ABSTRACT
The salt overly sensitive (SOS) pathway is the only mechanism known for Na+ extrusion in plant cells. SOS pathway activation involves Ca2+-sensing proteins, such as calcineurin B-like (CBL) proteins, and CBL-interacting protein kinases (CIPKs). In this signalling mechanism, a transit increase in cytosolic Ca2+ concentration triggered by Na+ accumulation is perceived by CBL (also known as SOS3). Afterward, SOS3 physically interacts with a CIPK (also known as SOS2), forming the SOS2/SOS3 complex, which can regulate the number downstream targets, controlling ionic homeostasis. For instance, the SOS2/SOS3 complex phosphorylates and activates the SOS1 plasmalemma protein, which is a Na+/H+ antiporter that extrudes Na+ out of the cell. The CBL-CIPK networking system displays specificity, complexity and diversity, constituting a critical response against salt stress and other abiotic stresses. In a study reported in the journal Plant and Cell Physiology, we showed that NH4+ induces the robust activation of transporters for Na+ homeostasis in root cells, especially the SOS1 antiporter and plasma membrane H+-ATPase, differently than does NO3−. Despite some studies having shown that external NH4+ ameliorates salt-induced effects on ionic homeostasis, there is no evidence that NH4+ per se or some product of its assimilation is responsible for these responses. Here, we speculate about the signalling role behind glutamine in CBL-CIPK modulation, which could effectively activate the SOS pathway in NH4+-fed stressed plants.
KEYWORDS: Na+/H+ antiporter, NH4+ nutrition, salt tolerance, SOS pathway, signal
Calcineurin B-Like (CBL) proteins are a family of small plant-specific peptides that function as Ca2+ sensors. Some CBL Ca2+ sensors, also referred to as SOS3-Like Calcium Binding Proteins (SCaBPs), are known to regulate ionic homeostasis.1 The CBL-interacting protein kinase (CIPK) family, also known as sucrose non-fermenting (SNF)-related kinases (SnRKs), includes a serine/threonine protein kinase (SOS2) belonging to the SNF1-related kinase 3 (SnRK3) family. CBL/CIPK (SOS3/SOS2) proteins not only act as essential components of the salt overly sensitive (SOS) pathway, activating the Na+/H+ SOS1 antiporter,2 but also may interact and positively regulate key components during salt tolerance responses. These components include vacuolar Na+/H+ antiporter (NHX) and H+-ATPase (V-ATPase) proton pumps, high-affinity K+ transporters (AtHKT1) and channels (AKT1) and H+-ATPase proton pumps of the plasma membrane.3,4,5,6
It is known that numerous CBL (11 genes in Arabidopsis) and CIPK (26 genes in Arabidopsis) protein isoforms are present in plants; therefore, different combinations of CBL-CIPK complexes could occur to mediate physiological and biochemical responses for both growth and developmental patterns and tolerance to environmental stresses.14,15 Thus, the exact flow of information will depend on the assembly of alternative and competing CBL-CIPK combinations.16 Some CBL and CIPK proteins have been functionally characterized,15 and the CBL4-CIPK24 (SOS3/SOS2) complex was revealed to specifically mediate salt stress signaling and tolerance.14 Interestingly, like CBL4-CIPK24 in Arabidopsis, many other members (CBL1, CBL9, PsCBL, CIPK1, CIPK2, CIPK25, CIPK26 and CIPK31) are also involved in the network-like signaling system to mediate response to salt stress, including ABA-dependent signaling cascades.16,17,18,19,20,21,22 However, although important progress has been achieved in understanding the downstream targets and physiological functions of some of these proteins, many queries remain to be explored.
To date, extensive studies have focused on identifying salt responsive regulators and focusing salt stress pathways at cellular, tissue and whole-plant level. Strictly, from salt exposure to SOS1 activation, stress signals always result in cytosolic Ca2+ alterations, which are unique and precisely translated by Ca2+ sensor proteins to relay the signaling network.1,23 Surprisingly, although 14-3-3 protein has been found to interact with SOS2 and constitute an important feature for salt tolerance,24 there is little or no evidence regarding of synergistic chemical inducers in inducing CBL/CIPK proteins under salinity, especially in roots.16
Some products of N assimilation might act as signalling molecules, activating specific members of the CIPK family in plants. It has been shown that AtCIPK8 and AtCIPK23 transcripts and N assimilation by-products accumulate to high levels under NO3− supply.7,8 Despite a relationship between NO3− nutrition and CIPK signalling factors having been demonstrated regarding both morphogenetic processes and NO3− uptake in roots, the roles of these factors in salt tolerance responses are still elusive. On the other hand, NH4+ supply also specifically activates other CIPK members, such as CIPK15, whose response is closely associated with the expression of N assimilation enzymes such glutamine synthetase (GS) and glutamate synthase (GOGAT).9
Recent studies have shown that glutamine functions as a signal molecule that induces several transcription factors responsible for plant defence against abiotic stress.10,11 In rice, exogenous glutamine increases the expression of some members of the CIPK family, which are responsible for amplifying and transducing the signal that activates pathways involved in the regulation of plant growth and stress responses. We previously reported that NH4+ supply severely restricts Na+ accumulation and transport to shoots through the efficient upregulation of SOS1 in sorghum roots, a response not observed for NO3− nutrition.12 Our data suggested that enhanced SOS1 antiporter activity may mostly involve post-transcriptional and/or post-translational regulation, as SOS1 transcript abundance was similar in salt-stressed plants treated with NH4+ or NO3−. Thus, we hypothesised that glutamine exerts a role as a signal molecule to activate the SOS2/SOS3 system and, subsequently, the SOS1 antiporter (Fig. 1). This idea is supported by high glutamine accumulations in NH4+-grown sorghum roots, as the accumulations were more prominent in response to salt stress.13
Figure 1.
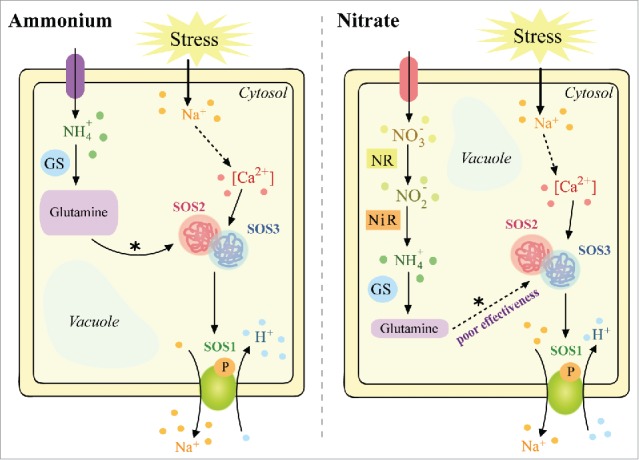
Putative model of CIPK modulation by glutamine. Under NH4+ supply, an over-accumulation of glutamine occurs in the root cells of sorghum, especially in salt-stressed plants. The signal from glutamine* is suggested to specifically modulate CIPK proteins at the allosteric, transcriptional or post-transcriptional level. Upon activation of CIPK proteins, the operation of the SOS pathway is augmented, activating Na+ extrusion by an SOS1 exchanger. Otherwise, NO3− nutrition may also modulate CIPK proteins, albeit at a lower intensity. Abbreviations: Ca2+, calcium; GS, glutamine synthetase; Na+, sodium; NH4+, ammonium; NiR, nitrite reductase; NO2−, nitrate; NO3−, nitrate; NR, nitrate reductase; SOS1, Salt Overly Sensitive 1, a plasma membrane-binding Na+/H+ antiporter; SOS2, Salt Overly Sensitive 2, also referred to as CBL-interacting protein kinase (CIPK); SOS3, Salt Overly Sensitive 3, also referred to as calcineurin B-like protein (CBL).
It is also plausible that activation of plasma membrane Na+/H+ antiporters by the CBL-CIPK complex occurs in NO3−-fed stressed plants, but this response was less intense than that of NH4+-fed stressed plants.12 Thus, this evidence supports the assumption that high levels of glutamine, as a consequence of enhanced GS activity, can stimulate the formation of CBL-CIPK-specific complexes acting on important mechanisms of ionic regulation during tolerance to salinity in NH4+-fed sorghum. Nevertheless, the roles of some complex combinations remain to be explored, including their interaction with specific signal molecules such as by-products of N metabolism under salt stress. For instance, specifically in sorghum, there is no evidence of a role for glutamine or NH4+ in activating the CBL4-CIPK24 complex, which is mostly found to function in Na+ extrusion in root cells under salt stress conditions.14,15 To clarify this mistake, much future effort should be addressed. Firstly, the crystal structures of CBL-CIPK complexes must be solved. Secondly, the CBL- and CIPK-interacting potential candidates should be studied with respect to antagonistic and synergistic effects. Finally, the physiological inputs and outputs of each CBL-CIPK combination should be investigated in order to complete understanding their biological roles.
The use of conventional plant physiology integrated with genetics and biochemical traits may constitute a reliable strategy for understanding the role of glutamine in CBL-CIPK signalling pathways. In this sense, the identification of glutamine as an element of the CBL-CIPK network emerges as a promising challenge for future studies to advance in knowledge and manipulation of signalling processes. Once enough mechanisms are elucidated, specific signalling pathways and cross-talk could be properly applied to crop management and plant breeding programmes in order to improve the capacity of plants to tolerate salt stress.
Acknowledgments
The authors are grateful for the financial support provided by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Instituto Nacional de Ciência e Tecnologia em Salinidade (INCTSal).
References
- 1.Ji H, Pardo JM, Batelli G, Oosten MJV, Bressan RA, Li X. The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol Plant. 2013;6:275–86. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- 2.Qiu QS, Guo Y, Dietrich MS, Shumaker KS, Zhu J. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. P Natl Acad Sci U S A. 2002;99:8436–41. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the SOS pathway. J Biol Chem. 2004; 279:207–15. doi: 10.1074/jbc.M307982200. [DOI] [PubMed] [Google Scholar]
- 4.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444:139–58. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Batelli G, Verslues PE, Agius F, Qiu QS, Fujii H, Pan S, Schumaker KS, Grillo S, Zhu JK. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol. 2007;27:7781–90. doi: 10.1128/MCB.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li R, Zhang J, Wei J, Wang H, Wang Y, Ma R. Functions and mechanisms of the CBL-CIPK signaling systems in plant response to abiotic stress. Prog Nat Sci. 2009;19:667–76. doi: 10.1016/j.pnsc.2008.06.030. [DOI] [Google Scholar]
- 7.Vidal EA, Tamayo KP, Gutierrez R. Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Syst Biol Med. 2010;2:683–93. doi: 10.1002/wsbm.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanin L, Zamboni A, Monte R, Tomasi N, Varanini Z, Cesco S, Pinton R. Transcriptomic analysis highlights reciprocal interaction of urea and nitrate for nitrogen acquisition by maize roots. Plant Cell Physiol. 2015;56:532–48. doi: 10.1093/pcp/pcu202. [DOI] [PubMed] [Google Scholar]
- 9.Yang S, Hao D, Song Z, Yang G, Wang L, Su Y. RNA-Seq analysis of differentially expressed genes in rice under varied nitrogen supplies. Gene. 2015;555:305–17. doi: 10.1016/j.gene.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 10.Kamada-Nobusada T, Makita N, Kojima M, Sakakibara H. Nitrogen-dependent regulation of de novo cytokinin biosynthesis in rice: the role of glutamine metabolism as an additional signal. Plant Cell Physiol. 2013;54:1881–93. doi: 10.1093/pcp/pct127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan CC, Chung TY, Juo YA, Hsieh MH. Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genomics. 2015;16:731. doi: 10.1186/s12864-015-1892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda RS, Mesquita RO, Costa JH, Alvarez-Pizarro JC, Prisco JT, Gomes-Filho E. Integrative control between proton pumps and SOS1 antiporters in roots is crucial for maintaining low Na+ accumulation and salt tolerance in ammonium-supplied Sorghum bicolor. Plant Cell Physiol. 2017;58:522–36. doi: 10.1093/pcp/pcw231. [DOI] [PubMed] [Google Scholar]
- 13.Miranda RS, Gomes-Filho E, Prisco JT, Alvarez-Pizarro JC. Ammonium improves tolerance to salinity stress in Sorghum bicolor plants. Plant Growth Regul. 2016;78:121–31. doi: 10.1007/s10725-015-0079-1. [DOI] [Google Scholar]
- 14.Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–45. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 15.Mao J, Manik SMN, Shi S, Chao J, Jin Y, Wang Q, Liu H. Mechanisms and physiological roles of CBL-CIPK networking systems in Arabidopsis thaliana. Genes. 2016;7:62. doi: 10.3390/genes7090062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Angelo C, Weinl S, Batistic O, Pandey GK, Cheong HY, Schultke S, Albrecht V, Ehlert B, Schulz B, Harter K, Luan S, Bock R, Kudla J. Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. The Plant Journal. 2006;48:857–872. doi: 10.1111/j.1365-313X.2006.02921.x. [DOI] [PubMed] [Google Scholar]
- 17.Cheong YH, Kim K-N, Pandey GK, Gupta R, Grant JJ, Luan S. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. The Plant Cell. 2001;15:1833–1845. doi: 10.1105/tpc.012393.genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey GK, Cheong YH, Kim K-N, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S. The calcium sensor calcineurin b-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. The Plant Cell. 2003;16:1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, Zhang J, Wu G, Wang H, Chen Y, Wei J. HbCIPK2, a novel CBL-interacting protein kinase from halophyte Hordeum brevisubulatum, confers salt and osmotic stress tolerance. Plant, Cell and Environment. 2012;35:1582–1600. doi: 10.1111/j.1365-3040.2012.02511.x. [DOI] [PubMed] [Google Scholar]
- 20.Luo Q, Wei Q, Wang R, Zhang Y, Zhang F, He Y, Zhou S, Feng J, Yang G, He G. BdCIPK31, a calcineurin B-like protein-interacting protein kinase, regulates plant response to drought and salt stress. Frontiers in Plant Science. 2017;8:1–16. doi: 10.3389/fpls.2017.01184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Lv F, Han X, Xia X, Yin W. The calcium sensor PeCBL1, interacting with PeCIPK24/25 and PeCIPK26, regulates Na+/K+ homeostasis in Populus euphratica. Plant Cell Reports. 2013;32:611–621. doi: 10.1007/s00299-013-1394-5. [DOI] [PubMed] [Google Scholar]
- 22.Meena MK, Ghawana S, Dwivedi V, Roy A, Chattopadhyay D. Expression of chickpea CIPK25 enhances root growth and tolerance to dehydration and salt stress in transgenic tobacco. Frontiers in Plant Science. 2015;6:1–11. doi: 10.3389/fpls.2015.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luan S. The CBL-CIPK network in plant calcium signaling. Trends in Plant Science. 2008;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Tan T, Cai J, Zhan E, Yang Y, Zhao J, Guo Y, Zhou H. Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant Molecular Biology. 2016;92:391–400. doi: 10.1007/s11103-016-0520-5. [DOI] [PubMed] [Google Scholar]


