Abstract
Histone deacetylases (HDACs) are negative regulators of transcription and have been shown to regulate specific changes in gene expression. In vertebrates, eighteen HDACs have thus far been identified and subdivided into four classes (I-IV). Key roles for several HDACs in bone development and biology have been elucidated through in vitro and in vivo models. By comparison, there is a paucity of data on the roles of individual HDACs in osteoclast formation and function. In this study, we investigated the gene expression patterns and the effects of suppressing individual class II (Hdac4, 5, 6, 9, and 10) and class IV (Hdac11) HDACs during osteoclast differentiation. We demonstrated that HDAC class II and IV members are differentially expressed during osteoclast differentiation. Additionally, individual shRNA-mediated suppression of Hdac4, 5, 9, 10 and 11 expression resulted in increased multinucleated osteoclast size and demineralization activity, with little to no change in the overall number of multinucleated osteoclasts formed compared with control shRNA-treated cells. We also detected increased expression of genes highly expressed in osteoclasts, including c-Fos, Nfatc1, Dc-stamp and Cathepsin K. These observations indicate that HDACs cooperatively regulate shared targets in a non-redundant manner.
Introduction
Bone is a dynamic tissue that is constantly remodeled through degradation by osteoclasts and renewal by osteoblasts [1]. Proper coordination between these cell types is essential for maintaining structural integrity of the skeleton throughout development. Osteoclasts are large, multinucleated cells derived from hematopoietic stem cells/monocyte precursors [1]. Osteoclast differentiation is governed mainly by two important cytokines: macrophage colony stimulating factor (M-CSF) and receptor activator of nuclear factor kappa B ligand (RANKL) [2–5]. RANKL stimulates the expression of key transcription factors such as NF-κB, c-Fos, MITF, and NFATc1 [5–8] that are necessary for osteoclast differentiation and maturation. Osteoclasts are needed for normal bone functions such as bone remodeling and fracture repair. However, uncontrolled osteoclast activity can lead to skeletal disorders such as osteoporosis. Therefore, it is important to determine mechanisms that regulate transcription of osteoclast genes. This knowledge may reveal key modulators of bone resorption that can be considered as therapeutic targets.
The existence of tissue-specific transcription factors alone is inadequate to control temporal gene expression; co-factors are needed for chromatin remodeling and recruitment of RNA polymerase II. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) control gene expression markings by being drafted to target genes through interactions with specific transcription factors [9]. Gene activation is linked to the recruitment of HATs by transcription factors. Conversely, HDACs bind to the same transcription factors and promote transcriptional repression. Thus, HATs and HDACs act as molecular switches for the functions of transcription factors. These processes have become essential mechanisms to examine regarding our understanding of bone physiology and diseases.
HDACs are negative regulators of transcription, and have been shown to induce specific changes to gene expression in various biological processes by deacetylating both histone and non-histone proteins [10–12]. Rooted in structural and functional similarities to yeast deacetylases, the 18 HDACs in the human genome are categorized into four classes: class I HDACs (HDAC1, 2, 3 and 8), class II HDACs (HDAC4, 5, 6, 7, 9 and 10), class III HDACs (Sirtuins1-7) and class IV HDACs (HDAC11) [13–19]. HDACs are emerging as important regulators of skeletal homeostasis [16]. Various in vitro studies have reported that HDAC inhibitors can repress osteoclast differentiation [20–22]. These inhibitors are broad-spectrum compounds that target multiple HDACs. However, the specific effects of individual HDACs on osteoclasts are largely unknown.
Previous work from our lab demonstrated that suppression of Hdac7 and Hdac3 have opposite effects on osteoclast differentiation in vitro [23]. Hdac7 suppression enhances differentiation, whereas suppression of Hdac3 inhibits osteoclast differentiation. Our lab [24] as well as Jin et al [25] demonstrated that HDAC7 inhibits osteoclastogenesis and bone resorption in vivo. We found that HDAC7 represses osteoclast differentiation through interacting with the transcription factor MITF [24], while Jin et al. showed that HDAC7 regulates NFATc1 activity [25]. Additionally, Jin et al [26] demonstrated that HDAC9 suppresses osteoclastogenesis through negatively regulating PPARγ. These findings suggest that each HDAC member can induce suppression of osteoclast differentiation through distinct and possibly multiple mechanisms. However, it is still unclear whether other HDAC members play separate and pivotal roles in osteoclastogenesis.
The goal of this study was to investigate the functions of class II and IV HDACs during osteoclastogenesis and to determine whether any redundant roles exist for the class II and IV HDACs. Our results revealed that suppression of individual HDACs enhance osteoclast differentiation, potentially through repression of different proteins within the same cellular pathways. We anticipate that characterization of changes in gene expression patterns due to specific suppression of each HDAC in osteoclasts will further our understanding of how class II and IV HDACs regulate osteoclastogenesis. This knowledge will lead to potential new therapies in treating resorption-mediated bone diseases.
Experimental procedures
Ethics
The use and care of the mice was reviewed and approved by the University of Minnesota Institutional Animal Care and Use Committee, IACUC protocol number 150732820A. Euthanasia was performed by CO2 inhalation. Isolation and culture of osteoclasts from mouse bone marrow cells, as well as virus generation and viral transduction of osteoclasts were performed under approval of the University of Minnesota Institutional Biosafety Committee, permit number 1506-32712H.
Primary osteoclast cell culture
Primary bone marrow macrophages (BMMs) were isolated from the femora and tibiae of C57BL/6 mice. The femora and tibiae were dissected out and adherent tissue was removed. The epiphyses of these long bones were removed, and the bone marrow was flushed from the diaphysis. Red blood cells were lysed from the flushed marrow using red blood cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.4), and the resulting cells were plated and cultured overnight in 100 mm tissue culture dishes (MidSci) in osteoclast media (phenol red-free alpha-MEM [Gibco] with 5% fetal bovine serum [Hyclone], 25 units/mL penicillin/streptomycin [Invitrogen], 400 mM L-Glutamine [Invitrogen], and supplemented with 1% CMG 14–12 culture supernatant containing M-CSF). CMG 14–12 cell line was obtained from Dr. Sunao Takeshita (Nagoya City University, Nagoya, Japan) [27]. The non-adherent cell population was then re-plated in 12-well plates (MidSci) at 2 x 105 cells/well in osteoclast media supplemented with 1% CMG 14–12 culture supernatant for 48 hours. Cells were then fed every two days with osteoclast media containing 1% CMG culture supernatant plus 10 ng/mL RANKL (R&D Systems) to stimulate osteoclastogenesis.
Tartrate resistant acid phosphatase (TRAP) staining
After five days of culture with 1% CMG 14–12 culture supernatant and 10 ng/ml RANKL, primary osteoclasts were rinsed in PBS and fixed with 4% paraformaldehyde for 20 minutes. Cells expressing TRAP were stained with Naphthol AS-MX phosphate and Fast Violet LB salt protocol as previously described [24]. The stained cells were then imaged and photographed with light microscopy and analyzed using NIH ImageJ to measure the number and size of TRAP-positive multinuclear cells.
Demineralization assay
BMMs were plated on calcium phosphate-coated plates (Corning) and cultured as above. After five days of stimulation with 1% CMG 14–12 culture supernatant and 10 ng/ml RANKL, plates were processed according to the manufacturer’s instructions, the demineralized area was photographed by dark field microscopy and analyzed using NIH ImageJ.
Lentiviral infection of osteoclasts
Lentiviral vectors (Open Biosystems) encoding shRNAs against Hdac4 (TRCN0000039249 and TRCN0000039252), Hdac5 (V2LMM 72835 and V3LMM 432047), Hdac6 (V2LMM 61798 and V2LMM 79557), Hdac9 (V3LMM 481592 and V3LMM 481594), Hdac10 (V3LMM 425386 and V3LMM 425387), Hdac11 (V2LMM 24029 and TRCN0000039224), or a control shRNA were used to produce replication-defective lentivirus according to the manufacturer’s protocols. Viral stocks were titrated by infection in HeLa cells. BMMs were isolated and cultured as described above. 48 hours after seeding the non-adherent population, lentiviruses were added at 10 MOI and incubated for 18 hours at 37°C in the presence of 1% CMG 14–12 culture supernatant. The following day cultures were stimulated with 1% CMG 14–12 culture supernatant and 10 ng/ml RANKL. Cells were used for either RNA extraction after 48 hours with 1% CMG 14–12 culture supernatant and RANKL treatment, or TRAP staining or demineralization assays after 5 days with 1% CMG 14–12 culture supernatant and RANKL.
RNA isolation and real-time PCR
RNA was harvested from cells plated in triplicate using TRIZOL Reagent (Ambion, Life Technologies) and quantified using UV spectroscopy. cDNA was then prepared from 1 μg of purified RNA using iScript cDNA Synthesis Kit (Bio-Rad) per the manufacture’s protocol. Quantitative real-time PCR (qRT-PCR) was performed in duplicate using CFX Connect Real-Time PCR system (Bio-Rad). Each 20 μl reaction mixture contained 1 μl cDNA, 10 μL iTaq Universal Sybr Green Supermix, and 500 nM forward and reverse primers. The PCR conditions were as follows: 95°C for 3 minute, and 40 cycles of 94°C for 15 seconds, 56°C for 30 seconds, and 72°C for 30 seconds, followed by melt curve analysis (95°C for 5 seconds, 65°C for 5 seconds, and then 65°C to 95°C with 0.5°C increase for 5 seconds). Experimental genes were normalized to Hprt. Primers amplified with equal efficiencies. Their sequences used are listed in S1 Table. All measurements were analyzed using the ΔΔCT method.
Immunoblot analysis
Protein cell lysates were harvested from primary osteoclasts in modified RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% IGEPAL, 0.25% sodium deoxycholate, 1 mM EDTA) supplemented with Halt Protease & Phosphatase Inhibitor Cocktail (Thermo Scientific). Lysates were cleared by centrifugation. Proteins were resolved by SDS-PAGE, transferred to PVDF membrane (Millipore), blocked, and blotted in primary antibody overnight at 4°C. The next day, blots were incubated with horseradish peroxidase conjugated secondary antibody (G.E. Healthcare) for 1 hour at room temperature. Antibody binding was detected using western blotting detection kit (Western Bright Quantum, Advansta). The following primary antibodies were all used at 1:1000 dilution: polyclonal HDAC4 produced using peptide corresponding to amino acids 14–28 of human HDAC4 –Sigma Aldrich (H9536, Antibody ID# AB477079); monoclonal HDAC5 produced using peptide corresponding to amino acids 371–443 of human HDAC5—Santa Cruz Biotechnology (SC-133225, Antibody ID# 2116791); polyclonal HDAC9 produced using peptide corresponding to amino acids 112–129 of HDAC9 –Thermo Scientific (PA5-23346, Antibody ID# AB2540870); polyclonal MITR produced from peptide against human HDAC9—Sigma Aldrich (SAB4503694, Antibody ID# AB10751345); polyclonal HDAC10 produced using peptide corresponding to amino acids 2–16 of human HDAC10 –Sigma Aldrich (H3413, Antibody ID# AB261940); polyclonal HDAC11 produced using peptide corresponding to amino acids 2–16 of human HDAC11 –Sigma Aldrich (H4539, Antibody ID# AB532246) and polyclonal ACTIN-Santa Cruz Biotechnology produced using carboxy terminus of human actin (SC-1616, Antibody ID# AB630836). The secondary antibodies used were all used at 1:10,000 dilution: anti-rabbit (NA-934, Antibody ID# AB772206) and anti-goat (SC-2020, Antibody ID# AB631728). All densitometry data was generated using the Image Lab Software (Bio Rad) following the manufacturer’s instructions for normalizing data to a housekeeping protein. The calculated volume intensity for HDAC expression was normalized to its corresponding actin from the same membrane.
Statistical analysis
All experiments were run in triplicate, performed three times, and results are expressed as mean ± standard deviation. Student’s t-test or one-way ANOVA analysis followed by a Tukey’s multiple comparison test were used to compare data using Graph-Pad Prism version 7.
Results
Expression of class II and IV HDACs during osteoclastogenesis
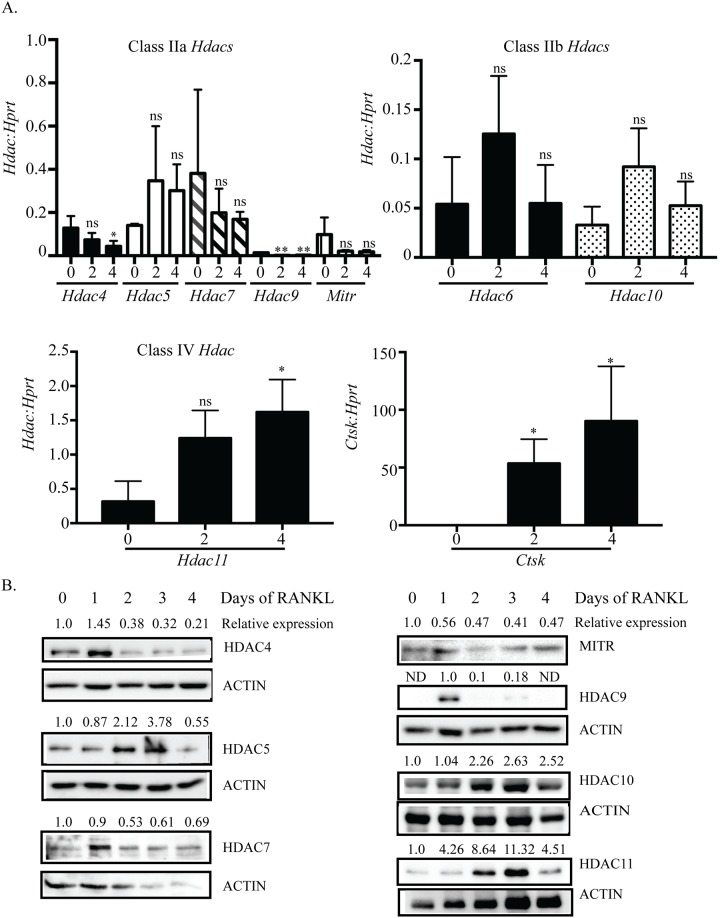
To investigate the roles of class II and IV HDACs during osteoclastogenesis we first examined their expression through the process of osteoclast differentiation. Non-adherent cells from C57BL/6 (wild-type) mouse bone marrow cells were cultured with 1% CMG for two days to promote bone marrow macrophages (BMMs) proliferation, and then treated with M-CSF plus RANKL for four more days to stimulate osteoclast differentiation. All Hdac members in class IIa, IIb and IV were expressed in both proliferating BMMs (day zero with M-CSF treatment) and differentiating osteoclasts (day one-day four with M-CSF plus RANKL treatment) (Fig 1A). Among these HDACs, the expression of Hdac4, Hdac7, Hdac9 and Histone deacetylase related protein (Hdrp/Mitr) were significantly down-regulated upon RANKL stimulation (day zero vs day two) and expression remained low during osteoclast differentiation (day one-day four, S1 Fig). Conversely, Hdac5, Hdac6, Hdac10 and Hdac11 expression increased with RANKL expression (Fig 1A, day zero vs day two). Jin et al [26] reported similar trends in HDAC mRNA expression. Hdac11, Hdac5 and Hdac7 were the most highly expressed Hdac RNAs during osteoclast differentiation. As expected, the expression of the osteoclast marker gene Cathepsin K (Ctsk) was significantly increased upon RANKL stimulation and served as a positive control for osteoclast differentiation (Fig 1A, bottom right panel). HDAC protein expression as measured by western blot demonstrated similar expression patterns to the mRNAs (Fig 1B, S2 Fig). These results show that HDAC class II and IV members are differentially expressed during osteoclast differentiation.
Fig 1. Class II and IV Hdac expression during osteoclast differentiation.
BMM cells from C57BL/6 mice were cultured in M-CSF only (day zero) or in M-CSF plus RANKL (day two and day four) to stimulate osteoclast differentiation. Class II and IV Hdac RNA expression (A) on day zero, day two and day four of osteoclast differentiation. qRT-PCR data shown are the mean of three independent experiments. * p <0.05 comparing vs. day zero, ns = not significant. Representative protein expression (B) of class II and IV HDACs on each day of osteoclast differentiation. Displayed relative expression values are HDAC expression relative to the normalized expression on day zero as calculated by Image Lab Software (BioRad). In the case of HDAC9 no band was identified for day zero so data is expressed relative to day one. The same lysates were analyzed for HDAC9 and MITR expression and therefore, have the same actin loading control.
Effect of shRNA suppression of Hdacs on osteoclast differentiation
Since we detected expression of all the examined Hdacs in osteoclast cultures, we next asked how suppression of each individual Hdac would affect osteoclast formation or function. To this end, we used lentiviral vectors encoding shRNAs against these Hdacs. Prior to beginning RANKL stimulation, BMMs from wild-type mice were infected with one of two lentiviral vectors encoding an shRNA against an Hdac or a control shRNA. For each Hdac, we obtained similar results with both shRNAs indicating that the outcomes were not due to off-target effects. Following infection with shRNAs, we confirmed target gene knockdown by qRT-PCR and examined changes in the expression of c-Fos, Nfatc1, Dc-stamp and Ctsk genes important for osteoclast formation or function. Because our experiments produced similar results in our analysis of osteoclast differentiation and activity assays using the two different Hdac shRNA, we grouped our PCR data together from the Hdac shRNA experiments for qRT-PCR. We employed TRAP staining and demineralization assays to measure osteoclast differentiation, fusion and activity.
Suppression of Hdac4 increases osteoclastogenesis
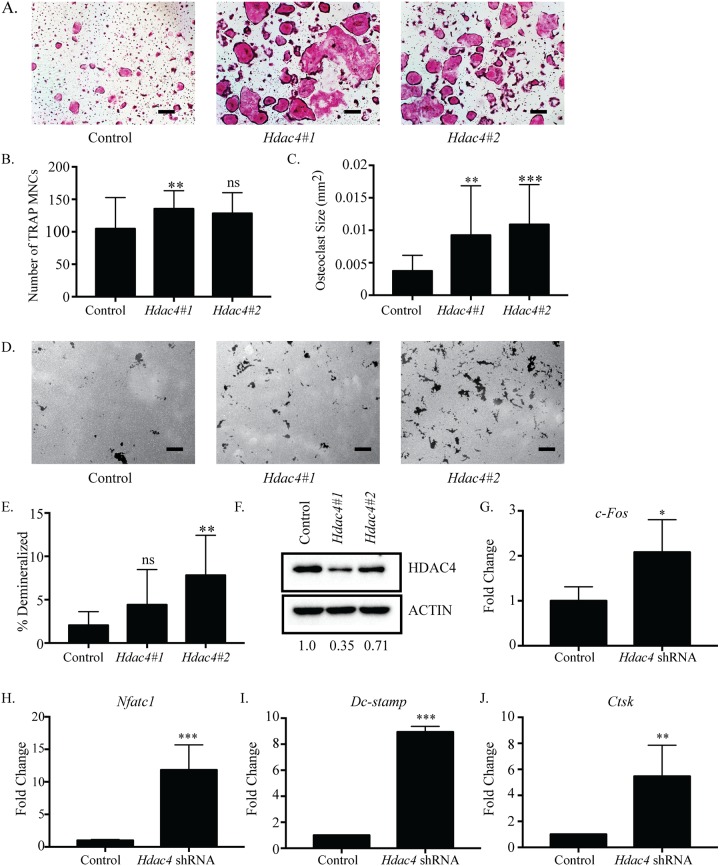
First, we investigated the effects of silencing Hdac4 on osteoclastogenesis. Cells infected with Hdac4 shRNA #1 or #2 showed enhanced osteoclast differentiation compared to control shRNA (Fig 2A). The observed TRAP-positive multinuclear cells (MNCs) from both Hdac4-shRNA cultures were more numerous, and their size significantly increased (3-fold) relative to the control (Fig 2B and 2C). Demineralization assays on calcium phosphate-coated plates demonstrated cells infected with Hdac4 shRNA #2 produced significantly increased total number of pits, average pit size, and total percent demineralized area compared to control shRNA infected cells. Hdac4 shRNA #1 infected cells showed similar trends (Fig 2D and 2E, S3 Fig). Moreover, BMMs infected with either Hdac4 shRNA showed Hdac4 RNA expression levels reduced by approximately 50% (S3 Fig), and Hdac4 protein expression levels reduced between 30–65% (Fig 2F). Hdac4 shRNA increased expression of c-Fos, Nfatc1, Dc-stamp and Ctsk compared to control shRNA (Fig 2G–2J). Taken together these results reveal that Hdac4 suppression increased osteoclast differentiation by producing larger and more numerous osteoclasts, suggesting HDAC4 inhibits osteoclastogenesis.
Fig 2. Accelerated osteoclast differentiation in Hdac4-suppressed osteoclasts.
Representative images of TRAP staining (A) of osteoclast cultures infected with control or Hdac4 shRNA-expressing lentivirus. Hdac4#1 represents Hdac4 shRNA #1and Hdac4#2 represents Hdac4 shRNA #2. Number (B) and size (C) of TRAP-positive MNCs. Representative images (D) and quantification (E) of demineralization activity of control and Hdac4 shRNA-expressing osteoclast cultures grown on calcium phosphate-coated plates. Scale bar represents 200 μm. Western blot (F) of control and Hdac4 shRNA-expressing cells with relative expression of shRNA expressing cells relative to control expressing cells indicated under the blots. Expression profile (G-J) of osteoclast genes c-Fos, Nfatc1, Dc-stamp and Ctsk. Data presented are the mean of three independent experiments. * p <0.05; ** p <0.01; *** p <0.001; ns = not significant compared to control infected cells.
Hdac5 is a negative regulator of osteoclastogenesis
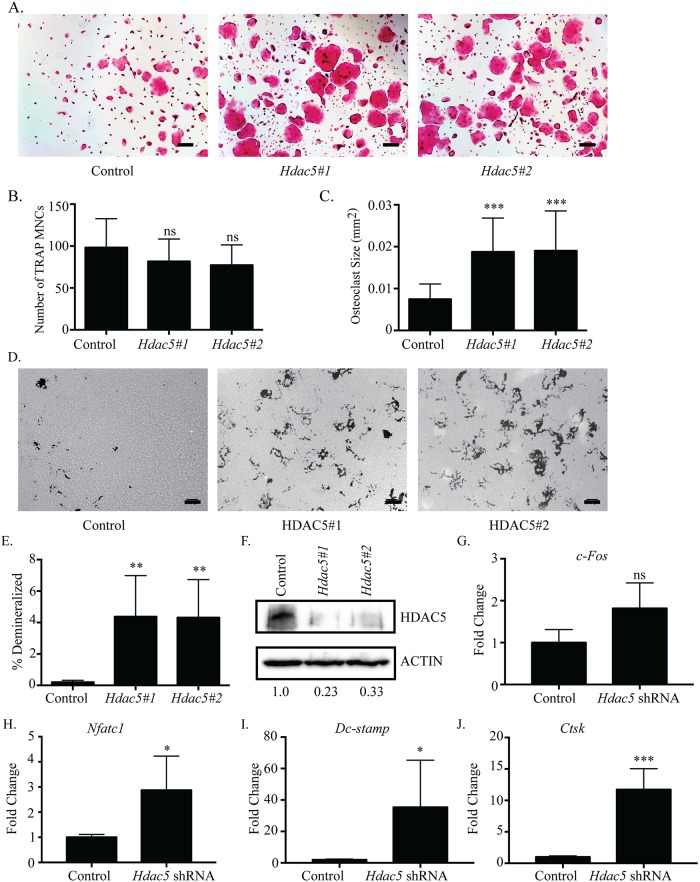
We investigated the effects of suppressing expression of the Class IIa deacetylase Hdac5. Osteoclast differentiation in either Hdac5-shRNA culture was enhanced compared to control shRNA (Fig 3A). The average size of TRAP-positive MNCs in Hdac5-shRNA cells was significantly increased (Fig 3B). However, the number of TRAP-positive MNCs per well in Hdac5-shRNA cells did not significantly change compared to control shRNA (Fig 3C). When osteoclast activity was determined, cells infected with each Hdac5-shRNA had significantly increased total number of pits, average pit size, and total percent demineralized area (Fig 3D and 3E, S4 Fig). The cultured osteoclasts also showed reduced Hdac5 mRNA expression levels (S4 Fig), and approximately 70–80% reduction in HDAC5 expression as measured by western blot (Fig 3F). Hdac5-suppressed osteoclasts showed an upward but not significant change in expression of c-Fos (Fig 3G); while expression of genes important for osteoclast formation or function including Nfatc1, Dc-stamp, and Ctsk (Fig 3H–3J) were significantly increased. These findings indicate that Hdac5 suppression enhances osteoclast differentiation through increased expression of genes such as Nfatc1, Dc-stamp, and Ctsk but independent of changes to c-Fos.
Fig 3. Suppression of Hdac5 enhances osteoclast differentiation.
Representative images of TRAP staining (A) of osteoclast cultures. Hdac5#1 represents Hdac5 shRNA #1and Hdac5#2 represents Hdac5 shRNA #2. Quantification of the number (B) and average size of TRAP-stained multinucleated osteoclasts (C). Representative photographs (D) and quantification (E) of demineralization activity of control and Hdac5 shRNA-expressing osteoclast cultures grown on calcium phosphate-coated plates. Scale bar represents 200 μm. Western blot (F) of control and Hdac5 shRNA-expressing cells with relative expression of shRNA expressing cells relative to control expressing cells indicated under the blots. Expression profile (G-J) of osteoclast genes c-Fos, Nfatc1, Dc-stamp and Ctsk. Data presented are the mean of three independent experiments. * p <0.05; ** p <0.01; *** p < 0.001; ns = not significant compared to control infected cells.
Hdac6 suppression does not affect osteoclast differentiation
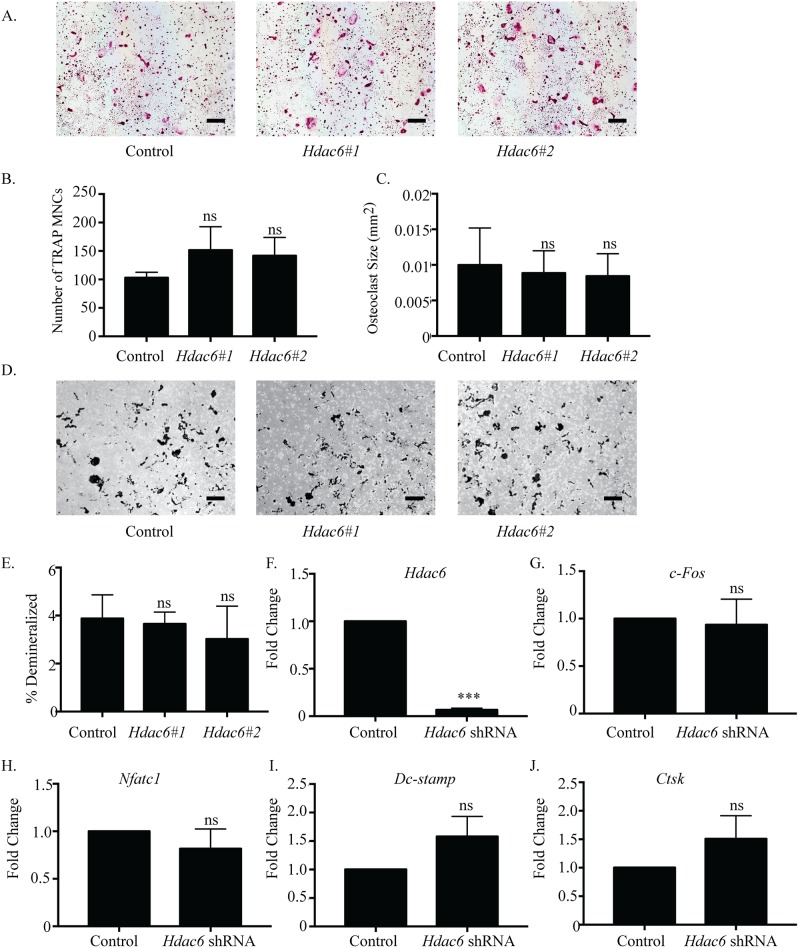
HDAC6, a member of class IIb HDAC family and tubulin deacetylase, has been shown to regulate tubulin dynamics and stability of the podosome belt in mature osteoclasts [28, 29]. However, the role of HDAC6 in early stages of osteoclast differentiation is unknown. There was no significant change in the number or size of TRAP-positive MNCs with Hdac6 suppression (Fig 4A–4C). Moreover, Hdac6-shRNA #1 showed no significant changes in the demineralization assay (Fig 4D and 4E, S5 Fig), but Hdac6-shRNA #2 had a significant decrease only in the average pit size (S4 Fig). Commercially verifiable antibodies to HDAC6 are not available; therefore, we verified both Hdac6 shRNAs significantly repressed Hdac6 expression by qPCR (Fig 4F). However, c-Fos, Nfatc1, Dc-stamp, and Ctsk were not significantly changed by Hdac6 suppression (Fig 4G–4J). These data show that knockdown of HDAC6 does not have any impact on osteoclast differentiation, at least within the parameters and genes analyzed.
Fig 4. Hdac6 suppression does not affect osteoclast differentiation.
Representative images of TRAP staining (A) of osteoclast cultures. Hdac6#1 represents Hdac6 shRNA #1and Hdac6#2 represents Hdac6 shRNA #2. Quantification of (B) number and (C) size of TRAP-positive MNCs. Representative photographs (D) and quantification (E) of demineralization activity of control and Hdac6 shRNA-expressing osteoclast cultures grown on calcium phosphate-coated plates. Scale bar represents 200 μm. qRT-PCR (F) of control and Hdac6 shRNA-expressing cells. Expression profile (G-J) of c-Fos, Nfatc1, Dc-stamp, and Ctsk. Data presented are the mean of three independent experiments. *** p < 0.001; ns = not significant compared to control infected cells.
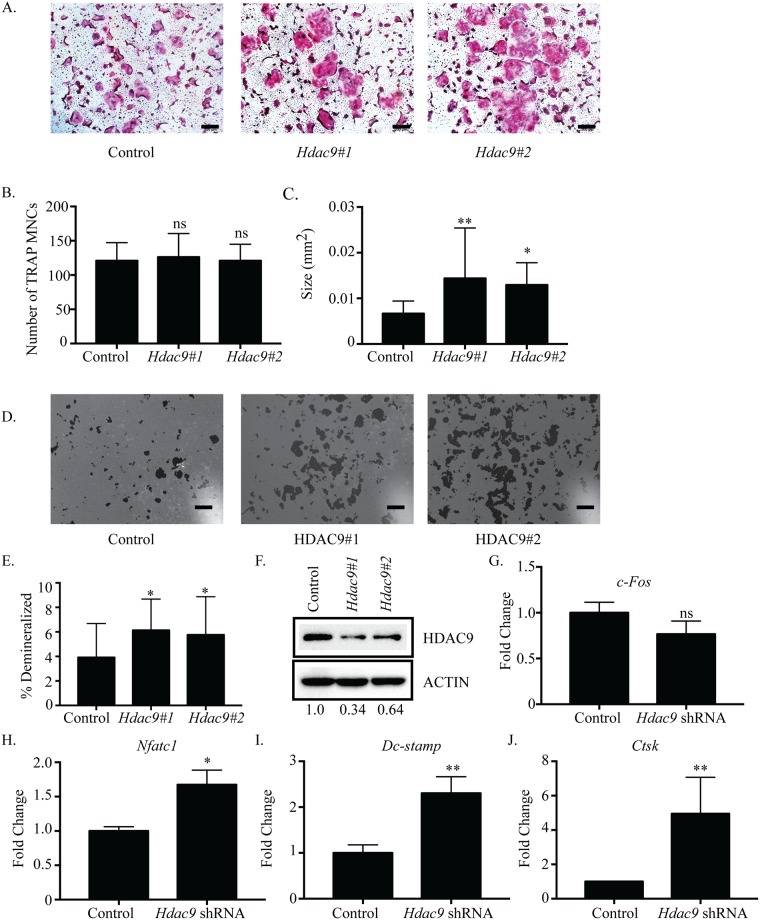
Suppression of Hdac9 inhibits osteoclastogenesis
We next used shRNAs to characterize the role of Hdac9 and Mitr in osteoclasts. Although our gene expression data (Fig 1) indicate that MITR is the predominant form of HDAC9 in osteoclasts, we were unable to identify shRNAs that distinguish between Mitr and full-length Hdac9. Consequently, both shRNAs used are predicted to target Hdac9 and Mitr. Suppression of Hdac9 increased the size of TRAP-positive MNCs, but had little effect on the number of osteoclasts formed (Fig 5A–5C). Demineralization assays showed that in Hdac9-shRNA infected cells, average pit size and demineralized area were significantly increased (Fig 5D and 5E, S6 Fig). As expected, in Hdac9-suppressed cells there was a reduction in Hdac9 protein expression (Fig 5F) as well as Hdac9 RNA (S6 Fig). Hdac9-suppressed cells revealed a slight but not significant reduction in c-Fos expression (Fig 5G), while expression of Nfatc1, Dc-stamp, and Ctsk was increased compared with control (Fig 5H–5J). These observations indicate that HDAC9/MITR inhibits osteoclastogenesis.
Fig 5. Suppression of Hdac9 inhibits osteoclast differentiation.
Representative images of TRAP staining (A) of osteoclast cultures infected with control or Hdac9 shRNA-expressing lentiviruses. Hdac9#1 represents Hdac9 shRNA #1and Hdac9#2 represents Hdac9 shRNA #2. Number (B) and size (C) of TRAP-positive MNCs. Representative photographs (D) and quantification (E) of demineralization activity of control and Hdac9 shRNA-expressing osteoclast cultures grown on calcium phosphate-coated plates. Scale bar represents 200 μm. Western blot (F) of control and Hdac9 shRNA-expressing cells with relative expression of shRNA expressing cells relative to control expressing cells indicated under the blots. Expression profile (G-J) of c-Fos, Nfatc1, Dc-stamp and Ctsk. Data presented are the mean of three independent experiments. * p <0.05; ** p <0.01; ns = not significant compared to control infected cells.
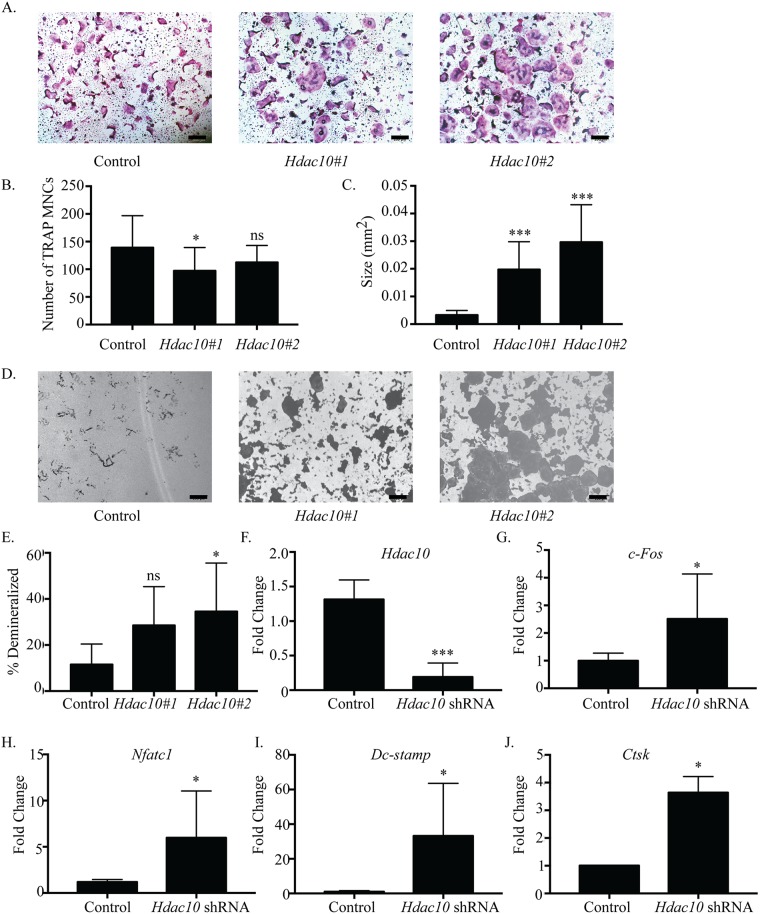
HDAC10 activity inhibits osteoclastogenesis
HDAC6 and HDAC10 are members of class IIb HDAC family. HDAC6 functions as a tubulin deacetylase, and its role as a disrupter of the actin belt in mature osteoclasts has been well established [30, 31], whereas the biological function(s) of HDAC10 remain largely unknown [32, 33]. To explore HDAC10’s role in osteoclast differentiation, we examined the effects of suppressing its expression (Fig 6A), finding that in Hdac10-suppressed cells showed a decrease in the mean number of TRAP-positive MNCs per well (Fig 6B). The average size of TRAP-positive MNCs in Hdac10-suppressed cells was significantly increased (Fig 6C). Demineralization assays show total number of pits, average pit size and percent demineralized area was increased with knockdown of Hdac10 (Fig 6D and 6E, S7 Fig). As expected, Hdac10-suppressed osteoclasts showed a reduction in Hdac10 mRNA expression compared to control shRNA (Fig 6F). We were unable to obtain a reliable western blot verifying HDAC10 protein reduction in shRNA expressing cells due to technical issues. Expression of c-Fos, Nfatc1, Dc-stamp, and Ctsk was significantly increased (Fig 6G–6J). These data indicate that reduction of Hdac10 expression enhanced osteoclast formation, suggesting that HDAC10, unlike HDAC6, may negatively regulate osteoclast differentiation.
Fig 6. Hdac10 suppression accelerates osteoclast differentiation.
Representative images of TRAP staining (A) of osteoclast cultures. Hdac10#1 represents Hdac10 shRNA #1and Hdac10#2 represents Hdac10 shRNA #2. Quantification of number (B) and size (C) of TRAP-positive MNCs. Representative images (D) and quantification (E) of demineralization activity of control and Hdac10 shRNA-expressing osteoclast cultures grow on calcium phosphate-coated plates. Scale bar represents 200 μm. qRT-PCR (F) of control and Hdac10 shRNA-expressing cells. Expression profile (G, H, I and J) of c-Fos, Nfatc1, Dc-stamp, and Ctsk. Data presented are the mean of three independent experiments. * p < 0.05; *** p <0.001; ns = not significant compared to control infected cells.
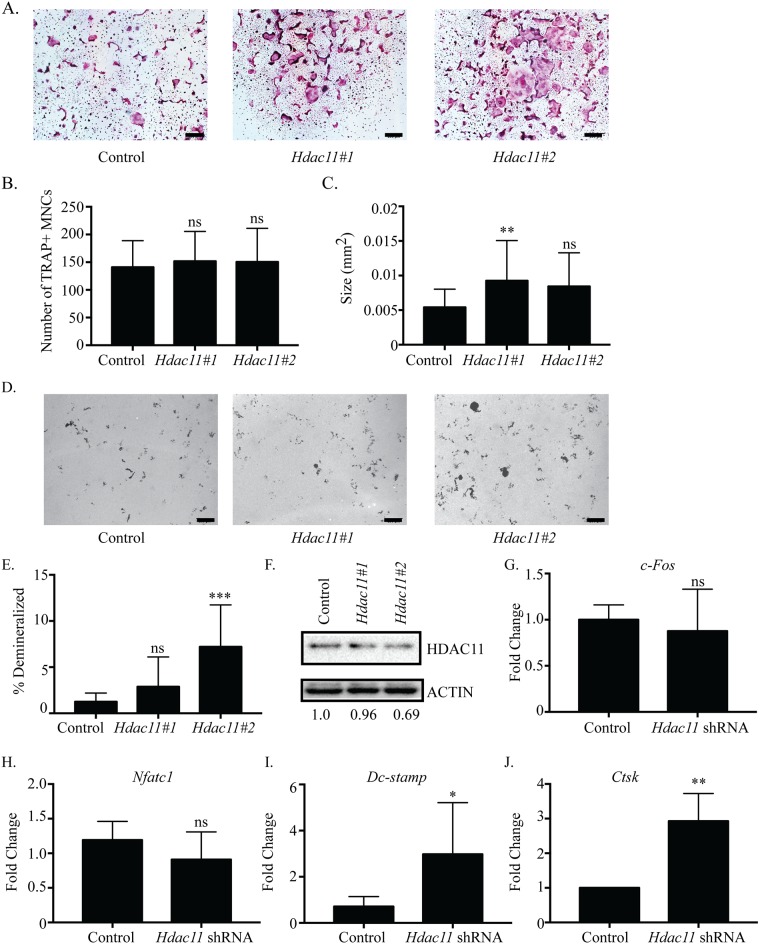
HDAC11 inhibits osteoclastogenesis
We examined the effects of suppressing Hdac11 on osteoclast differentiation. Cells infected with either Hdac11 shRNA had enhanced osteoclast differentiation (Fig 7A). Quantitative analysis of these cultures indicated that there was no significant difference in the average number of TRAP-positive MNCs formed in either Hdac11 shRNA cultures compared with control shRNA (Fig 7B). However, the average size of TRAP-positive MNCs was increased in both Hdac11 shRNA cultures (Fig 6C). Hdac11 shRNA #2 in demineralization assays increased the total number of pits, average pit size and total percent demineralized area significantly (Fig 7D and 7E, S7 Fig). Hdac11 shRNA #1 infected cells did not show a significant difference in average pit size but did show a difference in the total number of pits and total percent demineralized area (Fig 7D and 7E, S8 Fig). The Hdac11 shRNAs moderately reduced Hdac11 RNA (S8 Fig) and protein expression (Fig 7F), which led to increased Dc-stamp and Ctsk expression (Fig 7I and 7J) but no change in c-Fos (Fig 7G) and Nfatc1 expression (Fig 7H). These results reveal that Hdac11 suppression enhanced osteoclast formation, thus suggesting Hdac11 acts as an inhibitor of osteoclast differentiation.
Fig 7. Suppression of Hdac11 inhibits osteoclast differentiation.
Representative images of TRAP staining (A) of osteoclast cultures. Hdac11#1 represents Hdac11 shRNA #1and Hdac11#2 represents Hdac11 shRNA #2. Histomorphometric analysis of TRAP-stained osteoclasts (B-C). Representative photographs (D) and quantification (E) of demineralization activity of control and Hdac11 shRNA-expressing osteoclast cultures grown on calcium phosphate-coated plates. Scale bar represents 200 μm. Western blot (F) of control and Hdac11 shRNA-expressing cells with relative expression of shRNA expressing cells relative to control expressing cells indicated under the blots. Expression profile (G, H, I and J) of c-Fos, Nfatc1, Dc-stamp, and Ctsk. Data presented are the mean of three independent experiments. * p < 0.05; ** p <0.01; *** p <0.001; ns = not significant compared to control infected cells.
Discussion
To better understand how osteoclast differentiation and activity are regulated, it is necessary to investigate classes of transcriptional activators and repressors that function in osteoclasts. HDACs are a well-known class of transcriptional repressors that have been shown to be active in osteoclasts. Due to their high sequence homology and interactions with the same proteins in other cell types [14, 34], some HDACs may perform redundant roles in regulating osteoclast differentiation. However, recent studies demonstrated two class II HDACs, HDAC7 and HDAC9, repressed osteoclast differentiation using unique mechanisms. Deletion of either Hdac7 or Hdac9 in BMMs resulted in enhanced osteoclast differentiation in vitro, and increased bone resorption and osteopenia in mice in vivo [24, 26]. Our lab concluded HDAC7 interacted with and repressed the activity of MITF [24], another group demonstrated via reporter assays that HDAC7 might repress NFATc1 activity [25], and HDAC9 repressed PPAR-γ [26]. Though these mechanisms are different, it was still possible that the remaining Class II HDACs performed redundant roles in osteoclasts, in which case we hypothesized that loss of any one HDAC would be compensated for by the remaining proteins. To address this issue, we analyzed mRNA and protein expression, and the effects of shRNA-mediated knockdown of most class II (HDAC4, 5, 6, 9, and 10) and class IV (HDAC11) HDACs during osteoclast differentiation. To help identify how each HDAC was affecting osteoclasts, we used TRAP staining, demineralization assays, and qRT-PCR to evaluate changes in differentiation, activity, or gene expression due to Hdac knockdown. Individual knockdown of Hdac4, 5, 9, 10, and 11 produced similar phenotypes, which was observed as increased size of multinuclear TRAP-positive MNCs and percent demineralization of a calcium phosphate matrix, but little to no change in multinuclear TRAP-positive osteoclast number (Figs 2, 3 and 5–7). Additionally, temporal expression patterns of these HDACs were also different from each other, which is in agreement with a previous study [26]. The presence of similar osteoclast phenotypes when different HDACs are knocked down and their distinct expression profiles suggests that most HDACs have at least some unique targets rather than fully redundant roles.
Inhibiting differentiation in early stages of osteoclast differentiation
Hdac9 and the related Mitr are highly expressed in BMMs but drastically decrease in expression after RANKL stimulation. Hdac4 follows a similar trend but maintains a low level of expression throughout differentiation (Fig 1A and 1B). The initially high levels of HDAC9 and HDAC4 in BMMs which begin decreasing with the onset of RANKL stimulation could indicate that the presence of these HDACs is associated with maintaining proliferation rather than differentiation. Knockdown of Hdac4 showed a small but significant increase in osteoclast number (Fig 2B) and a trend towards increased c-Fos expression (Fig 2G). Supporting this, osteoclasts from Hdac4-KO mice showed increased c-Fos expression (data not shown, manuscript in preparation). Knockdown of Hdac9 demonstrated increased differentiation marker expression and osteoclast size but not increased osteoclast number or c-Fos expression, indicating HDAC9 may not be involved in proliferation through regulating c-Fos expression (Fig 5, [26]. Previous research on HDAC9 in osteoclasts showed both increased cell number and size of osteoclasts from global Hdac9-KO mice [26]. These contrasting results could be explained by the global deletion of Hdac9 affecting the BMM population at a stage before our transient shRNA studies began. Considering this, both previous and our work show upregulated expression of genes important for differentiation, implying knockdown of Hdac9 promotes a switch from proliferation to differentiation earlier than normal (Fig 5).
MITR is a truncated splice variant of HDAC9 that displays co-repressor activity despite lacking the C-terminal deacetylase catalytic domain. Specific suppression of Mitr but not of Hdac9 expression in neurons leads to cell death, demonstrating a necessary role for MITR separate from HDAC9 [35, 36]. It remains unknown whether MITR plays a distinct role in osteoclasts. The commercially available Hdac9 shRNAs we used are predicted to target both the full-length Hdac9 and Mitr transcripts (BLAST alignment and data not shown), and the global Hdac9-KO mice have reduced HDAC9 and MITR protein levels [37]. Future work should examine potential roles that are specific to MITR but separate from those of HDAC9.
Repressors of differentiation, fusion, and activity
Contrary to Hdac4, Hdac9, and Mitr’s early expression, Hdac5, Hdac10, and Hdac11 have expression levels that increase after RANKL treatment (Fig 1). Knockdown of any of these three HDACs results in increased osteoclast size and percent demineralization with little to no change in the number of osteoclasts (Figs 3, 6 and 7). Supporting this, knockdown of each HDAC increased Dc-stamp expression, which would lead to more fusion and, consequently, larger cells. However, knockdown did not affect c-Fos expression or cell number, indicating these HDACs mostly affect genes regulating differentiation, fusion, and possibly activity instead of proliferation (Figs 3, 6 and 7). Knockdown of either Hdac5 or Hdac10 showed a trend of increased Nfatc1 expression (Figs 3H and 6H). NFATc1 regulates expression of genes important for cell fusion (Dc-stamp) and resorption activity (Ctsk), as well as its own autoamplification after it is stably induced during differentiation [38–40]. Therefore, HDAC-mediated inhibition of Nfatc1 expression could reduce NFATc1 activity, concomitantly decreasing expression of genes that promote differentiation, fusion, and activity of osteoclasts. Indeed, Hdac5 shRNA treatment showed trends of increased Nfatc1 and Dc-stamp expression with significantly increased Ctsk expression (Fig 3). Supporting this, HDAC5 activity promotes deacetylation and, consequently, destabilization of NFATc1 [41]. Generally, class IIa HDACs rely on recruitment of class I HDACs, typically HDAC3, for deacetylation of targets [15]. Interestingly, HDAC10 interacts with all class I and IIa HDACs and could potentially act as an HDAC recruiter for the repression of targets [13]. Future HDAC5 and HDAC10 studies should concentrate on a potential role for their repression of NFATc1 either cooperatively or separately.
Suppression of Hdac11 resulted in a milder phenotype than either Hdac5- or Hdac10-shRNA treatment (Fig 7). This is a surprising result to us considering HDAC11 is more closely related to class I than II HDACs [13], and we have previously shown that loss of expression of Hdac3, a class I HDAC, inhibits osteoclast differentiation [23]. Distinct from Hdac5 and Hdac10, Hdac11 knockdown only increased Dc-stamp expression (Fig 7I). This potentially means HDAC11 specifically targets the Dc-stamp promoter or fusion genes without affecting the transcription of proteins such as NFATc1, which regulate both differentiation and fusion of osteoclasts. HDAC11 may interact with NFATc1 in a protein-protein interaction to regulate NFATc1’s ability to activate targets such as Dc-stamp. The mechanism(s) by which HDAC11 inhibits osteoclast differentiation will be explored in future work. HDAC11 has previously been shown to associate with and deacetylate the IL-10 promoter in bone marrow-derived antigen-presenting cells ([42]. Surprisingly, HDAC6 and HDAC11 physically interact and oppose each other in regarding to Il-10 expression; both HDAC6 and HDAC11 associate with the Il-10 promoter, but HDAC6 promotes expression while HDAC11 represses expression [43]. With the varied expression of different HDACs throughout differentiation, a similar mechanism may exist in osteoclasts. However, as explained in detail below, HDAC6 most likely is not exerting an opposing action on HDAC11 activity in osteoclasts.
The role of HDAC6
HDAC6 can be recruited to gene promoters to modulate gene expression [43, 44]. Though we successfully reduced Hdac6 expression, no observable effects on gene expression were seen (Fig 4). Since our four examined genes are far from an exhaustive characterization of altered gene expression by HDAC suppression, it is possible that HDAC6 does have some role in regulating osteoclast-specific gene expression that is outside of our examination or it does so in a redundant manner with another HDAC. Classically, HDAC6 is known to deacetylate α-tubulin, leading to changes in microtubule stabilization and dynamics [45, 46]. It may be that HDAC6’s role in regulating osteoclast differentiation is limited to effecting changes to microtubules but not gene expression. Further characterizing potential gene targets of HDAC6 in osteoclasts is beyond the scope of this work and may be further examined in the future. In vivo knockdown of HDAC6 in embryonic stem cells displayed hyperacetylated tubulin [47]. Supporting this, studies in osteoclasts demonstrated that inhibition of HDAC6 activity with chemical inhibitors led to hyperacetylated tubulin and stabilized podosomes [30, 48]. Microtubules are important in helping form and stabilize the podosome belt, which facilitates cell adhesion and matrix resorption by osteoclasts [49]. Knowing this, we predicted knockdown of Hdac6 to produce a significant effect in the demineralization assay; however, we did not observe any phenotype in terms of osteoclast size, number, or activity after Hdac6 depletion by shRNA (Fig 4). Though these results are surprising, they are not completely unexpected considering the conflicting results between HDAC6 knockdown and inhibition of activity in other cell types. Importantly, chemical inhibition of HDAC6 led to hyperacetylated tubulin and reduced microtubule dynamics, while siRNA-mediated knockdown of Hdac6 also showed hyperacetylated tubulin but did not affect microtubule dynamics [50, 51]. In this way, we would not expect Hdac6 knockdown to impact osteoclast size, number, or activity. Future HDAC6 research should concentrate on additional transcriptional regulatory roles or potential redundancy with Sirtuin 2, a class III HDAC known to also deacetylate α-tubulin [52].
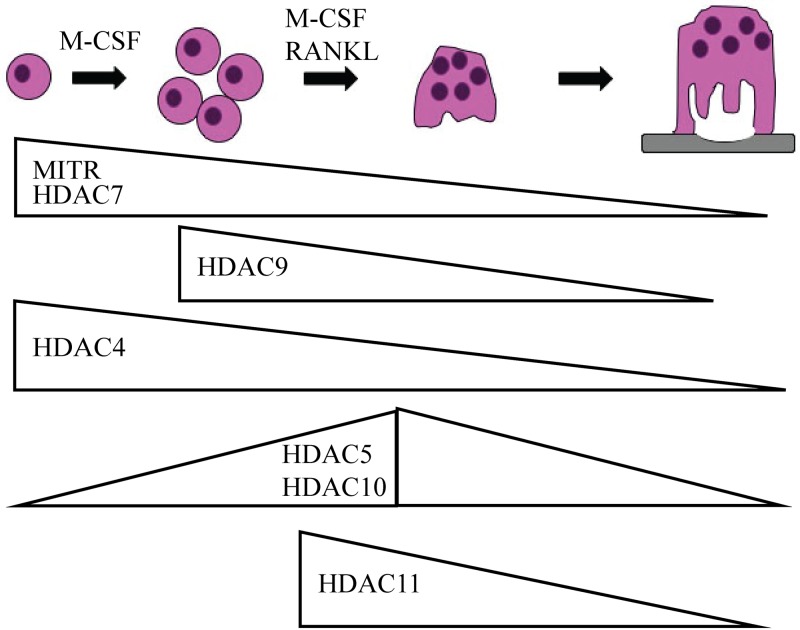
Overall, we have demonstrated for the first time the results of specifically knocking down Hdac4, 5, 6, 10, and 11 in osteoclasts. We found that HDACs differ in their temporal expression pattern during osteoclast differentiation (Fig 8). Except for HDAC6, our studies indicate that class II and IV HDACs have non-redundant roles during osteoclast differentiation. This study serves as an important starting block from which future work examining individual HDACs in osteoclasts should benefit.
Fig 8. Expression pattern of class II and IV HDACs during osteoclast differentiation.
Illustration of the stages of osteoclast differentiation with HDAC expression as determined by the western blots presented in Fig 1.
Supporting information
(DOCX)
BMMs were cultured in M-CSF only (day zero) or in M-CSF and RANKL (day one—day four) to stimulate osteoclast differentiation. qRT-PCR was performed to measure mRNA expression of Hdacs during osteoclast differentiation. Graphed data is the mean ± SD from three independent experiments. * p < 0.05; *** p <0.001; ns = not significant compared to day zero.
(TIF)
The western blots for HDAC10 and HDAC11 are labeled in the following order: 1) Bio-Rad Precision Plus Dual Color Protein Ladder 2) 293T protein lysates overexpressing HDAC 3) day zero 4) day one 5) day two 6) day three 7) day four osteoclast lysates. For HDAC5 full-length blot lanes are 2) 293T protein lysates overexpressing HDAC5 3) day zero 4) day one 5) day two 6) day three 7) day four osteoclast lysates. For HDAC4, HDAC7, HDAC9 and MITR lanes labeled are 3) day zero 4) day one 5) day two 6) day three 7) day four osteoclast lysates. Western blots analyzed for HDAC4 expression have two sets of osteoclast lysates on the blot, and those for HDAC9 and MITR expression have three different sets of lysates.
(TIF)
qPCR (A) of control and Hdac4 expressing cells and (B) full length western blot shown in Fig 2F. Number of pits (C) and average area of pits (D) of BMMs that were transduced with control or Hdac4 shRNAs (Hdac4#1 and Hdac4#2) and cultured on calcium phosphate-coated plates in the presence of M-CSF and RANKL. * p < 0.05; ** p <0.01, ns = not significant compared to control infected cells.
(TIF)
qPCR (A) of control and Hdac5 expressing cells and (B) full length western blot shown in Fig 3F. Number of pits (C) and average area of pits (D) made by BMMs that were infected with control or Hdac5 shRNAs (Hdac5#1 and Hdac5#2) and cultured on calcium phosphate-coated plates in the presence of M-CSF and RANKL. * p < 0.05; *** p <0.001 compared to control infected cells.
(TIF)
Number of pits (A) and average area of pits (B) of BMMs that were infected with control or Hdac6 shRNAs (Hdac6#1 and Hdac6#2) and cultured on calcium phosphate-coated plates in the presence of M-CSF and RANKL. ** p <0.01, ns = not significant compared to control infected cells.
(TIF)
qPCR (A) of control and Hdac9 infected cells and (B) full length western blot shown in Fig 5F. Number of pits (C) and average area (D) of pits of BMMs that were infected with control or Hdac9 shRNAs (Hdac9#1 and Hdac9#2) and plated on calcium phosphate-coated plates in the presence of M-CSF and RANKL. *** p <0.001, ns = not significant compared to control infected cells.
(TIF)
Number of pits (A) and average area of pits (B) made by BMMs infected with control or Hdac10 shRNAs (Hdac10#1 and Hdac10#2) and plated on calcium phosphate-coated plates in the presence of M-CSF and RANKL. * p < 0.05; ns = not significant compared to control infected cells.
(TIF)
qPCR (A) of control and Hdac11 infected cells and (B) full length western blot shown in Fig 7F. Number of pits (C) and average area of pits (D) of BMMs that were infected with control or Hdac11 shRNAs (Hdac11#1 and Hdac11#2) and plated on calcium phosphate-coated plates in the presence of M-CSF and RANKL.** p <0.01, *** p <0.001, ns = not significant compared to control infected cells.
(TIF)
Acknowledgments
We would like to thank Dr. Steve Bakke for his help in counting and measuring osteoclast TRAP and resorption cultures presented in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin, https://www.niams.nih.gov, R01AR061352 to KCM and EDJ and 2T32AR050938 to NCB. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–42. doi: 10.1038/nature01658 . [DOI] [PubMed] [Google Scholar]
- 2.Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20(3):345–57. doi: 10.1210/edrv.20.3.0367 . [DOI] [PubMed] [Google Scholar]
- 3.Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3:457–84. doi: 10.1146/annurev.pathmechdis.3.121806.151431 . [DOI] [PubMed] [Google Scholar]
- 4.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289(5484):1504–8. [DOI] [PubMed] [Google Scholar]
- 5.Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40(2):251–64. doi: 10.1016/j.bone.2006.09.023 . [DOI] [PubMed] [Google Scholar]
- 6.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4(8):638–49. doi: 10.1038/nrg1122 . [DOI] [PubMed] [Google Scholar]
- 7.Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet. 2000;24(2):184–7. doi: 10.1038/72855 . [DOI] [PubMed] [Google Scholar]
- 8.Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J Biol Chem. 2002;277(13):11077–83. doi: 10.1074/jbc.M111696200 . [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127 . [DOI] [PubMed] [Google Scholar]
- 10.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371 . [DOI] [PubMed] [Google Scholar]
- 11.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010 . [DOI] [PubMed] [Google Scholar]
- 12.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–49. doi: 10.1042/BJ20021321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9(1):45–57. . [DOI] [PubMed] [Google Scholar]
- 15.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104(44):17335–40. doi: 10.1073/pnas.0706487104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGee-Lawrence ME, Westendorf JJ. Histone deacetylases in skeletal development and bone mass maintenance. Gene. 2011;474(1–2):1–11. doi: 10.1016/j.gene.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21(7):993–1002. doi: 10.1359/jbmr.060415 . [DOI] [PubMed] [Google Scholar]
- 18.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–65. doi: 10.1146/annurev.biochem.74.082803.133500 . [DOI] [PubMed] [Google Scholar]
- 19.Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25(8):2873–84. doi: 10.1128/MCB.25.8.2873-2884.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantley MD, Fairlie DP, Bartold PM, Rainsford KD, Le GT, Lucke AJ, et al. Inhibitors of histone deacetylases in class I and class II suppress human osteoclasts in vitro. J Cell Physiol. 2011;226(12):3233–41. doi: 10.1002/jcp.22684 . [DOI] [PubMed] [Google Scholar]
- 21.Nakamura T, Kukita T, Shobuike T, Nagata K, Wu Z, Ogawa K, et al. Inhibition of histone deacetylase suppresses osteoclastogenesis and bone destruction by inducing IFN-beta production. J Immunol. 2005;175(9):5809–16. . [DOI] [PubMed] [Google Scholar]
- 22.Rahman MM, Kukita A, Kukita T, Shobuike T, Nakamura T, Kohashi O. Two histone deacetylase inhibitors, trichostatin A and sodium butyrate, suppress differentiation into osteoclasts but not into macrophages. Blood. 2003;101(9):3451–9. doi: 10.1182/blood-2002-08-2622 . [DOI] [PubMed] [Google Scholar]
- 23.Pham L, Kaiser B, Romsa A, Schwarz T, Gopalakrishnan R, Jensen ED, et al. HDAC3 and HDAC7 have opposite effects on osteoclast differentiation. J Biol Chem. 2011;286(14):12056–65. doi: 10.1074/jbc.M110.216853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stemig M, Astelford K, Emery A, Cho JJ, Allen B, Huang TH, et al. Deletion of histone deacetylase 7 in osteoclasts decreases bone mass in mice by interactions with MITF. PLoS One. 2015;10(4):e0123843 doi: 10.1371/journal.pone.0123843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z, Wei W, Dechow PC, Wan Y. HDAC7 inhibits osteoclastogenesis by reversing RANKL-triggered beta-catenin switch. Mol Endocrinol. 2013;27(2):325–35. doi: 10.1210/me.2012-1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Z, Wei W, Huynh H, Wan Y. HDAC9 Inhibits Osteoclastogenesis via Mutual Suppression of PPARgamma/RANKL Signaling. Mol Endocrinol. 2015;29(5):730–8. doi: 10.1210/me.2014-1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res. 2000;15(8):1477–88. doi: 10.1359/jbmr.2000.15.8.1477 . [DOI] [PubMed] [Google Scholar]
- 28.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–8. doi: 10.1038/417455a . [DOI] [PubMed] [Google Scholar]
- 29.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21(24):6820–31. doi: 10.1093/emboj/cdf682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118(Pt 13):2901–11. doi: 10.1242/jcs.02425 . [DOI] [PubMed] [Google Scholar]
- 31.Purev E, Neff L, Horne WC, Baron R. c-Cbl and Cbl-b act redundantly to protect osteoclasts from apoptosis and to displace HDAC6 from beta-tubulin, stabilizing microtubules and podosomes. Mol Biol Cell. 2009;20(18):4021–30. doi: 10.1091/mbc.E09-03-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer DD, Cai R, Bhatia U, Asselbergs FA, Song C, Terry R, et al. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J Biol Chem. 2002;277(8):6656–66. doi: 10.1074/jbc.M108055200 . [DOI] [PubMed] [Google Scholar]
- 33.Guardiola AR, Yao TP. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J Biol Chem. 2002;277(5):3350–6. doi: 10.1074/jbc.M109861200 . [DOI] [PubMed] [Google Scholar]
- 34.Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14(1):45–54. [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison BE, D'Mello SR. Polydactyly in mice lacking HDAC9/HDRP. Exp Biol Med (Maywood). 2008;233(8):980–8. doi: 10.3181/0802-RM-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison BE, Majdzadeh N, Zhang X, Lyles A, Bassel-Duby R, Olson EN, et al. Neuroprotection by histone deacetylase-related protein. Mol Cell Biol. 2006;26(9):3550–64. doi: 10.1128/MCB.26.9.3550-3564.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang CL, McKinsey TA, Chang SR, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110(4):479–88. doi: 10.1016/S0092-8674(02)00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim K, Lee SH, Ha Kim J, Choi Y, Kim N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP). Mol Endocrinol. 2008;22(1):176–85. doi: 10.1210/me.2007-0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202(9):1261–9. doi: 10.1084/jem.20051150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JH, Kim K, Jin HM, Song I, Youn BU, Lee SH, et al. Negative feedback control of osteoclast formation through ubiquitin-mediated down-regulation of NFATc1. J Biol Chem. 2010;285(8):5224–31. doi: 10.1074/jbc.M109.042812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JH, Kim K, Youn BU, Jin HM, Kim JY, Moon JB, et al. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem J. 2011;436(2):253–62. doi: 10.1042/BJ20110062 . [DOI] [PubMed] [Google Scholar]
- 42.Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10(1):92–100. doi: 10.1038/ni.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng F, Lienlaf M, Perez-Villarroel P, Wang HW, Lee C, Woan K, et al. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol Immunol. 2014;60(1):44–53. doi: 10.1016/j.molimm.2014.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–31. doi: 10.1016/j.cell.2009.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Peng L, Seto E, Huang S, Qiu Y. Modulation of histone deacetylase 6 (HDAC6) nuclear import and tubulin deacetylase activity through acetylation. J Biol Chem. 2012;287(34):29168–74. doi: 10.1074/jbc.M112.371120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Yang XJ. Tubulin acetylation: responsible enzymes, biological functions and human diseases. Cell Mol Life Sci. 2015;72(22):4237–55. doi: 10.1007/s00018-015-2000-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22(5):1168–79. doi: 10.1093/emboj/cdg115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akisaka T, Yoshida H, Takigawa T. Differential distribution of posttranslationally modified microtubules in osteoclasts. J Histochem Cytochem. 2011;59(6):630–8. doi: 10.1369/0022155411405334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duplan MB, Zalli D, Stephens S, Zenger S, Neff L, Oelkers JM, et al. Microtubule Dynamic Instability Controls Podosome Patterning in Osteoclasts through EB1, Cortactin, and Src. Molecular and Cellular Biology. 2014;34(1):16–29. doi: 10.1128/MCB.00578-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zilberman Y, Ballestrem C, Carramusa L, Mazitschek R, Khochbin S, Bershadsky A. Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. J Cell Sci. 2009;122(Pt 19):3531–41. doi: 10.1242/jcs.046813 . [DOI] [PubMed] [Google Scholar]
- 51.Asthana J, Kapoor S, Mohan R, Panda D. Inhibition of HDAC6 deacetylase activity increases its binding with microtubules and suppresses microtubule dynamic instability in MCF-7 cells. J Biol Chem. 2013;288(31):22516–26. doi: 10.1074/jbc.M113.489328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD(+)-dependent tubulin deacetylase. Molecular Cell. 2003;11(2):437–44. doi: 10.1016/S1097-2765(03)00038-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
BMMs were cultured in M-CSF only (day zero) or in M-CSF and RANKL (day one—day four) to stimulate osteoclast differentiation. qRT-PCR was performed to measure mRNA expression of Hdacs during osteoclast differentiation. Graphed data is the mean ± SD from three independent experiments. * p < 0.05; *** p <0.001; ns = not significant compared to day zero.
(TIF)
The western blots for HDAC10 and HDAC11 are labeled in the following order: 1) Bio-Rad Precision Plus Dual Color Protein Ladder 2) 293T protein lysates overexpressing HDAC 3) day zero 4) day one 5) day two 6) day three 7) day four osteoclast lysates. For HDAC5 full-length blot lanes are 2) 293T protein lysates overexpressing HDAC5 3) day zero 4) day one 5) day two 6) day three 7) day four osteoclast lysates. For HDAC4, HDAC7, HDAC9 and MITR lanes labeled are 3) day zero 4) day one 5) day two 6) day three 7) day four osteoclast lysates. Western blots analyzed for HDAC4 expression have two sets of osteoclast lysates on the blot, and those for HDAC9 and MITR expression have three different sets of lysates.
(TIF)
qPCR (A) of control and Hdac4 expressing cells and (B) full length western blot shown in Fig 2F. Number of pits (C) and average area of pits (D) of BMMs that were transduced with control or Hdac4 shRNAs (Hdac4#1 and Hdac4#2) and cultured on calcium phosphate-coated plates in the presence of M-CSF and RANKL. * p < 0.05; ** p <0.01, ns = not significant compared to control infected cells.
(TIF)
qPCR (A) of control and Hdac5 expressing cells and (B) full length western blot shown in Fig 3F. Number of pits (C) and average area of pits (D) made by BMMs that were infected with control or Hdac5 shRNAs (Hdac5#1 and Hdac5#2) and cultured on calcium phosphate-coated plates in the presence of M-CSF and RANKL. * p < 0.05; *** p <0.001 compared to control infected cells.
(TIF)
Number of pits (A) and average area of pits (B) of BMMs that were infected with control or Hdac6 shRNAs (Hdac6#1 and Hdac6#2) and cultured on calcium phosphate-coated plates in the presence of M-CSF and RANKL. ** p <0.01, ns = not significant compared to control infected cells.
(TIF)
qPCR (A) of control and Hdac9 infected cells and (B) full length western blot shown in Fig 5F. Number of pits (C) and average area (D) of pits of BMMs that were infected with control or Hdac9 shRNAs (Hdac9#1 and Hdac9#2) and plated on calcium phosphate-coated plates in the presence of M-CSF and RANKL. *** p <0.001, ns = not significant compared to control infected cells.
(TIF)
Number of pits (A) and average area of pits (B) made by BMMs infected with control or Hdac10 shRNAs (Hdac10#1 and Hdac10#2) and plated on calcium phosphate-coated plates in the presence of M-CSF and RANKL. * p < 0.05; ns = not significant compared to control infected cells.
(TIF)
qPCR (A) of control and Hdac11 infected cells and (B) full length western blot shown in Fig 7F. Number of pits (C) and average area of pits (D) of BMMs that were infected with control or Hdac11 shRNAs (Hdac11#1 and Hdac11#2) and plated on calcium phosphate-coated plates in the presence of M-CSF and RANKL.** p <0.01, *** p <0.001, ns = not significant compared to control infected cells.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.