ABSTRACT
Mitogen Activated Protein Kinases (MAPKs) are known to be important mediators of plant responses to biotic and abiotic stresses. In a recent report, we enlarged the understanding of the Arabidopsis thaliana MPK3 functions showing that the expression of a constitutively active (CA) form of the protein led to auto-immune phenotypes. CA-MPK3 plants are dwarf and display defense responses that are characterized by the accumulation of salicylic acid and phytoalexins as well as by the upregulation of several defense genes. Consistently with these data, we present here results demonstrating that, compared with wild type controls, CA-MPK3 plants are more resistant to the hemibiotrophic pathogen Pseudomonas syringae DC3000. Based on our previous work, we also discuss the mechanisms of robust plant immunity controlled by sustained MPK3 activity, focusing especially on the roles of disease resistance proteins.
KEYWORDS: Arabidopsis, MAPK, resistance protein, defense robustness, stress responses
CA-MPK3 plants are more resistant to pathogens
Activities of MPK3 and MPK6 are critical for the implementation of appropriate plant immunity. Rapid and transient MPK3/6 activities (from 5 minutes to 1 hour) are commonly associated with responses to Pathogen-Associated-Molecular-Patterns (PAMPs) in a process leading to PAMP-Triggered Immunity (PTI),1,2 while sustained MPK3/6 activities (from 3 to 12 hours) upon perception of a bacterial effector suggest their involvement in Effector-Triggered Immunity (ETI).3
Taking advantage of a gain-of-function approach,4 we revealed in a recent study that expression of a constitutively active (CA) form of Arabidopsis thaliana MPK3, driven by the endogenous MPK3 promoter, has a massive impact on the defense responses of plants.5 CA-MPK3 lines exhibit spontaneous cell death that is associated with the accumulation of reactive oxygen species, salicylic acid (SA), phytoalexins as well as with a transcriptional reprogramming of defense genes. However, the capacity of CA-MPK3 plants to resist to pathogen attacks was not known.
To test this, we challenged CA-MPK3 plants with the hemibiotrophic pathogen Pseudomonas syringae DC3000 and measured the levels of bacterial populations 4 dpi compared with those in Col-0 and in WT-MPK3 controls. As shown in Fig. 1, P. syringae DC3000 levels were significantly lower in the CA-MPK3 lines than in the Col-0 and WT-MPK3 lines. This result confirms that the defense induction observed in the CA-MPK3 plants can confer a higher resistance against pathogens.
Figure 1.
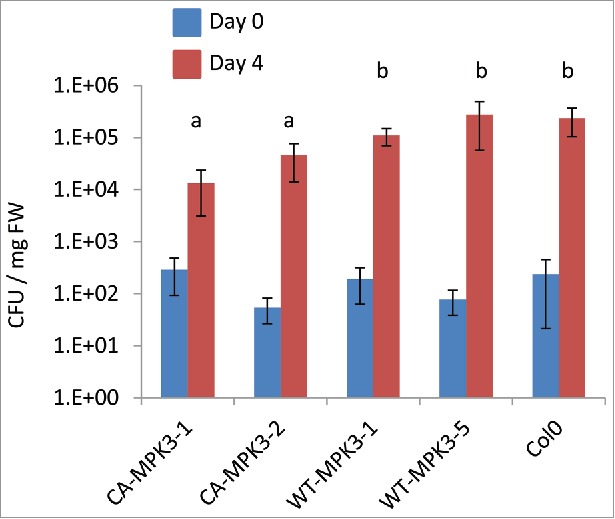
CA-MPK3 plants are more resistant to Pseudomonas syringae DC3000. Leaves of 1 month old plants grown in soil were infiltrated with P. syringae DC3000 solution (OD600 = 0,01). Bacterial populations were measured at day 0 (3 hpi) and at day 4. Letters indicate statistical differences at day 4 (Kruskal-Wallis test, followed by a post-hoc Tukey test; 3 < n < 5; p < 0,05). Experiments were repeated 3 times independendtly and gave similar results. CFU: colony-forming unit.
Expression of CA-MPK3 results in defense robustness
Defense robustness allows plants to cope with different pathogen strategies and to mount defense responses which are still effective even if some of their defense responses are impaired. For this reason robustness is considered as a feature of ETI rather than PTI.6 In line with other studies,7,3 we associated in our previous work the expression of CA-MPK3 proteins with defense robustness in plants. We notably showed that CA-MPK3/sid2 plants which do not accumulate SA anymore still retain most of the defense phenotypes of CA-MPK3, including the upregulation of some SA markers. This result comforts the notion that sustained MPK3 activity could allow plants to bypass SA signaling – for example, if SA signaling is inhibited by pathogen attack - and still be able to implement adequate defense responses.
Another result we obtained was that CA-MPK3/summ2, which is deficient in the disease resistance (R) protein SUMM2, exhibits a partial reversion of the CA-MPK3 phenotype. This finding demonstrates that some R proteins can act downstream of MPK3 activity. As SUMM2 is known to guard the MEKK1-MKK1/2-MPK4 pathway,8,9 we extrapolated that constitutive activity of MPK3 somehow interfered with this pathway to trigger SUMM2-mediated defense responses. More generally, an interesting hypothesis could be that, unlike transient activity, sustained activity of MPK3 (and/or other MAPKs) would target substrates which are guarded by different R proteins, each contributing to trigger partly overlapping and partly independent defense responses. This mechanism would confer defense robustness and account for the partial reversion of the phenotype in CA-MPK3/summ2. Figure 2 presents a model of these defense processes in which R proteins act both upstream and downstream of MAPKs. This model is also consistent with the fact that the CA-MPK3 phenotype is totally reverted by high temperature5 and the assumption that R proteins can function as rheostats, triggering defense responses in a temperature-dependent fashion.10
Figure 2.
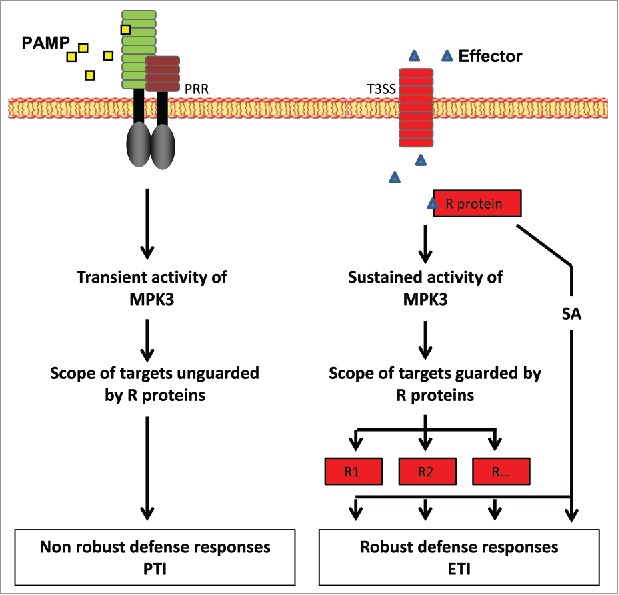
Model of defense robustness involving sustained MPK3 activity. Unlike transient activity in PTI, sustained MPK3 activity in ETI results in defense responses mediated by R proteins. These responses might be partly independent, partly redundant, thereby conferring defense robustness to the plant. PAMP: Pathogen-Associated Molecular Pattern; PRR: Pattern Recognition Receptor; T3SS: Type 3 Secretory System; R protein: Resistance protein, PTI: PAMPs-Triggered Immunity; ETI: Effector-Triggered Immunity.
When browsing the list of genes which are the most upregulated in CA-MPK3 plants compared with WT controls, we found several putative and annotated R genes (Table 1).5 Although increase in transcript abundance does not necessarily mean activation, it is known that overexpression of some R proteins can result in constitutive defense responses,11,12 strongly suggesting that there is indeed a correlation between expression levels and activation of R genes. Therefore the upregulated R genes could be promising candidates to further explore the link between defense robustness and sustained MPK3 activity. Since constitutive activation of the R proteins SNC113 and ADR114 leads to auto-immune phenotypes, the effects of snc1 and adr1 loss-of-function on the CA-MPK3 phenotype would be especially interesting to test our model. Furthermore, snc1 loss-of-function is already known to revert the auto-immune mkp1 phenotype that is characterized by increased basal activities of MPK3 and 6.15 Alternatively, crosses between CA-MPK3 and eds1 and/or ndr1 would be worth performing because these 2 genes are known to generally mediate defense responses initiated by R proteins.16,17
Table 1.
List of R genes upregulated in CA-MPK3. Data are from Genot et al. (2017). Fold Changes (FCh) are expressed in log2. Only genes with a FCh superior to 0,75 were considered.
| AGI code | CATMA annotation | FCh |
|---|---|---|
| AT2G32680 | “disease resistance family protein” | 1.96 |
| AT5G18350 | “disease resistance protein (TIR-NBS-LRR class), putative” | 1.85 |
| AT3G25020 | “disease resistance family protein” | 1.79 |
| AT3G24900 | “disease resistance family protein / LRR family protein” | 1.73 |
| AT1G47890 | “disease resistance family protein” | 1.69 |
| AT5G66890 | “disease resistance protein (CC-NBS-LRR class), putative” | 1.50 |
| AT3G25010 | “disease resistance family protein” | 1.49 |
| AT1G12290 | “disease resistance protein (CC-NBS-LRR class), putative” | 1.49 |
| AT4G11170 | “disease resistance protein (TIR-NBS-LRR class), putative” | 1.47 |
| AT4G19515 | “disease resistance family protein” | 1.44 |
| AT1G72920 | “disease resistance protein (TIR-NBS class), putative” | 1.42 |
| AT3G04220 | “disease resistance protein (TIR-NBS-LRR class), putative” | 1.35 |
| AT1G33560 | “ADR1 (ACTIVATED DISEASE RESISTANCE 1)” | 1.31 |
| AT1G57650 | “disease resistance protein (NBS-LRR class), putative” | 1.30 |
| AT2G15080 | “disease resistance family protein” | 1.27 |
| AT1G17615 | “disease resistance protein (TIR-NBS class), putative” | 1.23 |
| AT5G41750 | “disease resistance protein (TIR-NBS-LRR class), putative” | 1.19 |
| AT1G66090 | “disease resistance protein (TIR-NBS class), putative” | 1.19 |
| AT5G41740 | “disease resistance protein (TIR-NBS-LRR class), putative” | 1.14 |
| AT4G13810 | “disease resistance family protein / LRR family protein” | 1.12 |
| AT1G15890 | “disease resistance protein (CC-NBS-LRR class), putative” | 1.11 |
| AT5G58120 | “disease resistance protein (TIR-NBS-LRR class), putative” | 1.10 |
| AT4G16860 | “RPP4 (RECOGNITION OF PERONOSPORA PARASITICA 4)” | 1.10 |
| AT5G25910 | “disease resistance family protein” | 1.09 |
| AT1G57630 | “disease resistance protein (TIR class), putative” | 1.07 |
| AT1G50180 | “disease resistance protein (CC-NBS-LRR class), putative” | 1.07 |
| AT4G11170 | “disease resistance protein (TIR-NBS-LRR class), putative” | 1.05 |
| AT1G56510 | “disease resistance protein (TIR-NBS-LRR class), putative” | 0.83 |
| AT4G16890 | “SNC1 (SUPPRESSOR OF NPR1–1, CONSTITUTIVE 1)” | 0.79 |
Perspectives
CA-MPK3 plants exemplify the concept of trade-off between growth and immunity. On one hand the plants are more resistant to pathogens, but on the other hand they have a dwarf morphology. Campos et al. (2016)18 recently succeeded to obtain plants more resistant to pathogens without being affected in their development and showed that the trade-off between growth and immunity not necessarily relies on allocation costs but could also be due to genetic costs, i.e. to crosstalk in defense-growth signaling. Along the same path of ideas, a better understanding of the gene network involving sustained MPK3 activity and immune robustness, through notably epistatic studies as mentioned above, might unveil original mechanisms potentially allowing to uncouple defense and growth. This would open new perspectives to engineer plants with higher resistance to biotic stresses.
References
- 1.Nühse TS, Peck SC, Hirt H, Boller T. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK6. J Biol Chem. 2000;275:7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- 2.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 3.Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F. Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 2013;9:e1004015. doi: 10.1371/journal.pgen.1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berriri S, Garcia AV, Frei dit Frey N, Rozhon W, Pateyron S, Leonhardt N, Montillet JL, Leung J, Hirt H, Colcombet J. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell. 2012;24:4281–4293. doi: 10.1105/tpc.112.101253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genot B, Lang J, Berriri S, Garmier M, Gilard F, Pateyron S, Haustraete K, Van Der Streaten D, Hirt H, Colcombet J. Constitutively active Arabidopsis MAP Kinase 3 triggers defense responses involving salicylic acid and SUMM2 resistance protein. Plant Physiol. 2017;174(2):1238–1249. doi: 10.1104/pp.17.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, Tsuda K, Igarashi D, Hillmer RA, Sakakibara H, Myers CL, Katagiri F. Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe. 2014;15:84–94. doi: 10.1016/j.chom.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong Q, Qu N, Gao M, Zhang Z, Ding X, Yang F, Li Y, Dong OX, Chen S, Li X, et al.. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell. 2012;24:2225–2236. doi: 10.1105/tpc.112.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe. 2012;11:253–263. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Alcázar R, Parker JE. The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 2011;16:666–675. doi: 10.1016/j.tplants.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Stokes TL, Richards EJ. Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc Natl Acad Sci USA. 2002;99:7792–7796. doi: 10.1073/pnas.112040999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huh SU, Cevik V, Ding P, Duxbury Z, Ma Y, Tomlinson L, Sarris PF, Jones JDG. Protein-protein interactions in the RPS4/RRS1 immune receptor complex. PLOS Pathog. 2017;13:e1006376. doi: 10.1371/journal.ppat.1006376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 14.Grant JJ, Chini A, Basu D, Loake GJ. Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol Plant Microbe Interact. 2003;16:669–680. doi: 10.1094/MPMI.2003.16.8.669. [DOI] [PubMed] [Google Scholar]
- 15.Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R. MAP KINASE PHOSPHATASE1 and PROTEIN TYROSINE PHOSPHATASE1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21:2884–2897. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venugopal SC, Jeong RD, Mandal MK, Zhu S, Chandra-Shekara AC, Xia Y, Hersh M, Stromberg AJ, Navarre D, Kachroo A, et al.. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009;5:e1000545. doi: 10.1371/journal.pgen.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al.. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun. 2016;7:12570. doi: 10.1038/ncomms12570. [DOI] [PMC free article] [PubMed] [Google Scholar]


