Summary
During neuronal development, local mRNA translation is required for axon guidance and synaptogenesis, and dysregulation of this process contributes to multiple neurodevelopmental and cognitive disorders. However, regulation of local protein synthesis in developing axons remains poorly understood. Here, we uncover a novel role for the actin-regulatory protein Mena in the formation of a ribonucleoprotein complex that involves the RNA-binding proteins HnrnpK, and PCBP1, and regulates local translation of specific mRNAs in developing axons. We find that translation of dyrk1a, a Down Syndrome- and Autism Spectrum Disorders- related gene, is dependent on Mena, both in steady state conditions as well as upon BDNF stimulation. We identify hundreds of additional mRNAs that associate with the Mena complex, suggesting it plays broader role(s) in post-transcriptional gene regulation. Our work establishes a dual role for Mena in neurons, providing a potential link between regulation of actin dynamics and local translation.
Keywords: Mena, ENAH, Ena/VASP, developing axon, growth cone, axon guidance, local translation, ribonucleoprotein
In Brief
Vidaki et al, provide evidence that Mena, an actin regulator, forms a ribonucleoprotein complex in neurons, that involves translational repressors HnrnpK and PCBP1, and is required for local translation of dyrk1a and potentially other associated mRNAs in developing axons.
INTRODUCTION
During development, the exquisitely regulated process of axon guidance establishes the circuitry necessary for a properly functioning nervous system (NS) in the adult. Aberrant axonal navigation results in defective connectivity and multiple neurodevelopmental disorders including, among others, epilepsy, intellectual disabilities, autism and schizophrenia (McCandless, 2012; Sahin and Sur, 2015; Wegiel et al., 2010). The growth cone, a specialized structure at the tip of growing axons, must continuously sample the microenvironment for guidance cues and integrate this information rapidly into appropriate motility responses, frequently without sufficient time for transcriptional responses. Indeed, axons severed from their cell bodies can navigate correctly in vivo, and respond to guidance cues in vitro (Batista and Hengst, 2016; Campbell et al., 2001; Verma et al., 2005). Local mRNA translation is a key mechanism in such autonomous responses, and protein synthesis inhibitors block the ability of severed axons to respond to several guidance cues (Batista and Hengst, 2016; Jung et al., 2012). However, most of our understanding of regulated local protein synthesis is based on the characterization of individual mRNAs found in axons (Deglincerti and Jaffrey, 2012; Kim and Jung, 2015), with few details of the underlying molecular mechanism. Even in synapses, where local translation has been studied intensely, only a few proteins have been identified as key regulators of local mRNA translation (Bassell and Warren, 2008; Brown et al., 2001; Darnell et al., 2011; De Rubeis and Bagni, 2010; Deglincerti and Jaffrey, 2012; Fritzsche et al., 2013; Hutten et al., 2014; Kindler et al., 2012).
Mena (ENAH), a member of the Ena/VASP protein family, is highly expressed in the developing and adult NS, and is a known regulator of actin dynamics, integrin-mediated signaling, adhesion and cell motility (Bear and Gertler, 2009; Drees and Gertler, 2008; Gupton and Gertler, 2010; Gupton et al., 2012). Mena and its paralogs, VASP and EVL, are required for normal NS development during neurulation (Lanier et al., 1999; Menzies et al., 2004), neuritogenesis, migration (Dent et al., 2007; Kwiatkowski et al., 2007), axon guidance responses to both attractive and repulsive signals (Lebrand et al., 2004; Dent et al., 2011; Mcconnell et al., 2016), terminal axon branching (Dwivedy et al., 2007), dendritic morphology and synapse formation (Li et al., 2005; Lin et al., 2007). Of the three Ena/VASP proteins, Mena is the most abundant in the NS, and Mena-null animals exhibit clear defects in NS development, while VASP/ EVL double mutants exhibit no obvious NS phenotypes in animals with a wild type Mena allele (Kwiatkowski et al., 2007). Further, while Ena/VASP family proteins share a highly-conserved domain structure, Mena contains additional domains and alternatively-included sequences not found in VASP or EVL (Gertler and Condeelis, 2011).
Here we identify a ribonucleoprotein (RNP) complex containing Mena, known translation regulators and specific cytosolic mRNAs, including dyrk1a. Dyrk1a, a dual specificity kinase with multiple roles in neuronal development, has been implicated in the pathology and etiology of Down Syndrome, autism, intellectual disabilities, Alzheimer’s and Parkinson’s disease (Coutadeur et al., 2015; Di Vona et al., 2015; Krumm et al., 2014; O’Roak et al., 2012; Qian et al., 2013; Tejedor and Hämmerle, 2011; van Bon et al., 2015). We demonstrate a critical role for Mena in Dyrk1a protein synthesis, both under steady-state conditions and after BDNF stimulation.
RESULTS
Novel interactions of Mena with multiple RNA binding proteins in the developing brain
To explore the mechanisms underlying Mena function beyond its established roles in actin polymerization (Bear and Gertler, 2009) and in integrin-mediated signaling (Dent et al., 2007; Gupton et al., 2012; Gupton and Gertler, 2010) in the developing NS, we sought to identify its interactome. We performed mass spectrometry after immunoprecipitation (IP) of Mena from lysates of E15.5 mouse brains from Mena-wild type and Mena-deficient animals. Along with known Mena binding partners, including PFN2 and EVL (Giesemann et al., 2003; Riquelme et al., 2015), we identified additional Mena-associated proteins (Table S1). Surprisingly, multiple RNA-binding proteins (RBPs), including translation factors and mRNA transport proteins, were identified in the Mena IP (Figure S1A). To verify the specificity of these interactions, we performed co-IP experiments from E15.5 mouse brains and found that several of these RBPs co-IPed specifically with Mena, while FMR1, an RBP not identified by our proteomic screen did not (Figure 1A; multiple bands appearing on the Mena WB correspond to different Mena protein isoforms (Gertler et al., 1996; Lanier et al., 1999)). Several of the Mena-RBP interactions were also detected in N2A neuroblastoma cells and in mouse embryonic fibroblasts (MEFs) (Figure S1B and S1C). Interestingly, RNase treatment of brain lysates did not affect the recovery of RBPs in Mena co-IPs (data not shown), indicating that these Mena:RBP interactions do not depend on the presence of RNA.
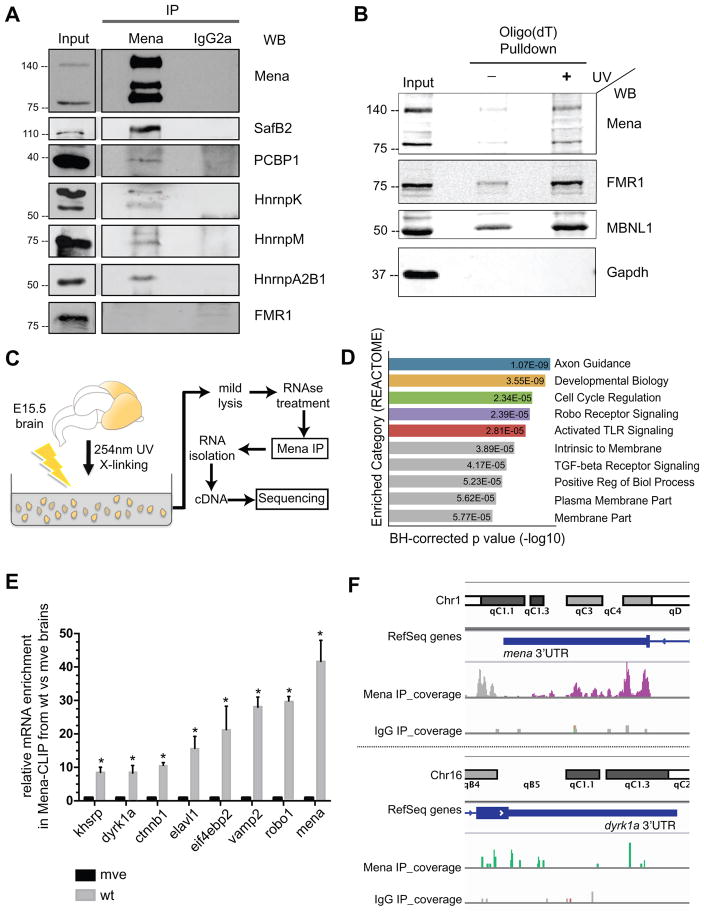
Figure 1. Mena interacts with RNA binding proteins and cytosolic mRNAs.
A. Panels show western blots, probed with antibodies to the indicated proteins, of Mena and IgG2a isotype control IPs and of 5% input lysate. B. Proteins enriched in Oligo(dT) pulldowns, analyzed by western blot probed with antibodies to Mena and to positive control RBPs, FMR1 and MBNL1, as indicated (Input is 5% of total lysate). C. Schematic representation of the modified HITS-CLIP protocol. E15.5 mouse brain tissues were triturated and UV-crosslinked to preserve RNP-complexes, and then homogenized in mild lysis buffer to generate lysates for Mena-IP. Co-IPed RNA was subsequently isolated and sequenced. D. Gene-Set-Enrichment-Analysis (REACTOME) of the mRNAs identified through Mena HITS-CLIP. E. qPCR validation of several mRNAs that specifically associated with Mena. Mena null brains were used as experimental controls. The graph represents relative mRNA enrichment of Mena-Associated mRNAs between the wt and mve samples ± StDEV (Student’s T test p*<0.05). F. Peaks in the 3’UTRs of mena and dyrk1a.
Mena associates with cytosolic mRNAs in vivo
Given that Mena associated with multiple RBPs in developing brain lysates, we wondered whether Mena-containing complexes were also associated with mRNA. To test this hypothesis, we used Oligo(dT) pulldown assays to capture polyadenylated mRNAs and mRNA-associated proteins from lysates prepared from brain tissue that was UV-crosslinked to preserve RNA-protein complexes (Figure S1D and Figure 1B). Western blot analysis of the captured proteins indicated that Mena was associated with mRNA (Figure 1B). To test whether this association is neuronal specific, we performed additional Oligo(dT) pulldown assays and found that in MEFs Mena is also in complex with cytosolic mRNAs (Figure S1E).
Identification of mRNAs associated with Mena in the brain
Mena can bind directly to a number of proteins, including ligands for its EVH1 (Ena/VASP Homology-1) domain, actin through its EVH2 domain and α5 integrin through its LERER domain, but lacks any known RNA binding sites (Bear and Gertler, 2009; Gertler et al., 1996; Gupton et al., 2012), raising the possibility that it associates with mRNA indirectly via one or more RBPs. To investigate this possibility, and to identify mRNAs in complex with Mena in neurons, we performed IP after crosslinking (CLIP) assays. UV-crosslinking was used to preserve RNA-protein complexes in E15.5 mouse brain tissues, followed by lysate preparation and Mena IP. The stringent lysis conditions typically used for CLIP diminished recovery of RNA associated with Mena by co-IP (data not shown), suggesting that the association of Mena with mRNA may be indirect. A modified CLIP protocol with mild lysis and IP conditions improved mRNA recovery in the Mena IP, allowing us to sequence the associated mRNA (Figure 1C: modified High-throughput RNA Sequencing after CLIP; HITS-CLIP, see Star Methods) (Darnell, 2010; Licatalosi et al., 2008). Binding peaks were identified by the presence of multiple sequence reads in the sample that exhibited more than 10 reads and that were at least 3-fold enriched in the Mena vs. control CLIP samples (Table S2). The majority of the peaks were distributed within exons (48%) or gene regions (47.8%), while 4.2% of peaks mapped to the 5’ or 3’ UTRs of mRNAs (Figure S1F and Table S2).
To identify potential biological processes controlled by the Mena-associated mRNAs, Gene Set Enrichment Analysis was performed using the Broad Institute platform (http://software.broadinstitute.org/gsea/index.jsp) (Figure 1D and Table S2). Several of the most enriched gene sets represented processes that involve Mena function (e.g. Axon Guidance, Robo signaling, etc) (Figure 1D). To confirm the specific association of selected mRNAs within each category with Mena, we performed quantitative RT-PCR after Mena CLIP assays from control and Mena-null E15.5 brains (referred to as mve throughout the text) (Figure 1E). Interestingly, some of the Mena-complex associated mRNAs encode proteins that have been functionally linked to Mena (e.g. Vamp2 (Gupton and Gertler, 2010), Robo1 (Bashaw et al., 2000; McConnell et al., 2016; Yu et al., 2002), Ctnnb1 (Najafov et al., 2012)), while others represent processes not previously associated with Mena (e.g. Khsrp, Elavl1, Eif4ebp2, Dyrk1a, etc). Two of the most prevalent mRNAs identified by sequencing were those of dyrk1a and mena itself, both demonstrating multiple sequencing peaks in their gene region and in their 3’UTR (Figure 1F).
Together, these data indicate that Mena can indirectly associate with specific mRNAs via its interacting RBPs, in a novel ribonucleoprotein (RNP) complex in the developing NS.
Mena associates with the mRNA of dyrk1a in neurons
The dyrk1a mRNA encodes a dual specificity kinase that has multiple functions in the NS (Barallobre et al., 2014; Hammerle et al., 2003; Hämmerle et al., 2008; Tejedor and Hämmerle, 2011). Dyrk1a inhibitors are being tested in Alzheimer’s disease treatment (Coutadeur et al., 2015; Janel et al., 2014), whereas in models of Parkinson’s disease Dyrk1a acts as a dopaminergic neuron survival factor (Barallobre et al., 2014). Moreover, Dyrk1a has been implicated in cases of autism and intellectual disability (Krumm et al., 2014; O’Roak et al., 2012; van Bon et al., 2015), and a Dyrk1a dosage-dependent role has been correlated with Down Syndrome etiology and pathology (Hammerle et al., 2003; Hämmerle et al., 2008; Tejedor and Hämmerle, 2011). Our HITS-CLIP data indicated that the Mena complex associated with the 3’UTR of dyrk1a mRNA (Figure 1F). As RBP binding to 3’UTRs can regulate cytosolic mRNA localization and translation (Szostak and Gebauer, 2013), we hypothesized that Mena might be important for dyrk1a mRNA dynamics in neurons.
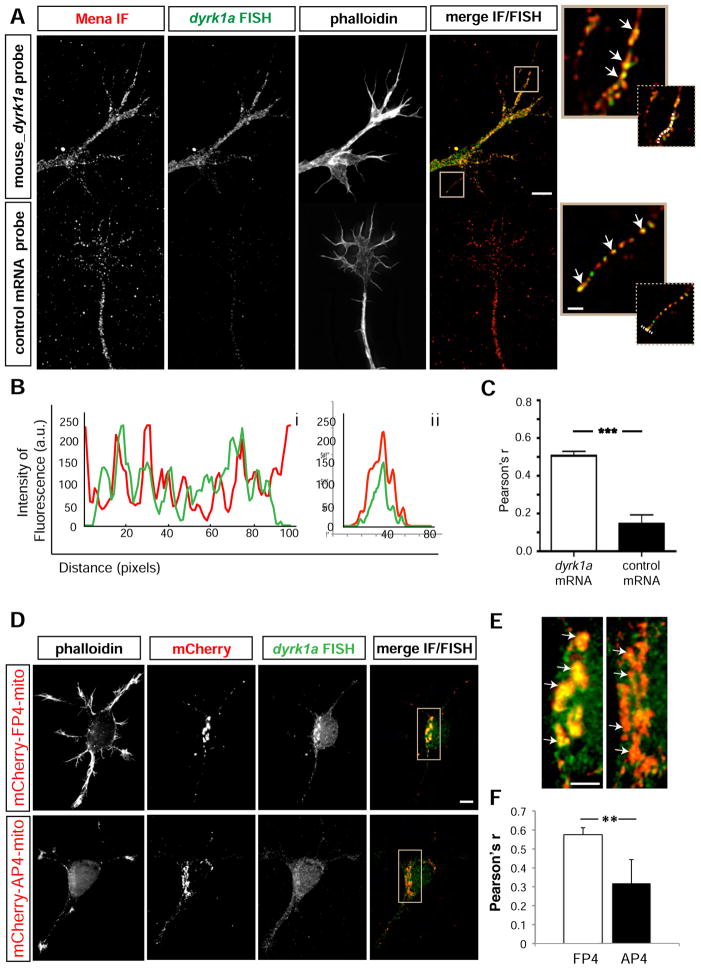
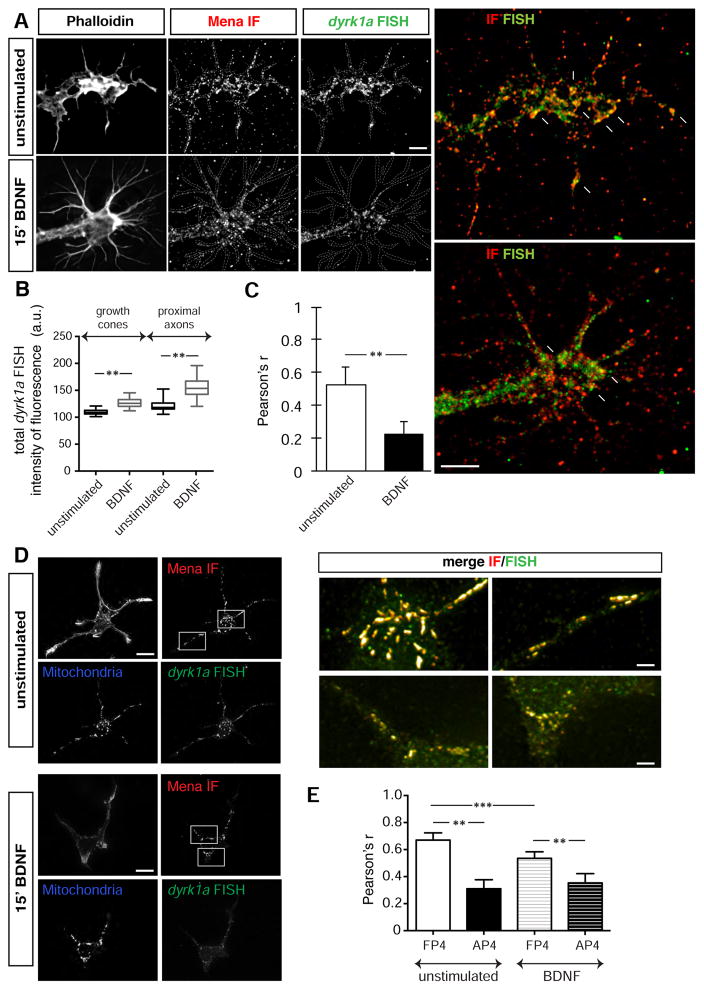
To detect dyrk1a mRNA and determine if it co-localizes with the Mena protein, we performed immunofluorescence (IF) for Mena along with Fluorescent In Situ Hybridization (FISH) for dyrk1a mRNA on cultured cortical neurons (Figure 2A: a’–d’), and observed extensive co-localization of the fluorescent signals along the axons and growth cones of neurons (Figure 2A: i & ii white arrows). Line scans and correlation-coefficient analyses (Bolte and Cordelieres, 2006) indicated that the distributions of Mena protein and dyrk1a mRNA were significantly correlated across growth cone filopodia (Figure 2Bi & ii and 2C). In contrast, IF for Mena and Dyrk1a proteins did not reveal significant overlap of the fluorescent signals (Figure S2A).
Figure 2. dyrk1a mRNA co-localizes with Mena in neuronal growth cones and axons A. Combined.
IF for Mena (a) and FISH of dyrk1a mRNA (b) on E15.5 + 2DIV cultured mouse cortical neurons revealed significant overlap of the two signals in axons and growth cones (d). In contrast, Mena (a’) and a control FISH probe (species-specific for human dyrk1a mRNA) (b’), fail to co-localize (d’). Ai, Aii. Higher magnification of filopodia showing co-localization between Mena and dyrk1a mRNA (white arrows). Phalloidin staining for F-actin (c,c’) was used to visualize morphology. B. Fluoresence intensity plots of protein and mRNA signals in scans along (i) or across (ii) stained filopodia (indicative dashed white lines depicted in Ai and Aii inserts). C. Pearson’s correlation coefficient for the protein and mRNA signals over the entire growth cone. Co-localization between Mena and dyrk1a mRNA is significantly higher than co-localization between Mena and a control mRNA probe (Student’s T test p***<0.001). The graph represents mean Pearson’s r ± StDEV. Scale bar for Aa-d and Aa’-d’: 5μm, Ai-ii: 1μm. D. An FP4-mito construct expressed in neurons (a-d) co-recruits the dyrk1a mRNA to the mitochondrial surface, in contrast to the control AP4-mito (a’-d’). Mena IF (b and b’), dyrk1a FISH (c & c’), F-actin staining and a merge of Mena IF+dyrk1a FISH (d,d’) are shown. E. Magnification of boxed inserts i and ii from D showing dyrk1a mRNA distribution with respect to the mitochondrial surface of FP4- and AP4-transfected neurons (white arrows in i and ii respectively). F. Pearson’s correlation coefficient for the mRNA and mitochondrial signal was assessed to verify the significant difference between AP4- and FP4-mito (Student’s T test p**<0.01). The graph represents mean Pearson’s r ± StDEV. Scale bar for Da-d and Da’-d’: 20 μm, Ei-ii: 5μm.
dyrk1a mRNA can be co-recruited to the mitochondrial surface along with Mena in a re-localization assay
To test whether Mena can affect the cytoplasmic localization of dyrk1a mRNA, we used a well-established mitochondrial sequestration assay (Bear et al., 2000) in which the expression of a construct with the high-affinity EVH1 domain-binding motif DFPPPPXDE fused to a mitochondrial targeting sequence (“FP4-mito”) re-localizes endogenous Ena/VASP proteins to the mitochondrial surface (Figure S2B). Expression of a control construct in which the EVH1-binding moiety is mutated to reduce affinity for EVH1 domains (“AP4-mito”) has only minimal effects on Ena/VASP re-localization. Using this assay, the vast majority of Mena (and its paralogs) is re-localized to the mitochondria, and in some cases, robustly interacting proteins are co-recruited (Gupton et al., 2012). We thus reasoned that any mRNA associated with a Mena-containing RNP complex might also be co-recruited with Mena to the mitochondria by FP4-mito. Using nucleofection, we expressed the FP4- and AP4-mito constructs in primary neurons from E15.5 mouse brains. 48hrs after nucleofection and plating, we performed FISH to detect the mRNA of dyrk1a in FP4- and AP4-mito-expressing neurons. Expression of the constructs did not affect the total levels of dyrk1a mRNA (Figure S2C). Interestingly, we found that a significant amount of dyrk1a mRNA was co-recruited with Mena to the mitochondrial surface by FP4-mito (Figure 2D: a–d & 2Ei), whereas dyrk1a mRNA localization was unaffected in AP4-expressing control neurons (Figure 2D: a’-d’ and 2Eii). The correlation between Mena and dyrk1a mRNA was significantly elevated on the mitochondrial surface of FP4- vs AP4-mito transfected neurons (Figure 2F). In contrast to the mRNA, IF analysis indicated that localization of the Dyrk1a protein was unaffected by FP4-Mito expression (Figure S2D).
Taken together, our data demonstrate that Mena and dyrk1a mRNA interact specifically and co-localize within neuronal axons and growth cones, and that this interaction is sufficient to relocalize the dyrk1a mRNA to mitochondria in an Ena/VASP-dependent manner.
Mena is necessary and sufficient to re-localize dyrk1a mRNA to the mitochondria
To test whether other members of the Ena/VASP family can also be found in complex with mRNAs, we performed VASP CLIP assays. Using RT-PCR we failed to detect Mena-associated mRNAs in complex with VASP (Figure S2E). We also failed to detect VASP associated with cytosolic mRNAs after Oligo(dT) Pulldown assays (data not shown), suggesting that the ability to associate with RNP complexes may be Mena-specific rather than a general property of Ena/VASP proteins. To test this hypothesis, we introduced FP4-mito into neurons isolated from Mena+/−;VASP+/+;EVL+/+ and from Mena−/−;VASP+/+;EVL+/+ E15.5 brains and analyzed the resulting effects on dyrk1a mRNA distribution. In the absence of Mena, both VASP (Figure S2F) and EVL (not shown) were recruited to the mitochondria by FP4-mito, as expected, but the mRNA of dyrk1a was not (S2G). Therefore Mena, unlike the other members of the Ena/VASP family, is necessary and sufficient to re-localize dyrk1a mRNA to the mitochondria, upon FP4-mito expression. As these data indicate that the ability to associate with dyrk1a and other mRNAs was Mena specific, we focused exclusively on characterizing the Mena-mRNA association in this study.
Candidate RBPs mediating the interaction between Mena and mRNAs
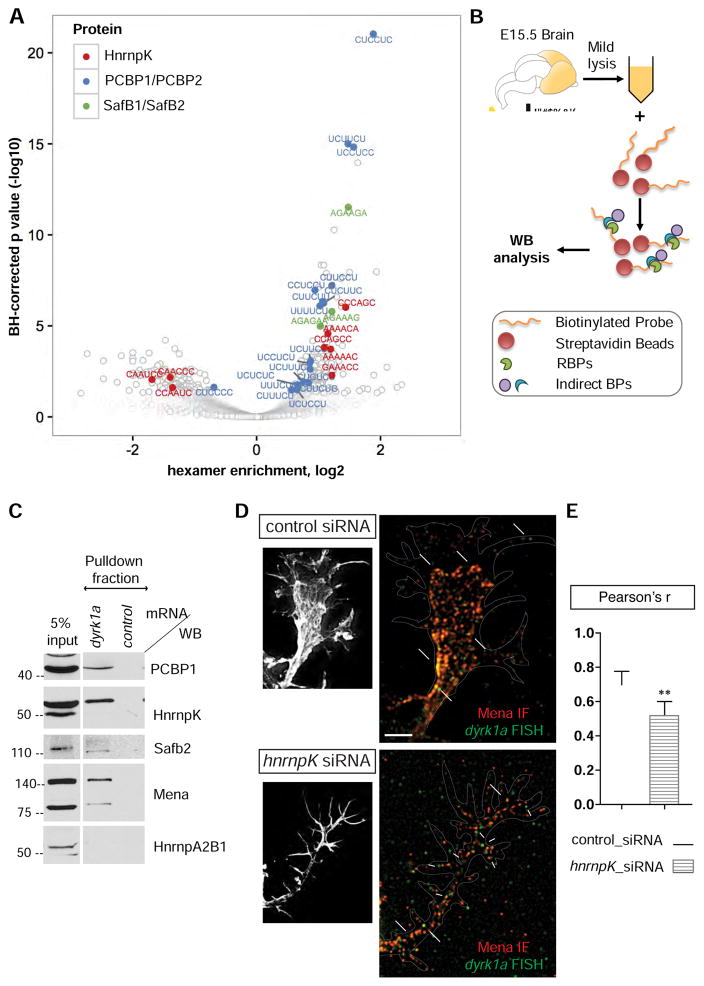
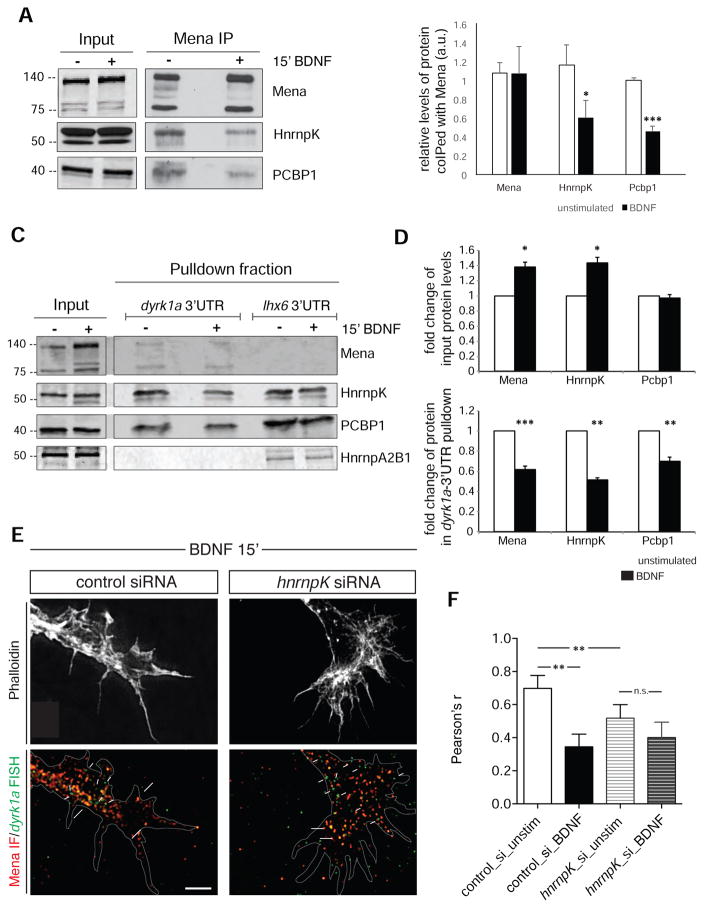
To understand the biological significance of the association between Mena and specific mRNAs, we investigated how RBPs mediate this indirect interaction. We used a custom script to perform an unbiased analysis to identify sequences enriched within the 3’UTRs of the Mena-associated mRNAs that could serve as potential RBP binding motifs. For simplicity, we searched only for hexamer sequences, as they have been previously identified as highly efficient kmer motifs with minimal contextual binding effects (i.e. secondary structure formation, etc) (Lambert et al., 2014). Interestingly, most of the enriched hexamers identified corresponded to binding motifs of RBPs that interact with Mena according to both our mass spectrometry data and co-IP validation experiments (Figure 3A). More specifically, the 3’UTR sequences derived from our Mena-HITS-CLIP data were enriched significantly for binding motifs of HnrnpK, PCBP1 (HnrnpE1), and Safb2, all of which were verified as interactors of Mena in brain lysates (Figure 1A & S1A).
Figure 3. RBPs mediating the interaction between Mena and dyrk1a 3’UTR.
A. Volcano plot of enriched hexamers within the Mena-associated 3’UTR sequences. Hexamers with a density higher in the Mena-HITS-CLIP compared to the control, have enrichments >1 (positive log values), whereas hexamers with densities lower in the Mena_HITS-CLIP than in the control, have enrichments <1 (negative log values). Some of the top hits correspond to RBPs associated with Mena, including HnrnpK, PCBP1 and Safb2. B. Schematic representation of the RNP-pulldown assay with the 3’UTR of dyrk1a mRNA as bait. C. Western blot analysis of the pulldown fraction revealed that Mena, Safb2, HnrnpK and PCBP1 can bind the 3’UTR of dyrk1a mRNA, unlike HnrnpA2B1, which was used as a negative control RBP. An RNA probe generated by in vitro transcription of λ-phage was used as a negative control bait. D. siRNA-mediated depletion of HnrnpK in neurons reduces signal overlap between Mena IF and dyrk1a FISH (large white arrows in ii), as opposed to control siRNAs (large white arrows in i). Smaller arrows in ii point to mRNA signal that does not overlap with Mena. E. Pearson’s coefficient correlation for the FISH and IF signal was assessed to verify the significant difference between neurons with control- and hnrnpK-siRNAs (Student’s T test p**<0.01). The graph represents mean Pearson’s r ± StDEV.
To test whether HnrnpK, SafB2 and PCBP1 could potentially mediate the indirect association of Mena with cytosolic mRNAs, we investigated whether they bind to the 3’UTR of the dyrk1a mRNA. We analyzed the sequences corresponding to the 3’UTR of dyrk1a in our Mena-HITS-CLIP data, using the RBPmap platform (http://rbpmap.technion.ac.il) (Akerman et al., 2009; Paz et al., 2010) and identified putative binding sites for HnrnpK, PCBP1 and Safb2, among others (Figure S3A and Table S3). To confirm that PCBP1, HnrnpK, Safb2 and Mena could associate with the 3’UTR of dyrk1a mRNA, we performed mRNA pulldown assays, using an in vitro transcribed biotinylated probe corresponding to the 3’UTR of dyrk1a mRNA, to capture interacting protein complexes from E15.5 brain lysates (Figure 3B). Western blot analysis of the captured fraction, revealed that all three candidate RBPs, as well as Mena, were enriched in the bound fraction associated with the 3’UTR of dyrk1a, but not with a control biotinylated RNA probe, consistent with our previous in silico predictions (Figure 3C). Further analysis using simultaneous dyrk1a FISH and IF for Mena and HnrnpK, revealed co-localization of the three fluorescent signals in the growth cones of primary neurons (Figure S3B), consistent with the existence of a Mena-RNP complex.
Taken together, our data indicated that the Mena-interacting RBPs HnrnpK, PCBP1, and Safb2 could mediate indirect association of Mena with dyrk1a, since all three can bind to the dyrk1a 3’UTR. We then asked if any of the candidate RBPs was required for the association of Mena with dyrk1a mRNA in neurons. To address this question, we sought to analyze the effects of depleting the RBPs on the extent of overlap between the Mena IF and dyrk1a FISH signals. We introduced siRNA pools for each RBP into primary neurons by co-nucleofection with a GFP plasmid to identify transfected neurons. As the nucleofection efficiency of siRNAs into primary neurons is low, and the resulting protein depletion is limited by the non-proliferative phenotype of primary neurons, we assessed the efficacy of depletion by each siRNA pool using IF for the proteins. Using this approach, we were able to generate convincing knockdown of HnrnpK, but not of either PCBP1 or Safb2 (Figure S3C and data not shown). We then asked whether co-localization between Mena and dyrk1a was sensitive to reduction in HnrnpK levels, and found that HnrnpK depletion significantly reduced the signal overlap between Mena IF and dyrk1a FISH (Figure 3D large white arrows), with more dyrk1a puncta lacking co-localized Mena signal (Figure 3D small white arrows). This result is consistent with our hypothesis that HnrnpK is involved in mediating the interaction between Mena protein and dyrk1a mRNA.
dyrk1a mRNA is locally translated in axons upon stimulation with BDNF
Based on the known roles of HnrnpK and PCBP1 in regulating cyotosolic mRNA localization and translation (Thiele et al., 2016; Torvund-jensen et al., 2014), we hypothesized that Mena-RNP complexes could facilitate transportation and/or local translation of the associated mRNAs, in axons and growth cones. To test this hypothesis, we stimulated neurons in culture with Brain Derived Neurotrophic Factor (BDNF) to elicit local translation (Jung et al., 2012; Santos et al., 2010; Schratt et al., 2004), and analyzed Dyrk1a protein abundance. We also tested the protein levels of Mena as our sequencing data indicated the mena mRNA associated with the protein. IF of primary cortical neurons showed that both Mena and Dyrk1a fluorescence intensity levels significantly increased in the growth cones and axons of BDNF stimulated vs. unstimulated cells (Figure S4A). This effect was blocked by addition of the translation inhibitor anisomycin, indicating that the increase in the Dyrk1a and Mena IF signal resulted from BDNF-elicited protein synthesis (Figure S4A).
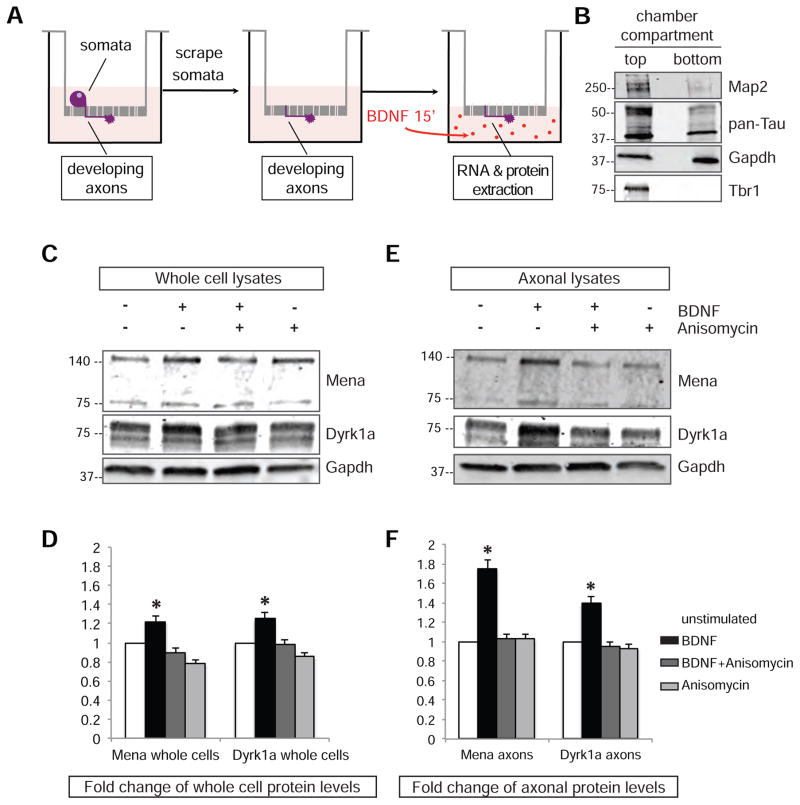
The BDNF-induced increase in axonal Mena and Dyrk1a proteins could arise from a global effect on their synthesis followed by protein trafficking into axons and growth cones, or, potentially, from local translation of axon-localized mRNAs. To investigate this possibility, we cultured cortical neurons on the top compartment of transwell chambers separated by filters with 1μm membrane pores (Figure 4A) that allow neuronal processes, but not neuronal cell bodies, to extend onto the bottom of the filter, and permit their physical fractionation from the soma. 36–48hrs after plating primary neurons on top of the filter, material harvested from the top and bottom compartments of the chamber was isolated and used to prepare lysates. Western blot analysis of lysates from the top and bottom compartment with known axon (pan-Tau), dendrite (Map2) and nuclear (Tbr1) markers, 36–48hrs after plating, verified that this assay successfully separates somata from neuronal processes (Figure 4B: absence of Tbr1 at the bottom compartment), and that the neuronal processes isolated from the bottom were primarily axons as opposed to dendrites (Figure 4B: enrichment of Tau and barely detectable levels of Map2), as anticipated based on the short time-window of the cultures. To assess protein synthesis using this system, neurons were allowed to grow for 36hours followed by starvation for 4hrs to minimize transcriptional and translational activity. Following starvation, the cell bodies were gently scraped off and removed from the top compartment of the filters, while the retained severed axons were stimulated with BDNF for 15’ along with controls in which the entire unscraped filter was stimulated, to measure local and global protein synthesis, respectively. To ensure we monitored de novo translational events, control experiments with anisomycin were performed. Western blot analysis revealed a significant increase in Mena and Dyrk1a protein levels upon BDNF stimulation compared to untreated and anisomycin-treated neurons, both globally, in whole cells, and even more evidently, locally in isolated axons that were stimulated (Figure 4C–D and 4E–F respectively). We also observed BDNF-elicited local translation of additional Mena-associated mRNAs using this assay (Figure S4B).
Figure 4. BDNF stimulation can induce local translation of Mena and Dyrk1a in axons.
A. Schematic representation of the assay for local translation. B. Western blot analysis of the top and bottom filter compartments C. Protein levels of Mena and Dyrk1a increase after BDNF stimulation in whole cell lysates. D. Quantification of Mena and Dyrk1a proteins in whole cells demonstrated elevated protein levels upon BDNF stimulation, but not when translation was blocked by anisomycin. The graph represents Mean ± StDEV (Two-Way Anova p<0.05). E. BDNF stimulation of axons only elicits a greater increase in the protein levels of both Mena and Dyrk1a in axonal lysates. F. Quantification of the proteins in isolated axonal preparations reveals significant changes upon BDNF stimulation. All values were normalized to loading controls (Gapdh) and then to the unstimulated protein levels to generate fold changes. The graph represents Mean ± StDEV (Two-Way Anova p*<0.05).
BDNF stimulation decreases the association between Mena and dyrk1a mRNA
Given that BDNF can induce translation of dyrk1a mRNA in developing neurons, we investigated whether and how the stimulation affects the association of Mena with dyrk1a. First, we asked whether the extent of Mena protein co-localization with dyrk1a mRNA in growth cones was altered upon stimulation. We performed IF for Mena and FISH for dyrk1a in cortical neurons with and without BDNF stimulation (Figure 5A). Notably, we observed an increase in the FISH signal of dyrk1a mRNA after BDNF stimulation, both in the growth cones and their proximal axon part (Figure 5B), indicating that increases in dyrk1a transcription, mRNA transport, or both occur upon BDNF stimulation. Interestingly, the overlap between the Mena IF and dyrk1a FISH signals was significantly decreased by BDNF treatment, raising the possibility that Mena:dyrk1a-containing complexes dissociate upon stimulation (Figure 5C). To test this hypothesis, we used the mitochondria re-localization assay described above and found that the amount of dyrk1a co-recruited with Mena to mitochondria in FP4-mito expressing neurons was reduced significantly by BDNF treatment (Figure 5D) compared to unstimulated neurons (Figure 5E).
Figure 5. BDNF stimulation reduces the association between Mena and the mRNA of dyrk1a A.
IF for Mena and FISH for dyrk1a before and after BDNF stimulation of cortical neurons in culture (b, c and b’, c’ respectively). Co-localization of the signal was reduced after the stimulation (white arrows in magnified panels d and d’). Scale bar: 5μm. B. Stimulation of neurons with BDNF resulted in a significant increase of total dyrk1a mRNA levels, both in the growth cones and in the proximal axon part (Student’s T test p**<0.01). The graph represents Mean ± StDEV. C. Pearson’s coefficient correlation for the FISH and IF signal was assessed before and after BDNF treatment, revealing significant decrease in co-localized of FISH and IF signal after stimulation (Student’s T test p<0.01). The graph represents Mean ± StDEV. D. Neurons expressing the FP4-mito construct were processed for Mena IF and dyrk1a FISH, before (a-f) and after BDNF stimulation (a’–f’). Scale bar a–d and a’–d’: 20μm; e-f and e’–f’: 5μm. E. Pearson’s coefficient correlation for the FISH and IF signal revealed significantly decreased mRNA signal co-recruited on the mitochondrial surface after BDNF stimulation (Student’s T test p**<0.01; p***<0.001). The graph represents Mean ± StDEV.
Overall, our results demonstrate that while BDNF stimulation increases total dyrk1a mRNA levels and local translation in axons, it reduces the association of Mena and dyrk1a.
BDNF stimulation results in partial dissociation of the Mena;dyrk1a RNP complexes
We next investigated the effects of BDNF stimulation on Mena:RNP complexes. We found that the levels of HnrnpK and PCBP1 recovered with Mena by coIP were significantly reduced in lysates of BDNF vs. unstimulated cultured primary neurons (Figure 6A and B). Taken together, these results suggest that BDNF stimulation induces Mena-RBP complex dissociation, which could lead to dissociation dyrk1a mRNA from with Mena. To test this hypothesis, we performed pulldowns using biotinylated dyrk1a 3’UTR from the lysates of neurons with or without BDNF stimulation. An irrelevant 3’UTR from lhx6, an mRNA not detected in the Mena-CLIPseq data (Table S2) was used as a negative control for the binding assay. Consistent with our previous findings, we observed that BDNF induced a significant increase in the protein levels of Mena and HnrnpK (Figure 6C input and 6D and Figure S4B), however, the amounts of Mena, HnrnpK and Pcbp1 pulled-down with the 3’UTR of dyrk1a mRNA were significantly decreased by BDNF stimulation (Figure 6C pulldown fraction and 6D). Therefore, the exogenous 3’UTR dyrk1a probe associated with the Mena-RBP complex more efficiently in the unstimulated condition than it did after stimulation. Together, these data indicate that, in addition to eliciting translation of dyrk1a, BDNF stimulation triggers dissociation of Mena from its interacting RBPs and from the dyrk1a mRNA.
Figure 6. The Mena-RNP complex is partially disassembled upon BDNF stimulation.
A. Western blot analysis of protein coIP after Mena-IP on unstimulated and BDNF-stimulated neurons in culture. Inputs and precipitated fractions are of different exposure times. B. Significantly reduced amounts of HnrnpK and PCBP1 coIP with Mena after 15’ of BDNF stimulation comparpared to unstimulated cells. Each precipitated protein value was normalized to its respective input and to the amount of precipitated Mena. The graph represents Mean ± StDEV (Student’s T test p*<0.05; p***<0.001). C. Western blot of biotinylated mRNA pulldown assays, before and after BDNF stimulation of neurons in culture (E15.5+2DIV). D. Quantification of the protein levels in the inputs used for the assay and in the mRNA pulldown fractions with and without BDNF stimulation. Input protein levels were normalized to the unstimulated lysate levels and the pulldown proteins were normalized to the respective input values. The graphs represent Mean ± StDEV (Student’s T test p*<0.05; p**<0.01; p***<0.001). E. IF for Mena and FISH for dyrk1a after BDNF stimulation of neurons that are HnrnpK-depleted. Overlap between the two signals is indicated by large white arrows in b and b’, whereas FISH signal not overlapping with Mena is shown by small white arrows. Scale bar: 5 μm. F. Pearson’s correlation coefficient for the FISH and IF signal revealed significantly decreased co-localization, both under steady-state conditions and after BDNF stimulation in the HnrnpK-depleted background (Student’s T test p**<0.01). The graph represents Mean ± StDEV.
Based on our results, we hypothesized that HnrnpK, which contributes to the association between Mena and dyrk1a, would be required to detect the BDNF-elicited reduction in their co-localization. We performed FISH for dyrk1a and IF for Mena in HnrnpK-depleted neurons (Figure 6E) and found that, as expected, the overlap between the fluorescence was significantly reduced compared to controls in the absence of HnrnpK in unstimulated cells (Figures 3D and 6F). Interestingly, HnrnpK-depleted neurons failed to exhibit any further significant reduction of Mena and dyrk1a co-localization after BDNF stimulation (Figure 6F)
Taken together, this set of results indicates that the association of dyrk1a mRNA with Mena depends, at least in part, on the presence of HnrnpK in unstimulated cells, suggesting that BDNF-elicited decreases in Mena:HnrnpK association could contribute to Mena dissociation from dyrk1a-containing RNPs.
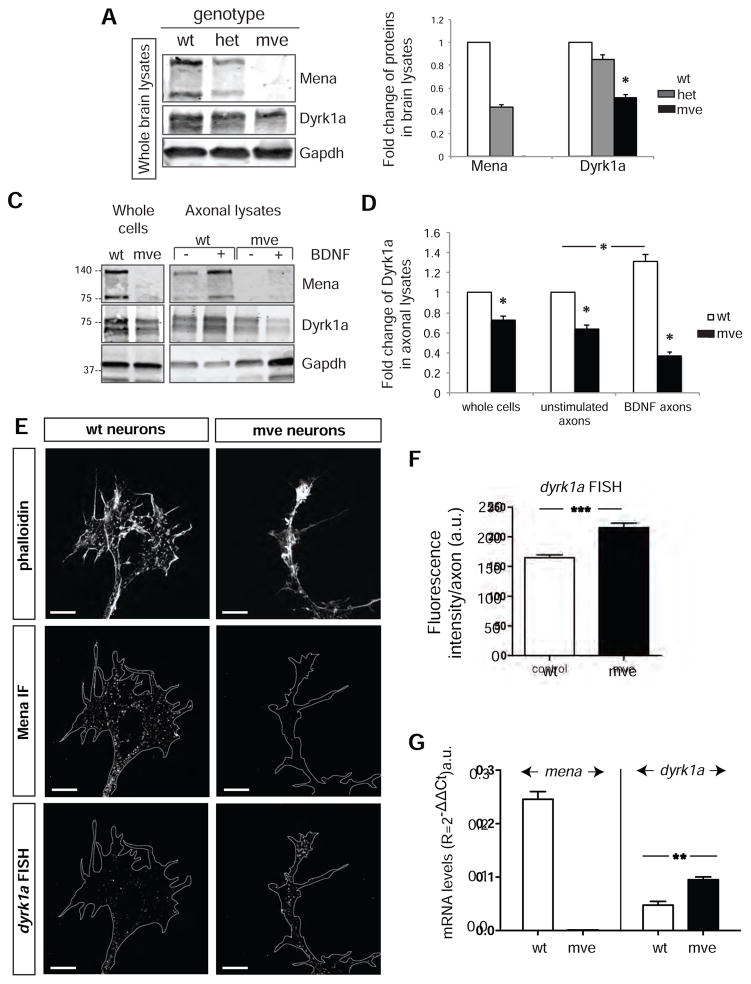
The absence of Mena disrupts Dyrk1a translation, but does not affect axonal targeting of the dyrk1a mRNA
Thus far, our data reveal an association between Mena and dyrk1a, through the formation of Mena-RNP complexes, and a potential role for those complexes in dyrk1a mRNA translation. We next investigated the requirement for Mena in dyrk1a localization and translation using material from Mena-deficient animals. Western blot analysis of E15.5 whole brain lysates revealed a significant decrease in Dyrk1a protein levels in Mena-deficient brains compared to control (Figure 7A and 7B).
Figure 7. The absence of Mena does not affect localization of dyrk1a mRNA, but significantly reduces both steady-state and BDNF-elicted increases in Dyrk1a protein levels.
A. Western blots of whole brain lysates of different Mena genotypes (wt: Mena+/+;VASP−/−;EVL−/−, het: Mena+/−;VASP−/−;EVL−/−, mve: Mena−/_;VASP−/−;EVL−/−). B. Quantification of A. Protein levels are normalized to the wt protein amount. The graph represents Mean ± StDEV (Student’s T test p<0.05). C. Western blot analysis of lysates from axotomy assays showing Dyrk1a in mve vs wt axons before and after BDNF stimulation. D. Dyrk1a protein levels were significantly decreased in mve axons and were not changed by BDNF stimulation. Values were normalized to the wt protein levels using the GAPDH loading controls. The graph represents Mean ± StDEV (Two-Way Anova p*<0.05). E. FISH for dyrk1a mRNA on cultured cortical neurons (E15.5+2DIV) from wt and mve brains. Scale bar: 5μm. F. Quantification of the fluorescence intensity in axons and growth cones of wt and mve neurons. The graph represents Mean ± StDEV (Student’s T test p***<0.001). G. Quantitative RT-PCR analysis of mRNA from wt and mve neurons. The graph represents Mean ± StDEV (Student’s T test p**<0.01).
To test whether local translation of the dyrk1a mRNA requires Mena-containing complexes, we examined the effect of BDNF stimulation on axons isolated from wt and mve primary cortical neurons. We observed that, compared to control, mve axons had significantly lower Dyrk1a protein levels under steady state conditions, and that Dyrk1a levels failed to increase upon BDNF stimulation (Figure 7C and 7D).
To verify that the Dyrk1a protein level reduction observed in mve neurons arose from defective translation rather than abnormal mRNA transport, we performed FISH for dyrk1a on mve neurons. While dyrk1a mRNA was normally targeted to axons and growth cones in both samples, dyrk1a mRNA levels were significantly increased in mve neurons, compared to control cells (Figure 7E and 7F). This could be explained by compensatory transport from the soma, and/or mRNA unmasking in the absence of Mena. Signal increases arising from mRNA unmasking can occur upon neuronal activation as mRNAs are released from protein complexes during translation or decay (Buxbaum et al., 2014). Pepsin treatment on wt neurons in culture as previously described (Buxbaum et al., 2014), followed by FISH, revealed a significant increase in the dyrk1a signal between untreated and pepsin-treated cells (Figure S5), indicating that part of dyrk1a RNA is masked by proteins in the absence of stimulation. To analyze the abundance dyrk1a mRNA in mve neurons in a protein complex-independent manner, we isolated total RNA and performed quantitative RT-PCR analysis for dyrk1a and observed significantly higher dyrk1a mRNA levels in mve vs. control neurons (Figure 7G). The increased abundance of dyrk1a mRNA in the absence of Mena likely arises as a consequence of elevated dyrk1a transcription, increased mRNA stability, or both, potentially triggered by impaired translation of dyrk1a mRNA.
DISCUSSION
We found an unanticipated role for Mena, but not for its paralogs VASP and EVL, as a key regulator of dyrk1a mRNA translation in neurons, and have identified a set of Mena-associated mRNAs that may be similarly regulated. Using BDNF to elicit protein synthesis (Santos et al., 2010; Schratt et al., 2004), we demonstrated that the mRNAs encoding Mena and Dyrk1a can be locally translated in axons severed form their cell bodies, and that this de novo protein synthesis is Mena-dependent. Our findings raise the intriguing possibility that Mena could act as a regulatory node that coordinates and balances actin polymerization and local protein synthesis in response to specific cues during neuronal development and, potentially, in adult neuroplasticity.
Interestingly, similar dual roles have been reported for CYFIPs, which can function either as a regulator of Arp2/3-mediated actin nucleation through the WAVE-complex, or as a local translation inhibitor in synaptic spines, via direct binding to the FMR1 RBP (De Rubeis et al., 2013), and for APC, which regulates microtubule dynamics, mRNA enrichment in filopodia (Mili et al., 2008), and axonal localization and translation of β2B-tubulin mRNA (Preitner et al., 2014). Notably, the mRNA set we have identified associated with Mena, is significantly different from mRNAs already known to be locally translated and associated with well-described RBPs, including FMR1, APC, Staufen, and Barentsz (Ascano et al., 2012; Balasanyan and Arnold, 2014; Brown et al., 2001; Fritzsche et al., 2013; Preitner et al., 2014), minus few exceptions (e.g. β-catenin (Baleriola and Hengst, 2014; Deglincerti and Jaffrey, 2012), suggesting that the Mena-containing complexes represent a novel RNP complex involved in localized mRNA translation in axons.
Mena associates indirectly with dyrk1a and other cytosolic mRNAs in an RNP containing the RBPs HnrnpK, PCBP1 and Safb2. Binding motifs for these three RBPs were enriched significantly in Mena-complex mRNAs, and they were all detected in pulldown assays with the dyrk1a 3’UTR. HnrnpK plays a critical role in linking Mena to mRNAs as HnrnpK depletion significantly reduced association between Mena and dyrk1a mRNA. Exactly how the Mena-RNP complex forms and connects Mena to specific mRNAs will require further investigation, though it is noteworthy that HnrnpK and Safb2 both contain LP4 motifs, which can mediate direct binding to the EVH1 domain of Ena/VASP proteins (Niebuhr et al., 1997), and that Safb2 (Townson et al., 2003) also contains a region of similarity to the LERER domain in Mena.
Two of the RBPs we found associated with Mena, HnrnpK and PCBP1, have varied roles in RNA metabolism, including regulation of mRNA translation (Gebauer and Hentze, 2004; Ostareck-lederer et al., 2002; Thiele et al., 2016; Torvund-jensen et al., 2014). Interestingly, HnrnpK and PCBP1 can form complexes that inhibit translation initiation when bound to the 3’UTRs of target mRNAs (Gebauer and Hentze, 2004). But how can Mena be associated with an mRNA and positively regulate its translation, when present in a complex that silences dyrk1a translation? Our results are consistent with the possibility that dyrk1a is translationally silenced by the HnrnpK and PCBP1 moieties in the Mena-RNP complex, and that de-repression of dyrk1a translation requires Mena. In Mena-deficient neurons, steady state levels of Dyrk1a protein are reduced, and BDNF stimulation fails to induce dyrk1a translation. Our data showing that BDNF stimulation disrupts Mena’s association with HnrnpK and PCBP1, as well as the recovery of Mena, HnrnpK and PCBP1 in pulldowns using the 3’UTR of dyrk1a, supports a speculative model in which dissociation of the Mena-RNP complex releases dyrk1a mRNA from its translationally-inhibited state.
The Mena-RNP complex is significantly enriched for many mRNAs encoding proteins involved in NS development and function, including dyrk1a. Dyrk1a is a dosage-sensitive, dual-specificity protein kinase that fulfills key roles during development and in tissue homeostasis, and its dysregulation results in multiple human pathologies (Chen et al., 2013; Hammerle et al., 2003; O’Roak et al., 2012; Qian et al., 2013; Tejedor and Hämmerle, 2011). Human Dyrk1a maps to chromosome 21, and it is overexpressed in Down syndrome (DS) individuals and DS mouse models. This alteration has been correlated with a wide range of the pathological phenotypes associated to DS, such as motor alterations, retinal abnormalities, osteoporotic bone phenotype, craniofacial dysmorphology, or increased risk of childhood leukemia (Arron et al., 2006; Kim et al., 2016; Malinge et al., 2012; Ortiz-Abalia et al., 2008; van Bon et al., 2015). In addition, a few cases of truncating mutations in one dyrk1a allele have been described in patients with general growth retardation and severe primary microcephaly (Van Bon et al., 2011), highlighting the extreme dosage sensitivity of this gene. Moreover, and as an indication of the pleiotropic activities of Dyrk1a, dysregulation of this kinase has also been linked to tumor growth and to pancreatic dysfunction (Fernandez-Martínez et al., 2015; Rachdi et al., 2014).
Like most of the mRNAs identified in this study, dyrk1a contains multiple binding sites for the Mena-complex in its 3’UTR, consistent with our data demonstrating that the Mena-RNP complex regulates local synthesis of Dyrk1a protein. Given the extreme dosage sensitivity of Dyrk1a and its implication in numerous neurodevelopmental disorders, our finding that Dyrk1a protein levels are regulated in a Mena-dependent manner in axons raises the intriguing possibility that dysregulation of the Mena-RNP complex may contribute to such disorders. Additional mRNAs that are associated with Mena, like the validated targets β-catenin and elavl1 (HuR) are also implicated in multiple developmental processes and pathophysiological conditions (Alami et al., 2014; Blanco et al., 2016; Holland et al., 2013; Krumm et al., 2014; Li et al., 2017; Lu et al., 2014; O’Roak et al., 2012; Wang et al., 2016). Therefore, the Mena-RNP complex may represent a target for the development of novel therapeutic strategies for multiple disease pathologies.
Interestingly, the Mena-RNP complex contains mena mRNA, which harbors multiple binding sites for the complex in its 3’UTR, raising the possibility that Mena regulates translation of its own mRNA. Mena-regulated translation of β-catenin could also affect mena mRNA abundance since β-catenin can regulate mena transcription (Najafov et al., 2012). These findings are consistent with the potential existence of regulatory feedback loops that control Mena protein abundance at the transcriptional and translational levels.
Neurons deficient for Ena/VASP proteins fail to respond properly to Netrin and Slit (Dent et al., 2011; Lebrand et al., 2004; Mcconnell et al., 2016), two axon guidance cues that require local translation (Campbell et al., 2001; Jung et al., 2012; Jung and Holt, 2011). Our results raise the possibility that, in addition to its established role in regulating filopodia dynamics in response to Netrin and Slit, Mena could contribute to local translation-dependent responses to these cues. Interestingly, both Mena and HnrnpK have been implicated in synapse formation and plasticity (Folci et al., 2014; Giesemann et al., 2003; Li et al., 2005; Lin et al., 2007; Proepper et al., 2011), raising the possibility that their synaptic functions involve regulated translation by the Mena-RNP complex. Future work will address how Mena function helps coordinate changes in cytoskeletal dynamics and protein synthesis in response to signals that regulate nervous system development and function.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and reagent requests may be directed to and will be fulfilled by the Lead Contact, Frank Gertler (fgertler@mit.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All experiments were performed according to the Guide for the Care and Use of Laboratory Animals and were approved by the National Institutes of Health, and the Committee on Animal Care at the Massachusetts Institute of Technology (Cambridge, MA, USA). Female pregnant mice were euthanized with CO2 and embryos were isolated and further dissected in 10mM Hepes and 1x HBSS (GIBCO/Invitrogen). Mice of the following strains were used: Swiss Webster, mixed background Mena+/+;VASP−/−;EVL−/−, Mena+/−;VASP−/−;EVL−/−, Mena−/−;VASP−/−;EVL−/− (mve), Mena+/+;VASP+/+;EVL+/+, and Mena−/−;VASP+/+;EVL+/+. The embryos were not assessed for their sex.
Primary neuron cultures
Cortical neurons from E15.5 mouse brains were plated on poly-D-lysine (PDL, SIGMA) or PDL and Laminin (Southern Biotech) and cultured for 2 days before treatments, unless otherwise indicated. Briefly, cortical tissue was dissected in 10mM Hepes and 1x HBSS, washed and trypsinized in the same buffer for 15min at 37°C. Tissues were then washed in DMEM 1x with 10% FBS to inactivate trypsin, and triturated in the same medium. Following trituration neurons were pelleted at 600g for 5 min, resuspended in serum-free Neurobasal medium (Invitrogen), supplemented with B27 (Gibco) and Pen/Strep (Gibco), and plated on PDL-coated coverslips or petri dishes.
Cell lines
MEFs and N2A cells were cultured at 37°C, 5% CO2, in Dulbecco’s modified Eagles’s medium (DMEM) supplemented with 10% fetal bovine serum and penicili/streptomycin (Pen/Strep).
METHOD DETAILS
Primary neuron stimulation with BDNF
Before stimulations with BDNF (50ng/ml; R&D systems) neurons were starved for 4 hrs in L15- Leibowitz medium (Invitrogen) and to block translation cells were incubated with 40μM Anisomycin (Sigma) in L15, for 30min prior to BDNF addition. BDNF was added for 15min. Where needed, neurons were transfected using Amaxa Nucleofector mouse neuron kit (LONZA) according to the manufacturer’s instructions. All experiments were repeated at least 3 times to eliminate technical and biological variations.
Primary neurons on transwell filters/Axotomy
Cortical neurons form E15.5 mouse brains were plated on the top compartment of 6-well hanging inserts with 1um membrane pores (Millipore, PET), coated on both sides of the membrane with PDL. The cells were cultured for 2 days in serum-free Neurobasal medium, supplemented with B27 and Pen/Strep. Prior to stimulation, neurons were starved as described above and the cell bodies were scraped from the top compartment of the filter, leaving the axons at the bottom. BDNF was added to the axons for 15min and after stimulation the bottom compartment was washed with ice cold PBS and lysed for protein or mRNA extraction. 40μM Anisomycin in L15, for 30min prior to BDNF addition, was used for translational inhibition. All experiments were repeated at least 3 times, to minimize technical and biological variability.
siRNA in primary neurons
siRNA smartpools against HnrnpK, Pcbp1 and Safb2 were obtained from Dharmacon and introduced in neurons with Amaxa Nucleofection, as per the manufacturer’s instructions. A GFP control plasmid provided with the mouse neuron nucleofection kit (LONZA) was co-transfected to visualize cells with the siRNAs. The knockdown efficiency was assessed by IF and microscopy.
Immunofluorescence (IF)
Coverslips were fixed for 20min at 37°C with 4% PFA in PHEM buffer (120mM Sucrose, 2mM MgCl2, 10mM EDTA, 25mM HEPES, 60mM PIPES), rinsed with PBS and then permeabilized with 0.3% Triton-X100 in PBS for 5min at RT. Blocking for 1hr in 10% serum in PBS was followed by incubation with primary antibodies diluted in blocking solution for 1hr at RT. After PBS rinses, secondary antibodies were added to the coverslips, diluted in blocking solution, for 45 min at RT. Phalloidin staining of F-actin was performed for 30 min at RT, followed by PBS rinses and mounting of the coverslips on slides with Fluoromount-G for imaging. Primary antibodies used: Ms-Anti-HnrnpK (1:50, SantaCruz) ms-anti-Mena (1:500) (Lebrand et al., 2004), Rb-anti-Mena (1:500) and Rb-anti-VASP (1:500) generated in the Gertler laboratory. All secondary antibodies used were from Jackson Laboratories, conjugated to -405, -488, -595, or -647 fluorophores and diluted 1:500.
RNA Fluorescent In Situ Hybridization (FISH)
RNA FISH was performed using custom Stellaris FISH probes, according to the manufacturer’s protocol. Briefly, cells on coverslips were fixed with 4% paraformaldehyde in PBS 1x for 15min at 37°C, and subsequently permeabilized with 0.3% Triton-X100 in PBS for 5min at RT. Coverslips were washed in 10% deionized Formamide, 2x Saline-sodium Citrate (SSC) (wash buffer) for 5min at RT and then hybridized in 10% Formamide, 2x SSC, 10% Dextran sulfate, 0,5 μg/ml Salmon Sperm DNA, 1mg/ml yeast tRNA, 1% BSA, and 125nM of RNA probe, in a dark humidified chamber at 37°C O/N. After hybridization the coverslips were washed in wash buffer at 37°C for 30min in the dark. Wherever IF was performed along with the FISH, the primary antibodies were diluted in the hybridization buffer with the probe and incubated simultaneously and the secondary antibodies were added to the post-hybridization wash (30min at 37°C). After the post-hybridization wash coverslips were incubated with phalloidin in PBS, 30min at RT, rinsed in PBS and mounted on slides with Fluoromount-G (Southern Biotech) for imaging. For the unmasking experiments, neurons were incubated with pepsin for 30sec after fixation, as previously described (Buxbaum et al., 2014). For the custom probes, the entire mRNA sequence of mouse and human dyrk1a was used on the Stellaris website. All experiments were repeated at least 3 times, and a minimum of 10 neurons per condition per experiment was imaged and used for quantifications.
Immunoprecipitation (IP)
Cortical neurons cultured for 3 days in vitro were lysed in lysis buffer (20 mM Tris ph 8.0, 200 mM NaCl, 2 mM MgCl2, 10% glycerol, 1% NP-40) supplemented with Roche complete EDTA-free protease and phosphatase inhibitor tablets) on ice for 20 minutes. For whole brain lysate, E15.5 mouse brains were homogenized in lysis buffer using a Dounce homogenizer chilled on ice. Collected lysates were cleared by centrifugation for 20 minutes 14k rpm at 4°C, and incubated overnight at 4°C with antibodies on magnetic protein G beads (incubated in PBS for 4hrs at 4°C. After IP, beads were washed three times with lysis buffer containing 0.4% NP40, and boiled in 2x SDS sample buffer for loading onto an acrylamide gel either for western blotting or for Mass Spectrometry.
HITS-CLIP Modification
E15.5 mouse brains were dissected, rinsed and triturated in PBS and UV-irradiated three times at 400 mJ/cm2 in a Stratalinker (254nm). The tissue suspension was collected by centrifugation and the pellet was lysed in 20 mM Tris ph 8.0, 200 mM NaCl, 2 mM MgCl2, 10% glycerol, 1% NP-40) supplemented with Roche complete EDTA-free protease and phosphatase inhibitor tablets. The lysate was subsequently treated with DNAseI (NEB, M0303L) and RNAseIF (NEB, M0243L) at 37°C and centrifuged for 10 min at 13000rpm. The cleared lysates were then used for a Mena IP. Following the IP, the samples were washed stringently three times with 1M NaCl in lysis/IP buffer and the beads were collected in TRIZOL reagent for RNA extraction, according to the manufacturer’s instructions. Purified RNA was subsequently used for the construction of libraries and sequencing on an ILLUMINA Platform (miSeq). The experiment was repeated twice, using 2 biological replicates per sample, per experiment, to eliminate technical and biological variability. Only the mRNAs that had more than 10 reads and 3-fold enrichment between the Mena and control IP samples were considered significant for subsequent analysis.
Oligo dT capture
E15.5 mouse brain lysates were prepared as described above for HITS-CLIP samples. The lysates were then treated with DNase for 5 min at 37°C and centrifuged 20 min at 14000 rpm at 4°C. Half of each lysate was treated with 1mg/ml RNaseA for 15’ at 37°C and all of the samples were finally heated at 65°C for 5min and kept on ice. OligodT Dynabeads (Pierce) were added to lysates for 12 min at RT and pelleted on a magnet. Following incubation, the beads were washed three times in 1M NaCl lysis buffer and 2xLaemli Buffer was used for Western blot analysis of each sample.
mRNA Pulldown assay
The sequence of the 3’UTR of the dyrk1a and lhx6 mRNA was cloned in a pBS KS vector and linearized. In vitro transcription was carried out on the linearized templates, using the Ampliscribe T7-Flash Biotin-RNA Transcription Kit, according to the manufacturer’s instructions (Epicentre), in order to generate biotinylated probes for the 3’UTR of dyrk1a mRNA. λ-phage DNA and the 3’UTR of the lhx6 gene was also used to generate a control, non-specific biotinylated RNA probe, by in vitro transcritpion. Following precipitation and reconstitution in H2O, the biotinylated probes were captured on Streptavidin Dynabeads (My Streptavidin T1 beads, Pierce) for 1hr at RT. E15.5 brains lysed in 20 mM Tris ph 8.0, 200 mM NaCl, 2 mM MgCl2, 10% glycerol, 1% NP-40 supplemented with Roche complete EDTA-free protease and phosphatase inhibitor tablets and RNAse Inhibitors (Ambion) were incubated with the beads, O/N at 4°C. The beads were subsequently washed in lysis buffer and processed for western blot analysis.
Western Blot
Protein samples were resolved by SDS-PAGE, transferred to nitrocellulose membranes and immunoblotted. Blocking was performed for 1 hour with 3% BSA in PBS at RT, and then the membranes were incubated with primary antibodies in PBS+0.1% Tween-20, O/N at 4°C. After thorough washes, the membranes were incubated with secondary HRP-conjugated antibodies at 1:5000 dilutions and they were visualized by enhanced chemiluminescence (SuperSignal West Pico Chemluminescent HRP substrate -ThermoFisher). Alternatively, fluorescent LICOR secondary antibodies were used at 1:10000, and the membranes were imaged on Odyssey imagers. Primary antibodies used: Rb-anti-PCBP1 (Abcam) 1:1000; Rb-anti-HnrnpK (Cell Signaling) 1:1000; Ms-anti-SafB2 (Abcam) 1:2000; Ms-anti-Dyrk1a (Abnova) 1:1000; Rb-anti-FMR1 (Cell Signaling) 1:1000; Ms-anti-beta Catenin (BD Biosciences) 1:2000; Rb-anti-HnrnpM (Bethyl Laboratories) 1:1000; Rb-anti-HnrnpA2B1 (Elabscience) 1:1000; Rb-anti-RRBP1 (Bethyl Laboratories) 1:1000; Ms-anti-MBNL1 (E. Wang) 1:1000; Ms-anti-tubulin (DM1A) 1:10000
Mass Spectrometry
We performed two technical replicates of this experiment, each replicate used 2 independent biological replicates for Mena IP from wild type brains, and 2 replicates for Mena IP from Mena-null brains (as a negative control for specificity). During IP for mass spectrometry the anti-Mena antibody (clone A351F7D9) was covalently crosslinked with DMP to protein G magnetic beads. Acrylamide gels were stained with Coomasie Brilliant Blue. After destaining with 40% ethanol/10% acetic acid, proteins were reduced with 20mM dithiothreitol (Sigma) for 1h at 56°C and then alkylated with 60mM iodoacetamide (Sigma) for 1h at 25°C in the dark. Proteins were then digested with 12.5ng/μl modified trypsin (Promega) in 50μl 100mM ammonium bicarbonate, pH 8.9 at 25°C overnight. Peptides were extracted by incubating the gel pieces with 50% acetonitrile/5% formic acid then 100mM ammonium bicarbonate, repeated twice followed by incubating the gel pieces with 100% acetonitrile then 100mM ammonium bicarbonate, repeated twice. Each fraction was collected, combined, and reduced to near dryness in a vacuum centrifuge. Per manufacturer’s instructions, each sample was labeled with a unique iTRAQ 4plex (ABSciex). Following the 1 h incubation, all samples were combined and concentrated to completion. The combined labeled peptides were desalted using Protea C18 spin tips and resuspended in 0.1% formic acid. Peptides were separated by reverse phase HPLC using an EASY- nLC1000 (Thermo) over a 140-minute gradient before nanoelectrospray using a QExactive mass spectrometer (Thermo). Mass spectrometry data were analyzed using Mascot (Matrix Science) and Proteome Discoverer (Thermo).
RT-PCR and Quantitative PCR
cDNA synthesis was performed using the Invitrogen Superscript III First Strand Synthesis for RT-PCR kit, with Random Hexamer primers, according to the manufacturer’s instructions. Quantitative PCR was performed using the Biorad iQ SYBR Green Supermix on a CFX96 Real Time PCR Detection System, with the following gene-specific primers: mouse GAPDH 5’-catgttccagtatgactccactc ; mouse GAPDH 3’-ggcctcaccccatttgatgt mouse Mena 5’-gggcagaaagattcaagacc ; mouse Mena 3’-gcgaagacattggcatcc mouse Dyrk1a 5’-caaacggagtgcaatcaaga ; mouse Dyrk1a 3’-agcacctctggagaccgata mouse Robo1 5’-catcaagaggatcagggagc ; mouse Robo1 3’-ggttgtcttcagctttcagtttc mouse Elavl1 5’-agccaatcccaaccagaac ; mouse Elavl1 3’-acaccagaaatcccactcatg mouse β-Ctnn 5’-ctatcccagaggctttatccaag ; mouse β-Ctnn 3’-ccagagtgaaaagaacggtagc mouse Khsrp 5’-gccaatcagactacaccaagg ; mouse Khsrp 3’-gccacttgtgttgcttcttg mouse Eif4ebp2 5’-ccatctgcccaatatccctg ; mouse Eif4ebp2 3’-tgtccatctcaaactgagcc mouse Vamp2 5’-aagttgtcggagctggatg ; mouse Vamp2 3’-cgcagatcactcccaagatg
Imaging
Imaging was performed using a Deltavision microscope (Applied Precision, Issaquah, WA), with a Coolsnap HQ camera (Photometrics); post-acquisition image processing was performed using SoftWoRx v.XX (Applied Precision). Maximum intensity projections of 2-4 optical sections were generated using ImageJ. Only growth cones that were not in contact with other cells or processes and had extended more than 0.3 mm away from the cell body were chosen for imaging. All fluorescence quantitation used original unprocessed image data, with no pixels at zero intensity or saturated. In panels displayed in the figures, for consistent visibility across the intensity range, contrast and brightness were adjusted uniformly within each experimental series.
QUANTIFICATION AND STATISTICAL ANALYSIS
All microscopy experiments were repeated at least 3 times with different biological samples, and at least 10 axons were analyzed per condition, per experiment. Quantified Signal intensities were normalized to cellular area within the region of interest. For colocalization studies, the JaCoP plugin of ImageJ was used to calculate Pearson’s Coefficient Corellation, including Coste’s Randomization. All biochemical assays were performed at least 3 times with two biological replicates each time per sample, to minimize variability. Statistical significance was assessed either in Excel, or Graphpad Prism6, using Student’s t test, non-parametric, or two-way ANOVA, specified in Figure legends for each experiment. All graphs represent mean values ± StDev.
Supplementary Material
TABLE S1 (Related to Figure 1 and S1): Proteins Interacting with Mena in developing mouse brains (excel file)
TABLE S2 (Related to Figure 1 and S1): mRNAs associated with Mena in developing mouse brains (excel file)
Highlights.
Mena is in complex with translational regulators HnrnpK and PCBP1 in neurons
The Mena-complex associates with multiple cytosolic mRNAs
dyrk1a is locally translated in axons in a Mena-dependent manner
Mena is potentially required for translational de-repression of dyrk1a
Acknowledgments
The authors thank Eliza Vasile for imaging assistance, Duanduan Ma for assistance with Bioinformatic Analysis, Jenny Tadros for technical support and Phil Sharp, Myriam Heiman, Russell McConnell and Guillaume Carmona for helpful discussion on the manuscript. This work was supported by NIH grant U01-CA184897 and funds from the Ludwig foundation (FBG), funds from the Koch Institute NCI core grant P30-CA14051.
Abbreviations
- RNP
ribonucleoprotein
- RBP
RNA-binding protein
- NS
nervous system
Footnotes
AUTHOR CONTRIBUTIONS
M.V. designed and performed experiments, analyzed data and wrote the paper. M.V. and F.D. performed qPCR analyses. M.V. and T.S. performed HITS-CLIP assays. E.L. performed FISH after unmasking. M.T. carried out bioinformatics analysis. A.T. performed qPCR analyses and gave intellectual support. M.V. and F.B.G. discussed the results and implications at all stages. F.B.G. designed experiments, gave technical and intellectual support and revised the manuscript.
DATA AND SOFTWARE AVAILABILITY
Mena protein interactions identified by IP-Mass Spectrometry are provided in Supplemental Table S1
mRNAs associated with Mena, as identified by CLIP-Sequencing, are provided in Supplemental Table S2
More information and access to the data are available upon request to the Lead Contact.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009:10. doi: 10.1186/gb-2009-10-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami NH, Smith RB, Carrasco Ma, Williams La, Winborn CS, Han SSW, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, Clare AJ, Badders NM, Bilican B, Chaum E, Chandran S, Shaw CE, Eggan KC, Maniatis T, Taylor JP. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–43. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–6. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasanyan V, Arnold DB. Actin and Myosin-dependent localization of mRNA to dendrites. PLoS One. 2014;9:e92349. doi: 10.1371/journal.pone.0092349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleriola J, Hengst U. Targeting Axonal Protein Synthesis in Neuroregeneration and Degeneration. Neurotherapeutics. 2014:57–65. doi: 10.1007/s13311-014-0308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barallobre MJ, Perier C, Bové J, Laguna a, Delabar JM, Vila M, Arbonés ML. DYRK1A promotes dopaminergic neuron survival in the developing brain and in a mouse model of Parkinson’s disease. Cell Death Dis. 2014;5:e1289. doi: 10.1038/cddis.2014.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–14. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista AFR, Hengst U. Intra-axonal protein synthesis in development and beyond. Int J Dev Neurosci. 2016 doi: 10.1016/j.ijdevneu.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative Regulation of Fibroblast Motility by Ena / VASP Proteins. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, Padhye S, Xu L, Weir SJ, Anant S, Meisner-Kober N, Brody JR, Dixon DA. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget. 2016:7. doi: 10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalisation analysis in light microscopy. J Microsc. 2006;224:13–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum Sa, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Wu B, Singer RH. Single -Actin mRNA Detection in Neurons Reveals a Mechanism for Regulating Its Translatability. Science (80- ) 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Regan AG, Lopez JS, Tannahill D, Harris WA, Holt CE. Semaphorin 3A Elicits Stage-Dependent Collapse , Turning , and Branching in Xenopus Retinal Growth Cones. 2001;21:8538–8547. doi: 10.1523/JNEUROSCI.21-21-08538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Lin JR, Tsai FC, Meyer T. Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle. Mol Cell. 2013;52:87–100. doi: 10.1016/j.molcel.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutadeur S, Benyamine H, Delalonde L, de Oliveira C, Leblond B, Foucourt A, Besson T, Casagrande AS, Taverne T, Girard A, Pando MP, Désiré L. A novel DYRK1A (Dual specificity tyrosine phosphorylation-regulated kinase 1A) inhibitor for the treatment of Alzheimer’s disease: effect on Tau and amyloid pathologies in vitro. J Neurochem. 2015;133:440–51. doi: 10.1111/jnc.13018. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–86. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Bagni C. Fragile X mental retardation protein control of neuronal mRNA metabolism: Insights into mRNA stability. Mol Cell Neurosci. 2010;43:43–50. doi: 10.1016/j.mcn.2009.09.013. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, Pasciuto E, Li KW, Fernández E, Di Marino D, Buzzi A, Ostroff LE, Klann E, Zwartkruis FJT, Komiyama NH, Grant SGN, Poujol C, Choquet D, Achsel T, Posthuma D, Smit AB, Bagni C. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79:1169–82. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A, Jaffrey SR. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:1–13. doi: 10.1098/rsob.120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The Growth Cone Cytoskeleton in Axon outgrowth and guidance.pdf. 2011 doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson Da, Gupton S, Van Veen JE, Furman C, Zhang J, Alberts AS, Mori S, Gertler FB. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9:1347–59. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- Di Vona C, Bezdan D, Islam ABMMK, Salichs E, López-Bigas N, Ossowski S, de la Luna S. Chromatin-wide profiling of DYRK1A reveals a role as a gene-specific RNA polymerase II CTD kinase. Mol Cell. 2015;57:506–20. doi: 10.1016/j.molcel.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Drees F, Gertler FB. Ena/VASP: proteins at the tip of the nervous system. Curr Opin Neurobiol. 2008;18:53–59. doi: 10.1016/j.conb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedy A, Gertler FB, Miller J, Holt CE, Lebrand C. Ena/VASP function in retinal axons is required for terminal arborization but not pathway navigation. Development. 2007;134:2137–2146. doi: 10.1242/dev.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folci A, Mapelli L, Sassone J, Prestori F, Angelo ED, Bassani S, Passafaro M. Loss of hnRNP K Impairs Synaptic Plasticity in Hippocampal Neurons. 2014;34:9088–9095. doi: 10.1523/JNEUROSCI.0303-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M, Doyle M, Bauer KE, Thomas S, Planyavsky M, Arn E, Bakosova A, Jungwirth K, Hörmann A, Palfi Z, Sandholzer J, Schwarz M, Macchi P, Colinge J, Superti-Furga G, Kiebler M. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5:1749–62. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. MOLECULAR MECHANISMS OF TRANSLATIONAL CONTROL. 2004:5. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler F, Condeelis J. Metastasis: tumor cells becoming MENAcing. Trends Cell Biol. 2011;21:81–90. doi: 10.1016/j.tcb.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/S0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Giesemann T, Nawrotzki R, Berho K, Rothkegel M, Schlu K, Schrader N, Schindelin H, Mendel RR, Kirsch J, Jockusch BM. Complex Formation between the Postsynaptic Scaffolding Protein Gephyrin , Profilin , and Mena3: A Possible Link to the Microfilament System. 2003;23:8330–8339. doi: 10.1523/JNEUROSCI.23-23-08330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. Integrin signaling switches the cytoskeletal and exocytic machinery that drives neuritogenesis. Dev Cell. 2010;18:725–736. doi: 10.1016/j.devcel.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Riquelme D, Hughes-Alford SK, Tadros J, Rudina SS, Hynes RO, Lauffenburger D, Gertler FB. Mena binds alpha5 integrin directly and modulates alpha5beta1 function. J Cell Biol. 2012;198:657–676. doi: 10.1083/jcb.201202079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle B, Carnicero a, Elizalde C, Ceron J, Martinez S, Tejedor FJ. Expression patterns and subcellular localization of the Down syndrome candidate protein MNB/DYRK1A suggest a role in late neuronal differentiation. Eur J Neurosci. 2003;17:2277–2286. doi: 10.1046/j.1460-9568.2003.02665.x. [DOI] [PubMed] [Google Scholar]
- Hämmerle B, Elizalde C, Tejedor FJ. The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur J Neurosci. 2008;27:1061–74. doi: 10.1111/j.1460-9568.2008.06092.x. [DOI] [PubMed] [Google Scholar]
- Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25:254–264. doi: 10.1016/j.ceb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Hutten S, Sharangdhar T, Kiebler M. Unmasking the messenger. RNA Biol. 2014;11:992–7. doi: 10.4161/rna.32091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janel N, Sarazin M, Corlier F, Corne H, de Souza LC, Hamelin L, Aka a, Lagarde J, Blehaut H, Hindié V, Rain JC, Arbones ML, Dubois B, Potier MC, Bottlaender M, Delabar JM. Plasma DYRK1A as a novel risk factor for Alzheimer’s disease. Transl Psychiatry. 2014;4:e425. doi: 10.1038/tp.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Holt CE. Local translation of mRNAs in neural development. Wiley Interdiscip Rev RNA. 2011;2:153–65. doi: 10.1002/wrna.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–24. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Jung H. Local protein synthesis in neuronal axons: why and how we study. BMB Rep. 2015;48:139–146. doi: 10.5483/BMBRep.2015.48.3.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee KS, Kim AK, Choi M, Choi K, Kang M, Chi SW, Lee MS, Lee JS, Lee SY, Song WJ, Yu K, Cho S. A chemical with proven clinical safety rescues Down-syndrome-related phenotypes in through DYRK1A inhibition. Dis Model Mech. 2016;9:839–48. doi: 10.1242/dmm.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Buzzi A, Di Marino D. Synaptic Plasticity. Advances in Experimental Medicine and Biology. 2012;970:285–305. doi: 10.1007/978-3-7091-0932-8. [DOI] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. E NA/VASP P ROTEINS: Regulators of the Actin Cytoskeleton and Cell Migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski AV, Rubinson Da, Dent EW, Edward van Veen J, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds Aa, Bronson RT, Mori S, Fässler R, Gertler FB. Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–55. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: Quantitative Assessment of the Sequence and Structural Binding Specificity of RNA Binding Proteins. Mol Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/S0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/S0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork K, Ramadoss S, Ding X, Li X, Wang CY. KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/β-catenin signalling. Nat Commun. 2017;8:15146. doi: 10.1038/ncomms15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li Y, Gao FB. Abelson, enabled, and p120 catenin exert distinct effects on dendritic morphogenesis in Drosophila. Dev Dyn. 2005;234:512–22. doi: 10.1002/dvdy.20496. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark Ta, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Lei YT, Hong CJ, Hsueh YP. Syndecan-2 induces filopodia and dendritic spine formation via the neurofibromin-PKA-Ena/VASP pathway. J Cell Biol. 2007;177:829–41. doi: 10.1083/jcb.200608121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zheng L, Si Y, Luo W, Dujardin G, Kwan T, Potochick NR, Thompson SR, Schneider DA, King PH. Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: Implications for amyotrophic lateral sclerosis. J Biol Chem. 2014;289:31792–31804. doi: 10.1074/jbc.M114.573246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinge S, Bliss-Moreau M, Kirsammer G, Diebold L, Chlon T, Gurbuxani S, Crispino JD. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of down syndrome. J Clin Invest. 2012;122:948–962. doi: 10.1172/JCI60455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandless DW. Epilepsy and Autism. 2012:1–18. doi: 10.1007/978-1-4614-0361-6. [DOI] [Google Scholar]
- McConnell RE, Van Veen JE, Vidaki M, Kwiatkowski AV, Meyer AS, Gertler FB. A requirement for filopodia extension toward Slit during Robo-mediated axon repulsion. J Cell Biol. 2016;213:261–274. doi: 10.1083/jcb.201509062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies AS, Aszodi A, Williams SE, Pfeifer A, Wehman AM, Goh KL, Mason CA, Fassler R, Gertler FB. Mena and Vasodilator-Stimulated Phosphoprotein Are Required for Multiple Actin-Dependent Processes That Shape the Vertebrate Nervous System. J Neurosci. 2004;24:8029–8038. doi: 10.1523/JNEUROSCI.1057-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–9. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafov A, Seker T, Even I, Hoxhaj G, Selvi O, Ozel DE, Koman A, Birgül-3yison N. MENA is a transcriptional target of the Wnt/beta-catenin pathway. PLoS One. 2012;7:e37013. doi: 10.1371/journal.pone.0037013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O’Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson Da, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Abalia J, Sahún I, Altafaj X, Andreu N, Estivill X, Dierssen M, Fillat C. Targeting Dyrk1A with AAVshRNA Attenuates Motor Alterations in TgDyrk1A, a Mouse Model of Down Syndrome. Am J Hum Genet. 2008;83:479–488. doi: 10.1016/j.ajhg.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-furga G, Hentze MW. c-Src-Mediated Phosphorylation of hnRNP K Drives Translational Activation of Specifically Silenced mRNAs. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, Akerman M, Dror I, Kosti I, Mandel-Gutfreund Y. SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010;38:W281–W285. doi: 10.1093/nar/gkq444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Quan J, Nowakowski DW, Hancock ML, Shi J, Tcherkezian J, Young-Pearse TL, Flanagan JG. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell. 2014;158:368–382. doi: 10.1016/j.cell.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proepper C, Steinestel K, Schmeisser MJ, Heinrich J, Steinestel J, Bockmann J, Liebau S, Boeckers TM. Heterogeneous Nuclear Ribonucleoprotein K Interacts with Abi-1 at Postsynaptic Sites and Modulates Dendritic Spine Morphology. 2011:6. doi: 10.1371/journal.pone.0027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Jin N, Shi J, Yin X, Jin X, Wang S, Cao M, Iqbal K, Gong CX, Liu F. Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) enhances tau expression. J Alzheimers Dis. 2013;37:529–38. doi: 10.3233/JAD-130824. [DOI] [PubMed] [Google Scholar]
- Rachdi L, Kariyawasam D, Aïello V, Herault Y, Janel N, Delabar JM, Polak M, Scharfmann R. Dyrk1A induces pancreatic ?? cell mass expansion and improves glucose tolerance. Cell Cycle. 2014;13:2221–2229. doi: 10.4161/cc.29250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme DN, Meyer AS, Barzik M, Keating A, Gertler FB. Selectivity in subunit composition of Ena/VASP tetramers. Biosci Rep. 2015;35:e00246–e00246. doi: 10.1042/BSR20150149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M, Sur M. Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science (80- ) 2015;350:aab3897. doi: 10.1126/science.aab3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gomez PF-MCZP. DYRK1A: the double-edged kinase as a protagonist in cell growth and tumorigenesis [WWW Document] [accessed 5.12.16];Mol Cell Oncol. 2015 doi: 10.4161/23723548.2014.970048. URL http://www.tandfonline.com/doi/pdf/10.4161/23723548.2014.970048. [DOI] [PMC free article] [PubMed]
- Santos AR, Comprido D, Duarte CB. Regulation of local translation at the synapse by BDNF. Prog Neurobiol. 2010;92:505–516. doi: 10.1016/j.pneurobio.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh Ea, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–77. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak E, Gebauer F. Translational control by 3’-UTR-binding proteins. Brief Funct Genomics. 2013;12:58–65. doi: 10.1093/bfgp/els056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor FJ, Hämmerle B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2011;278:223–35. doi: 10.1111/j.1742-4658.2010.07954.x. [DOI] [PubMed] [Google Scholar]
- Thiele B, Doller A, Ka T, Pregla R, Hetzer R, Regitz-zagrosek V. RNA-Binding Proteins Heterogeneous Nuclear Ribonucleoprotein A1 , E1 , and K Are Involved in Post-Transcriptional Control of Collagen I and III Synthesis. 2016 doi: 10.1161/01.RES.0000149166.33833.08. [DOI] [PubMed] [Google Scholar]
- Torvund-jensen J, Steengaard J, Reimer L, Fihl LB, Laursen LS. Transport and translation of MBP mRNA is regulated differently by distinct hnRNP proteins. 2014;2:1550–1564. doi: 10.1242/jcs.140855. [DOI] [PubMed] [Google Scholar]
- Townson SM, Dobrzycka KM, Lee AV, Air M, Deng W, Kang K, Jiang S, Kioka N, Michaelis K, Oesterreich S. SAFB2, a new scaffold attachment factor homolog and estrogen receptor corepressor. J Biol Chem. 2003;278:20059–20068. doi: 10.1074/jbc.M212988200. [DOI] [PubMed] [Google Scholar]
- van Bon BWM, Coe BP, Bernier R, Green C, Gerdts J, Witherspoon K, Kleefstra T, Willemsen MH, Kumar R, Bosco P, Fichera M, Li D, Amaral D, Cristofoli F, Peeters H, Haan E, Romano C, Mefford HC, Scheffer I, Gecz J, de Vries BBa, Eichler EE. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol Psychiatry. 2015:1–7. doi: 10.1038/mp.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bon BWM, Hoischen A, Hehir-Kwa J, De Brouwer APM, Ruivenkamp C, Gijsbers ACJ, Marcelis CL, De Leeuw N, Veltman JA, Brunner HG, De Vries BBA. Intragenic deletion in DYRK1A leads to mental retardation and primary microcephaly. Clin Genet. 2011;79:296–299. doi: 10.1111/j.1399-0004.2010.01544.x. [DOI] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–42. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ding N, Guo J, Xia J, Ruan Y. Dysregulation of TTP and HuR plays an important role in cancers. Tumor Biol. 2016;37:14451–14461. doi: 10.1007/s13277-016-5397-z. [DOI] [PubMed] [Google Scholar]
- Wegiel JJ, Kuchna I, Nowicki K, Imaki H, Marchi E, Ma SY, Chauhan A, Chauhan V, Bobrowicz TW, de Leon M, Louis La Saint, Cohen IL, London E, Brown WT, Wisniewski T. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119:755–70. doi: 10.1007/s00401-010-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Hao JC, Lim W, Tessier-Lavigne M, Bargmann CI. Shared receptors in axon guidance: SAX-3/Robo signals via UNC-34/Enabled and a Netrin-independent UNC-40/DCC function. Nat Neurosci. 2002;5:1147–54. doi: 10.1038/nn956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 (Related to Figure 1 and S1): Proteins Interacting with Mena in developing mouse brains (excel file)
TABLE S2 (Related to Figure 1 and S1): mRNAs associated with Mena in developing mouse brains (excel file)