Abstract
Background
The recent application of gene-sequencing technology has identified many new somatic mutations in patients with myelodysplastic syndromes (MDS). Among them, serine and arginine rich splicing factor 2 (SRSF2) mutations belonging to the RNA splicing pathway were of interest. Many studies have already reported the potential prognostic value of SRSF2 mutations in MDS patients, with controversial results. Therefore, a meta-analysis was performed to investigate their prognostic impact on MDS.
Methods
Databases, including PubMed, Embase and the Cochrane Library, were searched for relevant studies published up to 14 October 2016. Overall survival (OS) was selected as the primary endpoint, and acute myeloid leukemia (AML) transformation was the secondary endpoint. We extracted the corresponding hazard ratios (HRs) and their 95% confidence intervals (CIs) for OS and AML transformation from multivariate Cox proportional hazards models. The combined HRs with their 95% CIs were calculated using fixed or random effect models.
Results
A total of 10 cohort studies, covering 1864 patients with de novo MDS and 294 patients with SRSF2 mutations, were included in the final meta-analysis. Our results indicated that SRSF2 mutations had an adverse prognostic impact on OS (p<0.0001) and AML transformation (p = 0.0005) in the total population. Among the MDS patients with low or intermediate-1 risk defined according to the International Prognostic Scoring System (IPSS), SRSF2 mutations predicted a shorter OS (p = 0.009) and were more likely to transform to AML (p = 0.007).
Conclusions
This meta-analysis indicates an independent, adverse prognostic impact of SRSF2 mutations on OS and AML transformation in patients with de novo MDS. This also applies to the subgroup of low- or intermediate-1-IPSS risk MDS. The identification of mutations in SRSF2 can improve current risk stratification and help make treatment decisions.
Introduction
Myelodysplastic syndrome (MDS) is a group of clonal hematopoietic stem cell disorders characterized by ineffective hematopoiesis resulting in peripheral blood cytopenias and a risk of progression to acute myeloid leukemia (AML)[1]. The International Prognostic Scoring System (IPSS) and its revision (IPSS-R), depending mainly on bone marrow cytogenetics, marrow blast percentage and the presence of cytopenias, have been widely used as the standard for assessing prognosis and guiding treatment decisions for MDS patients[2,3]. Neither the IPSS nor the IPSS-R considers somatic mutations, though they play a significant role in the diagnosis and prognosis of leukemia. Recently, the application of whole-genome sequencing and whole-exome sequencing has led to the identification of new somatic mutations, which may provide important prognostic information in patients with MDS[4,5].
Through the above techniques, numerous mutations in genes encoding RNA splicing machinery, epigenetic modifiers, chromatin modifiers, transcription factors, signal transducers, RAS pathway members, cohesin complex members and DNA repair proteins have been identified in MDS[6]. Among them, gene mutations involved in the RNA splicing pathway are the most frequent molecular abnormalities[7,8]. RNA splicing is a process that generates mature mRNAs by excising introns and splicing exons from pre-messenger RNA (pre-mRNA); alteration of the excising and splicing of a single pre-mRNA transcript can be used to produce many different mRNAs and result in protein diversity[9,10]. The spliceosome mutations in MDS affect the major components of both the E and A splicing complexes in a mutually exclusive manner, leading to the impaired recognition of 3′-splice site recognition during pre-mRNA processing, inducing abnormally spliced mRNA species and compromising hematopoiesis[5].
One of the potential candidate genes involved in the RNA splicing pathway is serine and arginine rich splicing factor 2 (SRSF2). SRSF2, located on chromosome 17q25.2, encodes a protein belonging to the serine/arginine-rich (SR) splicing regulatory factor family[11]. SRSF2 plays a role in preventing exon skipping, ensuring the accuracy of splicing and regulating alternative pre-mRNA splicing[11]. Although the precise role of SRSF2 mutations in leukemogenesis remains elusive, many studies have already reported the potential prognostic value of SRSF2 mutations in MDS patients, with controversial results. Lin et al. [12] and Thol et al. [13] reported SRSF2 mutations were significantly correlated with poor survival in patients with MDS, while others [14,15] reported no prognostic impact of SRSF2 mutations. Hence, we performed a meta-analysis on data from related published studies to further explore the combined prognostic impacts of SRSF2 mutations for patients with de novo MDS.
Materials and methods
Literature search
We conducted a literature search of several databases, including PubMed, Embase and the Cochrane Library, for potentially relevant studies published up to October 14, 2016. The following terms were used to find eligible studies: “SRSF2” or “serine and arginine rich splicing factor 2” or “SC35” and “myelodysplastic syndrome” or “MDS” or “myelodysplasia” or “preleukemia”. No language limitations were added to the search strategy. References of eligible studies were manually searched to find other potentially relevant articles.
Inclusion and exclusion
Only papers that met all the following criteria were included: (1)The study focused on the prognostic impact of SRSF2 mutations in de novo MDS patients; (2)The study provided sufficient survival data for patients with SRSF2 mutations, at least on overall survival (OS); and (3)The study was published as a full article in English. Review articles, case reports and laboratory studies were excluded. If the same or overlapping data was presented in multiple studies, only the most recent or the highest quality study was included.
Two reviewers (Xue Zheng and Zhi Zhan) screened the database and identified the eligible studies, independently. Disagreements were resolved by discussion.
Data extraction
Two reviewers (Xue Zheng and Duolan Naren) independently extracted relevant information from each eligible study onto a spreadsheet. The data included the first author’s name, year of publication, country of origin, number of patients, age and gender distribution of patients, criteria for classification of MDS, MDS subtype, and the distribution of patients by IPSS classification. We selected OS as the primary endpoint and AML transformation as the secondary endpoint. Endpoints for OS were defined as either deceased (failure) or alive at last follow-up, and the time to AML transformation was calculated beginning from the time the patient entered the trial to the time of AML diagnosis. We extracted the corresponding hazard ratios (HRs) and their 95% confidence intervals (CIs) for OS and AML transformation from multivariate Cox proportional hazards models to evaluate the prognostic impact of SRSF2-mutated compared with unmutated patients with MDS. If the published paper did not report the required data for analysis, we contacted the corresponding authors to obtain missing data. Disagreements between reviewers regarding data abstraction were resolved through discussion.
Quality assessment
Two reviewers (Xue Zheng and Duolan Naren) independently evaluated the methodological quality of each included study. The quality of cohort studies was evaluated by the Newcastle-Ottawa quality assessment scale (NOS). The NOS includes a total of 9 points, with 4 points for selection, 2 points for comparability, and 3 points for exposure or outcome[16]. Cohort studies that scored six or more points were regarded to be of high quality[16]. Disagreements were resolved by discussion.
Statistical analysis
Review Manager version 5.3 software, following the recommendation of the Cochrane Collaboration (http://tech.cochrane.org/revman/download), was used to calculate the combined survival impact of SRSF2 mutations. The prognostic effect of SRSF2 mutations on OS and AML transformation was evaluated by calculation of the combined HRs and their 95% CIs with the generic inverse variance method. The result suggested statistical significance if the 95% CI did not overlap 1. Moreover, SRSF2 mutations contributed an adverse survival effect compared to unmutated patients when the HR was more than 1. The heterogeneity of the studies was evaluated through the chi-squared test, with significance set at a p-value of less than 0.10. The I2 statistic was used to quantify the heterogeneity. An I2 value of less than 25% was regarded as low heterogeneity, a value between 25 and 50% indicated moderate heterogeneity, and a value over 50% suggested high heterogeneity[17]. The random effect model was used if high heterogeneity was observed; otherwise, a fixed effect model was used for the meta-analysis.
We used sensitivity analysis and meta-regression analysis regarding the OS of the total population to evaluate the heterogeneity between studies. Sensitivity analysis was used to assess the influence of each study on the stability of the pooled results by sequential omission of one study at a time. Meta-regression analysis was used to evaluate the possible influence of publication year or study country on outcome when there were 10 or more studies including outcome. Funnel plots, Begg’s tests and Egger’s tests were used to screen for potential publication bias regarding the OS of the total population[18,19]. The calculations were carried out in Stata version 12.0 software (Stata Corp, College Station, TX, USA) with a p-value less than 0.05 being considered significant.
Results
Study identification and selection
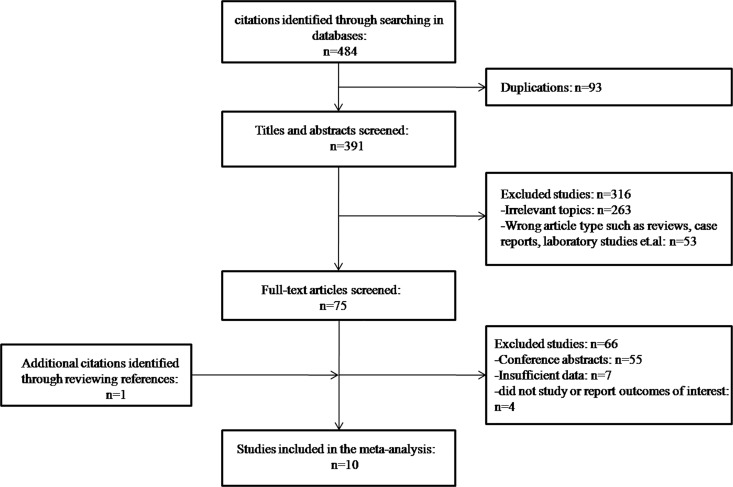
As shown in Fig 1, the initial search revealed 484 studies. After exclusion of 93 duplicates, 391 citations were further reviewed by reading the titles and abstracts, and 316 citations were then excluded for irrelevant article type or irrelevant subject. A total of 75 studies were left for full text review. Among them, 55 studies were excluded as conference abstracts, and 11 were further excluded because they did not evaluate outcome or did not provide sufficient data. During revision, one additional citation was reviewed and included in the final meta-analysis, leaving a total of 10 citations[12–15, 20–25](Fig 1).
Fig 1. Flow diagram of study selection.
Characteristics of included studies
The 10 included studies were all cohort studies and were published between 2012 and 2016 (Table 1). They were conducted in East Asia, Europe or North America. The studies included a total of 1864 patients with MDS, in which 294 patients harbored a mutation in SRSF2. For one study that included patients with both MDS and AML[23], only patients with MDS were studied in the meta-analysis. Occurrence of mutations of SRSF2 varied between 4.3% and 15% in MDS patients and were more frequent in chronic myelomonocytic leukemia (CMML) (29%-46%).
Table 1. Characteristics of studies included in the meta-analysis.
| MDS Subtype |
MDS | IPSS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Country | n/N | Age(years) | Sex(M/F) | RA/RCUD /RARS |
RAEB/ RAEB-t |
RCMD/ RCMD-RS |
CMML | Others | criteria | Low+Int1/Int2+ High/Unknown |
Adjustments | NOS |
| Wu | China | 13/304 | 57(11,89)* | 162/142 | 0/20/9 | 123/0 | 0/145 | 0 | 7 | WHO | 212/90/2 | 1,2,5,6,7 | 9 |
| 2016 | |||||||||||||
| Cui | China | 42/145 | 63(18,85)* | 98/47 | 0/0/0 | 0/0 | 0/0 | 145 | 0 | WHO | NR | NR | 8 |
| 2015 | |||||||||||||
| Kim | Korea | 5/52 | 52(18,73)* | 36/16 | 2/0/1 | 27/0 | 20/0 | 0 | 2 | WHO | 22/30/0 | 6,7 | 7 |
| 2015 | |||||||||||||
| Kang | Korea | 13/129 | 63.7±12.4# | 71/58 | 0/19/0 | 46/0 | 56/0 | 0 | 8 | WHO | NR | 1,2,4,7 | 8 |
| 2015 | |||||||||||||
| Karimi | Sweden | 15/100 | 72(32,88)* | 51/49 | 0/0/10 | 41/0 | 2/11 | 15 | 21 | WHO | NR | 1,2,4,7 | 8 |
| 2015 | |||||||||||||
| Lin | China | 5/108 | 60(20,86)* | 64/45 | 10/0/0 | 48/0 | 36/8 | 0 | 6 | WHO | 72/32/4 | 1,2,3,5,7 | 9 |
| 2014 | |||||||||||||
| Itzykson | France | 101/312 | 74(41,93)* | 210/102 | 0/0/0 | 0/0 | 0/0 | 312 | 0 | WHO | NR | 7 | 7 |
| 2013 | |||||||||||||
| Thol | Germany | 24/193 | NR(36,92)* | 119/74 | 38/0/20 | 53/0 | 21/9 | 0 | 52 | WHO | 96/51/43 | 1,3,7 | 8 |
| 2012 | |||||||||||||
| Wu | China | 34/233 | 66(18,95)* | 161/72 | 98 | 102 | 0/0 | 33 | 0 | FAB | 115/99/19 | 1,3,7 | 8 |
| 2012 | |||||||||||||
| Bejar | USA | 42/288 | 69(15,90)* | 203/85 | 173/0/41 | 71/3※ | 0/0 | 0 | 0 | FAB | 288/0/0 | 1,2,3,7 | 8 |
| 2012 |
Abbreviations: CMML, chronic myelomonocytic leukemia; FAB, French American British classification; IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndromes; n, number of patients with SRSF2 mutations; N, number of patients in total; NOS, Newcastle-Ottawa quality assessment scale; NR, not reported; RA, refractory anemia; RARS, RA with ringed sideroblasts; RAEB, RA with excess blasts; RAEB-t, RAEB in transformation; RCMD, refractory cytopenia with multilineage dysplasia; RCMD-RS, refractory cytopenia with multilineage dysplasia and ringed sideroblasts; RCUD, refractory cytopenia with unilineage dysplasia; WHO, World Health Organization.
*, median age (range)
#, mean age ± SD
※, the 3 patients with RAEB-t had circulating blasts but≤10% blasts in the bone marrow.
Main adjusted variables in multivariate Cox proportional hazards models: 1 = age; 2 = sex; 3 = IPSS; 4 = IPSS-R; 5 = neutrophils; 6 = hemoglobin; 7 = mutation status
In addition, we managed to contact the authors for some unpublished data. Kim et al. offered the HRs and their 95% CIs for OS after initial treatment of the MDS patients with SRSF2 mutations in their multivariate cox regression analysis. Similarly, Cui et al. and Bejar et al. also offered the HRs and their 95% CIs for OS in the same setting.
Quality assessment of included studies
As shown in Table 1, the mean overall NOS score was 8 (range 7–9), indicating that the quality of included studies was high.
Clinical characteristics of MDS patients with SRSF2 mutations
Patients carrying SRSF2 mutations were older than those with unmutated SRSF2[12,14,15,21,25], and SRSF2 mutations were significantly associated with old age (p<0.05)[14,15,21]. Patients with mutations in SRSF2 had an increased incidence of RUNX1, TET2, IDH1, IDH2 and ASXL1 mutations[13,15,21,24].
Analysis of outcomes
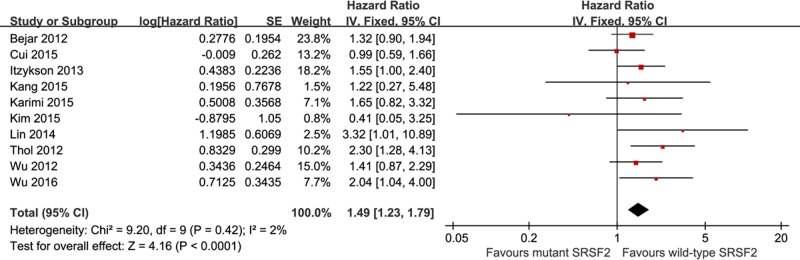
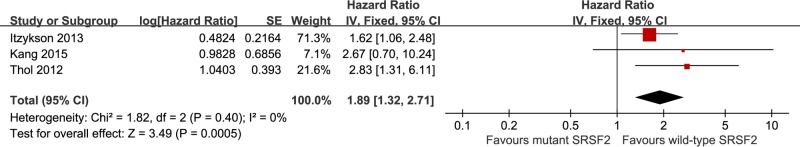
As shown in Fig 2, the meta-analysis of the effect of SRSF2 mutations on OS in MDS patients was performed for all cohort studies. Our results indicated that the presence of SRSF2 mutations was an independent adverse prognostic factor for OS (HR = 1.49, 95% CI: 1.23–1.79, with a p-value less than 0.0001) with a low heterogeneity (I2 = 2%). Three studies reported data on AML transformation, with 634 patients and 138 with SRSF2 mutations (Fig 3). The pooled HR for AML transformation was 1.89 (95% CI: 1.32–2.71, with a p-value of 0.0005, I2 = 0%) for SRSF2-mutated patients compared with unmutated patients, revealing that patients with SRSF2 mutations have a more rapid and frequent transformation to AML. There was no heterogeneity among the analyzed studies (I2 = 0%).
Fig 2. Forest plots of pooled HRs and 95% CIs for OS assessing the prognostic value of SRSF2 mutations in the cohort of MDS patients.
Fig 3. Forest plots of pooled HRs and 95% CIs for AML transformation assessing the prognostic value of SRSF2 mutations in the cohort of MDS patients.
Several studies focused on patients with low- or intermediate-1-IPSS risk MDS harboring SRSF2 mutations. The pooled HR for OS was 1.50 (95% CI: 1.11–2.03, with a p-value of 0.009, I2 = 0%), and the HR for AML transformation was 3.12 (95% CI: 1.36–7.11, with a p-value of 0.007, I2 = 0%) for patients with low- or intermediate-1-IPSS risk MDS with SRSF2 mutations, compared to SRSF2-unmutated patients (Fig 4), indicating that SRSF2 mutations predicted an independent unfavorable prognostic impact in terms of both OS and AML transformation. Our results showed no heterogeneity among these studies (I2 = 0%).
Fig 4.
Forest plots of pooled HRs and 95% CIs assessing the prognostic value of SRSF2 mutations in patients with low- or intermediate-1-IPSS risk MDS for: (A) OS and (B) AML transformation.
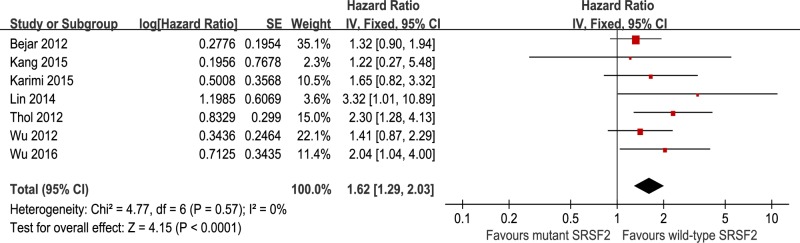
Among several studies, SRSF2 mutations were strongly associated with older age[14,15,21]. We further performed a subgroup analysis, examining whether SRSF2 mutations could still predict prognosis after adjusting for age. The meta-analysis of SRSF2 mutations after adjustment for age in COX multivariable models was carried out for 7 cohort studies, with 1355 patients and 146 with SRSF2 mutations (Fig 5). In this population, patients with SRSF2 mutations had a significantly shorter OS compared to unmutated patients (HR = 1.62, 95% CI: 1.29–2.03, with a p-value less than 0.0001), with no heterogeneity shown by I2 testing (I2 = 0%).
Fig 5. Forest plots of pooled HRs and 95% CIs for OS assessing the prognostic value of SRSF2 mutations in MDS patients after adjusting for age in COX multivariable models.
Sensitivity analysis, meta-regression, and publication bias
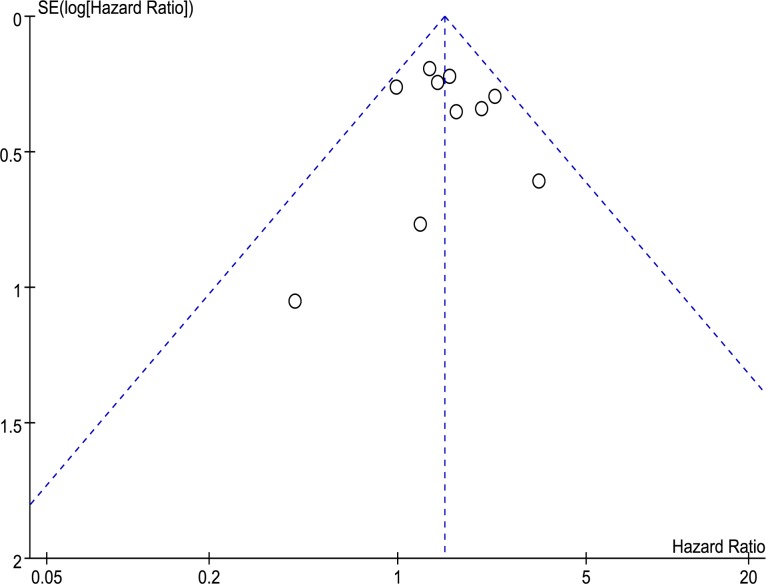
We conducted a sensitivity analysis by excluding one study at a time from the meta-analysis, examining the effect of individual studies on the combined HR. The result indicated that there were no significant effects of individual studies on the combined HR for OS in the total population. Meta-regression analysis showed no significant correlations between the publication year or the study country and OS in all patients. There was no significant publication bias in the funnel plot of OS of all population, which contained all 10 studies (Fig 6). The results of Begg’s test and Egger’s test also suggested no publication bias (p = 0.858 of Begg’s test and p = 0.774 of Egger’s test)
Fig 6. Funnel plots of publication bias for OS of all patients.
Discussion
The pooled meta-analysis of included studies demonstrated that patients with SRSF2 mutations had a worse outcome than unmutated patients in the total population, with a shorter OS and a more rapid and frequent transformation to AML. These mutations were also correlated with adverse outcomes in OS and AML transformation in the subgroup of low- or intermediate-1-IPSS risk patients. MDS is a heterogeneous disease with various clinical outcomes, thus, an accurate prediction of prognosis is crucial for selecting the appropriate treatment. Many patients with low- or intermediate-1-IPSS risk MDS have a more aggressive clinical course and an inferior OS than expected. This may be because neither the IPSS nor the IPSS-R takes into account gene mutations, which can provide prognostic information that is not captured by the IPSS and its revision. Our meta-analysis suggested that SRSF2 mutations were an independent molecular marker for shorter survival and AML transformation in the subgroup of low- or intermediate-1-IPSS risk MDS patients. A combination of SRSF2 status and IPSS assessment might improve prognostic evaluation for patients with low- or intermediate-1-IPSS risk MDS, and patients with SRSF2 mutations might benefit from more intensive treatment. However, this analysis is limited by the small number of included studies. More studies concerning SRSF2 mutations in this subgroup are needed to further verify or modify the results of the pooled analysis.
Increasing age is a well-known significant prognostic factor for survival in MDS patients[3], and our included studies demonstrated that MDS patents with SRSF2 mutations were of older age compared with unmutated patients. Wu et al. [15] found that the poor prognostic impact of SRSF2 mutations on OS might be explained by their close association with old age. Our meta-analysis suggested that SRSF2 mutations still had a significantly poor prognostic effect on OS after adjusting for the effect of age. Thus, mutations in SRSF2 provide independent prognostic information, which can not be attributed to old age.
Although the precise mechanisms of the contributions of SRSF2 mutations to the pathogenesis of MDS remain largely unknown, a growing number of studies have explained the mechanisms of SRSF2 mutations leading to the poor prognosis of MDS patients: (1) SRSF2 mutations occur early and are implicated as founder mutations in MDS[8,26], suggesting that they may play an important role in disease initiation. (2) SRSF2 mutations alter normal sequence-specific RNA binding activity, therefore altering the recognition of specific exonic splicing enhancer motifs to drive recurrent mis-splicing of key hematopoietic regulators[27]. Evidence from a study using an SRSF2 knock-in mouse model demonstrates that SRSF2 mutations can impair differentiation and increase apoptosis, resulting in peripheral cytopenias and morphologic dysplasia[27]. (3) SRSF2 has a critical role in maintaining genomic stability and regulating cell proliferation[28]. Depletion of SRSF2 can cause genomic instability, which may be a potential mechanism for acquiring other gene mutations during disease progression to induce AML transformation [28]. Yoshida et al. reported that approximately 20% of MDS cases have no known genetic changes, thus, SRSF2 mutations can not fully account for the pathogenesis of MDS[5]. Apart from the studies showing the above mechanisms, further studies are needed to sufficiently explain the contribution of SRSF2 mutations to MDS.
Our paper is the first meta-analysis concerning the controversial prognostic value of SRSF2 mutations in MDS patients. However, several limitations should be acknowledged. First, the adjusted factors from multivariate Cox proportional hazard models in each eligible study were different, which might represent methodological heterogeneity among the 10 studies. Second, there was much clinical heterogeneity between studies, such as the diverse treatment programs, age and gender distribution of patients, MDS subtype, cytogenetic and molecular abnormalities, time of follow-up, and the methods of gene sequencing, and this heterogeneity potentially effected clinical outcomes. Third, as SRSF2 mutations coexist with other mutations, such as mutations of RUNX1, TET2, IDH1, IDH2 and ASXL1, we could not evaluate the potential effects of gene-gene interactions on survival outcome. Finally, although Begg’s test and Egger’s test results suggested that no publication bias was observed, it still existed due to our using only published articles.
In conclusion, our meta-analysis indicates an independent, adverse prognostic impact of SRSF2 mutations on OS and AML transformation in patients with de novo MDS. This also applies to the subgroup of low- or intermediate-1-IPSS risk MDS. In addition, the inferior OS of patients with SRSF2 mutations could not be attributed to the close association of the mutation with old age. Mutations in SRSF2 can provide prognostic information that is not captured by the IPSS and might improve current risk stratification and treatment decisions. A better understanding of splicing mutations is leading to the development of spliceosome inhibitors, which could provide novel targeted therapies for patients[29]. Further studies using prospective randomized controlled trials are needed to confirm the prognostic impact of SRSF2 mutations, especially in patients with low- or intermediate-1-IPSS risk MDS.
Supporting information
(PDF)
(PDF)
Acknowledgments
We sincerely thank Pu Kuang and Jianli Zhao for advices of statistical analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by the Foundation of the Science & Technology Department of Sichuan Province (No. 2015SZ0234-5), Foundation of Administration of traditional Chinese medicine of Sichuan Province (No. 2014A038) and the Foundation of Science and Technology Bureau of Chengdu (No. 2016-HM01-00001-SF), http://www.scst.gov.cn/. YPG received these funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ades L, Itzykson R, Fenaux P. Myelodysplastic syndromes. Lancet. 2014; 383: 2239–2252. doi: 10.1016/S0140-6736(13)61901-7 [DOI] [PubMed] [Google Scholar]
- 2.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997; 89: 2079–2088. [PubMed] [Google Scholar]
- 3.Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Solé F, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012; 120: 2454–2465. doi: 10.1182/blood-2012-03-420489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter MJ, Shen D, Shao J, Ding L, White BS, Kandoth C, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013; 27: 1275–1282. doi: 10.1038/leu.2013.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011; 478: 64–69. doi: 10.1038/nature10496 [DOI] [PubMed] [Google Scholar]
- 6.Ganguly BB, Kadam NN. Mutations of myelodysplastic syndromes (MDS): An update. Mutat Res Rev Mutat Res. 2016; 769: 47–62. doi: 10.1016/j.mrrev.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 7.Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012; 119: 3211–3218. doi: 10.1182/blood-2011-12-400994 [DOI] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013; 122: 3616–3627. doi: 10.1182/blood-2013-08-518886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012; 37: 179–188. doi: 10.1016/j.tibs.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boultwood J, Dolatshad H, Varanasi SS, Yip BH, Pellagatti A. The role of splicing factor mutations in the pathogenesis of the myelodysplastic syndromes. Adv Biol Regul. 2014; 54: 153–161. doi: 10.1016/j.jbior.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 11.Maciejewski JP, Padgett RA. Defects in spliceosomal machinery: a new pathway of leukaemogenesis. Br J Haematol. 2012; 158: 165–173. doi: 10.1111/j.1365-2141.2012.09158.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J, Yang J, Wen XM, Yang L, Deng ZQ, Qian Z, et al. Detection of SRSF2-P95 mutation by high-resolution melting curve analysis and its effect on prognosis in myelodysplastic syndrome. PLoS One. 2014; doi: 10.1371/journal.pone.0115693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012; 119: 3578–3584. doi: 10.1182/blood-2011-12-399337 [DOI] [PubMed] [Google Scholar]
- 14.Kang MG, Kim HR, Seo BY, Lee JH, Choi SY, Kim SH, et al. The prognostic impact of mutations in spliceosomal genes for myelodysplastic syndrome patients without ring sideroblasts. BMC Cancer. 2015; doi: 10.1186/s12885-015-1493-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu SJ, Kuo YY, Hou HA, Li LY, Tseng MH, Huang CF, et al. The clinical implication of SRSF2 mutation in patients with myelodysplastic syndrome and its stability during disease evolution. Blood. 2012; 120: 3106–3111. doi: 10.1182/blood-2012-02-412296 [DOI] [PubMed] [Google Scholar]
- 16.Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
- 17.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003; 327: 557–560. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994; 50: 1088–1101. [PubMed] [Google Scholar]
- 19.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997; 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Song L, Xu L, Chang C, Xu F, Wu D, et al. Genetic landscape of recurrent ASXL1, U2AF1, SF3B1, SRSF2, and EZH2 mutations in 304 Chinese patients with myelodysplastic syndromes. Tumor Biol. 2016; 37: 4633–4640. [DOI] [PubMed] [Google Scholar]
- 21.Cui Y, Tong H, Du X., Li B, Gale RP, Qin T, et al. Impact of TET2, SRSF2, ASXL1 and SETBP1 mutations on survival of patients with chronic myelomonocytic leukemia. Exp Hematol Oncol. 2015; doi: 10.1186/s40164-015-0009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M, Yahng SA, Kwon A, Park J, Jeon YW, Yoon JH, et al. Mutation in TET2 or TP53 predicts poor survival in patients with myelodysplastic syndrome receiving hypomethylating treatment or stem cell transplantation. Bone Marrow Transplant. 2015; 50: 1132–1134. doi: 10.1038/bmt.2015.110 [DOI] [PubMed] [Google Scholar]
- 23.Karimi M, Nilsson C, Dimitriou M, Jansson M, Matsson H, Unneberg P, et al. High-throughput mutational screening adds clinically important information in myelodysplastic syndromes and secondary or therapy-related acute myeloid leukemia. Haematologica. 2015; 100: e223–e225. doi: 10.3324/haematol.2014.118034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013; 31: 2428–2436. doi: 10.1200/JCO.2012.47.3314 [DOI] [PubMed] [Google Scholar]
- 25.Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012; 30: 3376–3382. doi: 10.1200/JCO.2011.40.7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mian SA, Smith AE, Kulasekararaj AG, Kizilors A, Mohamedali AM, Lea NC, et al. Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome. Haematologica. 2013; 98: 1058–1066. doi: 10.3324/haematol.2012.075325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015; 27: 617–630. doi: 10.1016/j.ccell.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, et al. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007; 27: 5393–5402. doi: 10.1128/MCB.00288-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dehm SM. Test-firing ammunition for spliceosome inhibition in cancer. Clin Cancer Res. 2013; 19: 6064–6066. doi: 10.1158/1078-0432.CCR-13-2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.