Significance
Vibrational spectroscopy is an ideal technique for analysis of biofluids, as it provides a “spectral fingerprint” of all of the molecules present within a biological sample, thus generating a holistic picture of the sample’s status. Neurodegenerative diseases lack early and accurate diagnosis, and tests currently used for their detection are either invasive or expensive and time-consuming. This study used blood plasma to diagnose and differentiate various neurodegenerative diseases; the achieved sensitivities and specificities are equal to, or even higher than, the ones obtained by clinical/molecular methods. Herein, we show that spectroscopy could provide a simple and robust diagnostic test. Additional work should include asymptomatic individuals for an early screening test and exploration of neurodegenerative diseases at all stages of severity.
Keywords: Alzheimer’s disease, dementia with Lewy bodies, apolipoprotein E, differential diagnosis, spectroscopy
Abstract
The progressive aging of the world’s population makes a higher prevalence of neurodegenerative diseases inevitable. The necessity for an accurate, but at the same time, inexpensive and minimally invasive, diagnostic test is urgently required, not only to confirm the presence of the disease but also to discriminate between different types of dementia to provide the appropriate management and treatment. In this study, attenuated total reflection FTIR (ATR-FTIR) spectroscopy combined with chemometric techniques were used to analyze blood plasma samples from our cohort. Blood samples are easily collected by conventional venepuncture, permitting repeated measurements from the same individuals to monitor their progression throughout the years or evaluate any tested drugs. We included 549 individuals: 347 with various neurodegenerative diseases and 202 age-matched healthy individuals. Alzheimer’s disease (AD; n = 164) was identified with 70% sensitivity and specificity, which after the incorporation of apolipoprotein ε4 genotype (APOE ε4) information, increased to 86% when individuals carried one or two alleles of ε4, and to 72% sensitivity and 77% specificity when individuals did not carry ε4 alleles. Early AD cases (n = 14) were identified with 80% sensitivity and 74% specificity. Segregation of AD from dementia with Lewy bodies (DLB; n = 34) was achieved with 90% sensitivity and specificity. Other neurodegenerative diseases, such as frontotemporal dementia (FTD; n = 30), Parkinson’s disease (PD; n = 32), and progressive supranuclear palsy (PSP; n = 31), were included in our cohort for diagnostic purposes. Our method allows for both rapid and robust diagnosis of neurodegeneration and segregation between different dementias.
Neurodegenerative diseases are characterized by a common pathological mechanism of protein misfolding and aggregation and result in a progressive loss of neuronal function in the human brain/CNS and unavoidable death. Some of the common symptoms among them include problems with movement (e.g., parkinsonism) and mental functioning (e.g., dementia). Improvements in health care and social support have resulted in many elderly sufferers of neurodegenerative disease having prolonged lives with improved quality of life. However, these diseases remain incurable, and one of the main reasons for this is likely to be that their etiology is multifactorial. It is, therefore, crucial to develop new biomarkers to better comprehend the etiology of each disease and also, for screening and diagnosis, so that appropriate clinical intervention can begin as soon as possible. Surrogate markers of disease progression will also facilitate the development of new treatments, and this has led to an increased interest over recent years in the discovery of biomarkers for diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), dementia with Lewy bodies (DLB), vascular dementia, and frontotemporal dementia (FTD).
AD accounts for 60–70% of all types of dementia, with symptoms of memory loss; language problems; disorientation in identity, time, and place; cognitive decline; and eventually, death. Various pathological mechanisms contribute to brain damage, including amyloid deposition (1), neurofibrillary tangle formation (2), oxidative stress (3), inflammation (4, 5), and lipid dysregulation (6). A major genetic risk factor for AD is the ε4 allele of apolipoprotein E (APOE). APOE protein is involved in cholesterol distribution throughout the body, including brain, and exists in three different isoforms (E2, E3, E4) (7). Possession of one or more APOE ε4 alleles increases the risk of developing late-onset AD, reduces age of onset, and is associated with a higher cerebral amyloid plaque load and promotion of neurofibrillary tangle formation (8, 9).
Current approaches for diagnosing AD include measurement of amyloid-β (Aβ; specifically Aβ42) and total tau (T-tau) or phosphorylated tau (P-tau) levels in cerebrospinal fluid (CSF), structural neuroimaging techniques (MRI or computerized tomography), PET imaging of brain amyloid, or inflammation and batteries of neuropsychological tests (10). Usually, a combination of more than one test is necessary to provide a working diagnosis, while a definitive diagnosis can still only be given postmortem with histopathological examination (11). Brain scans present disadvantages, as they are expensive and time-consuming. CSF would be an optimal source for AD biomarkers, as it is in contact with the brain, allowing biochemical changes in the brain to be directly monitored. However, CSF collection is not universally obtainable, as it is invasive (lumbar puncture) and can be unpleasant for patients; therefore, not all clinicians are willing to take such samples routinely. Five hundred milliliters of CSF is discharged into the bloodstream every day, and therefore, alterations in CSF content can also facilitate discovery of blood-borne biomarkers of neurodegenerative disease (12).
Research has, therefore, focused on blood-based methods for diagnosis of neurodegenerative diseases. A major goal is the detection of AD at a very early stage, even before the patient becomes symptomatic (10, 13). Early diagnosis is crucial, as once the patient appears with clinical symptoms, the damage that has occurred in the brain may already be irreversible. However, if at-risk individuals are identified early, it may be possible to delay or even prevent additional progression of the disease by administration of drugs that decrease Aβ production or aggregation (preventing plaque formation) or tau protein aggregation (preventing tangle formation) (14). The diagnostic accuracy of existing biomarkers has been previously reported to vary from ∼70 to ∼95% (10, 13, 15, 16); however, most of the current tests cannot be used routinely because of their high cost and laborious sample processing.
Vibrational spectroscopy, including FTIR and Raman spectroscopy, has been widely used to discriminate and classify normal and pathological populations using cells, tissues, or biofluids (17–19). Readily accessible biofluids, such as blood plasma/serum, saliva, or urine, are considered ideal for clinical implementation because of routine methods of collection as well as their minimal sample preparation. Interrogation of these samples with spectroscopic techniques allows for the generation of a “spectral fingerprint,” which subsequently facilitates the discrimination of the different populations and identification of potential biomarkers. In the past few years, blood-based FTIR and Raman spectroscopy have been used for diagnosing, screening, or monitoring the progression/regression in a variety of diseases (20, 21). Spectroscopic techniques have many advantages over conventional molecular tests (e.g., ELISA), as they allow the investigation of a range of different molecules simultaneously instead of studying isolated molecules; thus, they are ideal for complicated, multifactorial diseases. They are also rapid, cost-effective, and nondestructive, which renders them a perfect candidate for translation to clinic.
In this study, attenuated total reflection FTIR (ATR-FTIR) spectroscopy was used to interrogate blood plasma samples from individuals with various neurodegenerative diseases. One of our goals was to differentiate AD as well as early AD patients from healthy control (HC) individuals and provide meaningful biomarkers that would assist in a more conclusive clinical diagnosis. We also took into consideration the APOE genotype and age of our participants as confounding factors. The second major objective of this study was to determine the spectral wavenumbers that allow segregation of AD from other dementias, such as DLB and FTD, where differential diagnosis—especially in early stages—can be challenging. The segregation of AD from DLB is of particular clinical importance, since accurate diagnosis is necessary for suitable medication. DLB is the second most common form of dementia (10–15% of all dementias) and shares characteristics with AD, leading to frequent misdiagnosis (22). Although AD remains the most common neurodegenerative disorder, other less common diseases, such as progressive supranuclear palsy (PSP) and PD, were also included for diagnostic purposes.
Results
AD Vs. HC.
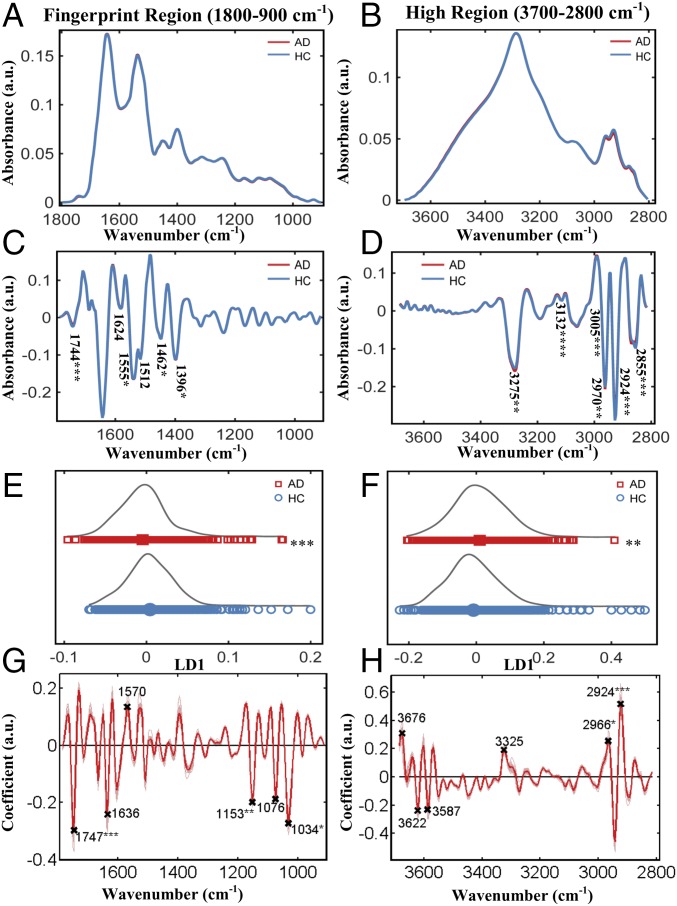
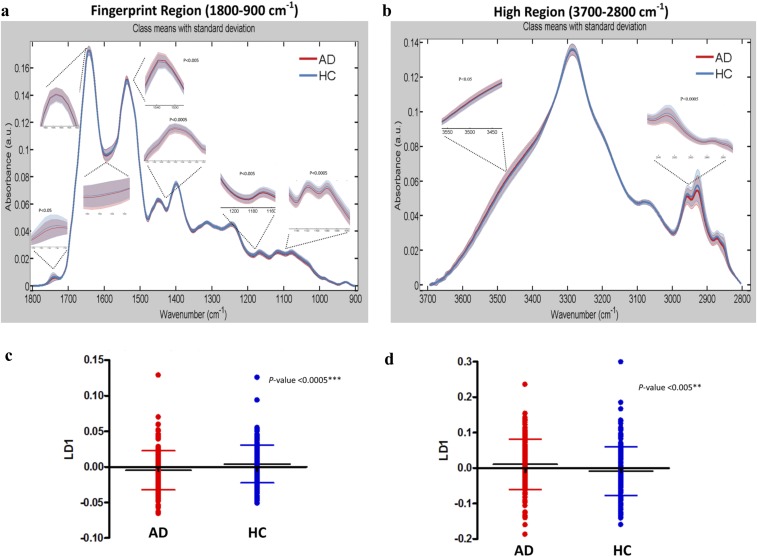
We primarily focused on the discrimination of AD patients from HCs; Fig. 1 A and B shows the preprocessed spectra (cut, rubber band baseline corrected, vector normalized) at the fingerprint region (1,800–900 cm−1) and high region (3,700–2,800 cm−1), respectively. At the fingerprint region (Fig. 1A and Fig. S1A), the AD group showed slightly higher peaks at 1,650–1,630 cm−1 [P = 0.095, 95% confidence interval (95% CI) = −0.005, 0.0004, stretching vibration (ν) of (C = O), Amide I, proteins] and 1,540–1,530 cm−1 [P = 0.003, 95% CI = −0.0031, −0.0007, bending vibration (δ) of (N-H), Amide II, proteins] as well as lower peaks at 1,750–1,735 cm−1 [P = 0.006, 95% CI = 0.0006, 0.0035, ν(C = O) of lipids], 1,590–1,580 cm−1 [P = 0.254, 95% CI = −0.0008, 0.003, δ(N-H) of Amide II, proteins], 1,470–1,430 cm−1 [P = 0.0002, 95% CI = 0.002, 0.006, δ(CH3) and δ(CH2) of lipids and proteins], 1,220–1,160 cm−1 [P = 0.0035, 95% CI = 0.0016, 0.0084, carbohydrates, asymmetric stretching (νas) of PO2− of DNA/RNA], and 1,150–1,040 cm−1 [P = 0.0002, 95% CI = 0.0057, 0.0185, carbohydrates and symmetric stretching (νs) of PO2− of DNA/RNA]. At the higher region (Fig. 1B and Fig. S1B), the AD group showed lower peaks at 2,950–2,850 cm−1 [P = 0.0004, 95% CI = 0.021, 0.017, νs(CH2) of lipids] and slightly higher peaks at 3,550–3,450 cm−1 [P = 0.0165, 95% CI = −0.048, −0.0047, ν of (–OH)]. The aforesaid areas of interest are shown enlarged in Fig. S1 A and B to make the differences more apparent.
Fig. 1.
AD (n = 164) vs. HC (n = 202). (A and B) Preprocessed spectra (cut, rubber band baseline corrected, vector normalized) at the fingerprint and high region. (C and D) Preprocessed spectra (cut, second-order differentiated, vector normalized) along with the wavenumbers responsible for differentiation. At the fingerprint region: 1,744 cm−1 (lipids), 1,624 cm−1 (Amide I), 1,555 cm−1 (Amide II), 1,512 cm−1 (Amide II), 1,462 cm−1 (lipids), and 1,396 cm−1 [δ(CH3) of proteins]. At the high region: 3,275 cm−1 (–OH stretching), 3,132 cm−1 [νs(N-H)], 3,005 cm−1 [νas (=C-H), lipids and fatty acids], 2,970 cm−1 [νas (CH3), lipids and fatty acids], 2,924 cm−1 [νas (CH2), lipids], and 2,855 cm−1 [νs(CH2), lipids]. (E and F) The 1D scores plot (LD1) after cross-validated PCA-LDA. (G and H) Loading plots showing the wavenumbers responsible for discrimination. At the fingerprint region: 1,747 cm−1 (lipids), 1,636 cm−1 (Amide I), 1,570 cm−1 (Amide II), 1,153 cm−1 (carbohydrate), 1,076 cm−1 [νs (PO2−)], and 1,034 cm−1 (glycogen). At the high region: 3,676 cm−1 (–OH stretching), 3,622 cm−1 (–OH stretching), 3,587 cm−1 (–OH stretching), 3,325 cm−1 (–NH stretching), 2,966 cm−1 (–CH stretching of lipids), and 2,924 cm−1 (–CH stretching of lipids). *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.00005.
Fig. S1.
AD (n = 164) vs. HC (n = 202). (A) Preprocessed spectra (cut, rubber band baseline corrected, vector normalized) at the fingerprint region 1,800–900 cm−1. The spectral regions of interest are magnified along with the P values for the regions that were statistically significant: 1,750–1,735 cm−1 (lipids; P = 0.006, 95% CI = 0.0006, 0.0035), 1,650–1,630 cm−1 (Amide I, proteins; P = 0.095, 95% CI = −0.005, 0.0004), 1,590–1,580 cm−1 (Amide II, proteins; P = 0.254, 95% CI = −0.0008, 0.003), 1,540–1,530 cm−1 (Amide II, proteins; P = 0.003, 95% CI = −0.0031, −0.0007), 1,470–1,430 cm−1 (lipids/proteins; P = 0.0002, 95% CI = 0.002, 0.006), 1,220–1,160 cm−1 (carbohydrates, DNA/RNA; P = 0.0035, 95% CI = 0.0016, 0.0084), and 1,150–1,040 cm−1 (carbohydrates, DNA/RNA; P = 0.0002, 95% CI = 0.0057, 0.0185). (B) Preprocessed spectra (cut, rubber band baseline corrected, vector normalized) at the high region 3,700–2,800 cm−1. The spectral regions of interest are magnified along with the P values for the regions that were statistically significant: 2,950–2,850 cm−1 (lipids; P = 0.0004, 95% CI = 0.021, 0.017) and 3,550–3,450 cm−1 (–OH stretching; P = 0.0165, 95% CI = −0.048, −0.0047). (C and D) The 1D scores plots (cross-validated PCA-LDA) comparing individuals rather than spectra. Significant differences were seen in both spectral regions (fingerprint region: P = 0.0007, 95% CI = 0.004, 0.014; high region: P = 0.003, 95% CI = −0.035, −0.007). Data are expressed as the mean ± SD. **P < 0.005; ***P < 0.0005.
The above-mentioned preprocessing method generates figures that allow for better visualization and gives an initial impression of the generic differences. However, spectra appeared to overlap to a great extent. This was expected, as the majority of blood components are common in all individuals; thus, it is difficult to detect differences associated with AD, and even when differences are present, they must be brought to light. For this reason, second-order differentiation was used to examine the spectra in more detail, as it resolves overlapping peaks. Fig. 1 C and D shows the mean spectra after second derivative analysis. For biomarker identification, we initially implemented the “difference-between-mean” (DBM) spectra approach with a peak detection algorithm (for six peaks) to extract wavenumbers that showed the largest differences between AD and HC. The selected wavenumbers for the fingerprint and high region are shown in Fig. 1 C and D, respectively.
Multivariate approaches were also implemented for additional examination of the spectra; Fig. 1 E–H was all generated after cross-validated principal component analysis followed by linear discriminant analysis (PCA-LDA). Fig. 1 E and F shows 1D scores plots showing the differences between the two classes; the “scores” here represent individual spectra. To investigate individuals rather than spectra, we averaged every 10 (because 10 spectra were collected per sample) before the statistical test. The generated figures (Fig. S1 C and D) represent the scores plots shown in Fig. 1 E and F, but this time, each score is an individual. The difference between AD and HC individuals was statistically significant in both regions (P = 0.0007, 95% CI = 0.004, 0.014 at the fingerprint region and P = 0.003, 95% CI = −0.035, −0.007 at the high region). To identify the spectral bands that mostly correlate to these differences, we then generated the loading plots along with the top six wavenumbers responsible for discrimination (Fig. 1 G and H).
Classification of AD and HC Before the Incorporation of APOE ε4 Genotype and Age Information.
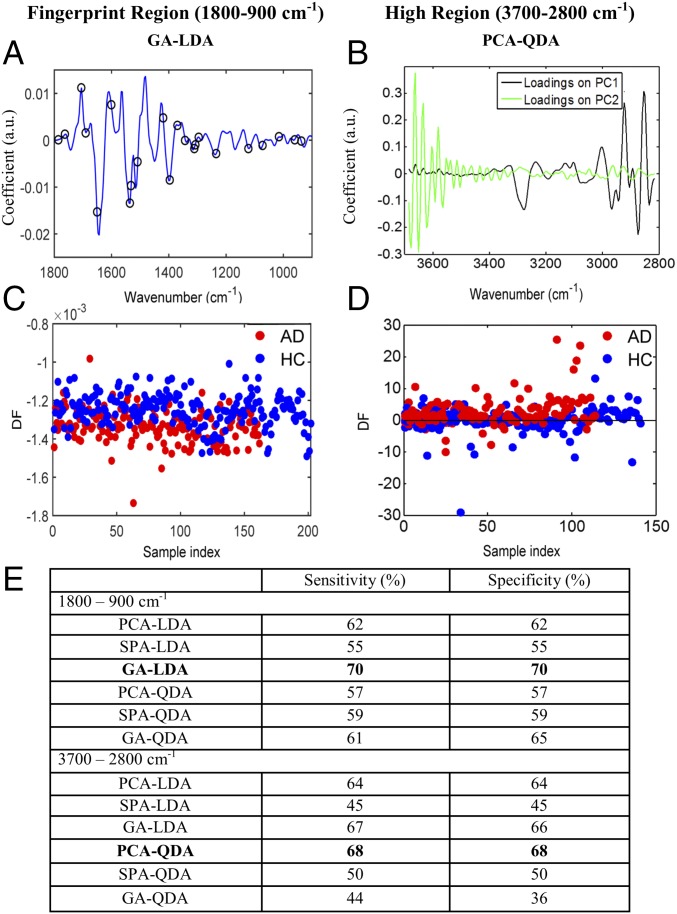
Classification of AD and HC was performed in both IR spectral regions of interest using PCA-LDA, principal component analysis followed by quadratic discriminant analysis (PCA-QDA) (Figs. S2 and S3), successive projection algorithm followed by linear discriminant analysis (SPA-LDA), successive projection algorithm followed by quadratic discriminant analysis (SPA-QDA) (Figs. S4 and S5), genetic algorithm followed by linear discriminant analysis (GA-LDA), and genetic algorithm followed by quadratic discriminant analysis (GA-QDA) (Figs. S6 and S7). The best classification ratios were obtained after GA-LDA for the fingerprint region (Fig. 2 A and C) and PCA-QDA for the high region (Fig. 2 B and D). GA-LDA used 23 wavenumbers (Fig. 2A) determined from the minimum cost function G (Fig. S6C), which identifies the optimal number of wavenumbers to be used. Fig. 2C is the scores plot illustrating classification by GA-LDA. Sensitivity and specificity achieved by GA-LDA, at the fingerprint region, were both equal to 70% (Fig. 2E). The loading and scores plots after PCA-QDA technique, at the high region, are shown in Fig. 2 B and D, respectively. In this case, the sensitivity and specificity were 68% (Fig. 2E).
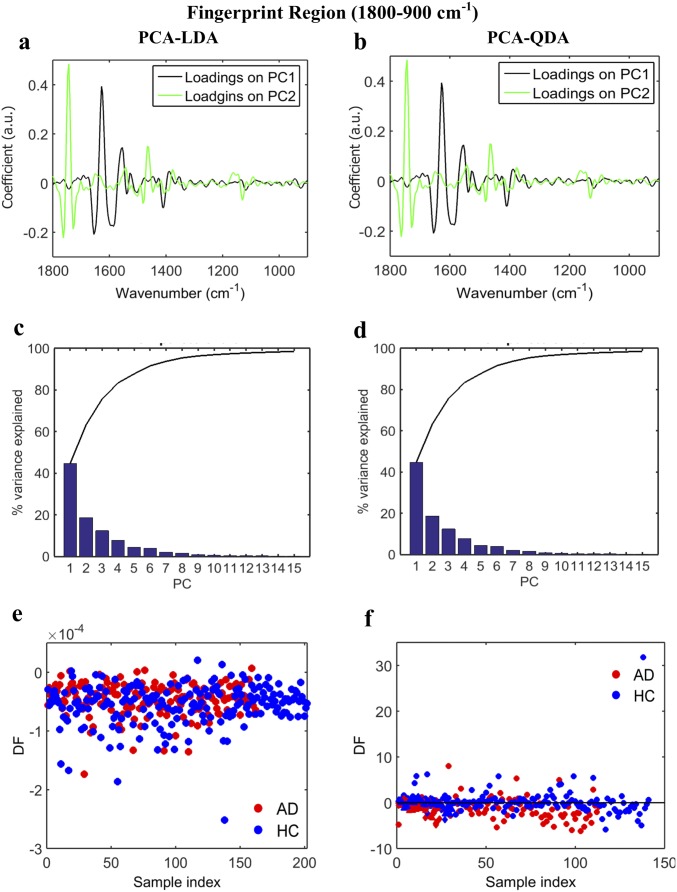
Fig. S2.
AD vs. HC using PCA-LDA/QDA at the fingerprint region (1,800–900 cm−1). (A and B) Loadings plots identifying the major discriminant wavenumbers after PCA-LDA and PCA-QDA, respectively. (C and D) The number of PCs used was optimized before PCA-LDA, using the Pareto function within the IRootLab toolbox, to prevent noise introduction. Ten PCs were used, representing >95% of the variance. (E and F) Scores plots illustrating classification by PCA-LDA and PCA-QDA, respectively. Sensitivity and specificity values are 62 and 62%, respectively, after PCA-LDA and 57 and 57%, respectively, after PCA-QDA (Fig. 2E). DF, discriminant function.
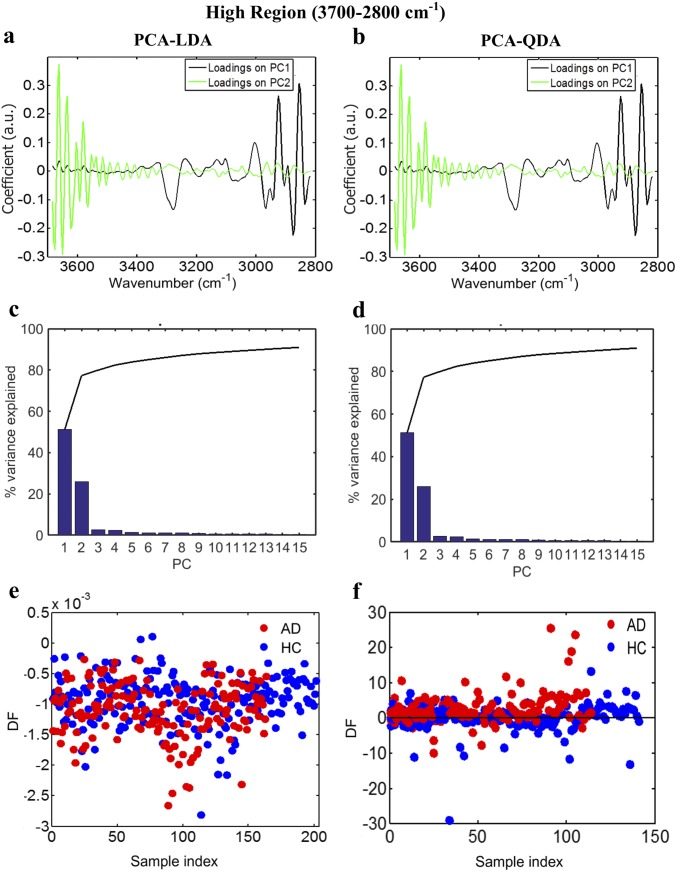
Fig. S3.
AD vs. HC using PCA-LDA/QDA at the high region (3,700–2,800 cm−1). (A and B) Loadings plots identifying the major discriminant wavenumbers after PCA-LDA and PCA-QDA, respectively. (C and D) The number of PCs used was optimized before PCA-LDA, using the Pareto function within the IRootLab toolbox, to prevent noise introduction. Ten PCs were used, representing >90% of the variance. (E and F) Scores plots illustrating classification by PCA-LDA and PCA-QDA, respectively. Sensitivity and specificity are 64 and 64%, respectively, after PCA-LDA and 68 and 68%, respectively, after PCA-QDA. DF, discriminant function.
Fig. S4.
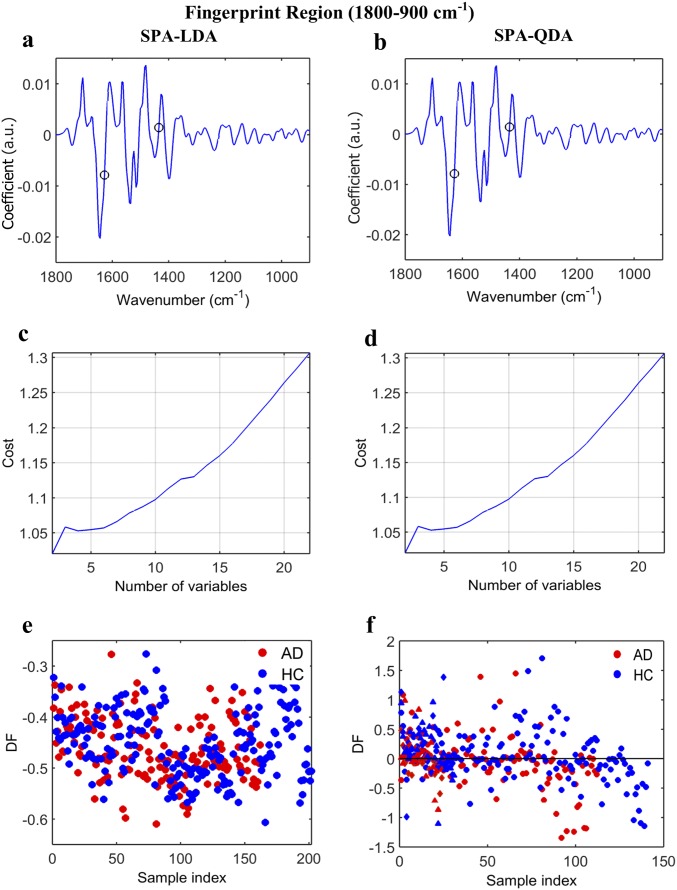
AD vs. HC using SPA-LDA/QDA at the fingerprint region (1,800–900 cm−1). (A and B) Wavenumber selection for SPA-LDA and SPA-QDA; the selected variables were 1,435 and 1,628 cm−1. (C and D) Cost/function plot identifying the optimal number of wavenumbers to be used for the SPA algorithm. (E and F) Scores plots illustrating classification by SPA-LDA and SPA-QDA, respectively. Sensitivity and specificity values are 55 and 55%, respectively, after SPA-LDA and 59 and 59%, respectively, after SPA-QDA (Fig. 2E). DF, discriminant function.
Fig. S5.
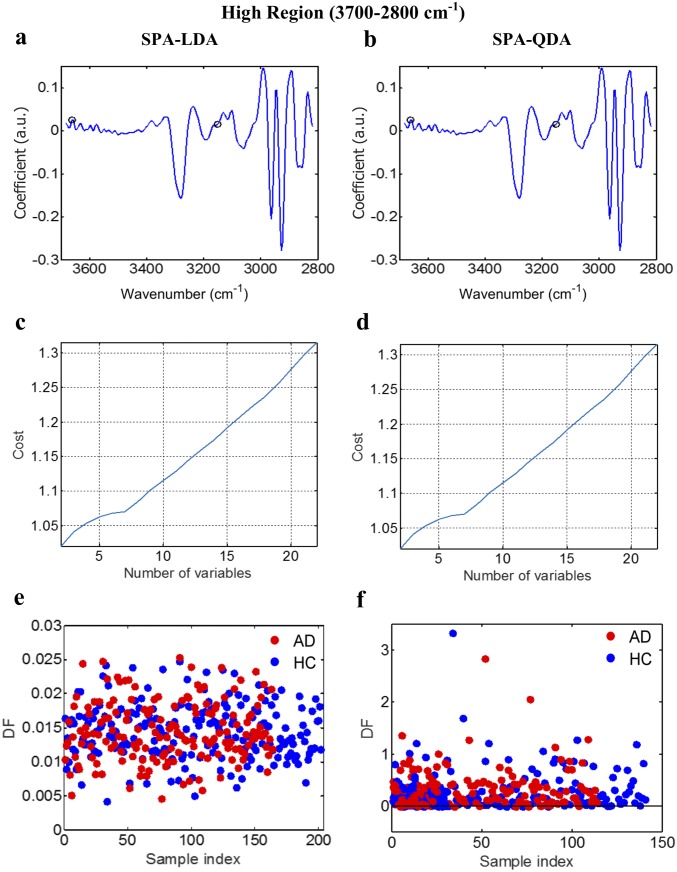
AD vs. HC using SPA-LDA/QDA at the high region (3,700–2,800 cm−1). (A and B) Wavenumber selection for SPA-LDA and SPA-QDA; the selected variables were 3,152 and 3,661 cm−1. (C and D) Cost/function plot identifying the optimal number of wavenumbers to be used for the SPA algorithm. (E and F) Scores plots illustrating classification by SPA-LDA and SPA-QDA, respectively. Sensitivity and specificity are 45 and 45%, respectively, after SPA-LDA and 50 and 50%, respectively, after SPA-QDA. DF, discriminant function.
Fig. S6.
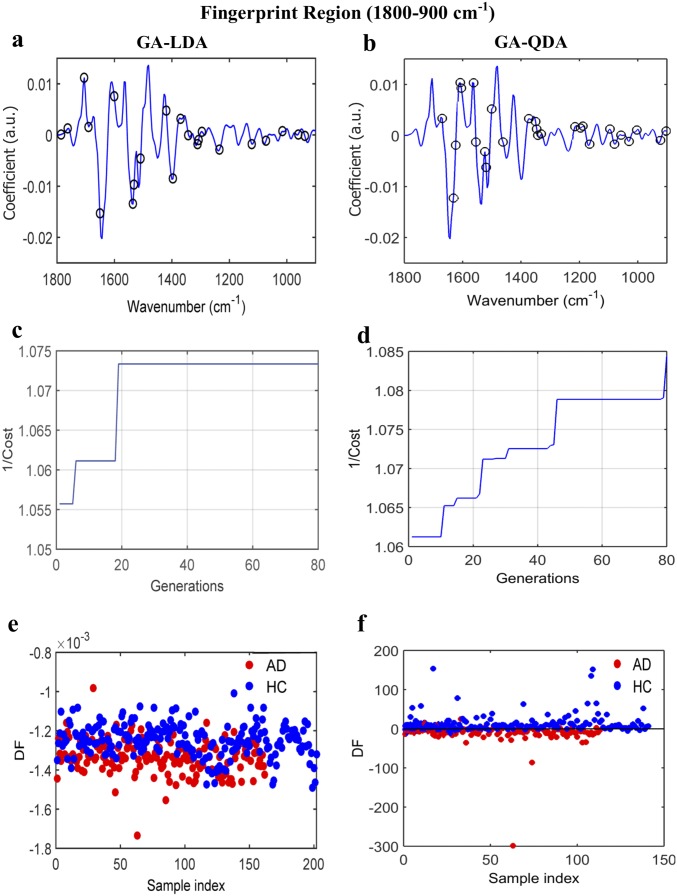
AD vs. HC using GA-LDA/QDA at the fingerprint region (1,800–900 cm−1). (A and B) Wavenumber selection for GA-LDA (900, 937, 960, 1,014, 1,072, 1,122, 1,234, 1,296, 1,308, 1,312, 1,342, 1,369, 1,396, 1,420, 1,508, 1,531, 1,535, 1,601, 1,651, 1,690, 1,705, 1,763, and 1,786 cm−1) and GA-QDA (903, 922, 1,003, 1,030, 1,057, 1,080, 1,096, 1,165, 1,188, 1,196, 1,215, 1,331, 1,342, 1,346, 1,350, 1,373, 1,462, 1,501, 1,520, 1,524, 1,555, 1,562, 1,605, 1,609, 1,624, 1,632, and 1,670 cm−1). (C and D) Cost/function plot identifying the optimal number of wavenumbers to be used for the GA algorithm. (E and F) Scores plots illustrating classification by GA-LDA and GA-QDA, respectively. Sensitivity and specificity are 70 and 70%, respectively, after GA-LDA and 61 and 65%, respectively, after GA-QDA. DF, discriminant function.
Fig. S7.
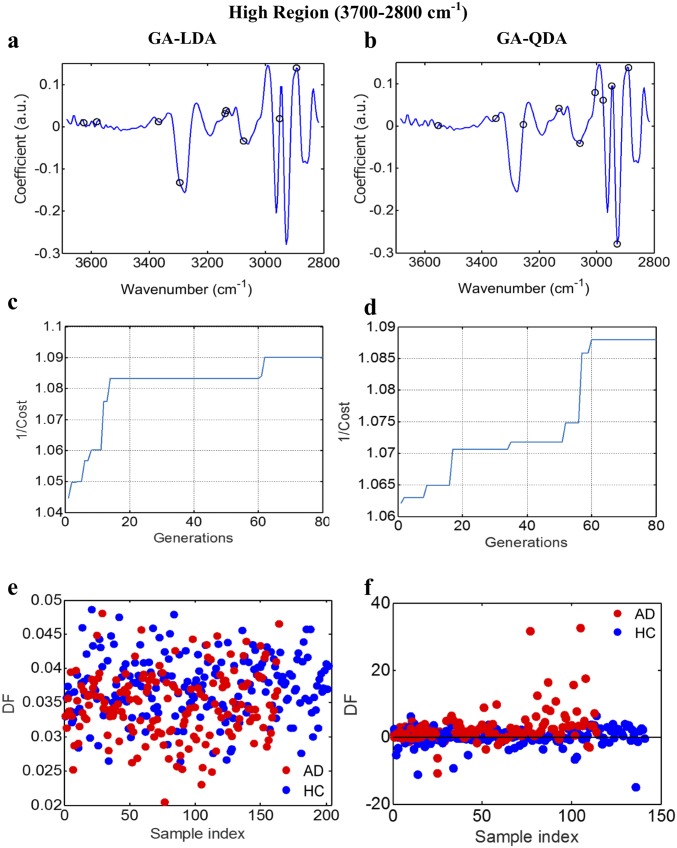
AD vs. HC using GA-LDA/QDA at the high region (3,700–2,800 cm−1). (A and B) Wavenumber selection for GA-LDA (2,893, 2,951, 3,074, 3,136, 3,140, 3,294, 3,368, 3,580, and 3,626 cm−1) and GA-QDA (2,889, 2,928, 2,947, 2,978, 3,005, 3,059, 3,132, 3,256, 3,352, and 3,553 cm−1). (C and D) Cost/function plot identifying the optimal number of wavenumbers to be used for the GA algorithm. (E and F) Scores plots illustrating classification by GA-LDA and GA-QDA, respectively. Sensitivity and specificity are 67 and 66%, respectively, after GA-LDA and 44 and 36%, respectively, after GA-QDA. DF, discriminant function.
Fig. 2.
Classification of AD before the incorporation of APOE ε4 genotype and age information. (A and B) The best classification techniques for the fingerprint region (GA-LDA) and high region (PCA-QDA). GA-LDA used 23 wavenumbers (900, 937, 960, 1,014, 1,072, 1,122, 1,234, 1,296, 1,308, 1,312, 1,342, 1,369, 1,396, 1,420, 1,508, 1,531, 1,535, 1,601, 1,651, 1,690, 1,705, 1,763, and 1,786 cm−1) for classification; PCA-QDA loadings plot depicts the most discriminant wavenumbers. (C and D) Scores plots illustrating classification by GA-LDA and PCA-QDA at the two regions. (E) Table showing the sensitivities and specificities achieved by all of the chemometric techniques used for both spectral regions. The ones shown in bold provided the best classification results. DF, discriminant function.
Classification of AD and HC After the Incorporation of APOE ε4 Genotype and Age Information.
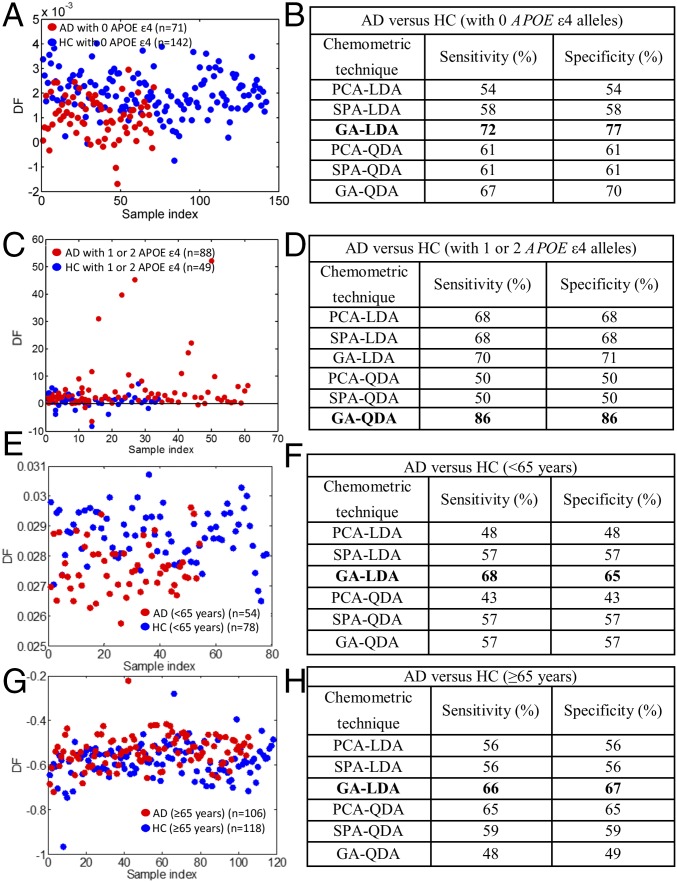
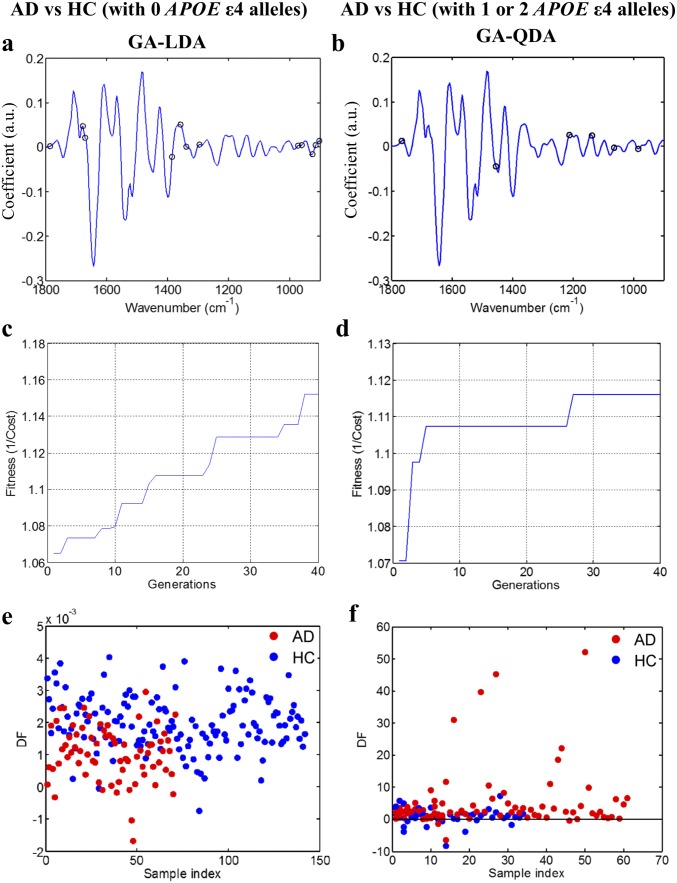
Similar to before, all of the available chemometric techniques were applied after incorporation of APOE ε4 and age information. GA-LDA provided higher accuracy than before, achieving sensitivity of 72% and specificity of 77% in individuals with no APOE ε4 alleles (Fig. 3 A and B and Fig. S8 A, C, and E). From the comparison of AD patients and controls with one or two APOE ε4 alleles, the best sensitivity and specificity were achieved by GA-QDA—each equal to 86% (Fig. 3 C and D and Fig. S8 B, D, and F).
Fig. 3.
Classification of AD after the incorporation of APOE ε4 genotype and age information. (A and B) Scores plot after GA-LDA, which provided the highest sensitivity (72%) and specificity (77%) for individuals with no APOE ε4 alleles. The different sensitivities and specificities achieved by all of the classification methods are also shown. (C and D) Scores plot after GA-QDA, which provided sensitivity and specificity of 86%, for individuals with one or two APOE ε4 alleles. (E and F) Scores plot after GA-LDA for the comparison of AD with HC (<65 y); sensitivity and specificity were 68 and 65%, respectively. (G and H) Scores plot after GA-LDA for the comparison of AD with HC (≥65 y old); sensitivity and specificity were 66 and 67%, respectively. DF, discriminant function.
Fig. S8.
Classification techniques for AD vs. HC after the incorporation of APOE ε4 genotype information. (A and B) Wavenumber selection using GA-LDA for individuals with no APOE ε4 alleles [AD (n = 71), HC (n = 142)]: 903, 914, 926, 960, 972, 1,296, 1,339, 1,358, 1,385, 1,670, 1,678, and 1,786 cm−1. Wavenumber selection using GA-QDA for individuals with one or two APOE ε4 alleles [AD (n = 88), HC (n = 49)]: 984, 1,065, 1,138, 1,211, 1,454, and 1,767 cm−1. (C and D) Cost/function plot identifying the optimal number of wavenumbers to be used for the GA algorithm. (E and F) Scores plots illustrating classification by GA-LDA and GA-QDA, respectively. The optimal sensitivities and specificities were 72 and 77%, respectively, for individuals with no APOE ε4 alleles and 86 and 86%, respectively, for individuals with one or two APOE ε4 alleles (Fig. 3 B and D). DF, discriminant function.
The same algorithms were applied to further investigate the effect of age on classification; GA-LDA provided the highest sensitivity and specificity for both comparison groups: AD vs. HC (<65 y old) and AD vs. HC (≥65 y old), respectively. In the comparison of the two classes of <65 y of age, sensitivity was 68%, and specificity was 65% (Fig. 3 E and F). In the ≥65-y-old age group comparison, sensitivity and specificity were equal to 66 and 67%, respectively (Fig. 3 G and H).
Classification of Early AD and HC.
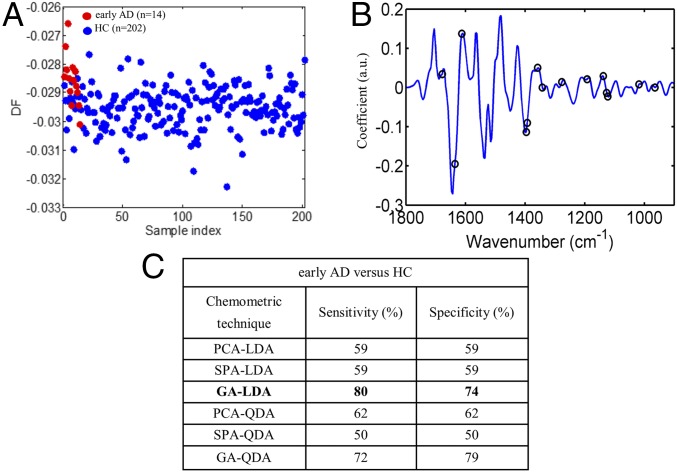
Our dataset included 14 cases of early AD; as previously shown, all of the classification algorithms were applied to compare early AD with HC (Fig. 4C). The chemometric technique that achieved the highest accuracy was GA-LDA, providing 80% sensitivity and 74% specificity. The selected wavenumbers for GA-LDA classification were 964, 1,018, 1,122, 1,126, 1,138, 1,192, 1,277, 1,342, 1,358, 1,393, 1,396, 1,612, 1,636, and 1,678 cm−1 (Fig. 4B).
Fig. 4.
Early AD (n = 14) vs. HC (n = 202). (A and B) Scores plot, illustrating classification by GA-LDA, and loadings plot, identifying the major discriminant variables by GA-LDA, which achieved the highest sensitivity and specificity of 80 and 74%, respectively. (C) Sensitivities and specificities achieved by all chemometric techniques. DF, discriminant function.
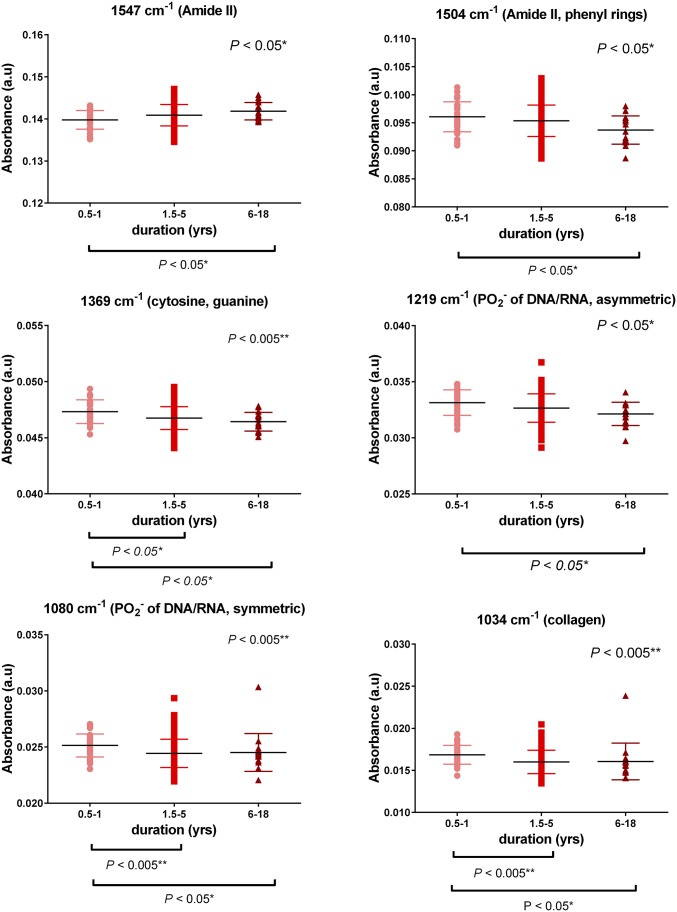
Duration of AD.
Patients were further classed and investigated according to disease duration to gain information on AD progression. Three different groups were examined based on duration: the first group (0.5–1 y) has 32 AD patients, the second group (1.5–5 y) has 111 patients, and the third group (6–18 y) has 17 patients. Relevant information was not available for four AD patients. Six spectral wavenumbers, corresponding to important biomolecules, showed substantial differences between the groups and therefore, were selected for additional exploration: 1,547 cm−1 (Amide II), 1,504 cm−1 (Amide II, phenyl rings), 1,369 cm−1 (cytosine, guanine), 1,219 cm−1 (νas of PO2− of DNA/RNA), 1,080 cm−1 (νs of PO2− of DNA/RNA), and 1,034 cm−1 (collagen). With increased duration, a decreasing trend was observed in most cases apart from the peak at 1,547 cm−1, where an increase was noted (Fig. 5). Statistical analysis was performed with data expressed as the mean ± SD. P values, for the groups that were found statistically different, are depicted on the respective graphs. Specifically, for the peak at 1,547 cm−1, significant differences were found between the first (0.5–1 y) and the third (6–18 y) group (P = 0.032, mean rank difference: −35.39). Peak at 1,504 cm−1 showed differences between the first group (0.5–1 y) and the third (6–18 y) group (P = 0.027, mean rank difference: 36.31). Peak at 1,369 cm−1 differentiated between the first (0.5–1 y) and second (1.5–5 y) groups (P = 0.03, mean rank difference: 24.1) as well as between the first (0.5–1 y) and the third (6–18 y) groups (P = 0.013, mean rank difference: 39.82). Peak at 1,219 cm−1 revealed differences between the first (0.5–1 y) and third (6–18 y) groups (P = 0.015, mean rank difference: 39.12). Peak at 1,080 cm−1 showed differences between the first (0.5–1 y) and second (1.5–5 y) groups (P = 0.0027, mean rank difference: 31.06) as well as between the first (0.5–1 y) and the third (6–18 y) groups (P = 0.038, mean rank difference: 34.65). Peak at 1,034 cm−1 showed differences between the first (0.5–1 y) and second (1.5–5 y) groups (P = 0.003, mean rank difference: 31.1) as well as between the first (0.5–1 y) and the third (6–18 y) groups (P = 0.015, mean rank difference: 39.06).
Fig. 5.
Duration of AD. The first group (0.5–1 y) includes 32 AD patients, the second group (1.5–5 y) includes 111 patients, and the third group (6–18 y) includes 17 patients. The most discriminant wavenumbers were 1,547 cm−1 (Amide II), 1,504 cm−1 (Amide II, phenyl rings), 1,369 cm−1 (cytosine, guanine), 1,219 cm−1 (νas of PO2− of DNA/RNA), 1,080 cm−1 (νs of PO2− of DNA/RNA), and 1,034 cm−1 (collagen). Data are expressed as mean (±SD). *P < 0.05; **P < 0.005.
Lipid-to-Protein, Phosphate-to-Carbohydrate, and RNA-to-DNA Ratios Related to Age.
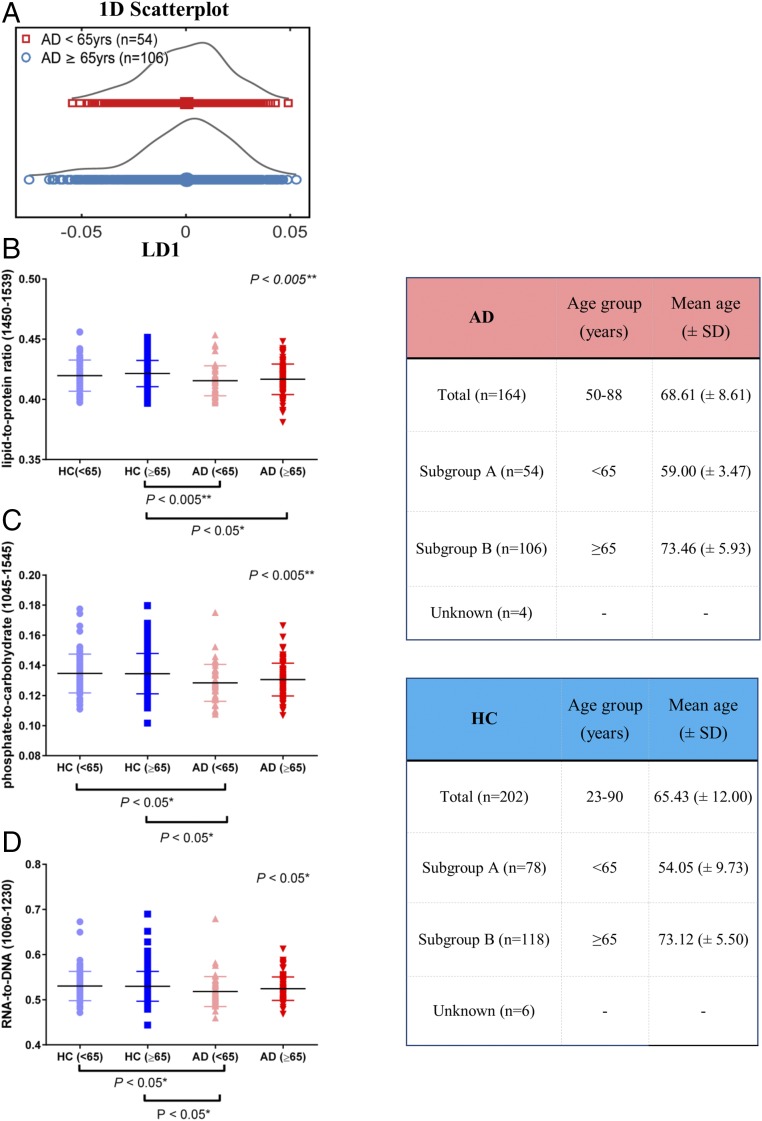
All individuals within the AD group were compared according to their age; patients were grouped into <65- and ≥65-y-old groups (Fig. 6A); people 65 y old and older are at higher risk of developing AD, and therefore, this age was used as a threshold value. Fig. 6A represents a 1D scores plot and was generated after cross-validated PCA-LDA (P = 0.6440, 95% CI = −0.0039, 0.0070).
Fig. 6.
Lipid-to-protein, phosphate-to-carbohydrate, and RNA-to-DNA ratios related to age. (A) Scores plot after cross-validated PCA-LDA showing differences and similarities of AD patients before and after the age of 65 y old; no significant differences were found after statistical analysis (P = 0.6440, 95% CI = −0.00391, 0.0070). (B–D) Intensity ratios of important spectral regions: lipid-to-protein (1,450/1,539 cm−1), phosphate-to-carbohydrate (1,045/1,545 cm−1), and RNA-to-DNA (1,060/1,230 cm−1), respectively. Data are expressed as mean (±SD). The two tables shown next to these graphs show the different subgroups and their mean age (±SD). *P < 0.05; **P < 0.005.
To further establish potential biomarkers and account for alterations between the different age groups of AD patients and HCs, we calculated the intensity ratio of six important spectral regions. Fig. 6 B–D represents the lipid-to-protein ratio (1,450/1,539 cm−1), phosphate-to-carbohydrate ratio (1,045/1,545 cm−1), and RNA-to-DNA ratio (1,060/1,230 cm−1), respectively. The two extra tables shown in Fig. 6 display the different subgroups for each class as well as their mean age (±SD). Subgroup A represents individuals of <65 y of age, and subgroup B represents individuals of ≥65 y of age.
No statistically significant differences were seen for the ratios within the HC or within AD groups. All of the different ratios, lipid-to-protein, phosphate-to-carbohydrate, and RNA-to-DNA, showed a decreasing trend for AD patients compared with HC. More specifically, in the lipid-to-protein ratio, statistically significant differences were seen after the comparison of HC (≥65 y old) vs. AD (<65 y old; P = 0.002, mean rank difference: 60.23) and HC (≥65 y old) vs. AD (≥65 y old; P = 0.014, mean rank difference: 42.05). Significant differences were also seen in the phosphate-to-carbohydrate ratio after the comparison of HC (<65 y old) vs. AD (<65 y old; P = 0.025, mean rank difference: 52.10) and HC (≥65 y old) vs. AD (<65 y old; P = 0.02, mean rank difference: 49.66). Lastly, the RNA-to-DNA ratio showed statistically significant differences for HC (<65 y old) vs. AD (<65 y old; P = 0.037, mean rank difference: 49.89) and HC (≥65 y old) vs. AD (<65 y old; P = 0.02, mean rank difference: 49.69).
Other Types of Neurodegenerative Diseases.
Another objective of this study was the identification of biomarkers that could be used for the diagnosis of less common neurodegenerative diseases (i.e., DLB, PD, FTD, and PSP); FTD and PSP patients were specifically selected for additional investigation, as the high number of subjects (>30) allowed for more robust conclusions. We also attempted to differentiate AD patients from patients with other types of dementia, such as DLB and FTD.
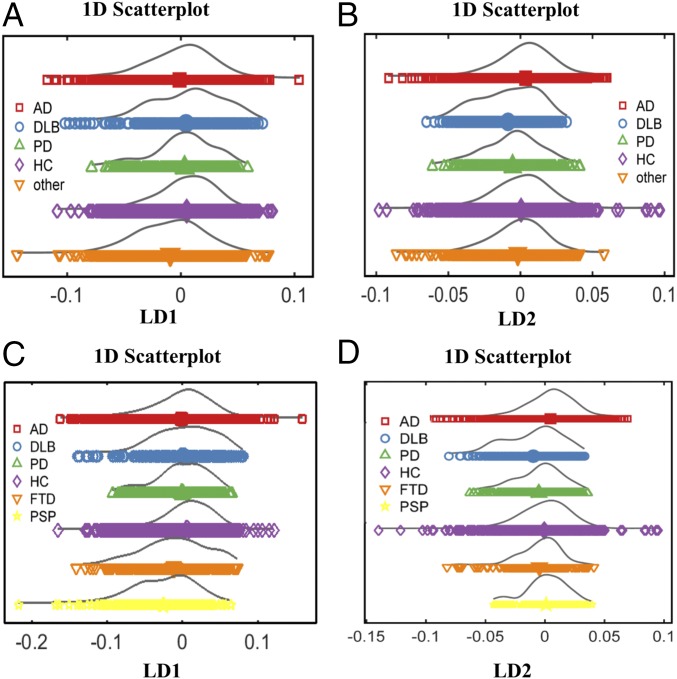
Initially, all of the different neurodegenerative pathologies were compared; the “other” group, containing less common neurodegenerative diseases, was also included to further validate our results and represent a pragmatic cohort. To visually account for similarities and differences between the various classes, we first generated the scores plots after cross-validated PCA-LDA (Fig. 7). With the use of linear discriminant 1 (LD1), the differentiation between the AD and HC was more prominent (Fig. 7A), while linear discriminant 2 (LD2) segregated better DLB and PD from HC (Fig. 7B). The other group was better separated from the HC when LD1 was used (Fig. 7A). LD1 was also responsible for the better segregation of FTD and PSP patients from HC (Fig. 7C). DLB and PD were not statistically different from HC when using LD1 (for the comparison between DLB and HC, P = 0.0521, 95% CI = −0.0353, 0.0002; for the comparison between PD and HC, P = 0.3315, 95% CI = −0.0282, 0.0089) (Fig. S9 A and B), whereas FTD and PSP were both statistically different from HC, with P = 0.0041, 95% CI = −0.0264, −0.0049 and P < 0.0001, 95% CI = −0.0439, −0.0201, respectively (Fig. S9 C and D).
Fig. 7.
Other types of neurodegenerative diseases. (A and B) The 1D scores plots after cross-validated PCA-LDA representing all of the different groups of neurodegenerative diseases [AD (n = 164), DLB (n = 34), PD (n = 32), HC (n = 202), other (n = 117)] using LD1 and LD2, respectively. (C and D) The 1D scores plots after cross-validated PCA-LDA after selection of FTD (n = 30) and PSP (n = 31) for additional analysis using LD1 and LD2, respectively.
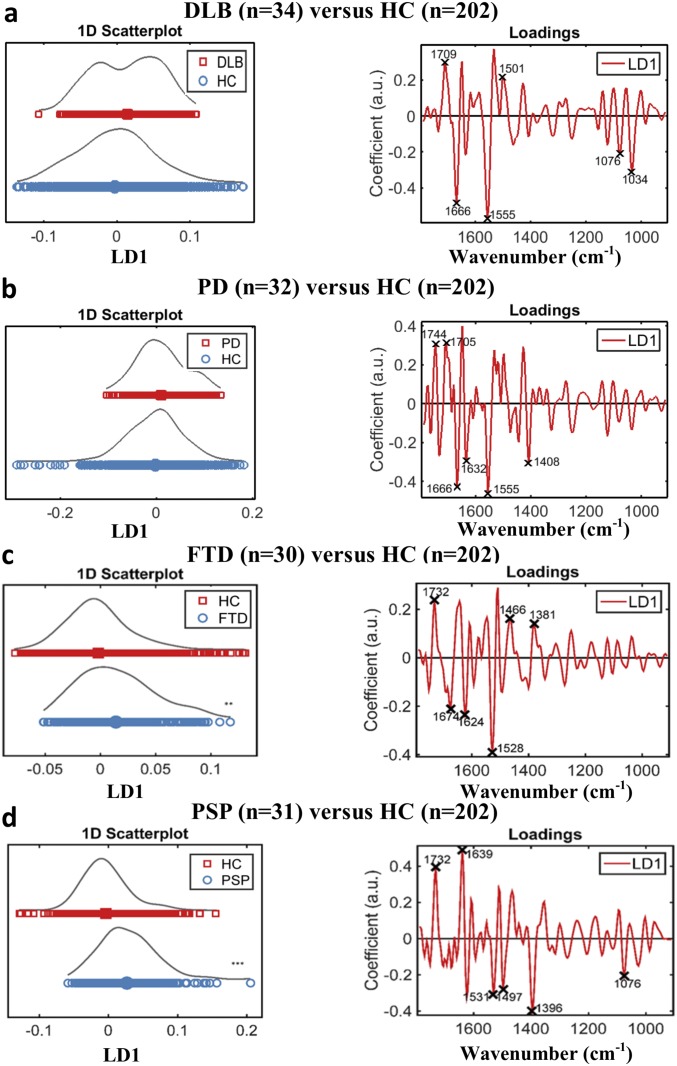
Fig. S9.
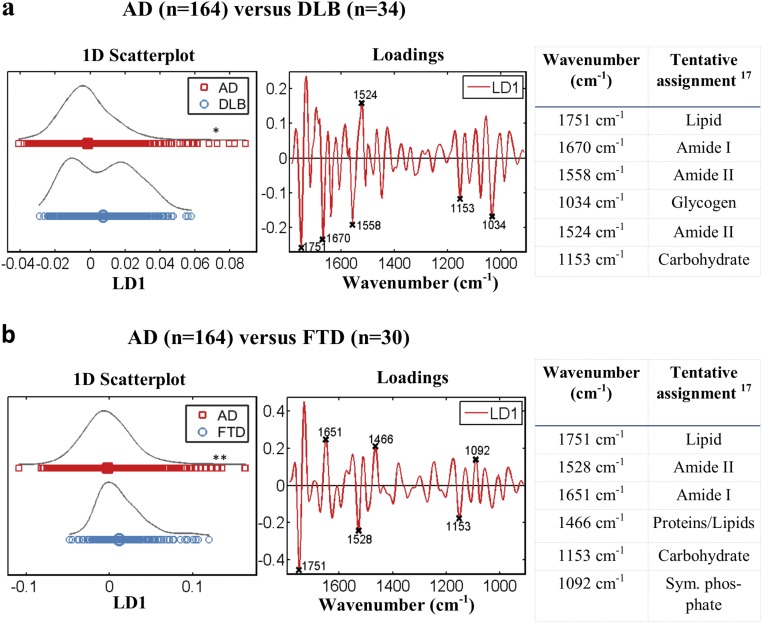
Diagnosis of other neurodegenerative diseases. (A) Scores and loading plots after cross-validated PCA-LDA for the diagnosis of DLB along with the six most discriminatory wavenumbers: 1,709 cm−1 (lipid), 1,666 cm−1 (Amide I), 1,555 cm−1 (Amide II), 1,501 cm−1 (Amide II), 1,076 cm−1 (symmetric stretching of PO2− of DNA/RNA), and 1,034 cm−1 (glycogen; P = 0.052, 95% CI = −0.0353, 0.0002). (B) Scores and loading plots after cross-validated PCA-LDA for the diagnosis of PD along with the six most discriminatory wavenumbers: 1,744 cm−1 (lipid), 1,705 cm−1 (lipid), 1,666 cm−1 (Amide I), 1,632 cm−1 (Amide I), 1,555 cm−1 (Amide II), and 1,408 cm−1 (–COO symmetric stretching of fatty/amino acids; P = 0.331, 95% CI = −0.0282, 0.0089). (C) Scores and loading plots after cross-validated PCA-LDA for the diagnosis of FTD along with the six most discriminatory wavenumbers: 1,732 cm−1 (lipid), 1,674 cm−1 (Amide I), 1,624 cm−1 (Amide I), 1,528 cm−1 (Amide II), 1,466 cm−1 (bending vibration of proteins/lipids), and 1,381 (–COO symmetric stretching of fatty/amino acids; P = 0.0041, 95% CI = −0.0264, −0.0049). (D) Scores and loading plots after cross-validated PCA-LDA for the diagnosis of PSP along with the six most discriminatory wavenumbers: 1,732 cm−1 (lipid), 1,636 cm−1 (Amide I), 1,531 cm−1 (Amide II), 1,497 cm−1 (Amide II), 1,396 cm−1 (–COO symmetric stretching of fatty/amino acids), and 1,076 cm−1 (symmetric stretching of PO2− of DNA/RNA); P < 0.0001, 95% CI = −0.0439, −0.0201). **P < 0.005; ***P < 0.0005.
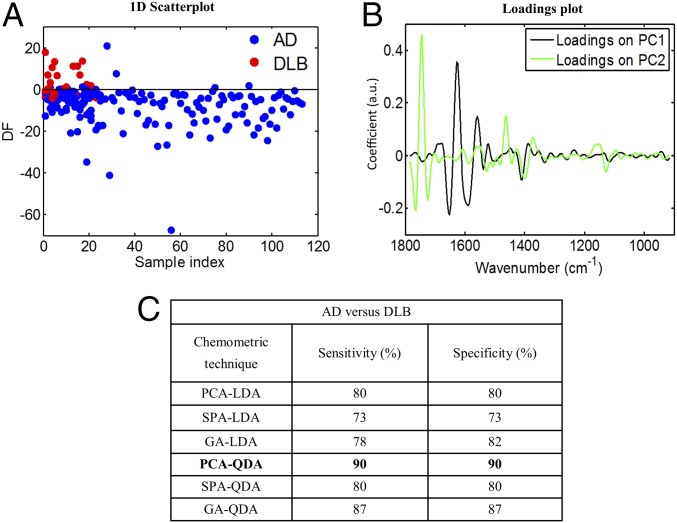
Of particular focus was the differentiation of AD and DLB. Chemometric techniques (PCA-LDA/PCA-QDA, SPA-LDA/SPA-QDA, and GA-LDA/GA-QDA) were applied, as previously, toward classification of AD vs. DLB (Fig. 8). The best classification ratios were achieved after PCA-QDA, giving sensitivity and specificity of 90%. Statistical analysis was also conducted after cross-validated PCA-LDA, and significant differences were found between the two groups (P = 0.178, 95% CI = 0.0012, 0.0157) (Fig. S10A). Finally, for the comparison of AD vs. FTD patients, significant differentiation was achieved after cross-validated PCA-LDA (P = 0.0040, 95% CI = 0.0041, 0.0216) (Fig. S10B).
Fig. 8.
AD (n = 164) vs. DLB (n = 34). (A and B) Scores plot, illustrating classification, and loadings plot, identifying the major discriminant wavenumbers, after PCA-QDA. (C) Sensitivity and specificity achieved for the segregation of AD from DLB; PCA-QDA achieved the best results, with 90% sensitivity and 90% specificity. DF, discriminant function.
Fig. S10.
AD vs. other types of dementia. (A) Scores and loading plots illustrating AD vs. DLB after cross-validated PCA-LDA along with the discriminatory wavenumbers and a table with their tentative assignments (P = 0.178, 95% CI = 0.0012, 0.0157). (B) Scores and loading plots illustrating AD vs. FTD after cross-validated PCA-LDA along with the discriminatory wavenumbers and a table with their tentative assignments (P = 0.0040, 95% CI = 0.0041, 0.0216). *P < 0.05; **P < 0.005.
Discussion
Using blood-based vibrational spectroscopy, we achieved results with significant clinical relevance at multiple levels. ATR-FTIR spectroscopy has been proven to be capable of detecting differences not only between patient and healthy groups but also, between various neurodegenerative dementias.
Overall, our results are in line with findings of previous studies, and most of the biomolecules that we identified, as contributors to the discrimination between AD and HC, have been previously described as potential biomarkers (10, 16, 23, 24). Generally, the IR region of 1,700–1,600 cm−1 is indicative of the protein secondary structure and could reflect protein conformational changes. A slight increase of the intensity at the Amide I region (1,650–1,630 cm−1; P = 0.1069, 95% CI = −0.005, 0.0005), observed in our AD patients, could presumably be caused by increased levels of Aβ plaques. In a recent study, using a more complex setting with a specialized, immuno-IR sensor, it was shown that the structural changes of Aβ peptide from α-helix to β-sheet, seen in AD, can be detectable by a significant downshift at the Amide I region (1,643 cm−1), and this further proves the importance of this region (13). Similar results were seen in an another study, which analyzed protein solutions containing soluble and aggregated forms of tau protein (random coil and β-sheet, respectively); in this case, a transition from a maximum peak at 1,650 cm−1 to a peak at 1,630 cm−1 was evident (25). However, in our study, statistically significant differences were not observed at this specific region. The increase at the Amide II region (1,540–1,530 cm−1; P = 0.003, 95% CI = −0.0031, −0.0007) could potentially be attributed to the overall accumulation of the above-mentioned proteins. The decrease in lipids (1,750–1,735 to 1,470–1,430 cm−1), comparing AD patients with HC, could be attributed to damaged cell membranes caused by free radicals; oxidative stress, implicated in the pathogenesis of AD, is the most likely reason for this increase in free radicals (26). The decrease seen at the DNA–RNA region could be explained by the loss of phosphoric acid and strand breaks also caused by hydroxyl radicals (27). The differences seen at the higher region could also occur because of disruption of phospholipid cell membranes (lower intensity at 2,950–2,850 cm−1) and free radicals (higher intensity at 3,550–3,450 cm−1). Multivariate approaches were further used to account for more detailed differences between the classes.
All of the six chemometric techniques used for classification between AD and HC identified differences that led to classification. The algorithms that achieved best classification were GA-LDA at the fingerprint region and PCA-QDA at the higher region. The sensitivity and specificity values found after the GA-LDA model at the fingerprint region (70%) were remarkable, considering that the spectral differences seen between AD and HC were very subtle. However, sensitivity and specificity at the higher region were slightly lower (68%), indicating the necessity of using the lower biofingerprint region toward the classification of AD samples rather than the higher region.
In a recent, large-scale study, which gathered information on 919 patients who had died and been autopsied, clinical diagnostic methods provided sensitivity ranging from 70.9 to 87.3% and specificity ranging from 44.3 to 70.8% (28). Having said that, an earlier study using 228 postmortem samples from the Manchester brain bank showed an overall agreement of 97% between clinical and pathological diagnoses of various neurodegenerative diseases. Clinical diagnosis of AD, specifically, was made with 97% sensitivity (four patients with pathologically confirmed AD had a different clinical diagnosis) and 100% specificity (a clinical diagnosis of AD was never wrong) (29). With regards to the subjects that we used for the specific study, it should be noted that, of the 2,450 samples so far accrued, 60 brains have been recovered; 14 thereof had a clinical diagnosis of AD, which was confirmed by pathological findings in 13 of them, whereas the other patient actually had corticobasal degeneration (CBD). Incidentally, there were four patients who had a clinical diagnosis of frontotemporal lobar degeneration—three with semantic dementia, one with FTD—and were identified as AD based on histopathology. However, none of these patient samples were available for a subgroup analysis in our spectroscopic study.
The performance of our chemometric analyses showed excellent agreement with these previous results. Although all patients with AD fulfilled relevant diagnostic criteria, it is well-known that there is clinical heterogeneity, with some patients showing disproportionate impairment of language, memory, frontal behavioral changes or orientation, and praxis. These may reflect topographic changes in the distribution of brain pathology in the early stages of the disease, partially in line with APOE genotype (30), and might influence the outcomes of the spectral analysis. Having said that, even if AD patients did show clinical uniformity overall, they could still have different phenotypes because of epigenetic alterations and/or environmental exposure that could influence the result (28, 31). Therefore, a clinical diagnosis would never reach 100% accuracy but rather, ∼80–90%. At the same time, some of the HC participants will inevitably have preclinical AD, since they are more or less age-matched to this group, also leading to rates lower than 100%. Unfortunately, preclinical diagnosis of these diseases is problematic, because there is, as yet, no really robust system to test for “prodromal” disease cases. Another possible explanation for the obtained classification values in this study could be the inclusion of all AD patients in one group without additional classification to different stages (mild, moderate, severe), as these were not fully recorded; this was also shown to adversely affect the results in a recent study, when AD patients of all stages were included in the same group (32). Nevertheless, in our AD dataset, we had 14 cases of early AD, and thus, we were able to conduct a subgroup analysis comparing them with the healthy subjects. The achieved sensitivity and specificity were 80 and 74%, respectively, implying that this spectroscopic technique may serve as a diagnostic tool for early AD as well; however, results should be interpreted with some caution because of the small number of early AD cases. In general, very high sensitivities/specificities should not be expected anyway, as the changes in the biochemical composition of blood plasma are relatively minor, compared with changes in CSF, for example, and do not necessarily reflect all possible alterations happening in the brain during AD progression (24).
APOE was also investigated, as it is a major risk factor for AD; homozygous ε4 individuals develop the disease 10–20 y earlier than the others (33), while APOE ε3 is considered “neutral,” and APOE ε2 is believed to serve a protective role (34). Furthermore, one ε4 allele increases the risk threefold, while two ε4 alleles increase the risk eightfold compared with the situation with no ε4 alleles. Different APOE genotypes would also cause differences in lipid composition and by extension, spectral information, and they could potentially “cloud” the issue when looking for a diagnostic. Therefore, after the incorporation of APOE ε4 information, the achieved classification results were noticeably higher, with sensitivity and specificity reaching 72 and 77% when the subjects had no APOE ε4 alleles as well as 86 and 86% when the subjects had one or two APOE ε4 alleles. After classifying AD and HC by age, lower values were achieved, suggesting that integration of APOE genotype is more important toward AD diagnosis. This could further support our aforementioned hypothesis, according to which differences in lipid deposition, caused by the presence of different APOE genotypes, could hide important spectral information; this would justify the higher classification values after classifying by genotype but not by age. In conclusion, these values for sensitivity/specificity, after consideration of APOE genotype information and along with the ease of sample collection and preparation, should ultimately allow the implementation of this test into a clinical setting.
From our analysis on AD duration, we found evidence of alterations being detectable, which correlate with duration. The increasing trend seen at 1,547 cm−1 is indicative of proteins predominantly in β-sheet conformation [Amide II, N-H bending, and C-N stretching (17)] and could potentially be attributed to an increased level of Aβ or more probably, tau proteins. One might presume that, by the time of clinical diagnosis, Aβ levels may well have peaked according to the amyloid cascade hypothesis, whereas tau pathology should still increase, spreading from temporal lobes into other neocortical regions. However, a decreasing trend is seen at 1,504 cm−1, which is again indicative of Amide II (proteins), but this time coming mainly from C-H bending vibrations of the proteins’ phenyl rings (17); this may be because of damage caused by reactive oxygen species (ROS) during oxidative stress (26), as phenyl rings are readily damaged by ROS. These peaks could provide information on progression and pathophysiology of AD, and a combined ratio with other important wavenumbers may serve as a potential biomarker. A decreasing trend is also seen at 1,369, 1,219, and 1,080 cm−1, which are assigned to cytosine/guanine and asymmetric and symmetric vibrations of phosphate of nucleic acids, respectively (17). We speculate that this general decrease in nucleic acids is caused by the increased levels of ROS that could lead to strand breaks and loss of phosphoric acid (27). In a relatively recent study, an elevated level of a specific type of collagen in the brain (collagen VI) was suggested to protect individuals against AD (35). The same study also pointed out that neurons of the brain were the source of this collagen VI. Therefore, this made us hypothesize that the progressive degradation of nerve cells coming with disease duration could potentially undermine their ability to produce collagen and that may explain the statistically significant decrease that we observe in individuals after 1 y of AD (i.e., peak at 1,034 cm−1).
After classification of the AD group into different age groups, the differences were not significant, suggesting that age difference alone is inadequate for discrimination when an individual has already developed the disease. Moreover, after calculation of different ratios of important spectral bands, potential biomarkers were established, and a classification process was facilitated by the generation of distinct patterns. Significant differences were seen after comparison of different age groups of HC and AD, and a combined ratio may serve as a more reliable biomarker.
In this study, we also incorporated a range of different neurodegenerative diseases, apart from AD, and achieved satisfactory segregation and classification results. The distinctive patterns seen between AD, DLB, and FTD may represent different pathological changes (36) and were mostly attributed to proteins and lipids. Accurate separation of DLB from AD is of high importance, as it would allow for appropriate medication/management and therefore, impact on outcomes; for instance, it is established that cholinesterase inhibitor treatment is more effective in DLB than AD patients (37). The high sensitivity and specificity (90%) achieved for distinguishing AD from DLB are outstanding and would provide an excellent diagnostic test toward clinical implementation. By reduction of misdiagnosed DLB cases and administration of appropriate drugs, many individuals would benefit by getting the appropriate clinical management. Potential biomarkers have also been suggested toward the diagnoses of DLB, PD, FTD, and PSP. It should be noted that PD and DLB did not show significant segregation from each other, which suggests common characteristics; the presence of Lewy bodies (i.e., α-synuclein), which is a hallmark of both diseases, is the main culprit for this similarity (38, 39).
A strength of this study is the large patient cohort, especially in the AD and HC groups, which to our knowledge, has not been previously repeated with similar techniques. However, some of the other diseases [motor neuron disease (MND), vascular dementia, etc.] are less common than AD; thus, the number of patients was limited. Additional exploration of these diseases using larger cohorts is required. Also, for prospective recruitment, patient characteristics should be recorded in more detail along with the stage of severity—mild (early stage), moderate (middle stage), and severe (late stage)—whenever possible.
Currently, there is no single definitive medical test for diagnosing AD. In terms of biological markers, the most widely studied molecules are undoubtedly Aβ and P-tau or T-tau. However, numerous studies attempting to diagnose AD using blood have shown controversial results (40). Specifically, plasma Aβ-42 and Aβ-40 levels have been seen to increase or decrease in AD patients compared with controls. Plasma total Aβ and Aβ-42 levels have shown inconsistent changes depending on whether an individual presented with familial or sporadic AD. Increase of Aβ-42 has been reported in women with mild cognitive impairment (MCI) but not in men. Other studies in plasma have found no differences at all between AD and controls; ratios of Aβ-42/Aβ-40 have also been conflicting, as some found that an elevated ratio predicts AD, while others found the opposite or no difference at all (41, 42). Several reasons have been implicated for these findings, such as the shift of Aβ levels over time and among individuals or the different stages of AD (42); this could potentially justify the nonsignificant increase (P = 0.1069, 95% CI = −0.005, 0.0005) that we observed at the Amide I region (1,650–1,630 cm−1) comparing AD patients with HCs. A recent metaanalysis of 231 studies concluded that plasma Aβ-42 and Aβ-40 were not strongly associated with AD and thus, should not be used in clinical practice (43).
Levels of tau protein in blood have also been investigated with the hope of diagnosing AD. A recent study reported elevated T-tau levels in AD patients compared with MCI and controls but found significant overlap between MCI and AD as well as no correlation between tau levels in CSF and plasma (44). Another study reported increased level of T-tau protein in AD, which when combined with the levels of Aβ-42, achieved improved sensitivities and specificities (80 and 82%, respectively) in differentiation of MCI from AD (45). The above-mentioned metaanalysis (43) also concluded that increased plasma level of T-tau was the only blood biomarker that discriminated AD from controls; however, it was pointed out that this finding would need additional verification in larger cohorts. Overall, there is still lack of optimal and reliable blood-based biomarkers that would allow screening of preclinical AD in a clinical setting (46). Most of the developed tests are for research use only, and this emphasizes the need to move on to new analytical approaches.
The available drugs for AD can only temporarily alleviate symptoms and do not treat the disease. The so far unsuccessful clinical trials, focusing on the reduction of Aβ and tau levels, suggest that there are more contributing factors/biomolecules to be explored. Biospectroscopy investigates a range of different biomolecules simultaneously, which could be advantageous in multifactorial diseases, like AD. In this study, we have achieved diagnosis of AD with an accuracy of ∼70–85% as well as differential diagnosis of AD and DLB with 90% sensitivity and specificity, which are outstanding considering the several overlapping features.
At the same time, biospectroscopy is rapid, cost-effective, and less laborious than other blood tests that require expensive antibodies/assays. For instance, a typical ELISA (96-well plate) has been calculated to cost approximately $1,000, with three kits needed to quantify three core biomarkers of AD disease: Aβ, T-tau, and P-tau (42, 43). Finally, this spectroscopic method could be used to further investigate individuals in combination with other clinically used neuropsychological tests and thus, help the clinician to decide whether to proceed to more specialized and expensive tests.
In summary, we have shown excellent diagnostic performance of our method for AD—especially after the APOE ε4 information–as well as for other neurodegenerative diseases. Our data suggest specific “fingerprint” regions capable of discriminating between pathological and healthy individuals as well as differentiating among the different types of dementia. Early AD cases were also segregated from HC with a high degree of sensitivity and specificity. Here, we would like to emphasize that, although we acknowledge the importance of diagnosing AD at an early stage, we also believe that there are some hurdles to overcome in diagnosing AD at any stage. Blood spectroscopy, as described in this study, would provide an alternative to current blood, CSF, or MRI tests. A prospective study, recruiting a larger number of individuals in healthy or predementia (MCI) state, would be required to confirm spectroscopy’s ability to perform as a screening test. By monitoring the latter individuals’ progression throughout the years, it would be possible to detect preclinical AD and establish early biomarkers implying the appearance of the disease. The ultimate aim would be to identify individuals destined to develop AD long before the symptoms occur and cease—or at least slow down—brain damage before it is too late.
Materials and Methods
Patients and Ethics.
Blood samples were collected at Salford Royal Hospital, with informed consent obtained before enrolment in accordance with Local Ethical Approval (05/Q1405/24 conferred by North West 10 Research Ethics Committee Greater Manchester North). This study included 549 individuals with various neurodegenerative diseases as well as age-matched healthy individuals, usually spouse controls. Patients with dementia were diagnosed according to a battery of psychological testing (Manchester Neuropsychology Inventory) performed at a specialist referral center (Cerebral Function Unit at the Greater Manchester Neurosciences Centre, Salford Royal Hospital). Patients with movement disorder but without apparent cognitive impairment were assessed in a specialist movement disorder clinic at Salford Royal Hospital, and diagnosis of PD, PSP, multiple system atrophy (MSA), or CBD was ascribed by consultant neurologists with expertise in this area. Also, patients with MND were assessed and diagnosed in Manchester MND Care Centre at Salford Royal Hospital. At time of diagnosis, patients were not receiving any medications, such as anticholinesterase treatments, which might affect the outcomes. Most patients receive MRI scans, but these were used only to support the neuropsychological outcomes.
The patient cohort was subgrouped in different classes depending on the type of neurodegenerative pathology; 164 patients with AD, 34 patients with DLB, 32 patients with PD, and a separate group named other containing 117 other less common neurodegenerative diseases. The other group consisted of 10 patients with CBD, 30 with FTD, 4 with MND, 5 with MSA, 20 with progressive nonfluent aphasia (PNFA), 31 with PSP, 14 with semantic dementia, and 3 patients with vascular dementia. The HC group was composed of 202 individuals who were recruited with informed consent as above. The information on clinical and demographic data is summarized in Table S1. A power test, using power of 80% (t test, confidence level of 95%) based on the training spectra set, indicated a number of 309 samples to be used for the comparison of AD with HC and 135 samples to be used for the comparison of AD with DLB. However, we have used 366 samples for AD vs. HC (power = 86%) and 198 samples for AD vs. DLB (power = 92%), covering larger data variability.
Table S1.
Clinical and demographic data for patient cohort
| Group (n) | Mean age (±SD, range) | APOE genotype | Gender | ||||||||
| ε2/2 | ε2/3 | ε2/4 | ε3/3 | ε3/4 | ε4/4 | n/a | Male | Female | n/a | ||
| AD (n = 164) | 68.61 (±8.61, 50–88) | — | 7 | 2 | 64 | 61 | 25 | 5 | 85 | 78 | 1 |
| DLB (n = 34) | 75.40 (±6.81, 63–88) | — | 4 | 3 | 13 | 11 | 3 | — | 27 | 5 | 2 |
| PD (n = 32) | 70.91 (±8.98, 50–81) | — | 1 | 1 | 20 | 10 | — | — | 19 | 13 | — |
| CBD (n = 10) | 75.3 (±6.60, 62–84) | — | 1 | — | 6 | 2 | 1 | — | 5 | 5 | — |
| FTD (n = 30) | 64.87 (±10.32, 42–78) | — | 5 | 1 | 15 | 8 | 1 | — | 18 | 12 | — |
| MND (n = 4) | 66.75 (±17.37, 47–85) | — | 1 | — | 2 | 1 | — | — | 1 | 3 | — |
| MSA (n = 5) | 64 (±11.79, 51–79) | — | — | — | 3 | 1 | 1 | — | 3 | 2 | — |
| PNFA (n = 20) | 72.75 (±8.52, 53–85) | — | 4 | 1 | 10 | 4 | — | 1 | 9 | 11 | — |
| PSP (n = 31) | 73.13 (±6.32, 60–85) | 1 | 2 | 1 | 20 | 5 | 1 | 1 | 21 | 10 | — |
| Semantic dementia (n = 14) | 68.43 (±6.71, 57–83) | — | 3 | — | 8 | 3 | — | — | 9 | 5 | — |
| Vascular dementia (n = 3) | 76.00 (±5.29, 72–84) | — | 1 | — | 1 | 1 | — | — | 3 | — | — |
| HC (n = 202) | 65.43 (±12, 23–90) | 2 | 20 | 10 | 120 | 37 | 2 | 11 | 75 | 124 | 3 |
n/a, not available.
Sample Collection and Preparation.
Whole-blood samples were collected into EDTA tubes and centrifuged at 1,130 × g at 4 °C for 10 min to separate erythrocytes from plasma. Plasma was collected in 0.5-mL clean plastic tubes, stored at −80 °C, and thawed at room temperature when necessary for experiments. After frozen samples were thawed, 50 μL from each were deposited on IR-reflective glass slides (MirrIR Low-E slides; Kevley Technologies) and left to air dry for ≈30 min before spectroscopic interrogation.
APOE Genotyping.
DNA was extracted by routine methods from blood samples of patients and control subjects; APOE alleles were determined by PCR (47). AD patients and HC individuals were classified for comparison into different categories depending on their APOE genotype (Table S1). Because of the established importance of APOE ε4 alleles in AD, we categorized our cohort into different groups to establish whether better classification is achieved. Both AD and HC individuals were categorized into two groups: one with no APOE ε4 alleles and another with one or two APOE ε4 alleles. AD patients with no APOE ε4 alleles (n = 71) were compared with HCs with no APOE ε4 alleles (n = 142), and similarly, patients with one or two APOE ε4 alleles (n = 88) were compared with HCs with one or two APOE ε4 alleles (n = 49).
ATR-FTIR Spectroscopy.
Spectra were obtained using a Tensor 27 FTIR spectrometer with Helios ATR attachment (Bruker Optics Ltd) operated by OPUS 5.5 software. The sampling area, defined by the internal reflection element, which was a diamond crystal, was ≈250 × 250 μm. Spectral resolution was 8 cm−1 with two times zero-filling, giving a data spacing of 4 cm−1 over the range 4,000–400 cm−1. Spectra were acquired from 10 different locations of each sample to minimize bias. The diamond crystal was cleaned with distilled water and dried every time before moving to the next sample; a background spectrum was also taken after the analysis of each sample to account for changes in ambient conditions.
Preprocessing of Spectra.
An in-house–developed IRootLab toolbox (trevisanj.github.io/irootlab/) was implemented within MATLAB R2012b software (MathWorks) for both preprocessing and computational analysis of the data. Preprocessing corrects problems associated with spectral data acquisition and increases the robustness of subsequent multivariate analyses. Spectra were preprocessed by two different methods and at two different regions of the mid-IR spectrum; the first region examined was the biochemical “fingerprint region” at 1,800–900 cm−1, and the second was a higher region at 3,700–2,800 cm−1, which is representative of –OH levels and lipid concentration. The first preprocessing method used rubber-band baseline correction followed by vector normalization and was included for better visualization. However, the second method, using Savitzky–Golay second-order differentiation (second-order polynomial and nine filter coefficients) and vector normalization, was the main approach used before additional multivariate analysis and classification. The second derivative was preferable as a preprocessing step, as it allows overlapping peaks to be resolved and accounts for differences in more detail (48).
The unsophisticated approach of DBM spectra was initially implemented to subtract the mean spectra of two different classes and account for differences; a peak-detecting algorithm was used to identify the most prominent peaks responsible for the segregation (49).
Computational Analysis and Classification.
Computational analysis of all spectra was performed using MATLAB with IRootLab and PLS Toolbox 7.9.3 (Eigenvector Research, Inc.). The multivariate analysis techniques used in this study were composed of PCA-LDA/PCA-QDA, SPA-LDA/SPA-QDA, and GA-LDA/GA-QDA.
PCA is an unsupervised technique that reduces the spectral data space to principal components (PCs) responsible for the majority of variance in the original dataset. This technique looks for inherent similarities/differences and provides a scores matrix representing the overall “identity” of each sample, a loadings matrix representing the spectral profile in each PC, and a residual matrix containing the unexplained data. The scores information can be used for exploratory analysis, providing possible classification between data classes. However, sometimes, PCA alone may not be enough, as it cannot distinguish whether the differences come from “within-class” or “between-class” variability (48, 50). For this reason, LDA, which is a supervised technique, is often applied after PCA to maximize the distance between the classes and minimize the distance within a class; consequently, PCA-LDA allows for better segregation.
For this study, the ∼230 variables (after the preprocessing) were reduced into 10 PCs that represented >95% of the variance. After mean-centering of the data, cross-validated PCA-LDA was performed to prevent overfitting of the data [with k-fold (k = 10) and leave-one-out validation]. After cross-validated PCA-LDA, information was visualized from 1D scores plots (using LD1 or LD2), while identification of specific wavenumbers accounting for separation was achieved from loading plots.
QDA is another supervised technique of discriminant analysis based, just like LDA, on the Mahalanobis distance between objects of classes. The main differences between the LDA and QDA are in the calculation of the classification score, in which QDA presents a more sophisticated approach accounting for different variance structures in the classes being analyzed. The LDA technique assumes that the classes have similar variance matrices, whereas QDA forms a separated variance model for each class (51). LDA and QDA classification scores are calculated as follows:
| [1] |
| [2] |
where is a measurement vector for sample , is the mean measurement vector of class , is the pooled covariance matrix, is the variance–covariance matrix, and is the prior probability of class (52).
SPA and GA are variable selection techniques used to reduce the spectral data space into few variables. SPA works by minimizing the variable collinearity (53), and GA works by simulating the data throughout an evolutionary process (54). The original space is maintained for both algorithms, and no transformation is made as in PCA. Therefore, the selected variables have the same meaning of the original ones (i.e., wavenumbers), and they are responsible for the region where there are more differences between the classes being analyzed or in other words, between the chemical changes.
For all classification models, samples were divided into training (70%), validation (15%), and prediction (15%) sets by applying Kennard–Stone uniform sampling selection algorithm (55). The training samples were used in the modeling procedure, whereas the prediction set was only used in the final classification evaluation using LDA and QDA discriminant approaches. The optimal number of variables for SPA and GA was determined with an average risk G of LDA/QDA misclassification. Such cost function is calculated in the validation set as
| [3] |
where is defined as
| [4] |
The numerator is the squared Mahalanobis distance between the object and the sample mean of its true class, and the denominator is the squared Mahalanobis distance between the object and the mean of the closest wrong class (56).
The GA calculations were performed during 80 generations with 160 chromosomes each. One-point crossover and mutation probabilities were set to 60 and 10%, respectively. GA is a nondeterministic algorithm, which can give different results by running the same equation/model. Therefore, the algorithm was repeated three times starting from random initial populations, with the best solution resulting from the three realizations of GA used.
Sensitivity (probability that a test result will be positive when disease is present) and specificity (probability that a test result will be negative when disease is not present) were given by the following equations:
| [5] |
| [6] |
where TP is defined as true positive, FN is defined as false negative, TN is defined as true negative, and FP is defined as false positive.
Data Sharing.
All data (raw spectra and preprocessed spectra) along with appropriate code identifiers will be uploaded onto the publicly accessible data repository Figshare.
Statistical Analysis.
Differences between two groups were assessed using a Student’s t test (two-tailed, nonparametric, Mann–Whitney test, 95% CI) in GraphPad Prism 6.0 (GraphPad Software). Statistical analysis was implemented at specific peaks pointed out after DBM spectra and cross-validated PCA-LDA as well as at scores plots as a whole. Before importing all of the information into GraphPad for additional statistical analysis, spectra were averaged every 10 (as 10 spectra were collected per individual) to account for differences between individuals rather than spectra. For the statistical analysis of three or more groups, one-way ANOVA was applied (nonparametric, Kruskal–Wallis test with a Dunn’s multiple comparison posttest). The data were expressed as the mean ± SD. A P value of 0.05 or less was considered significant in all statistical tests. Statistically significant differences were indicated with asterisks: *P < 0.05, **P < 0.005, ***P < 0.0005, or ****P < 0.00005.
Acknowledgments
We thank Mark Taylor, Kurimun Ismail, Yvonne Davidson, and Andrew Robinson for providing patient and sample information; Holly Butler for MATLAB tutorial; and Ketan Gajjar and Diane Halliwell for providing useful insights. M.P. was supported by the Rosemere Cancer Foundation. C.L.M.M. acknowledges Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Programa de Pós-Graduação em Química (PPGQ), and Universidade Federal do Rio Grande do Norte (UFRN) for the research grant. K.M.G.L. was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Grant 305962/2014-0.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data (raw spectra and preprocessed spectra) reported in this paper are available at the publicly accessible data repository Figshare (doi:10.6084/m9.figshare.5309797).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701517114/-/DCSupplemental.
References
- 1.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 2.Binder LI, Guillozet-Bongaarts AL, Garcia-Sierra F, Berry RW. Tau, tangles, and Alzheimer’s disease. Biochim Biophys Acta. 2005;1739:216–223. doi: 10.1016/j.bbadis.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Su B, et al. Oxidative stress signaling in Alzheimer’s disease. Curr Alzheimer Res. 2008;5:525–532. doi: 10.2174/156720508786898451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eikelenboom P, Zhan S-S, van Gool WA, Allsop D. Inflammatory mechanisms in Alzheimer’s disease. Trends Pharmacol Sci. 1994;15:447–450. doi: 10.1016/0165-6147(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 5.Heneka MT, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Paolo G, Kim T-W. Linking lipids to Alzheimer’s disease: Cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puglielli L, Tanzi RE, Kovacs DM. Alzheimer’s disease: The cholesterol connection. Nat Neurosci. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Q, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peila R, Rodriguez BL, Launer LJ. Honolulu-Asia Aging Study Type 2 diabetes, APOE gene, and the risk for dementia and related pathologies: The Honolulu-Asia Aging Study. Diabetes. 2002;51:1256–1262. doi: 10.2337/diabetes.51.4.1256. [DOI] [PubMed] [Google Scholar]
- 10.Mapstone M, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foy CML, et al. Diagnosing Alzheimer’s disease–non-clinicians and computerised algorithms together are as accurate as the best clinical practice. Int J Geriatr Psychiatry. 2007;22:1154–1163. doi: 10.1002/gps.1810. [DOI] [PubMed] [Google Scholar]
- 12.Hye A, et al. Proteome-based plasma biomarkers for Alzheimer’s disease. Brain. 2006;129:3042–3050. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- 13.Nabers A, et al. Amyloid-β-secondary structure distribution in cerebrospinal fluid and blood measured by an immuno-infrared-sensor: A biomarker candidate for Alzheimer’s disease. Anal Chem. 2016;88:2755–2762. doi: 10.1021/acs.analchem.5b04286. [DOI] [PubMed] [Google Scholar]
- 14.Sperling RA, Jack CR, Jr, Aisen PS. Testing the right target and right drug at the right stage. Sci Transl Med. 2011;3:111cm33. doi: 10.1126/scitranslmed.3002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rye PD, et al. A novel blood test for the early detection of Alzheimer’s disease. J Alzheimers Dis. 2011;23:121–129. doi: 10.3233/JAD-2010-101521. [DOI] [PubMed] [Google Scholar]
- 16.Carmona P, et al. Discrimination analysis of blood plasma associated with Alzheimer’s disease using vibrational spectroscopy. J Alzheimers Dis. 2013;34:911–920. doi: 10.3233/JAD-122041. [DOI] [PubMed] [Google Scholar]
- 17.Movasaghi Z, Rehman S, Rehman IU. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl Spectrosc Rev. 2008;43:134–179. [Google Scholar]
- 18.Baker MJ, et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat Protoc. 2014;9:1771–1791. doi: 10.1038/nprot.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler HJ, et al. Using Raman spectroscopy to characterize biological materials. Nat Protoc. 2016;11:664–687. doi: 10.1038/nprot.2016.036. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell AL, Gajjar KB, Theophilou G, Martin FL, Martin-Hirsch PL. Vibrational spectroscopy of biofluids for disease screening or diagnosis: Translation from the laboratory to a clinical setting. J Biophotonics. 2014;7:153–165. doi: 10.1002/jbio.201400018. [DOI] [PubMed] [Google Scholar]
- 21.Baker MJ, et al. Developing and understanding biofluid vibrational spectroscopy: A critical review. Chem Soc Rev. 2016;45:1803–1818. doi: 10.1039/c5cs00585j. [DOI] [PubMed] [Google Scholar]
- 22.Vann Jones SA, O’Brien JT. The prevalence and incidence of dementia with Lewy bodies: A systematic review of population and clinical studies. Psychol Med. 2014;44:673–683. doi: 10.1017/S0033291713000494. [DOI] [PubMed] [Google Scholar]
- 23.Burns DH, et al. Near-infrared spectroscopy of blood plasma for diagnosis of sporadic Alzheimer’s disease. J Alzheimers Dis. 2009;17:391–397. doi: 10.3233/JAD-2009-1053. [DOI] [PubMed] [Google Scholar]
- 24.Ryzhikova E, et al. Raman spectroscopy of blood serum for Alzheimer’s disease diagnostics: Specificity relative to other types of dementia. J Biophotonics. 2015;8:584–596. doi: 10.1002/jbio.201400060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Bergen M, Barghorn S, Biernat J, Mandelkow E-M, Mandelkow E. Tau aggregation is driven by a transition from random coil to beta sheet structure. Biochim Biophys Acta. 2005;1739:158–166. doi: 10.1016/j.bbadis.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Moreira PI, et al. Oxidative stress: The old enemy in Alzheimer’s disease pathophysiology. Curr Alzheimer Res. 2005;2:403–408. doi: 10.2174/156720505774330537. [DOI] [PubMed] [Google Scholar]
- 27.Peuchant E, et al. Infrared spectroscopy: A reagent-free method to distinguish Alzheimer’s disease patients from normal-aging subjects. Transl Res. 2008;152:103–112. doi: 10.1016/j.trsl.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Beach TG, Monsell SE, Phillips LE, Kukull W. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005–2010. J Neuropathol Exp Neurol. 2012;71:266–273. doi: 10.1097/NEN.0b013e31824b211b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snowden JS, et al. The clinical diagnosis of early-onset dementias: Diagnostic accuracy and clinicopathological relationships. Brain. 2011;134:2478–2492. doi: 10.1093/brain/awr189. [DOI] [PubMed] [Google Scholar]
- 30.Snowden JS, et al. Cognitive phenotypes in Alzheimer’s disease and genetic risk. Cortex. 2007;43:835–845. doi: 10.1016/s0010-9452(08)70683-x. [DOI] [PubMed] [Google Scholar]
- 31.Paraskevaidi M, Martin-Hirsch PL, Kyrgiou M, Martin FL. Underlying role of mitochondrial mutagenesis in the pathogenesis of a disease and current approaches for translational research. Mutagenesis. 2017;32:335–342. doi: 10.1093/mutage/gew058. [DOI] [PubMed] [Google Scholar]
- 32.Mordechai S, Shufan E, Porat Katz BS, Salman A. Early diagnosis of Alzheimer’s disease using infrared spectroscopy of isolated blood samples followed by multivariate analyses. Analyst. 2017;142:1276–1284. doi: 10.1039/c6an01580h. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prasad KN, Cole WC, Prasad KC. Risk factors for Alzheimer’s disease: Role of multiple antioxidants, non-steroidal anti-inflammatory and cholinergic agents alone or in combination in prevention and treatment. J Am Coll Nutr. 2002;21:506–522. doi: 10.1080/07315724.2002.10719249. [DOI] [PubMed] [Google Scholar]
- 35.Cheng JS, et al. Collagen VI protects neurons against Abeta toxicity. Nat Neurosci. 2009;12:119–121. doi: 10.1038/nn.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noe E, et al. Comparison of dementia with Lewy bodies to Alzheimer’s disease and Parkinson’s disease with dementia. Mov Disord. 2004;19:60–67. doi: 10.1002/mds.10633. [DOI] [PubMed] [Google Scholar]
- 37.McKeith I. Dementia with Lewy bodies. Dialogues Clin Neurosci. 2004;6:333–341. doi: 10.31887/DCNS.2004.6.3/imckeith. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spillantini MG, et al. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 39.El-Agnaf OM, et al. α-Synuclein implicated in Parkinson’s disease is present in extracellular biological fluids, including human plasma. FASEB J. 2003;17:1945–1947. doi: 10.1096/fj.03-0098fje. [DOI] [PubMed] [Google Scholar]
- 40.Kua EH, et al. The natural history of dementia. Psychogeriatrics. 2014;14:196–201. doi: 10.1111/psyg.12053. [DOI] [PubMed] [Google Scholar]
- 41.Cedazo-Minguez A, Winblad B. Biomarkers for Alzheimer’s disease and other forms of dementia: Clinical needs, limitations and future aspects. Exp Gerontol. 2010;45:5–14. doi: 10.1016/j.exger.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Humpel C. Identifying and validating biomarkers for Alzheimer’s disease. Trends Biotechnol. 2011;29:26–32. doi: 10.1016/j.tibtech.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsson B, et al. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: A systematic review and meta-analysis. Lancet Neurol. 2016;15:673–684. doi: 10.1016/S1474-4422(16)00070-3. [DOI] [PubMed] [Google Scholar]
- 44.Zetterberg H, et al. Plasma tau levels in Alzheimer’s disease. Alzheimers Res Ther. 2013;5:9. doi: 10.1186/alzrt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu M-J, et al. Combined plasma biomarkers for diagnosing mild cognition impairment and Alzheimer’s disease. ACS Chem Neurosci. 2013;4:1530–1536. doi: 10.1021/cn400129p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubois B, et al. Proceedings of the Meeting of the International Working Group (IWG) and the American Alzheimer’s Association on “The Preclinical State of AD”; July 23, 2015; Washington DC, USA Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria. Alzheimers Dement. 2016;12:292–323. doi: 10.1016/j.jalz.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson Y, et al. Apolipoprotein E epsilon4 allele frequency in vascular dementia. Dement Geriatr Cogn Disord. 2006;22:15–19. doi: 10.1159/000092960. [DOI] [PubMed] [Google Scholar]
- 48.Trevisan J, Angelov PP, Carmichael PL, Scott AD, Martin FL. Extracting biological information with computational analysis of Fourier-transform infrared (FTIR) biospectroscopy datasets: Current practices to future perspectives. Analyst. 2012;137:3202–3215. doi: 10.1039/c2an16300d. [DOI] [PubMed] [Google Scholar]
- 49.Trevisan J, et al. Syrian hamster embryo (SHE) assay (pH 6.7) coupled with infrared spectroscopy and chemometrics towards toxicological assessment. Analyst. 2010;135:3266–3272. doi: 10.1039/c0an00586j. [DOI] [PubMed] [Google Scholar]
- 50.Martin FL, et al. Distinguishing cell types or populations based on the computational analysis of their infrared spectra. Nat Protoc. 2010;5:1748–1760. doi: 10.1038/nprot.2010.133. [DOI] [PubMed] [Google Scholar]
- 51.Dixon SJ, Brereton RG. Comparison of performance of five common classifiers represented as boundary methods: Euclidean distance to centroids, linear discriminant analysis, quadratic discriminant analysis, learning vector quantization and support vector machines, as dependent on data structure. Chemometr Intell Lab Syst. 2009;95:1–17. [Google Scholar]
- 52.Wu W, et al. Comparison of regularized discriminant analysis linear discriminant analysis and quadratic discriminant analysis applied to NIR data. Anal Chim Acta. 1996;329:257–265. [Google Scholar]
- 53.Araújo MCU, et al. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemometr Intell Lab Syst. 2001;57:65–73. [Google Scholar]
- 54.Næs T, Isaksson T, Fearn T, Davies T. A User Friendly Guide to Multivariate Calibration and Classification. NIR Publications; Chicester, UK: 2002. [Google Scholar]
- 55.Kennard RW, Stone LA. Computer aided design of experiments. Technometrics. 1969;11:137–148. [Google Scholar]
- 56.Lima KMG, et al. Classification of cervical cytology for human papilloma virus (HPV) infection using biospectroscopy and variable selection techniques. Anal Methods. 2014;6:9643–9652. [Google Scholar]