SUMMARY
Antiviral responses are regulated by conjugation of ubiquitin (Ub) and interferon-stimulated gene 15 (ISG15) to proteins. Certain classes of viruses encode Ub- or ISG15-specific proteases belonging to the ovarian tumor (OTU) superfamily. Their activity is thought to suppress cellular immune responses, but studies demonstrating the function of viral OTU proteases during infection are lacking. Crimean-Congo hemorrhagic fever virus (CCHFV, family Nairoviridae) is a highly pathogenic human virus that encodes an OTU with both deubiquitinase and deISGylase activity as part of the viral RNA polymerase. We investigated CCHFV OTU function by inactivating protease catalytic activity or by selectively disrupting its deubiquitinase and deISGylase activity using reverse genetics. CCHFV OTU inactivation blocked viral replication independently of its RNA polymerase activity, while deubiquitinase activity proved critical for suppressing the interferon responses. Our findings provide insights into viral OTU functions and supports the development of therapeutics and vaccines.
Graphical Abstract
INTRODUCTION
The innate immune response is the first line of defense against viruses. They are sensed by pattern-recognition receptors (PRRs) such as Toll-like receptors and retinoic acid-inducible gene-I (RIG-I)-like receptors. PRR activation triggers the activation of intracellular signaling pathways that ultimately lead to the production of inflammatory cytokines and type I interferons (IFNs). Many cellular signaling pathways, including those involved in the innate immune response, are regulated by protein ubiquitination, a post-translational modification mechanism entailing the conjugation of a small 8 kDa ubiquitin protein (Ub). Ub is conjugated to either a lysine (K) side chain or the initiation methionine of the targeted protein, and can be conjugated as a single Ub moiety (mono-ubiquitin) or as elongated chains containing multiple Ub units (polyUb) (reviewed in (Heaton et al., 2015; Hu and Sun, 2016). Conversely, cellular deubiquitinases (DUBs) reverse this process by removing Ub from proteins.
Ubiquitination is critical for the regulation of type I IFN expression: various key signaling proteins such as RIG-I, MAVS, TRAF3, TBK1, and IRF3 require K63-linked ubiquitination for their activation. Conversely, K48-linked poly-ubiquitination generally serves as a degradation signal. Binding of secreted type I IFN activates the interferon-α/β receptor, resulting in the transcription of interferon-stimulated genes (ISGs) and induction of an antiviral state in infected and neighboring cells. To suppress these antiviral responses, viruses have adopted strategies to interfere with ubiquitination. Similar to cellular DUBs capable of forming a negative feedback system for the IFN responses (Kayagaki et al., 2007), virus-encoded DUBs of the ovarian tumor (OTU) superfamily are proposed to suppress the IFN responses. Viral OTU DUBs have been reported in both positive-strand (Tymoviridae and Arteriviridae) and negative-strand RNA viruses (Bunyaviridae and tenuiviruses) (Capodagli et al., 2013; Frias-Staheli et al., 2007; Honig et al., 2004; van Kasteren et al., 2012; Kinsella et al., 2004; Lombardi et al., 2013; Makarova, 2000; Zhang et al., 2007). In addition to their deubiquitinase activity, certain viral DUBs can also remove interferon-stimulated gene 15 (ISG15) modifications, ubiquitin-like and interferon-inducible proteins that are conjugated to target proteins in a manner similar to Ub. Viral DUB and deISGylation activity may be linked to host specificity, pathogenicity and disease severity. Ubiquitin is a highly conserved protein, whereas ISG15 displays considerable sequence variation among mammals. The role of ISG15 and protein ISGylation is complex and the outcome on virus replication is very unpredictable and even varies between host species. For instance, human ubiquitin-specific peptidase 18 (USP18) negatively regulates type-I IFN signaling by binding to the IFN receptor. Human ISG15 stabilizes USP18 and thus is part of this negative feedback, while no equivalent feedback exists in mice (Speer et al., 2016). ISG15 displays various antiviral functions, both as a secreted protein and upon conjugation. ISGylation can disrupt protein function, enhance activity or alter cellular localization. For example, ISGylation enhances antiviral responses by prolonging IRF3 activation (Shi et al., 2010), and can activate PKR in the absence of dsRNA (Okumura et al., 2013). In contrast, ISGylation can also negatively regulate RIG-I (Kim et al., 2008). ISGylation of viral proteins can interfere with their function; for example, ISGylation of human papillomavirus capsid interferes with particle assembly (Durfee et al., 2010). Furthermore, ISGylation of cellular factors blocks assembly and budding of several viruses, including Ebola virus, HIV and influenza (Okumura et al., 2006, 2008; Tang et al., 2010; Woods et al., 2011). ISG15 is not only important for intracellular signaling, as secreted ISG15 induces IFN-γ a cytokine critical for both innate and adaptive immunity (Recht et al., 1991).
Viral DUB immunosuppressive activity has almost exclusively been demonstrated using overexpression systems; few studies have assessed viral DUB activities in the course of a viral infection. In addition, data on viral DUB function during negative-strand RNA virus replication is completely lacking. Crimean-Congo hemorrhagic fever virus (CCHFV; order Bunyavirales, family Nairoviridae) possesses a negative-strand, tri-segmented genome, with the large (L) segment encoding an unusually large multifunctional protein (L). This protein contains the RNA-dependent RNA polymerase (RdRp) and an N-terminal OTU-like cysteine protease (OTU) domain, which is a distinctive feature of the genus Nairovirus. Other nairovirus OTUs appear to preferentially cleave either Ub or ISG15 moieties. For example, Dugbe virus displays a strong preference for Ub, whereas Erve virus displays a much higher activity towards ISG15 substrates (Capodagli et al., 2013). In contrast, the CCHFV OTU is capable of cleaving both Ub and ISG15 efficiently, and overexpression of the CCHFV OTU domain results in a general reduction of ubiquitinated and ISGylated protein levels. Furthermore, CCHFV OTU activity reduced the RIG-I/MAVS-mediated IFN response, as well as TNFα-mediated NF-κB (Frias-Staheli et al., 2007; van Kasteren et al., 2012). While current data support OTU activity as an immune evasion mechanism for CCHFV, these functions have yet to be investigated in the context of viral infection, where the complete L protein likely has a different spatiotemporal distribution than the overexpressed OTU domain. Here we investigate the deubiquitinase and deISGylase functions of the CCHFV OTU during viral infection using a reverse genetics approach. We found that simply inactivating the OTU by mutating the catalytic cysteine residue prohibits rescue of infectious CCHFV. Thus, to investigate the function of the OTU domain in the context of CCHFV infection, we generated recombinant CCHFV variants with altered OTU activities by disrupting its binding to Ub and ISG15. Using these CCHFV mutants, we confirmed that the OTU domain suppresses the cellular immune responses. Furthermore, we demonstrated that OTU DUB activity, rather than its deISGylase activity, is critical for suppressing the IFN responses. This study reveals critical functions of viral OTUs during infection.
RESULTS
OTU catalytic activity is required for recovery of infectious CCHFV and for efficient formation of virus-like particles
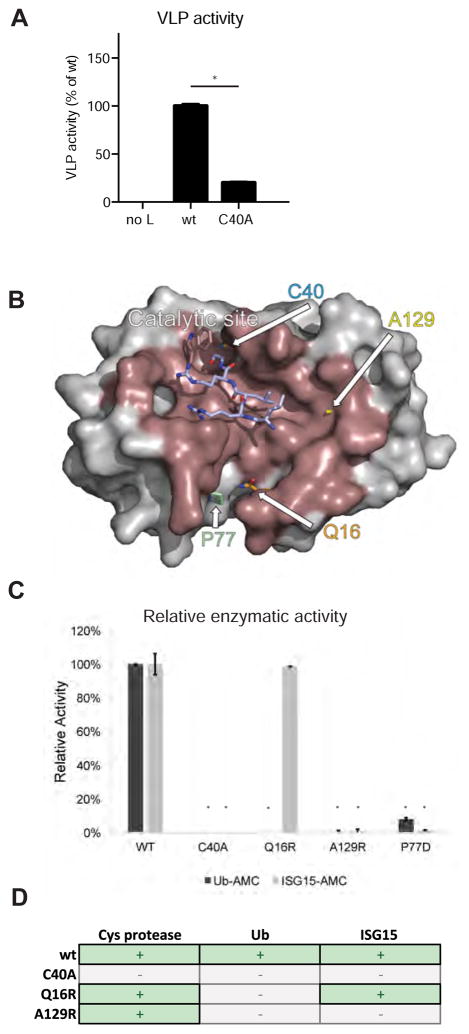
Many positive-strand RNA viruses that encode a DUB utilize the same protease to autoproteolytically process their viral polyproteins, a process required for replicase maturation. Therefore, inactivating the catalytic site of the viral protease prevents viral replication. Recently, viral DUB activity was separated from viral protein processing by reducing Ub binding affinities (van Kasteren et al., 2013). Although they are part of the viral polymerase, DUBs encoded by negative-strand RNA viruses (such as CCHFV) do not appear to utilize their protease to process viral proteins, and their activity is dispensable for RdRp function (Bergeron et al., 2010). Therefore, we expected to generate viable recombinant virus devoid of OTU proteolytic activity by mutating the OTU catalytic cysteine to alanine (C40A). Unexpectedly, multiple attempts to recover CCHFV with an inactive OTU failed. To explain this observation, we sought to identify if the formation of CCHF virus-like particles (VLPs) would be affected by the C40A mutation. Unlike what we previously observed with the minigenome system (Bergeron et al., 2010), an intact OTU domain was important for efficient formation of VLPs (Figure 1A), providing evidence that inactivating the OTU domain impairs CCHFV virion production and/or infectivity.
Figure 1. Design of OTU mutations and in vitro assessment of OTU activity.
(A) CCHF virus-like particles (VLPs) were generated by transfecting Huh7 cells with plasmids encoding CCHFV polymerase (L), nucleoprotein (NP), and glycoprotein (GPC), as well as a minigenome encoding luciferase and T7 polymerase. After 3 days, supernatants containing the VLPs were harvested and transferred to new Huh7 cells, and the luciferase signal was determined as a measure of VLP activity. Data are presented as mean ± SD of 3 biological replicates and * indicate p < 0.05 (B) A model of the CCHFV OTU in complex with ubiquitin (PDB entry 3PRP) was used to assist with the design of mutations that disrupt the OTU-Ub and OTU-ISG15 binding. The protease surface that forms the binding interface with ubiquitin is shaded in brown, with the residues targeted for mutation in this study indicated. The residues forming the shared consensus sequence of ubiquitin and ISG15, LRLRGG, are shown in purple. (C) OTU mutant activity profiles on 7-amino-4-methylcoumarin (AMC) fluorogenic substrates of monoubiquitin (Ub-AMC) and ISG15 (ISG-AMC) was assessed in vitro. Data are presented as mean of two biological replicates ± SD and * indicate p < 0.05 relative to WT (D) Overview of selected OTU mutations and their predicted effect on deubiquitinase and deISGylase activities. between the wild-type OTU or L protein and the individual mutants.
Mutating key residues in the OTU domain generates mutants with altered DUB and deISGylase activity
To study OTU function during replication, we mutated key residues within the domain to disrupt either Ub or ISG15 binding. The structure of the complete CCHFV L protein has not been elucidated, but that of the OTU domain was recently solved, providing insight into Ub and ISG15 binding specificity and allowing the design of OTU mutants unable to bind Ub and/or ISG15 (Akutsu et al., 2011; Capodagli et al., 2011; James et al., 2011) (Figure 1B). Mutations disrupting activity on ISG15 (P77D) or Ub (Q16R) were previously described (Akutsu et al., 2011), and we generated an additional OTU mutant predicted not to cleave Ub or ISG15 (A129R, Figure 1D).
To determine the functional activity of the OTU mutants, their capacity to cleave fluorogenic (AMC) substrates of Ub and ISG15 in vitro was analyzed (Figure 1C). The wild-type OTU efficiently cleaved Ub-AMC and ISG15-AMC. The Q16R mutation was designed to disrupt Ub binding and thus DUB activity, while retaining deISGylation activity. In contrast, the A129R mutant was designed to abrogate activity on both Ub and ISG15. The in vitro data confirmed that these OTU mutants indeed exhibited the predicted activities. An exception was the OTU-P77D mutant that was designed to exclusively disrupt deISGylase activity; the P77D mutation strongly affected DUB activity as well as deISGylase activity (Figure 1C). As further attempts to generate an Ub-specific OTU mutant remained unsuccessful, we employed the Q16R mutation, which retains deISGylase activity, and the A129R mutation, which displays minimal DUB and deISGylase activity, to study the OTU DUB function.
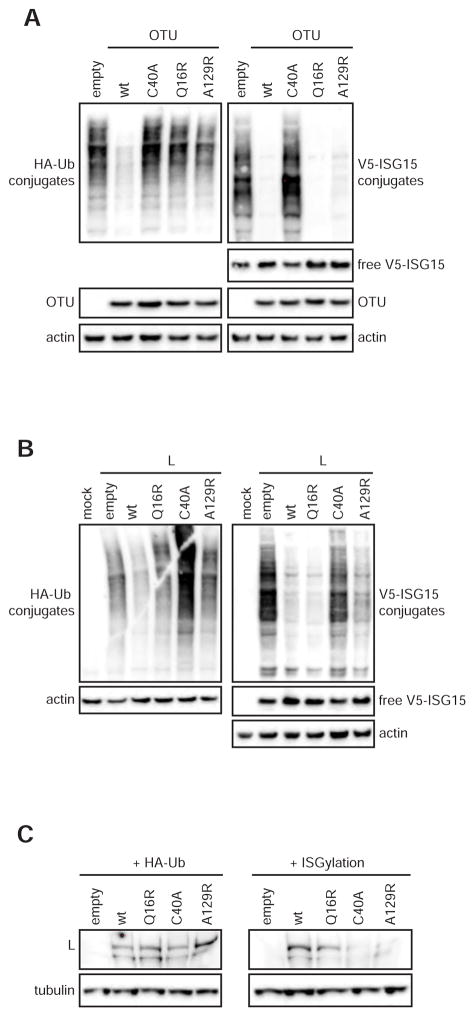
To further characterize the activity of the CCHFV OTU mutants, we assessed the OTU DUB and deISGylase activities in transfected cells. Plasmids encoding the OTU domain or complete CCHFV L protein were transfected with HA-Ub or V5-ISG15 in combination with plasmids expressing the enzymes required for ISG15 conjugation: Ube1L UbcH8, and HERC5. Co-transfection of the ISGylation enzymes is necessary as cellular levels of these ISGs are low in the absence of IFN stimulation. As expected, the wild-type L protein (L-wt) and OTU domain efficiently removed Ub and ISG15 conjugates upon overexpression, whereas the protease-inactive mutant C40A did not reduce levels of ubiquitinated or ISGylated proteins (Figure 2A, B). The Q16R mutants reduced levels of ISG15 conjugates, but did not affect HA-Ub conjugation. CCHFV L-A129R did not display detectable DUB activity, but deISGylase activity could be detected. In addition, overexpressed OTU-A129R showed deISGylase activity resembling that of the wild-type OTU. This is likely due to more efficient expression of the small OTU domain compared to the full-length L, and an apparently stronger affinity for ISG15 compared to Ub.
Figure 2. OTU activity in transfected cells.
To assess the DUB and deISGylase activity of the OTU mutants, Huh7 cells were co-transfected with plasmids expressing HA-Ub or V5-ISG15, Ube1L, UbcH8 and HERC5. Cell lysates were harvested 2 days post transfection, and proteins analyzed for Ub- or ISG15-conjugates. The OTU activities of both the single OTU domain (A) and the OTU in the context of the complete L protein were assessed by western blot (B). (C) The cellular levels of CCHFV L protein, co-expressed with HA-Ub or ISG15, were analyzed in Huh7 cells by western blot.
Strikingly, L-C40A and L-A129R protein levels were reduced by ISGylation, while it had no effect on L-wt or L-Q16R (Figure 2C and S1). As a result, L protein levels appear to correlate with the virus’s ability to remove ISG15 conjugates via their respective OTUs: L-wt and L-Q16R are able to remove ISG15 conjugates, whereas L-C40A and L-A129R cannot. This suggests that ISGylation destabilizes L or targets it for degradation, which under normal conditions would be countered by the OTU domain. Overall, we showed that the CCHFV OTU mutants generally displayed their expected activities both in vitro (Figure 1) and in transfected cells (Figure 2), and the inability of the OTU to counter ISGylation resulted in reduced L protein levels.
CCHFV OTU overexpression impacts the RIG-I mediated IFN-βresponse
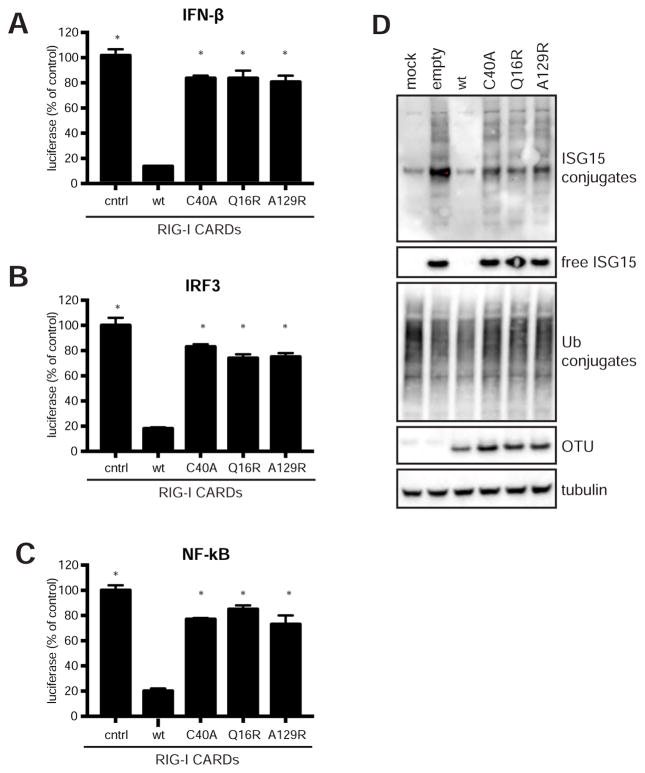
The RIG-I mediated IFN-β response is an important antiviral pathway activated by CCHFV. Overexpression of the OTU domain can block this response, presumably by removing Ub and/or ISG15 conjugates from RIG-I (van Kasteren et al., 2012; Spengler et al., 2015). To investigate directly if altering the OTU activity affects its ability to block the RIG-I mediated IFN-β response, we utilized a reporter assay in which IFN-β expression is induced by a plasmid expressing RIG-I CARDs. IFN-β expression was measured by co-transfecting a plasmid encoding firefly luciferase under control of the IFN-β promoter. As expected, expression of the wild-type OTU significantly decreased reporter activation (Figure 3A). The inactive OTU (C40A) was unable to block the RIG-I mediated reporter gene activation, indicating that CCHFV OTU catalytic activity is necessary for blocking RIG-I-induced IFN-β transcription. In addition, both the OTU-Q16R and OTU-A129R mutants were unable to block reporter gene activation, suggesting that OTU DUB activity is required and sufficient to block the RIG-I-mediated IFN-β response. This is in line with earlier observations that K63-linked poly-ubiquitination of RIG-I CARDs is required for RIG-I activation (Gack et al., 2007; Jiang et al., 2012), that overexpressed wild-type CCHFV OTU can remove Ub moieties from RIG-I CARDs (van Kasteren et al., 2012), and that CCHFV OTU has robust K63-linked polyUb activity (Capodagli et al., 2011). RIG-I activation results in a signaling cascade leading to phosphorylation of IRF3 and NF-κB, which are critical transcription factors activating the IFN-β promoter. To determine whether the CCHFV OTU blocks the activation of both IRF3 and NF-κB, we repeated this assay with the reporter plasmid containing repeats of DNA elements specifically recognized by IRF3 or NF-κB (Figure 3B, C). This resulted in similar outcomes, indicating that the wild-type OTU DUB activity likely blocks IFN-β activation by (directly or indirectly) suppressing both IRF3 and NF-κB activation. To investigate whether CCHFV OTU targets signaling proteins downstream of RIG-I, we overexpressed MAVS or TBK1 to activate the IFN-β signaling pathway; and these experiments produced similar results (Figure S2). This demonstrates that CCHFV OTU prevents activation of IRF3 and NF-κB signaling, either by directly interfering with IRF3 and NF-kB, or by interfering with activation of RIG-I, MAVS and/or TBK1, all of which are ubiquitinated and thus represent potential targets.
Figure 3. Effects of OTU mutations on immune suppression.
To assess the effect of OTU on the RIG-I-mediated IFN response, HEK293T cells were transfected with plasmids expressing the CCHFV OTU domain, the constitutively active RIG-I CARDs to induce IFN-β expression and a firefly luciferase reporter gene under control of the promoter of IFN-β (A), or containing binding sites for IRF3 (B) or NF-κB (C). Data are presented as mean ± SD of 3 biological replicates and * indicate statistical significance of p<0.05 between the wild-type OTU -and the individual mutants. OTU expression and activation of the RIG-I mediated immune response was confirmed by western blot analysis (D).
We confirmed OTU expression and CARD-mediated activation of the IFN-β response by visualizing endogenous ISG15 induction and conjugation (Figure 3D). The western blot data illustrate that wild-type OTU overexpression completely blocks the induction of ISG15 whereas ISGylation was not affected by the OTU mutants.
An active CCHFV OTU is important for efficient viral replication in IFN-competent cells
To further evaluate the function of the OTU mutants in viral infection, we generated the corresponding recombinant viruses and analyzed their growth kinetics (Figure 4). Unlike the catalytic cysteine mutant (C40A), CCHFV-Q16R and -A129R were easily rescued, enabling us to study OTU DUB and deISGylase function in the context of a CCHFV infection. The CCHFV-Q16R and -A129R mutants grew with similar kinetics compared to wild-type recombinant CCHFV (CCHFV-wt) in cells that produce no or little IFN in response to CCHFV infection (BSR-T7/5, SW13, and HAP1). In contrast, the CCHFV-Q16R and -A129R mutants were clearly attenuated in IFN-competent A549 cells, resulting in 2.5-log lower titers at 3 days post infection. The lower viral titers of the Q16R and A129R mutants were associated with more robust protein ISGylation and reduced levels of NP and viral RNA preceding the wild-type virus titer peak, suggestive of enhanced innate immune responses (Fig S3). Next, we assessed the effects of a mutated OTU domain on CCHFV growth kinetics in primary cells (monocyte-derived macrophages, human umbilical vein cells (HUVEC) and skin fibroblasts). CCHFV IbAr10200 does not efficiently infect macrophages (Peyrefitte et al., 2010) or fibroblasts, resulting in low titers. In monocyte-derived macrophages, we found that CCHFV-Q16R displayed similar growth kinetics to CCHFV-wt, whereas CCHFV-A129R was further attenuated. Both CCHFV-Q16R and –A129R replicated less efficiently than CCHFV-wt in primary fibroblasts, although none of these observations reached statistical significance. Although CCHFV infected HUVECs more efficiently, infection still resulted in low titers, and no differences in growth kinetics were observed (Figure 4). Therefore, significant differences in CCHFV OTU mutant growth kinetics are only observed in A549 IFN-competent cells, in which CCHFV-wt reached infectious titers of >108 TCID50/ml.
Figure 4. Growth kinetics of recombinant CCHFV OTU mutants.
Wild-type and mutant CCHFV growth kinetics were measured by TCID50. (A) A549, BSR-T7/5, Huh7, SW13, and HAP1 cells were infected with CCHFV at an MOI of 0.01. Monocyte-derived macrophages were infected with CCHFV (MOI 1). Data are mean ± SD of two independent experiments. * represent p < 0.05 between WT and OTU mutants. (B) HUVEC and primary skin fibroblast cells were infected at an MOI of 0.5. Data are mean ± SD of three biological replicates.
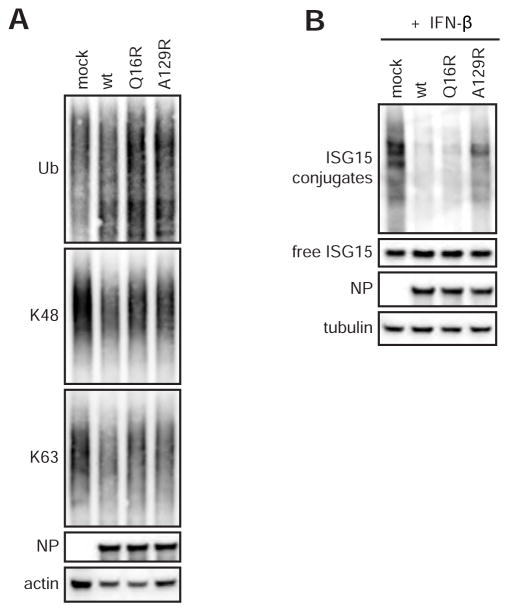
CCHFV OTU displays broad deISGylase activity and restricted DUB activity
As previously reported (Frias-Staheli et al., 2007; van Kasteren et al., 2012), we have shown that overexpression of the CCHFV OTU or complete L protein reduces the total levels of ubiquitinated and ISGylated proteins in cells co-transfected with tagged ISG15 and/or Ub. To investigate the function of the OTU on the level of ubiquitinated and ISGylated proteins in the context of infection, we infected cells with CCHFV-wt or CCHFV OTU mutants. We found that infected cells all displayed similar levels of ubiquitinated proteins to mock-infected cells, suggesting that CCHFV does not deconjugate Ub on a large scale during infection. Different ubiquitin linkages have variable and even opposite effects on proteins. Therefore, we addressed the levels of K48- and K63-linked polyubiquitin chains; the former promotes proteasomal degradation, while the latter is critical for IFN induction. Similar to total Ub, K48- or K63-linked Ub levels showed no apparent differences between CCHFV mutants (Figure 5A). Endogenous ISG15 was not detected under these experimental conditions. Therefore, to investigate the function of the OTU in inhibiting ISG15 induction and ISGylation, we treated CCHFV-infected cells with IFN to induce these responses. Infection with wild-type or mutant CCHFVs did not block IFN-induced ISG15, but both CCHFV-wt and CCHFV-Q16R reversed subsequent ISGylation (Figure 5B). In comparison, IFN-treated cells infected with CCHFV-A129R displayed robust protein ISGylation. While the OTU, when expressed both alone and within the full L protein, removes Ub conjugates in transfection experiments, it does not appear to extensively deconjugate Ub during viral infection (Figure 5B and S4A). Furthermore, CCHFV infection did not prevent the induction of IFN-stimulated genes such as ISG15, although strongly reduced levels of ISG15 conjugates were observed in cells infected with CCHFV possessing intact deISGylase activity (CCHFV-wt and –Q16R).
Figure 5. General levels of ubiquitinated/ISGylated proteins in CCHFV-infected cells.
(A) Western blot of ubiquitinated and ISGylated proteins in CCHFV-infected cells. Huh7 cells were infected with CCHFV at an MOI of 5. Cell lysates were harvested 24h post infection, separated by SDS-PAGE and probed for total Ub or K48- and K63-linked poly-Ub chains. (B) To assess protein ISGylation in infected cells, IFN β (1000 IU/ml) was added to the cell culture media 4 h post infection. Conjugation of ISG15 was visualized using western blotting.
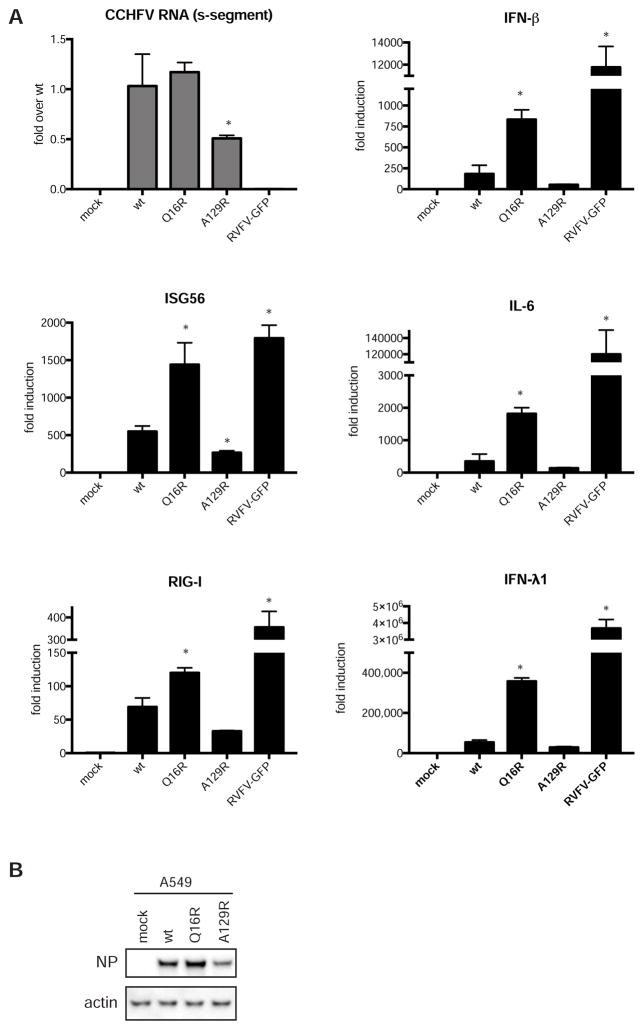
Blocking DUB activity enhances the cellular response to CCHFV infection
In transfected cells the wild-type OTU efficiently blocked induction of the innate immune responses, while the mutants could not. To investigate the function of the OTU on immune responses in cells infected with the CCHFV OTU mutants, IFN-competent A549 cells were infected with CCHFV, and RNA transcripts were analyzed. A Rift Valley fever virus (RVFV) vaccine strain expressing GFP instead of its IFN antagonist NSs served as a positive control for immune activation. Wild-type CCHFV induced various immune components, including IFN-β, IFN-λ1, ISG56, IL-6, and RIG-I, although not as efficiently as did the RVFV vaccine strain (Figure 6). Strikingly, infection with CCHFV-Q16R, in which Ub binding is disrupted, resulted in a strong increase in immune activation. In contrast, blocking OTU DUB/deISGylase activity (A129R) resulted in a similar immune response as observed with wild-type CCHFV. However, cells infected with CCHFV-A129R contained less viral RNA than cells infected with CCHFV-wt or -Q16R. These lower viral RNA levels may have contributed to the lower immune activation, despite the reduced DUB and deISGylase OTU activity of this mutant.
Figure 6. Immune responses in CCHFV-infected cells.
(A) A549 cells were infected with CCHFV or RVFV vaccine strain at an MOI of 3.3. RNA was isolated from samples harvested 24 h post infection, and transcripts of the indicated genes were quantified using RT-PCR. Data are mean ± SD of a representative experiment performed using 3 biological replicates, *p < 0.05. (B) CCHFV NP levels were determined by western blotting.
To verify that the relatively muted immune activation in response to CCHFV-A129R infection was not due to a negative effect of the OTU mutation on RdRp activity, we analyzed the mutants in a minigenome assay (Figure 7). Previously, we reported that an intact OTU cysteine protease was not required for minigenome activity (Bergeron et al., 2010). Importantly, the OTU mutants with impaired Ub (Q16R) or Ub/ISG15 (A129R) activity performed like wild-type or better in the minigenome assay. This indicates that the RdRp function of the L-Q16R and L-A129R mutants is not negatively affected, despite their low DUB activity (Figure 7).
Figure 7. Effect of OTU mutations on minigenome and VLP activity.
(A) Minigenome luciferase activity was measured 2 days post transfection. (see Experimental Procedures) (B) CCHF VLPs containing supernatants were harvested at 3 days post-transfection, and transferred to new Huh7 cells. The luciferase signal was determined as a measure for VLP activity. Data are mean ± SD of three biological replicates. *p < 0.05 indicate statistical significance between the wild-type L protein and the individual mutants.
An alternative explanation for the lower immune activation observed in cells infected with CCHFV-A129R may be found in OTU specificity. It is possible that CCHFV-A129R is more active at cleaving K63-linked polyubiquitin chains than CCHFV-Q16R. In this case, RIG-I activation and downstream immune signaling might be dampened, as K63-linked polyubiquitin chains are primarily required for immune activation (Gack et al., 2007). Therefore, we analyzed the relative efficiency with which the OTU mutants cleave K48- and K63-linked di-Ub in vitro (Figure S4B). These experiments showed that wild-type OTU efficiently cleaves both K48- and K63-linked di-Ub, whereas both Q16R and A129R display impaired activity on both types of linkage, demonstrating no preference for either Ub linkage. In contrast to the selective disruption of the DUB activity, disrupting both DUB and deISGylase activity did not result in enhanced immune responses.
To verify that the type I IFN response induced by the OTU mutants was also dependent on RIG-I as previously reported for wild-type CCHFV (Spengler et al., 2015), we knocked down RIG-I expression with siRNA (Figure S6). Similar to wild-type CCHFV, induction of IFN-β expression by the CCHFV OTU mutants was undetectable in RIG-I depleted cells. To verify if type I IFN contributes to the enhanced innate immune responses observed with CCHFV-Q16R, we blocked type I IFN signaling using a monoclonal antibody neutralizing IFNαβ receptor (IFNR) signaling in HUVECs (Figure S7). In CCHFV-wt and CCHFV-A129R infected cells, blocking IFN signaling resulted in decreased innate immune responses (IFN-β and ISG56 expression). In contrast, blocking IFNR signaling did not affect IFN-β expression in CCHFV-Q16R infected cells, and modestly reduced ISG56 expression. In agreement with the increase in IFN-β expression observed with CCHFV-Q16R, blocking IFNR signaling resulted in more viral RNA, but no increase in viral titers was observed. These results demonstrate that type I IFN signaling is important for IFN-β and ISG56 expression in CCHFV-wt and CCHFV-A129R infected cells, but not in CCHFV-Q16R infected cells. This suggests that the cellular immune response activated by CCHFV-Q16R is, in part, independent of IFN-β signaling.
DISCUSSION
IFN-induced innate immune responses are critical in controlling viral replication and disease outcomes. Using a set of recombinant CCHFVs with altered DUB/deISGylase activities, we demonstrated the IFN antagonist activity of CCHFV OTU and determined the relative contribution of OTU DUB and deISGylase activities on the suppression of IFN responses. Our study provides evidence that OTU activity of a negative-strand RNA virus suppresses IFN responses during viral infection and reverses protein ISGylation.
Viral DUBs are immune antagonists that are presumed to subvert the immune response by removing Ub and/or ISG15 moieties. To date, there are limited studies on viral deubiquitinase activity in the context of infection, and to our knowledge, no other report unequivocally demonstrates that virus-encoded deISGylases counter protein ISGylation in the context of a natural infection, such as CCHFV. Here we demonstrate that CCHFV OTU DUB activity is not as broad as previously anticipated. Studies on CCHFV OTU function using tagged Ub/ISG15 and a plasmid encoding the OTU domain reported dramatic reduction of tagged Ub- and ISG15-conjugated proteins ((Frias-Staheli et al., 2007; van Kasteren et al., 2012), Figure 2), suggesting a highly active and promiscuous protease. In contrast, CCHFV infection does not affect endogenous protein ubiquitination or levels of HA-Ub conjugates enough to demonstrate general reduction (Figure 3D, 5A, S4A), corroborating our observation that CCHFV OTU DUB activity is more selective than previously suggested by studies relying on overexpression systems (Holzer et al., 2011; van Kasteren et al., 2013). In contrast, infection with CCHFV-related Nairobi sheep disease virus (NSDV), resulted in reduced levels of HA-tagged ubiquitinated proteins (Holzer et al., 2011). However, it remains unclear whether this observation is attributable to NSDV OTU activity. If reverse genetics for NSDV become available, equivalent mutations to CCHFV OTU mutations would clarify whether NSDV DUB activity is responsible for the apparent general reduction in tagged-ubiquitinated proteins. Infection with equine arteritis virus (EAV) resulted in reduced levels of cellular ubiquitinated proteins, which could be reverted by disrupting the EAV-encoded DUB activity, suggesting that EAV DUB broadly targets ubiquitinated proteins (van Kasteren et al., 2013). This apparent difference between EAV and CCHFV might reflect the experimental set-up or fundamental differences between these unrelated viruses. Turnip yellow mosaic tymovirus displays a remarkably low activity towards Ub-AMC in vitro, despite cleaving the minimal recognition sequence (ZRLRGG-AMC) highly efficiently, suggesting that this tymovirus may target a select range of ubiquitinated proteins (Capodagli et al., 2013). Similarly, CCHFV DUB activity appears restricted to specific targets that have not yet been identified. Likely candidates include components of various antiviral-signaling pathways that require ubiquitination for their activation and regulation. IFN responses to CCHFV infection require the RIG-I-MAVS antiviral signaling axis, which included many signaling molecules regulated by ubiquitination. For example, RIG-I requires ubiquitination to activate MAVS, and activated MAVS in turn requires ubiquitination to initiate downstream signaling resulting in the activation of transcription factors IRF3 and NF-κB, and ultimately the production of type I IFNs and inflammatory cytokines. MAVS localizes both to the mitochondria and peroxisomes, and these pools of MAVS are respectively associated with production of IFN-β and IFN-λ (Dixit et al., 2010; Odendall et al., 2014). OTU DUB suppresses both IFN-β and IFN-λ1 suggesting that suppression occurs independently of MAVS localization. CCHFV OTU likely targets several ubiquitinated proteins in the RIG-I-MAVS antiviral signaling axis, and indeed, overexpression experiments demonstrated that the OTU could suppress pathway activation at multiple levels (RIG-I, MAVS, and TBK1). Future studies should be aimed at confirming bona fide substrates of CCHFV DUB. In contrast to ubiquitination, IFN-induced protein ISGylation was readily reversed in cells infected with wild-type CCHFV or a CCHFV mutant possessing intact deISGylase activity (Q16R, Fig 5B). Unlike CCHFV OTU DUB activity, its deISGylase broadly removes ISG15 moieties. Since ISGylation is induced by type I IFN, the effects of ISGylation on viral replication are expected to be most prominent at later stages of viral infection; indeed, disrupting OTU deISGylase activity can negatively affect CCHFV replication in macrophages (Figure 4), which constitutively express ISG15 and subsequent protein ISGylation (Kim et al., 2005). Also, A549 cells display a robust immune response upon CCHFV infection, including clear ISGylation, which results in growth attenuation of CCHFV-Q16R and –A129R (Figure S3, 4)
Co-expressing the ISGylation machinery with CCHFV L resulted in reduced levels of L proteins lacking deISGylase activity (L-C40A and L-A129R, Figure 2C), suggesting that ISGylation of the L protein leads to its degradation, unless ISGylation is countered by L protein’s OTU activity. Unfortunately, the lack of sensitivity of our L protein antibody prevented us from assessing L protein levels in infected cells. We expected the CCHFV mutant lacking deISGylase activity (A129R) to be most sensitive to IFN responses, including ISG15 conjugation. Consistent with this, we observed lower levels of viral RNA and nucleoprotein in IFN-producing cells infected with CCHFV-A129R, suggesting that deISGylase activity is required for optimal viral RNA synthesis in the presence of type I IFN. In contrast, CCHFV-A129R infection displays normal viral RNA and NP levels in cells that do not produce detectable levels of IFN-β upon CCHFV infection (Huh7, Figure S5A). Despite the lower CCHFV-A129R RNA levels observed in infected A549 cells compared to CCHFV-Q16R, both mutants have similarly attenuated growth kinetics compared to wild-type virus. This suggests that cellular viral RNA levels are not the limiting factor for the production of nascent virions.
Interpreting the effect of ISGylation on CCHFV infection and the effect of CCHFV-A129R on cellular immune responses is currently hindered by the unavailability of a CCHFV L protein mutant that selectively removes Ub, but not ISG15 conjugates. Previous studies performed in mice demonstrated a strong antiviral activity of ISG15 against a wide range of viruses. Recent studies have demonstrated that the immunological role of ISG15 in humans appears to be more complex. Free human ISG15 stabilizes USP18, a cellular deISGylase (Speer et al., 2016). Besides reversing ISGylation, USP18 also serves as a negative regulator of type I IFN signaling by binding to the IFN receptor and blocking downstream signaling (Malakhova et al., 2006). Thus, depletion of ISG15 would result in a reduction of USP18 and a loss of a negative regulator of IFN signaling. Later studies indicated that not only depletion of ISG15, but also of ISG15-conjugating enzyme Ube1L resulted in higher basal levels of various ISGs (Kim et al., 2008; Now and Yoo, 2016). Thus, studying ISGylation in human cell lines is hampered by the observation that depletion of ISG15 or Ube1L places the cells in an antiviral state. Conversely, studying the role of ISGylation on CCHFV replication in mice is hampered by the lack of an immunocompetent animal model for CCHFV. Currently, the only animal model for CCHF are mice lacking an intact type I IFN response (IFNAR, STAT-1 KO); consequently, induction of ISG15 by type I IFN is compromised in these animals.
Ubiquitination and ISGylation are intimately interconnected post-translational modification mechanisms that compete for the same lysine residues, and their conjugation can result in opposite effects. Cellular signaling is regulated by an intricate Ub/ISG15 balance, and the question remains to what extent CCHFV affects this balance. Deubiquitinase-deficient CCHFV-Q16R can only remove ISG15 moieties from their targets, which may effectively skew the balance towards the ubiquitinated state of these proteins. In contrast, CCHFV-A129R may induce CCHFV wild type-like immune responses because, like wild-type, the balance between ubiquitination and ISGylation states is not necessarily disrupted.
In conclusion, this study demonstrates that CCHFV suppresses the innate immune responses during infection and provides important insights into the mechanisms of this suppression by the CCHFV OTU. Structure-guided inactivation of the CCHFV OTU domain should be considered to boost immunogenicity of candidate CCHFV vaccines, and compounds targeting the OTU active site may form an attractive therapeutic strategy as they will likely block CCHFV replication and enhance innate immune responses.
EXPERIMENTAL PROCEDURES
VLP assay
The generation of CCHFV VLPs was described previously (Zivcec et al., 2015). Briefly, Huh7 cells were transfected with plasmids encoding the CCHFV strain IbAr10200 nucleoprotein (NP), codon-optimized glycoprotein precursor (GPC), codon-optimized L protein, T7 polymerase, and a CCHFV minigenome. Supernatants containing CCHFV VLPs were harvested 3 days post transfection and transferred to fresh Huh7 cells. VLP NanoLuc reporter activity was assessed the next day.
Cells
Huh7, HEK293T, A549, BSR-T7/5, SW13, and primary skin fibroblast cells were maintained in Dulbecco’s modified eagle media (DMEM) supplemented with 5–10% fetal calf serum, 1 mM sodium pyruvate, 1% non-essential amino acids and 100 U/ml of penicillin/streptomycin. BSR-T7/5 cells were a gift from K.K. Conzelmann (Ludwig-Maximilians-Universität, Munich, Germany), SW13 cells from P. Leyssen (Rega Instituut KU Leuven, Belgium). Primary skin fibroblasts were obtained from Coriell, HAP1 cells (Horizon Discovery) were maintained in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal calf serum and antibiotics. Monocyte-derived macrophages were obtained by isolating monocytes from whole blood pheresis products derived from a single healthy donor (Emory University hospital, Atlanta, GA, USA) as described previously (Zivcec et al. 2015). Human umbilical vein endothelial cells (HUVEC) were obtained from Lonza and maintained in EGM-plus media supplemented with the EGM BulletKit (Lonza).
Ethics statement monocyte-derived macrophages
The use of human blood products was approved by Emory University Institutional Review Board (IRB00045947). Under this protocol, no donor personal information was provided, and informed consent was given. All adult blood donors provided informed consent, and a parent or guardian of any child participant provided informed consent on the child’s behalf.
In vitro OTU activity
An OTU expression construct was created by synthesizing the first 169 amino acids from the CCHFV L protein (GenBank AAQ98866.2), codon-optimized for E. coli BL21 (Biobasic). OTU mutants C40A, Q16R, P77D and A129R were generated using QuikChange site-directed mutagenesis (Agilent Technologies). E. coli BL21 (DE3) cells were used for protein expression as previously published (Capodagli et al., 2013). Protein purification was conducted as previously described (Capodagli et al., 2011). In vitro OTU activity was analyzed as described previously (Capodagli et al., 2013). Each OTU mutant was assessed for activity against Ub- and ISG15-7-amido-4-methylcoumarin (AMC) substrates. Assays were conducted with 4 nM OTU and 1 μM substrate. All reactions were performed at 25°C in 100 mM NaCl, 50 mM HEPES [pH 7.5], 0.01 mg/mL bovine serum albumin, and 5 mM DTT in a 50 μL reaction volume. Turnover rates of AMC substrates were calculated based on the slope of fluorescence emitted by liberation of AMC from the substrate. Values were compared with the previously published turnover rate of wild-type CCHFV OTU (Capodagli et al., 2013).
OTU activity in transfected cells
Plasmids expressing the OTU domain (aa. 1–168) and various mutants were synthesized (Genscript) and cloned into pcDNA3.1 containing a C-terminal HA-tag. The OTU domain of full length L protein, was mutated using In Fusion cloning (Clontech). Huh7 cells were transfected with plasmids expressing the CCHFV L and pcDNA-HA-Ub (DUB assay) or plasmids required for IGSylation: pcDNA-Ube1L (E1), pcDNA-UbcH8 (E2), pcDNA-NTAP-HERC5 (E3) (J. Huibregtse, University of Texas at Austin), and pcDNA3.1-V5-hISG15 (Genscript). All plasmid transfections were performed with TransIT-LT1 (Mirus Bio) transfection reagent. Ubiquitin and ISG15 conjugation levels were assessed 48h post transfection using western blot analysis.
Western blot analysis and antibodies
Cell lysates were harvested in 2x Laemmli sample buffer, heated for 10 min at 95°C, and separated on 4–12% bis-tris SDS-PAGE gels. Proteins were transferred to nitrocellulose membranes using a semi-dry blotting system (Bio-Rad). Full-length CCHFV L protein was detected after separation on 4–8% tris-acetate gels with a rabbit antibody raised against a peptide corresponding to amino acids 1160–1174 (Genscript). HA-OTU and HA-Ub were visualized with rabbit-anti-HA (Life Technologies). CCHFV N was detected with hyperimmune mouse ascetic fluid (in house) or CCHFV anti-N antibody (IBT Bioservices). V5-hISG15 was detected with anti-V5 (Invitrogen) and endogenous ISG15 with anti-ISG15 (Santa Cruz). Endogenous Ub was detected with anti-Ub (Santa Cruz), FK2 (Enzo) or Cell Signaling antibodies (for K48 and K63 polyUb). B-actin or tubulin (Sigma) were used as loading control markers. SuperSignal West Dura Fast Western blotting kits (ThermoFisher Scientific) and a Bio-Rad ChemiDoc MP imaging system were used to reveal protein bands.
Minigenome assay (RdRp activity)
The CCHFV minigenome assay was performed as described previously (Zivcec et al., 2015), with the exception that Huh7 cells were used and minigenome reporter activity normalized to firefly transfection control (pGL3, Promega)
Luciferase reporter assay
The RIG-I-mediated IFN-β response was assessed by transfecting Huh7 or HEK293 cells with a plasmid encoding RIG-I CARD (S. Best, Rocky Mountain Laboratories, USA) and a reporter plasmid encoding firefly luciferase under control of the promoter of IFN-β (P125-luc), IRF3 (P55c1b-luc), or NF-κB (P55A2-luc), kindly provided by T. Fujita (Kyoto University, Japan). A plasmid constitutively expressing Renilla luciferase was used as a transfection control (pRL, Promega). Alternatively, plasmids encoding MAVS (pUNO-MAVS) or TBK1 (pUNO-TBK1, both from Invivogen) were used to activate the P125-luc reporter. Plasmids expressing the CCHFV OTU domain were co-transfected to assess the activity of OTU mutants. Cell lysates were harvested 16–24 h post transfection, and luciferase activities were determined using the Dual-Glo assay system (Promega). Renilla luciferase expression was used to normalize the firefly signal.
Di-ubiquitin cleavage assay
Activity of CCHFV OTU mutants were assessed for cleavage of K48- and K63-linked di-ubiquitin (di-Ub). The di-Ub substrates (10 μM) were incubated with each OTU (20 nM) at 37°C for 1 h in 100 mM NaCl, 5 mM HEPES [pH 7.5], and 2 mM DTT. Samples were taken at various time points and the reaction was terminated by mixing 9 μl of each reaction with 2x sodium dodecyl sulfate-tricine sample buffer and boiling for 5 min at 98°C. Samples were visualized on 10–20% tris-tricine gels by Coomassie staining.
Viruses
All CCHFV infections and rescue attempts were performed in BSL-4 facilities at the Centers for Disease Control and Prevention (Atlanta, GA, USA). Experiments involving cDNA encoding viral sequences were performed in accordance with approved Institutional Biosafety Committee (IBC) protocols. Recombinant CCHFV Ibar10200 was generated as described previously (Bergeron et al., 2015). Briefly, Huh7 cells were transfected with plasmids encoding the genomic segments L, M and S (T7-L, T7-M, and T7-S), helper plasmids expressing CCHFV NP and codon-optimized L, and the T7 polymerase. Viruses with mutated OTU were created by adding point mutations to the pT7-L plasmids using site-directed mutagenesis before virus rescue. After 3–5 days, supernatants were harvested for titration or passage on BSR-T7/5 cells. All recombinant viruses were sequence verified with deep sequencing technology using a MiniSeq System (Illumina).. Virus infectivity was measured by titration on BSR-T7/5 cells and calculated as the median tissue culture infective dose (TCID50). Infected cells were detected by immunofluorescence using a-CCHFV polyclonal mouse antiserum (HMAF) followed by an Alexa488-conjugated anti-mouse antibody (Invitrogen). The recombinant Rift Valley fever virus (RVFV) rZH501-deltaNSs:GFP-deltaNSm was described previously (Bird et al., 2008).
Quantitative RT-PCR
RNA was isolated using Magmax technology (Life Technologies) or trizol in combination with the direct-zol RNA miniprep kit (Zymo Research) and subjected to one-step qRT-PCR using the SuperScript III Platinum One-Step qRT-PCR Kit (Life Technologies). Cellular transcripts were amplified using various Taqman primer/probe sets (ThermoFisher). CCHFV S segment amplification was described previously (Bergeron et al., 2015). IPO8 was used for normalization purposes, and all reactions were performed on a Bio-Rad Thermal cycler Real-Time System.
RIG-I siRNA and antibody treatment
A549 cells were reverse-transfected with 10 nM siRNA targeting RIG-I (Santa Cruz) or control siRNA (Dharmacon) using Lipofectamine RNAiMAX (ThermoFisher Scientific). The siRNA-transfected cells were infected 2 days post transfection. Alternatively, cells were treated with 10 mg/ml Anti-Human IFNAR2 Antibody (PBL assay science) 1 h post infection.
Statistical analysis
Data were analyzed using GraphPad Prism version 7. The unpaired parametric two-sided Student’s t test was used to derive P values from all data presented and p < 0.05 defined as statically significant.
Supplementary Material
Acknowledgments
This study was supported by the CDC and a CDC foundation project funded by NIAID grant R01AI109008. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. We would like to thank Brian Hua and Tatyana Klimova for the critical review of this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
S.D.P., E.B. and F.E.M.S. conceived experiments. S.D.P., E.B., J.R.S. and F.E.M.S. wrote the manuscript. S.D.P. and E.B secured funding. E.B., F.E.M.S., M.D., M.Z, J.V.D., and S.R.W. performed experiments. M.Z. provided reagents. S.D.P., E.B., J.R.S., S.R.W, S.T.N, C.F.S. and M.Z. provided expertise and feedback.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc Natl Acad Sci U S A. 2011;108:2228–2233. doi: 10.1073/pnas.1015287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron E, Albariño CG, Khristova ML, Nichol ST. Crimean-Congo hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function. J Virol. 2010;84:216–226. doi: 10.1128/JVI.01859-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron É, Zivcec M, Chakrabarti AK, Nichol ST, Albariño CG, Spiropoulou CF. Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin. PLoS Pathog. 2015;11:e1004879. doi: 10.1371/journal.ppat.1004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird BH, Albariño CG, Hartman AL, Erickson BR, Ksiazek TG, Nichol ST. Rift valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J Virol. 2008;82:2681–2691. doi: 10.1128/JVI.02501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodagli GC, McKercher MA, Baker EA, Masters EM, Brunzelle JS, Pegan SD. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodagli GC, Deaton MK, Baker Ea, Lumpkin RJ, Pegan SD. Diversity of ubiquitin and ISG15 specificity among nairoviruses’ viral ovarian tumor domain proteases. J Virol. 2013;87:3815–3827. doi: 10.1128/JVI.03252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes Are Signaling Platforms for Antiviral Innate Immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 Conjugation System Broadly Targets Newly Synthesized Proteins: Implications for the Antiviral Function of ISG15. Mol Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Heaton SM, Borg NA, Dixit VM. Ubiquitin in the activation and attenuation of innate antiviral immunity. J Exp Med. 2015 doi: 10.1084/jem.20151531. jem.20151531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer B, Bakshi S, Bridgen A, Baron MD. Inhibition of Interferon Induction and Action by the Nairovirus Nairobi Sheep Disease Virus/Ganjam Virus. PLoS One. 2011;6:e28594–12. doi: 10.1371/journal.pone.0028594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig JE, Osborne JC, Nichol ST. Crimean–Congo hemorrhagic fever virus genome L RNA segment and encoded protein. Virology. 2004;321:29–35. doi: 10.1016/j.virol.2003.09.042. [DOI] [PubMed] [Google Scholar]
- Hu H, Sun SC. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457–483. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TW, Frias-Staheli N, Bacik JP, Levingston Macleod JM, Khajehpour M, García-Sastre A, Mark BL. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc Natl Acad Sci U S A. 2011;108:2222–2227. doi: 10.1073/pnas.1013388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam Ca, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:973–959. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren PB, Beugeling C, Ninaber DK, Frias-Staheli N, van Boheemen S, García-Sastre A, Snijder EJ, Kikkert M. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. J Virol. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren PB, Bailey-Elkin Ba, James TW, Ninaber DK, Beugeling C, Khajehpour M, Snijder EJ, Mark BL, Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci U S A. 2013;110:E838–47. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O’Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, et al. DUBA: A Deubiquitinase That Regulates Type I Interferon Production. Science (80-) 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- Kim KI, Malakhova OA, Hoebe K, Yan M, Beutler B, Zhang DE. Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling. J Immunol. 2005;175:847–854. doi: 10.4049/jimmunol.175.2.847. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Hwang SY, Imaizumi T, Yoo JY. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella E, Martin SG, Grolla A, Czub M, Feldmann H, Flick R. Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology. 2004;321:23–28. doi: 10.1016/j.virol.2003.09.046. [DOI] [PubMed] [Google Scholar]
- Lombardi C, Ayach M, Beaurepaire L, Chenon M, Andreani J, Guerois R, Jupin I, Bressanelli S. A Compact Viral Processing Proteinase/Ubiquitin Hydrolase from the OTU Family. PLoS Pathog. 2013;9:e1003560. doi: 10.1371/journal.ppat.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25:50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. Embo J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Now H, Yoo JY. AG490 and PF431396 Sensitive Tyrosine Kinase Control the Population Heterogeneity of Basal STAT1 Activity in Ube1l Deficient Cells. PLoS One. 2016;11:e0159453. doi: 10.1371/journal.pone.0159453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. Articles Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Okumura AJ, Uematsu K, Hatakeyama S, Zhang DE, Kamura T. Activation of double-stranded rna-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J Biol Chem. 2013;288:2839–2847. doi: 10.1074/jbc.M112.401851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrefitte CN, Perret M, Garcia S, Rodrigues R, Bagnaud A, Lacote S, Crance J, Vernet G, Garin D. Differential activation profiles of Crimean – Congo hemorrhagic fever virus- and Dugbe virus-infected antigen-presenting cells. J Gen Virol. 2010;91:189–198. doi: 10.1099/vir.0.015701-0. [DOI] [PubMed] [Google Scholar]
- Recht M, Borden EC, Knight E, Borden EC, Knight E. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J Immunol. 1991;147:2617–2623. [PubMed] [Google Scholar]
- Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, Zhu LH, Wang C. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer SD, Li Z, Buta S, Payelle-Brogard B, Qian L, Vigant F, Rubino E, Gardner TJ, Wedeking T, Hermann M, et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat Commun. 2016;7:11496. doi: 10.1038/ncomms11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler JR, Patel JR, Chakrabarti AK, Zivcec M, García-Sastre A, Spiropoulou CF, Bergeron E, Bergeron É. RIG-I Mediates an Antiviral Response to Crimean-Congo Hemorrhagic Fever Virus. J Virol. 2015;89:10219–10229. doi: 10.1128/JVI.01643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhong G, Zhu L, Liu X, Shan Y, Feng H, Bu Z, Chen H, Wang C. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J Immunol. 2010;184:5777–5790. doi: 10.4049/jimmunol.0903588. [DOI] [PubMed] [Google Scholar]
- Woods MW, Kelly JN, Hattlmann CJ, Tong JGK, Xu LS, Coleman MD, Quest GR, Smiley JR, Barr SD. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology. 2011;8:95. doi: 10.1186/1742-4690-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Yang J, Sun HR, Xin X, Wang HD, Chen JP, Adams MJ. Genomic analysis of rice stripe virus Zhejiang isolate shows the presence of an OTU-like domain in the RNA1 protein and a novel sequence motif conserved within the intergenic regions of ambisense segments of tenuiviruses. Arch Virol. 2007;152:1917–1923. doi: 10.1007/s00705-007-1013-2. [DOI] [PubMed] [Google Scholar]
- Zivcec M, Metcalfe MG, Albariño CG, Guerrero LW, Pegan SD, Spiropoulou CF, Bergeron É. Assessment of Inhibitors of Pathogenic Crimean-Congo Hemorrhagic Fever Virus Strains Using Virus-Like Particles. PLoS Negl Trop Dis. 2015;9:1–23. doi: 10.1371/journal.pntd.0004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.