Significance
UV tanning is a common social behavior, which increases melanin production and pigmentation of the skin. UV irradiation is a standard treatment of depigmenting diseases such as vitiligo. However, recurrent UV irradiation is genotoxic and facilitates skin aging and cancer. Here, we identified a method of inducing hyperpigmentation by inhibition of casein kinase 1α (CK1α). UV tanning is induced through activation of p53, via the Pomc/α-MSH/Mc1r/Mitf pathway, but both Pomc and Mc1r function can be compromised by aging or allelic polymorphism. In contrast, inhibition of CK1α activates a different pathway, p53/KitL/Kit, and raises protective eumelanin without the procarcinogenic pheomelanin. Inhibition of CK1α is therefore expected to be an effective strategy for skin protection from sunlight and for treating depigmenting diseases.
Keywords: casein kinase 1α, p53, Kit ligand, sunburn, melanocyte
Abstract
Casein kinase 1α (CK1α), a component of the β-catenin destruction complex, is a critical regulator of Wnt signaling; its ablation induces both Wnt and p53 activation. To characterize the role of CK1α (encoded by Csnk1a1) in skin physiology, we crossed mice harboring floxed Csnk1a1 with mice expressing K14–Cre–ERT2 to generate mice in which tamoxifen induces the deletion of Csnk1a1 exclusively in keratinocytes [single-knockout (SKO) mice]. As expected, CK1α loss was accompanied by β-catenin and p53 stabilization, with the preferential induction of p53 target genes, but phenotypically most striking was hyperpigmentation of the skin, importantly without tumorigenesis, for at least 9 mo after Csnk1a1 ablation. The number of epidermal melanocytes and eumelanin levels were dramatically increased in SKO mice. To clarify the putative role of p53 in epidermal hyperpigmentation, we established K14–Cre–ERT2 CK1α/p53 double-knockout (DKO) mice and found that coablation failed to induce epidermal hyperpigmentation, demonstrating that it was p53-dependent. Transcriptome analysis of the epidermis revealed p53-dependent up-regulation of Kit ligand (KitL). SKO mice treated with ACK2 (a Kit-neutralizing antibody) or imatinib (a Kit inhibitor) abrogated the CK1α ablation-induced hyperpigmentation, demonstrating that it requires the KitL/Kit pathway. Pro-opiomelanocortin (POMC), a precursor of α-melanocyte–stimulating hormone (α-MSH), was not activated in the CK1α ablation-induced hyperpigmentation, which is in contrast to the mechanism of p53-dependent UV tanning. Nevertheless, acute sunburn effects were successfully prevented in the hyperpigmented skin of SKO mice. CK1α inhibition induces skin-protective eumelanin but no carcinogenic pheomelanin and may therefore constitute an effective strategy for safely increasing eumelanin via UV-independent pathways, protecting against acute sunburn.
The epidermis, which is mainly composed of keratinocytes and melanocytes, is a highly sophisticated barrier tissue that protects the body against continuous external injuries such as UV radiation (hereafter, “UV”). UV can injure the skin both by indirect cellular damage via the generation of reactive oxygen species and by direct damage to the nucleotide structure in DNA, thereby causing an acute sunburn reaction and the development of skin cancers. Keratinocytes are sensitive to UV and are the major responders in the skin. They produce various paracrine factors in response to UV, which influence their microenvironment and activate adjacent melanocytes, forming a keratinocyte–melanocyte functional unit (1–3). Such paracrine factors produced by keratinocytes include α-melanocyte–stimulating hormone (α-MSH), adrenocorticosteroid hormone (ACTH), endothelin-1 (Edn1), and Kit ligand (KitL, also known as “stem cell factor”) (3–9). Skin hyperpigmentation, resulting from increased melanocyte density and/or melanin production with melanin distribution to neighboring keratinocytes, is important for UV protection. Melanin acts as a natural sunscreen that directly protects against UV and visible light radiation penetration to deep skin layers where proliferating cells reside (10) as well as acting as a potent antioxidant and free-radical scavenger. Individuals with darker skin have a reduced incidence of UV-induced skin cancers, whereas individuals with lighter skin are more prone to UV-induced damage and tumor formation and have weak tanning responses (11).
Casein kinase 1α (CK1α), encoded by the Csnk1a1 gene, is a component of the β-catenin degradation complex and is a critical regulator of the Wnt signaling pathway (12–14). CK1α phosphorylates β-catenin at Ser45, which primes it for subsequent phosphorylation by GSK-3β. GSK-3β destabilizes β-catenin by phosphorylating it at Ser33, Ser37, and Thr41, marking β-catenin for ubiquitination by SCFβ-TrCP E3 and proteasomal degradation. This CK1α-dependent phosphorylation functions as a molecular switch for the Wnt pathway (15). A homozygous deficiency of CK1α results in embryonic lethality, suggesting a fundamental role for CK1α in embryogenesis. In a study of murine intestine epithelium, CK1α deficiency was found to induce Wnt activation and DNA-damage response with robust p53 activation and cellular senescence in many types of tissues, including tissue stem cells (14, 16, 17). These facts suggest that CK1α plays important roles in cellular processes in various tissues that are at least partly coordinated with p53. p53, a well-known tumor-suppressor protein, is a transcription factor that plays a pivotal role in cellular responses to genotoxic stress and DNA damage (18). In the skin, p53 also acts as a central player against UV damage via the p53/proopiomelanocortin (POMC)/α-MSH/melanocortin 1 receptor (MC1R)/microphthalmia-associated transcription factor (MITF) skin-tanning pathway and through the DNA repair/cell-cycle arrest/apoptotic pathway (4, 19). As CK1α ablation is a robust means of activating p53 in many tissues, the physiological role of CK1α in the skin remains to be elucidated.
In this study, we aimed to clarify the effects of deleting CK1α in keratinocytes on skin physiology. We crossed mice with floxed Csnk1a1 (14) with mice expressing K14–Cre–ERT2 to generate mice in which tamoxifen induces the deletion of Csnk1a1 exclusively in keratinocytes. Phenotypically, we found that ablation of CK1α results mainly in skin hyperpigmentation accompanied by the activation of p53 in keratinocytes. We also generated K14–Cre–ERT2–CK1α/p53 double-knockout (DKO) mice to further address the role of p53 in the skin under CK1α ablation. Our study demonstrates that skin hyperpigmentation resulting from ablation of CK1α involves the p53/KitL/Kit signaling pathway, which is different from the pathway involved in UV tanning. This hyperpigmentation route has an important advantage compared with UV radiation; it is free of UV-related DNA damage yet successfully protects the skin from the UV-induced sunburn reaction.
Results
Ablation of CK1α in Keratinocytes Induces Skin Hyperpigmentation.
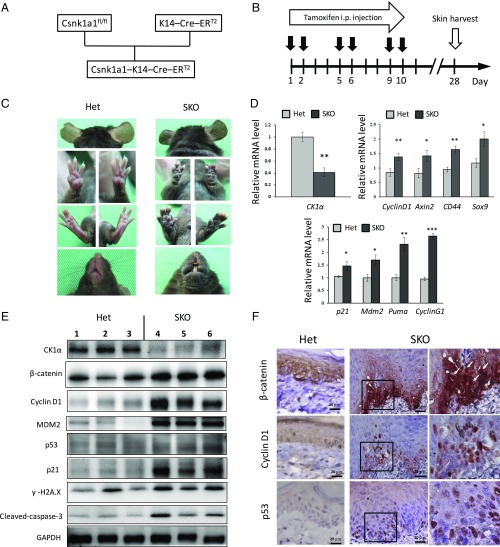
To characterize the role of CK1α in skin physiology, we developed Csnk1a1Δskin (CK1α-KO, hereafter “single-knockout,” “SKO”) mice by crossing mice in which Csnk1a1 was floxed with mice expressing K14–Cre–ERT2 (Fig. 1A). Expression of CK1α in keratinocytes was ablated by the i.p. injection of tamoxifen six times within 2 wk (Fig. 1B). Hyperpigmentation appeared on the ears, paws, tail, mouth, and trunk of SKO mice at 1 mo (Fig. 1C). CK1α ablation resulted in the stabilization and up-regulation of β-catenin (Fig. 1 E and F), which by itself may cause skin tumorigenesis (19–22), but we observed no skin tumor formation for at least 9 mo. Skin samples were then harvested, and mRNAs and proteins were extracted from the epidermis for analysis by RT-PCR and Western blot, respectively. The expression of CK1α mRNA was down-regulated in SKO mice compared with the control mice, as expected, and the mRNAs of Wnt-target genes (Cyclin D1, Axin 2, CD44, and Sox 9) were moderately up-regulated. At the same time, p53-target genes (p21, Mdm2, Puma, and Cyclin G1) were significantly up-regulated (Fig. 1D). Western blot analysis revealed that the level of CK1α protein was reduced in the epidermis of SKO mice compared with controls, which stabilized β-catenin and increased levels of Cyclin D1, p53, p21, and MDM2 (Fig. 1E). The CK1α ablation-induced DNA-damage response was demonstrated by increased γ-H2AX staining, and apoptosis was detected by cleaved-caspase 3 expression. Immunohistochemistry of the paw skin of SKO mice showed enhanced nuclear staining for β-catenin, cyclin D1, and p53 in the epidermis compared with the heterozygous control (Fig. 1F). This enhanced expression of protumorigenic Wnt-regulated proteins together with the tumor-suppressor p53-regulated proteins may explain the paucity of skin tumors in Csnk1a1Δskin, in contrast to the tumorigenic overexpression of stabilized β-catenin per se (20–23).
Fig. 1.
CK1α ablation in keratinocytes induces skin hyperpigmentation and activation of Wnt and p53 signaling pathways. (A) Scheme of SKO mice crossing. (B) CK1α is ablated in keratinocytes by i.p. tamoxifen injection. All samples were harvested at day 28. (C) SKO mice show skin hyperpigmentation of ears, paws, mouth, and trunk at 1 mo compared with heterozygous control (Het) mice. (D) RT-PCR analyses of the epidermis. Expression of CK1α mRNA is down-regulated in SKO mice. The mRNAs of Wnt-target genes and p53-target genes are up-regulated in SKO mice. *P < 0.05, **P < 0.01, and ***P < 0.001. (E) Western blot analyses of the epidermis. The reduced level of CK1α protein causes β-catenin stabilization and increases levels of cyclin D1, p53, p21, and Mdm2 in SKO mice. Increased levels of markers for DNA damage response (γ-H2AX) and apoptosis (cleaved-caspase 3) are also detected in SKO mice. (F) Immunohistochemistry of paw skin. Positive nuclear staining of β-catenin, Cyclin D1, and p53 in the basal layer of the epidermis of SKO mice. Panels in the Right column are 2.48× magnified on the square in the Middle column. (Scale bars, 20 μm.)
Ablation of CK1α in Keratinocytes Increases the Density of Epidermal Melanocytes and Eumelanin Deposits.
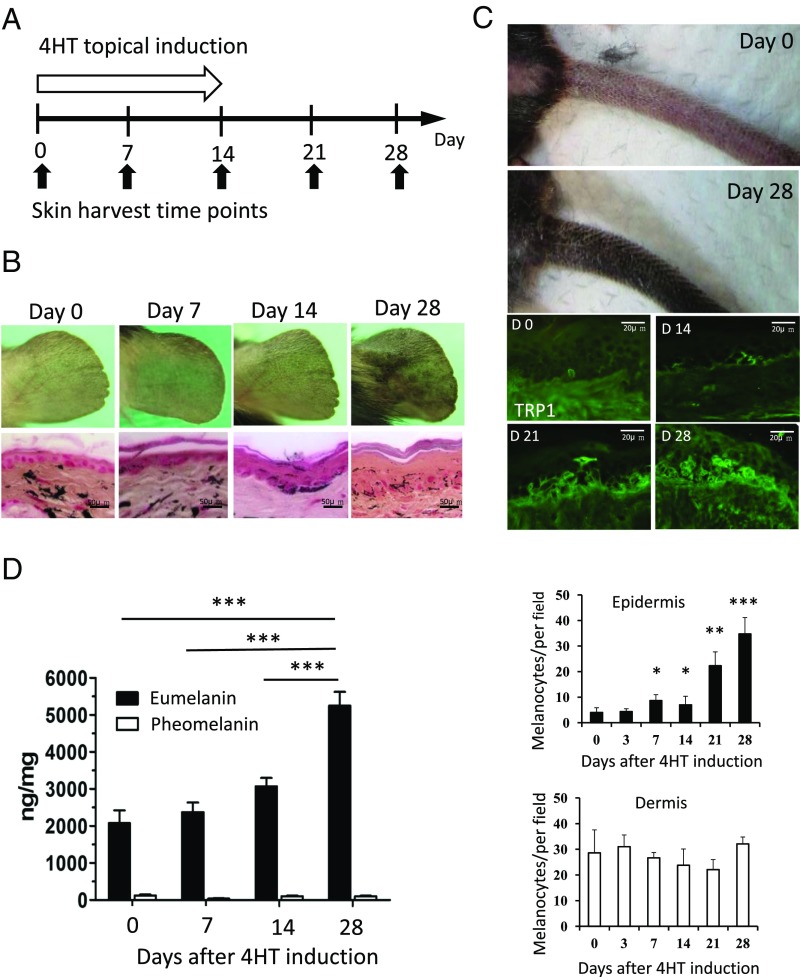
To investigate the mechanism of skin hyperpigmentation induced by the ablation of CK1α, we performed a time-course study of melanocyte density in the skin following topical 4-hydroxytamoxifen (4HT) induction on the ear and tail skin for 14 d (Fig. 2A). The resulting phenotypes were recorded using a digital camera and showed gradually increased pigmentation, especially after 3 wk. Ear and tail skin samples were harvested at different time points to produce paraffin-embedded and frozen sections. Fontana–Masson staining for melanin showed increased levels of melanin deposits in the epidermis of SKO mouse ears (Fig. 2B). Tyrosinase-related protein 1 (TRP1) staining to identify melanocytes demonstrated that the number of melanocytes in the SKO tail epidermis increased slowly during the first 14 d but increased rapidly thereafter. In contrast, the number of melanocytes in the dermis did not change significantly, implying that dermal melanocyte precursors were first activated and then migrated into the epidermis (Fig. 2C). Mice are generally considered to have a paucity of epidermal melanocytes in hair-bearing areas depending on the expression of KitL by basal keratinocytes. Ablation of CK1α in dorsal skin resulted in the maintenance of melanocytes in the interfollicular basal layer of epidermis with melanin deposition, compared with the controls (Fig. S1).
Fig. 2.
CK1α ablation in keratinocytes increased epidermal melanocyte number and eumelanin production. (A and B) Topical treatment with 4HT for 14 d induces skin hyperpigmentation on the ear. Fontana–Masson staining shows increased epidermal thickness and melanin content in the epidermis. (C) Topical treatment with 4HT for 14 d induces skin hyperpigmentation on the tail. TRP1 staining highlights the increased number of melanocytes in the tail epidermis. Quantitation of melanocyte staining shows that the number of dermal melanocytes was not significantly changed. (D) The amount of eumelanin is increased, but pheomelanin remains unchanged during CK1α inhibition. Data are shown as means ± SD; *P < 0.05, **P < 0.01, and ***P < 0.001.
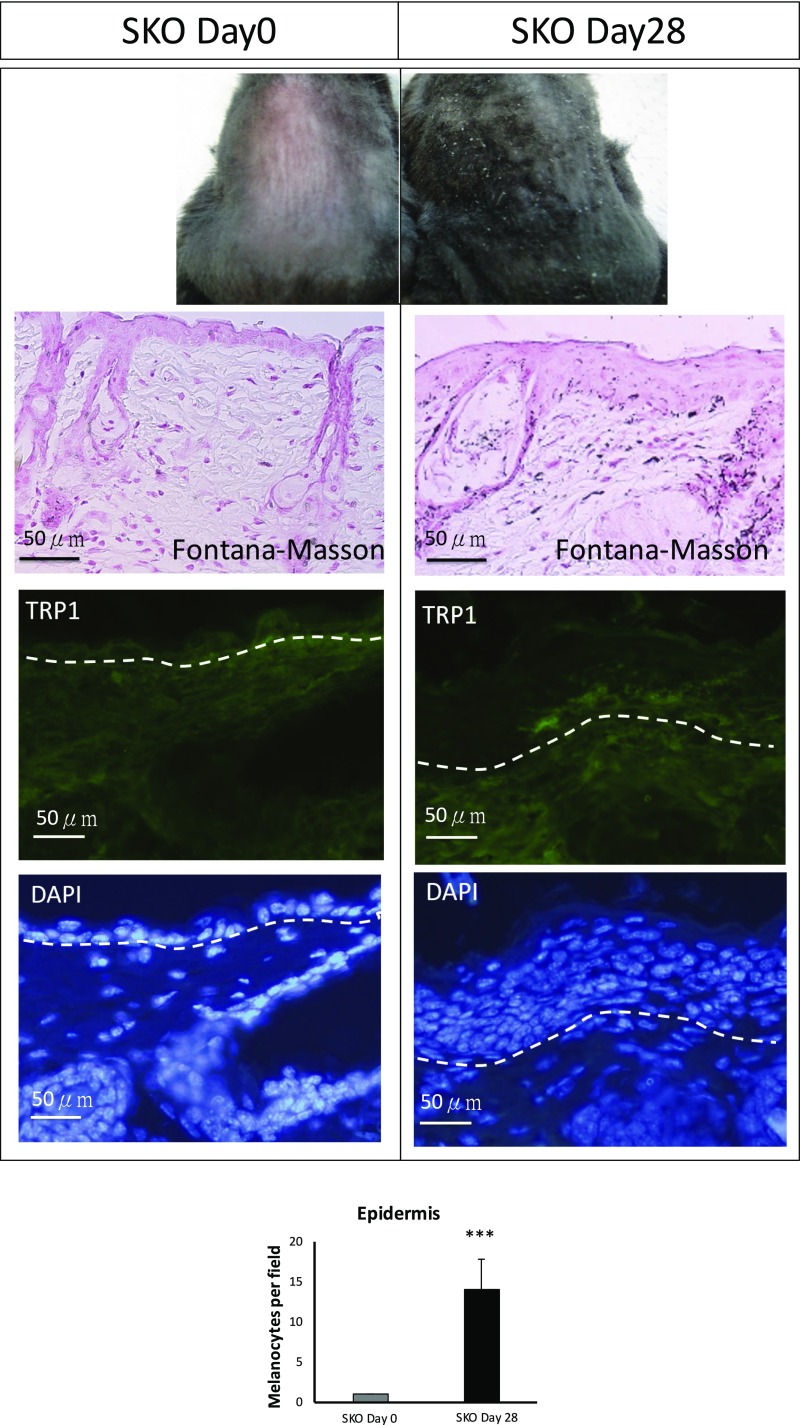
Fig. S1.
Phenotype examination and immunohistochemical staining of dorsal skin at days 0 and 28 after tamoxifen i.p. injection. SKO dorsal epidermis is hyperpigmented at day 28 compared with day 0. Melanin content and melanocyte density in the epidermis at day 28 are increased as assessed by Fontana–Masson staining and TRP1 staining. Data are shown as means ± SD; ***P < 0.001. (Scale bars, 50 μm.)
As CK1α depletion proceeded, melanocytes became larger and more dendritic, which indicates melanocytic differentiation (Fig. 2 B and C). Generally, melanocytes can produce two distinct types of melanin: brownish-black eumelanin and reddish-yellow pheomelanin. Two proteins, MC1R and α-MSH, encoded at the extension and Pomc1 loci, primarily control the relative amount of eumelanin and pheomelanin produced in melanocytes. To clarify the type of melanin that was increased by CK1α inhibition, we measured the amounts of both types of melanins in the epidermis and found that eumelanin was the main type of melanin increased by CK1α inhibition while pheomelanin remained at the same low level (Fig. 2D). The increase in the total amount of eumelanin may be mainly due to the expansion of the melanocyte population and may not be due to the melanin switch or increased synthesis of melanin per cell.
CK1α Ablation-Induced Skin Hyperpigmentation Is p53-Dependent.
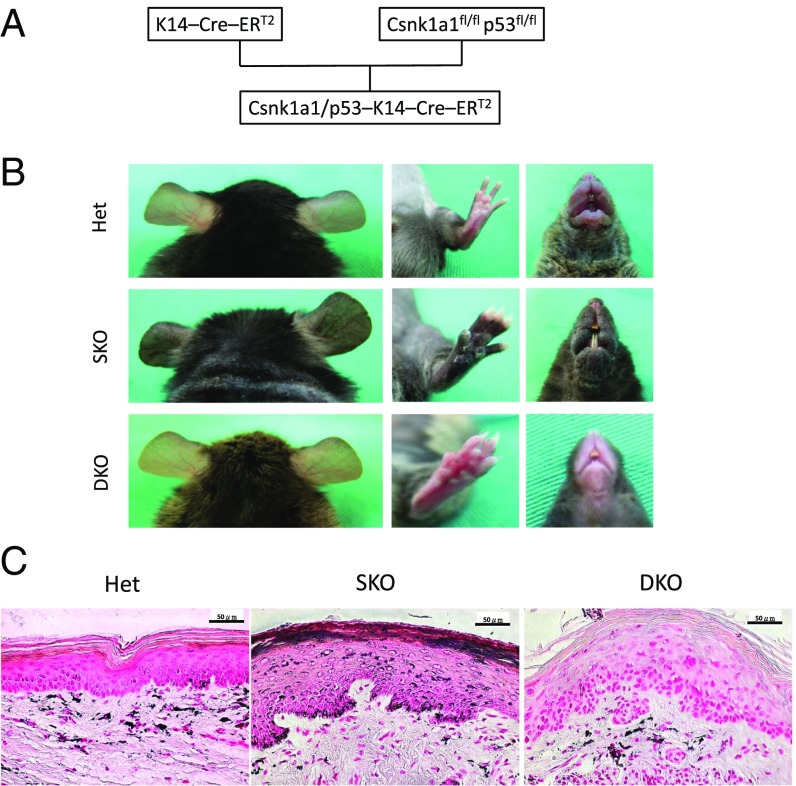
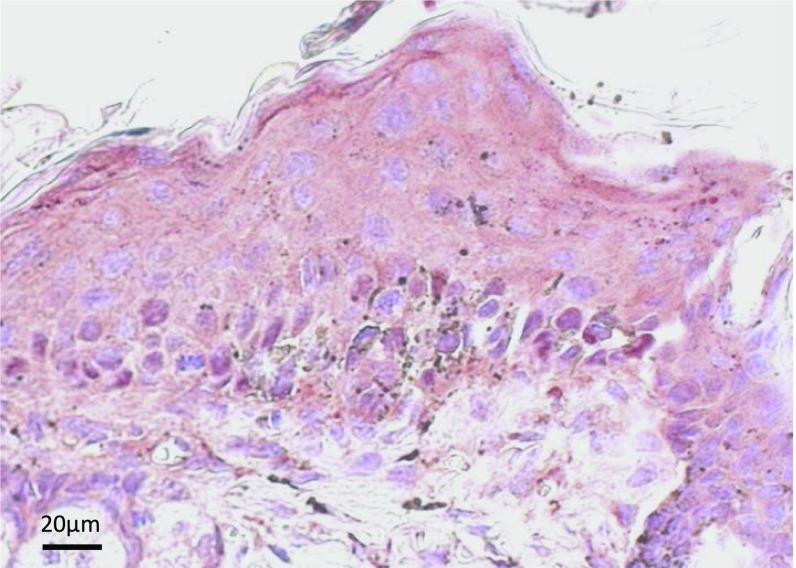
SKO mice developed hyperpigmented skin shortly after the ablation of CK1α in keratinocytes, while melanocytes should retain that gene. A previous study reported that UV-induced hyperpigmentation (UV tanning) is p53-dependent (4). As p53 is activated in keratinocytes following CK1α knockout in our mouse model (Fig. 1 D–F), we hypothesized that p53 transactivates one or more paracrine factor-related genes that influence melanocyte behavior. To test this hypothesis, we generated K14–Cre–ERT2–CK1α/p53 DKO mice and treated them i.p. with tamoxifen six times over 2 wk (Fig. 3A). Coablation of CK1α and p53 did not induce skin hyperpigmentation, indicating that the CK1α ablation-induced skin hyperpigmentation is p53-dependent (Fig. 3B). Fontana–Masson staining of paw skin sections demonstrated that in WT mice, melanin deposition was mainly in the dermis and less in the epidermis (Fig. 3C), whereas in SKO mice it was significantly increased in the epidermis. Remarkably, we observed epidermal hyperplasia in SKO (compared with heterozygous) mice and dysplasia in DKO mice, but melanin was mainly distributed in the dermis of DKO mice, similar to control heterozygous mice. In serial sections of SKO skin, increased numbers of melanocytes and melanin content matched the location of p53 expression (Fig. S2). Thus, histopathology further indicated that p53-regulated paracrine factors secreted by CK1α-ablated keratinocytes might have attracted dermal melanocytes to migrate into the epidermis and stimulate melanin deposits in the epidermis.
Fig. 3.
CK1α ablation in keratinocyte-induced skin hyperpigmentation is p53-dependent. (A) Scheme of the DKO mice crossing. (B) SKO mice develop skin hyperpigmentation, but DKO mice do not show skin hyperpigmentation. (C) Fontana–Masson staining of the paw skin. Melanin distribution in heterozygous control (Het) mice is mainly located in the dermis and less in the epidermis. In SKO mice, the epidermis becomes hyperplastic with increased melanin deposition. In DKO mice, the epidermis becomes dysplastic without melanin deposition. (Scale bars, 50 μm.)
Fig. S2.
Immunohistochemical staining for p53 of the epidermis in SKO mice 28 d after the initiation of tamoxifen i.p. treatment. Many dendritic melanocytes (indicated by black melanin pigment) are observed around the p53+ keratinocytes (red nuclear staining). (Scale bar, 20 μm.)
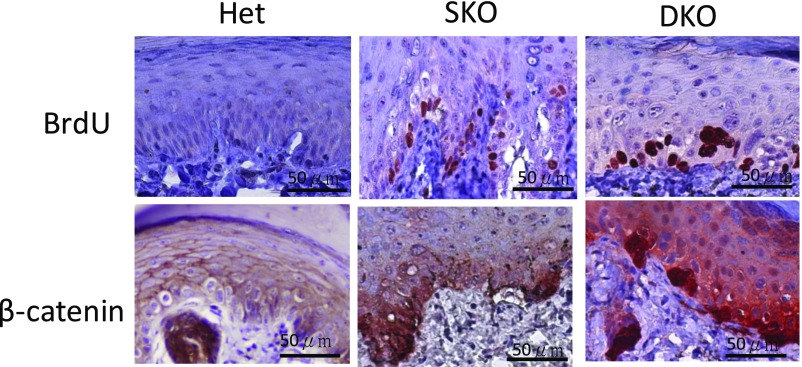
To address the mechanisms of hyperplasia or dysplasia in SKO and DKO skin, we performed histopathological staining for BrdU and β-catenin of heterozygous control, SKO, and DKO mice (Fig. S3). BrdU+ cells (red staining in the nuclei, indicating proliferating cells) and β-catenin nuclear staining are increased in the basal layer of the SKO epidermis, possibly contributing to the hyperplasia of the epidermis. BrdU+ cells and β-catenin nuclear staining likewise increased in the DKO epidermis, but most of the positive cells are multinucleated clumping cells, indicating dysfunctional mitoses associated with dysplasia of the epidermis. These data imply that the CK1α ablation-induced Wnt/β-catenin signaling pathway does not induce epidermal dysplasia unless WT p53 is lost, which is in line with the lack of development of skin tumors in SKO mice.
Fig. S3.
Immunohistochemistry for BrdU and β-catenin on paw skins of heterozygous (Het), SKO, and CK1α/p53-DKO mice 28 d after the initiation of tamoxifen i.p. treatment. BrdU+ cells (red staining in the nuclei, indicating proliferating cells) and β-catenin nuclear staining are increased in the basal layer of the SKO epidermis, which could contribute to the hyperplasia of the epidermis. In the CK1α/p53-DKO epidermis BrdU+ cells and β-catenin nuclear staining are increased, but most of the positive cells are multinucleated clumping cells. (Scale bars, 50 μm.)
The KitL/Kit Pathway Is Up-Regulated by p53 Activation in CK1α-Ablated Keratinocytes.
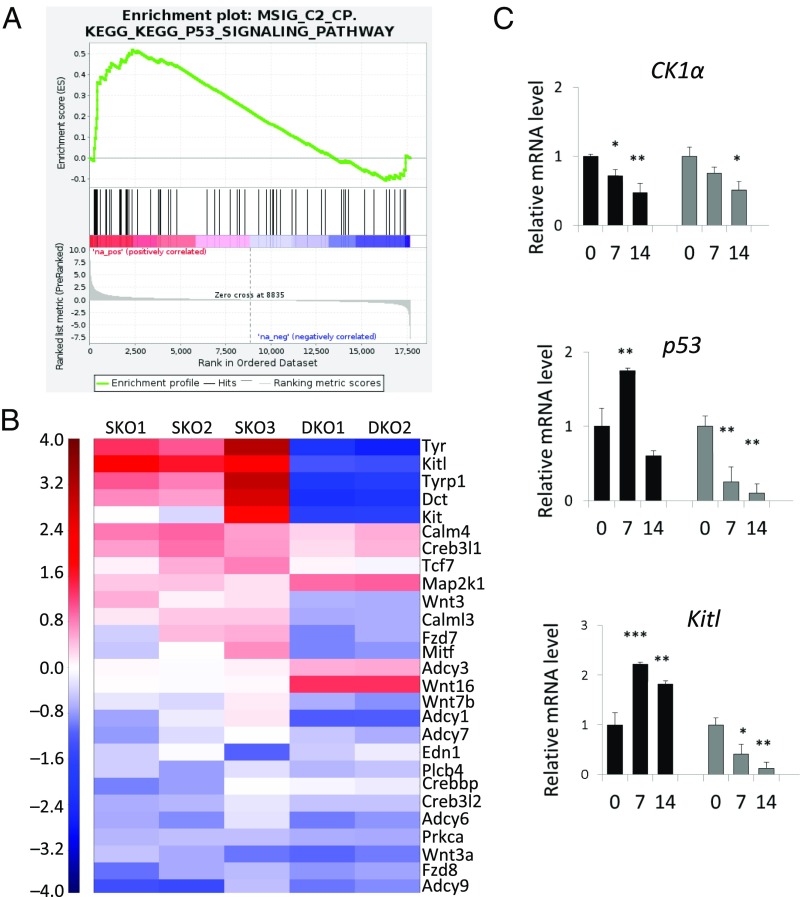
To clarify the paracrine factors induced by the ablation of CK1α in keratinocytes, we performed RNA-sequencing (RNA-seq) analysis on epidermal samples obtained from SKO mice, DKO mice, and heterozygous control mice at day 14 after the initiation of i.p. injections of tamoxifen. The cell populations in the epidermis include a major group of keratinocytes and a minor group of melanocytes. Consistent with the induction of p53 protein in the epidermis of SKO mice, we observed a significant enrichment of the Kyoto Encyclopedia of Genes and Genomes (KEGG) p53 signaling gene set in SKO versus controls (Fig. 4A and Dataset S1). We focused our transcriptome analysis further on all genes constituting the KEGG melanogenesis pathway. In line with the observed skin hyperpigmentation, the melanogenic enzymes Tyr (tyrosinase), Tyrp1 (tyrosinase-related protein 1), and Dct (dopachrome tautomerase) were consistently up-regulated in SKO samples compared with heterozygous and DKO controls (Fig. 4B). Remarkably, the most strongly and most significantly up-regulated gene was the secreted paracrine factor KitL [mean log2 fold change (FC) = 1.79; P value = 6.9 ×10−15]. In support of the observed p53 dependence, the induction of those genes was completely abolished by concomitant p53 loss. We further validated KitL expression levels in a time-course study after topical 4HT induction on the ears of SKO and DKO mice for 14 d. On day 7 of induction, the CK1α mRNA level in SKO mice was down-regulated around 0.7-fold, whereas the p53 mRNA level was up-regulated about 1.75-fold, indicating that p53 regulation is very sensitive to CK1α inhibition. KitL was up-regulated more than twofold in SKO mice compared with control mice within 7 d and then gradually declined to 1.8-fold at day 14. However, when p53 was lost, KitL was down-regulated rapidly to a very low level, which implies that the transactivation of KitL is p53-dependent (Fig. 4C).
Fig. 4.
Transcriptome analysis of CK1α-SKO and CK1α/p53-DKO mice in comparison with WT mice. Induction and skin sampling timing were as described in Fig. 2A. (A) Results of Gene Set Enrichment Analysis performed on the KEGG database indicates an enrichment of p53 target genes in SKO mice compared with WT mice. RNA was extracted from day 14 epidermis. (B) Among specific melanogenic genes of secreted paracrine factors, KitL is the most significantly up-regulated at day 14. (The entire gene list is given in Dataset S1.) (C) RT-PCR validation of epidermis at different time points. The CK1α ablation-induced p53 up-regulation is transient at day 7, and KitL up-regulation is sustained at days 7 and 14. Data are shown as means ± SD; *P < 0.05, **P < 0.01, and ***P < 0.001.
Skin Hyperpigmentation Induced by CK1α Ablation in Keratinocytes Is Abolished by Blocking the KitL/Kit Signaling Pathway.
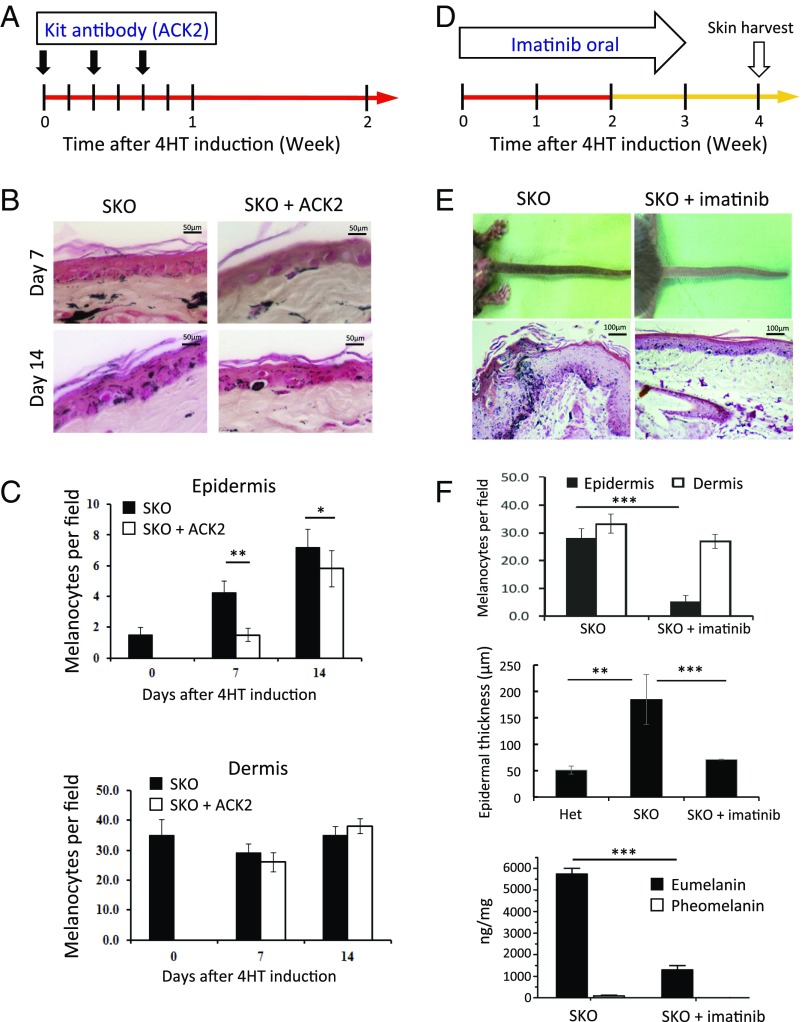
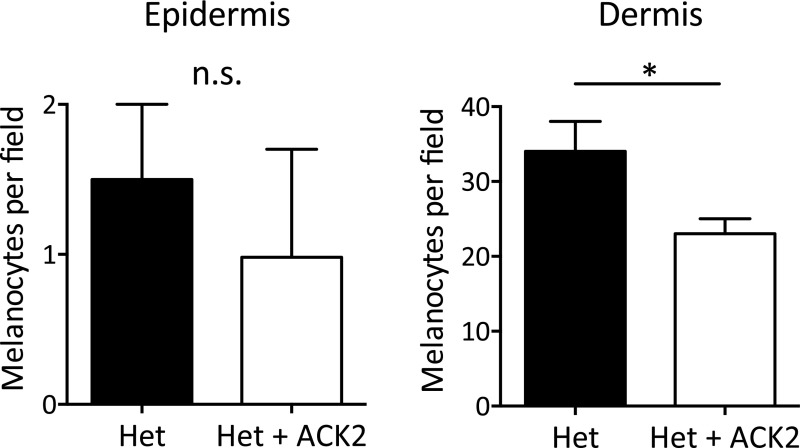
To further clarify the role of KitL in the skin hyperpigmentation of SKO mice, we blocked the KitL–Kit signaling pathway by injecting an anti-Kit antibody (ACK2) intradermally three times in 1 wk (Fig. 5A). SKO mice treated with ACK2 showed a decreased number of melanocytes in the epidermis but showed no significant change in the dermis compared with the noninjected control SKO mice (Fig. 5 B and C). However, the neutralization effect was transient, and the number of epidermal melanocytes had fully recovered by day 14 in the ACK2-treated SKO mice (Fig. 5C), unless the mice were given another injection. When we compared CK1α heterozygous mice and SKO mice, the ACK2 intradermal injection decreased the number of dermal melanocytes in CK1α heterozygous mice but did not affect the number of dermal melanocytes in SKO mice (Fig. 5C and Fig. S4). These findings suggest that (i) activation of the Kit receptor and the KitL–Kit signaling pathway are essential for melanocyte function and the migration of melanocytes from the dermis into the epidermis and (ii) blocking Kit using the ACK2 antibody can abolish the above effect of Kit signaling on dermal melanocytes; this effect is reversible, and melanocyte migration into the SKO epidermis resumes in the following week.
Fig. 5.
CK1α ablation in keratinocyte-induced skin hyperpigmentation can be prevented by blocking the KitL/Kit signaling pathway. (A–C) Intradermal injection of an anti-Kit antibody (ACK2) decreases the number of melanocytes in the epidermis of SKO mice at days 7 and 14 compared with vehicle-treated SKO mice. (D–F) Oral treatment with imatinib for 3 wk cancels the CK1α ablation-induced eumelanization and epidermal thickening. Phenotype examination reveals hypopigmentation in imatinib-treated mice compared with vehicle-treated mice at day 28. Histopathology shows thinning of the epidermis with decreased eumelanin and melanocytes in the epidermis in imatinib-treated mice at day 28. Data are shown as means ± SD; *P < 0.05, **P < 0.01, and ***P < 0.001. (Scale bars, 50 μm in B and 100 μm in E.)
Fig. S4.
Changes in melanocyte number after ACK2 intradermal injection in CK1α heterozygous (Het) mice 14 d after the 4HT administration. Dermal melanocytes were significantly reduced by ACK2 injection, whereas ACK2 did not affect epidermal melanocytes. Data are shown as means ± SD; *P < 0.05. n.s., not significant.
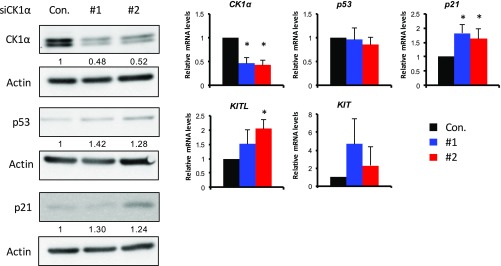
We then examined whether imatinib could cancel the CK1α ablation-induced hyperpigmentation. Imatinib is a potent inhibitor of receptor tyrosine kinases such as Kit, BCR-ABL, and the platelet-derived growth factor receptor (PDGFR) (24, 25). SKO mice treated with i.p. injections of tamoxifen were separated into two groups: one group treated with oral imatinib for 21 d and a control group treated with vehicle for 21 d (Fig. 5D). At day 28, phenotype examination revealed hypopigmentation in the imatinib-treated mice compared with the vehicle-treated mice, which were hyperpigmented as expected (Fig. 5E). Histopathology showed thinning of the epidermis with decreased melanin levels and melanocyte numbers in the epidermis of imatinib-treated mice compared with the vehicle-treated mice, which had epidermal hyperplasia and dense melanin deposits (Fig. 5 E and F). Eumelanin/pheomelanin analysis of the epidermis showed that eumelanin production was dramatically reduced after the imatinib treatment (Fig. 5F). These data indicate that skin pigmentation, as well as its thickening by CK1α ablation, is mainly mediated via the KitL–Kit signaling pathway, which is enhanced in SKO mice following p53 activation. This conclusion is further supported by analysis of human primary normal keratinocytes, in which siRNA-mediated depletion of CK1α significantly induced the expression of KITL (Fig. S5).
Fig. S5.
CK1α depletion in human primary keratinocytes induces p21 and KITL up-regulation. Human primary keratinocytes were transfected with CK1α siRNA. (Left) Two days after transfection, cell lysates were analyzed using Western blotting with antibodies as indicated. Total RNAs were isolated and reverse transcribed, and mRNA expression levels were analyzed by qPCR with primer pairs as indicated. (Right) mRNA levels were calculated relative to the scrambled control (Con.) with the housekeeping gene GAPDH as a reference. Data are shown as means ± SD; *P < 0.05.
CK1α Ablation-Induced Skin Hyperpigmentation Is Independent of the UV-Induced p53/Pomc/Mc1r Pathway.
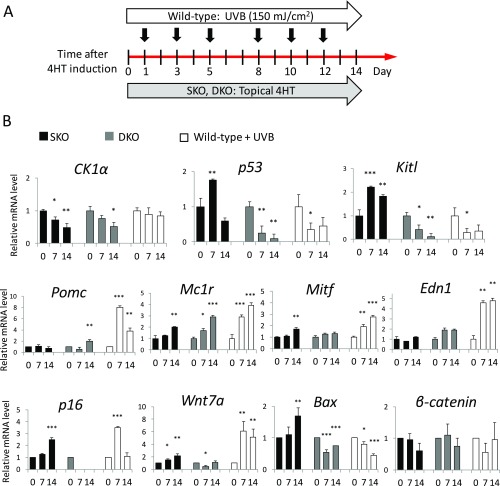
To compare the mechanisms of melanogenesis in the two types of p53-dependent hyperpigmentation, UV irradiation and CK1α inhibition, we designed an experiment using chronic UVB exposure (150 mJ/cm2 every other day, three times/wk) for 2 wk to compare with the topical induction of SKO and DKO mice with 4HT for 2 wk (Fig. S6). During the chronic UV irradiation, Pomc, a multicomponent precursor of α-MSH, was up-regulated about eightfold on day 7 in WT mice although p53 had passed the transient activation. Pomc was not activated by the ablation of CK1α. Other genes, including Mc1r, Mitf, Edn1, and Wnt7a, were up-regulated by UV irradiation much more intensely in WT mice than in SKO mice. The response of Wnt7a and Edn1, which are robustly induced by UV (8, 26), to SKO) was mild and delayed compared with the UV effect in WT mice. The CK1α ablation-induced up-regulation of KitL was sustained in SKO mice but not in DKO or UV-irradiated WT mice. Mitf up-regulation induced by CK1α ablation was delayed compared with the UV-induced Mitf up-regulation in WT mice. Taken together, our findings show that the genes influenced by CK1α ablation are entirely different from those affected by UV irradiation.
Fig. S6.
Comparison of signaling pathways between UV irradiation and CK1α ablation in keratinocytes. (A) Experimental design: WT mice under chronic UVB exposure and the topical 4HT induction of SKO and CK1α/p53-DKO mice. (B) RT-PCR analysis of epidermis at different time points (days 0, 7, and 14). Under chronic UV irradiation, Pomc is up-regulated eightfold on day 7. Pomc is not activated by CK1α ablation. Other genes, including Mc1r, Mitf, Edn1, and Wnt7a, are up-regulated by UV irradiation much more intensely than in the CK1α-ablated condition on days 7 and 14. The CK1α ablation-induced KitL up-regulation is sustained on days 7 and 14. The Mitf up-regulation induced by CK1α ablation is delayed compared with the UV-induced Mitf up-regulation. The genes influenced in response to CK1α ablation are different from those affected by UV irradiation. Data are shown as means ± SD; *P < 0.05, **P < 0.01, and ***P < 0.001.
Eumelanin Production Following CK1α Ablation Protects the Skin from Sunburn Damage.
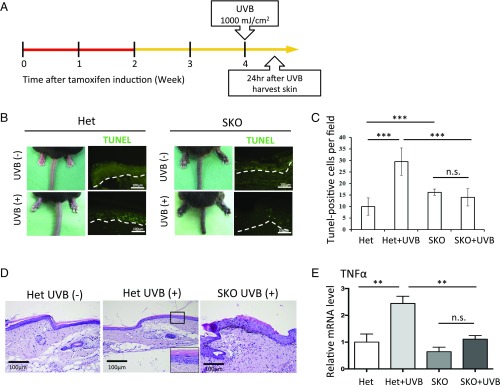
To determine whether the inhibition of CK1α could serve as a sun-protection strategy, we exposed CK1α heterozygous control mice and hyperpigmented SKO mice to a high, sunburn-inducing dose of UVB (1,000 mJ/cm2) and harvested skin samples 24 h later (Fig. 6A). Phenotype examination showed swelling of the tail, an acute sunburn damage response, in control mice after the sunburn dose of UVB (Fig. 6B). However, the hyperpigmented tails of SKO mice appeared normal in diameter, suggesting the sunburn-protective effect of CK1α-inhibition. TUNEL staining demonstrated that UVB exposure induced many sunburn cells (apoptotic cells) in the tail skin of control mice but induced many fewer apoptotic cells in SKO mice (Fig. 6 B and C). Inflammatory cells in the dermis were not remarkable either in control mice or in SKO mice by H&E staining (Fig. 6D). In contrast, the mRNA levels of TNFα, an inflammatory cytokine, were significantly up-regulated after UV radiation in control mice compared with SKO mice (Fig. 6E). These results indicate that CK1α inhibition successfully protects against acute UV reactions, including apoptosis of keratinocytes and skin swelling, and thus can be a potential strategy for sunburn protection.
Fig. 6.
CK1α ablation-induced eumelanization protects skin from sunburn damage. (A) Heterozygous control (Het) mice and eumelanized SKO mice were exposed to high-dose UVB (1 000 mJ/cm2). Skin samples were harvested 24 h after the UVB exposure. (B and C) Swelling of the tail, an acute type of sunburn damage, is observed in heterozygous mice after the sunburn dose of UVB exposure, whereas the eumelanized tails of SKO mice have a normal diameter. TUNEL staining demonstrated that the UVB exposure induced many sunburn cells (apoptotic cells) in the skin of heterozygous mice but induced many fewer apoptotic cells in SKO mice. (D) Inflammatory cells in the dermis are not remarkable either in heterozygous (±UVB) mice or in SKO (+UVB) mice by H&E staining. The Inset (3.6× magnified on the square) shows an apoptotic cell in the epidermis of Het mice after UVB exposure. (E) mRNA levels of TNFα, an inflammatory cytokine, were significantly up-regulated after UV radiation in heterozygous mice compared with SKO mice. Data are shown as means ± SD; **P < 0.01 and ***P < 0.001; n.s., not significant. (Scale bars, 100 μm.)
Discussion
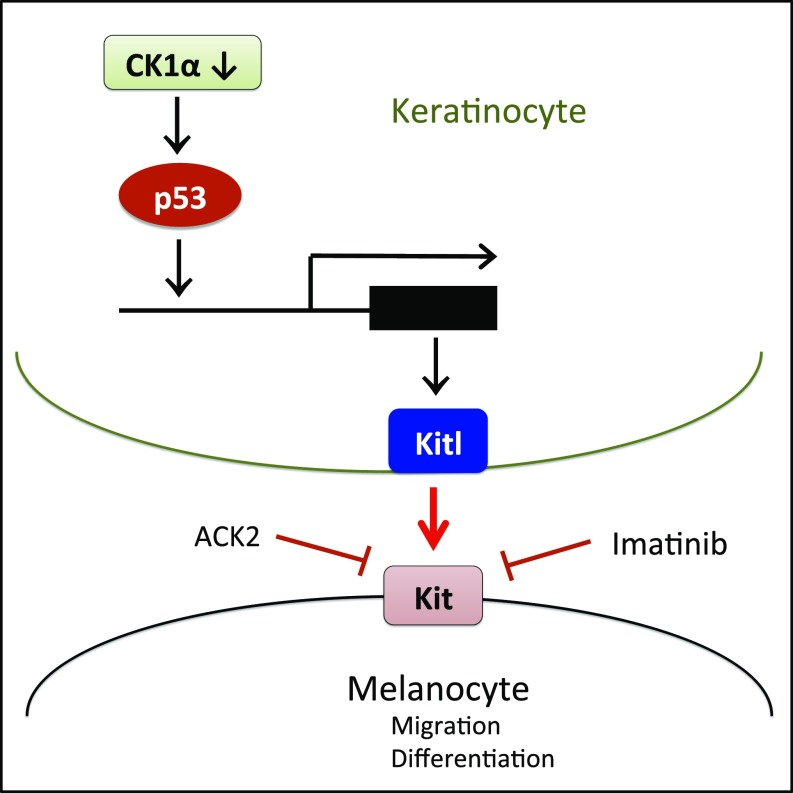
Here we demonstrate that activation of the p53/KitL/Kit signaling pathway by inhibition of CK1α in keratinocytes causes hyperpigmentation of the skin (summarized in Fig. 7) and successfully protects it from UV damage.
Fig. 7.
Schematic summary of CK1α inhibition-induced p53-dependent activation of the KitL/Kit signaling pathway in the keratinocyte/melanocyte unit.
The transcription factor p53 is a tumor-suppressor protein that triggers cell-cycle arrest, DNA repair mechanisms, and apoptosis and prevents the accumulation of damaged cells with potentially cancer-prone mutations (27). The mutational inactivation of p53 causes genome instability in human tumors that can mediate drug resistance. p53 has been shown to play a central role in the hyperpigmentation response to UV irradiation by regulating melanogenic paracrine cytokine networks in human epidermis. Previous studies have identified Pomc, KitL, and Edn1as p53-mediated melanogenic cytokines (4–8, 18).
Among the various signaling pathways that p53 governs, the major pathway regulating the synthesis of melanin is POMC/α-MSH/MC1R/MITF signaling. Once keratinocytes are exposed to UV, they increase their production of POMC, a multicomponent precursor of α-MSH and ACTH. α-MSH and ACTH are agonists of MC1R, the key receptor on melanocytes that ultimately leads to the generation of the secondary messenger cAMP signaling cascade and induces the transcription of enzymes necessary for melanin production. POMC and MC1R functions are thus important in regulating the amount of eumelanin pigment produced after UV exposure. However, the POMC and MC1R genes have a high number of polymorphic alleles. Individuals without the full function of POMC or MC1R typically have red/blond hair and fair skin and are unable to synthesize eumelanin after UV exposure (28, 29). Skin pigmentation stimulated in the absence of POMC/MC1R signaling can be a potential strategy to prevent UV damage and, consequently, the development of skin cancer, regardless of race or ethnicity. However, only a few such alternative methods have been suggested (30, 31).
Generally, melanocytes produce two distinct types of melanin: black-brown eumelanin that is prevalent in individuals with black and/or brown hair, and yellow-reddish pheomelanin that is primarily produced in individuals with red hair and freckles (3, 6, 32) but is also produced in the skin of individuals who do not have red hair and freckles (33). The beneficial effects of melanin are mainly due to the presence of eumelanin that absorbs most of the UV and scavenges the UV-generated free radicals (11), whereas pheomelanin is, in fact, carcinogenic (34), Therefore, the exclusive increase of eumelanin levels by SKO is a preferable means to prevent UV-induced DNA damage and skin cancers.
D’Orazio et al. (9) showed that forskolin, an agonist of cAMP, successfully induces eumelanization in MC1R-KO mice, without the need for UV. Since cAMP is located downstream of POMC/MC1R, cAMP is expected to activate MITF independently of POMC/MC1R function (9). Another p53-related pathway for eumelanization is Edn1. Keratinocyte expression of Edn1 is directly regulated by p53, and the in vivo disruption of keratinocyte-derived Edn1 signaling alters melanocyte proliferation and decreases epidermal and dermal melanocyte populations in both normal and UV-exposed mouse skin (8).
Distinct from those p53-regulated pathways, McGowan et al. (6) showed that ribosomal mutation-induced p53 stabilization in keratinocytes could stimulate KitL expression and epidermal melanocytosis via a paracrine mechanism. On this basis, it has been postulated that topical skin application of p53 activators may be used to induce a tanning response (30, 31).
Likewise, we now demonstrate that CK1α ablation in keratinocytes represents another approach to induce the p53/KitL/Kit cascade and epidermal melanocytosis. KitL secreted by keratinocytes is involved in regulating melanocyte function in normal skin as well as in UV-irradiated skin (35). Binding of KitL to its receptor Kit on melanocytes induces receptor dimerization and the autophosphorylation of specific tyrosine residues in its intracellular domain, leading to melanocyte precursor cell proliferation, differentiation, migration, and melanogenesis through the PI3K and Raf/Erk pathways (36). The KitL/Kit signaling pathway is also involved in regulating the migration of differentiated melanoblasts from melanocyte stem cells into the epidermis and their further differentiation into mature melanocytes (26, 37, 38). The loss of Kit receptor or KitL function blocks the migration and survival of melanocyte precursors at an early stage of melanocyte development (39). Treatment with the Kit-neutralizing antibody ACK2 is also known to inhibit melanocyte proliferation and in some instances to induce their apoptosis (40, 41). Furthermore, the forced expression of KitL in epidermal keratinocytes promotes epidermal pigmentation and maintains melanoblasts and melanocytes in the epidermis (42, 43). Constitutive expression of Kit ligand by epidermal keratinocytes in K14-Scf transgenic mice results in the maintenance of epidermal melanocytes in the interfollicular basal epidermal layer and subsequent epidermal pigmentation, primarily where devoid of melanocytes (9). Kit ligand-dependent pigmentation even can occur regardless of Tyr- or Mc1r-defective status (44). Mc1r-defective primary murine melanocytes have been difficult to culture in vitro, and conditioned supernatants containing stem cell factor derived from primary keratinocytes can facilitate their growth (45). These data indicate that KitL/Kit signaling plays a vital role in melanocyte precursor survival, differentiation, proliferation, and migration. Enhancing KitL/Kit signaling is therefore a potential means to induce skin pigmentation. CK1α ablation in keratinocytes produces the same melanocyte distribution pattern in the interfollicular epidermis, indicating KitL expression in basal keratinocytes plays an important role in maintaining melanocytes in epidermis (Fig. S1). It would be interesting to further test the effect of CK1α ablation in Mc1r-deficient mice to determine the possible value of CK1α inhibition-induced KitL/Kit activation and eumelanin production in the absence of Mc1r.
There is an increase in epidermal thickness following UV irradiation (46), which may play a role in avoiding DNA damage in the lower epidermis by the upper epidermis with increased melanin pigments. Epidermal thickening induced by CK1α-ablated keratinocytes together with hyperpigmentation may prevent sunburn damage. We have not detected any adverse effects of Wnt activation in the mouse skin, and even the hyperplasia thickening of the epidermis was apparently p53-KitL–mediated, as it was reversed by imatinib treatment, which has no effect on Wnt signaling.
In conclusion, we have demonstrated that skin hyperpigmentation is induced by the inhibition of CK1α in keratinocytes. This pigmentation response, similar to UV tanning, is p53-dependent, mediated by eumelanization, and associated with moderate epidermal thickening yet has a reduced secretion of inflammatory cytokines. Importantly, CK1α inhibition induces preferential p53 activation without skin tumor formation, suggesting that CK1α inhibition might be a safe approach for UV protection. The finding that the hyperpigmentation response prevented UV-induced DNA damage independently of the POMC/α-MSH/MC1R/MITF pathway suggests that CK1α inhibition might prove to be an effective strategy for sunburn protection in MC1R-deficient individuals.
Materials and Methods
Mice.
Csnk1a1 floxed and Csnk1a1/p53 double-floxed mice were reported previously (14). K14–Cre–ERT2 mice, purchased from the Jackson Laboratory, were used as a skin-specific deleter strain. Csnk1a1Δskin and Csnk1a1/p53Δskin mice were developed by crossing Csnk1a1 floxed or Csnk1a1/p53 floxed mice with K14–Cre–ERT2 mice. Mice were bred at the National Laboratory Animal Center, Tainan, Taiwan, and were maintained at a specific pathogen-free animal facility at the China Medical University. Mouse experiments were conducted with the approval of the China Medical University Animal Care Committee in accordance with legal and ethical standards.
Targeting Gene Activation.
For high-level recombination to ensure the deletion of CK1α expression, 6-wk-old K14–Cre–ERT2–CK1αfl/fl mice were treated six times with 120 mg/kg tamoxifen by i.p. injection (on days 1, 2, 5, 6, 9, and 10). Tamoxifen (T5648; Sigma) was dissolved in corn oil (C8267; Sigma). For the topical treatment to delete CK1α expression, 4HT (H6278; Sigma) was dissolved in 99.9% alcohol (32205; Sigma), and 1.5 mg/0.5 mL was administered to the dorsal part of both ears for 2 wk.
Histopathology, Immunostaining, and Fontana–Masson Staining.
Paraffin-embedded specimens were cut into 5-μm sections. After deparaffinization (tissue slides were soaked in xylene three times, each time for 5 min) and rehydration (slides were incubated in the following graded series of ethanol: 100, 100, 95, 90, and 70%, 5 min each), the slides were rinsed with distilled water for 5 min.
For Fontana–Masson staining (FMS-1-IFU; ScyTek), a freshly mixed ammoniacal silver solution was placed in a 58–60 °C water bath and allowed to equilibrate. The slides were incubated in warmed ammoniacal silver solution for 30–60 min or until the tissue sections became yellow/brown in color and were then rinsed with distilled water for 3–5 s. The slides were incubated in gold chloride solution (0.2%) for 30 s and were then rinsed with distilled water for 3–5 s. The slides were incubated in sodium thiosulfate solution (5%) for 1 min and were then rinsed with distilled water for 3–5 s. The slides were incubated in nuclear fast red solution for 5 min, rinsed for 1 min in running tap water, dehydrated in four changes of fresh absolute alcohol (95, 95, 100, and 100%) and xylene, and mounted with Histokitt (Assistent).
For histopathology, skin tissues were fixed overnight in 10% neutral buffered formalin at 4 °C and then transferred to 70% ethanol before being processed and embedded in paraffin. Paraffin sections were then stained with H&E.
For immunochemistry, paraffin sections were incubated in a humidity chamber for 15 min at 60 °C. Sections were deparaffinized in two changes of xylene for 5 min each and hydrated in two changes of 100% ethanol for 5 min each, then in 95% and 80% ethanol for 5 min each, and finally rinsed in distilled water. Antigen retrieval was enhanced by microwaving the slides in citrate buffer (DAKO; pH 6.0) for 20 min. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide in methanol. After blocking, sections were incubated overnight at 4 °C with antibodies to the following markers at a proper dilution: CK1α (C-19, 1/1,000; Santa Cruz), γ-H2AX (JBW301, 1/3,000; Millipore), p53 (CM5, 1/200; Leica Biosystems), p21 (F-5, 1/100; Santa Cruz), β-catenin (14 β-catenin, 1/100; BD Bioscience), cyclin D1 (SP4, 1/200; Thermo Fisher Scientific), cleaved caspase 3 (Asp175, 1/200; Cell Signaling Technology), and TRP1 (ab3312, 1/200; Abcam). Secondary antibodies used were HRP-polymer anti-mouse, anti-rabbit, anti-goat, and anti-rat (all from Nichirei). AEC chromogen (ScyTek) was used for detection, and hematoxylin was used as a counterstain. For melanin detection, Fontana–Masson staining (ScyTek) was performed according to the manufacturer’s instructions and was detected using a Carl Zeiss microscope.
Intradermal Injection of ACK2 into Mouse Dorsal Ear Skin.
ACK2 (anti-Kit antibody; eBioscience) was injected into the ear dorsal skin of 6-wk-old mice (20 μg/cm2) three times per week (on days 1, 3, and 5 during the topical treatment with 4HT). After the ACK2 antibody injection, the mice finished their 14 d of induced gene knockout with topical 4HT, and the samples were harvested 7, 14, and 28 d after the initiation of 4HT induction. The entire ear of each mouse was excised, washed and fixed with 4% paraformaldehyde, and processed for H&E and Fontana–Masson staining.
Administration of Imatinib.
Imatinib (270784; Sigma) was administered by oral gavage for 3 wk starting on the first day of induction at a dose of 100 mg/kg once daily. Placebo control mice were treated with sterile water alone.
Assays of Eumelanin and Pheomelanin.
Mice finished the 14 d of 4HT induction for SKO, and tail skins were obtained at day 28 for epidermal sheet separation. Skin samples were measured and rinsed with Ca2+-, Mg2+-free PBS (pH 7.4) to remove blood contaminants and were then treated with 0.25% trypsin (Difco; Becton Dickinson Microbiology Systems) in PBS (pH 7.2) for 16–18 h at 28 °C. The epidermis and dermis were separated and stored at −80 °C until use. For assays of melanin content, the tissues were minced with scissors and homogenized in 10 volumes of PBS at 28 °C. Samples of epidermis and dermis were processed for chemical analyses of eumelanin to detect the specific degradation product, pyrrole-2,3,5-tricarboxylic acid (PTCA) (47), and of pheomelanin to detect the specific degradation product, 4-amino-3-hydroxyphenylalanine (4-AHP) (48, 49). One nanogram of PTCA or 4-AHP corresponds to 50 ng of eumelanin or 9 ng of pheomelanin. The statistical significance of differences in the contents of eumelanin and pheomelanin was determined by Student’s t test for comparisons of groups of equal size.
TUNEL Staining.
Detection of apoptosis was conducted on 4-μM paraffin-embedded sections using a commercially available immunofluorescent TUNEL assay (Kit S7110, ApopTag; InterGen). Briefly, sections were deparaffinized with xylene and then were washed with two changes each of 100, 95, and 70% ethanol and PBS, each for 5 min. Tissues were pretreated with protein-digesting enzyme or Proteinase K (20 μg/ mL) at room temperature for 15 min and were then soaked in PBS for 5 min. Sections were then immediately incubated with working-strength Terminal deoxynucleotidyl Transferase (TdT) enzyme in a humidified chamber at 37 °C for 1 h. The sections were immersed in stop/wash buffer and were then gently rinsed with PBS. FITC-labeled anti-digoxigenin conjugate was then applied to each section and incubated at room temperature for 30 min in the dark. Slides were washed in PBS and mounted with Prolong Gold Antifade Reagent with DAPI (P36941; Life Technologies).
Real-Time RT-qPCR of Mouse Skin.
Total RNAs were extracted from epidermal sheets using TRIzol Reagent (Invitrogen). Complementary DNAs were synthesized using a reverse transcription kit (Thermo Scientific), and real-time RT-PCR was performed with a SYBR Advantage qPCR Premix kit, using the StepOnePlus real-time RT-PCR system (Applied Biosystems). Primer sequences are given in Table S1. Amplification was normalized to the housekeeping gene GAPDH or 18S ribosomal RNA, and differences between samples were quantified based on the ΔΔCt method.
Table S1.
PCR primers used in this study
| Primer | Forward | Reverse |
| Axin 2 | CTCCCCACCTTGAATGAAGA | ACTGGGTCGCTTCTCTTGAA |
| Bax | ATGCGTCCACCAAGAAGCTGA | AGCAATCATCCTCTGCAGCTCC |
| CD44 | AGAAAAATGGCCGCTACAGTATC | TGCATGTTTCAAAACCCTTGC |
| CK1α | GGTGCGGAAGATCGGATCT | TTCACTGCCACTTCCTCGC |
| Cyclin D1 | TTGACTGCCGAGAAGTTGTG | CCACTTGAGCTTGTTCACCA |
| Cyclin G1 | GCTGGCGCTATCTATCCTTG | GGTCAAATCTCGGCCACTTA |
| Edn1 | TCTCCGAGGGCGATGTGTA | CTTCTCCACCGAGGTCCGA |
| GAPDH | ACAACTTTGGCATTGTGGAA | GATGCAGGGATGATGTTCTG |
| KitL | TCCATCATAGACAAACTTGGGAAA | TTCTGGCCTCTTCGGAGATT |
| Mc1r | GACCGCTACATCTCCATCTTCT | AGGAGGAGGAAGAGGTTGAAGT |
| Mdm2 | TGTGTGAGCTGAGGGAGATG | CACTTACGCCATCGTCAAGA |
| Mitf | CAAATGGCAAATACGTTACCCG | CAATGCTCTTGCTTCAGACTCT |
| p21 | TCCACAGCGATATCCAGACA | AGACAACGGCACACTTTGCT |
| p53 | CCCCTGTCATCTTTTGTCCCT | AGCTGGCAGAATAGCTTATTGAG |
| Pomc | CCTTGGTTTGGGTGGAAGATG | GCGTTCTTGATGATGGCGTTC |
| Puma | CAAGAAGAGCAGCATCGACA | CTCCAGGATCCCTGGGTAAG |
| Sox9 | GGAGCTCAGCAAGACTCTGG | TGTAATCGGGGTGGTCTTTCT |
| TNFα | GCCAGGAGGGAGAACAGAAACTC | GGCCAGTGAGTGAAAGGGACA |
| Wnt7a | TCTCTGCTTTGGCGTCCTCTAC | GCCAGGCCAGGAATCTTGTTG |
| β-Catenin | ATGGAGCCGGACAGAAAAGC | TGGGAGGTGTCAACATCTTCTT |
RNA Extraction and Sequencing.
Mouse ear dorsal epidermis and tail epidermis were homogenized in QIAzol Lysis Reagent (Qiagen) with a TissueLyser (Qiagen) according to the manufacturer’s protocol. Following chloroform extraction, ethanol precipitation, and DNase digestion, RNAs were purified using an RNeasy Mini Kit (Qiagen). RNA integrity was assessed on an Experion StdSens RNA Chip (Bio-Rad).
RNA-seq libraries were prepared using a TruSeq Stranded mRNA Sample Prep kit (Illumina). Libraries were quantified on a Bioanalyzer (Agilent Technologies) and were sequenced on an Illumina HiSeq 1500 platform, rapid-run mode, single-read 50 bp (TruSeq Rapid SR Cluster Kit–HS, TruSeq Rapid SBS Kit–HS, 50 cycles) according to the manufacturer’s instructions.
For transcriptome analysis, sequenced reads were aligned to Ensembl v74 using STAR (version STAR_2.3.1z13_r470). Gene read counts were established as read count within merged exons of protein-coding transcripts (for genes with a protein gene product) or within merged exons of all transcripts (for noncoding genes). Fragments per kilobase of transcript per million mapped reads (FPKM) were calculated based on the total raw read count per gene and length of merged exons. Differential expression was assessed using DESeq (version 1.6.1) (50) based on raw read count data. P values and log2 FC values were derived from DESeq (after incrementing each raw read count by one to avoid undefined values). P values were corrected for multiple hypotheses testing using the Benjamini–Hochberg correction (51). Genes that did not yield a minimum raw read count of 50 and a minimum transcripts per million (TPM) of 5 in at least two samples were discarded due to insufficient coverage. Of the remaining genes, genes were considered differentially expressed if the absolute of the log2 FC was at least 1 (twofold induction/repression).
SI Materials and Methods
This section is related to Fig. S5.
Cell Culture of Human Primary Keratinocytes.
Human primary keratinocytes were purchased from the ATCC (no. PCS-200-010) and were maintained in keratinocyte basal medium supplemented with the corresponding keratinocyte supplements (Gibco) within a 37 °C humidified incubator containing 5% CO2.
siRNA Transfection.
All siRNAs were synthesized and annealed by Sigma-Aldrich. Transfection of siRNA was performed using Oligofectamine (Invitrogen). The sequences targeted by CK1α siRNA#1 and siRNA#2 are 5′-CCCGATATGCTAGCATCAA-3′ and 5′-GGGCTAAAGGCTGCAACAA-3′, respectively.
Real-Time RT-qPCR of Cultured Human Keratinocytes.
RNA samples were isolated using TRIzol extraction (Invitrogen) and reverse-transcribed (∼2 μg) using the Super-Script III reverse transcriptase (Invitrogen). qPCRs were performed using Power SYBR Green 2X Master Mix according to the manufacturer’s instructions (Applied Biosystems). Each sample was tested in duplicate. Three independent experiments were performed, and the results were normalized to the expression of GAPDH. Specific primer sequences used in this study were as follows: CK1α (Fwd, 5′-GTACTTCTCAGGACCCATCT-3′; Rev, 5′- TGCTAGCATATCGGGCAGTG-3′); p53 (Fwd, 5′-TGGGTCTTCAGTGAACCATT-3′; Rev, 5′-ACTACTTCCTGAAAACAACG -3′); p21 (Fwd, 5′- CTATGGACAGGGCATGCCAC -3′; Rev, 5′- TAGGTGGAGAAACGGGAACC -3′); KITL (Fwd, 5′- TCTGCAGGAATCGTGTGACT-3′; Rev, 5′-GACTTGGCAAAACATCCATCCC-3′); c-KIT (Fwd, 5′- CAAATCCATCCCCACACCCT-3′; Rev, 5′- TGTACTTCATACATGGGTTTCTGTA-3′); and GAPDH (Fwd, 5′ AGCACCAGGTGGTCTCCTCT 3′; Rev, 5′ TGAGGTCCACCACCCTGTTG 3′).
Western Blotting.
Cell lysates were collected for Western blotting. The antibodies used for immunoblotting were anti-actin (A2066; Sigma), anti-p53 (PAb 1801; Santa Cruz), and anti-p21 (sc-6246; Santa Cruz).
Supplementary Material
Acknowledgments
We thank Dr. Che-Hung Shen for his helpful discussion and assistance with Supporting Information preparation. This work was supported by Taiwan Ministry of Science and Technology Grants103-2314-B-037-065 and 104-2314-B-037-011-MY3; Taiwan National Health Research Institute Grant EX104-105-10402BI; Taiwan Academia Sinica Institute of Biomedical Sciences Grant CRC105-P04; the Israel Science Foundation Centers of Excellence; European Research Council Within the Seventh Framework Programme Grant 294390 PICHO; the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation; the Israel Cancer Research Fund; and Japan Society for the Promotion of Science Grants 26461705 and 15K09794.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw gene analysis data reported in this paper have been deposited in the ArrayExpress database (accession no. E-MTAB-5442).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702763114/-/DCSupplemental.
References
- 1.Natarajan VT, Ganju P, Ramkumar A, Grover R, Gokhale RS. Multifaceted pathways protect human skin from UV radiation. Nat Chem Biol. 2014;10:542–551. doi: 10.1038/nchembio.1548. [DOI] [PubMed] [Google Scholar]
- 2.Kondo S. The roles of keratinocyte-derived cytokines in the epidermis and their possible responses to UVA-irradiation. J Investig Dermatol Symp Proc. 1999;4:177–183. doi: 10.1038/sj.jidsp.5640205. [DOI] [PubMed] [Google Scholar]
- 3.Costin GE, Hearing VJ. Human skin pigmentation: Melanocytes modulate skin color in response to stress. FASEB J. 2007;21:976–994. doi: 10.1096/fj.06-6649rev. [DOI] [PubMed] [Google Scholar]
- 4.Cui R, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 5.Slominski A, Tobin DJ, Shibahara S, Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol Rev. 2004;84:1155–1228. doi: 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- 6.McGowan KA, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murase D, et al. The essential role of p53 in hyperpigmentation of the skin via regulation of paracrine melanogenic cytokine receptor signaling. J Biol Chem. 2009;284:4343–4353. doi: 10.1074/jbc.M805570200. [DOI] [PubMed] [Google Scholar]
- 8.Hyter S, et al. Endothelin-1 is a transcriptional target of p53 in epidermal keratinocytes and regulates ultraviolet-induced melanocyte homeostasis. Pigment Cell Melanoma Res. 2013;26:247–258. doi: 10.1111/pcmr.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Orazio JA, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 10.Kadekaro AL, et al. Cutaneous photobiology. The melanocyte vs. the sun: Who will win the final round? Pigment Cell Res. 2003;16:434–447. doi: 10.1034/j.1600-0749.2003.00088.x. [DOI] [PubMed] [Google Scholar]
- 11.Brenner M, Hearing VJ. The protective role of melanin against UV damage in human skin. Photochem Photobiol. 2008;84:539–549. doi: 10.1111/j.1751-1097.2007.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schittek B, Sinnberg T. Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis. Mol Cancer. 2014;13:231. doi: 10.1186/1476-4598-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Int J Biochem Cell Biol. 2011;43:465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Elyada E, et al. CKIα ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470:409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 15.Amit S, et al. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: A molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pribluda A, et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell. 2013;24:242–256. doi: 10.1016/j.ccr.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Schneider RK, et al. Role of casein kinase 1A1 in the biology and targeted therapy of del(5q) MDS. Cancer Cell. 2014;26:509–520. doi: 10.1016/j.ccr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogmundsdottir MH, Steingrimsson E. Selection, p53, and pigmentation. Pigment Cell Melanoma Res. 2014;27:154–155. doi: 10.1111/pcmr.12203. [DOI] [PubMed] [Google Scholar]
- 20.Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 21.Lo Celso C, Prowse DM, Watt FM. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–1799. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- 22.Watt FM, Collins CA. Role of beta-catenin in epidermal stem cell expansion, lineage selection, and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:503–512. doi: 10.1101/sqb.2008.73.011. [DOI] [PubMed] [Google Scholar]
- 23.Kretzschmar K, Weber C, Driskell RR, Calonje E, Watt FM. Compartmentalized epidermal activation of β-catenin differentially affects lineage reprogramming and underlies tumor heterogeneity. Cell Reports. 2016;14:269–281. doi: 10.1016/j.celrep.2015.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capdeville R, Silberman S. Imatinib: A targeted clinical drug development. Semin Hematol. 2003;40(2) Suppl 2:15–20. doi: 10.1053/shem.2003.50037. [DOI] [PubMed] [Google Scholar]
- 25.Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- 26.Yamada T, et al. Wnt/β-catenin and kit signaling sequentially regulate melanocyte stem cell differentiation in UVB-induced epidermal pigmentation. J Invest Dermatol. 2013;133:2753–2762. doi: 10.1038/jid.2013.235. [DOI] [PubMed] [Google Scholar]
- 27.Huart AS, MacLaine NJ, Narayan V, Hupp TR. Exploiting the MDM2-CK1α protein-protein interface to develop novel biologics that induce UBL-kinase-modification and inhibit cell growth. PLoS One. 2012;7:e43391. doi: 10.1371/journal.pone.0043391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller AJ, Tsao H. New insights into pigmentary pathways and skin cancer. Br J Dermatol. 2010;162:22–28. doi: 10.1111/j.1365-2133.2009.09565.x. [DOI] [PubMed] [Google Scholar]
- 29.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 30.Oren M, Bartek J. The sunny side of p53. Cell. 2007;128:826–828. doi: 10.1016/j.cell.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 31.Barsh G, Attardi LD. A healthy tan? N Engl J Med. 2007;356:2208–2210. doi: 10.1056/NEJMcibr071775. [DOI] [PubMed] [Google Scholar]
- 32.Prota G. The role of peroxidase in melanogenesis revisited. Pigment Cell Res. 1992;(Suppl 2):25–31. doi: 10.1111/j.1600-0749.1990.tb00344.x. [DOI] [PubMed] [Google Scholar]
- 33.Thody AJ, et al. Pheomelanin as well as eumelanin is present in human epidermis. J Invest Dermatol. 1991;97:340–344. doi: 10.1111/1523-1747.ep12480680. [DOI] [PubMed] [Google Scholar]
- 34.Mitra D, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature. 2012;491:449–453. doi: 10.1038/nature11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hachiya A, Kobayashi A, Ohuchi A, Takema Y, Imokawa G. The paracrine role of stem cell factor/c-kit signaling in the activation of human melanocytes in ultraviolet-B-induced pigmentation. J Invest Dermatol. 2001;116:578–586. doi: 10.1046/j.1523-1747.2001.01290.x. [DOI] [PubMed] [Google Scholar]
- 36.Monsel G, Ortonne N, Bagot M, Bensussan A, Dumaz N. c-Kit mutants require hypoxia-inducible factor 1alpha to transform melanocytes. Oncogene. 2010;29:227–236. doi: 10.1038/onc.2009.320. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura EK, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 38.Rabbani P, et al. Coordinated activation of Wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration. Cell. 2011;145:941–955. doi: 10.1016/j.cell.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wehrle-Haller B, Weston JA. Soluble and cell-bound forms of steel factor activity play distinct roles in melanocyte precursor dispersal and survival on the lateral neural crest migration pathway. Development. 1995;121:731–742. doi: 10.1242/dev.121.3.731. [DOI] [PubMed] [Google Scholar]
- 40.Ito M, et al. Removal of stem cell factor or addition of monoclonal anti-c-KIT antibody induces apoptosis in murine melanocyte precursors. J Invest Dermatol. 1999;112:796–801. doi: 10.1046/j.1523-1747.1999.00552.x. [DOI] [PubMed] [Google Scholar]
- 41.Botchkareva NV, Khlgatian M, Longley BJ, Botchkarev VA, Gilchrest BA. SCF/c-kit signaling is required for cyclic regeneration of the hair pigmentation unit. FASEB J. 2001;15:645–658. doi: 10.1096/fj.00-0368com. [DOI] [PubMed] [Google Scholar]
- 42.Kunisada T, et al. Murine cutaneous mastocytosis and epidermal melanocytosis induced by keratinocyte expression of transgenic stem cell factor. J Exp Med. 1998;187:1565–1573. doi: 10.1084/jem.187.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunisada T, et al. Transgene expression of steel factor in the basal layer of epidermis promotes survival, proliferation, differentiation and migration of melanocyte precursors. Development. 1998;125:2915–2923. doi: 10.1242/dev.125.15.2915. [DOI] [PubMed] [Google Scholar]
- 44.Vanover JC, et al. Stem cell factor rescues tyrosinase expression and pigmentation in discreet anatomic locations in albino mice. Pigment Cell Melanoma Res. 2009;22:827–838. doi: 10.1111/j.1755-148X.2009.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott TL, Wakamatsu K, Ito S, D’Orazio JA. Purification and growth of melanocortin 1 receptor (Mc1r)- defective primary murine melanocytes is dependent on stem cell factor (SFC) from keratinocyte-conditioned media. In Vitro Cell Dev Biol Anim. 2009;45:577–583. doi: 10.1007/s11626-009-9232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearse AD, Gaskell SA, Marks R. Epidermal changes in human skin following irradiation with either UVB or UVA. J Invest Dermatol. 1987;88:83–87. doi: 10.1111/1523-1747.ep12465094. [DOI] [PubMed] [Google Scholar]
- 47.Ito S, et al. Usefulness of alkaline hydrogen peroxide oxidation to analyze eumelanin and pheomelanin in various tissue samples: Application to chemical analysis of human hair melanins. Pigment Cell Melanoma Res. 2011;24:605–613. doi: 10.1111/j.1755-148X.2011.00864.x. [DOI] [PubMed] [Google Scholar]
- 48.Wakamatsu K, Ito S. Advanced chemical methods in melanin determination. Pigment Cell Res. 2002;15:174–183. doi: 10.1034/j.1600-0749.2002.02017.x. [DOI] [PubMed] [Google Scholar]
- 49.Wakamatsu K, Ito S, Rees JL. The usefulness of 4-amino-3-hydroxyphenylalanine as a specific marker of pheomelanin. Pigment Cell Res. 2002;15:225–232. doi: 10.1034/j.1600-0749.2002.02009.x. [DOI] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.