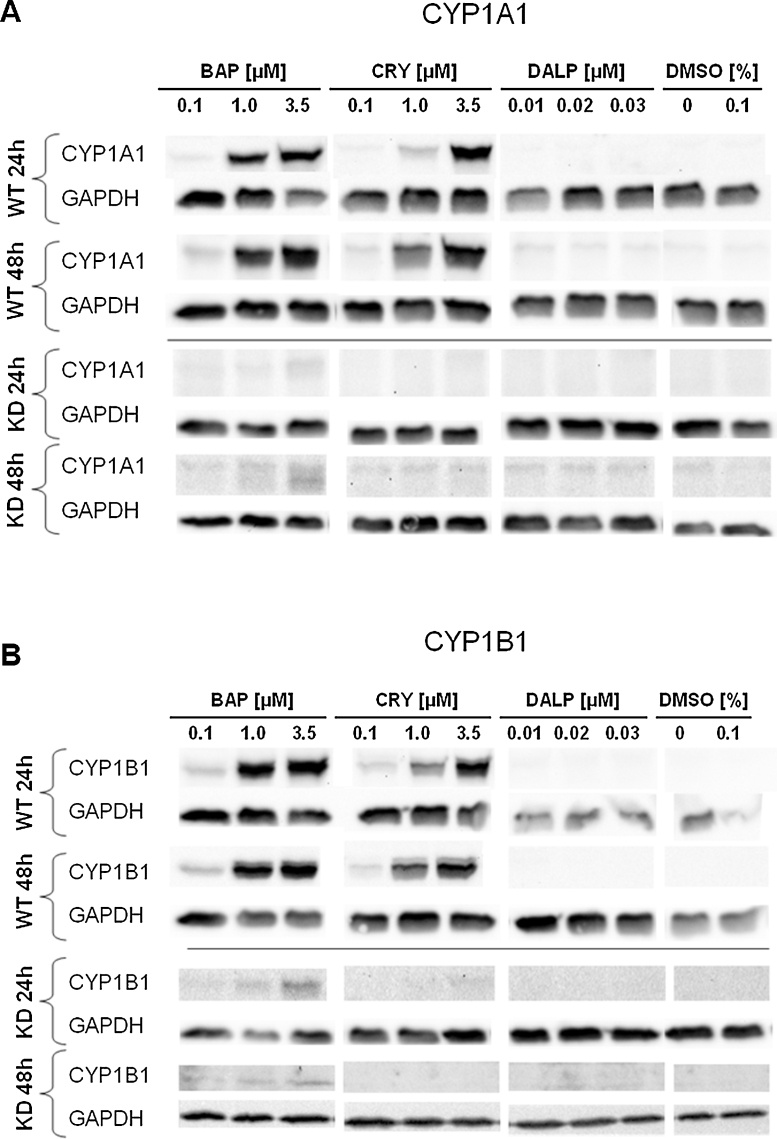
To indirectly assess AHR activation by these compounds, induction of the downstream genes CYP1A1 and CYP1B1 was examined at different concentrations and at both the mRNA (Fig. 2) and the protein level (Fig. 3).
Fig. 2.
Effect of BAP, CRY and DALP on the AHR target genes CYP1A1 and CYP1B1 in HaCaT WT and AHR-KD cells. Cells were exposed for 24 h and 48 h to (A) BAP (0.1 μM, 1.0 μM, 3.5 μM), (B) CRY (0.1 μM, 1.0 μM, 3.5 μM), and (C) DALP (0.01 μM, 0.02 μM, 0.03 μM), respectively. Controls contained 0.1% DMSO (vehicle) only. Relative gene expression of CYP1A1 and CYP1B1 was subsequently quantified with SYBR Green using real-time PCR. Target gene expression was normalized to the transcription level of an endogenous gene (HPRT). The data represent the mean of two independent experiments (±SD).
Fig. 3.
Effect of BAP, CRY and DALP on the CYP1A1 (A) and CYP1B1 (B) protein levels of the in HaCaT WT and AHR-KD cells. Cells were treated for 24 h and 48 h with BAP (0.1 μM, 1 μM, 3.5 μM), CRY (0.1 μM, 1 μM, 3.5 μM), and DALP (0.01 μM, 0.02 μM, 0.03 μM), respectively. Controls remained either untreated or were treated with 0.1% DMSO (vehicle). Western-blot analysis of total protein lysates was performed as described in the Section 2. Data shown are representative for two biological replicates.
After selection of appropriate concentrations and time-points corresponding cellular metabolites were analyzed using a targeted metabolomics platform.
3.1. Cytotoxicity
Cell viability was only weakly affected in both cell lines after treatment with up to 20 μM BAP for 48 h (Fig. 1A). In HaCaT WT cells viability only decreased to about 80% at the highest concentration applied. In contrast, CRY caused strong cytotoxic effects in both cell lines (calculated EC50 values of 3.8 μM for WT and 2.0 μM for AHR-KD cells; Fig. 1B). Treatment with DALP also led to enhanced cytotoxicity in WT cells with a calculated EC50 value of 0.035 μM (Fig. 1C). On the other side, the viability of AHR-KD cells remained unaffected at DALP concentrations of up to 20 μM.
3.2. CYP induction on mRNA level
To investigate the activation of AHR by BAP, CRY or DALP, induction of the downstream genes CYP1A1 and CYP1B1 was measured at the mRNA level. Both CYP enzymes are involved in the metabolism of the three PAHs [13]. As expected, BAP caused a very strong and concentration-dependent induction of CYP1A1 in WT but only a slight elevation of transcription levels in AHR-KD cells after 24 h of incubation (Fig. 2A). For CYP1B1 induction a clear chemical-related effect could be observed as well albeit to a much lower extent. Compared to BAP, CRY was also found inducing both genes in WT cells but less strongly. There was again no significant induction in AHR-KD cells (Fig. 2B). Conversely, DALP only weakly increased the transcription level of CYP1A1 and those of CYP1B1 were even less affected (Fig. 2C). Although this finding is matched by another study where DALP showed similar results in the skin of mice [24], it nevertheless seems kind of surprising and also pointed to the possibility that the time-points picked might have been too late to assess this end-point adequately. Therefore, we also tested earlier time-points (2 and 8 h) and confirmed that no significant induction of CYP1A1 or CYP1B1 occurred upon treatment of HaCaT cells with DALP concentrations of up to 1 μM (data not shown).
3.3. CYP induction on protein level
Western blots confirmed the qPCR data on the protein level. Both CYP1A1 and CYP1B1 showed a strong concentration-dependent induction by BAP, a moderate induction by CRY and no induction by DALP in HaCaT WT cells (Fig. 3). For AHR-KD cells faint signals were detected for BAP only.
Based on the results obtained in cytotoxicity tests, qPCR and western blots, concentrations of 1 μM (BAP and CRY) and 0.01 μM (DALP) were selected to be applied in the subsequent metabolomics study. HaCaT cells (WT and AHR-KD) were exposed to each chemical for 48 h. This experimental setting uses a sub-toxic dose for each PAH sufficient to activate the AHR in HaCaT WT cells. Exposure for 48 h was regarded to be suitable for the detection of metabolomic alterations. So, this time-point represents a scenario after maximal AHR activation and CYP mRNA induction at which downstream CYP protein levels were still found elevated.
3.4. Metabolomics approach: identification of metabolomics biomarkers
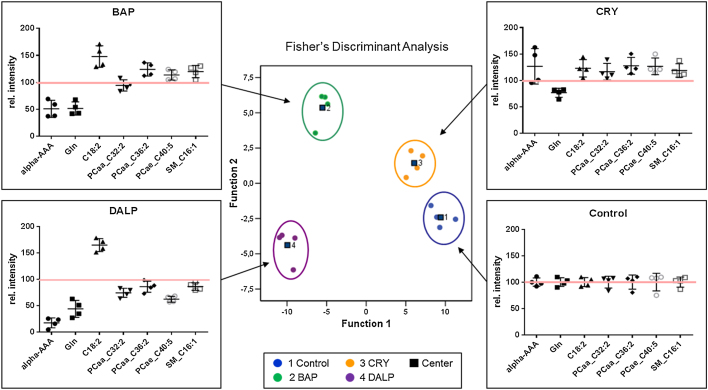
In total, 166 metabolites were detected at all exposure scenarios and analyzed using the AbsoluteIDQ platform. For HaCaT WT and AHR-KD cells 126 and 45 metabolites, respectively, were selected for a multivariate model since they showed significant group differences (p ≤ 0.05). PCA was performed to achieve dimension reduction of the datasets and the resulting factors were used for a post-hoc Fisher’s discriminant analysis resulting in group separation as described elsewhere [36]. For HaCaT WT cells, this discriminant model resulted in 6 factors accounting for 100% of the observed variance in the system. Evaluation of the PCA factors that contributed most to discriminant functions F(×1) and F(×2) revealed a high contribution (90% explanation of the overall variance observed) of PCA factors 1–3 to the separation. Interestingly, mainly PCae’s and PCaa’s were loading high on factor 1, acylcarnitines on factor 2 and a mix of metabolite classes including the amino acid glutamine (Gln) and α-aminoadipic acid (alpha-AAA) were loading high on factor 3. For further data reduction the following thresholds were applied to the loading factors: for factor 1 ≥ 0.970, for factor 2 ≥ 0.950 and for factor 3 ≥ 0.330, respectively. This methodology revealed 24 metabolites (see Table 1) that could be used to achieve complete separation of all exposure groups and controls in the model applied (Fig. 4).
Table 1.
List of 24 metabolites revealing with significant changes between the different treatment groups of HaCaT WT cells (p-values result from MANOVA; metabolite concentrations in nmol/g pellet weight ± SD). Abbreviations used: BAs = biogenic amines (alpha-AAA = a-aminoadipic acid); ACs = acylcarnitines (Cx:y = acylcarnitines, Cx:y-OH = hydroxyacylcarnitines, Cx:y-DC = dicarboxyacylcarnitines); AAs = amino acids (Gln = glutamine); PCs = phosphatidylcholines (PCaa_Cx:y = diacyl, PCae_Cx:y = acyl-alkyl); SMs = sphingomyelines (SM_Cx:y); x = number of carbon atoms in the lipid chain, y = number of double bonds.
| Metabolite | Category | Control | BAP | CRY | DALP | P value |
|---|---|---|---|---|---|---|
| alpha-AAA | BAs | 11.2 ± 0.9 | 5.7 ± 1.8 | 14.2 ± 3.8 | 2.0 ± 1.1 | 0.000 |
| C10:2 | ACs | 0.7 ± 0.1 | 1.0 ± 0.1 | 0.9 ± 0.1 | 1.1 ± 0.1 | 0.001 |
| C12:1 | 0.9 ± 0.1 | 1.2 ± 0.1 | 1.0 ± 0.2 | 1.4 ± 0.1 | 0.000 | |
| C12-DC | 2.9 ± 0.3 | 3.8 ± 0.2 | 3.3 ± 0.6 | 4.2 ± 0.2 | 0.001 | |
| C18:1-OH | 0.7 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 | 1.0 ± 0.0 | 0.001 | |
| C6 (C4:1-DC) | 1.7 ± 0.3 | 2.3 ± 0.2 | 1.9 ± 0.3 | 2.5 ± 0.2 | 0.004 | |
| C18:2 | 0.7 ± 0.1 | 1.0 ± 0.1 | 0.8 ± 0.1 | 1.1 ± 0.1 | 0.000 | |
| C4 | 2.0 ± 0.3 | 2.5 ± 0.3 | 2.3 ± 0.5 | 2.9 ± 0.3 | 0.029 | |
| C4:1 | 0.8 ± 0.1 | 1.1 ± 0.1 | 0.9 ± 0.2 | 1.3 ± 0.1 | 0.001 | |
| Gln | AAs | 149.4 ± 12.4 | 77.1 ± 18.2 | 114.3 ± 12.8 | 65.7 ± 23.9 | 0.000 |
| PCaa_C32:2 | PCs (aa) | 109.8 ± 12.4 | 103.6 ± 11.5 | 127.8 ± 17.3 | 81.9 ± 9.5 | 0.003 |
| PCaa_C36:2 | 1644.0 ± 221.7 | 2039.2 ± 198.8 | 2097.1 ± 262.1 | 1417.7 ± 173.7 | 0.002 | |
| PCae_C32:1 | PCs (ae) | 196.3 ± 29.9 | 198.8 ± 17.1 | 231.2 ± 36.0 | 164.7 ± 17.4 | 0.029 |
| PCae_C34:2 | 224.4 ± 36.1 | 238.2 ± 23.8 | 275.4 ± 48.2 | 170.7 ± 25.4 | 0.009 | |
| PCae_C34:3 | 24.4 ± 3.0 | 26.8 ± 3.1 | 29.7 ± 5.0 | 20.8 ± 3.3 | 0.031 | |
| PCae_C36:2 | 309.1 ± 43.7 | 354.1 ± 28.2 | 385.0 ± 53.6 | 235.3 ± 25.3 | 0.001 | |
| PCae_C36:3 | 96.2 ± 15.0 | 108.6 ± 10.5 | 120.5 ± 17.9 | 65.2 ± 8.9 | 0.001 | |
| PCae_C38:1 | 23.7 ± 4.2 | 29.7 ± 2.2 | 29.9 ± 5.0 | 18.7 ± 1.2 | 0.002 | |
| PCae_C38:2 | 62.5 ± 9.7 | 74.9 ± 6.3 | 78.0 ± 13.3 | 50.7 ± 5.2 | 0.004 | |
| PCae_C38:3 | 27.3 ± 3.6 | 33.8 ± 3.9 | 34.7 ± 5.8 | 18.5 ± 1.8 | 0.000 | |
| PCae_C38:4 | 58.2 ± 7.5 | 69.0 ± 7.3 | 70.5 ± 9.1 | 38.1 ± 4.0 | 0.000 | |
| PCae_C40:4 | 10.1 ± 1.5 | 11.7 ± 1.5 | 12.8 ± 2.0 | 7.6 ± 0.5 | 0.002 | |
| PCae_C40:5 | 26.3 ± 4.4 | 30.0 ± 2.5 | 33.3 ± 4.2 | 16.5 ± 1.5 | 0.000 | |
| SM_C16:1 | SMs | 29.5 ± 2.8 | 35.3 ± 3.4 | 35.0 ± 3.8 | 25.5 ± 2.1 | 0.002 |
Fig. 4.
Fisher’s discriminant analysis of targeted metabolomics data obtained in HaCaT WT cells. Cells were treated with BAP (1 μM), CRY (1 μM) or DALP (0.01 μM) for 48 h in 4 replicates. Control samples contain 0.1% DMSO only. Metabolites of total cell extracts were analyzed using the p180 Kit from BIOCRATES. In HaCaT WT cells 24 metabolites contributed to a group separation between the three different PAH exposure groups and the control. Cross-validation of the data revealed a re-grouping of 100% using the leave-one-out system. Scatter dot plots for each exposure group (BAP, CRY and DALP) show the metabolic patterns of selected metabolites picked from the group of 24 metabolites that contributed to group separation. The short black lines refer to the mean values ± SD. The long red lines refer to the relative mean values of untreated controls which were set to 100%.
The performance of the discriminant models was subsequently verified by applying the “leave-one-out” cross-validation formalism [37], [38] resulting in 100% accuracy. In the case of HaCaT AHR-KD cells, Fisher’s discriminant analysis of the selected 45 metabolites led to a separation of the exposure groups by including 7 factors, but cross-validation revealed only 81.3% of accuracy. One case of each exposure group was falsely assigned: one BAP-exposed sample was predicted as control, one CRY-treated sample as DALP-treated sample and one DALP-treated sample as CRY-treated sample. Accordingly, no reliable separation of exposure groups in HaCaT AHR-KD cells could be achieved. In HaCaT WT cells, the 24 identified metabolites that were used in the multivariate model to distinguish between different treatment groups comprise 6 different metabolite classes (BAs, AAs, ACs, PCaa’s, PCae’s and SMs, see Tbl. 1): the two polar metabolites alpha-AAA (BAs) and the amino acid Gln, two PCaa's of medium chain length (i.e., PCaa_C32:2 and PCaa_C36:2), several PCae's of medium and long chain length as well as SM_C16:1. Scatter dot plots (Fig. 4) show examples of metabolites of each class providing insight in the metabolomics patterns altered by the three PAHs that were essential for group separation. All PAHs show a down-regulation of Gln. Furthermore, metabolomics patterns of BAP and DALP show similarities in a strong down-regulation of alpha-AAA and a strong up-regulation of carnitines (e.g. C18:2). In contrast, PCaa’s, PCae’s and the SM_C16:1 are lowered in DALP-treated cells and increased in those treated with BAP. For CRY-treated cells a slight elevation of 23 metabolites was observed; here only Gln levels were down-regulated.
3.5. Metabolomics approach: alterations of metabolomics profiles by each PAH
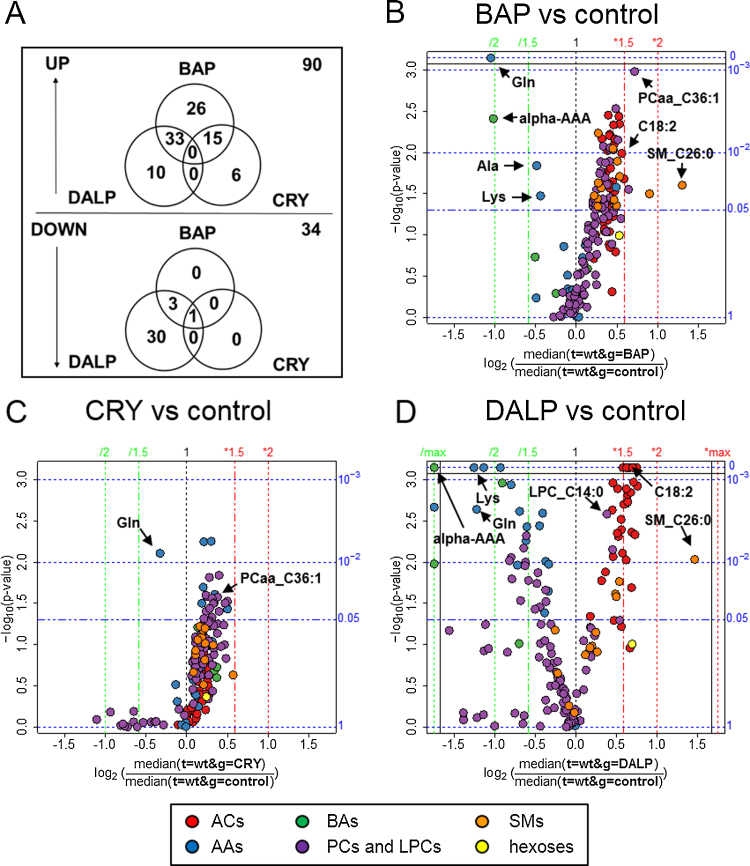
We used the described multivariate model to examine which metabolites contribute to the separation of the three PAHs. In an additional statistical analysis we evaluated alterations, as caused by each PAH in comparison to non-treated controls, of the complete metabolomics pattern consisting of a total of 166 metabolites. In Venn diagrams and volcano plots the metabolomics changes are visualized (Fig. 5).
Fig. 5.
Differential alterations of metabolomics profiles caused by BAP, CRY and DALP in HaCaT WT cells. Venn diagrams of up- and down-regulated levels of metabolites (A). Volcano plots of metabolites influenced by each single PAH compared to control; BAP (B), CRY (C) and DALP (D). Fold changes were calculated as the ratio between the median of the two groups compared. The significance expressed as p-values is calculated by a two-sided, unpaired t-test. (B–D) The x-axes represent the fold changes as log2 values of the ratio between the median of the treated cells compared to untreated controls. The y-axes represent the −log10 values of the determined p-values via t-test. Axes on the right side represent the p-values (p < 0.05 is regarded as significant). On the top axis, fold changes are presented. Abbreviations used: AAs, amino acids; ACs, acylcarnitines; BAs, biogenic amines; PCs, phosphatidylcholines; LPCs, lysophosphatidylcholines; SMs, sphingomyelins.
The plots depict the effects on the measured metabolites caused by each PAH in HaCaT WT cells. Venn diagrams (Fig. 5A) summarizing all significant changes show the differential alterations caused by the three compounds. Most of the changes were noticed as up-regulations of the levels of metabolites mainly caused by the two potent carcinogens BAP and DALP. These two compounds share a third of all up-regulated metabolites. Still 26 metabolites are uniquely elevated upon BAP exposure. BAP is also sharing certain up-regulated metabolites with CRY, while DALP and CRY do not have any metabolite changes in common. Down-regulation of metabolites was mainly caused by DALP with 30 uniquely altered metabolites compared to none by BAP and CRY. Only Gln has been consistently found down-regulated in all treatment groups (BAP p = 0.001, CRY p = 0.008, DALP p = 0.002). Therefore we verified relative Gln levels by an enzymatic assay. As seen in the targeted metabolomics approach, Gln was downregulated approximately two-fold in HaCaT WT cells treated with BAP and DALP (Supplementary Fig. 4). However, this orthogonal method could not confirm the effect of CRY on Gln levels. In AHR-KD cells, only BAP provoked a decrease of Gln concentration in accordance with the LC–MS/MS measurements. Metabolomics profile changes caused by BAP (Fig. 5B) and DALP (Fig. 5D) show similarities in up-regulation of ACs (e.g., C18:2 BAP p = 0.01; DALP p ≤ 0.003), SMs (e.g., SM_C26:0; BAP p = 0.025; DALP p = 0.009) and LPCs (e.g., LPC_C14:0; BAP p = 0.008; DALP p = 0.007), as well as down-regulation of AAs (e.g., Lys; BAP p = 0.034; DALP p ≤ 0.0005) and one BA (i.e., alpha-AAA; BAP p = 0.004; DALP p ≤ 0.0005) (Supplementary Tbl. 1). Treatment with BAP specifically induces a significant up-regulation of 9 SMs and SM(OH)s, for example SM(OH)_C14:1 (p = 0.006) and SM_C24:0 (p = 0.009), which are not altered in DALP- or CRY-treated cells (Supplementary Tbl. 1). In the case of DALP 37 ACs are significantly up-regulated. Twenty eight among them are also found up-regulated in cells treated with BAP. Conversely, treatment of WT cells with CRY did not affect its AC levels. The treatment of cells with DALP is further down-regulating 11 PCae’s that remain unaffected upon exposure to BAP and CRY. Twenty PCaa’s were found increased in cells after BAP exposure, 12 of which are also up-regulated after CRY exposure. Treatment with DALP caused a strong down-regulation of 15 AAs and 3 BAs (i.e., alpha-AAA, sarcosine, taurine). By contrast, treatment of cells with CRY and BAP resulted in the down-regulation of only 1 (Gln) and 3 (Ala, Gln and Lys) AAs, respectively. In CRY-exposed cells, many PCaa’s and PCae’s were up-regulated similarly to BAP-exposed cells. On the other side, SM, AC, LPC and BA concentrations remained completely unaffected in HaCaT WT cells upon treatment with CRY when compared to control cells. The results obtained for AHR-KD cells are presented in Supplementary Fig. 1 and Supplementary Tbl. 2.
4. Discussion
Carcinogenic PAHs occur in crude oil fractions used for the production of rubber, certain plastics and elastomers. Since keratinocytes are potential target cells for PAH exposure we studied the influence of BAP, CRY and DALP on 166 endogenous metabolitesof HaCaT cells. HaCaT cells represent a non-tumorigenic permanent epithelial cell line from adult human skin [39] which is known to be metabolically competent [40]. Hence, exposure of these cells against PAHs may lead to PAH biotransformation and the activation of certain toxicological signaling cascades involved in the expression of an adverse outcome.
4.1. PAH-mediated cytotoxicity and influence on CYP1A1 and CYP1B1 expression
Even though BAP is metabolized in HaCaT WT cells to its ultimate carcinogen (—)-anti-BAP-7,8-diol-9,10-epoxide (BPDE) [40] and has been shown to cause metabolism-independent pro-apoptotic effects [41], viability tests revealed only little cytotoxicity of BAP in HaCaT WT or AHR-KD cells after 48 h at the concentrations applied. Compared to the other two PAHs investigated, BAP caused a strong induction of CYP1A1 and CYP1B1 on both mRNA and protein level (Fig. 1, Fig. 2). This is in accordance with its high potency to bind and activate the AHR [42]. However, besides CYP induction, AHR activation may also promote defensive mechanisms such as, e.g., the synthesis of glutathione S-transferases (GST) [5], which then—in turn—may detoxify BAP-derived electrophiles such as BPDE [43]. This mechanism might contribute to the high tolerance of HaCaT cells against BAP. On the other side, the tetracyclic PAH CRY revealed with some cytotoxicity in HaCaT cells irrespective of its AHR expression status and most likely due to the induction of oxidative stress [44]. Conversely, DALP showed very strong cytotoxicity only in HaCaT WT cells (EC50: approx. 0.035 μM) without inducing CYP1A1 or CYP1B1. Since the strong cytotoxicity of DALP was observed in HaCaT WT without significant induction of CYP enzymes, but completely disappeared in HaCaT AHR-KD cells, AHR-dependent toxicity of DALP in this particular cell line seems rather mediated through other mechanisms than diol-epoxide-DNA adduct formation. Notwithstanding, formation of DALP-11,12-diol-13,14-epoxides (DALPDE) has been already shown to contribute to DALP-mediated cytotoxicity in certain cell models such as mouse embryo fibroblasts [45].
4.2. Effects of BAP, CRY and DALP on metabolites of HaCaT WT and AHR-KD cells
For the first time a metabolomics analysis was conducted in HaCaT cells and the differential influences of the three selected PAHs were recorded and considered as individual signatures of toxicity. Applying a multivariate model, the 166 metabolites analyzed allowed no separation of the different exposure groups in HaCaT AHR-KD cells while HaCaT WT cells revealed strongly altered and differentiable metabolomics profiles (Fig. 4, Fig. 5). This result points to differences of AHR signaling dependent on the particular PAH used. Since CYP1A1 and CYP1B1 expression was found significantly increased in WT cells treated with BAP and CRY (Fig. 2, Fig. 3), metabolites formed through enzyme-mediated catalysis most likely contributed to the overall metabolomics alterations observed with these two PAHs. On the other hand, DALP-treated WT cells showed a lack of considerable CYP induction. In addition, metabolite levels remained virtually unaffected in AHR knockdown cells treated with DALP (Supplementary Fig. 1). Aiming to explain these results for DALP, we considered three possible reasons: Firstly, AHR-KD cells might become insensitive against PAH effects and differentiate differently due to the lack of AHR. Contrasting this assumption, however, on the metabolite levels only minor changes between AHR-KD cells and WT cells could be detected in untreated controls (Supplementary Fig. 2). Secondly, albeit extremely low, basal CYP1B1 levels in WT cells could be sufficient for the formation of genotoxic metabolites of DALP. We detected 4-fold lower basal CYP mRNA levels in AHR-KD cells compared to WT cells (Supplementary Fig. 3). Also Topinka et al. found a negligible CYP induction at DALP concentrations that nevertheless caused high levels of DNA adducts [46]. A 10,000 fold reduction of basal CYP1B1 levels has been shown after knockout of AHR in MCF-7 cells [47]. As CYP1B1 is mainly responsible for DALP’s metabolic activation [48], a decrease in the cellular level of this enzyme might explain the insensitivity of AHR-KD cells against the potential toxicity of this compound. Thirdly, the AHR-dependent toxicity of DALP could also be caused by non-genotoxic AHR signaling alone. Although not investigated yet DALP might function as a selective AHR modulator (SAHRM), thereby triggering alternative AHR responses without xenobiotic response element (XRE) activation [49].
4.3. Alterations in metabolite levels of amino acids and biogenic amines
The levels of the amino acid Gln were found strongly lowered upon exposure of HaCaT WT cells to BAP and DALP and weakly upon CRY exposure (Fig. 4, Fig. 5). Conversely, in AHR-KD cells Gln levels were only decreased compared to controls upon treatment with BAP, but not in cells treated with CRY or DALP (Supplementary Figs. 1, 4). Significantly depleted levels of Gln have been reported in the literature in lung-derived A549 cells treated with different PAH quinones [50]. Since BAP quinones can easily be generated via both CYP-mediated monooxygenation followed by non-enzymatic oxidation and peroxidase-mediated one-electron oxidation [5], in the absence of a functional AHR the contribution of the latter pathway might become more relevant in the generation of PAH quinones.
The strong depletion in the levels of Gln points to a PAH-mediated impairment of both glycolysis and the tricarboxyclic acid (TCA) cycle whose intermediates provide anabolic precursors for the biosynthesis of fatty acids, nucleic acids, and proteins. This is further supported by significantly reduced levels of alpha-AAA, an intermediate in lysine biosynthesis, which has also been found depleted in HaCaT cells after exposure to BAP or DALP (Fig. 5, Supplementary Tbl. 1). Altogether, the levels of 15 AAs were decreased after DALP exposure (Supplementary Tbl. 1). Three of them (Ala, Gln and Lys) were also lowered in HaCaT cells after exposure to BAP. In particular, Glu and Gly are necessary in the biosynthesis of glutathione (GSH), a tripeptide which acts as protective agent against reactive PAH metabolites such as DALPDE and thus can prevent DNA adduct formation [43]. Therefore, depletion of these AAs could be also due to enhanced GSH synthesis.
4.4. Alterations in the lipid metabolite profile caused by the three different PAHs
Upon conversion of long-chain acyl-CoAs into ACs, these products are actively transported across the inner mitochondrial membrane in exchange for free carnitine. Then, through catalysis of carnitine acyltransferase II, acyl-CoA is being recovered within the mitochondrial matrix and directed towards β-oxidation [51]. Recently, Westmann et al. showed that fatty acid β-oxidation in chicken embryo liver was significantly reduced in response to the treatment with a mixture of 16 different PAHs [52]. In transgenic mice activation of AHR inhibited hepatic fatty acid β-oxidation along with acyl-CoA oxidase 1, the rate limiting enzyme in peroxisomal β-oxidation [53]. Down-regulation of acyl-CoA synthetase was shown after BAP exposure of marine diatoms, thus providing evidence that PAHs may interfere with the regulation of lipid metabolism [54]. In our experiments significant increases of AC levels have been detected in HaCaT WT cells after exposure to BAP or DALP (Fig. 5, Supplementary Tbl. 1). A more than 3-fold increase of carnitine palmitoyltransferase 1 (CPT1), which is localized in the outer mitochondrial membrane, and is responsible for the formation of ACs by catalyzing the transfer of the acyl group of a long-chain acyl-CoA to carnitine, was reported in the liver of rats upon treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) [55], [56]. This might explain why ACs start to accumulate in cells that have been challenged by the carcinogens BAP and DALP. TCDD is known as a strong AHR inducer, thus the detected changes may be linked to AHR activation. While the exact molecular mechanisms of this interference between these carcinogenic PAHs and AC degradation remain obscure, lowered rates of ß-oxidation and subsequent TCA cycle activity can explain why AA profiles were found strongly decreased under the very same conditions (Fig. 5, Supplementary Tbl. 1).
The phospholipid groups of PCs and SMs are important components of cell membranes and involved in regulation of cell function, membrane protein trafficking and inflammation [57]. PAHs are highly lipophilic compounds that can be directly incorporated into the cell membrane thereby disturbing the biological membrane function [58]. Perturbation of membrane’s integrity and the ultimate loss of functionality have been shown in diatoms upon BAP exposure [54] and might also be causative for the strong PC levels changes observed in our study (Supplementary Tbl. 1). Additionally, inducing canonical AHR signaling can be assumed to play a role in the phospholipid pattern changes of HaCaT cells exposed to BAP or CRY, but not to DALP. A recent study predicted genes like PPARδ and Lpin involved in lipid biosynthesis to be direct AHR targets [59]. An activation of PPARδ was also found in a recent proteomics study using Hepa1c1c7 cells exposed to BAP [60]. Yet, this interference of AHR signaling with lipid metabolism is still poorly understood. For instance, the PCaa_C36:2 levels of cells were only increased after BAP and CRY exposure and therefore identified as a potential biomarker for canonical AHR activation after PAH exposure. PCaa_C36:2 was also detected as a possible biomarker to distinguish smokers from non-smokers in the KORA (Cooperative Health Research in the Region of Augsburg, Germany) study [61]. As cigarette smoke contains a mixture of carcinogenic PAHs including BAP [62], our data provide further evidence that this particular lipid can serve as biomarker indicating the exposure of cells against AHR activating PAHs. By contrast, DALP, that did not induce AHR downstream signaling in HaCaT cells (i.e., induction of CYP enzymes), indeed caused a measurable but insignificant decrease in the level of PCaa_C36:2 (Tbl. 1).
The increases of the levels of certain SMs (i.e., SM(OH)_C24:1, SM_C18:0, SM_C26:0, SM_C26:1) found in HaCaT WT cells after treatment with BAP or DALP can be associated with the onset of apoptosis. Sphingolipids represent a complex group of biomolecules that function as transducers in eukaryotic cells, modulating cell growth, differentiation and cell death [63]. SMs can be hydrolyzed by sphingomyelinase (SMase) to ceramides. After hydrolysis of SMs, ceramides can function as second messengers activating various cytokines such as TNF-α that are involved in the signaling of apoptosis [64]. Therefore, the increases of SM levels found in cells after exposure to the strong carcinogens BAP and DALP are likely a result of the pro-apoptotic effects of these compounds. Exposure of HaCaT WT cells against BAP at concentrations known to activate the AHR revealed a strong enhancement of CD95 (Fas)-mediated apoptosis [41], which can similarly be triggered by ceramides [65].
5. Conclusion
Applying a multivariate model we have identified altogether 24 biomarkers in HaCaT WT cells that revealed sufficient to discriminate between exposure groups after treatment with three different PAHs. In previous PAH toxicity studies, long known cellular endpoints such as CYP1A1 induction and DNA adduct formation have been repeatedly proposed to be used in the risk assessment of these environmental pollutants. The data presented complement this set of rather historical biomarkers by novel metabolites originating from lipid metabolism and turnover. The alterations in the metabolomics profiles measured suggest influences of PAHs on the energy and lipid metabolism in the cells investigated.
This study is a proof of principle work using single compound exposure only. As PAH exposure might lead to interactions if such compounds occur in mixtures, in future experiments binary mixtures as well as multi PAH mixtures should be studied and the resulting metabolomic pattern changes compared. We would suggest experiments using PAH concentrations with identical potency in combination with experiments investigating co-exposure scenarios, where one of the investigated chemicals is at low dose in order to identify additive or synergistic effects. Further mechanistic insights should be gained by experiments that relate metabolic changes to toxicological pathways. By specific inhibition of steps in relevant signaling pathways (e.g., AHR, p53 or Nrf2) resulting metabolomic marker sets could be related to those pathway steps. This way metabolomic biomarkers indicative for inhibition or activation of relevant signaling pathways could be identified [35]. All this information would finally help to develop in vitro risk assessment strategies for PAH mixtures.
Acknowledgement
The authors acknowledge intramural funding at BfR, SFP grant #1322-434.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.toxrep.2016.09.003.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Lithner D., Damberg J., Dave G., Larsson Å. Leachates from plastic consumer products- screening for toxicity with Daphnia magna. Chemosphere. 2009;74:1195–1200. doi: 10.1016/j.chemosphere.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 2.Lau O.W., Wong S.K. Contamination in food from packaging material. J. Chromatogr. A. 2000;882:255–270. doi: 10.1016/s0021-9673(00)00356-3. [DOI] [PubMed] [Google Scholar]
- 3.Lewtas J. Air pollution combustion emissions: characterization of causative agents and mechanisms associated with cancer, reproductive, and cardiovascular effects. Mutat. Res. 2007;636:95–133. doi: 10.1016/j.mrrev.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Lijinsky W. The formation and occurrence of polynuclear aromatic hydrocarbons associated with food. Mutat. Res. 1991;259:251–261. doi: 10.1016/0165-1218(91)90121-2. [DOI] [PubMed] [Google Scholar]
- 5.Luch A., Baird W.M. Carcinogenic polycyclic aromatic hydrocarbons. In: McQueen C.A., editor. Compr. Toxicol. 2nd ed. Elsevier, Inc.; Amsterdam: 2010. pp. 85–123. [Google Scholar]
- 6.Lassen P., Hoffmann L., Thomsen M. PAHs in toys and childcare products. Dan. Environ. Prot. Agency. 2011;114:5–41. [Google Scholar]
- 7.Tsai P.-J., Shieh H.-Y., Hsieh L.-T., Lee W.-J. The fate of PAHs in the carbon black manufacturing process. Atmos. Environ. 2001;35:3495–3501. [Google Scholar]
- 8.Galea K.S., Davis A., Todd D., Maccalman L., Mcgonagle C., Cherrie J.W. Dermal exposure from transfer of lubricants and fuels by consumers. J. Expo. Sci. Environ. Epidemiol. 2014;24:665–672. doi: 10.1038/jes.2014.41. [DOI] [PubMed] [Google Scholar]
- 9.Ng K.M.E., Chu I., Bronaugh R.L., Franklin C.A., Somers D.A. Percutaneous absorption and metabolism of pyrene, benzo[a]pyrene, and di(2-ethylhexyl) phthalate: comparison of in vitro and in vivo results in the hairless guinea pig. Toxicol. Appl. Pharmacol. 1992;115:216–223. doi: 10.1016/0041-008x(92)90326-n. [DOI] [PubMed] [Google Scholar]
- 10.Paschke M., Hutzler C., Brinkmann J., Henkler F., Luch A. Polycyclic aromatic hydrocarbons in newspaper inks: migration, metabolism, and genotoxicity in human skin. Polycycl Aromat. Compd. 2015;35:32–40. [Google Scholar]
- 11.Bartsch N., Heidler J., Vieth B., Hutzler C., Luch A. Skin permeation of polycyclic aromatic hydrocarbons: a solvent-based in vitro approach to assess dermal exposures against benzo[a]pyrene and dibenzopyrenes. J. Occup. Environ. Hyg. 2016 doi: 10.1080/15459624.2016.1200724. ahead of print. [DOI] [PubMed] [Google Scholar]
- 12.Boffetta P., Jourenkova N., Gustavsson P. Cancer risk from occupational and environmental exposure to polycyclic aromatic hydrocarbons. Cancer Causes Control. 1997;8:444–472. doi: 10.1023/a:1018465507029. http://www.jstor.org/stable/3552702 [DOI] [PubMed] [Google Scholar]
- 13.Shimada T., Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab. Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- 15.Hall M., Grover P.L. Polycyclic Aromatic Hydrocarbons: Metabolism, Activation and Tumour Initiation. In: Cooper C.S., Grover P.L., editors. Chem. Carcinog. Mutagen. I. Springer; Berlin: 1990. pp. 27–372. [Google Scholar]
- 16.Schwarz D., Kisselev P., Cascorbi I., Schunck W.-H., Roots I. Differential metabolism of benzo[a]pyrene and benzo[a]pyrene-7 8-dihydrodiol by human CYP1A1 variants. Carcinogenesis. 2001;22:453–459. doi: 10.1093/carcin/22.3.453. [DOI] [PubMed] [Google Scholar]
- 17.Buters J.T.M., Mahadevan B., Quintanilla-Martinez L., Gonzalez F.J., Greim H., Baird W.M., Luch A. Cytochrome P450 1B1 determines susceptibility to dibenzo[a,l]pyrene-induced tumor formation. Chem. Res. Toxicol. 2002;15:1127–1135. doi: 10.1021/tx020017q. [DOI] [PubMed] [Google Scholar]
- 18.Cavalieri E.L., Rogan E.G. Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25:677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- 19.Penning T.M., Ohnishi S.T., Ohnishi T., Harvey R.G. Generation of reactive oxygen species during the enzymatic oxidation of polycyclic aromatic hydrocarbon trans-dihydrodiols catalyzed by dihydrodiol dehydrogenase. Chem. Res. Toxicol. 1996;9:84–92. doi: 10.1021/tx950055s. [DOI] [PubMed] [Google Scholar]
- 20.Xue W., Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol. Appl. Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Denison M.S., Nagy S.R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 22.Murray I.A., Patterson A.D., Perdew G.H. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marston C.P., Pereira C., Ferguson J., Fischer K., Hedstrom O., Dashwood W.-M., Baird W.M. Effect of a complex environmental mixture from coal tar containing polycyclic aromatic hydrocarbons (PAH) on the tumor initiation, PAH-DNA binding and metabolic activation of carcinogenic PAH in mouse epidermis. Carcinogenesis. 2001;22:1077–1086. doi: 10.1093/carcin/22.7.1077. [DOI] [PubMed] [Google Scholar]
- 24.Siddens L.K., Larkin A., Krueger S.K., Bradfield C.A., Waters K.M., Tilton S.C., Pereira C.B., Löhr C.V., Arlt V.M., Phillips D.H., Williams D.E., Baird W.M. Polycyclic aromatic hydrocarbons as skin carcinogens: comparison of benzo[a]pyrene, dibenzo[def, p]chrysene and three environmental mixtures in the FVB/N mouse. Toxicol. Appl. Pharmacol. 2012;264:377–386. doi: 10.1016/j.taap.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.IARC Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monogr. Eval. Carcinog. Risks Hum. 2010;92:1–853. [PMC free article] [PubMed] [Google Scholar]
- 26.Nordqvist M., Thakker D.R., Vyas K.P., Yagi H., Levin W., Ryan D.E., Thomas P.E., Conney A.H., Jerina D.M. Metabolism of chrysene and phenanthrene to bay-region diol epoxides by rat liver enzymes. Mol. Pharmacol. 1981;19:168–178. http://molpharm.aspetjournals.org/content/19/1/168.long [PubMed] [Google Scholar]
- 27.Hodgson R.M., Weston A., Grover P.L. Metabolic activation of chrysene in mouse skin: evidence for the involvement of a triol-epoxide. Carcinogenesis. 1983;4:1639–1643. doi: 10.1093/carcin/4.12.1639. [DOI] [PubMed] [Google Scholar]
- 28.Levin W., Thakker D.R., Wood A.W., Chang R.L., Lehr R.E., Jerina D.M., Conney A.H. Evidence that benzo[a]anthracene 3,4-diol-1,2-epoxide is an ultimate carcinogen on mouse skin. Cancer Res. 1978;38:1705–1710. http://cancerres.aacrjournals.org/content/38/6/1705.long [PubMed] [Google Scholar]
- 29.Ralston H.H.S., Seidel A., Luch K.L., Platt Baird W.M. The potent carcinogen dibenzo[a,l]pyrene is metabolically activated to fjord-region 11,12-diol 13,14-epoxides in human mammary carcinoma MCF-7 cell cultures. Cancer Res. 1994;54:887–890. http://cancerres.aacrjournals.org/content/54/4/887 [PubMed] [Google Scholar]
- 30.Pushparajah D.S., Plant K.E., Plant N.J., Ioannides C. Synergistic and antagonistic interactions of binary mixtures of polycyclic aromatic hydrocarbons in the upregulation of CYP1 activity and mRNA levels in precision-cut rat liver slices. Environ. Toxicol. 2016 doi: 10.1002/tox.22276. [DOI] [PubMed] [Google Scholar]
- 31.Barlow Y., Pye R.J. Keratinocyte culture. In: Walker J.M., Pollard J.W., editors. Anim. Cell Cult. Humana Press; 1990: 2016. pp. 83–97. [Google Scholar]
- 32.Kalmes M., Neumeyer A., Rio P., Hanenberg H., Fritsche E., Blömeke B. Impact of the arylhydrocarbon receptor on eugenol- and isoeugenol-induced cell cycle arrest in human immortalized keratinocytes (HaCaT) Biol. Chem. 2006;387:1201–1207. doi: 10.1515/BC.2006.148. [DOI] [PubMed] [Google Scholar]
- 33.Jourdan C., Petersen A.-K., Gieger C., Döring A., Illig T., Wang-Sattler R., Meisinger C., Peters A., Adamski J., Prehn C., Suhre K., Altmaier E., Kastenmüller G., Römisch-Margl W., Theis F.J., Krumsiek J., Wichmann H.-E., Linseisen J. Body fat free mass is associated with the serum metabolite profile in a population-based study. PLoS One. 2012;7:e40009. doi: 10.1371/journal.pone.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.US Department of Health Human Services . Center for Veterinary Medicine (CVM), Guidance for Industry, Bioanalytical Method Validation; 2001. Food and Drug Administration, Center for Drug Evaluation and Research (CDER) [Google Scholar]
- 35.Jungnickel H., Potratz S., Baumann S., Tarnow P., von Bergen M., Luch A. Identification of lipidomic biomarkers for coexposure to subtoxic doses of benzo[a]pyrene and cadmium: the toxicological cascade biomarker approach. Environ. Sci. Technol. 2014;48:10423–10431. doi: 10.1021/es502419w. [DOI] [PubMed] [Google Scholar]
- 36.Ami D., Natalello A., Mereghetti P., Neri T., Zanoni M., Monti M., Doglia S.M., Redi C.A. FT-IR spectroscopy supported by PCA −LDA analysis for the study of embryonic stem cell differentiation. Spectroscopy. 2010;24:89–97. [Google Scholar]
- 37.Stone M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. 1974;36:111–147. www.jstor.org/stable/2984809 [Google Scholar]
- 38.Dillon W.R., Goldstein M. Wiley; New York: 1984. Multivariate Analysis. Methods and Applications. [Google Scholar]
- 39.Boukamp P., Petrussevska R.T., Breitkreutz D., Hornung J., Markham A., Fusenig N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marie C., Maître A., Douki T., Gateau M., Tarantini A., Guiraud P., Favier A., Ravanat J.-L. Influence of the metabolic properties of human cells on the kinetic of formation of the major benzo[a]pyrene DNA adducts. J. Appl. Toxicol. 2008;28:579–590. doi: 10.1002/jat.1306. [DOI] [PubMed] [Google Scholar]
- 41.Stolpmann K., Brinkmann J., Salzmann S., Genkinger D., Fritsche E., Hutzler C., Wajant H., Luch A., Henkler F. Activation of the aryl hydrocarbon receptor sensitises human keratinocytes for CD95L- and TRAIL-induced apoptosis. Cell Death Dis. 2012;3:e388. doi: 10.1038/cddis.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nebert D.W., Roe A.L., Dieter M.Z., Solis W.A., Yang Y., Dalton T.P. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- 43.Sundberg K., Dreij K., Seidel A., Jernström B. Glutathione conjugation and DNA adduct formation of dibenzo[a,l]pyrene and benzo[a]pyrene diol epoxides in V79 cells stably expressing different human glutathione transferases. Chem. Res. Toxicol. 2002;15:170–179. doi: 10.1021/tx015546t. [DOI] [PubMed] [Google Scholar]
- 44.Ali D., Verma A., Mujtaba F., Dwivedi A., Hans R.K., Ray R.S. UVB-induced apoptosis and DNA damaging potential of chrysene via reactive oxygen species in human keratinocytes. Toxicol. Lett. 2011;204:199–207. doi: 10.1016/j.toxlet.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 45.Yoon J.-H., Besaratinia A., Feng Z., Tang M., Amin S., Luch A., Pfeifer G.P. DNA damage, repair, and mutation induction by (+)-syn- and (—)-anti-dibenzo[a, l]pyrene-11, 12-diol-13, 14-epoxides in mouse cells. Cancer Res. 2004;64:7321–7328. doi: 10.1158/0008-5472.CAN-04-1094. [DOI] [PubMed] [Google Scholar]
- 46.Topinka J., Marvanová S., Vondrácek J., Sevastyanova O., Nováková Z., Krcmár P., Pencíková K., Machala M. DNA adducts formation and induction of apoptosis in rat liver epithelial stem-like cells exposed to carcinogenic polycyclic aromatic hydrocarbons. Mutat. Res. 2008;638:122–132. doi: 10.1016/j.mrfmmm.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Ahmed S., Wang A., Celius T., Matthews J. Zinc finger nuclease-mediated knockout of AHR or ARNT in human breast cancer cells abolishes basal and ligand-dependent regulation of CYP1B1 and differentially affects estrogen receptor α transactivation. Toxicol. Sci. 2014;138:89–103. doi: 10.1093/toxsci/kft274. [DOI] [PubMed] [Google Scholar]
- 48.Luch A., Coffing S.L., Tang Y.M., Schneider A., Soballa V., Greim H., Jefcoate C.R., Seidel A., Greenlee W.F., Baird W.M., Doehmer J. Stable expression of human cytochrome P450 1B1 in V79 chinese hamster cells and metabolically catalyzed DNA adduct formation of dibenzo[a,l]pyrene. Chem. Res. Toxicol. 1998;11:686–695. doi: 10.1021/tx970236p. [DOI] [PubMed] [Google Scholar]
- 49.Murray I.A., Morales J.L., Flaveny C.A., DiNatale B.C., Chiaro C., Gowdahalli K., Amin S., Perdew G.H. Evidence for ligand-mediated selective modulation of aryl hydrocarbon receptor activity. Mol. Pharmacol. 2010;77:247–254. doi: 10.1124/mol.109.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurbani D., Bharti S.K., Kumar A., Pandey A.K., Ana G.R.E.E., Verma A., Khan A.H., Patel D.K., Mudiam M.K.R., Jain S.K., Roy R., Dhawan A. Polycyclic aromatic hydrocarbons and their quinones modulate the metabolic profile and induce DNA damage in human alveolar and bronchiolar cells. Int. J. Hyg. Environ. Health. 2013;216:553–565. doi: 10.1016/j.ijheh.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Minnich A., Tian N., Byan L., Bilder G. A potent PPARα agonist stimulates mitochondrial fatty acid β-oxidation in liver and skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2001;280:270–279. doi: 10.1152/ajpendo.2001.280.2.E270. http://ajpendo.physiology.org/content/280/2/E270.long [DOI] [PubMed] [Google Scholar]
- 52.Westman O., Nordén M., Larsson M., Johansson J., Venizelos N., Hollert H., Engwall M. Polycyclic aromatic hydrocarbons (PAHs) reduce hepatic β-oxidation of fatty acids in chick embryos. Environ. Sci. Pollut. Res. Int. 2013;20:1881–1888. doi: 10.1007/s11356-012-1418-7. [DOI] [PubMed] [Google Scholar]
- 53.Lee J.H., Wada T., Febbraio M., He J., Matsubara T., Lee M.J., Gonzalez F.J., Xie W. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 2010;139:653–663. doi: 10.1053/j.gastro.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carvalho R.N., Lettieri T. Proteomic analysis of the marine diatom Thalassiosira pseudonana upon exposure to benzo[a]pyrene. BMC Genomics. 2011;12:159. doi: 10.1186/1471-2164-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fletcher N., Wahlström D., Lundberg R., Nilsson C.B., Nilsson K.C., Stockling K., Hellmold H., Håkansson H. 2 3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the mRNA expression of critical genes associated with cholesterol metabolism, bile acid biosynthesis, and bile transport in rat liver: a microarray study. Toxicol. Appl. Pharmacol. 2005;207:1–24. doi: 10.1016/j.taap.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Forgacs A.L., Kent M.N., Makley M.K., Mets B., DelRaso N., Jahns G.L., Burgoon L.D., Zacharewski T.R., Reo N.V. Comparative metabolomic and genomic analyses of TCDD-elicited metabolic disruption in mouse and rat liver. Toxicol. Sci. 2012;125:41–55. doi: 10.1093/toxsci/kfr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gault C.R., Obeid L.M., Hannun Y.A. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv. Exp. Med. Biol. 2011;688:1–23. doi: 10.1007/978-1-4419-6741-1_1. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3069696/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiménez M., Aranda F.J., Teruel J.A., Ortiz A. The chemical toxic benzo[a]pyrene perturbs the physical organization of phosphatidylcholine membranes. Environ. Toxicol. Chem. 2002;21:787–793. [PubMed] [Google Scholar]
- 59.Michaelson J.J., Trump S., Rudzok S., Gräbsch C., Madureira D.J., Dautel F., Mai J., Attinger S., Schirmer K., von Bergen M., Lehmann I., Beyer A. Transcriptional signatures of regulatory and toxic responses to benzo[a]pyrene exposure. BMC Genomics. 2011;12:502. doi: 10.1186/1471-2164-12-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalkhof S., Dautel F., Loguercio S., Baumann S., Trump S., Jungnickel H., Otto W., Rudzok S., Potratz S., Luch A., Lehmann I., Beyer A., Von Bergen M. Pathway and time-resolved benzo[a]pyrene toxicity on Hepa1c1c7 cells at toxic and subtoxic exposure. J. Proteome Res. 2015;14:164–182. doi: 10.1021/pr500957t. [DOI] [PubMed] [Google Scholar]
- 61.Wang-Sattler R., Yu Y., Mittelstrass K., Lattka E., Altmaier E., Gieger C., Ladwig K.H., Dahmen N., Weinberger K.M., Hao P., Liu L., Li Y., Wichmann H.E., Adamski J., Suhre K., Illig T. Metabolic profiling reveals distinct variations linked to nicotine consumption in humans − first results from the KORA study. PLoS One. 2008;3:e3863. doi: 10.1371/journal.pone.0003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hecht S.S. Tobacco carcinogens, their biomarkers and tobacco-induced cancer, Nat. Rev. Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 63.Andrieu-Abadie N., Levade T. Sphingomyelin hydrolysis during apoptosis. Biochim. Biophys. Acta. 2002;1585:126–134. doi: 10.1016/s1388-1981(02)00332-3. [DOI] [PubMed] [Google Scholar]
- 64.Wymann M.P., Schneiter R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 65.Cremesti A., Paris F., Grassmé H., Holler N., Tschopp J., Fuks Z., Gulbins E., Kolesnick R. Ceramide enables fas to cap and kill. J. Biol. Chem. 2001;276:23954–23961. doi: 10.1074/jbc.M101866200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.