Abstract
Isoniazid (INH), recommended by WHO (World Health Organization) in the treatment of tuberculosis (TB), is metabolized primarily by the genetically polymorphic N-acetyltransferase 2 (NAT2) enzyme. The human population is divided into three different phenotypic groups according to acetylation rate: slow, intermediate, and fast acetylators. The objective of this study was to explore the relationship between NAT2 genotypes and the serum concentrations of INH. Blood samples from 96 patients with TB were taken for the analysis. NAT2 polymorphisms on coding region were examined by polymerase chain reaction (PCR) direct sequencing; the acetylation status was obtained by measuring isoniazid (INH) and its metabolite, acetylisoniazid (AcINH) in plasma was obtained by using the liquid chromatography coupled to mass spectrometry. TB patients were distributed into two groups of fast and slow acetylators according to the acetylation index calculated based on the plasma concentration of INH in the 3rd hour (T3) after an oral dose. Our PCR analysis identified several alleles, where NAT2*4, NAT2*5A, NAT2*6A, and NAT2*13A were the most important. The concentrations of INH varied between 1.10 mg/L and 13.10 mg/L at the 3rd hour and between 0.1 and 9.5 mg/L at the 6th hour. The use of the acetylating index I3 allowed the classification of tested patients into two phenotypic groups: slow acetylators (44.3% of TB patients), and rapid acetylators (55.7%). Patient’s acetylation profile provides valuable information on their therapeutic, pharmacological, and toxicological responses.
Chemical compounds studied in this article: Isoniazid (PubChem CID: 3767), Acethylisoiazid (PubChem CID: 71602), Beta hydroxyethyl theophylline (PubChem CID: 1892), Formic acid (PubChem CID: 284), Ammonium formate (PubChem CID: 2723923), Acetronitrile (PubChem CID: 6342), Methanol (PubChem CID: 887), Ethidium bromide (PubChem CID: 14710)
Keywords: NAT2, Genotyping, Isoniazid, Acetylation, Toxicity
1. Introduction
The concept of personalized medicine has emerged as a means of improving therapeutic response and to minimize adverse drug reactions. Significant functional changes and several polymorphic enzymes involved in drug metabolism have now been identified with an influence on the systemic concentration of the drug. N-Acetyltransferase 2 (NAT2) carries out an important metabolic biotransformation of isoniazid (INH), and was first recognized among tuberculosis (TB) patients treated with this antibiotic [1], [2].
More than 50 years after its introduction, isoniazid remains one of the basic drugs used to treat tuberculosis, administered either alone or as a prophylactic agent. Tuberculosis is now re-emerging and is been classified as a major public health issue, causing approximately 1.5 million deaths worldwide in 2014 (890 000 men, 480 000 women and 140 000 children) [3]. In Senegal, about 12,256 new cases of TB infections are reported yearly: 76% have pulmonary infection and 3% are deadly [4]. Thanks to the use of highly efficient treatment protocols, such as directly observed therapy (DOT) and the free care treatment for all patients without exception, Senegal reported a therapeutic success rate that increased from 76% in 2006 to 85% in 2009 [4].
Despite multi drug treatment régimes (pyrazinamide, isoniazid, ethambutol and rifampicin), patients have a wide variation in plasma concentrations and adverse drug reactions such as hepatitis are often observed [5], [6]. Patients who are slow acetylators are known to be at risk of most drug-induced toxicity, whereas rapid acetylators may experience treatment failure. Therefore, studies of the influence of NAT2 genotypes on the ability to metabolize isoniazid can help personalize and optimize the treatments régimes; making them therapeutically more efficient while simultaneously minimizing the side effects associated with the use of this drug [7].
Therefore, the absence of data in INH acetylation in Senegal constitute a toxicity risk factor in the therapeutic treatment of TB. Our aim is to alleviate that with a profiling of the INH acetylation.
In this study, we will investigate the various mutations of the NAT2 found among patients with tuberculosis and associate the kinetics of INH metabolism to the acetylation profile in these patients in order to provide a treatment regime that can be used to better manage the disease and to prevent the risk of INH toxicity.
2. Material and methods
2.1. Patients
TB patients were recruited, with their written consent after authorization from the ethics committee, from the pneumophtisiology service of the university hospital of Fann, located in the city of Dakar. The unit takes care of TB patients as well as respiratory diseases patients. Our study was composed of 96 newly diagnosed TB patients, positive with Koch’s bacillus with treatment-naive. Patients were well representative of the different regions of Senegal and were composed of 57% women and 43% men, with age ranging from 17 to 74 years. Blood samples were taken by venepuncturewith ethylenediamine tetraacetic acid (EDTA) tube at T0 time zero for genotyping of NAT2, and with heparinised tubes at T3 (after three hours) and T6 (after 6 h) after an oral administration of 5 mg of isoniazid per kg of body weight (Isoniazid Tablets BP100 mg Macleods Pharmaceuticals Ltd.) for the dosage of INH. Patients with hepatic or renal insufficiency who were likely to alter the metabolism or elimination of the drug were excluded. The renal and hepatic functions have been monitored by clinicians with serum transminase testing as well as urine protein testing. However, results from such monitoring were not included; patients were selected by clinicians themselves. Whole blood collected and the serum (collected by centrifugation immediately after sampling) were stored at −80 °C until required.
2.2. NAT2 genotyping
A 5- or 10-mL blood sample from all 96 participants was collected by the venojet system, employing a Vacutainer with EDTA anticoagulant. After collection, blood samples were frozen at −20 °C until analysis. Genomic deoxyribonucleic acid (DNA) was isolated from peripheral blood leucocytes using the Nucleon BACC3 kit (GE Healthcare, Saclay, France) or the NucleonSpin kit (Macherey-Nagel Eurl, Hoerdt, France), according to the available volume of blood and the manufacturer's instructions. The quality and quantity of DNA were determined using an UV–vis (UV) spectrophotometer NanodropND (Thermo Scientific, Courtaboeuf, France). After extraction, DNA samples were stored at −80 °C.
2.2.1. Genomic DNA isolation
A 873-bp fragment covering the coding region of NAT2 was amplified by PCR using a pair of specific primers (NAT2F: 5′-GTTTTTCTTGCTTAGGGGATC-3′ and NAT2R: 5′-ATTAGTGAGTTGCGTGATACATA-3′). The amplification reaction was performed in a final volume of 25 μL in the presence of 200 ng DNA, 20 mM Tris-HCl buffer pH 8.4, 50 mM KCl, 1.5 mM MgCl2, 1.25 U Taq DNA polymerase, 0.2 mM of each dNTP and 0.4 μM of each primer. The PCR thermal profile included an initial denaturation step at 94 °C for 2 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and extension at 72 °C for 1 min. The last cycle was followed by a 7 min extension step at 72 °C. Size and specificity of PCR fragments were controlled on a 2% agarose ethidium bromide-stained gel.
2.2.2. Sequencing analysis
After purification with a MultiScreen 384-PCR Filter Plate (Milipore, Billerica, MA, USA), the amplicons were sequenced on both strands with the aforementioned primers using the Big-Dye® Terminator version 3-1 cycle sequencing kit (Applied Biosystems, Villebonsur Yvette, France) and an ABI 3730 XL DNA sequencer (Applied BioSystems). The sequence analysis and SNP identification were performed using SeqScape v.2.5 software (Applied BioSystems) using EnsemblGenom Browser Accession Number ENST00000286479 (http://www.ensembl.org/index.html) as the NAT2 genomic reference sequence.
2.2.3. Data analysis
The allele characterization and designation were performed using the NAT2 allele nomenclature consensus published in 1995 and updated in April 2016 [http://nat.mbg.duth.gr/human/nat2/allele].
The Chi-Square analysis (χ2-test) was used to determine whether the genotype distribution for each single-nucleotide polymorphisms (SNP) was in Hardy–Weinberg equilibrium and the significance was set at p < 0.05 [8].
2.3. Study of the kinetics of isoniazid metabolism
Plasma isoniazid concentrations were measured for 79 of the 96 tuberculosis patients. Venous blood samples were collected into heparin anticoagulant at 3 and 6 h after administration of a single oral dose of isoniazid 5 mg/kg in the morning. Plasma was separated from blood cells by centrifugation and stored at −80 °C until analysis.
2.3.1. Measurement of isoniazid and acetylisoniazid concentrations
Estimation of isoniazid (INH) and its metabolite, acetylisoniazid (AcINH) in plasma was carried out using liquid chromatography coupled to mass spectrometry (UPLC–MS/MS).
The chromatographic separation was performed on a liquid chromatograph Ultra Performance (UPLC) Acquity (Waters Corporation, Milford, Ma, USA) equipped with an HSS column C18 2.1 × 150 mm, 1.8 μm (Waters Corporation) maintained at 50 °C. The UPLC was coupled to a triple quadrupole mass spectrometer Xevo TQD (Waters Corporation) via an electrospray interface. The control software of the system is MassLynx V4.1 software (Waters Corporation).
All chemicals were of high performance liquid chromatography (HPLC) grade. INH, AcINH, the internal standard I.S. (β-hydroxyethyl theophylline), formic acid, ammonium formate, acetonitrile and deionized water Biosolve were purchased from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Methanol was obtained from VWR Prolabo (Fontenay-sous-Bois, France).
To 100 μL aliquots of each standard, QC sample and blank plasma were deproteinized by adding 350 μL of acetonitrile in the presence of 50 μL of internal standard. After briefly mixing on a vortex shaker for 30 s protein was removed by centrifugation for 5 min at 10000 rounds per minute (rpm). A volume of 100 μL of the supernatant was then transferred to an autosampler vial and diluted in 900 μL of mobile phase. Then, 10 μL of this final mixture were injected into the UPLC–MS/MS. All plasma samples needed to be processed immediately once they were thawed at room temperature.
The mobile phase was a mixture of ammonium formate 5 mM at pH 3 (A) and acetonitrile 0.1% in formic acid (B) (95:5, v/v). The flow rate was 0.4 mL/min and the gradient profile used began with a isocratic elution of (A:B) 95:5 v/v for 5 min, followed by a gradual linear decrease of A to (A:B) 95:5 v/v until 2 min and from the 7th min, return to initial conditions (A:B) 95:5 v/v.
The Electrospray Ionization (ESI) source was operated in positive mode and the final optimized conditions were as follows: capillary voltage, 3 kV; cone voltage, 30 V for INH and AcINH, 35 V for I.S.; extractor voltage, 3 V; collision energy, 12 eV for INH, 14 eV for AcINH, 19 eV for I.S.; source temperature, 140∘C; desolvation temperature, 500∘C; cone gas flow, 50 L/h; desolvation gas flow, 750 L/h and collision gas (argon) flow 0.16 mL/min. Quantification was achieved under multiple reaction monitoring (MRM) mode using the following transitions: INH m/z 138.0–121.0, AcINH m/z 180.0–138.0 and I.S. m/z 225.0–181.0.
2.3.2. Determination of the index acetylation among tuberculosis patients
To distribute the TB patients into two groups of fast and slow acetylators, we used the method of Vivien et al. [9]. This method involves calculating the acetylation index based on the plasma concentration of INH in the 3rd hour (T3) after an oral dose. Vivien et al. showed that there is a linear correlation between the dose of INH (oral) and the plasma concentration of INH: the dose/concentration ratio is constant for each patient; the slope of the straight dose/concentration has a different value for each patient: this is the index acetylation I3:
Median values of I3 were used for results analysis: I3 < 0.45: fast acetylator; I3 = 0.45–0.65: intermediate acetylator; I3 > 0.65: slow acetylator.
The index of acetylation will determine the daily dose of INH; a recommended dosage of 1.5 μg/mL INH at the 3rd hour.
3. Results
3.1. NAT2 alleles and genotypes
Twenty one different haplotypes that were observed are presented in Table 1. The major alleles found were NAT2*4, NAT2*5A, NAT2*6A, and NAT2*13A followed by NAT2*5B, NAT2*6C and NAT2*12A. Some alleles were observed at frequencies of <1% (NAT2*5M, NAT2*6B, NAT2*6L, NAT2*7A, NAT2*7B, NAT2*12H).
Table 1.
Observed frequencies of NAT2 haplotypes in the Senegalese tuberculosis patients (N = 96).
| NAT2 haplotype | SNP | Observed frequency (%) |
|---|---|---|
| NAT2*4 | None | 10.9 |
| NAT2*5A | T341C, C481T | 17.2 |
| NAT2*5B | T341C, C481T, A803G | 6.3 |
| NAT2*5D | T341C | 2.6 |
| NAT2*5M | T341C, C481T, A803G, G838A | 0.5 |
| Total NAT2*5 | 26.6 | |
| NAT2*6A | C282T, G590A | 18.8 |
| NAT2*6B | G590A | 0.5 |
| NAT2*6C | C282T, G590A, A803G | 5.2 |
| NAT2*6J | C282T, G590A, G857A | 1.6 |
| NAT2*6L | C282T, C345T, G590A | 0.5 |
| Total NAT2*6 | 26.6 | |
| NAT2*7A | G857A | 0.5 |
| NAT2*7B | C282T, G857A | 0.5 |
| NAT2*7C | C282T, A803G, G857A | 2.1 |
| Total NAT2*7 | 3.1 | |
| NAT2*12A | A803G | 4.7 |
| NAT2*12B | C282T, A803G | 3.6 |
| NAT2*12C | C481T, A803G | 3.1 |
| NAT2*12H | C403G, A803G | 0.5 |
| Total NAT2*12 | 12 | |
| NAT2*13A | C282T | 10.4 |
| NAT2*14A | G191A | 2.6 |
| NAT2*14B | G191A, C282T | 3.6 |
| NAT2*14E | G191A, A803G | 3.6 |
| Total NAT2*14 | 9.9 | |
| New allele (838) | G838A | 0.5 |
The distribution of the NAT2 genotype frequencies and predicted phenotypes are summarized in Table 2. The most frequent slow acetylator genotype observed was NAT2*5/*6 (16.7%), followed by NAT2*5/*14 (10.4%), and NAT2*6/*6 (7.3%). Among rapid acetylators, NAT2*5/*12, NAT2*4/*6, NAT2*4/*13 were the most frequent genotypes, respectively. No differences were observed between the slow and rapid acetylator groups of tuberculosis patients, while 2 individuals remained unclassified (unknown phenotype) as they were carriers of one slow-acetylator allele and one allele with unknown functional effect (c.838G > A and allele NAT2*12H).
Table 2.
Distribution of NAT2 genotypes and predicted acetylator phenotype (n = 96).
| Genotype | Number of subjects | Observed frequency (%) | Predicted phenotype |
|---|---|---|---|
| NAT2*5/*5 | 5 | 5.2 | Slow |
| NAT2*5/*6 | 16 | 16.7 | Slow |
| NAT2*5/*7 | 4 | 4.2 | Slow |
| NAT2*5/*14 | 10 | 10.4 | Slow |
| NAT2*6/*6 | 7 | 7.3 | Slow |
| NAT2*6/*7 | 1 | 1 | Slow |
| NAT2*6/*14 | 4 | 4.2 | Slow |
| Total Lent | 47 | 49 | |
| NAT2*4/*4 | 1 | 1 | Rapid |
| NAT2*4/*5 | 1 | 1 | Rapid |
| NAT2*4/*6 | 7 | 7.3 | Rapid |
| NAT2*4/*12 | 3 | 3.1 | Rapid |
| NAT2*4/*13 | 6 | 6.3 | Rapid |
| NAT2*4/*14 | 2 | 2.1 | Rapid |
| NAT2*5/*12 | 9 | 9.4 | Rapid |
| NAT2*6/*12 | 4 | 4.2 | Rapid |
| NAT2*6/*13 | 4 | 4.2 | Rapid |
| NAT2*7/*13 | 1 | 1 | Rapid |
| NAT2*12/*12 | 1 | 1 | Rapid |
| NAT2*12/*13 | 4 | 4.2 | Rapid |
| NAT2*13/*13 | 1 | 1 | Rapid |
| NAT2*13/*14 | 3 | 3.1 | Rapid |
| Total Rapide | 47 | 49 | |
| NAT2*6/*new (838) | 1 | 1 | Unknown |
| NAT2*5/*12H | 1 | 1 | Unknown |
| Total Unknowna | 2 | 2 | |
| Total | 96 | 100 | |
Two out of the 96 tested subjects had an unknown acetylator phenotype as they were composite heterozygotes for a slow-acetylator allele and an allele with unknown functional effect.
3.2. NAT2 genotypes and plasma isoniazid concentration
Predicted phenotypes were deduced from genotypes based on a bimodal distribution of the acetylation of isoniazid [10]. Patients were grouped as slow or rapid acetylators if a genotype comprised two slow alleles or one or two fast alleles respectively. Plasma isoniazid concentrations were measured in all 79 patients and the levels compared between slow and rapid acetylators.
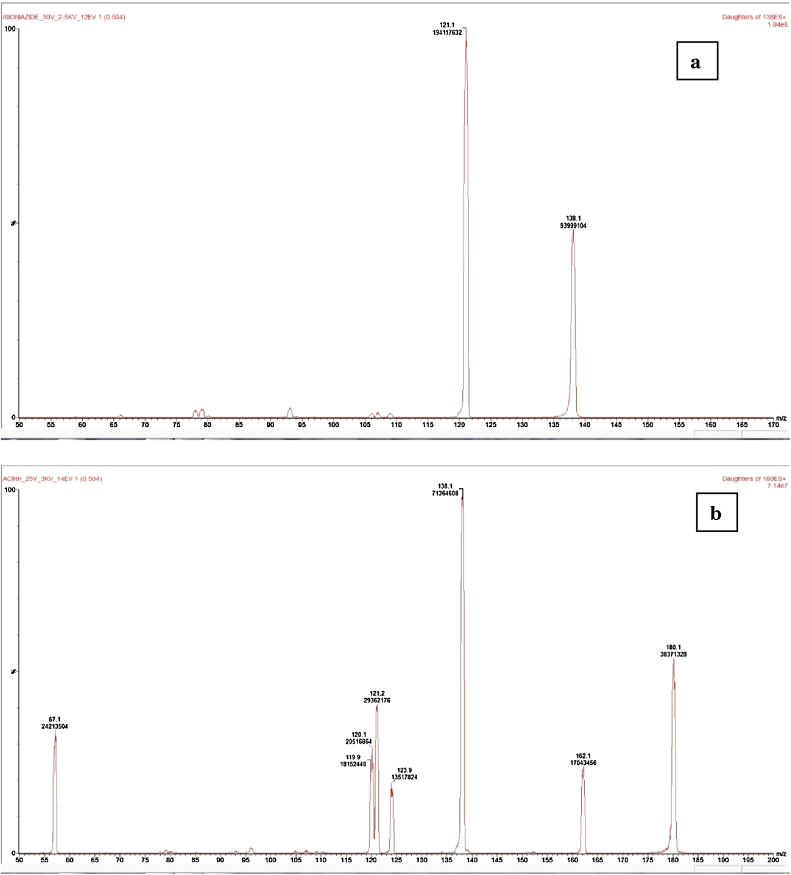
The retention times in UHPLC of INH, AcINH and the I.S. were 0.9, 0.98. and 3.54 min, respectively. The calibration curve was linear over a concentration range of 0.1–20.0 mg/L and the limit of quantification (LOQ) is the lowest calibration point of 0.1 mg/L. The acquisition of the mass spectrum with a 30 V cone allowed the forming of the protonated molecular ion [MH]+ of m/z = 138 for isoniazid and m/z = 180 for AcINH (Fig. 1).
Fig. 1.
Representative product ion mass spectra of (a) INH, (b) AcINH.
Table 3 shows the overall mean (95% CI) isoniazid plasma concentrations observed at 3 and 6 h. The concentrations of INH obtained after oral administration of 5 mg/kg body weight varied between 1.10 mg/L and 13.10 mg/L with a mean of 4.78 + 3.03 mg/L for isoniazid at the 3rd hour and between 0.1 and 9.5 mg/L at the 6th hour. While, the concentration of acetylisoniazid varied between 0.1 and 3.7 at the 3rd hour and between 0.1 and 2.2 at the 6th hour (Table 3).
Table 3.
Plasma Isoniazid and Acetylisoniazid concentrations in TB patients (N = 79) after 3 and 6 h from the administration of a dose (5 mg/kg body weight) of INH.
| 3 h (mg/L) | 6 h (mg/L) | |
|---|---|---|
| Isoniazid | 4.78 + 3.03 (1.10–13.10) | 2.60 + 1.97 (0.1–9.5) |
| Acetylisoniazid | 1.34 + 0.96 (0.1–3.7) | 0.56 + 0.5 (0.1–2.2) |
The use of the acetylating index I3 allowed tested patients to be classified into two phenotypic groups: slow acetylators, 44.3% of TB patients, and rapid acetylators 55.7%. For a better correlation of genotyping results, intermediate acetylators were classified in fast groups.
Table 4 gives the concentrations of INH and AcINH at 3 h (T3) and 6 h (T6) in TB patients according to their acetylation profile. At 3 h, INH concentrations varied between 2.8 and 13.1 mg/L in slow acetylators and between 1.3 and 3.5 mg/L in rapid acetylators. The concentrations of the metabolite, AcINH ranged from 0.2 to 3.7 mg/L, depending on the acetylation profile.
Table 4.
Plasma Isoniazid and Acetylisoniazid concentrations in slow and rapid acetylator TB patients (N = 79) after 3 and 6 h from the administration of a dose (5 mg/kg body weight) of INH.
| Slow Acetylators |
Rapid Acetylators |
|||
|---|---|---|---|---|
| 3 h (mg/L) | 6 h (mg/L) | 3 h (mg/L) | 6 h (mg/L) | |
| Isoniazid | 6.79 + 2.70 (2.8–13.10) |
3.5 + 2.25 (0.1–9.5) |
2.37 + 0.6 (1.3–3.5) |
1.53 + 0.67 (0.5–3.1) |
| Acétylisoniazid | 1.52 + 0.60 (0.2–3.7) |
0.77 + 0.6 (0.1–2.2) |
1.13 + 0.8 (0.2–3.1) |
0.41 + 0.32 (0.1–1.2) |
At 6 h, INH concentrations varied between 0.1 and 2.2 mg/L in slow acetylators and between 0.1 and 1.2 mg/L in rapid acetylators. Similarly, AcINH concentrations ranged from 0.1 and 2.2 mg/L in slow acetylators and between 0.1 and 1.2 mg/L in rapid acetylators.
3.3. Genetic polymorphism correlation of NAT2—kinetics of INH metabolism
Table 5 gives the joint proportions of the genotyping and phenotyping results in 77 patients with tuberculosis. The kappa coefficient, κ is 58% between phenotyping using the acetylation index method and genotyping by the PCR method; with two undetermined genotyping (slow phenotyping) that have not been taken into account.
Table 5.
Proportional distribution of genotype and phenotype within the population of TB patients (N = 77).
| Phenotype | Slow | 35 | 7 | 42 |
| Rapid | 9 | 26 | 35 | |
| Total | 44 | 33 | 77 | |
| Genotype |
||||
| Slow | Rapid | Total | ||
bTwo cases were not been taken into account because of slow phenotyping and undetermined genotyping.
4. Discussion
The distribution of TB patients was made according to the index of acetylation, with 44.3% of rapid acetylators and 55.7% of slow acetylators. This distribution is similar to that observed in healthy Senegalese subjects [8]. In South Africa, a similar study in children with TB gave 39% of slow acetylators and 61% of fast acetylators [11]. In that study, the author used a dose of 10 mg/kg body weight, based on international recommendations stating that the child should receive more drugs for extra precaution and to ensure that a child with rapid acetylator capacity has an adequate dose of INH to properly impregnate all the body compartments [12]. In our study, children under 15 years old were excluded as we consider them potentially to group differently.
The kinetics of INH metabolism following an oral administration of 5 mg/kg body weight was used to calculate the mean concentrations of INH and AcINH after 3 and 6 h of exposure. The optimum point to calculate the index of acetylation is at 3 h after administration (T3), as this corresponds to the time at which the ratio of the metabolite concentration and that of the parent molecule stabilizes for slow acetylators [13], [14], [15]. In this study, we have not calculated the values of kinetic parameters (Cmax, Tmax, T0.5, AUC and Cmax) because of insufficient time-points for this purpose. However, the study of the mean concentrations in slow acetylators and fast acetylators indicates whether the obtained results are located in the zone of active INH concentrations. However, the study average concentrations in slow acetylators and fast acetylators allows us to see if the concentration of INH obtained based on acetylation profiles is located in the zone of active INH concentrations.
The decrease in concentration of INH in the blood is not directly proportional to the increase of the plasma concentration of AcINH. This is due to the first pass hepatic effect of the drug and its metabolite with a linear index of the activity of the enzyme. This allowed the use of the AcINH/INH ratio for population classification according to the rate of acetylation [16], [6]. Thus, several researchers have used the ratio [AcINH]/[INH] as a parameter of choice for determining the acetylation phenotype [17], [18], [19], [20].
Studies have shown that a concentration of INH between 3 and 5 mg/L is desirable at 2 h after drug administration [22], [23], [21], although it has also been suggested that a concentration of 1.5 mg/L at 3 h is an optimum effective amount of drug against mycobacteria [24], [25], [26]. But, these values can be dependent on the co-administration of combinations with other drugs such as rifampicin. Indeed, in a multi-drug therapy regime, demonstrated differences in slow and fast phenotypes in INH are more likely to originate from an effect of toxicity rather than from a therapeutic origin [27], [28]. However, in cases of improper drug administration [29], drug malabsorption [30], or widely spaced intermittent therapy [31], [32], treatment failure may be well justified in fast acetylators. Thus, an efficient dose of INH must take into account the possibility of drug toxicity based on the profile of acetylation of NAT2.
The results obtained in this study show concentrations above the recommended values at 3 h for both slow and fast acetylators. It is therefore possible that a dose lower than 5 mg/kg body weight should be considered in tuberculosis treatment in Senegal to avoid toxicity related mainly to the slow metabolizing phenotype.
According to previous studies of TB patients within the literature, a dose of 3 mg/kg may be proposed for slow acetylators versus a dose of 6 mg/kg for fast acetylators; this in order to achieve bactericidal concentrations of INH and simultaneously avoid cases of liver toxicity and peripheral neuropathy [33], [34], [35]. Hence, a priori knowledge of the acetylation rate of a patient is important to administer an appropriate therapy.
In our study, the correlation between genotype and phenotype through the plasma concentration of INH and the metabolic ratios at 3 h post-dose, gave a concordance coefficient kappa (κ) of 58%; showing a moderate agreement between the two techniques. This suggests that the plasma concentrations of INH are:
-
•
either low, due to the increase of acetylation of the drug, and thus its metabolites, in fast acetylator phenotype;
-
•
or there is a discrepancy between the plasma concentration of INH and the metabolic ratio related to poor drug absorption.
The debate has always been complicated after it became clear that there were significant differences between individuals in their ability to eliminate INH [36], [37]. Because of such considerable differences in response to INH exposure, it is urgent for each country to conduct a profiling study from its own population, for proper follow up to possible therapeutic and toxic consequences linked to the use of certain drugs or combinations of drugs. We have demonstrated that an acetylation profile allows an optimized treatment of TB with a net reduction of the INH toxicity.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
We are particularly grateful to Dr. Richard Robins, Director of Research of the French National Centre for Scientific Research (CNRS), from the CEISAM Laboratory in Nantes, for his useful suggestions and careful rereading of the article; to Mr. Christophe Hallaert and Ms Isabelle Szuster Department of Genopathie of Centre de Biologie Pathologie de Lille2 for their help and technical assistance in performing a confirmatory sequencing of NAT2.
Contributor Information
A. Toure, Email: amitoure@hotmail.com, aminatatr@gmail.com.
M. Cabral, Email: tildacabral@yahoo.fr.
A. Niang, Email: niangpneumo@gmail.com.
C. Diop, Email: cheikhkoki@hotmail.com.
A. Garat, Email: anne.garat-2@univ-lille2.fr.
L. Humbert, Email: luc.humbert@chru-lille.fr.
M. Fall, Email: madoufal@gmail.com.
A. Diouf, Email: amdiouf@refer.sn.
F. Broly, Email: f-broly@chru-lille.fr.
M. Lhermitte, Email: m.lhermitte@wanadoo.fr.
D. Allorge, Email: dallorge@univ-lille2.fr.
References
- 1.Evans D.A.P., Manley K.A., Mc Kusick V.A. Genetic control of isoniazid metabolism in man. Br. Med. J. 1960;13:485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubota R., Ohno M., Hasunuma T., Iijima H., Azuma J. Dose-escalation study of isoniazid in healthy volunteers with the rapid acetylator genotype of arylamine N-acetyltransferase 2. Eur. J. Clin. Pharmacol. 2007;63:927–933. doi: 10.1007/s00228-007-0333-1. [DOI] [PubMed] [Google Scholar]
- 3.WHO . 20th edition. 2015. Global Tuberculosis Report 2015. 204p. Available from URL: http://www.who.int/tb/publications/global_report/en/ (accessed 26.02.16.) [Google Scholar]
- 4.Programme National de Lutte contre la Tuberculose (PNT) au Sénégal: Rapport annuel 2012; ministère de la Santé et de l’Action Sociale du Sénégal: 109p.
- 5.Huang Y.S., Chern H.D., Su W.J., Wu J.C., Lai S.L., Yang S.Y., Chang F.Y., Lee S.D. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology. 2002;35(4):883–889. doi: 10.1053/jhep.2002.32102. [DOI] [PubMed] [Google Scholar]
- 6.Zabost A., Brzezinska S., Kozinska M., Blachnio M., Jagodzinski J., Zwolska Z., Augustynowicz-Kope T.E. Correlation of N-acetyltransferase 2 genotype with isoniazid acetylation in Polish tuberculosis patients. BioMed Res. Int. 2013;2013:853602. doi: 10.1155/2013/853602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinshilboum R. Inheritance and drug response. N. Engl. J. Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 8.Touré A., Diop C., Cabral M., Fall M., Lhermitte M., Diouf A., Broly F., Allorge D. Study of NAT2 genetic polymorphism in West African subjects: example of an healthy non-smoker Senegalese population. Mol. Biol. Rep. 2012;39:10489–10496. doi: 10.1007/s11033-012-1931-2. [DOI] [PubMed] [Google Scholar]
- 9.Vivien J.N., Thibier R., Lepeuple A. La pharmacocinétique de l’isoniazide dans la race blanche. Rev. Mal. Respir. 1973;1:753–773. [Google Scholar]
- 10.Parkin D.P., Vandenplas S., Botha F.J., Vandenplas M.L., Seifart H.I., van Helden P.D., van der Walt B.J., Donald P.R., van Jaarsveld P.P. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am. J. Respir. Crit. Care Med. 1997;155(5):1717–1722. doi: 10.1164/ajrccm.155.5.9154882. [DOI] [PubMed] [Google Scholar]
- 11.Schaaf H.S., Parkin D.P., Seifart H.I., Werely C.J., Hesseling P.B., van Helden P.D., Maritz J.S., Donald P.R. Isoniazid pharmacokinetics in children treated for respiratory tuberculosis. Arch. Dis. Child. 2005;90:614–618. doi: 10.1136/adc.2004.052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Academy of Pediatrics . Tuberculosis. In: Pickering L.K., Baker C.J., Kimberlin D.W., Long S.S., editors. Red Book: 2009 Report of the Committee on Infectious Diseases. 28th edition. American Academy of Pediatrics; Elk Grove Village, IL: 2009. pp. 680–701. [Google Scholar]
- 13.Hutchings A., Routledge P.A. A simple method for determining acetylator phenotype using isoniazid. Br. J. Clin. Pharm. 1986;22:343–345. doi: 10.1111/j.1365-2125.1986.tb02897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mashimo M., Suzuki T., Abe M., Deguchi T. Molecular genotyping of N-acetylation polymorphism to predict phenotype. Hum. Genet. 1992;60:139–143. doi: 10.1007/BF00210758. [DOI] [PubMed] [Google Scholar]
- 15.Smith C.A., Wadelius M., Gough A.C., Harrison D.J., Wolf C.R., Rane A. A simplified assay for the arylamine N-acetyltransferase 2 polymorphism validated by phenotyping with isoniazid. J. Med. Genet. 1997;34(9):758–760. doi: 10.1136/jmg.34.9.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson P.R., Tucker G.T., Lennard M.S., Woods H.F. Polymorphic drug oxidation: pharmacokinetic basis and comparision of experimental indices. Br. J. Clin. Pharm. 1986;22:541–550. doi: 10.1111/j.1365-2125.1986.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matar K.M., Mayet A.Y., Ayoola E.A., Bawazir S.A., Al-Faleh F.Z., Al-Wazzan A. Isoniazid acetylation phenotyping in Saudi Arabs. J. Clin. Pharm. Ther. 2004;29(5):443–447. doi: 10.1111/j.1365-2710.2004.00588.x. [DOI] [PubMed] [Google Scholar]
- 18.Ng K.Y., Zhou H., Zhang Y.L., Hybertson B., Randolph T., Christians U. Quantification of isoniazid and acetylisoniazid in rat plasma and alveolar macrophages by liquid chromatography–tandem mass spectrometry with on-line extraction. J. Chromatogr. B. 2007;847:188–198. doi: 10.1016/j.jchromb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Singh N., Dubey S., Chinnaraj S., Golani A., Maitra A. Study of NAT2 gene polymorphisms in an indian population. association with plasma isoniazid concentration in a cohort of tuberculosis patients. Mol. Diagn. Ther. 2009;13(1):49–58. doi: 10.1007/BF03256314. [DOI] [PubMed] [Google Scholar]
- 20.Kayhan S., Akgüneş A. Therapeutic monitoring of isoniazid, rifampicin, ethambutol and pyrazinamide serum levels in the treatment of active pulmonary tuberculosis and determinants of their serum concentrations. Afr. J. Pharm. Pharmacol. 2011;5(17):2015–2041. [Google Scholar]
- 21.Alsultan A., Peloquin C.A. Therapeutic drug monitoring in the treatment of tuberculosis: an update. Drugs. 2014;74(8):839–854. doi: 10.1007/s40265-014-0222-8. [DOI] [PubMed] [Google Scholar]
- 22.Peloquin C.A. Therapeutic drug monitoring in the treatment of tuberculosis. Drugs. 2002;62(15):2169–2183. doi: 10.2165/00003495-200262150-00001. [DOI] [PubMed] [Google Scholar]
- 23.Babalik A., Babalik A., Mannix S., Francis D., Menzies D. Therapeutic drug monitoring in the treatment of active tuberculosis. Can. Respir. J. 2011;18(4):225–229. doi: 10.1155/2011/307150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moussa L.A., Khassouani C.E., Soulaymani R., Janab M., Cassanasc G., Alricd R., Hüed B. Therapeutic isoniazid monitoring using a simple high-performance liquid chromatographic method with ultraviolet detection. J. Chromatogr. B. 2002;766(1):181–187. doi: 10.1016/s0378-4347(01)00434-0. [DOI] [PubMed] [Google Scholar]
- 25.Kinzig-Schippers M., Tomalik-Scharte D., Jetter A., Scheidel B., Jakob V., Rodamer M., Cascorbi I., Doroshyenko O., Sörgel F., Fuhr U. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob. Agents Chemother. 2005;49(5):1733–1738. doi: 10.1128/AAC.49.5.1733-1738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azuma J., Ohno M., Kubota R., Yokota S., Nagai T., Tsuyuguchi K., Okuda Y., Takashima T., Kamimura S., Fujio Y., Kawase I. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics-based therapy. Eur. J. Clin. Pharmacol. 2013;69:1091–1101. doi: 10.1007/s00228-012-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellard G.A. The potential clinical significance of isoniazid acetylator phenotype in the treatment of pulmonary tuberculosis. Tubercle. 1984;65:211–227. doi: 10.1016/0041-3879(84)90079-5. [DOI] [PubMed] [Google Scholar]
- 28.Tostmann A., Boeree M.J., Aarnoutse R.E., de Lange W.C., van der Ven A.J., Dekhuijzen R. Antituberculosis drug-induced hepatotoxicity: concise up-to-date review. J. Grastroenterol. Hepatol. 2008;23:192–202. doi: 10.1111/j.1440-1746.2007.05207.x. [DOI] [PubMed] [Google Scholar]
- 29.Mitchison D.A. How drug resistance emerges as result of poor compliance during short course chemotherapy for tuberculosis. Int. J. Tuberc. Lung Dis. 1998;2:10–15. [PubMed] [Google Scholar]
- 30.Peloquin C.A., Nitta A.T., Burman W.J., Brudney K.F., Miranda-Massari J.R., McGuinness M.E., Berning S.E., Gerena G.T. Low antituberculosis drug concentrations in patients with AIDS. Ann. Pharmacother. 1996;30(9):919–925. doi: 10.1177/106002809603000901. [DOI] [PubMed] [Google Scholar]
- 31.Weiner M., Burman W., Vernon A., Benator D., Peloquin C.A., Khan A., Weis S., King B., Shah N., Hodge T., Tuberculosis Trials Consortium Low isoniazid concentrations and outcome of tuberculosis treatment with once-weekly isoniazid and rifapentine. Am. J. Respir. Crit. Care Med. 2003;167:1341–1347. doi: 10.1164/rccm.200208-951OC. [DOI] [PubMed] [Google Scholar]
- 32.Weiner M., Benator D., Burman W., Peloquin C.A., Khan A., Vernon A., Jones B., Silva-Trigo C., Zhao Z., Hodge T., Tuberculosis Trials Consortium Association between acquired rifamycin resistance and the pharmacokinetics of rifabutin and isoniazid among patients with HIV and tuberculosis. Clin. Infect. Dis. 2005;40:1481–1491. doi: 10.1086/429321. [DOI] [PubMed] [Google Scholar]
- 33.Ohno M., Yamaguchi I., Yamamoto I., Fukuda T., Yokota S., Maekura R., Ito M., Yamamoto Y., Ogura T., Maeda K., Komuta K., Igarashi T., Azuma J. Slow N-acetyltransferase 2 genotype affects the incidence of isoniazid and rifampicin-induced hepatotoxicity. Int. J. Tuberc. Lung Dis. 2000;4:256–261. [PubMed] [Google Scholar]
- 34.Hiratsuka M., Kishikawa Y., Takekuma Y. Genotyping of the N-acetyltransferase 2 polymorphism in the prediction of adverse drug reactions to isoniazid in Japanese patients. Drug Metab. Pharmacokinet. 2002;17:357–362. doi: 10.2133/dmpk.17.357. [DOI] [PubMed] [Google Scholar]
- 35.Ben Mahmoud L., Ghozzi H., Kamoun A., Hakim A., Hachicha H., Hammami S., Sahnoun Z., Zalila N., Makni H., Zeghal K. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatotoxicity in Tunisian patients with tuberculosis. Pathol. Biol. 2012;60(5):324–330. doi: 10.1016/j.patbio.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Bönicke R., Reif W. Enzymatische inaktiviering von isonikotinsaure hydrazide in menslichen und tierschen organismus. Arch. Exp. Pathol. Pharmakol. 1953;220:321–333. [PubMed] [Google Scholar]
- 37.Hughes H.B. On the metabolic fate of isoniazid. J. Pharmacol. Exp. Ther. 1953;109:444–454. [PubMed] [Google Scholar]