Significance
Age-associated increased marrow adiposity is often coupled with increased bone loss, presenting a significant age-associated osteoporosis health problem. Here, we examined the mechanisms underlying how transcription factors regulate mesenchymal stem cell lineage determination and showed that Cbfβ plays a critical role in stimulating osteogenesis and inhibiting adipogenesis through activating Wnt10b/β-catenin signaling and inhibiting adipogenesis regulatory gene (c/ebpα) expression. The insights resulting from this study will fill an important knowledge gap and may facilitate the development of novel bone loss therapeutics that minimize the adverse side effects on bone homeostasis.
Keywords: Cbfβ, transcription factor, osteoblast, Wnt/β-catenin, bone−fat interaction
Abstract
The mechanism underlying how transcription factors regulate mesenchymal stem cell lineage commitment remains unclear. To determine the role of core-binding factor subunit beta (Cbfβ) in osteoblast lineage commitment, we generated three mouse models by deleting Cbfβ at different osteoblast lineage stages. We demonstrated that the Cbfβf/fPrx1-Cre, Cbfβf/fCol2α1-Cre, and Cbfβf/fOsx-Cre mice exhibited severe osteoporosis with substantial accumulation of marrow adipocytes resembling aged bone from enhanced adipogenesis, indicating that mesenchymal stem cells and osteoblasts can be programed and reprogramed, respectively, into adipocytes. Consistently, Cbfβ-deficient calvarial cells and bone marrow mesenchymal stem cells displayed strong adipogenic potential, with 5- to ∼70-fold increased adipocyte gene expression, which can be rescued by Cbfβ overexpression. Canonical Wnt signaling was impeded in the Cbfβ-deficient cells, with ∼80% decrease of Wnt10b expression. Accordingly, ChIP and luciferase assays demonstrated that Cbfβ/RUNX2 binds to Wnt10b promoter driving Wnt10b expression. Furthermore, Wnt3a suppressed adipogenesis but did not rescue osteoblastogenesis in Cbfβ-deficient cells. Notably, mixing culture of Cbfβ-deficient with normal cells demonstrates that Cbfβ functions not only through WNT paracrine pathway but also through endogenous signaling. Further analysis shows that Cbfβ/RUNX2 inhibits c/ebpα expression at transcriptional level. Our results show that, besides its osteogenic role, Cbfβ governs osteoblast−adipocyte lineage commitment both cell nonautonomously through enhancing β-catenin signaling and cell autonomously through suppressing adipogenesis gene expression to maintain osteoblast lineage commitment, indicating Cbfβ may be a therapeutic target for osteoporosis.
Age-associated osteoporosis is a major health problem, which is characterized by an imbalance in bone remodeling and metabolism due to reduced osteoblast-mediated bone formation and increased bone marrow adiposity associated with aging. Bone marrow mesenchymal stem cells (MSCs) are multipotent progenitors that give rise to osteoblasts, adipocytes, and chondrocytes upon specific stimulation for cell differentiation. Notably, the majority of conditions associated with bone loss (e.g., aging, glucocorticoid treatment, and thiazolidinedione treatment) stem from a decreased number of osteoblasts and an increased number of adipocytes in the bone marrow (1–4). The preference of MSC differentiation into osteoblast or adipocyte has particular relevance to the maintenance of normal bone homeostasis. It is believed that, under pathologic or aging conditions, switch of MSC commitment promotes increased adipocytes at the expense of osteoblast differentiation in the bone marrow (5).
Differentiation of MSCs into adipocyte or osteoblast is driven by different transcriptional factors and orchestrated by many signaling pathways (3). Notably, peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer binding protein α (C/EBPα) can promote adipocyte differentiation (3, 6, 7), whereas Runx2 and Dlx5 direct osteoblast differentiation (8). Wnt ligands comprise a large family of secreted cysteine-rich and highly hydrophobic glycoproteins that regulate a wide variety of cellular functions (9). Disruption of the canonical Wnt signaling was shown to reprogram myoblast and preosteoblast cells into the adipocyte lineage (10, 11). Despite the recent insights, the mechanisms underlying how transcriptional factors regulate β-catenin activity to determine MSC fate, how β-catenin is activated to support healthy bone density and quality, and whether and how different stages of osteoblasts can be reprogramed to adipocytes are unknown.
Cbfβ is a non-DNA-binding partner of Runt-related transcription factors (Runx1, Runx2, and Runx3) (12–14). We have recently reported that Cbfβ is involved in skeletal development and osteoblast and chondrocyte differentiation as well as fracture healing (15–18). Nevertheless, the function of Cbfβ in osteoblast−adipocyte lineage commitment and maintenance has not yet been determined. To address these questions, we deleted the Cbfβ gene at different osteoblast lineage stages, i.e., MSCs, osteochondroprogenitors, and osteoblast using the Prx1-Cre, Col2α1-Cre, and Osx-Cre mouse lines, respectively (19–21). Our Cbfβ conditional deletion revealed that Cbfβ is critical for osteoblast lineage commitment and maintenance through promoting Wnt10b expression and inhibiting the expressions of c/epbα.
Results
Loss of Cbfβ in MSC, Osteochondroprogenitor, or Early Osteoblast Causes Increased Bone Marrow Adiposity Accompanied by Decreased Bone Mass.
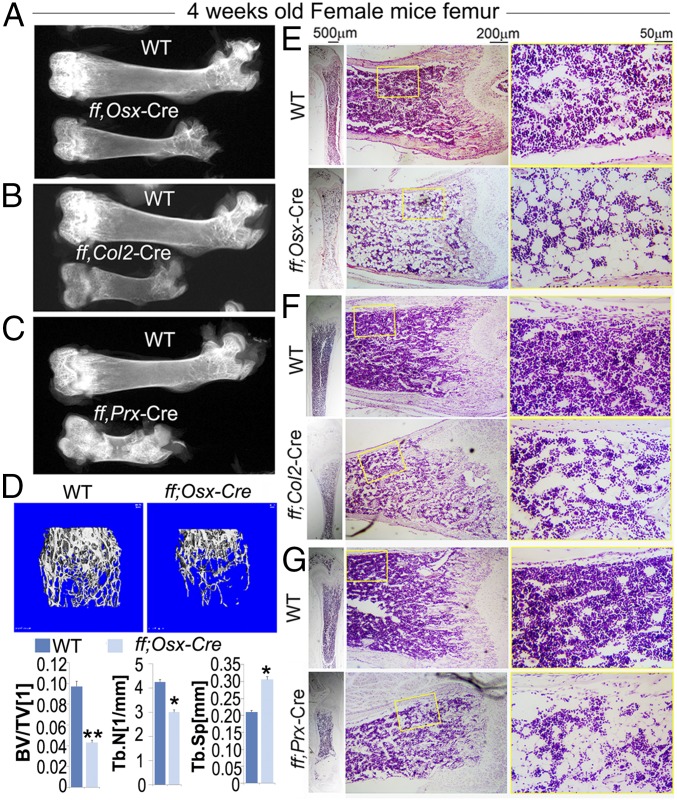
Previous mouse model with Cbfβ deficiency (CbfβGFP/GFP knock-in mice, Cbfβ−/− embryos rescued by Tek-GFP/Cbfβ and Gata1-Cbfβ transgenes, and Cbfβf/fTwist-Cre mice) exhibited skeletal dysplasia and died around birth. To investigate the role of Cbfβ in osteoblast−adipocyte lineage commitment in vivo, we sought to delete Cbfβ in skeletal cells at various differentiation stages by generating the Cbfβf/fPrx1-Cre, Cbfβf/fCol2α1-Cre, and Cbfβf/fOsx-Cre conditional knockout mice. Cbfβf/fPrx1-Cre and Cbfβf/fOsx-Cre mice survived to adulthood, and Cbfβf/fCol2α1-Cre survived to about 3 wk to 4 wk of age (15–18). Interestingly, radiographic analysis showed that postnatal Cbfβf/fPrx1-Cre, Cbfβf/fCol2α1-Cre, and Cbfβf/fOsx-Cre mice had reduced bone density compared with their wild-type (WT) littermates (Fig. 1 A–C). Microcomputed tomography (μCT) analysis of the distal femora of 4-wk-old mouse femurs further confirmed a reduction in bone volume and trabecular bone number, and an increase in trabecular bone separation in the Cbfβf/fOsx-Cre mice compared with WT controls (Fig. 1D). Femoral sections from Cbfβf/fPrx1-Cre, Cbfβf/fCol2α1-Cre, and Cbfβf/fOsx-Cre mice were subsequently subjected to further histological analysis (Fig. 1 E–G). Hematoxylin and eosin (H&E) staining showed that trabecular bone number was largely reduced in the Cbfβf/fPrx1-Cre (Fig. 1G), Cbfβf/fCol2α1-Cre (Fig. 1F), and Cbfβf/fOsx-Cre mice (Fig. 1E). While performing femoral bone histology of the Cbfβ-deficient mice and their WT littermates, we unexpectedly observed dramatic change in the bone marrow fat content. As shown in the high-magnification images, adipocyte-like vacuoles accumulated in the bone marrow of Cbfβf/fPrx1-Cre, Cbfβf/fCol2α1-Cre, and Cbfβf/fOsx-Cre mice but not WT mice (Fig. 1 E–G, Right).
Fig. 1.
Cbfβ deficiency reduced bone density. (A−C) X-ray of mouse femurs of 4-wk-old (A) Cbfβf/fOsx-Cre mice, (B) Cbfβf/fCol2α1-Cre mice, and (C) Cbfβf/fPrx1-Cre mice. (D) The μ-CT analysis of 4-wk-old Cbfβf/fOsx-Cre and WT mouse femurs. Quantification data indicate bone volume/tissue volume (BV/TV), trabecular number (Tb.N), and trabecular separation (Tb.Sp). (E−G) (Left) H&E staining of mouse femurs from 4-wk-old (E) WT and Cbfβf/fOsx-Cre mice, (F) WT and Cbfβf/fCol2α1-Cre mice, and (G) WT and Cbfβf/fPrx1-Cre mice. (Right) Images with higher magnification. Data were presented as mean ± SEM, n = 3. *P ≤ 0.05; **P ≤ 0.01.
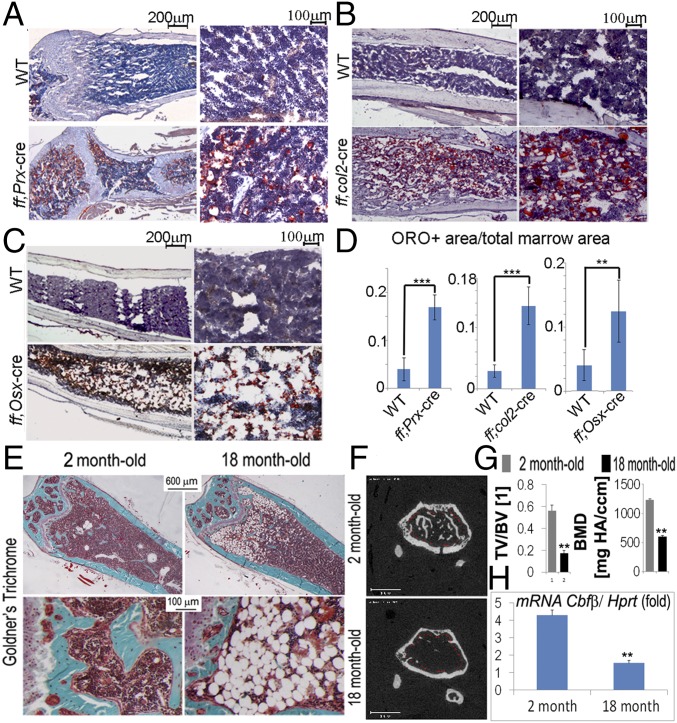
To further confirm that the adipocyte-like vacuoles observed in H&E staining slides are adipocytes, we performed Oil Red O staining using femoral frozen sections. Consistently, more adipocytes (red color) were present in the bone marrow of Cbfβ-deficient mice compared with that of WT (Fig. 2 A–D). Oil Red O+ area per tissue area was increased by about threefold in Cbfβf/fPrx1-Cre mice (Fig. 2 A and D), about threefold in Cbfβf/fCol2α1-Cre mice (Fig. 2 B and D), and about twofold in Cbfβf/fOsx-Cre mice (Fig. 2 C and D). Our histomorphological analysis of femoral sections of 18-wk-old WT mice by Goldner’s trichrome staining revealed that aged mice showed a dramatic decrease in mineralized tissue and an increase in marrow adiposity compared with 2-mo-old controls (Fig. 2E). The reduced bone density due to aging was further confirmed by μCT analysis (Fig. 2 F and G). Interestingly, Cbfβ mRNA expression levels in 18-mo-old WT mice were also drastically reduced compared with that of the 2-mo-old group (Fig. 2H). The results indicate that deficiency of Cbfβ in MSC, osteochondroprogenitor, or early osteoblast leads to high bone marrow adiposity and low bone mass, resembling the bone phenotype in aged WT mice.
Fig. 2.
Cbfβ-deficient mice had accumulated marrow adipocyte. Oil Red O staining of 2-wk-old WT and (A) Cbfβf/fPrx1-Cre mice, (B) Cbfβf/fCol2α1-Cre mice, and (C) Cbfβf/fOsx-Cre mice. (D) Quantification of adipocyte area per bone marrow area in A−C. (E) (Top) Goldner’s Trichrome analysis of WT femurs at 2 mo and 18 mo of age. (Bottom) Higher magnification images. (F) The μ-CT analysis of WT mouse femurs at 2 and 18 mo. (G) Quantification for F is shown as TV/BV and bone mineral density (BMD). (H) The qRT-PCR to assess Cbfβ mRNA in 2-mo-old and 18-mo-old mice. Data were presented as mean ± SEM, n = 30. **P ≤ 0.01; ***P ≤ 0.005.
Cbfβ-Deficient Calvarial Cells Transdifferentiated into Adipocytes.
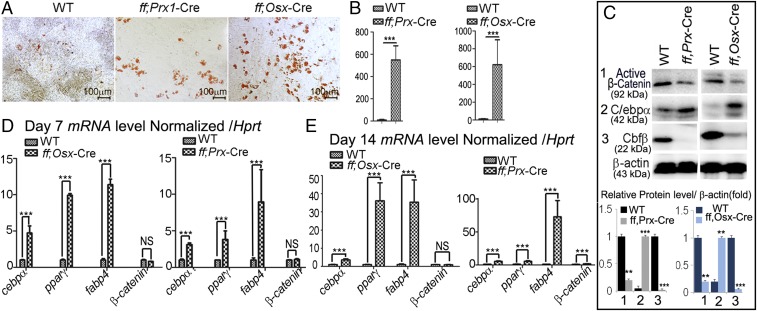
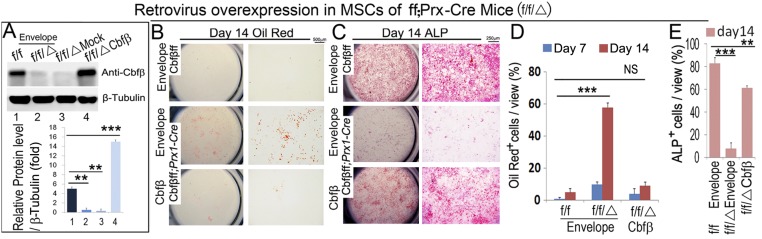
The increased bone marrow adiposity caused by knockout of Cbfβ in MSCs, osteochondroprogenitors, or early osteoblasts indicates that the absence of Cbfβ favors adipogenesis. To determine whether Cbfβ-deficient calvarial cells transdifferentiate into adipocytes in vitro, calvarial osteoblast precursor cells derived from WT, Cbfβf/fPrx1-Cre, and Cbfβf/fOsx-Cre newborn mice were maintained in the osteogenic medium for 14 d, and adipocyte formation was detected by Oil Red O staining (Fig. 3A). A significantly higher number of Cbfβf/fPrx1-Cre and Cbfβf/fOsx-Cre cells were committed to adipocyte lineage (Fig. 3B). Efficient deletion of Cbfβ in Cbfβf/fPrx1-Cre and Cbfβf/fOsx-Cre cells was confirmed by Western blot (Fig. 3C). On the contrary, retrovirus-mediated overexpression of Cbfβ in Cbfβf/fPrx1-Cre cells partially rescued osteoblast differentiation (Fig. S1 C and E) and, meanwhile, completely reduced adipocyte formation rates (Fig. S1 B and D). The deletion and overexpression of Cbfβ were confirmed by Western blot (Fig. S1A). Interestingly, active β-catenin (nonphosphorylated β-catenin) protein was decreased greatly in the Cbfβ-deficient cells whereas c/ebpα expression was significantly up-regulated (Fig. 3C). We further examined the expression of adipogenesis-related genes with quantitative real time PCR (qPCR), using cells maintained in the osteogenic medium for 7 d (D7) and 14 d (D14) (Fig. 3 D and E). At D7 and D14, c/ebpα expression was increased by fivefold in Cbfβf/fOsx-Cre cells, and threefold in Cbfβf/fPrx1-Cre cells, compared with that in WT cells (Fig. 3 D and E). Furthermore, PPARγ expression was increased by 10-fold in D7 Cbfβf/fOsx-Cre cells, 32-fold in D14 Cbfβf/fOsx-Cre cells, fourfold in D7 Cbfβf/fPrx1-Cre cells, and fivefold in D14 Cbfβf/fPrx1-Cre cells, compared with the WT cohorts (Fig. 3 D and E). Fabp4 expression was also increased dramatically (more than 30-fold in D7, more than 50-fold in D14) in the mutant cells (Fig. 3 D and E). Unexpectedly, despite the reduced expression level of active β-catenin protein (Fig. 3C), β-catenin mRNA expression did not change in the WT, Cbfβf/fPrx1-Cre, and Cbfβf/fOsx-Cre cells (Fig. 3 D and E).
Fig. 3.
Cbfβ-deficient cells had increased adipocyte formation with decreased β-catenin proteins. (A) Oil Red O staining of WT, Cbfβf/fPrx1-Cre, and Cbfβf/fOsx-Cre calvarial cells cultured in osteogenic medium. (B) Quantification of adipocyte number and area of A. (C) Western blot and quantification of protein level of active β-catenin, C/ebpα, and Cbfβ in WT, Cbfβf/fPrx1-Cre, and Cbfβf/fOsx-Cre cells. (D and E) Expression of C/ebpα, Pparγ, Fabp4, and β-catenin mRNA in (D) D7 and (E) D14 WT, Cbfβf/fPrx1-Cre, and Cbfβf/fOsx-Cre cells. Data were presented as mean ± SEM, n = 15. **P ≤ 0.01; ***P ≤ 0.005; NS, not significant.
Fig. S1.
Overexpression of Cbfβ rescues osteoblast differentiation and inhibits adipocyte differentiation in Cbfβ-deficient cells. (A) Deletion and overexpression of Cbfβ confirmed by Western blot. Quantification of protein level is copresented in the lower panel. (B and C) Cbfβf/f MSCs, Cbfβf/fPrx1-Cre MSCs, and Cbfβf/fPrx1-Cre MSCs overexpressing Cbfβ mediated by retrovirus were cultured in osteogenic medium for 14 d, followed by (B) Oil Red O staining and (C) ALP staining to detect adipocyte and osteoblast formation, respectively. (D) Quantification of Oil Red O+ cell number in B. (E) Quantification of ALP+ cell number in C. The data were presented as mean ± SEM, n = 3. **P ≤ 0.01; ***P ≤ 0.005; NS, not significant.
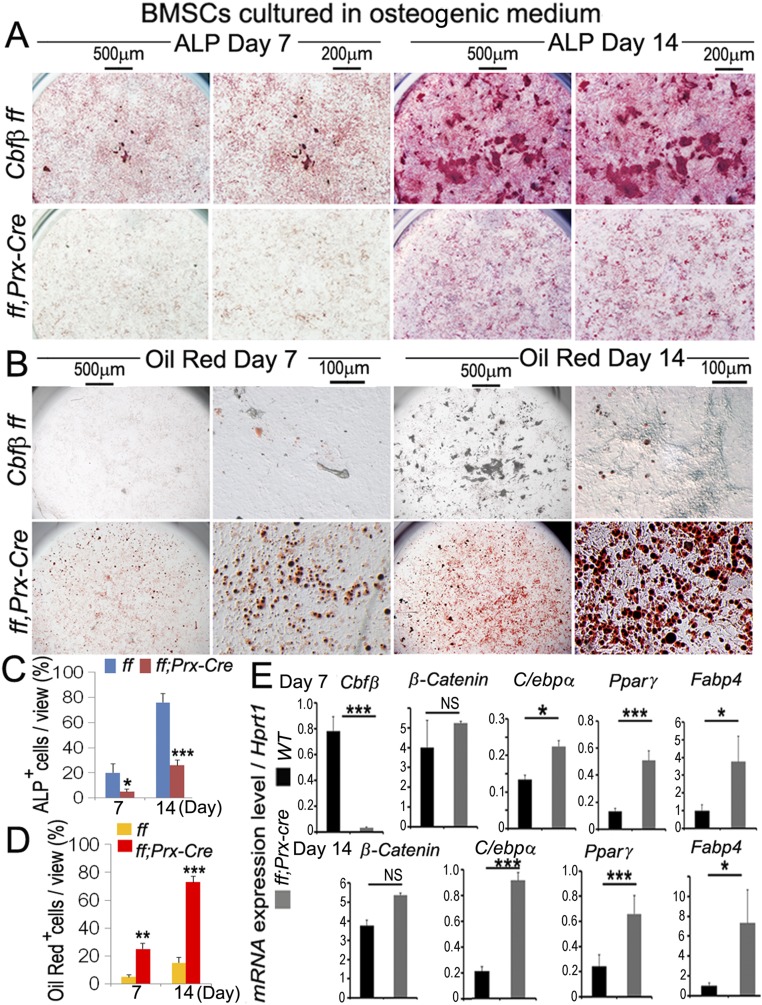
Cbfβ Deficiency Promotes Adipogenesis and Inhibits Osteoblastogenesis in Marrow MSC.
MSCs derived from WT and Cbfβf/fPrx1-Cre mice were maintained in the osteogenic medium for 7 d and 14 d. Osteoblast formation was detected by alkaline phosphatase (ALP) staining (Fig. S2 A and C). Adipocyte formation was detected by Oil Red O staining (Fig. S2 B and D). Cbfβ-deficient MSCs had approximately fourfold more adipocyte (Oil Red O+ cells) formation (Fig. S2 B and D) and approximately threefold less osteoblast (ALP+ cells) formation (Fig. S2 A and C). Efficient deletion of Cbfβ in Cbfβf/fPrx1-Cre cells was confirmed by qPCR (Fig. S2D). Similar to calvarial cells, expression of adipocyte markers (c/ebpα, Fabp4, and Pparγ) was increased in the Cbfβf/fPrx1-Cre cells, whereas β-catenin mRNA expression was not changed (Fig. S2D) in both D7 and D17.
Fig. S2.
Cbfβ-deficient MSC has decreased osteogenic potency with increased adipocyte formation. (A and B) Cbfβf/f and Cbfβf/fPrx1-Cre bone marrow MSCs were cultured in osteogenic medium for 7 d and 14 d, followed by (A) ALP staining and (B) Oil Red staining to show osteoblast and adipocyte formation, respectively. (C) Quantification of ALP+ cell number in A. (D) Quantification of Oil Red O+ cell number in B. (E) Real-time PCR to show Cbfβ, β-catenin, c/ebpα, PPARγ, and Fabp4 expression in day 7 and day 14 cells. The data were presented as mean ± SEM, n = 3. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.005; NS, not significant.
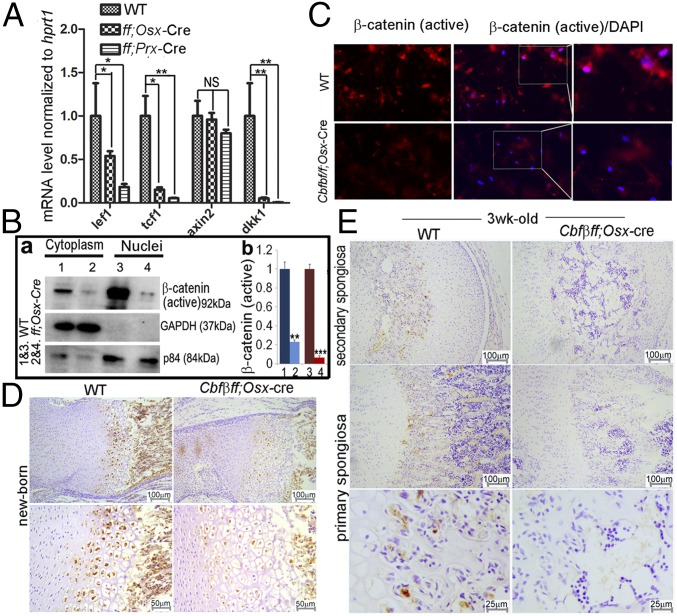
Cbfβ Inhibits β-Catenin Signaling in Vivo and in Vitro.
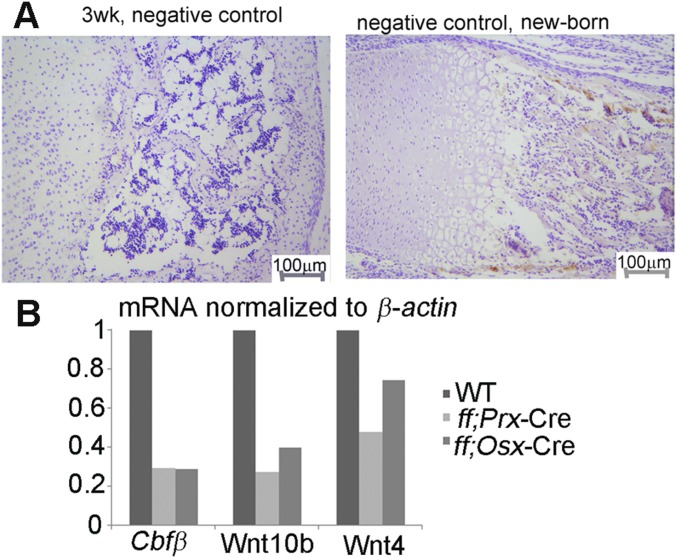
In deciphering the molecular basis of Cbfβ’s roles in osteoblast−adipocyte lineage allocation, mRNA were harvested from WT, Cbfβf/fPrx1-Cre, and Cbfβf/fOsx-Cre calvarial cells, which were maintained in the osteogenic medium for 7 d. Genes whose expression has been reported to be regulated by β-catenin (lef1, tcf1, axin2, and dkk1) were analyzed by quantitative real-time (qRT)-PCR. The expression of lef1, tcf1, and dkk1 was down-regulated significantly in both Cbfβf/fPrx1-Cre and Cbfβf/fOsx-Cre cells (Fig. 4A). The β-catenin needs to translocate into the nuclei to truly exert its transcriptional function; thus we also examined the expression of active β-catenin in the cytoplasm and nuclei, separately. We found a lower level of active β-catenin in both cytoplasm and nuclei of Cbfβ-deficient osteoblasts (Fig. 4B). The down-regulation of active β-catenin in the whole cell of the mutant cells was confirmed by cellular immunofluorescent (IF) staining (Fig. 4C). We also detected active β-catenin expression in the newborn and 3-wk-old WT and Cbfβf/fOsx-Cre femurs (Fig. 4 D and E; negative controls are presented in Fig. S3A). We found that the active β-catenin protein level was down-regulated in the hypertrophic chondrocytes of newborn Cbfβf/fOsx-Cre femurs (Fig. 4D), and both primary spongiosa and secondary spongiosa of 3-wk-old Cbfβf/fOsx-Cre femurs (Fig. 4E), compared with that of the WT littermates. Taken together, our data indicate that β-catenin activity was influenced in the absence of Cbfβ.
Fig. 4.
Cbfβ-deficiency inhibited β-catenin protein signaling in vivo and in vitro. (A) The qRT-PCR analysis of lef1, tcf1, axin2, and dkk1 expression normalized by hprt1. (B) Immunoblotting analysis and quantification of β-catenin protein level was reduced in both (a) cytoplasm and (b) nuclei of Cbfβ-deficient cells compared with WT cells. (C) IF staining of active β-catenin in Cbfβ-deficient and WT cells. (D and E) Immunostaining of active β-catenin in newborn and 3-wk-old Cbfβ-deficient mice femurs compared with WT femurs. The data were presented as mean ± SEM, n = 15. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.005; NS, not significant.
Fig. S3.
Mouse IgG isotype negative control and qRT-PCR analysis of gene expression at the transcriptional level. (A) Mouse IgG isotype negative control for the immunostaining in Fig. 4 D and E confirms the specificity of the staining, n = 6. (B) Wnt4, Wnt10b, and Cbfβ mRNA expression in WT, Cbfβf/ fPrx-Cre, and Cbfβf/ fOsx-Cre newborn mice long bone tissues, normalized to hprt1.
Cbfβ Regulates Wnt10b Expression at the Transcriptional Level.
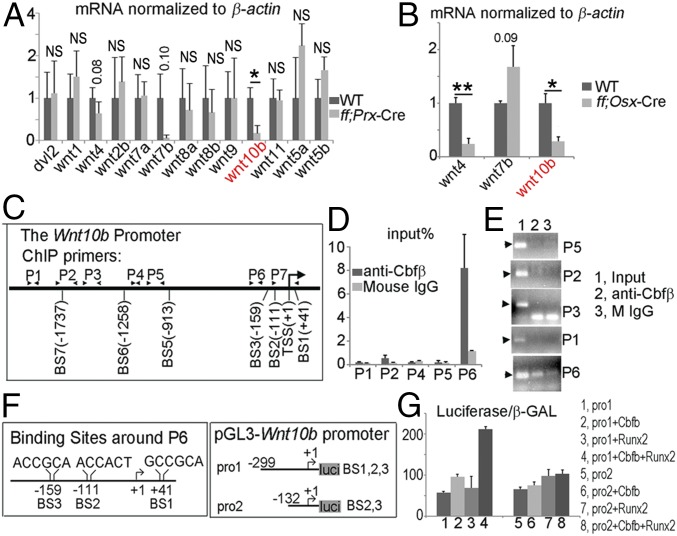
Abrogated β-catenin activity might be caused by altered expression of canonical Wnts. To address this question, we performed qRT-PCR to detect Wnts expression in the WT, Cbfβf/fPrx1-Cre, and Cbfβf/fOsx-Cre preosteoblasts. We found that, although expression of most Wnts (Wnt1, Wnt7b, Wnt7a, Wnt5a, Wnt11, etc.) was not drastically changed, Wnt4 expression is significantly reduced in Cbfβf/fOsx-Cre cells but not Cbfβf/fPrx1-Cre cells. Importantly, the expression of Wnt10b was significantly down-regulated in both Cbfβf/fPrx1-Cre and Cbfβf/fOsx-Cre cells (Fig. 5 A and B). Consistently, Wnt10b expression is also down-regulated in the Cbfβf/fPrx1-Cre and Cbfβf/fOsx-Cre newborn long bone (Fig. S3B). Using an online transcription factor binding site predictive tool (alggen.lsi.upc.es), we identified six binding sites on the Wnt10b promoter for Cbfβ/Runx complex (+41, −111, −159, −913, −1,258, and −1,737) (Fig. 5C). ChIP assay showed that Cbfβ/Runx complex potentially binds to the Wnt10b promoter on the binding sites around 200 bp (predicted binding sites −159, −111, +41) but is not likely to bind to the predicted binding sites −913, −1,258, and −1,737 (Fig. 5 C–E). Then we constructed the pGL3-Wnt10b promoter containing −159, −111, and +41 binding sites (pro1), and pGL3-Wnt10b promoter containing −111 and +41 binding sites (pro2) (Fig. 5F). Luciferase analysis revealed that Cbfβ with Runx2 enhances pro1-driven luciferase activity but did not change pro2-driven luciferase activity (Fig. 5G). Taken together, Cbfβ/Runx2 regulates Wnt10b expression at transcriptional level, by binding to the Wnt10b promoter at the binding sites −159.
Fig. 5.
Cbfβ promotes Wnt10b expression at the transcriptional level. (A) The qRT-PCR analysis of dvl2 and Wnts mRNA expression in WT and Cbfβf/fPrx1-Cre cells. (B) The qRT-PCR analysis of Wnt4, Wnt7b, and Wnt10b mRNA expression in WT and Cbfβf/fOsx-Cre cells. (C) Schematic illustration of the Wnt10b promoter. Transcriptional start site (TSS), predicted Cbfβ/Runx binding site (BS), and ChIP primers (P1 through P7) were indicated in the promoter. (D) ChIP qPCR assay using anti-Cbfβ antibody and primers indicated in C. (E) Agarose gel image using ChIP qPCR products in D. (F) Schematic illustration of the predicted Cbfβ/Runx binding site 1,2,3 on the Wnt10b promoter, and the construction of pGL3-Wnt10b promoter vectors. (G) Cotransfection of pGL3-Wnt10b promoter with Runx2 and/or Cbfβ expressing vector into C3H10T1/2 cell line. β-GAL expressing vector was also cotransfected. Promoter activity was measured by luciferase normalized to β-GAL. The data were presented as mean ± SEM, n = 20. *P ≤ 0.05 **P ≤ 0.01; NS, not significant.
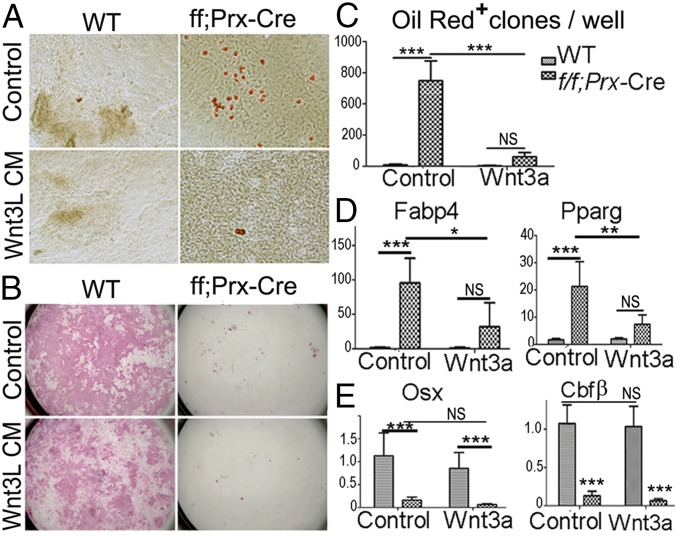
Wnt3a Suppresses Lineage Switch of Cbfβ-Deficient Calvarial Osteoblast.
As we showed in Cbfβ Inhibits β-Catenin Signaling in Vivo and in Vitro and Cbfβ Regulates Wnt10b Expression at the Transcriptional Level, Wnt signaling plays an important role in Cbfβ-mediated lineage commitment. Thus, we sought to suppress the adipogenesis from Cbfβ-deficient calvarial cells by potentiating Wnt signaling using Wnt3L conditional medium (Fig. 6). Wnt3L conditional medium suppressed adipogenesis of Cbfβ-deficient cells (Fig. 6 A and C) but did not help rescue osteoblast formation (Fig. 6B). Consistently, Wnt3a dramatically repressed adipocyte-specific gene expression (i.e., fabp4 and pparg) in Cbfβ-deficient cells, rendering their expression level similar to those in WT cells (Fig. 6D). However, Osx (an osteoblast marker) remained down-regulated in Cbfβ-deficient cells under the treatment of Wnt3L conditional medium (Fig. 6E). Meanwhile, expression of Cbfβ itself was not changed by Wnt3L conditional medium (Fig. 6E).
Fig. 6.
Canonical Wnt inhibits excessive adipocyte formation in Cbfβ-deficient calvarial cells. (A) Oil Red O staining and (B) ALP staining of WT and Cbfβf/fPrx1-Cre calvarial cells cultured in osteogenic medium or osteogenic medium supplemented with Wnt3L conditional medium. (C) Quantification of adipocyte number in A. (D) Fabp4 and PPARg and (E) Osx and Cbfβ mRNA levels in WT and Cbfβf/fPrx1-Cre calvarial cells cultured in osteogenic medium or osteogenic medium supplemented with Wnt3L conditional medium, normalized to hprt1. The data were presented as mean ± SEM, n = 10. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.005; NS, not significant.
Cbfβ Suppresses Adipocyte Formation Through both Cell-Autonomous Gene Regulation Pathway and Nonautonomous Wnt10b Paracrine/Autocrine Pathway.
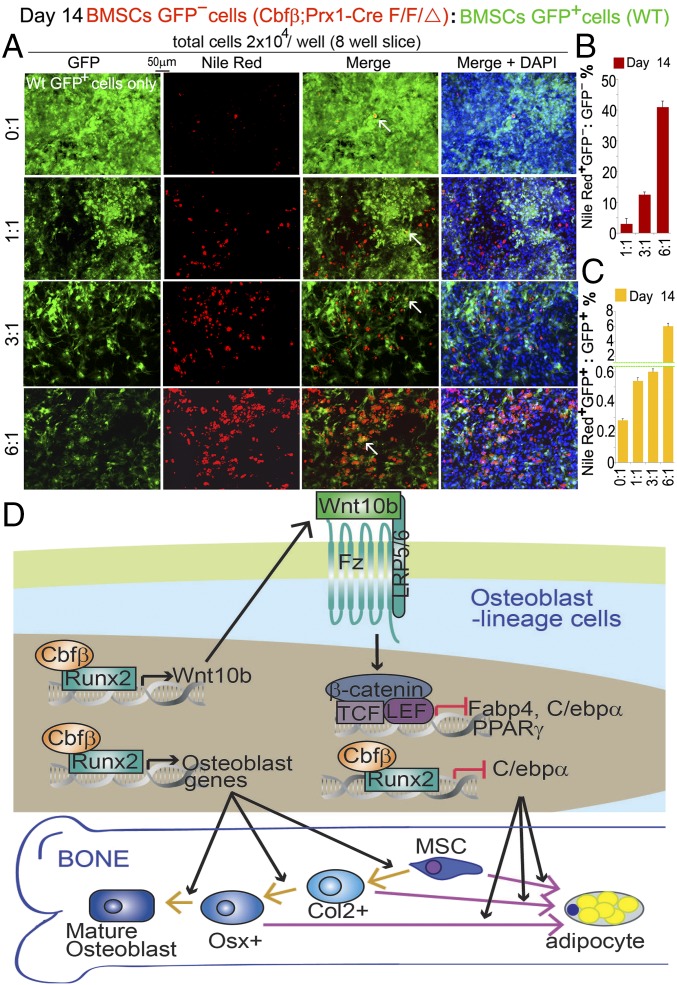
To further test whether Cbfβ antagonizes adipocyte differentiation through Wnt10b paracrine/autocrine pathway, we combined GFP−;Cbfβ-deficient MSCs with GFP+ WT MSCs at different ratios and cultured them in osteogenic medium for 14 d. Then adipocyte formation was stained by Nile Red, and counterstained with DAPI. The mixing culture experiment confirmed that Cbfβ-deficient MSCs were prone to forming more adipocyte; as more Cbfβ-deficient MSCs were present in the mixing culture, more adipocytes are formed (Fig. 7A). The results showed that mixing culture with Cbfβ-deficient MSCs increased the adipocyte formation rate of GFP+ normal cells (Nile Red+GFP+/GFP+ ratios 0.28%, 0.55%, 1.5%, and 5.9%, based on GFP-:GFP+ ratios 0:1, 1:1, 3:1, and 6:1, respectively) (Fig. 7C). Higher ratio of Cbfβ-deficient MSC in the mixing culture also increased the adipocyte formation rate of GFP–Cbfβ-deficient cells (Nile Red+GFP−/GFP− ratios 2.5%, 12%, and 40%, based on GFP-:GFP+ ratios 1:1, 3:1, 6:1, respectively) (Fig. 7B). From GFP-:GFP+ ratio 3:1 to 6:1, adipocyte formation rate of GFP–Cbfβ-deficient cells increases by 2.3-fold (Fig. 7B). Adipocyte formation rate was 4.55-, 8-, and 6.78-fold higher in Cbfβ-deficient cells compared with WT type GFP+ cells (Fig. 7 B and C) based on GFP-:GFP+ ratios 1:1, 3:1, and 6:1, respectively. This observation indicates that, besides autocrine/paracrine signaling, Cbfβ also regulates adipocyte formation through cell-autonomous pathway.
Fig. 7.
Cbfβ-deficient and normal cells reciprocally regulate their respective adipocyte potency. Cbfβf/fPrx1-Cre;GFP- and GFP+ bone marrow MSCs were mixed together in different ratios and cultured in osteogenic medium. (A) Adipocytes were labeled by Nile Red and couterstained by DAPI on D14. Quantification of (B) NileRed+GFP-/GFP- and (C) NileRed +GFP+/GFP+ ratios in A. The data were presented as mean ± SEM, n = 3. (D) Working model for Cbfβ controlling osteoblast−adipocyte commitment through Wnt10b/β-catenin signaling.
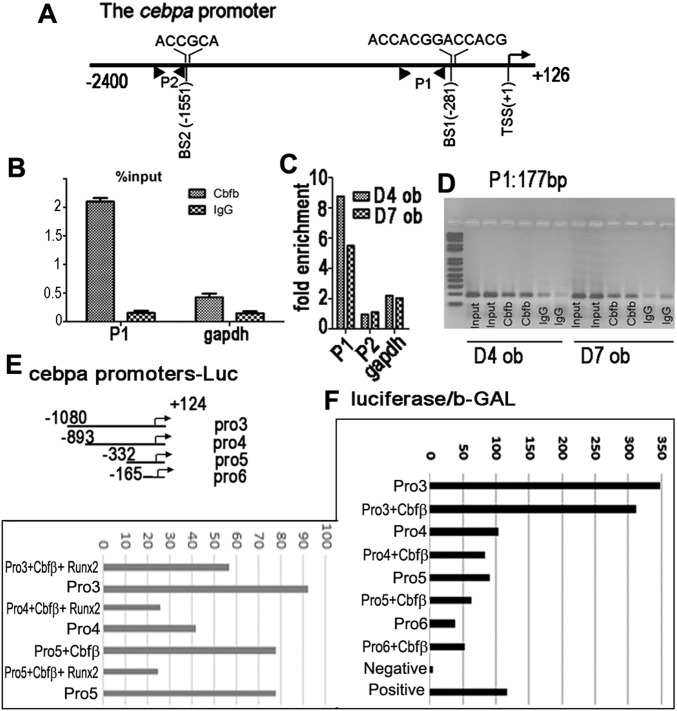
Using the online transcription factor binding site predictive tool (alggen.lsi.upc.es), we identified two binding sites on the c/ebpα promoter for Cbfβ/Runx complex (−281, −2,400) (Fig. S4A). ChIP assay showed that Cbfβ/Runx complex binds to the Wnt10b promoter on the binding sites around 300 bp (predicted binding site −281) but is not likely to bind to the predicted binding site −2,400 (Fig. S4 B–D). Then we constructed the pGL3-c/ebpα promoter with −281 binding site (pro3,4,5) and without −281 binding site (pro6) (Fig. S4E). Luciferase analysis revealed that pro3,4,5-driven luciferase activity, but not pro6-driven luciferase activity, is inhibited by Cbfβ overexpression (Fig. S4F). Furthermore, pro3,4,5-driven luciferase activity is more notably inhibited by Cbfβ/Runx2 cotransfection (Fig. S4F). Taken together, Cbfβ/Runx2 regulates c/ebpα expression at transcriptional level, by binding to the c/ebpα promoter at the binding site −281. Besides Wnt signaling, Cbfβ-mediated c/ebpα also contributes to lineage commitment.
Fig. S4.
Runx2/ Cbfβ binds to the cebpa promoter to inhibit its transcription. (A) Schematic illustration of the mouse cebpa promoter (−2,400... +126). P1 and P2, BS, and TSS were indicated in the promoter. (B) ChIP qPCR assay using D4 osteoblast, anti-Cbfβ antibody, and P1 indicated in A; the result is presented in the form of percent input. (C) ChIP qPCR assay using D4 and D8 osteoblast, anti-Cbfβ antibody, and primers indicated in A; the result is presented in the form of fold enrichment. (D) Agarose gel image using ChIP qPCR products in C. (E) Schematic illustration of the construction of pGL3-cebpa promoter vectors. (F) Cotransfection of pGL3-cebpa promoters with Runx2 and/or Cbfβ expressing vector into C3H10T1/2 cell line. β-GAL expressing vector was also cotransfected. Promoter activity was measured by luciferase normalized to β-GAL. The data were presented as mean ± SEM, n = 3.
Discussion
Taken together, our data demonstrate that Cbfβ plays a critical role in the maintenance of osteoblast lineage, and the switch from osteoblastogensis to adipogenesis can occur at any stage of osteoblast differentiation (Fig. 7D). Importantly, Cbfβ suppresses adipogenesis through Wnt signaling and c/ebpα expression, whereas it regulates osteoblast differentiation through different mechanisms (Fig. 7D).
Cbfβ Is Essential to Preserve Osteoblast Lineage Commitment and Maintenance.
Using the genetic dissection tool of conditional gene deletion by the Cre/LoxP system, we discovered that Cbfβ deficiency from the Cbfβf/fPrx1-Cre, Cbfβf/fCol2α1-Cre, and Cbfβf/fOsx-Cre mouse models result in bone marrow adipocyte accumulation in vivo, resembling the conditions associated with osteoporosis in aging human bone (Figs. 1–3). Our data show that, besides MSCs, Cbfβ-deficient osteoblasts lineage cells at different stages of osteoblastogenesis can be reprogramed into adipocytes. This finding indicates the importance of Cbfβ in osteoblast lineage commitment and maintenance as well as in adipogenesis in aging bone marrow, and that the switch from osteoblastogensis to adipogenesis can occur at any stage of osteoblast differentiation (Fig. 7D).
Role of Cbfβ in MSC Differentiation and Osteoporosis.
Our previous studies also showed that Cbfβ deficiency inhibited chondrocyte proliferation and hypertrophy, and impaired growth plate development due to disrupted Ihh−PTHrP regulatory loop (19–21), which in turn regulated endochondral bone formation. Our previous and current studies showed that Ihh and β-catenin signaling plays a role at distinct differentiation stages (19). Although β-catenin signaling is a critical regulator of bone marrow adipocyte (10, 11, 22), adipogenic role of Ihh is unclear. Thus, regarding Cbfβ’s role in osteoblast−adipocyte lineage commitment, canonical Wnt might be more critical than Ihh pathway.
Runx/Cbfβ Complex Promotes Wnt10b/β-Catenin Signaling and Inhibits c/ebpa Expression to Inhibit Lineage Switch into Adipocytes.
Wnt/β-catenin directs mesenchymal cell commitment to the osteogenic lineage and prevents adipogenic differentiation, resulting in increased new bone formation (10, 11, 22). Overexpression of canonical Wnt ligand Wnt10b induces higher bone mass in mice, and Wnt10b inhibits differentiation of preadipocytes and blocks adipose tissue development (23, 24). Interestingly, Wnt10b is down-regulated in Cbfβ-deficient cells and tissues, and ChIP plus promoter activity assay showed that Wnt10b is up-regulated by Cbfβ at the transcriptional level (Fig. 5). In addition, adipocyte differentiation can be suppressed but osteoblast differentiation cannot be rescued in the presence of Wnt3L conditional medium containing Wnt3a (Fig. 6). Those observations indicate that the Runx/Cbfβ complex inhibits programing and reprograming of MSCs and osteoblast into adipocytes through Wnt10b/β-catenin signaling, whereas regulation of osteoblast differentiation by Runx/Cbfβ is independent of this pathway (Fig. 7D).
In the experiment of mixing culture of Cbfβ-deficient and GFP+ normal cells, in the presence of higher ratio of Cbfβ-deficient cells, both Cbfβ-deficient and GFP+ WT cells have higher adipocyte formation rate, confirming that Cbfβ inhibits adipocyte formation through paracrine Wnt10b signaling (Fig. 7 A–C). However, adipocyte formation rate is always much higher in the Cbfβ-deficient cells regardless of mixing ratios, indicating that Cbfβ also inhibits adipocyte differentiation cell-autonomously (Fig. 7 A–C). Further studies using ChIP and luciferase assay prove that Cbfβ/Runx2 complex inhibits the expression of c/ebpa, an important adipogenic transcriptional factor, at transcriptional level (Fig. S4). Taken together, our data show that Cbfβ regulates osteoblast differentiation from both endogenous and paracrine signaling (Fig. 7D).
Cbfβ May Be Essential for Prevention of Human Age-Associated Osteoporosis Resulting from Elevated Adipogenesis.
Our results showed, along with reduced bone mineral density in aged WT mice, Cbfβ expression was dramatically reduced in aged mice, indicating that low Cbfβ expression might be a major cause of age-related osteoporosis with increased adipogenesis (Fig. 2). The fact that lineage switch from osteoblastogensis to adipogenesis can occur at multiple stages of osteoblast differentiation in the Cbfβ-deficient cells (Figs. 1 and 2) demonstrates that Cbfβ may be essential for prevention of human age-associated osteoporosis due to elevated adipogenesis. Further understanding the mechanism underlying osteoblast−adipocyte lineage allocation will fill an important knowledge gap and may facilitate the development of novel bone loss therapeutics while minimizing the adverse side effects on bone hemostasis.
Materials and Methods
The study was approved by the University of Alabama at Birmingham (UAB) Animal Care and Use Committee, conformed to National Institutes of Health guidelines, and followed all recommendations of Animal Research: Reporting in Vivo Experiments guidelines. For more detailed description, please refer to SI Materials and Methods. For primer sequences please refer to Tables S1–S3.
Table S1.
Primers used for qPCR
| Gene | Forward primers | Reverse primers |
| Cbfβ | CCGCGAGTGCGAGATTAAGTA | GTTCTGGAAGCGTGTCTGG |
| Cebpα | CAAGAACAGCAACGAGTACCG | GTCACTGGTCAACTCCAGCAC |
| Pparγ | GGAAGACCACTCGCATTCCTT | GTAATCAGCAACCATTGGGTCA |
| fabp4 | AGCACCATAACCTTAGATGGGG | CGTGGAAGTGACGCCTTTCA |
| β-catenin | TGACGTTGACATCCGTAAAGAC | TGCTAGGAGCCAGAGCAGTAA |
| lef1 | TGAGTGCACGCTAAAGGAGA | GCTGTCATTCTGGGACCTGT |
| tcf1 | ACATGAAGGAGATGAGAGCCA | CTTCTTCTTTCCGTAGTTATC |
| axin2 | ATGTCCTGTCTGCCAGCGTTC | CAAGCACTAGCCAGTGGGTCAA |
| dkk1 | AAGATGAGGAGTGCGGCTCTG | GGCGGCGTTGTGGTCATTAC |
| β-actin | GTACGACCAGAGGCATACAGG | GATGACGATATCGCTGCGCTG |
| Hprt | GGTGGAGATGATCTCTCAACTTTAA | AGGAAAGCAAAGTCTGCATTGTT |
| Dvl2 | GCTTCCACATGGCCATGGGC | AGGCACTGCTGGTGAGAGTCACAG |
| Wnt1 | ACATAGCCTCCTCCACGAAC | GGAGGTGATTGCGAAGATG |
| Wnt4 | CGTAGCCTTCTCACAGTCCTTT | CGTCAAACTTCTCCTTTAGCG |
| Wnt2b | TCGTCTATGCTATCTCGTCAGC | ACAGCGGTTGTTGTGTAAGTTC |
| Wnt7a | ACGCCATCATCGTCATAGG | TTGCTTCTCCTTGTCGCA |
| Wnt7b | TCCAAGGTCAACGCAATG | GGGAAGGGTGTCCTCAAATAG |
| Wnt8a | GCGTGTGGTGAGCAGATACTAC | TCGTGGAAGGTCTACAGGCTA |
| Wnt8b | GTCCAAAGGCTTACCTGGTCT | CACAGTTGTCAAAGTCTCCGAG |
| Wnt9 | GTGTGGCTTTCGTGAGCAT | GGTATTCCAGCACTGAACAATG |
| Wnt10b | TTGCTCGGATTTCTGTCTAGG | ACTCCACACAATGCCTGCTA |
| Wnt11 | TGAATCAGACGCAACACTGTAA | CTCTCCAGGTCAAGCAGGTAGT |
| Wnt5a | CTGCAGCACAGTGGACAATAC | TAGCGTGGATTCGTTCCC |
| Wnt5b | CCTGACTACTGCTTGCGTAATG | ACATACTGGTCCACAACCTCG |
Table S3.
Primers used for subcloning
| Gene | Primer |
| wnt10bproF1 | AG GGTACC ACCGTCAGTAACCCTAAGGTCGT |
| wnt10bproF2 | AG GGTACC TGGAGTGGGGCTAATCGTGATCC |
| wnt10bproR | CCC AAGCTT CTAACTCACGGGTGGTTCTGCTC |
Generation of Cbfβ CKO Mice.
Cbfβf/f mice and mice with tissue-specific promoter-driven Cre were crossed to generate heterozygous mice (15–18).
Histology and tissue preparation were performed as described previously (25).
Immunohistochemistry (IHC) and IF staining were performed using a commercial kit (CAT#BMK-2202; Vector Laboratories) (25–27).
Cell Culture and Osteoblast Function.
Osteoblastogenesis from primary calvarial cells and bone marrow MSC was performed as described (15, 28).
Gene expression analyses (Western blot and qRT-PCR) were performed as previously described (25, 26).
Nile Red Staining.
Differentiated cells were stained by 25 ng/mL Nile Red and counterstained by DAPI.
ChIP was performed as described using primary osteoblast lysates and MAGnify ChIP system (Cat#492024; Invitrogen) (15).
Promoter Luciferase Assay.
Promoter assay-related plasmids were transfected into C3H10T1/2 cells. Luciferase was detected and analyzed (15).
Statistical Analysis.
The number of animals to be used in this study was determined in accordance with power analysis and our previous studies (25, 26). Detailed materials and methods can also be retained in SI Materials and Methods.
SI Materials and Methods
Generation of Cbfβ CKO Mice.
Cbfβf/f, Prx1-Cre, and Osx-Cre mice were purchased from Jackson Laboratory. Col2α1-Cre mice were kindly given by Rosa Sera. Cbfβf/f mice and mice with tissue-specific promoter-driven Cre were crossed to generate heterozygous mice, which were intercrossed to obtain homozygous CKO (conditional knockout) mice (15–18). To confirm the generation of CKO mice, the genotypes of the mice were determined by PCR and Western blot using both tissues of mesenchymal lineage (i.e., osteoblast, cartilage) and nonspecific tissues (i.e., heart, lung, liver). All mice were maintained under a 12-h light–dark cycle with ad libitum access to regular food and water at the UAB Animal Facility. Both male and female mice of each strain were randomly selected into groups of five animals each. The investigators were not blinded during allocation, animal handling, and endpoint measurements. The study was approved by the UAB Animal Care and Use Committee, conformed to National Institutes of Health guidelines, and followed all recommendations of Animal Research: Reporting in Vivo Experiments guidelines.
Histology and Tissue Preparation.
Histology and tissue preparation were performed as described previously (25). Murine femurs and tibiae were harvested, skinned, and eviscerated before fixing in 4% paraformaldehyde (PFA) in 1× PBS overnight. Samples were then dehydrated in ethanol and decalcified in 10% EDTA for 1 wk. For paraffin sections, samples were dehydrated in ethanol, cleared in xylene, embedded in paraffin, sectioned at 6 μm with a Leica microtome, and then mounted on Superfrost Plus slides (Fisher). For frozen sections, samples were infiltrated in 30% sucrose, embedded in optimum cutting temperature (OCT) compound, sectioned at 8 µm with a freezing microtome, and then mounted on Superfrost Plus slides (Fisher).
H&E Staining.
H&E staining was performed as described previously (25). Mice were skinned and eviscerated, and then fixed in 4% PFA overnight. Specimens were dehydrated in ethanol and embedded in paraffin. Sections were cut at a thickness of 6 μm with a microtome and then mounted on Superfrost Plus slides (Fisher). Sections were deparaffinized and hydrated through a xylene and graded ethanol series, rinsed in hematoxylin, in 1% acid alcohol and ammonia-H2O, and then in eosin. Slides were dehydrated in graded ethanol and xylene.
IHC.
The 6-μm paraffin sections were deparaffined, and antigens retrieval was achieved by heat treatment with a commercial reagent (Abcam AB970). Immunohistochemistry staining was performed using a commercial kit (Mouse on Mouse Basic kit CAT#BMK-2202; Vector Laboratories) and mouse anti-active β-catenin antibody (05-665; Millipore) as previously described (25, 27). The procedure followed the manufacturer’s instructions. Slides were counterstained by hematoxylin.
IF.
Calvarial cells cultured on cover slides were maintained in osteogenic medium and then fixed in 10% formalin. IF staining was performed using a commercial kit (Mouse on Mouse Basic kit CAT#BMK-2202; Vector Laboratories), mouse–anti-active–β-catenin antibody (05-665; Millipore), and Rhodamine Red-conjugated Streptavidin (016-290-084; Jackson Immunoresearch Inc.), following the manufacturer’s instructions as previously described (25, 26). Slides were counterstained by Hoechst 33342 (H3570; Life Technology).
Oil Red O Staining.
The 8-μm frozen sections were used for Oil Red O staining. The slides were fixed in 10% formalin, counterstained by hematoxylin and then stained in Oil Red O staining solution (0.08% oil Red O in 40% isopropanol solution). The stained slides were mounted with 50% glycerol/PBS and visualized by light microscope.
Cell Culture and Osteoblast Function.
Primary calvarial osteoblasts were isolated from newborn mice and seeded in culture at 3 × 103 cells per square centimeter as described (15). Subsequently, cells were induced using osteogenic medium and BGJb medium (12591; Gibco) supplemented with 10% (vol/vol) FBS, 50 μg/mL l-ascorbic acid (A4544; Sigma-Aldrich), and 5 mM glycerolphosphate (G9891; Sigma-Aldrich). Bone marrow MSCs were isolated as described (28). Cells were passaged and osteoblastogenesis were induced using osteogenic medium (α-MEM medium supplemented with 50 μg/mL l-ascorbic acid, 5 mM β-glycerolphosphate, and Dexamethasone). Osteoblastogenesis was analyzed by ALP staining according to manufacturer’s manual (A2356; Sigma-Aldrich). Osteoblast mineralization was examined by Von Kossa staining.
Wnt3L Conditional Medium.
Wnt3L conditional medium is prepared from L Wnt-3A (ATCC CRL-2647) cells according to manufacturer’s instruction.
Western Blot Analysis.
Protein samples were prepared from calvaria-derived osteoblasts in protein lysis buffer as described (25, 26). Nuclei and cytoplasmic fractions were separated using a commercial kit (NE-PER Nuclear and Cytoplasmic Extraction Reagents; Fisher Inc.) following manufacturer’s instructions. Proteins were resolved on SDS/PAGE and electrotransferred on nitrocellulose membranes. Cbfβ, active–β-catenin, and C/ebpα protein levels were analyzed using the following primary antibodies: rabbit–anti-Cbfβ (sc-56751; Santacruz), rabbit–anti-C/ebpα (sc-61; Santacruz), mouse–anti-active–β-catenin (05-665; Millipore), and mouse–anti–β-tubulin (E7; DSHB). Horseradish peroxidase-linked anti-rabbit IgG (7074) and Horseradish peroxidase-linked anti-mouse IgG (7076) were from Cell Signaling.
mRNA Preparation from 2- and 18-mo-Old Mice.
Femoral bone was isolated from 2- and 18-mo-old mice, and bone marrow was flushed out. The prepared samples were then transferred to a tube containing beads (Nextadvance Inc.) and homogenized using a Blender (Bullet BlenderH; Nextadvance). RNA extraction was carried out under standard procedures using TRIzol reagent (Invitrogen).
qRT-PCR Analysis.
Total RNA was isolated from cultured cells at day 7 and day 14 (as indicated) with TRIzol reagent (15596018; Life Technologies). Mouse cDNA was reverse-transcribed from 0.5 g total RNA with SuperScript VILO Master Mix (11755050; Life Technologies). The qRT-PCR was performed using the one-step RT-PCR System as previously described (25, 26). Primer sequences are presented in the Table S1.
Nile Red Staining.
Differentiated cells are fixed by paraformaldehyde, rinsed by PBS, stained by 25 ng/mL Nile Red, counterstained by DAPI, and visualized by fluorescent microscope.
ChIP.
ChIP was performed as described using primary osteoblast lysates (15). After immunoprecipitation using monoclonal anti-Cbfβ antibody (sc-20693 X) and DNA extraction, quantitative PCR was performed using the primers in the promoter region of mouse Wnt10b genes (primer sequences are presented in the Table S2).
Table S2.
Primers used for ChIP assay
| Gene | Forward primer | Reverse primer |
| Wnt10b pro4 | 5′- CCGAGGCAGACCCTTACTTCCTT -3′ | 5′- AGAAACCTTGCGCAGTTCTATGC -3′ |
| Wnt10b pro4 | 5′- CAGCGAGGGGAAGGAGTTCTCTA -3′ | 5′- AAAGCACTGGTGTGCTCTGACTC -3′ |
| Wnt10b pro2 | 5′- AGTGAGGTCATTAGGGAGACGGG -3′ | 5′- CCTGGGGTCACAGATGGAGGTAT -3′ |
| Wnt10b pro3 | 5′- AGGGAACTGTCTTTGCTGCCATT -3′ | 5′- GCTGAATGTAAGTGGATGGGCCT -3′ |
| Wnt10b pro1 | 5′- TCCCGGAACAGAAAATGAAGCCT -3′ | 5′- AGGACCCTGAGGTTTTAACAGGGA -3′ |
| Wnt10b pro6 | 5′- ACCGTCAGTAACCCTAAGGTCGT -3′ | 5′- ATCTGGTCCCAATGACAGGGTCT -3′ |
| Wnt10b pro7 | 5′- GTGAGTGGAACTCCGACCACTTG -3′ | 5′- CTAACTCACGGGTGGTTCTGCTC -3′ |
| Cebpa P1 | GCCTTTTAGAAATCCGGGTGGGA | TCTAAGTCACCCACTTCCAGCCA |
| Cebpa P2 | GCTCTGCACCGAAAATGAAGCAG | AGAACCGACTTCGTCTGAAGGAC |
Promoter Luciferase Assay.
The promoter region (−) and (+) of the mouse Wnt10b gene was amplified by PCR using Wnt10b Bac clone (cat#CH29-27K23; CHORI). Primer sequences are available in Table S3. Then the promoter regions were inserted into the pGL3-basic vector to construct the pGL3-Wnt10b promoter 1 vector (pro-1) and pGL3-Wnt10b promoter 2 vector (pro-2). The insertions of the constructs were confirmed by sequencing. C3H10T1/2 cells were cultured in 24-well plates, and were transiently transfected with a DNA mixture containing the pGL3-Wnt10b construct (0.3 μg) and β-GAL-expressing plasmids (0.03 μg), with or without Runx2 expressing vector (pcDNA3.1-Runx2, 0.3 μg) or Cbfβ-expressing vector (psport6-CMV-Cbfβ, 0.3 μg) using Lipofectamine and Plus reagents. Luciferase was detected using Glo Luciferase Assay System (Promega) 48 h posttransfection as described (15). The β-GAL activity of the cell lysates was analyzed using β-Galactosidase Enzyme Assay System (E2000; Promega). The level of luciferase activity was normalized to the level of β-GAL activity.
Statistical Analysis.
The number of animals used in this study was determined in accordance with power analysis and our previous studies (25, 26). In brief, our study used five mice per group per experiment. Data are presented as mean ± SEM (n ≥ 6). Statistical significance was assessed using Student’s t test. Values were considered statistically significant at P < 0.05. Results are representative of at least four individual experiments. Figures are representative of the data.
Acknowledgments
We thank Ms. Diep N. Nguyen for her excellent assistance with the manuscript. We appreciate the assistance of the Center for Metabolic Bone Disease at the UAB (P30 AR046031). We are also grateful for the assistance of the Small Animal Phenotyping Core, Metabolism Core, and Neuroscience Molecular Detection Core Laboratory at UAB (P30 NS0474666). This work was supported by National Institutes of Health Grants AR-044741 (to Y.-P.L.), DE-023813 (to Y.-P.L.), and AR-070135 (to W.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619294114/-/DCSupplemental.
References
- 1.Kirkland JL, Tchkonia T, Pirtskhalava T, Han J, Karagiannides I. Adipogenesis and aging: Does aging make fat go MAD? Exp Gerontol. 2002;37:757–767. doi: 10.1016/s0531-5565(02)00014-1. [DOI] [PubMed] [Google Scholar]
- 2.Duque G. Bone and fat connection in aging bone. Curr Opin Rheumatol. 2008;20:429–434. doi: 10.1097/BOR.0b013e3283025e9c. [DOI] [PubMed] [Google Scholar]
- 3.Muruganandan S, Roman AA, Sinal CJ. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: Cross talk with the osteoblastogenic program. Cell Mol Life Sci. 2009;66:236–253. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonough AK, Rosenthal RS, Cao X, Saag KG. The effect of thiazolidinediones on BMD and osteoporosis. Nat Clin Pract Endocrinol Metab. 2008;4:507–513. doi: 10.1038/ncpendmet0920. [DOI] [PubMed] [Google Scholar]
- 5.Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- 6.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 7.Lin FT, Lane MD. CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Yang S, Shao J, Li YP. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front Biosci. 2007;12:3068–3092. doi: 10.2741/2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takada I, Kouzmenko AP, Kato S. Molecular switching of osteoblastogenesis versus adipogenesis: Implications for targeted therapies. Expert Opin Ther Targets. 2009;13:593–603. doi: 10.1517/14728220902915310. [DOI] [PubMed] [Google Scholar]
- 10.Ross SE, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 11.Song L, et al. Loss of wnt/β-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res. 2012;27:2344–2358. doi: 10.1002/jbmr.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa E, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa E, et al. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, et al. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, et al. Cbfβ deletion in mice recapitulates cleidocranial dysplasia and reveals multiple functions of Cbfβ required for skeletal development. Proc Natl Acad Sci USA. 2014;111:8482–8487. doi: 10.1073/pnas.1310617111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, et al. Deletion of core-binding factor β (Cbfβ) in mesenchymal progenitor cells provides new insights into Cbfβ/Runxs complex function in cartilage and bone development. Bone. 2014;65:49–59. doi: 10.1016/j.bone.2014.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M, et al. Chondrocyte-specific knockout of Cbfβ reveals the indispensable function of Cbfβ in chondrocyte maturation, growth plate development and trabecular bone formation in mice. Int J Biol Sci. 2014;10:861–872. doi: 10.7150/ijbs.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian F, et al. Core binding factor beta (Cbfbeta) controls the balance of chondrocyte proliferation and differentiation by upregulating Indian hedgehog (Ihh) expression and inhibiting parathyroid hormone-related protein receptor (PPR) expression in postnatal cartilage and bone formation. J Bone Miner Res. 2014;29:1564–1574. doi: 10.1002/jbmr.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 20.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16:1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, et al. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88:904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takada I, Kouzmenko AP, Kato S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol. 2009;5:442–447. doi: 10.1038/nrrheum.2009.137. [DOI] [PubMed] [Google Scholar]
- 23.Longo KA, et al. Wnt10b inhibits development of white and brown adipose tissues. J Biol Chem. 2004;279:35503–35509. doi: 10.1074/jbc.M402937200. [DOI] [PubMed] [Google Scholar]
- 24.Bennett CN, et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci USA. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W, et al. C/EBPα regulates osteoclast lineage commitment. Proc Natl Acad Sci USA. 2013;110:7294–7299. doi: 10.1073/pnas.1211383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu M, et al. Gα13 negatively controls osteoclastogenesis through inhibition of the Akt-GSK3β-NFATc1 signalling pathway. Nat Commun. 2017;8:13700. doi: 10.1038/ncomms13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YP, Chen W, Liang Y, Li E, Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet. 1999;23:447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]
- 28.Soleimani M, Nadri S. A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat Protoc. 2009;4:102–106. doi: 10.1038/nprot.2008.221. [DOI] [PubMed] [Google Scholar]