Abstract
Cell reprogramming of somatic cells into pluripotent states and subsequent differentiation into certain phenotypes has helped progress regenerative medicine research and other medical applications. Recent research has used viral vectors to induce this reprogramming; however, limitations include low efficiency and safety concerns. In this review, we discuss how biomaterial methods offer potential avenues for either increasing viability and downstream applicability of viral methods, or providing a safer alternative. The use of non-viral delivery systems, such as electroporation, micro/nanoparticles, nucleic acids and the modulation of culture substrate topography and stiffness have generated valuable insights regarding cell reprogramming.
Keywords: biomaterials, cell reprogramming, non-viral delivery, biophysical cues
TOC image
Improving cell reprogramming methods is vital to regenerative medicine efforts. This review discusses advances in reprogramming using biomaterial approaches.
1. Introduction
Both embryonic (ESC) and induced pluripotent stem cells (iPSC) act as templates for differentiation into desired cell phenotypes and ultimately engineered tissues [1, 2]. This is beneficial for regenerative medicine because it advances the possible medical interventions used to repair or replace damaged tissue [3]. This is limited, however, by lack of patient specificity and ethical concerns of ESCs [1]. Thus, more emphasis on iPSC usage has been conducted in recent years as they counteract the limitations of ESCs through patient specificity and cell proliferative properties from somatic cells. Using cell reprogramming with defined transcription factors to change somatic cells to iPSCs for repeated tissue development is vital to expand regenerative medicine [4]. Increasing the efficiency of cell reprogramming and subsequent differentiation into particular cell phenotypes benefit cell therapy and disease modeling research by providing easier to construct tissues that mitigate any ethical concerns [5].
Biomaterials are another technology used in regenerative medicine that has gained attention in recent years. Advancements in material sciences have characterized a plethora of substances for their bulk and surface properties and determined their biocompatibility [6]. Naturally, research has begun to combine the benefits of biomaterial methods with stem cell developments for more efficient and viable human cell and tissue engineering [7]. As more materials compatible with stem cells are discovered, characterizing properties such as charge, surface features, and potential for cell adhesion, apoptosis, or tissue necrosis have been elucidated to determine how these biomaterials can be used as scaffolds for tissue engineering [8, 9]. Biomaterials can affect the efficiency of cell reprogramming and differentiation by providing both genetic and epigenetic influences to cells as either a niche or delivery mechanism.
In this review, we will briefly discuss the current state of cell reprogramming and differentiation practices, their limitations, and how various biomaterial methods are a potential avenue to improve the impact of this technology. This includes harnessing the effects of biochemical and biophysical cues critical for cell development, the use of methods that do not rely on viral vectors while increasing cell phenotype specificity, and engineering culture substrates to benefit cell reprogramming and differentiation.
2. Cell reprogramming
With the landmark study on establishing pluripotency in somatic cells from Yamanaka and Takahashi [10], cell reprogramming has developed into an innovative area of biomedical research. This phenomenon of actively converting one cell type to another is particularly effective with stem cells, which are proliferative and have the capacity to be easily modified. Stem cells in the adult human body can regenerate the specific tissues from which they originate, yet they lack the pluripotency that immature stem cells have [11]. Human iPSCs (hiPSCs) can be studied for genetic defects and stability, be directly differentiated into specific tissues, and be used for disease modeling or potential implantation into patients. The supply of iPSCs is less scarce than ESCs and provides less risk of immune rejection since these cells can be patient specific [5]. Induced pluripotent stem cells can be developed by introducing transcription factors to somatic cells like skin fibroblasts for research studies and blood or adipose cells for possible clinical usage [4, 12]. These factors include Oct4, Sox2, Myc, and Klf4 [10]. Other factors and small molecules display pluripotency promotion in somatic cells as a more controlled replacement [13, 14, 15]. The knowledge of mechanisms to pluripotency was limited until one group sought to map the genomic characterization of fibroblasts to pluripotency. This study indicated the specific gene expressions and epigenetic signatures that lead to reprogramming to an iPSC state. This data outlines a molecular description of the reprogramming routes and is now publicly accessible [16]. Their findings suggest that mouse embryonic fibroblasts (MEFs) follow two different paths to pluripotency dependent on histone loss and reacquisition, transgene dependency, and trimethylation within the genome. By mapping the genomic pathways of cells, cell reprogramming can become more controlled. While our understanding of cell pathways grows, more emphasis is made on reprogramming methods.
Cell reprogramming is predominantly achieved via viral infection. Retroviruses, lentiviruses, and non-integrating viruses induce pluripotency by the introduction of genes that drive the adoption of a pluripotent state [17]. Direct reprogramming or transdifferentiation to specific cell types has also been reported using viral induction. Inserting reprogramming factors into fibroblasts to bypass pluripotency and transdifferentiate to specific cell phenotypes is currently inefficient but has the potential to optimize controlled cell development [18]. Examples of this method have focused on cardiomyocyte development from fibroblasts by expression of particular transcription factors during viral induction [19, 20, 21]. The variety of different factors and exposure to environments not conducive to iPSC production in these studies provide a plethora of methods specific to cell phenotypes needed for reprogramming. The current issues with these methods are long term functionality of reprogrammed cells, overcoming reprogramming inefficiency using transcription factors in vitro, and translation to in vivo conditions within humans.
Viral delivery systems are not ideal as there are implant safety concerns with tumor development, despite increased efficiency in pluripotency [17]. Mitigating oncogenic cell behavior through other induction methods, while improving proliferative and reprogramming efficiency, has been a focus of current stem cell research [11]. Replacing viruses with alternative extracellular chemicals or delivery systems provides another alternative to reduce tumor formation [17]. Factors in the microenvironment have been studied that promote forms of stemness, such as low attachment substrates that form 3D spheres [22]. Observations of stem cells have concluded that they will aggregate into spherical clusters and thus developing substrates that promote this behavior in somatic cells can increase cell reprogramming effectiveness. These types of studies guide the focus on creating microenvironments that promote specific cell proliferation during reprogramming through cell signaling and biophysical properties.
Techniques using matrix manipulation and novel delivery systems can improve cell reprogramming and more accurately model in vivo conditions to better elucidate mechanisms by which cell programming occurs (Figure 1). By using biomaterials in conjunction with stem or somatic cells, more control over cell reprogramming and differentiation can be attained. Using known biophysical and biochemical cues to create synthetic systems that replicate the optimal conditions needed for reprogramming will help to alleviate the lack of efficiency in cell specific growth and maturation (Figure 2). Developing systems that do not require viral induction will mitigate the mutation risk with injecting foreign genetic material into cells. Examples include electroporation, micro- or nanoparticles, and nucleic acid complexation. To manipulate microenvironments, factors like topography, substrate stiffness, and stretching have been used to study their effects on cell proliferation and differentiation. These methods seek to either improve current viral methods or to mitigate risk by using non-viral systems with the possibility of increasing reprogramming efficiency (Table 1).
Figure 1.
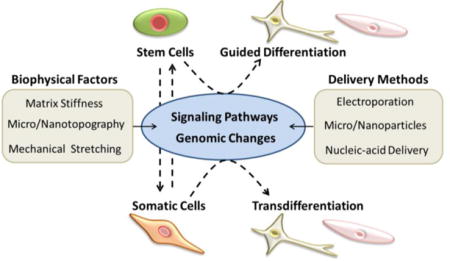
Pathways to differentiation and reprogramming to pluripotency or direct reprogramming: Pluripotent and somatic cells are exposed to genomic changes and micro-environmental cues to promote reprogramming or differentiation into the appropriate phenotype. Once cells have differentiated into a somatic form (i.e. skin fibroblasts), they can be reprogrammed using these same techniques into stem cells or convert directly to certain phenotypes. Manipulating the extracellular matrix/substrate that cells develop on by adjusting stiffness, topographical cues, or stretching are among common biomaterial techniques attempting to use microenvironments to control reprogramming and differentiation. Delivering genomic markers into cells via carrier systems like nucleic acids and nanoparticles is another way to actively adjust stem cell fate or convert from a somatic cell to pluripotency or induced phenotypes.
Figure 2.
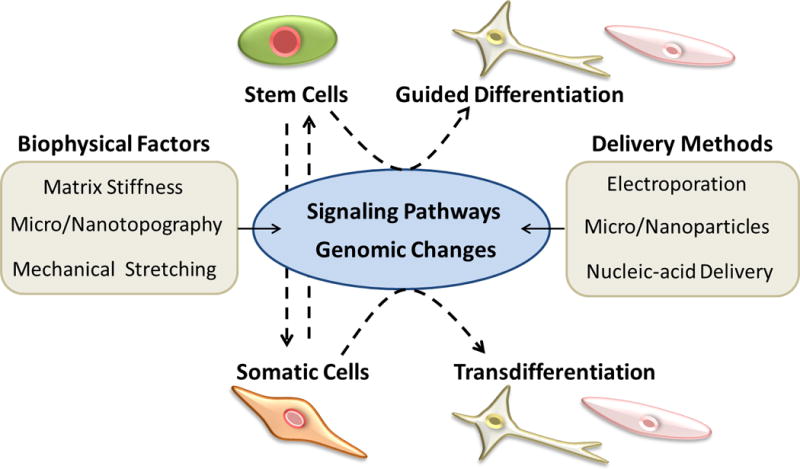
Different methods of cell manipulation that lead to reprogramming and differentiation. This includes (A) topographical cues that signals cytoskeletal organization and thus intracellular pathways, (B) varying stiffness that promotes cell phenotype specific reprogramming through intercellular signaling, and (C) addition of soluble factors to cell culture to activate cell pathways towards direct reprogramming. Reproduced with permission from ref. 57. Copyright (2009) Elsevier, from ref. 55. Copyright (2013) American Chemical Society, and from ref. 27. Copyright (2016) Wiley Periodicals, Inc.
Table 1.
Cellular reprogramming methods, specific phenotypes, and factors
| Reprogramming Method | Original Phenotype | Reprogrammed Phenotype | Viral or non-viral | Reprogramming Factor | Ref. |
|---|---|---|---|---|---|
| Electroporation | Fibroblasts | iPSC | Non-viral | mRNA | [30] |
| Nucleofection | Adipose SC | iPSC | Non-viral | minicircle DNA | [32] |
| Nanoparticles | iPSC | Hepatocyte | Non-viral | FMSN | [37] |
| Fibroblasts | iPSC | Non-viral | Plasmid DNA magnetic nanoparticle | [38] | |
| Fibroblasts | iPSC | Non-viral | Chitosan nanoparticle | [42] | |
| hMSC | Osteoblasts | Non-viral | PAMAM dendrimer | [43] | |
| hMSC | Osteoblasts | Non-viral | Gold nanoparticle | [44] | |
| Complexation | aRPE cell | aRPE progenitor | Non-viral | PU-PEI plasmid complex | [50] |
| BM-MNC | iPSC | Non-viral | MicroRNA:DOTAP copolymer | [51] | |
| HUMSC | iPSC | Non-viral | CPEPS encapsulated plasmid | [53] | |
| Topography | Fibroblast | Induced neuron | Viral | Micrograting/post | [66] |
| Cardiac progenitor | Cardiomyocyte | Viral | Microgroove | [69] | |
| Fibroblast | Cardiomyocyte | Viral | Microgroove | [70] | |
| ESC/iPSC | Pancreatic cell | Non-viral | Nanopore | [72] | |
| Fibroblast | Neurons | Viral | Nanogrooves | [81] | |
| Stiffness | Fibroblast | iPSC | Viral | 0.1 kPa stiffness | [86] |
| hMSC | Adipocyte | Non-viral | 0.5 kPa stiffness | [88] | |
| hMSC | Osteoblast | Non-viral | 64 kPa stiffness | [88] | |
| Stretching | hMSC | Smooth muscle cell | Non-viral | Cyclic uniaxial stretch | [102] |
| hMSC | Tenocyte | Non-viral | Cyclic stretch | [104] |
3. Non-viral methods for reprogramming to pluripotency and differentiation
Over the last decade, viral reprogramming approaches have shown a potential for inducing pluripotency from somatic cells [23, 24, 25, 26]. However, viral methods for cell reprogramming have inherent drawbacks including genomic integration and insertional mutagenesis. Moreover, as previously mentioned, tumor development is another major concern for viral reprogramming approaches due to unintended oncogene activation. To overcome these limitations, non-viral methods have greatly attracted the attention of many researchers in the field of cell reprogramming [27, 28]. Non-viral methods include electroporation of cell membranes, delivering genes or proteins in nano-particulated form and complexation of genes with lipid or cationic polymers (Figure 3). In this section, recent advances in each non-viral approach for cell reprogramming and differentiation are discussed along with the advantages and disadvantages of each strategy.
Figure 3.
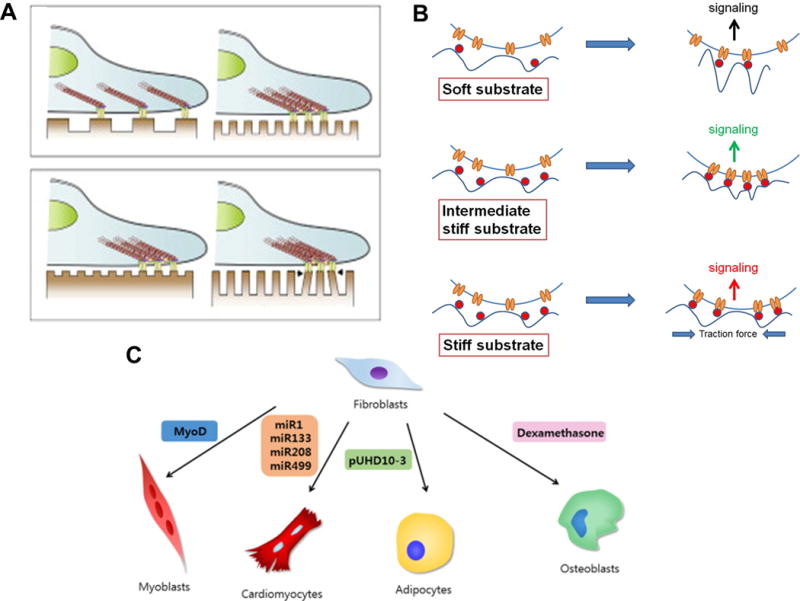
Examples of non-viral approaches (A) Schematic of cell reprogramming and myogenic commitment of human chondrocytes using electroporation and induced factors. (B) Schematic representation of the entry of encapsulated cationic bolaamphiphile-protein complex into the cell and their transformation of the cells to iPSCs. The cationic bolaamphiphile-protein complex enters the cell via the cell membrane (1). Dissociation and release of proteins into the cytoplasm (2). Released proteins get translocated to nucleus (3). Proteins bind to their respective response elements and trigger the transcription of the genes associated with reprogramming (4). (C) Schematic of arginine-glycine-aspartic (Arg-Gly-Asp, RGD) peptide-modified dendrimer-entrapped gold nanoparticles (Au DENPs) specific gene delivery to stem cells. Reproduced with permission from ref. 27. Copyright (2016) Wiley Periodicals, from ref. 41. Copyright (2013) Elsevier, and from ref. 43. Copyright (2015) ACS Publications.
3.1. Electroporation and nucleofection
In order to avoid rapid clearance and degradation in the extracellular environment when introducing naked nucleic acids for expressing reprogramming factors, electroporation has been adapted to deliver nucleic acids directly to the cytoplasm. Due to the highly charged nature of nucleic acids, it is physically impossible for them to penetrate the hydrophobic core of cell membranes. For the electroporation of foreign genes to the cells, an electrostatic field is applied to the cells in suspension. This causes the lipid molecules comprising the membrane to shift their position and create pores, thus resulting in an increased permeability of the cell membrane [29]. This allows foreign substances to enter the membrane, independent of their charge. It has been shown that electroporation works both in vitro and in vivo, and works for hard-to-transfect cells as well.
Electroporation was utilized for delivering mRNA to express Yamanaka factors and SV40 large T in human fibroblast cells to activate pluripotency (Figure 4A) [30]. Along with mRNA, the fibroblasts were also treated with small molecules (valproic acid, BIX01294 and 59-aza-29-deoxycytidine) to help increase the degree of reprogramming. In this study, activation of pluripotency was achieved with high transfection efficiency, and they showed that only a transient expression of reprogramming factors using mRNAs is sufficient to generate iPSCs without causing genetic modification. However, mRNA-based reprogramming approaches still need to overcome the innate immune response activated by introducing a large amount of long RNAs [31]. It was also reported that multiple rounds of electroporation resulted in massive cell death [30]. As shown in the study, electroporation can cause thermal damage. This leads to low cell viability after gene electrotransfer, even though transfection efficiency might be high in leftover viable cells. Moreover, it is a labor-intensive procedure compared to other non-viral approaches, and is not considered a universal method since it requires a specific device, leading researchers to seek alternative approaches.
Figure 4.
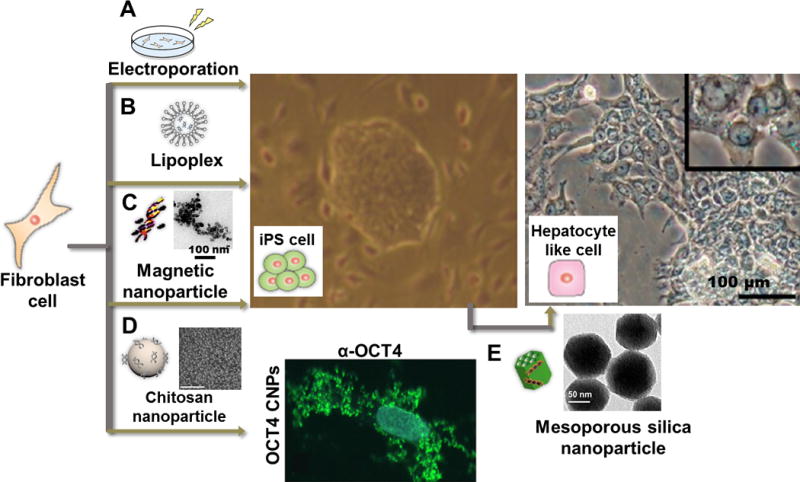
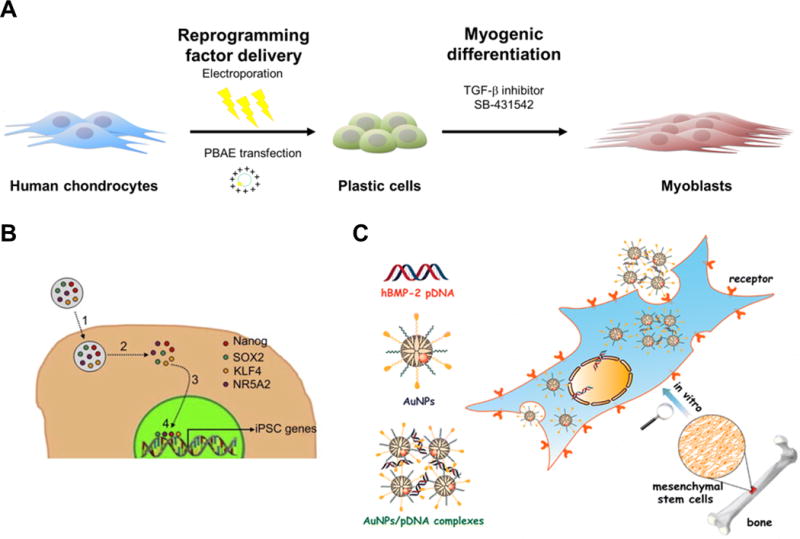
Schematic illustrations of non-viral cellular reprogramming approaches and cellular reprogramming results from ref. 37, 40 and 42. (A) mRNA for expressing Yamanaka factors can be introduced by electroporation. (B) Cationic lipid-mediated approach to activate pluripotency in fibroblast cells. (C) Magnetofection of pDNA for reprogramming factors complexed with magnetic nanoparticles (PolyMag) can also be used for generating iPSCs from human fibroblast cells. (TEM image of pDNA-Polymag complexes is also presented). (D) Cellular uptake assay of soluble Oct4 and Oct4 loaded CNPs towards human primary fibroblasts indicates CNPs can be utilized as delivery agent for reprogramming factors. The cells were stained with Oct4 antibody (α-Oct4; green) and Hoechst33258 for nuclear DNA (blue). (SEM image of CNPs loaded with protein is also presented). (E) Introduction of pDNA-loaded FMSNs to iPSCs resulted in differentiation to hepatocyte-like cells (TEM image of FMSNs is also presented). Reproduced with permission from ref. 37. Copyright (2013) American Chemical Society, from ref. 40. Copyright (2011) Elsevier, and from ref. 42. Copyright (2016) Impact Journals, LLC.
As a modified manner, nucleofection has been exploited for introduction of genes to cells. Nucleofection shares a physical basis with electroporation, but the main difference is that nucleofection involves complexation of genes with cell specific reagents prior to electroporation. It provides a great advantage when transfecting non-dividing cells or resting red blood cells and primary cells which are known to be difficult to transfect. Nucleofection was applied to deliver minicircle DNA that contained one cassette of four reprogramming factors (Oct4, Sox2, Lin28, Nanog) with the green fluorescent protein (GFP) reporter [32]. In between the gene encoding each protein, self-cleavage peptide 2A sequences were placed to separate each gene. This study demonstrated generation of transgene-free hiPSCs from adult human adipose stem cells with the advantage of higher transfection efficiency from nucleofection of minicircle DNA due to lower activation of exogenous silencing mechanisms. However, it had an overall transfection efficiency of ~0.005%, which is still significantly lower compared to that with viral approaches.
As described, both electroporation and nucleofection of genes for reprogramming show higher transfection efficiency compared to other approaches. However, cell damage caused by the process and potential cytotoxicity still remains problematic.
3.2. Nanoparticles
Nanoparticles made from a range of materials, including metals, polymers and lipids, have been extensively studied in fields of drug delivery, catalysts, diagnostics and therapeutics [33]. In particular, it has attracted much attention in the area of gene delivery, mainly due to its suitable size for cell penetration, large surface area for loading molecular cargo and protection of loaded substances from external threats [34, 35]. Nano-particulated forms of gene or protein carriers have been applied for cell reprogramming to achieve stable gene expression from the delivered reprogramming factors and thus improve reprogramming efficiency.
One example is mesoporous silica nanoparticles (MSNs) which have the advantages of tunable charge, low cytotoxicity and enhanced delivery efficiency [36]. Furthermore, MSNs are known to have a highly porous internal structure, which provides large surface area for loading high amounts of molecular cargo within the nanoparticle. MSNs were advantageously used for the rapid differentiation of miPSCs to definitive lineage cells (Figure 4D) [37]. To be specific, non-viral vectors encoding hepatocyte nuclear factor 3β (pHNF3β) were electrostatically adsorbed to positively charged FITC-labeled MSNs (FMSNs). By delivering a FMSN-pHNF3β complex to miPSCs, it was confirmed that the complex could improve miPSC differentiation toward hepatocyte-like lineage with mature liver function. Moreover, it has been demonstrated that FMSNs could be used for labeling miPSCs without affecting pluripotency of the cells.
Calcium phosphate nanoparticles (CaP-NPs) were also adapted for Yamanaka factors to human umbilical cord mesenchymal stem cells (HUMSCs) [38]. In this study, 20–50 nm sized pOSKM (plasmid DNAs encoding Yamanaka factors) loaded CaP-NPs were synthesized by forming ionic complexes via co-precipitation of calcium chloride and disodium hydrogen phosphate. This synthesis was in the presence of pOSKM within water-in-oil microemulsions. The benefits of calcium phosphates are their material properties, including good biodegradability and biocompatibility. This system resulted in reprogramming efficiencies of 0.049%, which was higher than the results from previous studies.
Magnetic nanoparticles were also employed for nanofection of iPS genes, which involved a magnetic field to promote transfection (Figure 4C). This approach, termed ‘magnetofection’, has been utilized for non-viral gene delivery with high efficiency for over a decade [39]. For example, magnetic nanoparticles made from biodegradable iron oxide with an overall size of 10–20 nm were coated with a cationic polymer, polyethylenimine (PEI), prior to forming complexes with plasmid DNA for reprogramming factors [40]. Then, by applying a magnetic field to the cells, they are allowed to rapidly get in contact with the complexes. Since the magnetofection process significantly increased the local concentration of delivering genes on the cell membrane, high transfection efficiency along with high reprogramming efficiency in MEFs could be achieved in this study, suggesting this system may serve as a potential efficient reprogramming tool in future.
For the delivery of reprogramming factors in nanoparticle form, protein-encapsulated cationic bolaamphiphile sub-micron sized particles were also introduced for generating hiPSCs [41]. Cationic 1,12-aminododecane based bolaamphiphile has a central hydrophobic core for formation of stable structures by hydrophobic interaction between carrier and protein cargo. In addition, the bolaamphiphile has a primary amine for facilitating electrostatic interactions and hydrogen bonding between carrier and protein, and secondary amine for enhancing endosomal escape of the complexes. This study demonstrated hiPSC generation from human foreskin fibroblasts by delivering a protein-bolaamphiphile complex bearing four reprogramming factors (KLF4, Nanog, NR5A2 and Sox2). The colonies appeared between 15 and 20 days with a reprogramming efficiency of 0.05% which is similar to that of viral reprogramming approaches. Although the mechanism of releasing protein cargo from the carrier is yet to be elucidated, this approach has shown potential to be utilized in hiPSC generation.
More recently, chitosan nanoparticles (CNPs) were employed to deliver recombinant Oct4 to human fibroblasts, demonstrating a potential of CNPs to be a promising tool for the generation of transgene-free hiPSCs (Figure 4E) [42]. A nanoparticle formulation provides protection for the loaded recombinant Oct4, while naked recombinant Oct4 is known to become rapidly degraded in cell culture conditions. Furthermore, it was shown that Oct4 encapsulated in CNPs efficiently entered fibroblasts without compromising its biological function, while soluble Oct4 protein was unable to enter the cells. While these protein induced pluripotent stem cells (piPSCs) are expected to be safer compared to iPSCs using viral vectors, a further development of piPSCs is limited due to its poor reproducibility [15].
Another example is a polyethylene glycol (PEG)-conjugated poly(amidoamine) (PAMAM) dendrimer conjugated gene delivery system [43]. The dendrimers were conjugated with the RGD peptide for facilitating cell adhesion and subsequent entry, as well as with pDNA encoding human bone morphogenetic protein-2 (hBMP-2) and reporters (plasmid DNA carrying both the enhanced green fluorescent protein and the luciferase; pEGFPLuc), resulting in nanoparticles with overall size of few hundred nanometers. In the assembly process, gold nanoparticles (AuNPs) with the size of 2 nm were also entrapped within the nanoparticles, providing the AuNP-entrapped dendrimer carrier with an improved biocompatibility due to reduced dendrimer amine functionality and potential use for X-ray imaging or photothermal therapy. Using this dendrimer complex for gene delivery, the expression of hBMP-2 in human mesenchymal stem cells (hMSCs) was achieved, which could then lead to hMSC differentiation down an osteoblastic lineage.
Functionalized AuNPs have been employed to study cellular responses on osteogenesis of bone marrow-derived hMSCs recently [44]. This study found that AuNPs functionalize with carboxyl group induced up-regulation of growth factors FGF-2 and TGF-β in hMSCs, which can lead to cell proliferation and bone expansion.
Moreover, another study reported that magnetic core-shell nanoparticles conjugated with multiple targeting genes and gene repression molecules could be utilized for gene repression in stem cells, to initiate enhanced differentiation of neural stem cells into functional neurons [45]. Although these studies were not for generating iPSCs or inducing stem cell differentiation, gaining knowledge of how different nanomaterials affect cells is expected to accelerate development of technology for stem cell reprogramming and regenerative medicine in the future.
Reprogramming with nanoparticle-based approaches allows compact packing of genes to deliver and provides the genes with extra functionalities such as targeting ability. Although nanoparticles-based reprogramming approach is still not a general method applied as much as it has been for other therapeutic and clinical purposes, it is expected to be developed further with the help of advances in synthetic approaches of nanoparticles and further understanding of nanoparticles in reprogramming.
3.3. Nucleic acid-based complexation
To introduce cells with foreign genes, liposome-based approaches have been applied in the field of gene delivery. There are quite a few liposomal carriers that are commercially available, and they are recognized as a safe carrier to the cell when compared to viral carriers. Lipofection is carried out by fusion with cell membrane made up of phospholipid bilayer to deliver molecular cargo to the cells (Figure 4B). For instance, cationic lipid-mediated introduction of mRNAs encoding Yamanaka factors was reported in 2010 and 2011 [46, 47]. A proof-of-concept study determined that fibroblasts converted to less mature cardiomyocytes via direct reprogramming using lipid based miRNA methods as well [48]. This is particularly beneficial to heart regeneration and proves to independently influence cell reprogramming.
Due to the negatively charged phosphate backbone of nucleic acids, cationic polymers are widely used for synthesizing polyplexes for efficient gene delivery. Polyplexes can protect the cargo from nuclease-mediated degradation while having small sizes for entering target cells. Moreover, a polymer-based gene delivery system is less harmful to the recipient cell, compared to viral gene delivery [49]. In addition, positively charged polymers are believed to induce a “proton sponge effect”, which causes a burst release of the cargo in the endosome, therefore enhancing the efficiency of gene delivery. Although there are still issues regarding biocompatibility of some polymers used to fabricate polyplexes, it is considered an effective strategy for condensing genes and effectively delivering genes to cells for cell reprogramming.
Plasmids encoding Oct4 and SirT1 complexed with cationic polyurethanes-short branch PEI (PU-PEI) were used for overexpression of genes in aging cells [50]. In this study, it was hypothesized that upregulation of these two factors may attribute to bring aged retinal pigmented epithelium (aRPE) cells back into a rejuvenated state. It was found that overexpression of Oct4 and SirT1 in aRPE cells was achieved by utilizing PU-PEI as non-cytotoxic carrier with high transfection efficiency. The two overexpressed factors enhanced antioxidant enzymatic activities of the cells, and the cells were reprogrammed into retinal progenitor-like cells without being in iPSC-like states.
Polyketal copolymer PK3 was used to complex with mixtures of microRNA:DOTAP ion-pairs to induce pluripotency from mouse bone marrow-derived mononuclear cells (BM-MNCs) [51]. PK3 remains stable at pH 7.4 for several weeks, while it is rapidly degraded at pH 4.5. Therefore, miRNA payloads could be protected from serum nuclease-mediated digestion at neutral pH, then miRNA:DOTAP ion-pairs could be released from the complex in endosomes due to rapid hydrolysis of PK3 at endosomal pH. This approach has been applied for other gene delivery systems like siRNA delivery to macrophages as well [52].
More recently, positively charged Pleurotus eryngii polysaccharides (CPEPS) were employed for delivering plasmid DNA encoding two of the Yamanaka factors (Oct4 and Sox2) with miR302-367 to replace oncogenic Klf4 and c-Myc [53]. The microRNA and CPEPS were self-assembled by electrostatic interaction, resulting in 40 to 100 nm sized nanoparticles. By using naturally occurring polysaccharide with biodegradability and biocompatibility, the nanoparticles showed lower cytotoxicity compared to commercially available liposomal carrier, Lipofectamin2000, while successfully carrying out hiPSC generation from HUMSCs with high reprogramming efficiency (0.044%).
As exemplified above, lipoplexes and polyplexes have been facilitated for efficient non-viral gene delivery to induce pluripotency or differentiation. By selecting an appropriate kind of lipid or polymer for complexation of genes, the encapsulated genes can be provided with extrinsic properties, including physical, chemical and physicochemical properties, which could lead to boosted gene delivery and reprogramming efficiency.
4. Biophysical regulation of cell reprogramming
Mechanisms of extracellular matrix (ECM) manipulation on cell fate have been used as a means to investigate the role that substrates can play on cell reprogramming and differentiation. Biomaterials similar to the in vivo ECM in stiffness, topography, or material may guide specified differentiation of iPSCs [54, 55]. For example, the discovery that naturally occurring ECM molecules like collagen can influence differentiation through their presence and degradation has led to studies of biomaterial influences on cell lineage [56]. This is based on substrates sequestering and activating signals with cells, highlighting the effects of topographical cues in the substrate. Developing engineered substrates that manipulate both intrinsic and extrinsic factors such as cell shape and intracellular pathways provide a controlled environment for tissue engineering and regenerative medicine [55, 57, 58, 59].
There are many different approaches to be taken when incorporating biomaterials in cell structures that affect the intricate signaling pathways cells induce [60]. Examples include using proteins like collagen or laminin in conjunction with polymers like agarose and alginate to promote the xenogeneic proliferation of stem cells. Synthetic polymers, on the other hand, can have highly controlled properties and be easily screened for biomimetic behavior. However, they are lacking in biological activity and cell control [61]. Synthetic biomaterials like graphene promote fibroblast reprogramming into a pluripotent and proliferative state through activation of mesenchymal to epithelial transition epigenetic changes. This property enhanced the ability to reprogram cells virally from fibroblasts to iPSCs [62].
Given the cues that microenvironments provide to cells, it should be expected that the combination of biochemical and biophysical changes provides a synergistic effect on proliferation and programming (Figure 5). Studies using PEG hydrogels with Yamanaka factors [63] or nanotopography with biofunctional growth factors [64] are proving that cell reprogramming can be enhanced via these engineered cues. In this section, factors such as substrate topography, stiffness, and stretching that have potential for more efficient cell reprogramming are discussed.
Figure 5.
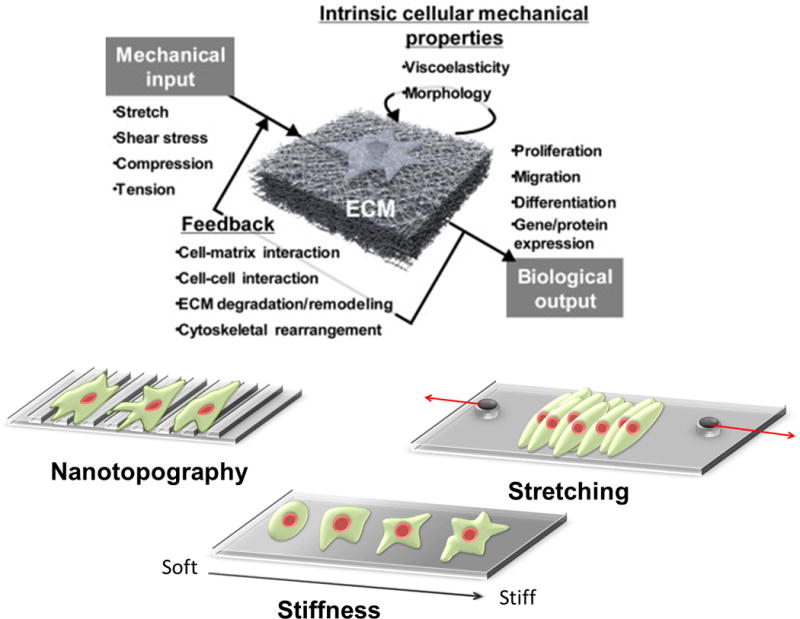
Mechanotransduction is the process by which cells convert mechanical inputs into biological responses. Mechanotransduction often involves a feedback process, and their mechanical environment is dynamic and complex. Biophysical factors that induce these responses include nanotopography, stiffness, and stretching. Modified with permission from ref. 84. Copyright (2017) Elsevier.
4.1 Micro/nanotopography
The major effects of topography on cells are cell adhesion, proliferation, morphological changes, migration, and organization, which ultimately helps determine cell fate [65]. The ranges of topographical cues investigated span from the micro to the nano scale. Initially, studies focused on microtopography as a means of reprogramming and differentiation, but nanotopography has garnered more attention in recent years due to their increased similarities to in vivo ECM structure.
On the micro scale, one study used both microgratings and microposts to investigate effects on cell reprogramming of MEFs to induced neurons (iN) using overexpression of transcription factors (Figure 6B) [66]. These designs both promoted iN outgrowth, as well as neuronal gene expression and regulation compared to flat substrates. Neurites showed a difference in cell shape, furthering the claim that controlled niches and cell shapes are coupled in defining cell pathways. Microtopographic scaffolds using electrospun fibers have been used as another platform for mouse neuronal reprogramming and development [67]. The three dimensional thick fibre neuronal scaffolds improved iN growth onto hippocampal brain slices. The emphasis of this design is that it also focused on subtype specific neurons working together in a 3D microniche, suggesting that the scaffold promoted specificity and biomimicry. These studies move closer to controlling niches for more efficient cell reprogramming into a neuronal lineage by actively modeling in vivo conditions.
Figure 6.
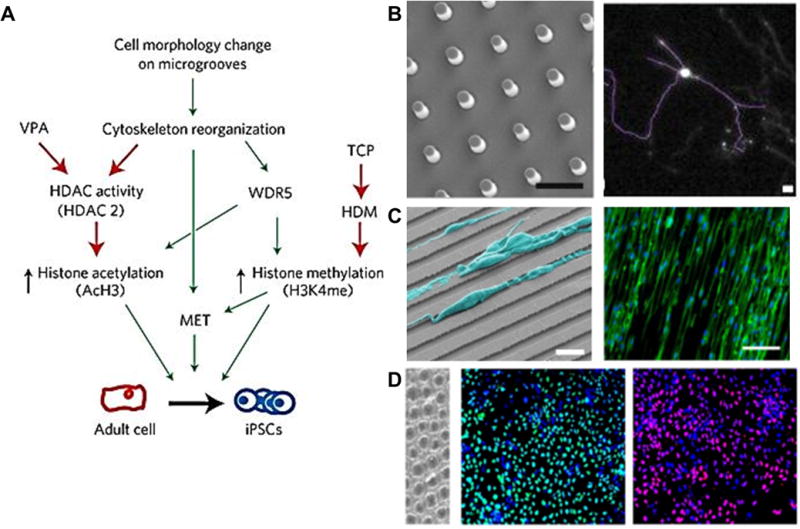
Topographical cues acting as micro-environments for cell adhesion and in vivo mimicry from ref. 66, 69, 71, and 74. (A) A summary of microtopographical regulation of histone modifications and cell reprogramming in adult mouse fibroblasts. HDM, histone demethylase. (B) Images of substrate topographies with micron-sized features fabricated by micro-imprinting. Scale bar 5 μm. Neurites of induced neurons on micropost substrate display more branching. Scale bar 100 μm. (C) SEM image of cardiac stem cells aligned on grooves (Bar: 20 microns), immunofluorescent imaging showing overlaid alignment of cardiac stem cells on grooved surface stained for cytoskeletal actin (Phallodin, green) and nuclei (DAPI, blue). (D) Immunofluorescence image of human ES-cell-derived endoderm differentiated by stimulating cells with growth factors on nanopit surfaces and stained against markers FOXA2 and SOX17. Scale bar: 100 μm. Reproduced with permission from ref. 71. Copyright (2013) Nature Materials. Reproduced with permission from ref. 66. Copyright (2014) Elsevier. Reproduced with permission from ref. 69. Copyright (2015) Elsevier. Reproduced with permission from ref. 74. Copyright (2016) American Chemical Society.
Various micropattern topographies have been explored on multiple cell types based on respective cell niches monitored in vivo. Human adult renal progenitor/stem cells (ARPCs) have been differentiated into a tubular form using micropatterns [68]. Micropatterns containing ECM proteins (i.e. fibronectin) were designed and ARPCs were interfaced on these structures to promote differentiation without the use of exogenous chemicals. The results showed viable differentiated cells that lasted up to 20 days. An important factor of this study is that the micropattern was able to control cell shape via elongation. As the surfaces decreased patterning width (50 μm to 5 μm), cells also began to lose their stemness, suggesting that stricter topographies promote differentiation.
Another example using parallel microgrooves enhanced mouse cardiac progenitor cells into cardiomyocyte like cells (Figure 6C) [69]. Differentiation was recognized through biochemical factors, and proved that viral induction of stem cells combined with the anisotropy of microgrooves provided a more organized, biomimetic atmosphere. This excludes the intervention of chemical inducers to create a simpler method for more efficient cell manipulation.
A more recent study goes further by reprogramming fibroblasts to cardiomyocytes on microgrooves while exploring other biophysical cues. A new mechanism for controlled cell reprogramming in which Mkl1 activity was regulated on microgrooves was introduced. This regulation and the presence of a sarcomeric structure were only partially shown with a biochemical approach compared to the microgrooves [70]. Microtopography promotes a mesenchymal-to-epithelial transition in adult mouse fibroblasts as well. Microgrooves have been observed to affect histone acetylation activity, suggesting that changes in cell morphology play a major role in epigenetic states (Figure 6A) [71].
Shrinking to the nanotopographical cue has led to more interesting results such as being used for disease modeling and drug screening. Through development of nanotopographical substrates, a novel disease model for Duchenne muscular dystrophy has been discovered [72, 73]. By using patient specific hiPSCs, this particular model can more accurately test disease phenotypes without the effort of in vivo testing. Using nanotopographical substrates as a biomimetic tool, distinctions in disease models and cell differentiation can more effectively be researched.
Furthermore, pancreatic differentiation and regulation of pancreatic transcriptional factors has been demonstrated from both hESCs and hiPSCs using 200 nm nanopore patterned surfaces (Figure 6D) [74]. Pancreatic endocrine cells that produce insulin and other chemicals were enhanced in the presence of biochemical factors. By providing a specific biochemical and biophysical environment through nanotopography, these endocrine cells were more easily controlled and thus could be differentiated with more efficiency.
Anisotropy has become a pinnacle part of differentiating cells as it provides an even more accurate representation of the cell niche in vivo. Being able to control anisotropy spatio-temporally has been a goal for some experiments [75]. Anisotropy and its relation to cell behavior have been investigated through cardiac pathways for stem cells. Substrate nanogroove width has been optimized to the 700–1000 nm range and combined with a coated peptide which further influenced human cardiomyocyte development [76]. Such substrates could be optimized for specific cells and their appropriate niches and thus push the boundary of drug screening, disease modeling, or other physiologically relevant studies.
Investigation into the cytoskeletal behavior and mechanical properties of hMSCs on different substrates with nanotopography has proven interdependence with stiffness, however. This suggests that two different substrates could change the mechanics of an attaching cell and its subsequent signaling pathways [77]. Thus, it becomes important to consider material mechanical properties and not just biocompatibility.
Comparing micro to nanopattern designs in recent research suggests favorability towards the latter [78], yet there are studies that try to implement both these patterns into a single gradient mold [79, 80]. As previously mentioned, nanotopography does well at aligning cells along its specific pattern, more so than microtopography. Mouse fibroblasts have been shown to translate into neurons more effectively using nanogrooves compared to microgrooves [78]. Synthetic 350 nm nanogratings increase proliferation and differentiation of hMSCs into a neuronal lineage. By promoting elongation around the grating axis, intracellular neuronal markers were subsequently upregulated and further enhance with exposure to biochemical cues. The emphasis here is that extracellular cues can synergistically promote reprogramming of cells to controlled phenotypes [81]. This type of research has shown the possibility for different approaches to activate cellular pathways by enabling proper cell organization and exposure to bioactive surface molecules that can enhance differentiation. By tuning nanotopographical cues, cell behavior can be studied in a dynamic medium that further represents the matrix in which these cells could be applied [82, 83, 84].
4.2 Matrix stiffness
Some studies have coupled surface chemistry and stiffness together as one factor for cell differentiation, but groups had begun to realize the significance of stiffness as its own variable for cell niches. One group sought to reconcile challenges between stiffness and surface chemistry. Their results suggested that when stem cells were exposed to PEG hydrogels with arginine-glycine-aspartate (RGD) peptide nanoarrays on the surface, that both factors equally and independently played a role in cell differentiation [85]. The PEG stiffness could be correlated to the focal adhesion of cells to substrates. Stiffness promotes cell tension and thus activation of chemical pathways that lead to reprogramming into appropriate phenotypes. Softer substrates as low as 0.1 kPa made of polyacrylamide hydrogel have been reported to promote iPSC induction using mesenchymal-to-epithelial activation or regulation of stemness factors [86, 87]. This proves that stiffness control can help to control iPSC production for future applications.
Matrix elasticity is thus directly correlated with stem cell differentiation. In particular, as substrates get stiffer (up to 100 kPa), the closer they are to osteogenic differentiation [88]. Softer matrices promote cell roundness and increasing stiffness develops branching, spindle, and polygonal shapes linked with other cell phenotypes [89]. Depending on substrate material, stiffness thresholds will overlap and thus differentiation into two different phenotypes will occur without the presence of other biochemical factors. For example, adipocytes reside in tissues around 1–10 kPa [88] which overlaps with that of cardiac muscles [90]. This knowledge has been used to create hydrogels seeded with ECM proteins (fibronectin and collagen) and tunable stiffness to direct stem cells towards specific differentiation pathways [91]. Tuned stiffness in conjunction with the appropriate coated proteins on substrates signals cells to the desired phenotypes. Investigations of cardiomyocytes derived from ESCs on stiffness ranging from 1 to 50 kPa emphasize that increasing stiffness spread cells and promotes stress fiber formation [92]. This effect influences the idea that anisotropy plays a major role in cell fate, much like research pertaining to topography.
Research has not been limited to synthetic polymer gels. A soft collagen gel substrate displayed hard substrate characteristics in cells [93]. Fibroblasts exhibited morphology and cytoskeletal cues like spreading and monolayer organization. A collagen gel is reminiscent of ECM conditions in that collagen is one of the representative proteins existent in vivo. However, some synthetic polymers like a co-polymer system of n-octyl methacrylate crosslinked with diethylene glycol dimethacrylate (DEGDMA/nOM) prove ideal for differentiation studies looking for independent effects of substrate stiffness without the inconsistency of surface chemistry [94]. This co-polymer system elucidated the mechanosensitivity of cells to their substrates by revealing that after a certain stiffness threshold, gene expression does not increase but rather re-organizes corresponding to the formation of osteoblastic markers which are known to associate with higher stiffness. Since surface chemistry is often coupled with substrate stiffness, careful examination into consistent surface chemistry provides clearer insights into substrate reprogramming and differentiating efficiency.
Expanding to varying stiffness is the next step to discovering how cells adapt to their surroundings. Creating stiffness gradients and applying hMSCs to this stimuli shows preferential migration to stiffer substrates [95]. Stem cells will retain plasticity during this durotaxis phenomenon. The challenge here is relating the soft hydrogel substrates often used in vitro with the fibrillary nature of in vivo ECM. The importance of aligned tissue has been established in research pertaining to topographical cues and thus coupling knowledge in stiffness gradients and topographical cues could promote a better understanding of in vivo processes.
New mechanisms for cell regulation involving substrate stiffness have recently been discovered and prove that cytoskeletal activity is only part of the equation. Substrate stiffness dictates hMSC differentiation through the MIF-mediated AKT/YAP/RUNX2 pathway. Once a stem cell contacts a stiffer substrate, intracellular migration inhibitory factor (MIF) increases and starts a pathway towards AKT/YAP signaling inside the nucleus which promotes osteogenic development (Figure 7) [96]. Other regulation mechanisms such as the p190RhoGAP pathway have been characterized proving the intricacies and intertwining intracellular signaling that occurs once a cell comes into contact with a substrate with certain rigidity. This particular pathway activates cell-cell communications and cell-substrate signaling simultaneously to adjust cell formation and proliferation to a differentiated phenotype [97]. Linking these two mechanisms illustrates the growing complexity of cell signaling required to adjust to microenvironments. Thus, being able to tune these matrices for highly controlled reprogramming and subsequent differentiation becomes a bigger challenge for future studies.
Figure 7.

Substrate stiffness activates intracellular pathways and cytoskeletal organization to influence cell behavior. (A) Confocal laser scanning microscope images of immunofluorescent-labeled YAP (green) and DAPI-labeled nuclei (blue) in hMSCs cultured on fibrous substrates with (75PLLA) and without (PLLA) annealing. Arrows indicate the location of cell nuclei. Scale bar = 200 μm. (B) Illustration of an identified molecular mechanism elucidating how substrate stiffness directs hMSCs toward the bone-specific lineage. Reproduced with permission from ref. 96. Copyright (2016) Elsevier.
4.3 Mechanical stretching
Matrix mechanical stress has been proposed as another marker for biomimicry. Stem cells need to differentiate into correct phenotypes under stressed conditions to actively replace damaged cells or create the tissue structure as a whole. Since particular phenotypes experience mechanical stress as part of their natural environment, mimicking that attribute has become an investigation for stem cell differentiation. Most studies investigating mechanical stress focus on specific phenotypes such as cardiomyocytes and osteoblasts [98, 99, 100, 101].
Looking at vascular smooth muscle growth and maturation, mechanical stress has induced activation of these markers from hMSCs [102]. Other studies have linked induced stress to spontaneous ligament directed differentiation [100]. The direction of stretch contributes to cell behavior as well. Comparing uniaxial to equiaxial or radial stress on hMSCs suggests that cell orientation and subsequent cell signaling became more important when stress was uniaxial [99]. These results concluded that uniaxial strain is better suited for differentiation towards smooth muscle cells; pending cell orientation can be controlled. Also, MSCs can sense the subtle difference between different types of mechanical loading and respond accordingly. Overall, no matter the amount of mechanical loading, hMSCs will lose their stemness and gain the factors towards osteoblastic or tendon/ligament differentiation [98, 101]. The cell orientation will convert to a direction perpendicular to stress orientation, providing a tissue response that seeks to balance the stress it is exposed to [94]. For example, a recent study developed a probe to investigate cell actin stress and its role in cell reprogramming and differentiation, which could elucidate how cellular mechanics can influence cell fate [103]. By measuring actin mechanics directly during reprogramming experiments, the correlation between environmental cues and intracellular signaling can be justified and more controlled in future research.
One aspect of cell differentiation in response to mechanical cues in the environment that is paramount when inducing stress is the release of ECM producing factors and biochemical signals [102]. Mechanical stress enhances ECM components to be developed and thus contributes to tissue construction and inherent cell differentiation in the process. By producing ECM components, cells change their environment, and in contrast, are influenced by their environment to do such an action. Xu et al proposes a signal network involving chemicals associated with recognizing other environmental indicators. They examined the cytoskeletal organization of hMSCs and their relation to the Rho/ROCK pathway linked with the intracellular chemical FAK. By inhibiting each of these processes, this network subsequently could not be seen in vitro, proving that this signaling network is the key component to cell differentiation via environmental change [104]. Other studies suggest that mechanical stretching can even regenerate skin tissue [105]. Stretching could upregulate gene expression related to cell proliferation, vascularization, etc. This idea highlights the important regenerative properties of stem cells and how that could be advantageous.
Depending on in vitro conditions, mechanical loading can take precedence as a factor of cell manipulation. One study tested the threshold in which mechanical loads can change morphology and direction of 3T3 cells grown using nanotopographical cues [106]. Cells pre-aligned by micro/nanostructures could be altered by cyclic in-plane strain, regardless of the structure size. However, this time dependent study also found that over time and depletion of mechanical stress, cells would re-orient themselves. This suggests that mechanical stress plays an active role only during application and after effects can be mitigated by other factors. Isolating mechanical stretching as a factor helps to understand the mechanisms by which cells behave. Yet, combining it with nanotopography synergistically enhances stem cell differentiation [107]. In converting mouse fibroblasts to a pluripotent state, mechanical stretching not only re-oriented cells perpendicularly as previous studies showed, but actually lowered reprogramming efficiency (Figure 8) [70]. This suggests that mechanical stretching can influence cytoskeletal response to microenvironments and cell fate.
Figure 8.

Mechanical stretching induces perpendicular alignment and reduction of reprogramming efficiency. (A) Transduced TTF with and without cyclical stretch fixed and stained on day 10. Scale bar denotes 50 μm. (B) Yield of GFP + cells, normalized by initial number of cells seeded, quantified on day 10 after stretching at various frequencies. (*p < 0.005, Bonferroni test, compared to static. n = 5.) Reproduced with permission from ref. 70. Copyright (2016) Elsevier.
Stimulation on cell-seeded tendon slices have shown promise in promoting tenogenic differentiation through upregulation of Type I collagen, decorin, and tenomodulin while maintaining proper strength and stiffness needed [101]. Mechanical stress becomes more valuable in cell phenotypes that endure those stresses in vivo. Optimizing stretching for that purpose proves to be effective in creating proper tissue constructs that replicate those environments.
The effects of this mechanism are restricted to two dimensional cultures and remain a challenge to exploit in three dimensional cultures. As some studies combine topography and stretching, tissue engineering research is one step closer to moving to broader constructs that invoke the proper anisotropy needed to be applied in vivo. Incorporating topography such as ridges that remain intact for long periods of time with mechanical stretching has produced more stable biomaterials that promote long-term tissue regeneration [109]. Much like stiffness gradients, strain gradients in three dimensional hydrogels align cells towards the direction of strain and confirm that focal adhesions affect cell behavior [108]. Gradient biomaterials can be used to manipulate cell behavior towards niches that actively represent specific ECM properties for reprogramming and differentiation. Previously mentioned topography [79, 80], stiffness [95], and strain [107] gradient biomaterials have shown that cells can be controlled using these active niches for reprogramming purposes.
5. Conclusions and perspectives
Manipulation of both biophysical cues and genetic makeup has profound effects on cell behavior and can be optimized for future medical applications. By creating biomaterial systems that deliver specific genetic markers and transcription factors while mitigating harmful effects, and exposing cells to biomaterial scaffolds that accurately represent in vivo conditions, controlled efficient cell reprogramming can be accomplished. Finding alternatives or additions to viral induction of somatic or stem cells has been the goal of recent reprogramming research that employs these biomaterial delivery systems, and successful routes have been discovered using plasmids and particles that can enter cells. Increasing differentiation and proliferation efficiency has been an opportunity that has been tackled by developing biomaterial scaffolds that are finely tuned to mimic in vivo conditions by using topographical, stiffness, and stretching cues. For biomaterial systems, electroporation and nucleofection can be effective methods for transfection of genes and better reprogramming compared to viral approaches, but it risks significant damage to cells in the process. Nanoparticle systems, however, is in its relative infancy for reprogramming purposes and requires optimization for reproducibility despite potential for targeted gene transfer and cell manipulation. Nucleic acid based complexes have been proven a safer carrier for genes compared to viruses and increased proficiency in cell reprogramming. Yet, some of these methods require intracellular manipulation of cell pathways that could be influenced by extracellular matrices. Developing biomaterial scaffolds with tunable nanotopography, stiffness, and stress can provide this route of cell reprogramming without directly changing the cell genome. These methods still rely on viral approaches of factor introduction to somatic and stem cells however and require more control.
Future research will look to further explore the pathways associated with these biomaterial systems and their effect on cell behavior, and optimize the efficiency of cell reprogramming by synergistically combining delivery and matrix systems. In order to provide better outcomes in biomedical research via more efficient patient specific treatments, being able to control cells on demand is required. The major milestones for achieving this outcome include elucidating the requirements of chemical signaling necessary to optimize the efficiency of reprogramming to pluripotency from a somatic state, developing a dynamic or gradient biomaterial platform that adapts to cell behavior, incorporating biochemical and biophysical cues synergistically to yield specific tissue phenotypes with precision, and exploring the effects of three dimensional cell niches and their possible enhancement of reprogramming [110]. Recognizing what cues and factors take priority in how cells react and then being able to tune these accordingly would provide more cell reprogramming control and specificity. The current challenge is to develop niche-mimicking biomaterials as a system where stem cells will be activated either manually or autonomously to a specific cell phenotype on demand [111]. Therefore, using all the known microenvironmental factors to engineer better cell niches will prove to be advantageous.
Acknowledgments
This work was supported by the National Institute of Health R21EB02013201 and R01NS09438801 (to D.-H.K.). This work was also supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea (NRF) funded by the Korean Government (NRF-2016M3A9C6917402).
Biographies
Deok-Ho Kim is currently a Professor in the Department of Bioengineering at the University of Washington. He received his PhD in Biomedical Engineering from The Johns Hopkins University. His current research interests cover multiscale biomimetic materials/devices, functional tissue engineering, microscale stem/tumor cell niche engineering, and cell mechanobiology. He has published over 140 peer reviewed journal articles and referenced conference proceedings, book chapters and patent applications, and given over 100 national and international invited lectures.
Jong Bum Lee is currently a Professor in the Department of Chemical Engineering at the University of Seoul. He received his Ph.D. in Biological and Environmental Engineering from the Cornell University. His current research interests cover multiscale biomaterials engineering, nucleic acid engineering and theranostics. He has published over 50 peer-reviewed journal articles, book chapters and patents, and given over 100 national and international invited lectures.
References
- 1.Rippon HJ, Bishop AE. Cell Proliferation. 2004;37:23–34. doi: 10.1111/j.1365-2184.2004.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Jiang W. BioScience. 2015;65:468–475. [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Nishikawa SI, Goldstein RA, Nierras CR. Nat Rev Mol Cell Biol. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- 6.Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 7.Tong Z, Solanki A, Hamilos A, Levy O, Wen K, Yin X, Karp JM. EMBO J. 2015;34:987–1008. doi: 10.15252/embj.201490756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neuss S, Apel C, Buttler P, Denecke B, Dhanasingh A, Ding X, Grafahrend D, Groger A, Hemmrich K, Herr A, Jahnen-Dechent W, Mastitskaya S, Perez-Bouza A, Rosewick S, Salber J, Wöltje M, Zenke M. Biomaterials. 2008;29:302–313. doi: 10.1016/j.biomaterials.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Martino S, D’angelo F, Armentano I, Kenny JM, Orlacchio A. Biotechnol Adv. 2012;30:338–351. doi: 10.1016/j.biotechadv.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Wu SM, Hochedlinger K. Nat Cell Biol. 2011;13:734–734. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry FP, Murphy J. Int J Biochem Cell Bio. 2004;36:568–584. doi: 10.1016/j.biocel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Jopling C, Boue S, Belmonte JCI. Nat Rev Mol Cell Bio. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 14.Tanabe S. World J Stem Cells. 2015;7:992–998. doi: 10.4252/wjsc.v7.i7.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi A, Ling QD, Kumar SS, Munusamy MA, Alarfaj AA, Chang Y, Kao SH, Lin KC, Wang HC, Umezawa A. Lab Invest. 2015;95:26–42. doi: 10.1038/labinvest.2014.132. [DOI] [PubMed] [Google Scholar]
- 16.Hussein SMI, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL, Mosbergen R, Li M, Lee DS, Cloonan N, Wood DLA, Munoz J, Middleton R, Korn O, Patel HR, White CA, Shin JY, Gauthier ME, Cao KAL, Kim JI, Mar JC, Shakiba N, Ritchie W, Rasko JEJ, Grimmond SM, Zandstra PW, Wells CA, Preiss T, Seo JS, Heck AJR, Rogers IM, Nagy A. Nature. 2015;523:626–626. doi: 10.1038/nature14606. [DOI] [PubMed] [Google Scholar]
- 17.O’doherty R, Greiser U, Wang W. BioMed Res Int. 2013;2013:1–6. doi: 10.1155/2013/705902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JX, Krane M, Deutsch MA, Wang L, Rav-Acha M, Gregoire S, Engels MC, Rajarajan K, Karra R, Abel ED, Wu JC, Milan D, Wu SM. Circ Res. 2012;111:50–55. doi: 10.1161/CIRCRESAHA.112.270264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Nat Cell Bio. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 21.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, Bassel-Duby R, Olson EN. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su G, Zhao Y, Wei J, Han J, Chen L, Xiao Z, Chen B, Dai J. Biomaterials. 2013;34:3215–3222. doi: 10.1016/j.biomaterials.2013.01.044. [DOI] [PubMed] [Google Scholar]
- 23.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 24.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Belmonte JCI. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 25.Tsai SY, Clavel C, Kim S, Ang YS, Grisanti L, Lee DF, Kelley K, Rendl M. Stem Cells. 2010;28:221–228. doi: 10.1002/stem.281. [DOI] [PubMed] [Google Scholar]
- 26.Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, Nishikawa SI. Proc Natl Acad Sci USA. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee ES, Kim SHL, Lee H, Hwang NS. J Biomed Mater Res Part B. 2016;104:686–697. doi: 10.1002/jbm.b.33601. [DOI] [PubMed] [Google Scholar]
- 28.Deng XY, Wang H, Wang T, Fang XT, Zou LL, Li ZY, Liu CB. Curr Stem Cell Res Ther. 2015;10:153–158. doi: 10.2174/1574888X09666140923101914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarmush ML, Golberg A, Serša G, Kotnik T, Miklavčič D. Annu Rev Biomed Eng. 2014;16:295–320. doi: 10.1146/annurev-bioeng-071813-104622. [DOI] [PubMed] [Google Scholar]
- 30.Plews JR, Li JL, Jones M, Moore HD, Andrews PW, Na J. PLoS One. 2010;5:e14397. doi: 10.1371/journal.pone.0014397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drews K, Tavernier G, Demeester J, Lehrach H, Smedt SCD, Rejman J, Adjaye J. Biomaterals. 2012;33:4059–4068. doi: 10.1016/j.biomaterials.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 32.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li A, Robbins RC, Kay MA, Longaker MT, Wu JC. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petros RA, DeSimone JM. Nat Rev Drug Discovery. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 34.Muthiah M, Park IK, Cho CS. Expert Opin Drug Delivery. 2013;10:1259–1273. doi: 10.1517/17425247.2013.798640. [DOI] [PubMed] [Google Scholar]
- 35.Williford JM, Wu J, Ren Y, Archang MM, Leong KW, Mao MQ. Ann Biomed Eng. 2014;16:347–370. doi: 10.1146/annurev-bioeng-071813-105119. [DOI] [PubMed] [Google Scholar]
- 36.Mamaeva V, Sahlgren C, Lindén M. Adv Drug Delivery Rev. 2013;65:689–702. doi: 10.1016/j.addr.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Tasi PH, Hung Y, Chiou SH, Mou CY. ACS Nano. 2013;7:8423–8440. doi: 10.1021/nn401418n. [DOI] [PubMed] [Google Scholar]
- 38.Cao X, Deng W, Qu R, Yu Q, Li J, Yang Y, Cao Y, Gao X, Xu X, Yu J. Adv Funct Mater. 2013;23:5403–5411. [Google Scholar]
- 39.Scherer F, Anton M, Schillinger U, Henke J, Bergemann C, Kru¨ger A, Gänsbacher B, Plank C. Gene Ther. 2002;9:102–109. doi: 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- 40.Lee CH, Kim JH, Lee HJ, Jeon K, Lim HJ, Choi HY, Lee ER, Park SH, Park JY, Hong SH, Kim SH, Cho SG. Biomaterials. 2011;32:6683–6691. doi: 10.1016/j.biomaterials.2011.05.070. [DOI] [PubMed] [Google Scholar]
- 41.Khan M, Narayanan K, Lu HF, Choo Y, Du C, Wiradharma N, Yang YY, Wan ACA. Biomaterials. 2013;34:5336–5343. doi: 10.1016/j.biomaterials.2013.03.072. [DOI] [PubMed] [Google Scholar]
- 42.Tammam S, Malak P, Correa D, Rothfuss O, Azzazy HM, Lampecht A, Schulze-Osthoff K. Oncotarget. 2016;7:37728–37739. doi: 10.18632/oncotarget.9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong L, Alves CS, Hou W, Qiu J, Möhwald H, Tomás H, Shi X. ACS Appl Mater Interfaces. 2015;7:4833–4843. doi: 10.1021/am508760w. [DOI] [PubMed] [Google Scholar]
- 44.Li JJ, Kawazoe N, Chen G. Biomaterials. 2015;54:226–236. doi: 10.1016/j.biomaterials.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Patel S, Chueng STD, Yin PT, Dardir K, Song Z, Pasquale N, Kwan K, Sugiyama H, Lee KB. Angew Chem Int Ed. 2015;54:11983–11988. doi: 10.1002/anie.201504902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavernier G, Wolfrum K, Demeester J, De Smedt SC, Adjaye J, Rejman J. Biomaterials. 2012;33:412–417. doi: 10.1016/j.biomaterials.2011.09.062. [DOI] [PubMed] [Google Scholar]
- 48.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 50.Peng CH, Cherng JY, Chiou GY, Chen YC, Chien CH, Kao CL, Chang YL, Chien Y, Chen LK, Liu JH, Chen SJ, Chiou SH. Biomaterials. 2011;32:9077–9088. doi: 10.1016/j.biomaterials.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 51.Sohn YD, Somasuntharam I, Che PL, Jayswal R, Murthy N, Davis ME, Yoon YS. Biomaterials. 2013;34:4235–4241. doi: 10.1016/j.biomaterials.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim H, Noh J, Kim Y, Kim H, Kim J, Khang G, Lee D. Biomacromolecules. 2013;14:240–247. doi: 10.1021/bm301669e. [DOI] [PubMed] [Google Scholar]
- 53.Deng W, Cao X, Chen J, Zhang Z, Yu Q, Wang Y, Shao G, Zhou J, Gao X, Yu J, Xu X. ACS Appl Mater Interfaces. 2015;7:18957–18966. doi: 10.1021/acsami.5b06768. [DOI] [PubMed] [Google Scholar]
- 54.Wasik AM, Grabarek J, Pantovic A, Cieślar-Pobuda A, Asgari HR, Bundgaard-Nielsen C, Rafat M, Dixon IM, Ghavami S, Łos MJ. Int Rev Cell Mol Biol. 2014:167–203. doi: 10.1016/B978-0-12-800097-7.00005-1. [DOI] [PubMed] [Google Scholar]
- 55.Higuchi A, Ling Q-D, Chang Y, Hsu S-T, Umezawa A. Chem Rev. 2013;113:3297–3328. doi: 10.1021/cr300426x. [DOI] [PubMed] [Google Scholar]
- 56.Murphy WL, Mcdevitt TC, Engler AJ. Nat Mater. 2014;13:756–756. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higuchi A, Ling QD, Kumar SS, Chang Y, Alarfaj AA, Munusamy MA, Murugan K, Hsu ST, Umezawa A. J Mater Chem B. 2015;3:8032–8058. doi: 10.1039/c5tb01276g. [DOI] [PubMed] [Google Scholar]
- 59.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higuchi A, Kumar SS, Ling QD, Alarfaj AA, Munusamy MA, Murugan K, Hsu ST, Benelli G, Umezawa A. Prog Polym Sci. 2016 [Google Scholar]
- 61.Prewitz M, Seib FP, Pompe T, Werner C. Macromol Rapid Commun. 2012;33:1420–1431. doi: 10.1002/marc.201200382. [DOI] [PubMed] [Google Scholar]
- 62.Yoo J, Kim J, Baek S, Park Y, Im H, Kim J. Biomaterials. 2014;35:8321–8329. doi: 10.1016/j.biomaterials.2014.05.096. [DOI] [PubMed] [Google Scholar]
- 63.Smith AW, Hoyne JD, Nguyen PK, McCreedy DA, Aly H, Efimov IR, Rentschler S, Elbert DL. Biomaterials. 2013;34:6559–6571. doi: 10.1016/j.biomaterials.2013.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boroujeni SM, Mashayekhan S, Vakilian S, Ardeshirylajimi A, Soleimani M. J Biomed Mater Res Part A. 2016;104:1610–1621. doi: 10.1002/jbm.a.35686. [DOI] [PubMed] [Google Scholar]
- 65.Park S, Im G. J Biomed Mater Res Part A. 2014;103:1238–1245. doi: 10.1002/jbm.a.35236. [DOI] [PubMed] [Google Scholar]
- 66.Kulangara K, Adler AF, Wang H, Chellappan M, Hammett E, Yasuda R, Leong KW. Biomaterials. 2014;35:5327–5336. doi: 10.1016/j.biomaterials.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carlson AL, Bennett NK, Francis NL, Halikere A, Clarke S, Moore JC, Hart RP, Paradiso K, Wernig M, Kohn J, Pang ZP, Moghe PV. Nat Commun. 2016;7:10862. doi: 10.1038/ncomms10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sciancalepore AG, Portone A, Moffa M, Persano L, Luca MD, Paiano A, Sallustio F, Schena FP, Bucci C, Pisignano D. Biomaterials. 2016;94:57–69. doi: 10.1016/j.biomaterials.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 69.Morez C, Noseda M, Paiva MA, Belian E, Schneider MD, Stevens MM. Biomaterials. 2015;70:94–104. doi: 10.1016/j.biomaterials.2015.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sia J, Yu P, Srivastava D, Li S. Biomaterials. 2016;103:1–11. doi: 10.1016/j.biomaterials.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 71.Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Macadangdang J, Guan X, Smith AST, Lucero R, Czerniecki S, Childers MK, Mack DL, Kim DH. Cell Mol Bioeng. 2015;8:320–332. doi: 10.1007/s12195-015-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith AS, Davis J, Lee G, Mack DL, Kim DH. Drug Discovery Today. 2016;21:1387–1398. doi: 10.1016/j.drudis.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim JH, Kim HW, Cha KJ, Han J, Jang YJ, Kim DS, Kim JH. ACS Nano. 2016;10:3342–3355. doi: 10.1021/acsnano.5b06985. [DOI] [PubMed] [Google Scholar]
- 75.Mengsteab PY, Uto K, Smith AS, Frankel S, Fisher E, Nawas Z, Macadangdang J, Ebara M, Kim DH. Biomaterials. 2016;86:1–10. doi: 10.1016/j.biomaterials.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carson D, Hnilova M, Yang X, Nemeth CL, Tsui JH, Smith AS, Jiao A, Regnier M, Murry CE, Tamerler C, Kim DH. ACS Appl Mater Interfaces. 2016;8:21923–21932. doi: 10.1021/acsami.5b11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yim EK, Darling EM, Kulangara K, Guilak F, Leong KW. Biomaterials. 2010;31:1299–1306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoo J, Noh M, Kim H, Jeon NL, Kim BS, Kim J. Biomaterials. 2015;45:36–45. doi: 10.1016/j.biomaterials.2014.12.049. [DOI] [PubMed] [Google Scholar]
- 79.Klymov A, Bronkhorst EM, Riet JT, Jansen JA, Walboomers XF. Acta Biomater. 2015;16:117–125. doi: 10.1016/j.actbio.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 80.Kim J, Bae WG, Choung HW, Lim KT, Seonwoo H, Jeong HE, Suh KY, Jeon NL, Choung PH, Chung JH. Biomaterials. 2014;35:9058–9067. doi: 10.1016/j.biomaterials.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 81.Yim EK, Pang SW, Leong KW. Exp Cell Res. 2007;313:1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith AS, Macadangdang J, Leung W, Laflamme MA, Kim DH. Biotechnology Advances. 2017;35:77–94. doi: 10.1016/j.biotechadv.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pioner JCACAM, Racca AW, Klaiman JM, Yang K-C, Guan X, Pabon L, Muskheli V, Zaunbrecher R, Macadangdang J, Jeong MY, Mack DL, Childers MK, Kim D-H, Tesi C, Poggesi C, Murry CE, Regnier M. Stem Cell Reports. 2016;6:885–896. doi: 10.1016/j.stemcr.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uto K, Tsui JH, Deforest CA, Kim DH. Progress in Polymer Science. 2017;65:53–82. doi: 10.1016/j.progpolymsci.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye K, Wang X, Cao L, Li S, Li Z, Yu L, Ding J. Nano Lett. 2015;15:4720–4729. doi: 10.1021/acs.nanolett.5b01619. [DOI] [PubMed] [Google Scholar]
- 86.Choi B, Park KS, Kim JH, Ko KW, Kim JS, Han DK, Lee SH. Macromol Biosci. 2015;16:199–206. doi: 10.1002/mabi.201500273. [DOI] [PubMed] [Google Scholar]
- 87.Higuchi S, Watanabe TM, Kawauchi K, Ichimura T, Fujita H. J Biosci Bioeng. 2014;117:749–755. doi: 10.1016/j.jbiosc.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 88.Vertelov G, Gutierrez E, Lee SA, Ronan E, Groisman A, Tkachenko E. Sci Rep. 2016;6:33411. doi: 10.1038/srep33411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 90.Zhao W, Li X, Liu X, Zhang N, Wen X. Mater Sci Eng: C. 2014;40:316–323. doi: 10.1016/j.msec.2014.03.048. [DOI] [PubMed] [Google Scholar]
- 91.Macrí-Pellizzeri L, Pelacho B, Sancho A, Iglesias-García O, Simón-Yarza AM, Soriano-Navarro M, González-Granero S, García-Verdugo JM, De-Juan-Pardo EM, Prosper F. Tissue Eng Part A. 2015;21:1633–1641. doi: 10.1089/ten.TEA.2014.0251. [DOI] [PubMed] [Google Scholar]
- 92.Jacot JG, Kita-Matsuo H, Wei KA, Chen HV, Omens JH, Mercola M, Mcculloch AD. Ann N Y Acad Sci. 2010;1188:121–127. doi: 10.1111/j.1749-6632.2009.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ali MY, Chuang CY, Saif MTA. Soft Matter. 2014;10:8829–8837. doi: 10.1039/c4sm01602e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coombs KE, Leonard AT, Rush MN, Santistevan DA, Hedberg-Dirk EL. J Biomed Mater Res Part A. 2016 doi: 10.1002/jbm.a.35864. [DOI] [PubMed] [Google Scholar]
- 95.Tse JR, Engler AJ. PLoS ONE. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yuan H, Zhou Y, Lee MS, Zhang Y, Li WJ. Acta Biomater. 2016;42:247–257. doi: 10.1016/j.actbio.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hubbi KME, Ahn EH, Downey J, Afzal J, Kim D-H, Rey S, Chang C, Kundu A, Semenza GL, Abraham RM, Levchenko A. Sci Signaling. 2012;5:ra41. doi: 10.1126/scisignal.2003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen YJ, Huang CH, Lee IC, Lee YT, Chen MH, Young TH. Connect Tissue Res. 2008;49:7–14. doi: 10.1080/03008200701818561. [DOI] [PubMed] [Google Scholar]
- 99.Park JS, Chu JS, Cheng C, Chen F, Chen D, Li S. Biotechnol Bioeng. 2004;88:359–368. doi: 10.1002/bit.20250. [DOI] [PubMed] [Google Scholar]
- 100.Altman GH, Horan RL, Martin I, Farhadi J, Stark PR, Volloch V, Richmond JC, Vunjak-Novakovic G, Kaplan DL. FASEB J. 2002;16:270–272. doi: 10.1096/fj.01-0656fje. [DOI] [PubMed] [Google Scholar]
- 101.Qin TW, Sun YL, Thoreson AR, Steinmann SP, Amadio PC, An KN, Zhao C. Biomaterials. 2015;51:43–50. doi: 10.1016/j.biomaterials.2015.01.070. [DOI] [PubMed] [Google Scholar]
- 102.Lee IC, Wang JH, Lee YT, Young TH. Biochem Biophys Res Commun. 2007;352:147–152. doi: 10.1016/j.bbrc.2006.10.170. [DOI] [PubMed] [Google Scholar]
- 103.Guo J, Wang Y, Sachs F, Meng F. Proc Natl Acad Sci U S A. 2014;111:E5252–5261. doi: 10.1073/pnas.1411683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu B, Song G, Ju Y, Li X, Song Y, Watanabe S. J Cell Physiol. 2012;227:2722–2729. doi: 10.1002/jcp.23016. [DOI] [PubMed] [Google Scholar]
- 105.Liang X, Huang X, Zhou Y, Jin R, Li Q. Stem Cells Transl Med. 2016;5:960–969. doi: 10.5966/sctm.2015-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Q, Huang H, Wei K, Zhao Y. Biotechnol Bioeng. 2016;113:2191–2201. doi: 10.1002/bit.25981. [DOI] [PubMed] [Google Scholar]
- 107.Gu SR, Kang YG, Shin JW, Shin JW. J Biosci Bioeng. 2016 doi: 10.1016/j.jbiosc.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 108.Wang L, Li Y, Chen B, Liu S, Li M, Zheng L, Wang P, Lu TJ, Xu F. ACS Appl Mater Interfaces. 2015;7:15088–15097. doi: 10.1021/acsami.5b04450. [DOI] [PubMed] [Google Scholar]
- 109.Wang ZY, Lim J, Ho YS, Zhang QY, Chong MSK, Tang M, Hong MH, Chan JKY, Teoh SH, Thian ES. J Biomed Mater Res Part A. 2013;102:2197–2207. doi: 10.1002/jbm.a.34899. [DOI] [PubMed] [Google Scholar]
- 110.Caiazzo M, Okawa Y, Ranga A, Piersigilli A, Tabata Y, Lutolf MP. Nat Mater. 2016;15:344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 111.Anderson HJ, Sahoo JK, Ulijn RV, Dalby MJ. Front Bioeng Biotechnol. 2016;4:38. doi: 10.3389/fbioe.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]


