ABSTRACT
In Arabidopsis thaliana, six acyl-CoA-binding proteins (ACBPs), designated as AtACBP1 to AtACBP6, have been identified to function in various events related to plant stress and development. The 10-kDa AtACBP6 is the smallest in this protein family, and recombinant AtACBP6 interacts with lipids in vitro by binding to acyl-CoA esters and phosphatidylcholine. Using anti-AtACBP6 antibodies in immunoelectron microscopy, we have localized AtACBP6 in the Arabidopsis phloem. The detection of immunogold grains in the plasmodesmata suggested that AtACBP6 could move from the companion cells to the sieve elements via the plasmodesmata. As AtACBP6 has been identified in a membrane-based interactome analysis to be a potential protein partner of Plasmodesmata-Localized Protein, PDLP8, AtACBP6-PDLP8 interaction was investigated herein utilizing isothermal titration calorimetry, as well as pull-down and bimolecular fluorescence complementation assays (BiFC). Notably, BiFC data revealed that AtACBP6-PDLP8 interaction occurred at the plasma membrane, which was unexpected as AtACBP6 has been previously identified in the cytosol. AtACBP6 expression was generally higher than PDLP8 in β-glucuronidase (GUS) assays on transgenic Arabidopsis transformed with AtACBP6 or PDLP8 promoter-driven GUS, consistent with qRT-PCR and microarray results. Furthermore, western blot analysis using anti-AtACBP6 antibodies showed a reduction in AtACBP6 expression in the pdlp8 T-DNA insertional mutant, suggesting that PDLP8 may possibly influence AtACBP6 accumulation in the sieve elements, probably in the plasmodesmata, where PDLP8 is confined and to where AtACBP6 has been immunodetected.
KEYWORDS: Acyl-CoA-binding protein, arabidopsis thaliana, PDLP8, phloem, plasma membrane, plasmodesmata, protein-protein interaction
Introduction
In higher plants, the plasmodesmata serve as vital structures in cell walls for intercellular transport of water and nutrients, to facilitate molecular movement within the symplastic space.1,2 They also are important in development and pathogenesis.3,4 In Arabidopsis, an eight-membered family of Plasmodesmata-Located Proteins, PDLPs (PDLP1-PDLP8), have been reported to be involved in virus translocation through plasmodesmal tubular-guided movement.5 These PDLPs, predicted to range between 30 to 35 kD, were identified based on sequence homology to Arabidopsis PDLP1 and had been characterized by proteomic analysis of the cell wall.5,6
Recent studies on PDLPs demonstrated their localization in the plasma membrane within the plasmodesmatal channels, and their function in the regulation of cell-to-cell movement of viruses and macromolecules.5,6,7 A reduction in cell-to-cell movement of fluorescent molecules and viral movement proteins occurred in transgenic plant lines overexpressing PDLP1 or PDLP5.5,6,7 Furthermore, PDLP5, as a regulator of plasmodesmal gating, was responsive to virulent Pseudomonas syringae and salicylic acid (SA)-dependent callose deposition at the plasmodesmata.8,9
The six members of the Arabidopsis thaliana acyl-CoA-binding protein (AtACBP) family share a conserved acyl-CoA-binding domain and they have been reported to function in plant stress and development.10,11 Ankyrin repeat-containing AtACBPs, AtACBP1 and AtACBP2, are membrane-associated proteins that can facilitate protein-protein interactions12-16 as well as mediate heavy metal stress responses.17-19 They have also been shown to exert overlapping functions in embryo development.20,21 AtACBP3 has been identified as an extracellularly-targeted protein which functions in pathogen defense and autophagy-mediated leaf senescence.22,23,24 Both cytosolic AtACBP4 and AtACBP5 contain kelch-motifs that are potentially capable of protein-protein interactions and they play complementary roles in floral development.25-28 The smallest (10-kDa) member, AtACBP6, is also a cytosolic protein and conferred freezing tolerance upon its ectopic expression in transgenic Arabidopsis rosettes and flowers.29-30 More recently, immunoelectron microscopy using anti-AtACBP6 in 5-week-old Arabidopsis revealed that AtACBP6 was detected in the companion cells, sieve elements and the plasmodesmata.28
Using a membrane-based interactome analysis, Jones et al. (2014) identified a panel of interactors, including a number of putative AtACBP6 protein interactors.31 Amongst those predicted AtACBP6 interactors, PDLP8 (At3g60720) ignited our interest as its plasmodesmal localization coincided with recent observation that AtACBP6 could be transported from the companion cells to the sieve elements via the plasmodesmata.28 While PDLP8 has been hypothesized to function in intercellular movement of molecules,5 phloem-localized AtACBP6 has been designated a role in lipid transport through the plasmodesmata.28 Herein, the interaction of PDLP8 and AtACBP6 was verified using isothermal titration calorimetry (ITC), as well as pull-down and bimolecular fluorescence complementation (BiFC) assays. The relationship between PDLP8 and AtACBP6 was further explored using qRT-PCR, phloem exudate analysis on the pdlp8 mutant, and GUS assays on PDLP8pro::GUS and AtACBP6pro::GUS transgenic Arabidopsis lines.
Results
Interaction of AtACBP6 and PDLP8 at the plasma membrane
Some candidates of putative “protein partners” for AtACBP6 was identified using membrane-based interactome analysis (Table 1). More than 100 protein partners were predicted to interact with AtACBP6.31 Among them, PDLP8 showed the highest interaction scores with AtACBP6 (www.associomics.org; Table 1). To further evaluate putative AtACBP6-PDLP8 interaction, computational prediction was performed prior to experimental verification. As shown in the ribbon diagram (Fig. 1A), two α-helices, four β-sheets and a visible loop appeared in the 3-D structure of PDLP8 when generated using 4XRE32 as template in SWISS MODEL. A model on AtACBP6-PDLP8 interaction was predicted by docking using Patchdock/Firedock (http://bioinfo3d.cs.tau.ac.il/PatchDock/) (Fig. 1B). Our results suggested possibility of AtACBP6-PDLP8 interaction, with the α1 and α2 helices of AtACBP6 predicted to interact with the β3 and β4 sheets of PDLP8 (Fig. 1B).
Table 1.
Putative protein interactors of AtACBP6.
| Locus | Interaction Score | Gene description |
|---|---|---|
| AT3G53510 | ++++ | ABC-2 type transporter family protein |
| AT1G51460 | ++++ | ABC-2 type transporter family protein |
| AT1G70520 | ++++ | CRK2, cysteine-rich RLK (RECEPTOR-like protein kinase) 2 |
| AT1G72300 | ++++ | Leucine-rich receptor-like protein kinase family protein |
| AT3G05360 | ++++ | AtRLP30, RLP30, receptor like protein 30 |
| AT3G60720 | ++++ | PDLP8, plasmodesmata-located protein 8 |
| AT4G37220 | ++++ | Cold acclimation protein WCOR413 family |
| AT2G42770 | ++++ | Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein |
| AT5G40420 | ++++ | OLEO2, OLE2, oleosin 2 |
| AT5G47530 | ++++ | Auxin-responsive family protein |
The list was selected from 153 candidate proteins that scored the highest from database reported by Jones et al. (2014) Science 344:711–716 (www.associomics.org).31
Figure 1.
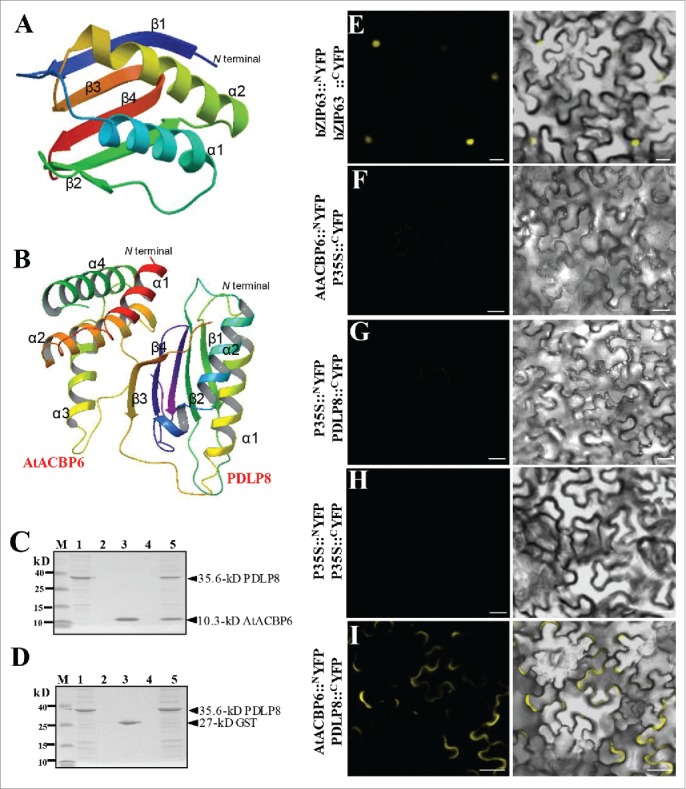
PDLP8 interacts with AtACBP6 at the plasma membrane. (A-B) The predicted 3-D structure of PDLP8 interacting with AtACBP6. An overall ribbon diagram showing the fold of PDLP8 (A), with α-helices colored yellow and light blue and β-sheets colored dark blue, green, orange and red. There are two helices and three β-sheets in this protein, with only one visible loop. The model were generated from 4XRE as template by using SWISS MODEL (www.swissmodel.expasy.org). The model consists of 2 α-helices and 4 β-sheets. The interaction between PDLP8 and AtACBP6 by docking via patchdock/firedock. Secondary structure is marked to show 4 α-helices in AtACBP6 as well as the 2 α-helices and 4 β-sheets in PDLP8 (B). The α1 and α2 helices of AtACBP6 are predicted to interact with the β3 and β4 sheets in PDLP8. Best model is represented with a binding affinity of -12.80 (ΔG). (C-D) The interaction between (His)6-PDLP8 and GST-tagged AtACBP6 (with the GST tag cleaved) was visualized by 15% SDS-PAGE. M, low-range rainbow marker; Lane 1, recombinant (His)6-PDLP8; Lane 2, flow-through after recombinant (His)6-PDLP8 is bound to the Ni-NTA magnetic agarose beads; Lane 3, recombinant GST-tagged AtACBP6 (with the GST tag cleaved); Lane 4, flow-through after incubation of recombinant (His)6-PDLP8 and recombinant AtACBP6 on the Ni-NTA magnetic agarose beads following washing to remove excess recombinant AtACBP6; Lane 5, elution from Ni-NTA magnetic agarose beads using buffer B. Arrows indicate the various recombinant proteins. (E-I) The interaction of PDLP8 and AtACBP6 was confirmed by bimolecular fluorescence complementation (BiFC). The interaction of BiFC assays using Agrobacterium-infiltrated tobacco leaves to examine the interaction between AtACBP6 with PDLP8 in vivo. (E) bZIP63-YFPN and bZIP63-YFPC, the positive controls, were expressed in the nucleus. (F-I) The combination of P35S::NYFP and 35S::cYFP, AtACBP6::NYFP and 35S::cYFP as well as P35S::NYFP and PDLP8::CYFP did not show any interaction, while AtACBP6::NYFP and PDLP8::CYFP showed signals of interaction in the plasma membrane (I). Bar = 20 µm.
Next, pull-down assays (Fig. 1C, D) were carried out to confirm the binding of PDLP8 with AtACBP6, using recombinant AtACBP6 and PDLP8 proteins (SDS-PAGE analysis in Figs. S1). As (His)6-PDLP8 (35.6 kD) and GST-AtACBP6 (37.4 kD) are close in molecular size (Fig. S1), PreScission Protease was used to remove the GST tag to distinguish the cleaved recombinant AtACBP6 (10.3 kD) protein from (His)6-PDLP8 (35.6 kD). Both fractions of (His)6-PDLP8 and AtACBP6 were detected in the eluant from Ni-NTA magnetic agarose beads (Fig. 1C), validating in vitro binding of PDLP8 and AtACBP6. Furthermore, ITC data revealed that PDLP8 could bind AtACBP6 with a dissociation constant (Kd) ranging from 11.4 uM to 3.7 uM (Table 2). Binding affinity between AtACBP6 and PDLP8, though modest, was thus confirmed.
Table 2.
ITC binding constant and thermodynamic parameters for (His)6-PDLP8 and (His)6-AtACBP6 interaction.
| Binding | n | Kd (μM) | ΔH (kcal/mol) | -TΔS (kcal/mol) |
|---|---|---|---|---|
| 250 uM AtACBP6 vs 25 uM PDLP8 | 1.91 | 6.8 ± 3.1 | 1.78 | −8.8 |
| 250 uM PDLP8 vs 25 uM AtACBP6 | 2.19 | 7.6 ± 3.8 | 1.44 | −8.4 |
Experiments were carried out at 25°C and reported values are the mean of at least two independent titrations. n, number of binding sites (n = ligand/receptor); Kd, the dissociation constant; ΔH, the enthalpy changes; ΔS, the entropy changes. The binding entropy and enthalpy determination ligand binding.
As PDLP8 is a transmembrane protein and AtACBP6 protein is localized in the cytosol,5,29 an investigation on where and how these two proteins could interact subcellularly was pursued. In BiFC assays (Fig. 1E-I), transient co-expression of AtACBP6::CYFP and PDLP8::NYFP in N. benthamiana (tobacco) leaf cells showed YFP fluorescence signals at the plasma membrane (Fig. 1I), indicating interaction of AtACBP6 and PDLP8 at the plasma membrane. The nuclear basic leucine zipper (bZIP) transcription factor bZIP63 fused to the N/C-terminal of YFP (bZIP63::NYFP and bZIP63::CYFP) combination served as our positive controls,33 and florescent signals were detected in the nucleus (Fig. 1E). The negative control pairs (P35S::NYFP + P35S::CYFP, AtACBP6CYFP + P35S::NYFP and P35S::CYFP + PDLP8::NYFP) did not show any fluorescence signal (Fig. 1F-H).
Comparison of PDLP8 and AtACBP6 expression in Arabidopsis tissues
The spatial expression pattern of PDLP8 and AtACBP6 was compared using GUS assay on transgenic Arabidopsis harboring PDLP8pro::GUS (Fig. 2A-F) and AtACBP6pro::GUS (Fig. 2G-L). PDLP8pro::GUS showed GUS expression in buds (Fig. 2D) after 24 h, while only weak GUS signals could be detected in the vasculature of 3-week-old rosette (Fig. 2F). In comparison, the expression of AtACBP6pro::GUS was stronger with GUS signals detected after merely 2-h GUS staining (Fig. 2I). GUS expression was not observed in the vector control (pB101.3)-transformed plants at similar reaction periods (Fig. 2M-R).
Figure 2.
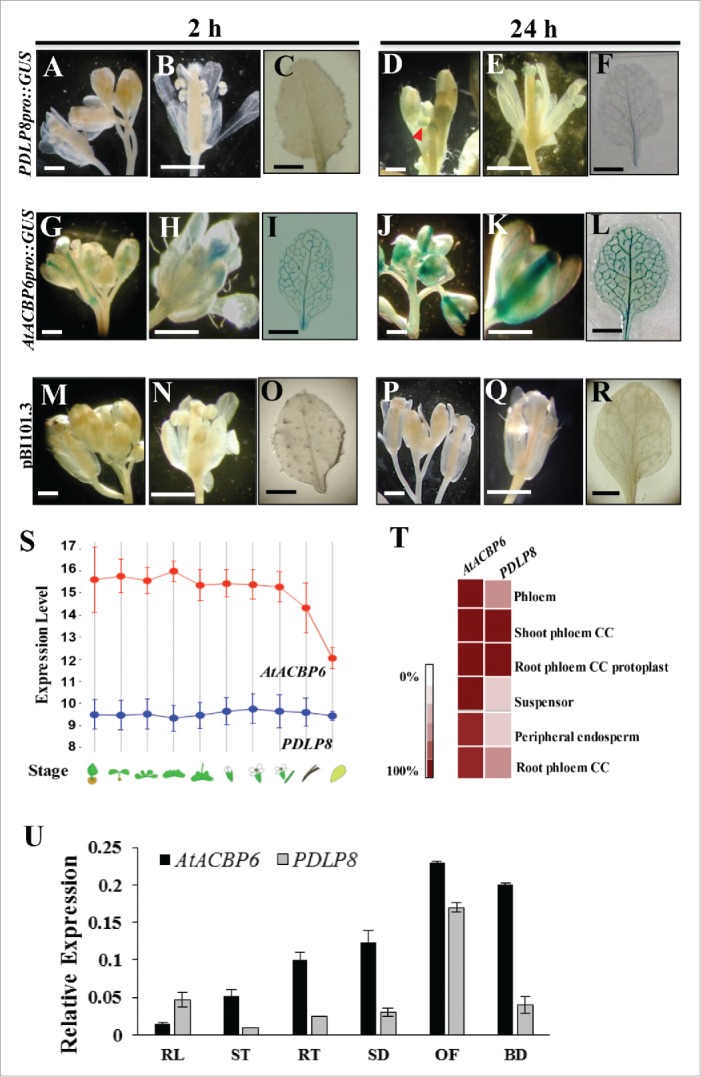
Analysis in the expression pattern of AtACBP6 and PDLP8. Histochemical staining shows GUS expression of PDLP8pro::GUS (A-F) and AtACBP6pro::GUS (G-L), 8-week-old inflorescence, 10-DAF mature flowers and 3-week-old rosette leaves. Different GUS staining times are shown (2 h in A-C and G-I; 24 h in D-F and J-L). GUS expression was not detected in vector (pB101.3)-transformed control plants (M-R). (S-T) A comparison in expression of AtACBP6 and PDLP8 as carried out by analysis of microarray data from Genevestigator (https://genevestigator.com/gv/). (S) Expression of AtACBP6 and PDLP8 in different developmental stages. The red line indicates the expression of AtACBP6 and the blue represents the expression of PDLP8. (T) Heat map of the expression of AtACBP6 and PDLP8 in the phloem. CC, companion cells. (U) Quantitative real-time PCR analysis of PDLP8 and AtACBP6 expression in 4-week-old rosette leaves (RL), 4-week-old stems (ST), 4-week-old stems roots (RT), 1-week-old seedlings (SD), buds (BD) and 8-week-old open flowers (OF). Results were normalized against the expression of ACTIN2. Each bar represents a mean value of six repeats from two independent biological samples ± SD.
By comparing the microarray data obtained from Genevestigator, the expression level of AtACBP6 was indeed much higher than PDLP8 (Fig. 2S). Furthermore, both AtACBP6 and PDLP8 appeared to have higher relative expression values in shoot and root phloem companion cells (Fig. 2T). Also, the mRNA expression level of AtACBP6 and PDLP8 in various Arabidopsis tissues was measured by qRT-PCR analysis (Fig. 2U). Our results showed that the expression of PDLP8 mRNA was relatively higher in the reproductive tissues (flowers and buds) than in vegetative organs (leaves and stems) and the expression pattern of PDLP8 was similar to AtACBP6 (Fig. 2). However, in terms of absolute expression level, AtACBP6 appeared to be much higher (Fig. 2). Collectively, we demonstrated that PDLP8 and AtACBP6 exhibited similar expression patterns in Arabidopsis, with AtACBP6 expression exceeding PDLP8.
Reduced AtACBP6 accumulation in pdlp8 phloem exudates
To further understand the interaction between PDLP8 and AtACBP6, the pdlp8 mutant (SALK_089929C) was characterized. The presence of a T-DNA insert in the pdlp8 mutant was confirmed by gene-specific primers ML2427 and ML2428 as well as a T-DNA border primer (LBa1) as shown in Fig. 3A. In wild-type Arabidopsis (Fig. 3B, lanes 1 and 2), a 0.65-kb band was amplified by ML2427/ML2428, while no such band was detected following PCR using primer pair Lba1/ML2428. Except for lane 6 (Fig. 3B), all the other SALK_089929C samples did not show any bands after PCR using primer pair ML2427/ML2428 (unlike Lba1/ML2428), suggesting that the samples in lanes 3, 4, 5, 7 and 8 were homozygous. When qRT-PCR analysis was carried out to analyze PDLP8 mRNA expression in SALK_089929C in comparison to the wild type, the results indicated an approximately 10-fold decrease in PDLP8 expression in SALK_089929C (Fig. 3C).
Figure 3.
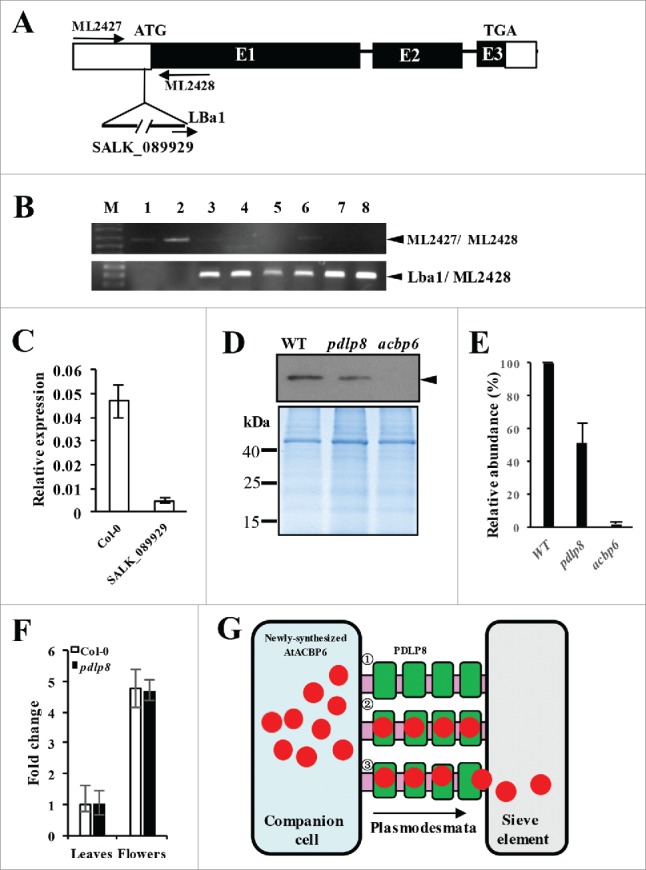
Characterization of the pdlp8 mutant by PCR, qRT-PCR, and western blot analysis on its phloem exudates using anti-AtACBP6 antibodies. (A) Location of the T-DNA insertion in the PDLP8 gene. (B) Specificity of primer combinations, ML2427 and ML2428 (top gel) and LBa1 and ML2428 (bottom gel) in PCR to identify pdlp8 (homozygous) lines. Lanes 1 and 2 are from wild type 4-week-old rosette leaf DNA. Lanes 3 to 8 are from pdlp8 mutant plant DNA. The 0.65-kb ML2427/ML2428, and 0.3-kb Lba1/ML2428 PCR bands are denoted by arrowheads. (C) qRT-PCR analysis of PDLP8 expression in Col-0 and the SALK_089929 (pdlp8 mutant). Relative expression level of PDLP8 was normalized against ACTIN2. (D) Western blot analysis on 5-week-old phloem exudates of Col-0, pdlp8 and acbp6 using anti-AtACBP6 antibodies. The 10.4-kDa AtACBP6 band is indicated by an arrowhead. Coomassie Blue-stained gel identically loaded with proteins as in western blot analysis displayed below. (E) The immunoreactive bands of the total phloem extract proteins were densitometrically quantified. Each value represents the mean of three replicates (± SE). (F) qRT-PCR analysis on AtACBP6 mRNA expression in 5-week-old leaves and flowers from Arabidopsis Col-0 and pdlp8. The AtACBP6 mRNA expression in 5-week-old leaves from Arabidopsis Col-0 was normalized as 1, and fold changes in AtACBP6 mRNA expression in pdlp8 were compared. (G) Previous studies have demonstrated the AtACBP6 protein could be transported from companion cells to sieve elements via the plasmodesmata.28 A model indicating the role for AtACBP6-PDLP8 interaction in influencing AtACBP6 transport from the companion cell (blue) to the sieve element (grey) is proposed.1 PDLP8 (green) is localized to the plasmodesmata (pink).2 AtACBP6 (red) presumably interacts with PDLP8.3 PDLP8 (green) may facilitate AtACBP6 transport from the companion cells (blue) to the sieve element (grey), by unloading AtACBP6 (red) to the sieve element.
To investigate whether PDLP8 could affect AtACBP6 uptake into the sieve elements, phloem exudates from pdlp8 mutant plants were collected for western blot analysis using anti-AtACBP6-specific antibodies with Col-0 and acbp6 as controls. The phloem exudates from Col-0 showed a strong cross-reacting band while acbp6 phloem exudates did not show any bands as expected (Fig. 3D). A weaker band was visualized in the pdlp8 phloem exudates, suggesting that AtACBP6 accumulation was reduced in the absence of PDLP8 in the phloem (Fig. 3D, E). In terms of mRNA expression, since AtACBP6 mRNA could not be detected in the Col-0 phloem exudates,28 we were not able to determine whether AtACBP6 mRNA had declined in the pdlp8 mutant. The AtACBP6 mRNA expression level of leaves and flowers, however, showed non-significant differences between 5-week-old Col-0 and pdlp8 (Fig. 3F). As the phloem exudates originated from the sieve elements, we concluded that the absence of PDLP8 diminished AtACBP6 accumulation in the sieve elements.
Discussion
Plasmodesmata are plant specific cell-to-cell junctions that connect adjacent cells via a cytosolic sleeve and contiguous endoplasmic reticulum, allowing the symplastic movement of signals and macromolecules.9,34 It has been widely acknowledged that these channels regulate a number of developmental and stress-related processes in all plant species.35,36 In the phloem, companion cells are connected to sieve elements by the plasmodesmata which facilitate the loading and unloading of materials into and out of the phloem.37 In conjunction with the prediction that PDLP8 interacts with AtACBP6,31 this study verifies their interaction and supports AtACBP6 movement in the phloem. Among the protein partners of AtACBP6, PDLP8 is unusual because membrane-bound PDLP8 interacts with cytosolic AtACBP6 only when AtACBP6 is translocated through the plasmodemata. BiFC results confirmed their interaction at the plasma membrane (Fig. 1). In a previous proteomic study on cell suspension cultures, AtACBP6 was detected in the plasma membrane fraction,38 suggesting that small amounts of AtACBP6 may adhere to the plasma membrane. It has been observed that PDLP1 and its homologs are distributed on specific subdomains as punctate patterns on the plasma membrane,7 which slightly differs from the PDLP8-AtACBP6 interaction as observed in this study. Whether PDLP8 is located on the plasmodesmata as well as the plasma membrane, or AtACBP6 interaction would alter PDLP8 localization remains to be further investigated.
Analysis in expression profiles of PDLP8 in different tissues of Arabidopsis by qRT-PCR showed its high expression in seedling and flowers (Fig. 2), similar to some extent to the distribution of AtACBP6 as reported by Hsiao et al. (2015),26 although variation in their expression levels was evident. The results of qRT-PCR analysis was supported by GUS assays of higher AtACBP6pro::GUS expression than PDLP8pro::GUS in flowers and 3-week-old rosette leaves (Fig. 2). As PDLP8 belongs to the PDLP family and is presumed to be specifically targeted to the plasmodesmata,5 it was not surprising that lower PDLP8pro::GUS expression occurred given its confinement to the plasmodesmata. In the Arabidopsis PDLP family, PDLP8 showed conservation in amino acid sequence to other members, with highest identity (41.67%) to PDLP6 (Fig. S2; Table S1). Recent studies have revealed that PDLP1 or PDLP5 overexpression in transgenic Arabidopsis enhanced systemic acquired resistance (SAR) during pathogen infections.39 It has also been reported that PDLP1- and PDLP5-overexpressing lines exhibited reduced DIR1 translocation and SAR manifestation.39 Previously, acbp6 plants were observed to be compromised in fungal (Botrytis cinerea and Colletotrichum higginsianum) and Pseudomonas syringae infections.40 Given the amino acid sequence similarity shared by the PDLP family members, it would be interesting in the future to explore whether AtACBP6-PDLP8 interaction affects SAR.
Interestingly, the pdlp8 mutant had displayed a decrease in AtACBP6 expression in the phloem exudates (Fig. 3D, E), implying that PDLP8 expression does affect AtACBP6 accumulation in the phloem. However, the AtACBP6 mRNA level did not decrease in 5-week-old pdlp8 leaves (Fig. 3F), suggesting that PDLP8 may not affect AtACBP6 expression. Given that AtACBP6 mRNA was not detected in the phloem exudate, and AtACBP6 has been hypothesized to move from the companion cells to the sieve elements via the plasmodesmata,28 PDLP8 possibly facilitates AtACBP6 movement in the plasmodesmata as shown in the model (Fig. 3G). PDLP8, when anchored at the plasmodesmata, may guide AtACBP6 protein movement from the companion cells to the sieve elements. Correspondingly it would be of no surprise if AtACBP6 exceeds PDLP8 levels; which would explain for the discrepancy in their distinct absolute expression levels as observed in GUS assays and qRT-PCR analysis.
Computational analysis on the membrane-based interactome database (www.associomics.org) had predicted a number of putative “interactors” of PDLP8 including cytosolic and non-cytosolic proteins, despite PDLP8 being anchored in the plasmodesmata. As the absence of PDLP8 in pdlp8 did not affect AtACBP6 mRNA expression (Fig. 3F), PDLP8 is more likely to function in the transport of protein from the companion cells to the sieve elements. It is worth mentioning that as the pdlp8 mutant showed ∼50% (but not 100%) decrease in AtACBP6 amount in phloem exudates when compared with Col-0 (Fig. 3D, E), it may be possible that other proteins facilitate AtACBP6 into the sieve elements besides PDLP8. Owing to the conservation PDLP8 shares with other PDLP members, AtACBP6 may also interact with the remaining untested PDLPs (PDLP1-PDLP7) when trafficked through the plasmodesmata. This hypothesis needs to be verified experimentally in a future study. Collectively, these findings confirm an interaction between AtACBP6 and PDLP8, and AtACBP6 reduction in pdlp8 phloem exudates is consistent with the absence of its interactor, PDLP8. This study has provided an insight on AtACBP6 transport, most likely mediated by PDLP8, from the companion cells to the sieve elements via the plasmodesmata. It further provides basis for further investigations on PDLP8 function in the symplastic movement of AtACBP6.
Materials and methods
Plant materials and growth conditions
The acbp6 and pdlp8 T-DNA insertion mutants identified from the SALK collection (SALK_104339 and SALK_089929C, respectively; http://signal.salk.edu) were purchased from the Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org) and the acbp6 mutant (SALK_104339) was previously characterized.29 Seeds were surface-sterilized in a solution containing 20% bleach and 1% Triton X-100 for 30 minutes, followed by several times of double-distilled water (ddH2O). Seeds were sowed on Murashige and Skoog (MS) medium agar plates. They were stratified for two days at 4°C in darkness before germination. Plates were incubated in a tissue culture room under continuous light for two weeks and subsequently potted in soil at 23°C/21°C (16 h day/8 h night) cycles. For phloem exudate extraction, wild type, acbp6 and pdlp8 Arabidopsis were grown in a growth chamber maintained at 21°C under 16 h of light and 8 h of dark. Transgenic lines expressing PDLP8pro::GUS or AtACBP6pro::GUS were also grown in a growth chamber maintained at 21°C under 16 h of light and 8 h of dark.
Identification of the pdlp8 mutant
The T-DNA insertion in pdlp8 mutant (SALK_089929C) was identified using the T-DNA left border primer LBa1 and ML2428 together with the PDLP8-specific forward (ML2427) and reverse (ML2428) primer pair (Table S2). Upon identification of the T-DNA insertional site, individual homozygous T-DNA mutant plants were verified by PCR.
Expression and purification of (His)6-tagged AtACBP6, GST-tagged AtACBP6 and (His)6-PDLP8
To generate the construct for expression and purification of recombinant (His)6-PDLP8, an 852-bp PDLP8 cDNA from wild-type Arabidopsis was amplified by primers ML2459 and ML2426 (Table S2). The PCR product was digested with BamHI and PstI and cloned into similar restriction sites on (His)6-tagged expression vector pRSET B (Invitrogen, Carlsbad, CA, USA) to yield plasmid pAT787. To generate the construct for expression and purification of recombinant Glutathione S-transferase (GST)-AtACBP6, plasmid pAT505 which contains a 0.53-kb AtACBP6 BamHI-EcoRI cDNA fragment was cloned into the pGEX-6p-1 vector (Promega) to express tagged GST-AtACBP6 recombinant protein. Expression and purification of Escherichia coli BL21(DE3)pLysS transformed with plasmid pAT505 was carried out as described previously.41
The recombinant (His)6-PDLP8 protein was purified from the inclusion body as previously described,41 with 2% (w/v) sarkosyl added to maintain its protein reactivity.42 GST-AtACBP6 and (His)6-AtACBP6 were analyzed using 15% SDS-PAGE while 10% SDS-PAGE was used for detecting (His)6-PDLP8. All above purified recombinant proteins were subjected to dialysis and refolding in a refolding buffer (50 mM HEPES sodium salt, 200 mM NaCl, 2 mM MgCl2, 5 mM EDTA, 10% glycerol, 0.005% (v/v) Tween-20, pH 7.9) at 4°C, and they were finally concentrated using a Centricon-10 (Amicon) spin-column and subsequently used for in vitro binding assays.
Pull-down assays
Ten micrograms of the recombinant (His)6-PDLP8 protein was immobilized to the Ni-NTA magnetic agarose beads and incubated with 10 μg of the GST-tagged AtACBP6 at room temperature for 1 h.43 After washing several times with washing buffer (20 m mM Tris-HCl, pH 7.5, 0.3 M NaCl, 50 mM imidazole), the PDLP8-AtACBP6 protein complex was eluted from the Ni-NTA magnetic agarose beads with elution buffer (20 mM Tris-HCl, pH 7.5, 0.3 M NaCl, 300 mM imidazole). Purified protein expressed from the GST-tagged blank vector was used as a negative control. The elution fractions were loaded into an SDS-PAGE gel and stained with Coomassie Blue.
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) analysis was carried out using a MicroCal iTC200 system (GE Healthcare, Piscataway, NJ, USA). (His)6-AtACBP6 protein was purified as described previously29 and (His)6-PDLP8 was purified in this study. The interaction of (His)6-AtACBP6 and (His)6-PDLP8 was performed according to Pierce et al. (1999).44 Raw data sets were included and analyzed using ORIGIN7 (OriginLab, Northampton, MA, USA) that was provided alongside the instrument.
Bimolecular fluorescence complementation (BiFC)
BiFC assays were carried out by transient transfection of Nicotiana benthamiana (tobacco) leaves.33 The plasmid containing the AtACBP6 coding region on vector pSPYCE::35S was used to generate the pSPYCE::35SAtACBP6 construct. Similar methods were adopted for amplifying the PDLP8 coding region to yield pSPYNE-35SPDLP8.33 The PCR primers for cloning each fragment following PCR-amplification are listed in Table S2. Agrobacterium tumefaciens GV3101 transformation was carried out.45 A. tumefaciens GV3101 derivatives containing with fluorescence-tagged construct were incubated overnight with shaking at 28°C and 3-week-old tobacco leaves were infiltrated. Two days after infiltration, epidermal cell layers were examined for fluorescence under a Zeiss LSM 510 META microscope using a 505 to 530 nm emission filter.
Quantitative real-time polymerase chain reactions (qRT-PCR)
Various tissues from wild-type Arabidopsis were used to extract RNA using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) followed by DNase (Promega, Madison, WI) treatment. First-strand cDNA was synthesized using the Superscript First-Strand Synthesis System (Invitrogen). Subsequently, qRT-PCR was carried out using the FastStart Universal SYBR Green Master Mix (Roche, www.roche.com) on a StepOne Plus Real-Time PCR System (Applied Biosystems, www.appliedbiosystems.com). Relative gene expression was normalized to the expression of ACTIN2 and gene-specific primers used are listed in Table S2. Three independent biological repeats were performed.
Generation of PDLP8pro::GUS transgenic lines
A 0.65-kb fragment of the AtPDLP8 5′-flanking region was amplified by PCR with primer pair ML2427 and ML2428 using wild-type A. thaliana Col-0 genomic DNA as template. The primers used are listed in Table S2. Fragments were purified and cloned into pGEM-T Easy (Promega) to yield plasmid pAT783. The BamHI-SmaI fragment was subcloned into the corresponding restriction sites on the binary vector pBI101.3 (Clontech) to generate AtPDLP8pro::GUS fusion plasmid pAT788.
The plasmid pAT788 was utilized to generate transgenic Arabidopsis by the floral dip approach.45 Transformants were selected on Murashige and Skoog plates containing 50 mgL−1 of kanamycin. T1 transformants were confirmed by PCR, and seedlings which segregated in a 3:1 Mendelian ratio were potted to yield the T2 generation. Seeds from T2 lines were germinated, and the resultant T3 lines that displayed 100% kanamycin-resistant segregation were deemed homozygous and were subject to further analysis.
β-glucuronidase (GUS) histochemical assays
Histochemical GUS assays were performed on transgenic Arabidopsis expressing PDLP8pro::GUS and AtACBP6pro::GUS. Different tissues from transgenic PDLP8pro::GUS and AtACBP6pro::GUS Arabidopsis T2 lines were stained with GUS staining solution (100 mM sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide and 1 mg ml−1 5-bromo-4-chloro-3-indolyl-β-D-glucuronide). Transgenic samples and the vector-transformed control plants were vacuum-infiltrated in GUS staining buffer for 1 h, followed by a 2-h incubation at room temperature. Chlorophyll was removed using 70% ethanol as described by Zheng et al. (2012).24
Western blot analysis
Phloem exudate proteins were extracted from 5-week-old Arabidopsis for western blot analysis as previously described.28 Protein concentration was determined by the Bradford assay and 20 µg of total protein were loaded for SDS-PAGE, followed by blotting to Hybond-C membrane (Amersham) with the Trans-Blot Cell (BioRad). The blots were incubated with rabbit polyclonal antibodies raised against a synthetic peptide (VEGKSSEEAMNDY) corresponding to amino acids 63–75 of AtACBP6.29 Detection of immunoreactive signals was carried out using the ECL™ Western Blotting Detection Reagents (Amersham) according to the manufacturer's instructions. The intensities of immunoreactive bands were densitometrically quantified using the gel-analyzer function of ImageJ software v.1.46 (National Institutes of Health, USA).
Supplementary Material
Disclosure of potential conflicts of interest
The authors declare that there is no conflict of interest.
Acknowledgements
We thank Julie Chu for expression of recombinant (His)6-tagged PDLP8 and Geoffrey Kong for help in expression of recombinant GST-tagged AtACBP6 used in pull-down assays. This work was supported by the Wilson and Amelia Wong Endowment Fund and the Research Grants Council of Hong Kong (HKU765813M). ZWY was supported by a University Postgraduate Fellowship and QFC was supported by a Postgraduate Scholarship.
Author contributions
This study was designed, directed and coordinated by MLC and ZWY. MLC provided the conceptual and technical guidance throughout this project. ZWY planned and performed the bimolecular fluorescence complementation assays, protein expressions, pull-down assays, GUS assays, qRT-PCR analysis and western blot analysis. QFC constructed the GST-tagged plasmid and optimized the protein expression condition. The manuscript was written by ZWY and MLC.
Abbreviations
- ACBP
Acyl-CoA-binding protein
- BiFC
Bimolecular fluorescence complementation
- GUS
β-Glucuronidase
- ITC
Isothermal titration calorimetry
- PDLP
Plasmodesmata-Located Protein
- qRT-PCR
Quantitative real-time polymerase chain reaction
- MS
Murashige and Skoog
- SA
Salicylic acid
- SAR
Systemic acquired resistance
References
- 1.Epel BL. Plasmodesmata: Composition, structure and trafficking. Plant Mol Biol. 1994;26:1343–56. doi: 10.1007/BF00016479. [DOI] [PubMed] [Google Scholar]
- 2.Roberts AG, Oparka KJ. Plasmodesmata and the control of symplastic transport. Plant Cell Environ. 2003;26:103–124. doi: 10.1046/j.1365-3040.2003.00950.x. [DOI] [Google Scholar]
- 3.Lucas WJ, Lee JY. Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol. 2004;5:712–726. doi: 10.1038/nrm1470. [DOI] [PubMed] [Google Scholar]
- 4.Lee JY, Lu H. Plasmodesmata: The battleground against intruders. Trends Plant Sci. 2011;16:201–210. doi: 10.1016/j.tplants.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Amari K, Boutant E, Hofmann C, Schmitt-Keichinger C, Fernandez-Calvino L, Didier P, Lerich A, Mutterer J, Thomas CL, Heinlein M, et al.. A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog. 2010;6:e1001119. doi: 10.1371/journal.ppat.1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 2008;6:e7. doi: 10.1371/journal.pbio.0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al.. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell. 2011;23:3353–73. doi: 10.1105/tpc.111.087742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Sager R, Cui W, Zhang C, Lu H, Lee JY. Salicylic acid regulates plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell. 2013;25:2315–29. doi: 10.1105/tpc.113.110676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JY. New and old roles of plasmodesmata in immunity and parallels to tunneling nanotubes. Plant Sci. 2014;221-222:13–20. doi: 10.1016/j.plantsci.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao S, Chye ML. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol Biochem. 2009;47:479–484. doi: 10.1016/j.plaphy.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Xiao S, Chye ML. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog Lipid Res. 2011;50:141–151. doi: 10.1016/j.plipres.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Chye ML, Huang BQ, Zee SY. Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J. 1999;18:205–214. doi: 10.1046/j.1365-313X.1999.00443.x. [DOI] [PubMed] [Google Scholar]
- 13.Chye ML, Li HY, Yung MH. Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14C palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol Biol. 2000;44:711–721. doi: 10.1023/A:1026524108095. [DOI] [PubMed] [Google Scholar]
- 14.Li HY, Chye ML. Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Mol Biol. 2003;51:483–492. doi: 10.1023/A:1022330304402. [DOI] [PubMed] [Google Scholar]
- 15.Li HY, Chye ML. Arabidopsis Acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol Biol. 2004;54:233–243. doi: 10.1023/B:PLAN.0000028790.75090.ab. [DOI] [PubMed] [Google Scholar]
- 16.Du ZY, Chye ML. Interactions between Arabidopsis acyl-CoA-binding proteins and their protein partners. Planta. 2013;238:239–245. doi: 10.1007/s00425-013-1904-2. [DOI] [PubMed] [Google Scholar]
- 17.Xiao S, Chye ML. Arabidopsis ACBP1 overexpressors are Pb(II)-tolerant and accumulate Pb(II). Plant Signal Behav. 2008;3:693–694. doi: 10.4161/psb.3.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao W, Li HY, Xiao S, Chye ML. Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J. 2010;62:989–1003. doi: 10.1111/j.1469-8137.2008.02631.x. [DOI] [PubMed] [Google Scholar]
- 19.Gao W, Xiao S, Li HY, Tsao SW, Chye ML. Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with heavy-metal-binding farnesylated protein AtFP6. New Phytol. 2009;181:89–102. [DOI] [PubMed] [Google Scholar]
- 20.Chen QF, Xiao S, Qi W, Mishra G, Ma J, Wang M, Chye ML. The Arabidopsis acbp1acbp2 double mutant lacking acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytol. 2010;186:843–855. doi: 10.1111/j.1469-8137.2010.03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du ZY, Chen MX, Chen QF, Xiao S, Chye ML. Arabidopsis acyl-CoA-binding protein ACBP1 participates in the regulation of seed germination and seedling development. Plant J. 2013;74:294–309. doi: 10.1111/tpj.12121. [DOI] [PubMed] [Google Scholar]
- 22.Xiao S, Chye ML. Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiol. 2011;156:2069–81. doi: 10.1104/pp.111.176933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, Ma J, Wang M, Welti R, Chye ML. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22:1463–82. doi: 10.1105/tpc.110.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng SX, Xiao S, Chye ML. The gene encoding Arabidopsis acyl-CoA-binding protein 3 is pathogen inducible and subject to circadian regulation. J Exp Bot. 2012;63:2985–3000. doi: 10.1093/jxb/ers009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye ZW, Chye ML. Plant cytosolic Acyl-CoA-binding proteins. Lipids. 2016;51:1–13. doi: 10.1007/s11745-015-4103-z. [DOI] [PubMed] [Google Scholar]
- 26.Hsiao AS, Yeung EC, Ye ZW, Chye ML. The arabidopsis cytosolic acyl-CoA-binding proteins play combinatory roles in pollen development. Plant Cell Physiol. 2015;56(2):322–333. doi: 10.1093/pcp/pcu163. [DOI] [PubMed] [Google Scholar]
- 27.Ye ZW, Xu J, Shi J, Zhang D, Chye ML. Kelch-motif containing acyl-CoA binding proteins AtACBP4 and AtACBP5 are differentially expressed and function in floral lipid metabolism. Plant Mol Biol. 2017;93:209–225. doi: 10.1007/s11103-016-0557-5. [DOI] [PubMed] [Google Scholar]
- 28.Ye ZW, Lung SC, Hu TH, Chen QF, Suen YL, Wang M, Hoffmann-Benning S, Yeung E, Chye ML. Arabidopsis acyl-CoA-binding protein ACBP6 localizes in the phloem and affects jasmonate composition. Plant Mol Biol. 2016;92:717–730. doi: 10.1007/s11103-016-0541-0. [DOI] [PubMed] [Google Scholar]
- 29.Chen QF, Xiao S, Chye ML. Overexpression of the Arabidopsis 10-kgdalton acyl-coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiol. 2008;148:304–315. doi: 10.1104/pp.108.123331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao P, Chen QF, Chye ML. Transgenic Arabidopsis flowers overexpressing acyl-CoA-binding protein ACBP6 are freezing tolerant. Plant Cell Physiol. 2014;55(6):1055–71. doi: 10.1093/pcp/pcu037. [DOI] [PubMed] [Google Scholar]
- 31.Jones AM, Xuan Y, Xu M, Wang RS, Ho CH, Lalonde S, You CH, Sardi MI, Parsa SA, Smith-Valle E, et al.. Border control-a membrane-linked interactome of Arabidopsis. Science. 2014;344:711–716. doi: 10.1126/science.1251358. [DOI] [PubMed] [Google Scholar]
- 32.Miyakawa T, Mizushima H, Ohtsuka J, Oda M, Kawai F, Tanokura M. Structural basis for the Ca2+-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190. Appl Microbiol Biotechnol. 2015;99:4297–4307. doi: 10.1007/s00253-014-6272-8. [DOI] [PubMed] [Google Scholar]
- 33.Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al.. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 34.Lee JY, Cho SK, Sager R. Plasmodesmata and noncell autonomous signaling in plants. The plant plasma membrane. Berlin Heidelberg: Springer; 2011. p. 87–107. [Google Scholar]
- 35.Burch-Smith TM, Stonebloom S, Xu M, Zambryski PC. Plasmodesmata during development: Re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma. 2011;248:61–74. doi: 10.1007/s00709-010-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burch-Smith TM, Zambryski PC. Plasmodesmata paradigm shift: Regulation from without versus within. Annu Rev Plant Biol. 2012;63:239–260. doi: 10.1146/annurev-arplant-042811-105453. [DOI] [PubMed] [Google Scholar]
- 37.Turgeon R, Wolf S. Phloem transport: Cellular pathways and molecular trafficking. Annu Rev Plant Biol. 2009;60:207–221. doi: 10.1146/annurev.arplant.043008.092045. [DOI] [PubMed] [Google Scholar]
- 38.Benschop JJ, Mohammed S, O'Flaherty M, Heck AJ, Slijper M, Menke FL. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Carella P, Isaacs M, Cameron RK. Plasmodesmata-located protein overexpression negatively impacts the manifestation of systemic acquired resistance and the long-distance movement of Defective in Induced Resistance1 in Arabidopsis. Plant Biol. 2015;17:395–401. doi: 10.1111/plb.12234. [DOI] [PubMed] [Google Scholar]
- 40.Xia Y, Yu K, Gao QM, Wilson EV, Navarre D, Kachroo P, Kachroo A. Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Frontiers Plant Sci. 2012;3:224. doi: 10.3389/fpls.2012.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leung KC, Li HY, Xiao S, Tse MH, Chye ML. Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta. 2006;223:871–881. doi: 10.1007/s00425-005-0139-2. [DOI] [PubMed] [Google Scholar]
- 42.Massiah MA, Wright KM, Du H. Obtaining soluble folded proteins from inclusion bodies using sarkosyl, triton X-100, and CHAPS: Application to LB and M9 minimal media. Curr Protoc Protein Sci. 2016;84:1–6. doi: 10.1002/0471140864.ps0613s84. [DOI] [PubMed] [Google Scholar]
- 43.Yuan S, Chu H, Singh K, Zhao H, Zhang K, Kao RY, Chow BKC, Zhou J, Zheng BJ. A novel small-molecule inhibitor of influenza A virus acts by suppressing PA endonuclease activity of the viral polymerase. Sci Rep. 2016;6:22880. doi: 10.1038/srep22880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 45.Clough SJ, Bent AF. Floral dip: A simpliied method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


