Abstract
Background
It is unclear whether the transcriptional subtypes of high grade serous ovarian cancer (HGSOC) apply to high grade clear cell (HGCCOC) or high grade endometrioid ovarian cancer (HGEOC). We aim to delineate transcriptional profiles of HGCCOCs and HGEOCs.
Methods
We used Agilent microarrays to determine gene expression profiles of 276 well annotated ovarian cancers (OCs) including 37 HGCCOCs and 66 HGEOCs. We excluded low grade OCs as these are known to be distinct molecular entities. We applied the prespecified TCGA and CLOVAR gene signatures using consensus non-negative matrix factorization (NMF).
Results
We confirm the presence of four TCGA transcriptional subtypes and their significant prognostic relevance (p<0.001) across all three histological subtypes (HGSOC, HGCCOC and HGEOCs). However, we also demonstrate that 22/37 (59%) HGCCOCs and 30/67 (45%) HGEOCs form 2 additional separate clusters with distinct gene signatures. Importantly, of the HGCCOC and HGEOCs that clustered separately 62% and 65% were early stage (FIGO I /II), respectively. These finding were confirmed using the reduced CLOVAR gene set for classification where most early stage HGCCOCs and HGEOCs formed a distinct cluster of their own. When restricting the analysis to the four TCGA signatures (ssGSEA or NMF with CLOVAR genes) most early stage HGCCOCs and HGEOC were assigned to the differentiated subtype.
Conclusions
Using transcriptional profiling the current study suggests that HGCCOCs and HGEOCs of advanced stage group together with HGSOCs. However, HGCCOCs and HGEOCs of early disease stages may have distinct transcriptional signatures similar to those seen in their low grade counterparts.
Keywords: Ovarian cancer, molecular subtypes, endometrioid, clear cell and high grade serous histologies
Introduction
Microarray-based gene expression studies demonstrate that ovarian cancer (OC) is both a clinically diverse and molecularly heterogeneous disease, comprising subtypes with distinct gene expression patterns that are each associated with statistically significant different clinical outcomes. A gene expression analysis of high-grade serous and endometrioid OCs as part of the Australian Ovarian Cancer Study identified distinct molecular subtypes that have been designated with neutral descriptors (C1, C2, C4, and C5) (1). The four molecular subtypes were validated in 489 high grade serous ovarian cancer (HGSOC) cases using 1,500 intrinsically variable genes for consensus non-negative matrix factorization (NMF) clustering and were termed immunoreactive, differentiated, proliferative and mesenchymal on the basis of gene expression in the clusters (2). These four molecular subtypes have been independently validated and have been shown to be of independent prognostic relevance (3). Using the TCGA ovarian cancer data set, Verhaak et al. recently confirmed the four molecular subtypes of high grade serous ovarian cancer (HGSOC) using a reduced subtype gene expression signature, named “Classification of Ovarian Cancer” (CLOVAR) (4). This reduced CLOVAR gene signature is composed of a 100 genes capable of predicting the ovarian cancer subtypes (4). Validation studies in independent data sets demonstrated that the CLOVAR signature classifies HGSOC with small error rates, making implementation using medium-throughput expression profiling platforms feasible (4).
The main objective of a molecular classification of OC into subtypes with distinct gene expression patterns is to develop robust biomarker signatures that will allow clinicians to identify women likely to benefit from a given therapy. These evolving subgroups are thought to have distinct biologic features that can translate into different therapeutic implications. Epithelial ovarian cancer is a heterogeneous disease consisting of tumors with different histology and grade. The most common OC types are the serous tumors followed by endometrioid and clear-cell cancers which represent 50%–60%, 25% and 4% of all ovarian tumors, respectively (5). Importantly, however, the evolving molecular classification using the four main subtype signatures have almost exclusively been studied and applied to HGSOC (2,3,4). Although some early gene expression studies have included endometrioid and clear cell ovarian cancers (6–10) these studies were limited by their small sample size and the use of early generation microarrays. Nevertheless these studies did suggest that clear cell and endometrioid ovarian cancers may be distinguished from serous ovarian cancers based on their gene expression profiles (6–10). However, many of these early studies included well differentiated tumors (G1) known to be distinct molecular entities (11). To date it is unclear if the evolving signatures which have been used to successfully classify HGSOC into four molecular subtypes could also be used to classify these less common epithelial ovarian cancer histologies. Although clear cell carcinomas and endometrioid carcinomas have been previously shown to be in part driven by pathways distinct from those driving progression of HGSOC we wanted to investigate whether high grade clear cell ovarian cancers (HGCCOCs) or high grade endometrioid ovarian cancers (HGEOCs) may nevertheless in part share gene signatures that have been described in HGSOCs. For instance, we thought it would be important to know if gene signatures characterizing an immunoreactive or mesenchymal subtype can also be found in HGEOCs or HGCCOCs because the evolving molecular signatures are becoming increasingly clinically relevant.
In the present study we, therefore, examined the transcriptional profiles of 276 ovarian cancer cases including 37 HGCCOCs, 66 HGEOCs and 173 previously published HGSOCs using Agilent Whole Human Genome 4×44K Expression Arrays (3). All low grade tumors were excluded from this study as they are known to represent distinct biologic entities (11). We applied the pre-specified TCGA gene expression signatures and the reduced CLOVAR gene signatures to this cohort of 276 well annotated OCs from Mayo Clinic. Moreover, we also performed single sample gene set enrichment analysis (ssGSEA) which calculates separate enrichment scores for each sample and allows the assignment to the nearest TGCA subgroup.
Materials and methods
Patient cohort
Fresh frozen tumors were collected from a series of 276 consecutive women with high grade serous, clear cell and endometrioid ovarian, primary peritoneal or fallopian tube cancer who underwent surgery by a gynecologic oncologist at Mayo Clinic between 1994 and 2005. All patients signed an Institutional Review Board approved consent for bio-banking, clinical data extraction, and molecular analysis. Clinical data were abstracted from medical records and tumor registry. Thirteen patients (7.5%) were included in the TCGA study.
Sample processing and gene expression profiling
Samples were collected during surgery, snap frozen within 30 minutes, and stored at −80°C until RNA extraction. Samples were reviewed by a pathologist specialized in gynecologic oncology (G.K.) and selected to have >70% tumor cell content. RNA was isolated using RNeasy (Qiagen Inc., Valencia, CA) and quantified using a Nanodrop Spectrophotomer (Agilent Technologies, Santa Clara, CA). Gene expression profiles were established using Agilent Whole Human Genome 4×44K Expression Arrays. Total RNA (750ng) with RNA Integrity Number >8.0 was labeled with cyanine 5-CTP or cyanine 3-CTP using the Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies), purified on RNeasy Mini columns (Qiagen Inc.), and hybridized to expression arrays (using a mixed reference containing equal amounts of each of 106 ovarian tumor samples). Slides were scanned using the Agilent 2565BA Scanner and data were exported by the Agilent Feature Extraction Software (version 7.5.1) into Rosetta Resolver (Rosetta Inpharmatics LLC, Cambridge, MA). Log ratios of signal from individual tumor to signal from the reference mix were used for analysis. The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number: GSE73614.
Non negative matrix factorization (NMF) and class prediction
Molecular classification was determined blinded to demographic and clinical information. To identify genes associated with TCGA subtypes, we analyzed expression and molecular subtype data of TCGA cases (https://tcga-data.nci.nih.gov/docs/publications/ov_2011/). Next, we mapped these signature genes to corresponding Agilent probe-set IDs. We selected 1844 probes matching the TCGA signature gene set. Subclasses were computed by reducing the dimensionality of the expression data from thousands of genes to a few metagenes using a consensus NMF clustering method (12). This method computes multiple k-factor factorization decompositions of the expression matrix and evaluates the stability of the solutions using a cophenetic coefficient. The same analysis was repeated using 161 probes representing 100 genes that represented the CLOVAR subtype signature derived by Verhaak and colleagues (4).
Gene set enrichment analysis
Gene set activation scores for each of the subtype expression signatures were generated using single sample Gene Set Enrichment Analysis (ssGSEA) (13) using Bioconductor package GSVA downloadable at http://www.bioconductor.org (14). Raw enrichment scores were expressed as relative z-scores. Subtype assignment of each tumor sample was determined using a z-score cut-off 0.6.
Statistical analysis of molecular subtypes and patient outcome
Subgroup assignments were compared by use of the chi-square test. Overall survival is depicted according to the method of Kaplan and Meier, and the curves were compared with use of the log rank test. All statistical tests were two-sided.
Results
We used Agilent microarrays to determine gene expression profiles of 276 well annotated OCs including 37 HGCCOCs and 66 HGEOCs and 173 HGSOCs. Patient and disease characteristics for the entire cohort and the histologic subtypes are shown in Table 1. Median patient age was 63 years (range 24–89). In the overall cohort of 276 patients 56 (21%) of the patients had early stage (FIGO I or II) disease, while 220 (79%) were diagnosed with advanced stage disease (FIGO III or IV). In the cohort of HGEOC, HGCCOC, and HGSOC 28/66 (33%), 20/37 (54%) and 8/173 (5%) of the patients had early stage disease, respectively. All tissue specimens were centrally reviewed by a pathologist specialized in gynecologic oncology (G.K.) for confirmation of tissue morphology and histologic grade. All samples were of high grade (12% G2 and 88% G3) (Table 1). In the cohort of HGEOC, HGCCOC, and HGSOC 26/66 (39%), 3/37 (8%) and 2/173 (2%) of the patients had grade 2 disease, respectively (Table 1).
Table 1.
Patient and disease characteristics for 276 high grade epithelial ovarian cancer patients.
| HGEOC | HGCCOC | HGSOC | All | |
|---|---|---|---|---|
| N = 66 | N = 37 | N = 173 | N = 276 | |
| Age at diagnosis | ||||
| Median | 58 | 64 | 64 | 63 |
| Range | 38–86 | 41–88 | 24–89 | 24–89 |
| Grade | ||||
| 2 | 26 (39%) | 3 (8%) | 4 (2%) | 33 (12%) |
| 3 | 40 (61%) | 34 (92%) | 169 (98%) | 143 (88%) |
| FIGO Stage | ||||
| 1 | 19 (29%) | 13 (35%) | 0 | 32 (12%) |
| 2 | 9 (14%) | 7 (19%) | 8 (5%) | 24 (9%) |
| 3 | 31 (47%) | 15 (41%) | 124 (72%) | 170 (61%) |
| 4 | 7 (10%) | 2 (5%) | 41 (23%) | 50 (18%) |
| Debulking Status | ||||
| Optimal | 58 (88%) | 26 (70%) | 122 (70%) | 206 (75%) |
| Suboptimal | 5 (7%) | 0 | 48 (28%) | 53 (19%) |
| Unknown | 3 (5%) | 11 (30%) | 3 (2%) | 17 (6%) |
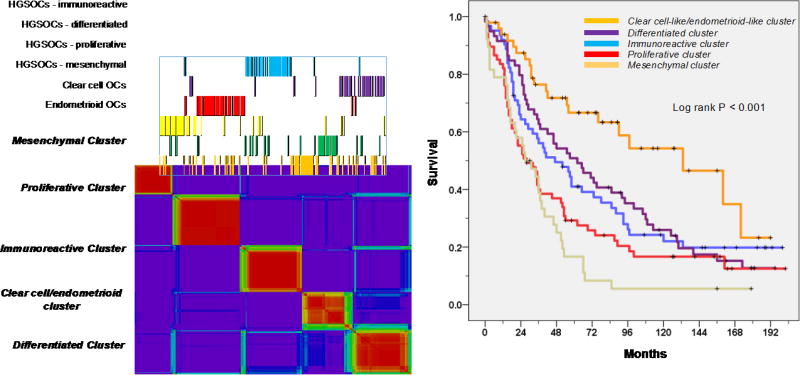
A gene set representing the pre-specified TCGA signatures was used to classify 276 arrays representing HGSOC (n=173) and HGCCOC (n=37) and HGEOC (n=66). We confirmed the presence of the immunoreactive, differentiated, proliferative and mesenchymal transcriptional subtypes and their prognostic relevance (p<0.001) across all three histological subtypes (HGSOC, HGCCOC and HGEOCs) (Figure 1). However, we also demonstrated that 22/37 (59%) HGCCOCs and 30/67 (45%) HGEOCs formed two additional separate clusters with distinct gene signatures (Supplemental Figures 1 and 2). Importantly, of the HGCCOC and HGEOCs that clustered separately 62% and 65% were of early FIGO stage (FIGO I /II), respectively (Table 2). Analysis of grade distribution across the transcriptional subtypes also demonstrated that of the HGCCOC and HGEOCs that clustered separately 21% and 50% were grade 2 tumors, respectively (Table 2).
Figure 1.
Unsupervised clustering (non-negative matrix factorization) using 1500 TCGA genes depicting six clusters; overall survival according to transcriptional subtypes.
Table 2.
Cross tabulation assignment of high grade clear cell ovarian cancers and endometrioid ovarian cancers to the molecular subtypes (TCGA 6 clusters; CLOVAR 5 and 4 clusters) according to their FIGO stage (FIGO I/I versus III/IV) and grade (2 versus 3).
| FIGO Stage | Grade | |||||||
|---|---|---|---|---|---|---|---|---|
| I/II | III/IV | 2 | 3 | |||||
| N | 103 | 48 | 55 | P value |
29 | 74 | P value |
|
| TCGA 6* | Immunoreactive | 12 (100%) | 4 (33%) | 8 (67%) | 0.001 | 2 (17%) | 10 (83%) | 0.019 |
| Differentiated | 15 (100%) | 1 (7%) | 14 (93%) | 2 (13%) | 13 (87%) | |||
| Proliferative | 12 (100%) | 3 (25%) | 9 (75%) | 3 (25%) | 9 (75%) | |||
| Mesenchymal | 6 (100%) | 3 (50%) | 3 (50%) | 0 | 6 (100%) | |||
| Clear cell-like | 24 (100%) | 15 (62%) | 9 (38%) | 5 (21%) | 19 (79%) | |||
| Endometrioid-like | 34 (100%) | 22 (65%) | 12 (35%) | 17 (50%) | 17 (50%) | |||
| CLOVAR 5# | Immunoreactive | 15 (100%) | 3 (20%) | 12 (80%) | 0.001 | 1 (7%) | 14 (93%) | 0.022 |
| Differentiated | 19 (100%) | 6 (32%) | 13 (68%) | 4 (21%) | 15 (79%) | |||
| Proliferative | 15 (100%) | 4 (27%) | 11 (73%) | 4 (27%) | 11(73%) | |||
| Mesenchymal | 7 (100%) | 2 (29%) | 5 (71%) | 0 | 7 (100%) | |||
| Clear cell-like/Endometrioid-like | 47 (100%) | 33 (70%) | 14 (30%) | 20 (43%) | 27 (57%) | |||
| CLOVAR 4# | Immunoreactive | 18 (100%) | 5 (28%) | 13 (72%) | 0.037 | 1 (6%) | 17 (94%) | 0.005 |
| Differentiated | 56 (100%) | 33 (59%) | 23 (41%) | 23 (41%) | 33 (59%) | |||
| Proliferative | 20 (100%) | 8 (40%) | 12 (60%) | 5 (25%) | 15 (75%) | |||
| Mesenchymal | 9 (100%) | 2 (22%) | 7 (78%) | 0 | 9 (100%) | |||
Assignment using unsupervised clustering with the 1500 most differentially expressed genes described by the TCGA.
Assignment using unsupervised clustering with the reduced gene set CLOVAR genes described by Verhaak et al.
Survival was significantly better for patients with tumors that demonstrated an endometrioid-like or a clear cell-like signature when compared to those patients with tumors that displayed one of the established TCGA signatures (Figure 1). However, the good median survival in the endometrioid-like and clear cell-like cluster of 133 and 160 months, respectively, was clearly driven by early stage disease. When performing a survival analysis separately for stage III/IV patients the median survival of the endometrioid-like and clear cell-like cluster was reduced from 133 to 90 months and from 160 to 23 months, respectively. The survival analysis that was performed separately for stage III/IV patients still showed improved outcome for the endometrioid-like cluster, however, the survival for the clear cell-like cluster was comparable with the remaining poor outcome groups of HGSOC when limiting the analysis to stage III/IV patients. These results underscore the notion that improved survival of the endometrioid-like or clear cell-like cluster was due to the increased rate of stage I/II diagnosis.
These finding were confirmed using the reduced CLOVAR gene set for classification into 5 subgroups where most early stage HGCCOCs and HGEOCs formed a distinct combined cluster of their own (Figure 2). Using the 100 CLOVAR genes we confirm the presence of the immunoreactive, differentiated, proliferative and mesenchymal transcriptional subtypes and their prognostic relevance across all three histological subtypes (HGSOC, HGCCOC and HGEOCs) (Figure 2, p<0.001). In addition, 47/103 (46%) of HGCCOCs and HGEOCs form an additional fifth separate cluster. Importantly, of these HGCCOC and HGEOCs that clustered separately 70% were of early stage (FIGO I/II), and 43% were grade 2, respectively (Table 2). Survival was significantly better for patients with tumors that express the endometrioid-like/clear cell-like signature when compared to those patients with tumors that express one of the TCGA signatures but when the analysis was restricted to stage III/IV patients it again became apparent that this was due to the increased rate of stage I/II diagnosis.
Figure 2.
Unsupervised clustering (non-negative matrix factorization) using the 100 CLOVAR genes depicting five clusters; overall survival according to transcriptional subtypes.
Furthermore, using the 100 CLOVAR genes described by Verhaak and colleagues for consensus NMF we were able to demonstrate stable clustering of all cases into four transcriptional subtypes (Figure 3 and Supplementary Figure 1). Consensus matrices and sample correlation matrices are shown for k = 2 to k = 8 and depict stable clustering into four groups (Cophrenic coef. 0.994) (Supplementary Figure 1). Overall survival analysis confirmed the prognostic significance of these four TCGA transcriptional subtypes (Figure 3, p=<0.001). Patients whose tumors expressed the differentiated signature had the best overall survival. This appears to contrast with earlier reports where patients whose tumors expressed the immunoreactive signature demonstrated the best overall survival (Verhaak 2013, Konecny 2014). However, this may be explained by the fact that in the present study 56/103 HGCCOC and HGEOC cases were assigned to the differentiated subtype when using the four group assignment which is expected to positively impact outcome of the differentiated subgroup (Table 2).
Figure 3.
Unsupervised clustering (non-negative matrix factorization) using the 100 CLOVAR genes depicting four clusters; overall survival according to transcriptional subtypes.
Single-sample Gene Set Enrichment Analysis (ssGSEA) calculates separate enrichment scores for each pairing of a sample and gene set. Each ssGSEA enrichment score represents the degree to which the genes in a particular gene set are coordinately up- or down-regulated within a sample and it allows the assignment of an individual sample to the nearest or closest transcriptional subtype. In the present analysis we used the CLOVAR gene set for ssGSEA. Consistent with the stable clustering into four groups using NMF classification, ssGSEA allowed the assignment of each HGCCOC or HGEOC to one of the four known TCGA subgroups which showed a high concordance with the consensus NMF clustering into four groups (p<0.001, data not shown). Similar to the results obtained by consensus NMF, ssGSEA assigned 47/103 HGCCOC and HGEOC cases to the differentiated subtype.
Discussion
We tested the hypothesis whether gene expression based molecular subtyping of epithelial ovarian cancer developed for high grade serous histology could be applied to high grade endometrioid and clear cell histologies. Using a gene set representing the1500 genes described by the TCGA that defined the immunoreactive, differentiated, proliferative and mesenchymal subtypes, consensus NMF revealed six distinct expression subtypes. In addition to the four TCGA subtypes we describe an endometrioid-like and a clear cell-like expression subtype when using the 1500 most variable genes described by the TCGA (2). When using the smaller CLOVAR gene set for consensus NMF it is possible that more subtle difference between HGCCOC and HGEOC, which may require a bigger number of genes to be detected, may not be captured by the reduced gene list. Nevertheless, the CLOVAR signature did group many HGEOCs and HGCCOCs into one new distinct group that was enriched for cases with early stage disease. Using transcriptional profiling the current study suggests that many HGCCOCs and HGEOCs of advanced stage group together with HGSOCs. However, HGCCOCs and HGEOCs of early disease stages may have distinct transcriptional signatures more similar to those seen in their low grade counterparts. Finding from previous studies support this hypothesis (10,15–17). Wu and colleagues identified two distinctive subgroups of endometrioid carcinoma, based on variance in their global gene expression patterns (15). One of these subgroups was highly similar to serous carcinoma and tended to be of higher grade. Genetic annotation also revealed that p53 mutations were common among those endometrioid carcinomas with a serous-like gene expression profile (15). Moreover, deregulated β-catenin signaling and defects in the PI3K–PTEN pathway were shown to be typical among those endometrioid carcinomas that did not share gene expression homology to serous carcinoma (15). In a further analysis, WT1 gene expression was associated with those endometrioid ovarian cancers with a serous like expression profile (16). Further studies will be necessary to understand whether these molecular differences can also be identified in the current cohort of HGEOCs. These molecular features may ultimately help us to better delineate serous from endometrioid histologies as histopathological diagnosis of high-grade endometrioid and serous cancer of the ovary is poorly reproducible under the current morphology based classification system, especially for anaplastic, high grade tumors.
Previous studies suggest that clear cell cancer of the ovary is a unique ovarian cancer subtype (18). Recent work has shown that a subset of clear cell cancers evolve from endometriosis (19). Molecular alterations within clear cell cancers include unique cytokine expression patterns and c-met amplification as well as mutations in ARID1A and the PI3K/PTEN pathway (20–22). Clear cell-specific clinical trials based upon these biologic discoveries have been designed and are presently active (23). Furthermore, a recent study suggests that clear cell ovarian cancers have distinct molecular characteristics especially with regards to IL6-STAT3-HIF signaling and possible susceptibility to angiogenesis inhibition when compared to high grade serous and endometrioid histologies (10). In our data set, early stage clear cell cancer of high grade had a distinct gene expression profile, confirming earlier reports on the distinct features of clear cell ovarian cancer. However, we were also able to show that many HGCCOCs of advanced stage grouped together with HGSOCs. This may in part be explained by the fact that the TCGA expression signatures may reflect disease processes that are more critical for advanced ovarian cancer than for early disease stages. While it is evident that clear cell carcinomas are in part driven by pathways distinct from those driving progression of HGSOC, they may, nevertheless, in part share gene signatures that have been described in HGSOCs. As the evolving molecular signtures in HGSOC are becoming increasingly clinically relevant it is important to understand whether therapeutically relevant gene signatures like the immunoreactive or mesenchymal signature can also be found in HGCCOCs.
set Consensus NMF and ssGSEA using the reduced CLOVAR gene set both likewise demonstrated stable clustering of all OC cases into four subgroups. Importantly we were able to show that HGCCOC and HGEOC cases of early stage and G2 tended to fall into the differentiated expression subgroup and that HGCCOCs and HGEOCs of advanced stage more likely group together with the three remaining HGSOCs transcriptional subtypes. Importantly, however, before any of the proposed classifications can be converted to clinical use, further clinical validation of their predictive importance will be key in moving either of them forward. Associations between subtype signatures and treatment responses will help to clarify which signatures can be deemed to be the most appropriate to help classify OC. Only these studies will allow us to ultimately determine which subtype signatures are most appropriate to select patients for any given therapy. Possibly these evolving ovarian cancer transcriptional signatures will be used as predictive signatures similar to how gene expression based classifiers are currently being used to guide treatment decisions in breast cancer (24). Results from a recent retrospective study support this notion. Using NMF to assign the gene expression profiles of 359 formalin fixed paraffin embedded tumor samples from patients treated in a phase III frontline ovarian cancer trial, which assessed the efficacy of bevacizumab, into the four molecular subtypes demonstrated a preferential treatment effect of bevacizumab in the proliferative and mesenchymal subtypes (25). These preliminary data underscore the potential clinical utility of transcriptional signatures across all advanced ovarian cancer types including HGCCOCs and HGEOCs. Our study, however, is not without limitations as neither additional genomic nor immunohistochemical analyses have been performed to further delineate the biologic underpinnings of the distinct transcriptional signatures seen between advanced and early stage HGCCOCs and HGEOCs. Nevertheless, a strength of our study is the fact that we were able to profile a relatively large number of rare high grade epithelial ovarian cancers which allowed us to demonstrate the existence of the immunoreactive, differentiated, proliferative and mesenchymal transcriptional subtypes across advanced ovarian cancer cases irrespective of their specific histology. Importantly, however, rare high grade epithelial ovarian cancers of early disease stages may have distinct transcriptional signatures similar to those seen in their low grade counterparts. These findings could aid in the implementation of medium-throughput expression profiling platforms to guide treatment decisions in advanced epithelial ovarian cancer and may support their use across different histologies.
Supplementary Material
Highlights.
TCGA gene signatures are present in high grade ovarian cancers of rare histology.
Early stage high grade ovarian cancers of rare histology cluster separately from advanced stages.
TCGA gene signatures may help to stratify patients with rare high grade ovarian cancer.
Acknowledgments
This work supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Thelma L. Culverson Endowed Cancer Research Fund, and the Stranahan Foundation for Translational Cancer Research and Advanced Clinical Cancer Research. Funding was also provided by the Fred C. and Katherine B. Andersen Foundation, the US National Institute of Health (P50 163393, R01 CA122443) and Mayo Clinic Ovarian Cancer SPORE (P50 CA136393).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that there are no conflicts of interest.
References
- 1.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–1. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014;106(10) doi: 10.1093/jnci/dju249. pii: dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–25. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei W, Dizon D, Vathipadiekal V, Birrer MJ. Ovarian cancer: genomic analysis. Ann Oncol. 2013;24(Suppl 10):7–15. doi: 10.1093/annonc/mdt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14(11):4376–86. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62(16):4722–9. [PubMed] [Google Scholar]
- 8.Zorn KK, Bonome T, Gangi L, Chandramouli GV, Awtrey CS, Gardner GJ, et al. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin Cancer Res. 2005;11:6422–30. doi: 10.1158/1078-0432.CCR-05-0508. [DOI] [PubMed] [Google Scholar]
- 9.Marquez RT, Baggerly KA, Patterson AP, Liu J, Broaddus R, Frumovitz M. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin Cancer Res. 2005;11(17):6116–26. doi: 10.1158/1078-0432.CCR-04-2509. [DOI] [PubMed] [Google Scholar]
- 10.Anglesio MS, George J, Kulbe H, Friedlander M, Rischin D, Lemech C, et al. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res. 2011;17(8):2538–48. doi: 10.1158/1078-0432.CCR-10-3314. [DOI] [PubMed] [Google Scholar]
- 11.Oswald AJ, Gourley C. Low-grade epithelial ovarian cancer: a number of distinct clinical entities? Curr Opin Oncol. 2015;27(5):412–9. doi: 10.1097/CCO.0000000000000216. [DOI] [PubMed] [Google Scholar]
- 12.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, Akyol A, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/β-catenin and PI3K/Pten signaling pathways. Cancer Cell. 2007;11(4):321–333. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Madore J, Ren F, Filali-Mouhim A, Sanchez L, Köbel M, Tonin PN, et al. Characterization of the molecular differences between ovarian endometrioid carcinoma and ovarian serous carcinoma. J Pathol. 2010;220(3):392–400. doi: 10.1002/path.2659. [DOI] [PubMed] [Google Scholar]
- 17.Pamuła-Piłat J, Rubel T, Rzepecka IK, Olbryt M, Herok R, Dansonka-Mieszkowska A. Gene expression profiles in three histologic types, clear-cell, endometrioid and serous ovarian carcinomas. J Biol Regul Homeost Agents. 2014;28(4):659–74. [PubMed] [Google Scholar]
- 18.Del Carmen MG, Birrer M, Schorge JO. Clear cell carcinoma of the ovary: a review of the literature. Gynecol Oncol. 2012;126:481–490. doi: 10.1016/j.ygyno.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Pearce CL, Templeman C, Rossing MA, Lee A, Near AM, Webb PM, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case–control studies. Lancet Oncol. 2012;13(4):385–94. doi: 10.1016/S1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanaihara N, Anglesio MS, Ochiai K, Hirata Y, Saito M, Nagata C, et al. Cytokine gene expression signature in ovarian clear cell carcinoma. Int J Oncol. 2012;41:1094–1100. doi: 10.3892/ijo.2012.1533. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita Y, Akatsuka S, Shinjo K, Yatabe Y, Kobayashi H, Seko H, et al. Met is the most frequently amplified gene in endometriosis-associated ovarian clear cell adenocarcinoma and correlates with worsened prognosis. PLoS ONE. 2013;8:e57724. doi: 10.1371/journal.pone.0057724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones S, Wang TL, Shih IeM, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penson RT1, Dizon DS, Birrer MJ. Clear cell cancer of the ovary. Curr Opin Oncol. 2013;25(5):553–7. doi: 10.1097/CCO.0b013e328363e0c7. [DOI] [PubMed] [Google Scholar]
- 24.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2000. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 25.Winterhoff B, Kommoss S, Konecny G, Wang C, Riska S, Oberg A, et al. Bevacizumab may Differentially Improve Survival for Patients with the Proliferative and Mesenchymal Molecular Subtype of Ovarian Cancer. J Clin Oncol. 2014;32 doi: 10.1158/1078-0432.CCR-16-2196. (suppl; abstr 5509) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.