Abstract
The design and synthesis of compounds that target mixed, AT/GC, DNA sequences is described. The design concept connects two N-methyl-benzimidazole-thiophene single GC recognition units with a flexible linker that lets the compound fit the shape and twist of the DNA minor groove while covering a full turn of the double helix.
Graphical Abstract
Mixed-sequence-binding, rationally-designed heterocyclic dications specifically recognize a full turn of the DNA minor groove.
Small, synthetic molecules that selectively target biological macromolecules in cells and induce specific responses, such as changes in gene expression, are a central goal of biomolecular compound design and synthesis research as well as therapeutic development.1–8 While there is significant progress in this area with proteins, there is limited progress in the variety of compound designs to target mixed base pair (bp) DNA sequences. Approaches for compounds that can target the DNA component of DNA-protein complexes, for example, would help remove this block to progress and provide an important step forward in the area.9–12 In order to effectively target DNA in cells, compounds with appropriate physical properties must be designed along with the development of synthetic approaches that are reasonable in cost and effort.
Heterocyclic-cation, minor-groove binders that can selectively bind to DNA have become a valuable resource in therapeutics, and compounds that successfully act on specific cancers, parasitic organisms and bacteria are now in animal and clinical testing as well as therapeutic use.1,13–15 These successful agents have provided proof of concept that selective and functional recognition of the DNA minor groove with a set of modules, combined in different ways for different sequences, is possible. The relatively simple agents available, however, primarily recognize only A·T bp sequences and have off-target effects. 16
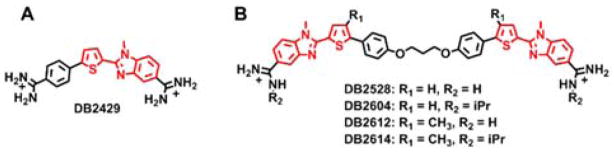
Our approach to overcome this limitation is design and preparation of modular minor-groove, sequence-specific compounds that can recognize G·C in addition to A·T bp in longer DNA sequences than most current agents. The design strategy starts with heterocyclic cationic molecular modules that have effective DNA binding affinity in AT sequences, cell and nuclear uptake, reasonable syntheses, relatively low human toxicity and a range of physical properties to increase the chances of finding active compounds for the treatment of a variety of diseases.11 Using this approach a thiophene-N-methylbenzimidazole (thiophene-N-MeBI) compound (DB2429, Figure 1A) was designed according to the “σ-hole” preorganizing principle for macromolecular targeting.17 This effect is based on the presence of low-lying C–S σ* orbitals on the thiophene S that possess positive electrostatic potential for interaction with electron donating atoms such as the unsubstituted N in N-MeBI.18,19 Such rational control of the conformation of small molecules is a key concept of optimized molecular design for targeting macromolecules and can lead to significantly improved binding affinity and specificity that is independent of compound–DNA contacts. Compounds with furan or pyridine in place of the thiophene in DB2429 bind with less affinity and selectivity in support of the role of σ-hole interactions with the thiophene.17
Fig. 1.
A) The compound DB2429 with best binding affinity and selectivity for a single G·C bp sequence found in our initial research. B) Chemical structure of new compounds designed to link two GC recognition modules. Red: G·C bp recognition module.
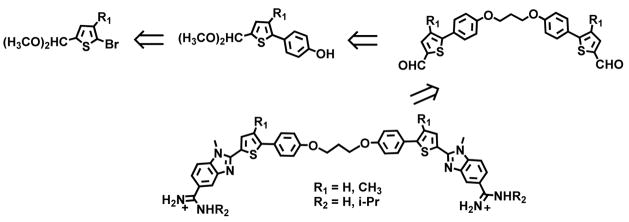
The critical question at this point, which is addressed in this report for the first time, is how to extend relatively simple compounds, such as DB2429, to selectively recognize longer, more complex DNA sequences with additional G·C bp. Our approach combines modular GC recognition units to prepare agents that can selectively recognize two G·C bps in DNA sequences such as (A/T)3-(G/C)-(A/T)n-(G/C)-(A/T)3 where “n” can be 0–5 bps (Figure 1). For the initial test of this approach, a flexible linker, –O-(CH2)3-O-, for DB2429-type GC recognition molecular modules was used. The design strategy includes enough flexibility to match the shape and curvature of the DNA minor groove and an appropriate length with modules spaced to bind to the GAAAC DNA sequence and a full turn of the DNA helix.
Our first transcription factor (TF)-DNA complex for targeting was the PU.1 TF that is of critical importance in the development of acute myeloid leukemia (AML). Compounds that bind AT or single G·C bp sequences have been successful at targeting the PU.1 promoter and inhibiting AML in cells.20 To increase the effectiveness of the TF-promoter DNA targeting approach, we wish to target additional TFs, such as the Blimp-1 TF protein that impairs T cell function and is correlated with AML progression.21 Blimp-1 binds to a consensus GAAAC/G promoter sequence,22,23 and agents that bind strongly to that sequence have the potential to inhibit the TF and provide new anti-AML activity. Scheme 1 shows a retrosynthetic analysis which allows modular assembly of small molecules designed to bind to DNA sequences with two G·C bps. The target bis-amidino-N-methylbenzimidazolethiophenes are prepared from amidino-N-methylphenylenediamines by coupling with the appropriate bis-thiophenecarboxaldehydes in the presence of selected oxidizing agents. The required bis-aldehydes are obtained by linking the protected 5-(4-hydroxyphenyl) thiophene-2-carboxaldehdyes with 1, 3-dibromopropane employing standard Williamson Ether synthesis methodology. The protected 5-(4-hydroxyphenyl) thiophene-2-carboxaldehdyes are readily accessible using standard Suzuki coupling reactions between various substituted bromothiophenes and 4-hydroxyphenylboronic acid. A set of related compounds prepared using this approach are shown in Figure 1B. Their synthesis routes are shown in Supporting Information, Scheme S1. The resulting agents are particularly striking because relatively simple changes in chemical groups in AT specific compounds, the addition of specific H-bond accepting groups for G·C bp recognition, converts them into compounds (Figure 1B) that strongly and specifically recognize mixed bps sequences of DNA much more strongly than the original AT sequences. This is the first example where heterocyclic cations linked in this manner have been systematically designed and prepared to recognize G·C bp in complex DNA sequences.
Scheme 1.
Retrosynthetic analysis for bis-amidine-N-Methylbenzimidazole-thiophene compounds from bis-thiophenecarboxaldehydes that are prepared by linking arylthiophenecarboxaldehydes. Full synthetic details are in Supplementary Materials.
Thermal melting (Tm) provides a rapid, evaluation screen of compounds with selected DNA sequences (Supporting Information, Table S1, S2).24,25 Binding to DNA gives an increase in the melting temperature that is related to binding affinity. Hairpin DNA oligomers, which have monomolecular melting transitions, were chosen for the screen. DB2429 (Figure 1A) shows our approach of design of single G recognition compounds and for linking these modular units to recognize more complex sequences. DB2528 and analogs are the first test of this concept and Tm results are shown in Table S2. DB2528, which combines two DB2429 units with a flexible –O-(CH2)3-O-linker (Figure 1B), does not bind well to a single G segment (AAAAGTTTT, ΔTm = 2°C) but binds with a ΔTm of 9°C with the target two G·C bps sequence, GAAAC. It has lower Tm with longer, GAAAAC and GAAAAAC sequences (6°C). It is very encouraging that it shows no significant binding with an all AT sequence (ΔTm = 1°C), and closer G·C bps (GC, GAC, GAAC), all with ΔTm <2°C (Table S1, S2, Figure S1). The ΔTm for GAAAC and CAAAG are essentially identical but the less symmetrical sequence GAAAG gives a 3°C lower ΔTm (Figure S1). Modification of the amidine (DB2604 and 2614) or thiophene (DB2612 and 2614) groups can enhance interactions with the minor groove in cases where self-association limits DNA interactions, but the additions did not enhance binding with DB2528 (Figure 1B, Table S2). The replacement of N-MeBI in DB2528 with benzimidazole (BI) gives a more classical minor groove binder that shows strong binding with the AT sequences and weaker binding to the G sequences (not shown). This result clearly confirms the crucial importance of N-MeBI to the G·C bp recognition. Circular dichroism (CD) results with DB2528 and the GAAAC sequence show a strong positive induced CD peak at the compound absorption wavelength (Figure S2). This result clearly indicates that the compound is a minor groove binder. Addition of a pure AT sequence to DB2528 results in very little spectral change in agreement with quite weak binding to pure AT sequence.
Biosensor-SPR methods provide an excellent way to quantitatively evaluate the interaction of small molecules with immobilized biomolecules.26 SPR provides sensitive, real-time progress of the binding reaction with binding affinity, kinetics, and stoichiometry of complex formation.27,28 Based on the Tm results, the interactions of DB2528 were evaluated by SPR with G·C bp sequences (Table S3). As can be seen in Figure 2, DB2528 binds strongly with GAAAC and global kinetics fitting yielded a single binding site with a KD of 5 nm, a rapid on-rate (ka = 2.83±0.5×106 M−1s−1) and a very slow off-rate (kd = 1.7±0.4×10−2 s−1). The association and dissociation of DB2528 with the shorter GAAG sequence are quite fast with a high KD = 149 nM. Pure AT and single G sequences were also studied by SPR, and it is encouraging that DB2528 has no detectable binding with either of them. These results indicate excellent selectivity and strong binding for a two G sequence that has a specific distance between the two G·C bps. As a test of the SPR binding results, the KD was determined by a fluorescence anisotropy titration of the compound with the GAAAC DNA, and the KD value is in agreement with the results from SPR (Table S3, and Figure S3) and validates the SPR results for binding affinity. Clearly, the combination of thiophene-N-MeBI and a flexible –O-(CH2)3-O-linker creates an effective module for strong two G·C bps specific recognition of a full turn of the DNA helix.
Fig. 2.
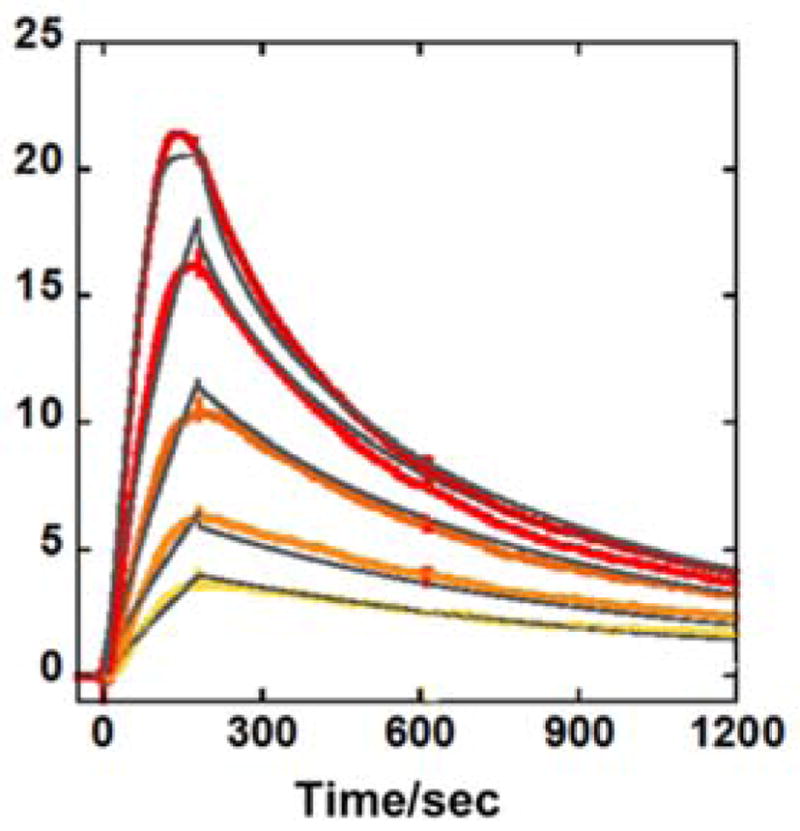
Representative SPR sensorgrams for DB2528 in the presence of GAAAC DNA, concentrations of DB2528 from bottom to top are 10, 15, 30, 50, and 100 nM. The solid black lines are best-fit values for the global kinetic fitting of the results with a single site function.
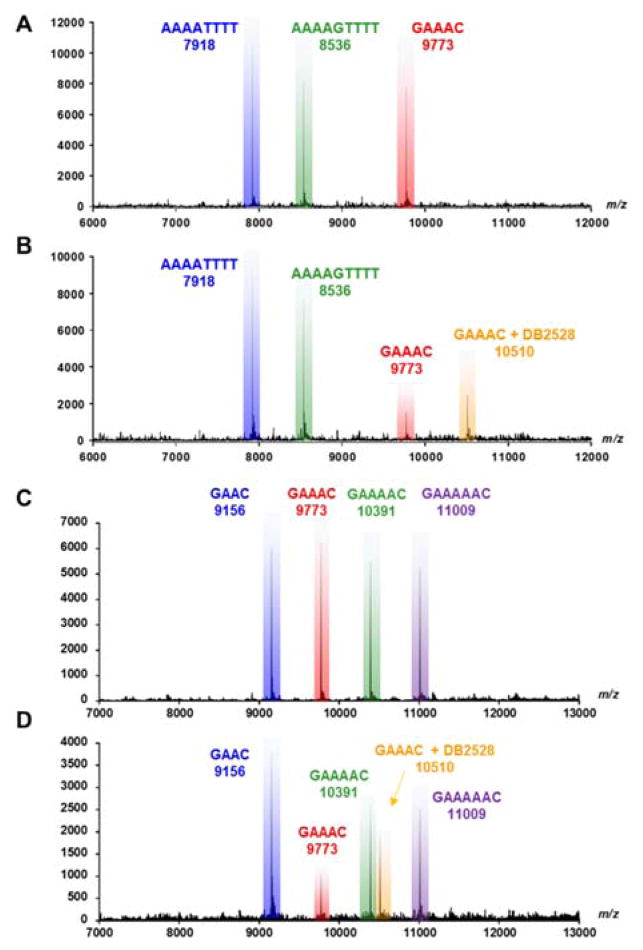
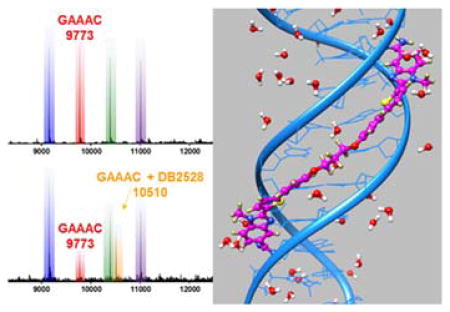
Competition ESI-mass spectrometry (MS) provides a direct analysis of relative binding affinity, stoichiometry and specificity for small molecule DNA complexes.29,30 The use of several DNA sequences simultaneously mixed with a compound creates a competitive binding environment for direct comparison of DNA interactions. The competition ESI-MS analysis results of DB2528 with DNA sequences AAAATTTT, AAAAGTTTT, and GAAAC, are shown in Figure 3A, B. The upper plot shows three peaks for the three DNA sequences. On addition of DB2528, the peak for GAAAC (9773) decreases with the simultaneous appearance of a new peak at m/z=10510 that is characteristic of a 1:1 GAAAC-DB2528 complex (Figure 3B). There is no appearance of any complex peak with the other DNA sequences. Results for competition ESI-MS analysis of DB2528 with sequences GAAC, GAAAC, GAAAAC, and GAAAAAC are shown in Figure 3C, D to test the sequence specificity. The upper plot shows four peaks for the four sequences. With the addition of DB2528, only the peak for GAAAC (9773) decreases with the simultaneous appearance of a new peak at m/z=10510 for a 1:1 GAAAC-DB2528 complex. There is no significant complex peak with any of the other tested sequences. In agreement with the binding results, the competition ESI-MS results clearly indicate that DB2528 binds with GAAAC very strongly and specifically in a 1:1 complex.
Fig. 3.
ESI-MS negative mode spectra of the competition binding of sequences A), B) AAAATTTT, AAAAGTTTT and GAAAC(10 μM each) ; C), D) GAAC, GAAAC, GAAAAC and GAAAAAC (10 μM each); with 40 μM DB2528 in buffer (50 mm ammonium acetate with 10% methanol(v/v), pH 6.8). A), C) The ESI-MS spectra of free DNA mixture. B), D) The ESI-MS spectra of DNA mixture with DB2528. The ESI-MS results shown here are deconvoluted spectra and molecular weights are shown with each peak. Full DNA sequences are in Table S1.
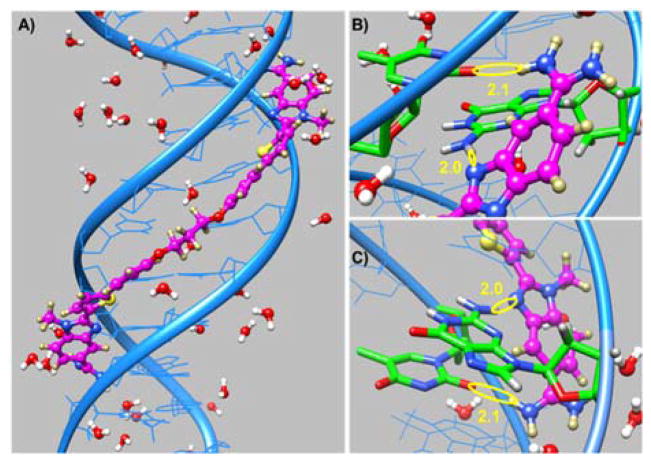
DB2528 was designed to H-bond with two G·C bps separated by three A·T bp and for terminal amidines H-bonds with AT. The compound binds strongly and specifically to this sequence but how does it fit to the minor groove and A·T/G·C bps? To answer this question, the molecular dynamics structures for the DB2528 complex with the minor groove of AGAAACT (Figure 4) were determined with the AMBER 14 software suite and the ff99 force field. Force constants for DB2528 were added and the structure determined as previously described.31–33 The full view in Figure 4A and Figure S4A show DB2528 is able to match the curvature of the DNA minor groove and can cover a full turn of the double helix. The same view is shown in space filling model in Figure S4B and shows the excellent contacts and van der Waals interactions that the compound makes with the minor groove molecular walls. A view of the complex structure at the upper part of the model is shown in Figure 4B. This view shows strong H-bond between the N of N-MeBI and the G-NH that points out into the groove. There is also an H-bond between the amidine-NH that points to the floor of the groove and a T=O. The view in Figure 4C shows the same H-bond pattern with the other end of the complex. The amidine-N-MeBI-thiophene-phenyl-O-can easily be seen tracking along the minor groove. Again, there are strong H-bonds from the G-NH to the N of N-MeBI and from the amidine to a T=O. As can be seen, both amidines are strongly hydrated and this certainly contributes to the energetics of binding and the fit to the groove (Figure 4A and Figure S4). The compound curves around the groove and there is an exact indexing of two N-MeBI-G-NH and amidine-A·T bp.
Fig. 4.
A) Minor groove view of the DB2528-DNA complex with proximal water molecules. B) Hydrogen bonding the upper G-NH and an opposite strand T-O2 with N of N-MeBI and NH of an amidine of DB2528 respectively are shown in yellow circles (in Å). C) View of hydrogen bonding of the lower G-NH and an opposite strand T-O2 with N of N-MeBI and NH of an amidine of DB2528, respectively is shown in yellow circles. Upper and lower are with respect to the model in A. DNA is represented in ribbon style (in blue) whereas G and T involved in H-bonding are shown in stick representation (B and C in green). DB2528 is shown in the ball and stick representation (B and C in magenta).
In conclusion, the initial goal of this work was to link previously designed single G·C bp binding modules with a flexible linker for recognition of two G·C bps in the core sequence AGAAACT that has functional significance. With optimized design, synthesis and analysis a linked heterocyclic-diamidine has been obtained that has an excellent match to the DNA minor groove shape and registry of the compound with DNA functional groups. These results show, for the first time, what can be accomplished with modular compounds of this type in selective targeting of complex DNA. The compound has synthesis that is reasonable in cost and time and offers a promising route to develop a broad array of modular agents for control of gene expression.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM111749 to WDW and DWB.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Neidle S, Thurston DE. Nat Rev Cancer. 2005;5(4):285–296. doi: 10.1038/nrc1587. [DOI] [PubMed] [Google Scholar]
- 2.Howell LA, Searcey M. ChemBioChem. 2009;10(13):2139–2143. doi: 10.1002/cbic.200900243. [DOI] [PubMed] [Google Scholar]
- 3.Mantaj J, Jackson PJM, Rahman KM, Thurston DE. Angew Chem Int Ed. 2017;56(2):462–488. doi: 10.1002/anie.201510610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaires JB. Biopolymers. 2015;103(9):473–479. doi: 10.1002/bip.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savreux-Lenglet G, Depauw S, David-Cordonnier MH. Int J Mol Sci. 2015;16(11):26555–26581. doi: 10.3390/ijms161125971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett MP, Gemmell CG, Suckling CJ. Pharmacol Ther. 2013;139(1):12–23. doi: 10.1016/j.pharmthera.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Dardonville C, Martínez JJN. Curr Med Chem. 2017:24. doi: 10.2174/0929867324666170623091522. [DOI] [PubMed] [Google Scholar]
- 8.Aikawa H, Yano A, Nakatani K. Org Biomol Chem. 2017;15(6):1313–1316. doi: 10.1039/c6ob02273a. [DOI] [PubMed] [Google Scholar]
- 9.Mosquera J, Jiménez-Balsa A, Dodero VI, Vázquez ME, Mascareñas JL. Nat Commun. 2013;4:1874. doi: 10.1038/ncomms2825. [DOI] [PubMed] [Google Scholar]
- 10.He G, Tolic A, Bashkin JK, Poon GMK. Nucleic Acids Res. 2015;43(8):4322–4331. doi: 10.1093/nar/gkv267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munde M, Wang S, Kumar A, Stephens CE, Farahat AA, Boykin DW, Wilson WD, Poon GMK. Nucleic Acids Res. 2014;42(2):1379–1390. doi: 10.1093/nar/gkt955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millan CR, Acosta-Reyes FJ, Lagartera L, Ebiloma GU, Lemgruber L, Nué Martínez JJ, Saperas N, Dardonville C, de Koning HP, Campos JL. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang JS, Dervan PB. Q Rev Biophys. 2015;48(4):453–464. doi: 10.1017/S0033583515000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X, Gray PJ, Von Hoff DD. Cancer Treat Rev. 2009;35(5):437–450. doi: 10.1016/j.ctrv.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Lin C, Mathad RI, Zhang Z, Sidell N, Yang D. Nucleic Acids Res. 2014;42(9):6012–6024. doi: 10.1093/nar/gku219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanjunda R, Wilson WD. Curr Protoc Nucleic Acid Chem. 2012;Chapter 8(Unit8.8) doi: 10.1002/0471142700.nc0808s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo P, Paul A, Kumar A, Farahat AA, Kumar D, Wang S, Boykin DW, Wilson WD. Chem Eur J. 2016;22(43):15404–15412. doi: 10.1002/chem.201603422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beno BR, Yeung KS, Bartberger MD, Pennington LD, Meanwell NA. J Med Chem. 2015;58(11):4383–4438. doi: 10.1021/jm501853m. [DOI] [PubMed] [Google Scholar]
- 19.Tilly D, Chevallier F, Mongin F. Synthesis. 2016;48(2):184–199. [Google Scholar]
- 20.Antony-Debré I, Paul A, Leite J, Mitchell K, Kim HM, Carvajal LA, Todorova TI, Huang K, Kumar A, Farahat AA, Bartholdy B, Narayanagari S-R, Chen J, Ambesi-Impiombato A, Ferrando AA, Mantzaris I, Gavathiotis E, Verma A, Will B, Boykin DW, Wilson WD, Poon GMK, Steidl U. J Clin Invest. 2017 doi: 10.1172/JCI92504. In Press, Manuscript No.: 92504-JCI-RG-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu L, Kong Y, Zhang J, Claxton DF, Ehmann WC, Rybka WB, Palmisiano ND, Wang M, Jia B, Bayerl M, Schell TD, Hohl RJ, Zeng H, Zheng H. J Hematol Oncol. 2017;10(124):1–13. doi: 10.1186/s13045-017-0486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo TC, Calame KL. J Immunol. 2004;173(9):5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 23.Deng S, Yuan T, Cheng X, Jian R, Jiang J. Mol Biol Rep. 2010;37(8):3747–3755. doi: 10.1007/s11033-010-0028-z. [DOI] [PubMed] [Google Scholar]
- 24.Munde M, Kumar A, Peixoto P, Depauw S, Ismail MA, Farahat AA, Paul A, Say MV, David-Cordonnier MH, Boykin DW, Wilson WD. Biochemistry. 2014;53(7):1218–1227. doi: 10.1021/bi401582t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi X, Chaires JB. Nucleic Acids Res. 2006;34(2):e14. doi: 10.1093/nar/gnj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nanjunda R, Munde M, Liu Y, Wilson W. In: Methods for Studying Nucleic Acid/Drug Interactions. Wanunu Y, editor. 2011. pp. 92–122. [Google Scholar]
- 27.Liu Y, Kumar A, Depauw S, Nhili R, David-Cordonnier MH, Lee MP, Ismail MA, Farahat AA, Say M, Chackal-Catoen S, Batista-Parra A, Neidle S, Boykin DW, Wilson WD. J Am Chem Soc. 2011;133(26):10171–10183. doi: 10.1021/ja202006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor RD, Asamitsu S, Takenaka T, Yamamoto M, Hashiya K, Kawamoto Y, Bando T, Nagase H, Sugiyama H. Chem Eur J. 2014;20(5):1310–1317. doi: 10.1002/chem.201303295. [DOI] [PubMed] [Google Scholar]
- 29.Laughlin S, Wang S, Kumar A, Farahat AA, Boykin DW, Wilson WD. Chem Eur J. 2015;21(14):5528–5539. doi: 10.1002/chem.201406322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul A, Nanjunda R, Kumar A, Laughlin S, Nhili R, Depauw S, Deuser SS, Chai Y, Chaudhary AS, David-Cordonnier MH, Boykin DW, Wilson WD. Bioorg Med Chem Lett. 2015;25(21):4927–4932. doi: 10.1016/j.bmcl.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Athri P, Wilson WD. J Am Chem Soc. 2009;131(22):7618–7625. doi: 10.1021/ja809249h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laughlin-Toth S, Carter EK, Ivanov I, Wilson WD. Nucleic Acids Res. 2017;45(3):1297–1306. doi: 10.1093/nar/gkw1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Špačková N, Cheatham TE, Ryjáček F, Lankaš F, van Meervelt L, Hobza P, Šponer J. J Am Chem Soc. 2003;125(7):1759–1769. doi: 10.1021/ja025660d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.