ABSTRACT
Polyamines (PA) in plant play roles in growth and development and in responses to environmental stresses. The family of polyamine oxidases (PAO) contributes to a balanced homeostasis of PAs catalyzing two different reactions, terminal catabolic (TC) and back-conversion (BC) pathway, in PA catabolism. From the seven PAOs encoded by the rice genome (OsPAO1 – OsPAO7) OsPAO6 could so far not be characterized due to failure in obtaining the coding cDNA based on accessions in the genomic databases. We report cloning and characterization of the correct OsPAO6 cDNA with a length of 1,742 bp. The 1,491 bp long open reading frame codes for a 497-amino acid protein from nine exons. The protein which has 92% identity to OsPAO7 localizes to plasma membrane.
KEYWORDS: Polyamines, polyamine oxidase, rice, Oryza sativa, coding region, cDNA, catabolism
Polyamines (PAs) are aliphatic compounds with low molecular masses occurring in all living organisms.1,2 They play roles not only in growth, organogenesis, development and aging process but also in coping with various environmental stresses.3,4,5 The major common PAs in plant are the diamine, putrescine (Put), the triamine, spermidine (Spd), and two tetraamines, spermine (Spm) and thermospermine (T-Spm).6,7 Some plants contain norspermidine (NorSpd) and norspermine (NorSpm) as minor uncommon PAs.8 PAs are derived from the amino acids ornithine, arginine and methionine. Put is synthesized from ornithine by ornithine decarboxylase (ODC) or from arginine by three sequential enzyme reactions catalyzed by arginine decarboxylase (ADC), agmatine iminohydrolase (AIH) and N-carbamoylputresine amidohydrolase (CPA). Spd synthesis from Put is catalyzed by Spd synthase which requires another substrate, decarboxylated S-adenosylmethionine (dcSAM). The latter is synthesized from methionine by two enzyme reactions; S-adenosylmethionine synthase (SAMS) and S-adenosylmethionine decarboxylase (SAMDC). Spm and T-Spm are derived from Spd by Spm synthase (SPMS) and T-Spm synthase (ACL5 or T-SpmS), respectively. SPDS, SPMS and ACL5 are collectively called ‘aminopropyl transferase’ because they transfer an ‘aminopropyl residue’ from dcSAM to Put or Spd. As described above, the PA biosynthetic pathway in plant is well established. On the other hand, plant PA catabolic pathway(s) were only recently explored. Two enzymes, copper-dependent amine oxidase (CuAO: EC 1.4.3.6) and flavin adenine dinucleotide (FAD)-associated polyamine oxidase (PAO: EC 1.5.3.11), are involved in PA catabolism. The former forms homodimers of 70∼90 kDa subunits, and each subunit contains a single copper ion and a 2,4,5-trihydroxyphenylalanine quinone (TPQ) cofactor.9,10 In contrast, PAO is a monomeric enzyme. The first characterized apoplastic maize PAO and barley PAOs oxidize PAs in a terminal catabolic (TC) pathway. 1,3-diaminopropane (DAP), produced by TC-type PAO, is converted to NorSpd and then NorSpm by the aminopropyl transferases with broad specificity.8 The family of PAO encoding genes from Arabidopsis comprises five members, AtPAO1 to AtPAO5. All the Arabidopsis PAO enzymes were charecterized and shown to oxidize PAs in an alternative pathway, the back-conversion (BC) pathway.11,12,13 This type of PAO reaction converts Spm and T-Spm to Spd, and/or further to Put, along with the production of 3-aminopropanal and H2O2. The O. sativa genome contains seven PAO genes termed OsPAO1 to OsPAO7, respectively. Phylogenetic analysis on angiosperm PAOs revealed that they are at least classified into four clades, I-IV. OsPAOs are distributed to clade II (OsPAO2, OsPAO6 and OsPAO7), clade III (OsPAO1) and clade IV (OsPAO3, OsPAO4 and OsPAO5), respectively, but no member is found in clade I.14 Characterization of the different PAO enzymes in terms of substrate and product specificity, cellular localization and gene expression is of great importance for further investigating polyamine function. Five of the seven OsPAOs were so far partially characterized by cloning and expression of the recombinant proteins. OsPAO3, OsPAO4 and OsPAO5 localize in peroxisomes and catalyze BC-type reactions,14 OsPAO1 resides in cytoplasm and catalyzes BC-type reaction, whereas OsPAO7 localizes in apoplast and catalyzes TC-type reactions.15,16 To complete characterization of all OsPAOs the cDNAs of OsPAO2 and OsPAO6 were still missing and could so far not be amplified and cloned based on the DNA sequence available from public databases.
Here we report that the presumed coding region for OsPAO6 derived from the genomic DNA sequence (Acc. No. NM_001069545) was incorrect and could be replaced by the newly assigned OsPAO6 cDNA sequence (Acc. No. LC107620). According to the public database (National Center for Biotechnology Information), the coding region of OsPAO6 cDNA comprised 1,623 bp. Furthermore, 8 EST (Expressed sequence tag) clones of OsPAO6 were reported (http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Os&CID=73758). Based on this information we designed seven specific oligonucleotide primers for OsPAO6 covering the open reading frame (ORF) and the 3´-untranslated region (3´-UTR) (Fig. 1A). Expression analysis by reverse transcription (RT)-polymerase chain reaction (PCR) with the primer combination F3 + R2 showed that the OsPAO6 gene is highly expressed in 7-day-old rice seedlings treated for 6 h with 100 µM of the phytohormone jasmonic acid (Fig. 1B). Therefore, RNA isolated from JA-treated seedlings was used for RT-PCR with several primer combinations. With F2 and R2 primer combination, a single DNA fragment of approximately 1,200 bp in size was amplified. DNA sequencing revealed that the fragment corresponds to the nucleotide positions (np) 450 to 1,623 of the provisional 1,623 bp-ORF. However, using the forward primer F1, in combinations with each of the reverse primers no PCR products could be detected (Fig. 1C), suggesting that the 5’ region of the full-length cDNA is different from the reported cDNA sequence. The correct 5´-end could be determined by rapid amplification of cDNA ends (RACE), 5’- RACE. The resulting full-length OsPAO6 cDNA of 1,742 bp (Accession number, LC107620) encodes an ORF of 1,491 bp starting at np 39 and ending at the np 1,529, which encodes a 497-amino acid protein from 9 exons (E1 to E9) while the previous OsPAO6 cDNA sequence was derived from 12 exons (E1 to E12) (Fig. 2). Alignment of the previous and new OsPAO6 amino acid sequences showed, as expected, that the amino-terminal regions were quite different and the newly assigned OsPAO6 showed high identity (92%) to OsPAO7 (Suppl. Fig. 1). With the SignalP ver 4.1 program (http://www.cbs.dtu.dk/services/SignalP/) a signal peptidase recognition site between amino acid positions 27 and 28 was predicted within the OsPAO6 sequence (data not shown). Thus, subcellular localization of OsPAO6 in plant cells was tested using two green fluorescent protein (GFP)-fusion constructs. The one covers the full-length ORF region of OsPAO6 fused to the GFP gene (FL), the other contains the signal peptide (SP) and transmembrane domain (TD) region of OsPAO6 fused to GFP gene (SP+TD). DNA fragments encoding FL OsPAO6 and the hypothetical SP and TD portions of OsPAO6 were amplified using the respective primer pairs and cloned into plasmid pBI221:GFP, resulting in pBI221:OsPAO6-GFP (FL) and pBI221:SP+TD-GFP (SP +TD).14 Those constructs were delivered to onion epidermal cells by particle bombardment (PDS-1000/He, Bio-Rad) together with the positive plasma membrane-localized marker construct, pBI221:DsRED-OsPIP2;1.17 The result indicated that both OsPAO6-GFP fusion proteins (FL and SP+TD) localized in plasma membrane (Fig. 3). Therefore, it is likely that OsPAO6 targets to the apoplastic space via its signal peptide. Same localization has been shown for closely related OsPAO7 which is a TC-type PAO enzyme specifically expressed during anther development.16 Having the correct OsPAO6 coding region in hands it will now be possible to complete enzymatic characterization of all rice PAOs. It is of interest whether OsPAO6 significantly differs from OsPAO7 in reaction type (TC or BC) and in substrate and product specificity. Also differences in expression in response to various environmental stresses and in the diverse plant organs should be investigated. Furthermore, the here described example of OsPAO6 shows that detection and correction of putative coding sequence annotation in genomes needs fine tuning as recently described.18
Figure 1.

Cloning of OsPAO6 cDNA. (A) Schematic illustration of OsPAO6 cDNA available in the public database and the position and orientation of the primers. (B) RT-PCR result using F3 and R2 primers which showed that OsPAO6 was responsive to JA. (C) RT-PCR results using the other primer combination.
Figure 2.

The genome assignment of the new OsPAO6 transcript. The upper figure is derived from the NCBI information. The provisional assignment indicates that OsPAO6 is consisting of 12 exons and 11 introns, whereas the newly isolated OsPAO6 is consisting of 9 exons and 8 introns. The E2 to E9 totally correspond to the old E5 to E12, respectively. White box and black box indicate the untranslated region and the coding region, respectively. The newly identified E1 corresponds to the region between the previous E4 and E5 which is highlighted by the dotted lines.
Figure 3.
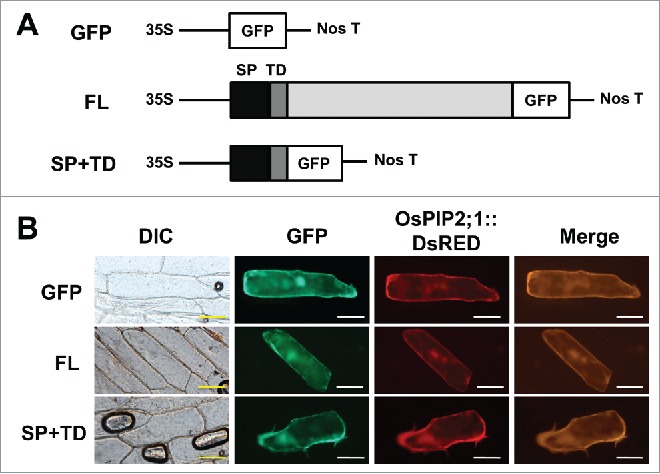
Subcellular localization of OsPAO6 in plant cells. (A) Schematic drawing of the OsPAO6::GFP fusion constructs. OsPIP2;1 encodes a plasma membrane intrinsic protein (a form of aquaporin), is a plasma membrane-localized marker. FL: the full-length region of OsPAO6 was fused to the GFP; SP+SD: the signal peptide region of OsPAO6 was fused to the GFP. 35S, Cauliflower mosaic virus 35S promoter; Nos T, the terminator region derived from the nopaline synthase gene. (B) Subcellular localization of OsPAO6 in onion epidermal cells. Bar corresponds to 10 μm.
Supplementary Material
Disclosure of potential conflicts of interests
No potential conflicts of interest were disclosed.
Acknowledgments
GS acknowledges support by JSPS postdoctoral fellowship for foreign researchers and by German Academic Exchange Service (DAAD) program Research Stays for University Academics and Scientists.
Funding
This work was supported by grants from the Japan Society for the Promotion of Science (JSPS) toTK (15K14705, 2604081).
Abbreviations
- PA
polyamine
- PAO
polyamine oxidase
- RT-PCR
reverse transcription polymerase chain reaction
References
- 1.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 2.Cohen SS. A guide to the polyamines. Oxford University Press: Oxford; 1998. [Google Scholar]
- 3.Alcázar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, Carrasco P, Tiburcio AF. Polyamines: Molecules with regulatory functions in plant abiotic stress tolerance. Planta. 2010;231:1237–1249. doi: 10.1007/s00425-010-1130-0. [DOI] [PubMed] [Google Scholar]
- 4.Minocha R, Majumdar R, Minocha SC. Polyamines and abiotic stress in plants: A complex relationship. Front. Plant Sci. 2014;5:175. doi: 10.3389/fpls.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berberich T, Sagor GHM, Kusano T. Polyamines in Plant Stress Response – In: Kusano T, Suzuki H, editors. Polyamines, A Universal Molecular Nexus for Growth, Survival, and Specialized Metabolism. Tokyo: Springer; 2015. p. 155–168. [Google Scholar]
- 6.Kusano T, Berberich T, Tateda C, Takahashi Y. Polyamines: Essential factors for growth and survival. Planta. 2008;228:367–381. doi: 10.1007/s00425-008-0772-7. [DOI] [PubMed] [Google Scholar]
- 7.Tiburcio AF, Altabella T, Bitrian M, Alcazar R. The roles of polyamines during the lifespan of plants: from development to stress. Planta. 2014;240:1–18. doi: 10.1007/s00425-014-2055-9. [DOI] [PubMed] [Google Scholar]
- 8.Fuell C, Elliot KA, Hanfrey CC, Franceschetti M, Michael AJ. Polyamine biosynthetic diversity in plants and algae. Plant Physiol. Biochem. 2010;48:513–520. doi: 10.1016/j.plaphy.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Kusano T, Kim DW, Liu T, Berberich T. Polyamine catabolism in plants in: Polyamine: A universal molecular Nexus for growth, survival and specialized metabolism. In: Kusano T, Suzuki H, editors. Tokyo: Springer; 2015. p. 77–88. [Google Scholar]
- 10.Medda R, Padiglia A, Flores G. Plant copper-amine oxidases. Phytochemistry. 2010;39:1–9. doi: 10.1016/0031-9422(94)00756-J. [DOI] [Google Scholar]
- 11.Cona A, Rea G, Angelini R, Federico R, Tavladoraki P. Functions of amine oxidases in plant development and defence. Trends Plant Sci. 2006;11:80–88. doi: 10.1016/j.tplants.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Angelini R, Cona A, Federico R, Fincato P, Tavladoraki P, Tisi A. Plant amine oxidases “on the move:” an update. Plant Physiol Biochem. 2010;48:560–564. doi: 10.1016/j.plaphy.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Moschou PN, Wu J, Cona A, Tavladoraki P, Angelini R, Roubelakis-Angelakis K A. The polyamines and their catabolic products are significant players in the turnover of nitrogenous molecules in plants. J Exp Bot. 2012;63:5003–5015. doi: 10.1093/jxb/ers202. [DOI] [PubMed] [Google Scholar]
- 14.Ono Y, Kim DW, Watanabe K, Sasaki A, Niitsu M, Berberich T, Kusano T, Takahashi Y. Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids. 2012;42:867–876. doi: 10.1007/s00726-011-1002-3. [DOI] [PubMed] [Google Scholar]
- 15.Liu T, Kim DW, Niitsu M, Berberich T, Kusano T. Polyamine oxidase 1 from rice (Oryza sativa) is a functional ortholog of Arabidopsis Polyamine oxidase 5. Plant Sign Behavior. 2014a;9:e29773. doi: 10.4161/psb.29773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Kim DW, Niitsu M, Maeda S, Watanabe M, Kamio Y, Berberich T, Kusano T. Polyamine oxidase 7 is a terminal catabolism-type enzyme in Oryza sativa and is specifically expressed in anther organ. Plant Cell Physiol. 2014;55:1110–1122. doi: 10.1093/pcp/pcu047. [DOI] [PubMed] [Google Scholar]
- 17.Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005;46:1568–77. doi: 10.1093/pcp/pci172. [DOI] [PubMed] [Google Scholar]
- 18.Prasad TS, Mohanty AK, Kumar M, Sreenivasamurthy SK, Dey G, Nirujogi RS, Pinto SM, Madugundu AK, Patil AH, Advani J, et al.. Integrating transcriptomic and proteomic data for accurate assembly and annotation of genomes. Genome Res. 2017;27:133–144. doi: 10.1101/gr.201368.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


