Significance
Campylobacter jejuni is naturally transformable but is selective in the DNA used in transformation. Natural transformation allows for rapid adaptation and provides a means for DNA repair, both of which enhance bacterial fitness. Our work demonstrates that the methylation status of C. jejuni DNA is critical for transformation; methylation of a single 6-bp sequence in proximity to the transforming DNA is sufficient to allow nontransforming DNA to transform C. jejuni. Our data argue against previously described means for DNA discrimination; specifically, there is no evidence that a restriction enzyme recognizes the unmodified 6-bp sequence, and no evidence of a typical DNA uptake sequence. Therefore, we have identified a distinct DNA discrimination mechanism in Campylobacter.
Keywords: DNA discrimination, Campylobacter jejuni, natural transformation, DNA methyltransferase, competence
Abstract
Campylobacter jejuni, a leading cause of bacterial gastroenteritis, is naturally competent. Like many competent organisms, C. jejuni restricts the DNA that can be used for transformation to minimize undesirable changes in the chromosome. Although C. jejuni can be transformed by C. jejuni-derived DNA, it is poorly transformed by the same DNA propagated in Escherichia coli or produced with PCR. Our work indicates that methylation plays an important role in marking DNA for transformation. We have identified a highly conserved DNA methyltransferase, which we term Campylobacter transformation system methyltransferase (ctsM), which methylates an overrepresented 6-bp sequence in the chromosome. DNA derived from a ctsM mutant transforms C. jejuni significantly less well than DNA derived from ctsM+ (parental) cells. The ctsM mutation itself does not affect transformation efficiency when parental DNA is used, suggesting that CtsM is important for marking transforming DNA, but not for transformation itself. The mutant has no growth defect, arguing against ongoing restriction of its own DNA. We further show that E. coli plasmid and PCR-derived DNA can efficiently transform C. jejuni when only a subset of the CtsM sites are methylated in vitro. A single methylation event 1 kb upstream of the DNA involved in homologous recombination is sufficient to transform C. jejuni, whereas otherwise identical unmethylated DNA is not. Methylation influences DNA uptake, with a slight effect also seen on DNA binding. This mechanism of DNA discrimination in C. jejuni is distinct from the DNA discrimination described in other competent bacteria.
Natural transformation, the ability of bacterial cells using dedicated machinery to transport DNA into the cytosol and recombine it into the genome, is a widespread trait. As a mechanism of horizontal gene transfer, it may allow bacteria to acquire new genes to better adapt to their environment. More than 65 species of both Gram-negative and Gram-positive bacteria, and some archaea, are known to be naturally competent, expressing protein complexes that span all cellular compartments to acquire exogenous DNA and transport it into the cell for recombination with the chromosome (1, 2).
One such microbe is the Gram-negative bacterium Campylobacter jejuni, a leading cause of bacterial gastroenteritis worldwide. C. jejuni often colonizes the avian intestinal tract, explaining why poultry is a common source of human infection (3). Horizontal gene transfer in vivo has been demonstrated during experimental infection of chicks, a natural host for this pathogen (4).
C. jejuni is selective in the DNA that it incorporates during natural transformation, using DNA originating in only closely related strains (5). Other bacteria are also selective with respect to the DNA incorporated during transformation, often through a mechanism involving DNA uptake sequences (DUS), which are 10- to 12-bp sequences repeated frequently throughout the genome (6–8). DUS have not been identified in Campylobacter, and evidence indicates that DNA discrimination is not solely sequence-based, because an identical piece of DNA transforms C. jejuni when the DNA is isolated from C. jejuni, but not when it is isolated from E. coli (9).
DNA methyltransferases (MTases) play many roles in biology; in bacteria, a key role is in restriction-modification (RM) systems, where they aid discrimination of foreign DNA, including DNA from infecting phages. DNA MTases methylate specific sequences of bacterial DNA to protect the host from its own restriction endonucleases (REases), although a subset of REases have no paired MTase and digest DNA only when it is methylated in the target sequence (10). In either case, incoming phage DNA lacks the methylation pattern of its host, thereby enabling host REases to selectively degrade that DNA, in a process known as restriction.
RM systems play a role in transformation in Helicobacter pylori, a close relative of C. jejuni that contains more than 20 RM systems (11). The HpyAXII RM system degrades unmethylated plasmid and chromosomal DNA during natural transformation (12). Other RM systems degrade incoming DNA lacking the proper methylation motifs, while also decreasing the length of integrated DNA (13, 14). Similarly, increasing numbers of NlaIV restriction sites have an inhibitory effect on transformation in Neisseria meningitidis (15). A negative correlation between RM systems and natural transformation has been demonstrated in Pseudomonas stutzeri as well (16). In all of these cases, incoming DNA lacking the appropriate methylation motifs is then degraded and thus cannot complete the transformation process by recombining onto the chromosome. In C. jejuni strain 11168, the RM system encoded by Cj1051c plays a role in transformation, as cells with mutations in this gene are transformed to higher levels with DNA derived from other Campylobacter strains; however, these mutants cannot be transformed by E. coli-derived plasmids (17). Although this system is important in C. jejuni strain 11168, it is not widely distributed among Campylobacter strains and so cannot completely explain their universal discrimination against plasmid DNA derived from E. coli. Therefore, Cj1051c is unlikely to be a major determinant of DNA discrimination in C. jejuni.
Numerous DNA MTases have been identified in different C. jejuni strains. Active DNA MTases of C. jejuni, and the sequences that they methylate, were identified using single-molecule real-time (SMRT) sequencing (18–23). With this approach, one methylation motif—RAm6ATTY—was present in all four strains tested to date. We hypothesized that by recognizing methylation at this site, Campylobacter might distinguish DNA of close relatives from that of other species. We examined the role of this single methylation motif in strain DRH212, a streptomycin-resistant derivative of the human disease isolate 81–176. This methylation mark is conferred by a single DNA MTase, CJJ81-176_0240, which is conserved in Campylobacter species but not in other closely related genera (18). This MTase lacks a cognate restriction enzyme, putting it into the class of orphan methyltransferases, similar to the dam enzyme of E. coli and others. We hypothesized this MTase is important for DNA discrimination in natural transformation.
This study demonstrates that the RAm6ATTY motif functions effectively as an uptake sequence in C. jejuni. DNA purified from a Cjj81-176_0240 mutant isolate, which lacks methylation at this motif, transforms C. jejuni at levels orders of magnitude lower than DNA carrying the methyl mark. Thus, we term this MTase Campylobacter transformation system methyltransferase (CtsM). DNA methylated in vivo or in vitro at the RAATTY site is required for wild-type (WT) levels of transformation into C. jejuni. Furthermore, methylation of this motif enables E. coli- or PCR-derived DNA to serve as a higher-efficiency substrate for transformation compared with similar DNA lacking the RAATTY methylation mark. Experiments using radiolabeled DNA demonstrate that methylation influences DNA uptake, with a slight effect seen on DNA binding as well.
Results
One Methylation Motif Is Conserved Among All SMRT-Sequenced Strains of Campylobacter.
Campylobacter species contain numerous annotated DNA MTases (24). The introduction of SMRT sequencing has enabled easy identification of all adenosine methylation motifs, as well as some cytosine methylation motifs (23). These methylation motifs can be paired with their corresponding MTase genes using bioinformatics and genetic analysis. To date, 10 Campylobacter strains have been sequenced using this method, including four C. jejuni isolates (18–22). These strains include two laboratory isolates, one ovine isolate, and one human isolate. (Table 1). Of the 34 methylation motifs identified, only one methylation motif—and its cognate DNA MTase—is conserved among the 10 strains (Table 1). Five motifs were found in more than one strain, but the majority of motifs were only found in one strain (Table S1; not included in Table 1). These data demonstrate that methylation patters differ even within the same Campylobacter species.
Table 1.
Conserved methylation motifs in Campylobacter
| Recognition sequence | C. jejuni laboratory strains | C. jejuni human isolate | C. jejuni sheep isolate | C. coli | C. peloridis | C. subantarcticus | UPTC NCTC | Campylobacter spp. | ||
| 81–176 (18) | 11168 (18) | F38011 (19) | IA3902 (20) | BfR-CA-9557 (21) | (22) | LMB 24374 (22) | LMG 24377 (22) | 11845 (22) | RM16704 (22) | |
| RAATTY | + | + | + | + | + | + | + | + | + | + |
| GATC | − | − | − | − | + | + | + | + | + | − |
| TAAYN5TGC | + | + | − | + | +/− | − | − | − | − | − |
| CAAGAA | − | − | − | − | + | + | − | − | − | − |
| GCAAGG | + | +/− | +/− | + | − | − | − | − | − | − |
| GAGN5GT | − | + | − | +/− | − | − | − | − | − | − |
Methylation motifs were determined using SMRT sequencing. Ambiguous nucleotides are denoted using IUPAC ambiguity codes. +/− indicates that the motif contains one nucleotide difference between the two strains. Underlining indicates the methylated base. Only motifs found to be methylated in more than one strain are included here. Numbers in parentheses refer to citations.
Table S1.
Methylation motifs in Campylobacter isolates
| Recognition sequence | Laboratory strains | Human isolate F38011 (19) | Sheep isolate IA3902 (20) | C. coli BfR-CA-9557 (21) | C. peloridis (22) | C. subantarcticus | UPTC NCTC 11845 (22) | Campylobacter spp. RM16704 (22) | ||
| 81–176 (18) | 11168 (18) | LMB 24374 (22) | LMG 24377 (22) | |||||||
| RAATTY | + | + | + | + | + | + | + | + | + | + |
| GATC | − | − | − | − | + | + | + | + | + | − |
| TAAYN5TGC | + | + | − | + | +/− | − | − | − | − | − |
| CAAGAA | − | − | − | − | + | + | − | − | − | − |
| GCAAGG | + | +/− | +/− | + | − | − | − | − | − | − |
| GAGN5GT | − | + | − | +/− | − | − | − | − | − | − |
| CAAYN6ACT | + | − | − | − | − | − | − | − | − | − |
| GGRCA | + | − | − | − | − | − | − | − | − | − |
| GAGN5RTG | − | − | + | − | − | − | − | − | − | − |
| AN4CGNAAATTY | − | − | + | − | − | − | − | − | − | − |
| CTAN8TAYC | − | − | + | − | − | − | − | − | − | − |
| GAAGAA | − | − | − | + | − | − | − | − | − | − |
| RCATC | − | − | − | − | + | − | − | − | − | − |
| GGGTDA | − | − | − | − | + | − | − | − | − | − |
| DACATTGB | − | − | − | − | + | − | − | − | − | − |
| TAAAN5TTTA* | − | − | − | − | + | − | − | − | − | − |
| CACN5TTTA* | − | − | − | − | + | − | − | − | − | − |
| CAAYN7TTYG† | − | − | − | − | + | − | − | − | − | − |
| CRAAN7RTTG† | − | − | − | − | + | − | − | − | − | − |
| GGRAT | − | − | − | − | − | + | − | − | − | − |
| TAAYN6RTTG | − | − | − | − | − | + | − | − | − | − |
| ACCTA‡ | − | − | − | − | − | + | − | − | − | − |
| ATCN7TCC | − | − | − | − | − | − | + | − | − | − |
| GAYN7TYTC | − | − | − | − | − | − | + | − | − | − |
| GNCGAA | − | − | − | − | − | − | + | − | − | − |
| AACCA | − | − | − | − | − | − | + | − | − | − |
| GGAAY‡ | − | − | − | − | − | − | + | − | − | − |
| GAGG | − | − | − | − | − | − | − | + | − | − |
| GTRGAG | − | − | − | − | − | − | − | + | − | − |
| CCAAT | − | − | − | − | − | − | − | + | − | − |
| GWCAT | − | − | − | − | − | − | − | + | − | − |
| GCGAA | − | − | − | − | − | − | − | − | + | − |
| CAYN6RTG | − | − | − | − | − | − | − | − | + | − |
| GANTC | − | − | − | − | − | − | − | − | − | + |
| GCATC | − | − | − | − | − | − | − | − | − | + |
| CGAN8ACA | − | − | − | − | − | − | − | − | − | + |
Methylation motifs were determined using SMRT sequencing. Ambiguous nucleotides are denoted using IUPAC ambiguity codes. +/− indicates that the motif contains one nucleotide difference between the two strains. Underlining indicates the methylated base.
and † indicate double-stranded motifs where the methylation occurs in different spots on each strand.
Indicates that ≈50% of DNA was methylated.
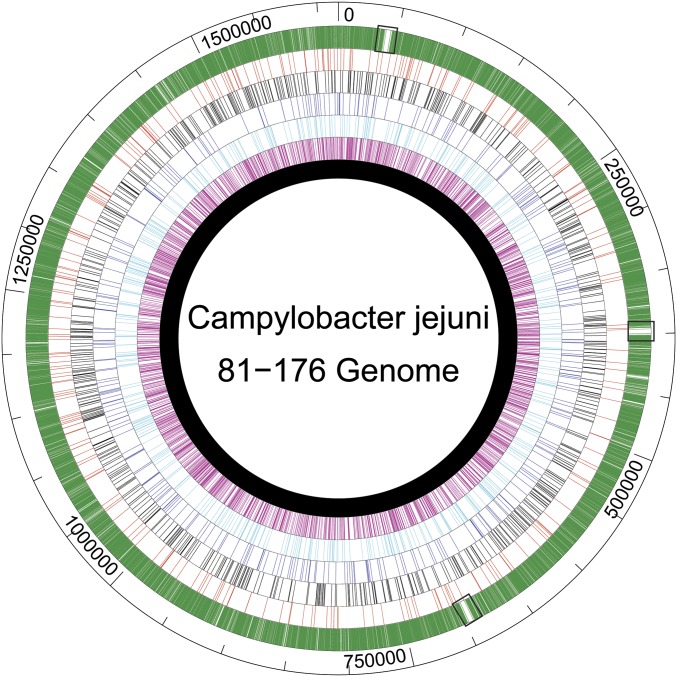
The single universally conserved methylation motif among these strains is RAm6ATTY. Not only is this motif methylated in all SMRT-sequenced strains of Campylobacter, it is overrepresented in the C. jejuni genome, and nearly all of the available sites are evidently methylated (26,609 out of 26,876) (18). Based on the genome size of C. jejuni, there is a RAATTY motif approximately every 60 bp, fourfold more frequent than predicted for a 6-base motif with the same degenerate base composition. This is significantly more frequent than the other methylation motifs identified, including another identified 6-bp recognition sequence, GCAAGG, for which 684 out of 700 total sites were methylated (Fig. 1) (18). We hypothesized that recognition of methylation of this highly conserved RAATTY motif would enable Campylobacter to favor DNA from close relatives over DNA from other species during transformation.
Fig. 1.
Distribution of all possible methylation motifs in 81–176. CtsM (RAATTY) sites are indicated in green. For comparison (in order toward the center), EcoRI (GAATTC) sites are in red, M.CjeFIII (GCAAGG) sites are in black, M.CjeFII (CAAYN6ACT) sites are in dark blue, M.CjeFIV (TAAYN5TGC) sites are in light blue, and M.CjeFV (GGRCA) sites are in fuschia. The chromosome is in black. Small black boxes indicate the ribosomal gene clusters.
In strain 81–176, this methylation mark is conferred by CJJ81176_0240, which we term ctsM (18). This N6 adenine-specific DNA MTase is an orphan, lacking a cognate restriction enzyme. Using a BLAST search, we determined that the gene is highly conserved among Campylobacter species, but has more limited distribution outside the genus. The top 100 BLASTp hits were for either C. jejuni or Camphylobacter coli, with identities >80%, while a BLASTp hit from H. pylori found only 30% identity. We hypothesized that CtsM plays a role in DNA discrimination during transformation, for two reasons. First, its high degree of conservation within Campylobacter implies an important role in a variety of habitats, while its lack of conservation outside the genus suggests that it might help discriminate Campylobacter DNA from non-Campylobacter DNA. Second, its lack of a cognate restriction enzyme as an orphan DNA MTase indicates that it might play a unique role in the biology of this bacterium outside of restriction-modification per se.
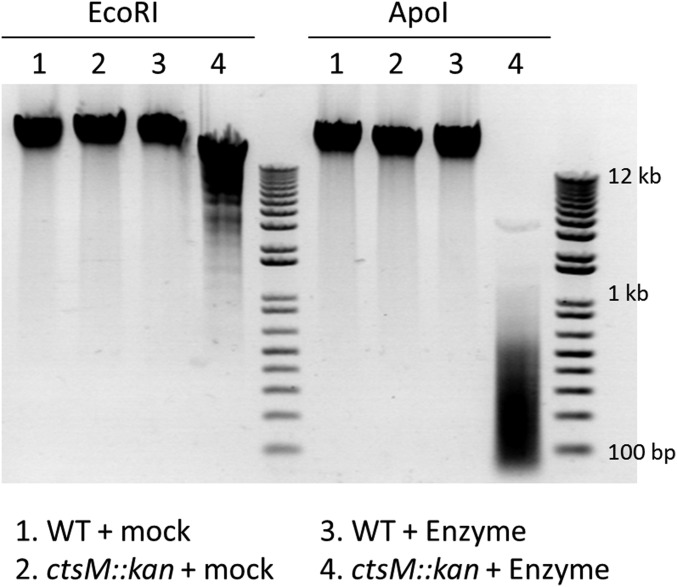
We constructed a ctsM::kan insertion/deletion strain, and assayed its methylation status by testing the sensitivity of DNA from the mutant strain to restriction by the REases EcoRI or ApoI. The recognition site for each REase includes the methylation substrate of CtsM (specifically, GAATTC for EcoRI and RAATTY for ApoI), and neither enzyme cleaves DNA when that site is methylated at the position acted on by CtsM. EcoRI and ApoI were able to digest genomic DNA (gDNA) from the mutant strain, but not gDNA from the WT strain (Fig. 2), confirming that loss of CtsM in the cell alters methylation of the DNA at RAATTY.
Fig. 2.
DNA from ctsM::kan is degraded by ApoI and EcoRI. gDNA was purified from either WT C. jejuni strain DRH153 (kanR) or ctsM::kan mutants. The DNA was then either mock-digested (lanes 1 and 2) or digested (lanes 3 and 4) with the restriction enzymes EcoRI (Left) or ApoI (Right). The DNA was then electrophoresed on an agarose gel and visualized using ethidium bromide. ApoI (RAATTY) should cut approximately four times as frequently as EcoRI (GAATTC).
DNA Not Methylated at RAm6ATTY Is a Poor Transformation Substrate.
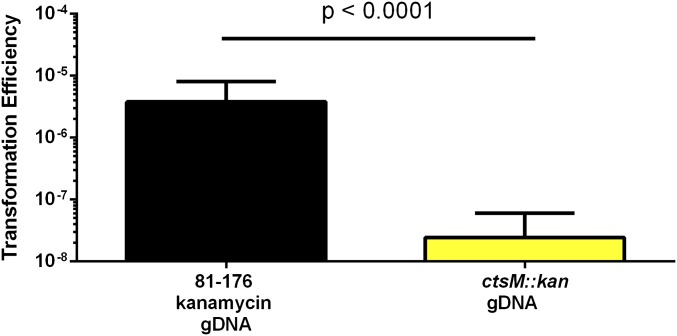
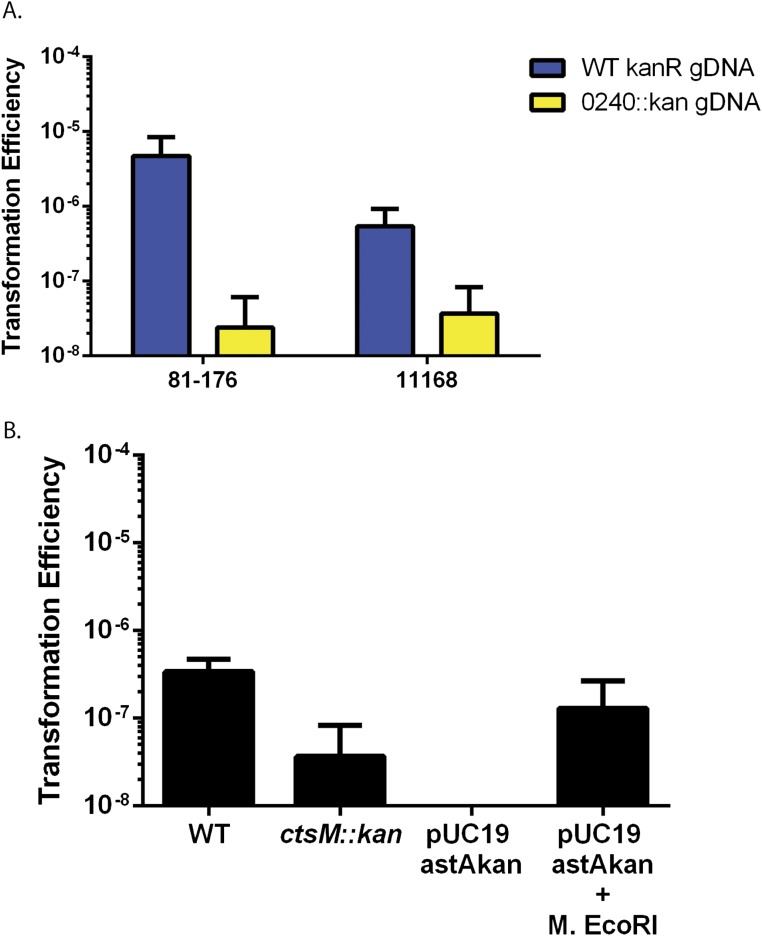
Having confirmed the disruption of DNA methylation in the ctsM::kan mutant strain, we tested whether methylation at RAATTY affects the transformation frequency of DNA. gDNA was purified from a strain carrying a marker for kanamycin resistance in the astA gene (CtsM+) or from the ctsM::kan (CtsM−) mutant strain. Both were used in separate transformation assays to transform WT C. jejuni strain DRH212. The mutant gDNA from the CtsM− cells, lacking methylation at the RAATTY site, transformed to ∼200-fold lower levels than gDNA from the CtsM+ cells, indicating that the RAm6ATTY methylation mark is critical for the ability of gDNA to serve as a substrate in transformation (Fig. 3).
Fig. 3.
DNA lacking CtsM methylation is a poor substrate for transformation. DNA was purified from either DRH153 or the ctsM::kan mutant. This DNA was then used to transform WT cells. Transformation efficiency was calculated as described in Materials and Methods. Results are the compilation of three biological replicates, each of which included three technical replicates. Error bars indicate SD. Significance was determined using the Mann–Whitney U test.
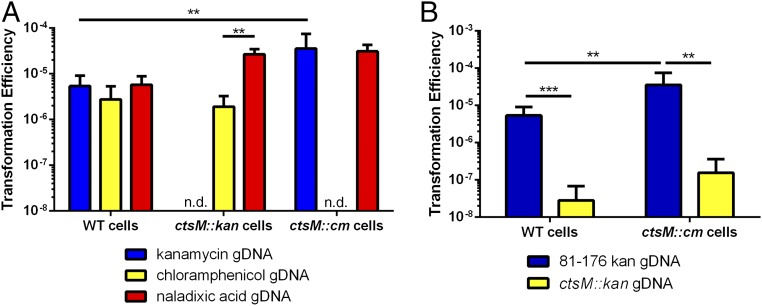
We tested the ability of the ctsM mutant strain itself to be transformed, using gDNA from cells containing different antibiotic resistance marker cassettes or alleles (kanamycin and chloramphenicol cassettes at the astA locus, or a gyrA allele encoding resistance to naladixic acid). Purified gDNA from these different antibiotic resistance mutants was used in transformation assays with WT and ctsM mutant C. jejuni. No significant difference from WT in transformation frequency of the ctsM mutants was detected when using these gDNAs as substrates (Fig. 4A). This indicates that the MTase does not affect the ability of the cell to be transformed; rather, it is just required for the DNA to act efficiently as a substrate in transforming other cells. Similar to WT cells, the ctsM mutants were severely deficient in their ability to be transformed with gDNA lacking methylation at RAATTY (Fig. 4B). Collectively, our data suggest that ctsM is required for methylation at the RAATTY site and, while not required for cells to be competent for transformation, is required for DNA to serve as an efficient transformation substrate.
Fig. 4.
ctsM mutants are transformed with WT frequency by WT gDNA, but not by their own gDNA. (A) WT DNA with varying antibiotic resistance [kanamycin (DRH153), chloramphenicol (DRH154), and naladixic acid (JMB202)] was purified and used to transform WT, ctsM::kan, and ctsM::cm cells. Transformation efficiency was calculated as described previously. n.d. indicates that transformation efficiency was not determined because the cells already contained the antibiotic resistance used to transform cells. (B) DRH154 cells or ctsM::cm cells were transformed with either WT kanR gDNA or DNA from the ctsM::kan gDNA, and transformation efficiency was measured as described previously. Data represent three biological replicates, each of which contained three technical replicates. Error bars indicate SD. **P < 0.01; ***P < 0.001, Mann–Whitney U test.
In Vitro Methylation Enables Transformation of DNA from a ctsM::kan Mutant.
The ctsM mutant strain produces gDNA that (i) serves as a poor substrate for natural transformation and (ii) is more readily digested by enzymes (EcoRI and ApoI) that are inhibited by methylation at the site modified by this methyltransferase. To test more directly whether the methylation mark provided by CtsM is required for transformation, we took advantage of the fact that EcoRI MTase (M.EcoRI) recognizes a subset of RAATTY sites, GAATTC. We used commercially obtained M.EcoRI to methylate gDNA from the ctsM::kan strain. As controls, we used two other commercially available MTases, M.TaqI and M.HpaII, which methylate the sites TCGm6A and Cm5CGG, respectively. Given the position of the methylated bases, the HpaII MTase would interfere with neither of the REases that we used, even though its target could overlap the CtsM sequence (e.g., RAATTC5mCGG). M.TaqI methylation would block 1/8 ApoI sites, because ApoI REase is sensitive to methylation of the first A (TCGm6AATTY) (25), although it would not affect EcoRI REase, because methylation of the first A (TCGm6AATTC) does not block cleavage (26).
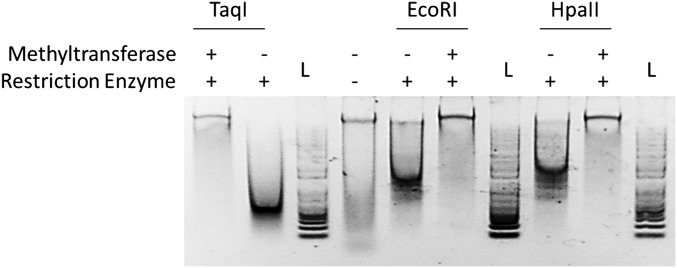
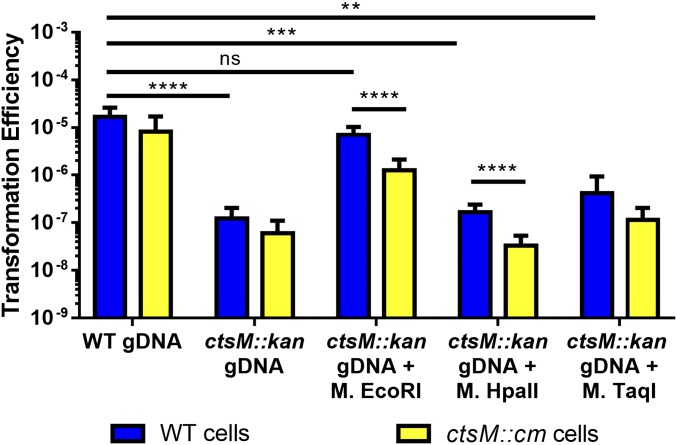
After treatment with the MTases, purified gDNA from ctsM::kan was protected from digestion by each cognate restriction enzyme, confirming successful methylation of the gDNA (Fig. S1). We then measured the transformation efficiency of ctsM+ C. jejuni with these methylated gDNAs as substrates, compared with transformation with gDNA from the ctsM+ strain (Fig. 5, blue bars). The gDNA methylated by M.EcoRI transformed as well as gDNA from ctsM+ C. jejuni, with a frequency ≈100-fold greater than that of gDNA isolated from the ctsM mutant that had not been methylated by M.EcoRI or that had been methylated by M.TaqI or M.HpaII.
Fig. S1.
Digest of gDNA treated with methyltransferases. ctsM::kan gDNA was treated as indicated. The DNA was then electrophoresed on an agarose gel and visualized using Gel Red. L, marker ladder.
Fig. 5.
In vitro methylation at the CtsM site RAATTY, but not at other sites, significantly enhances gDNA substrate capability. DNA was purified from either DRH153 (kanr) or ctsM::kan cells. ctsM::kan DNA was then either mock-methylated or in vitro methylated with EcoRI MTase, TaqI MTase, or HpaII MTase. EcoRI MTase methylates GAm6ATTC, TaqI methylates TCG m6A, and HpaII methylates C m6CGG. WT cells (blue) or ctsM::cm cells (yellow) were transformed with the methylated DNA. Transformation efficiency was calculated as described previously. Data represent three biological replicates, each of which contained three technical replicates. Error bars indicate SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Kruskal–Wallis test with Dunn’s multiple-comparisons test. ns, not significant.
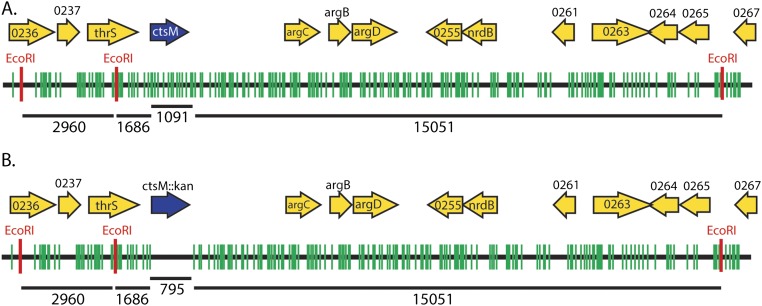
These results indicate that methylation of the RAATTY site is required for WT levels of transformation. Furthermore, while the M.EcoRI site overlaps with and methylates the same site as CtsM, the site is more specific (GAATTC vs. RAATTY) than the site recognized by the Campylobacter enzyme. Given that fewer overall sites will be methylated by M.EcoRI than by CtsM (∼550 vs. 26,609), our data also suggest that not every genomic RAm6ATTY site must be methylated for transformation to occur. As shown in Fig. S2B, there are no EcoRI sites in the kanamycin cassette. The nearest EcoRI site is 1,686 bp upstream of the insertion; the closest site 3′ is >15 kb away. While M.EcoRI has some “star” activity that results in modification of sequences differing at one position from GAATTC, that off-site activity requires high enzyme concentrations and buffer conditions that were avoided here (27). This indicates that the methylated RAATTY site can be at least 1,600 bases away from the DNA that we know recombines (i.e., the antibiotic cassette) and still allow for transformation.
Fig. S2.
Chromosome location of ctsM with methylation RAATTY and GAATTC sites. (A) WT chromosome with RAATTY sites (green) and EcoRI sites (red) shown for DNA surrounding ctsM. ORFs are indicated by yellow arrows; those smaller than 200 aa are not shown. ctsM is shown in blue. Numbers under chromosomes indicate bases. (B) RAATTY sites and EcoRI sites in a ctsM::kan mutant.
In Vitro Methylation by M. EcoRI Is Sufficient to Allow E. coli Plasmids and PCR Products to Transform Campylobacter.
Relative to gDNA purified from WT C. jejuni, DNA from laboratory strains of E. coli or derived in vitro by PCR is a poor substrate for Campylobacter transformation. The mechanism for this discrimination is unclear. After showing that methylation of RAATTY is necessary for transformation, we next wanted to determine whether this methylation is also sufficient for transformation.
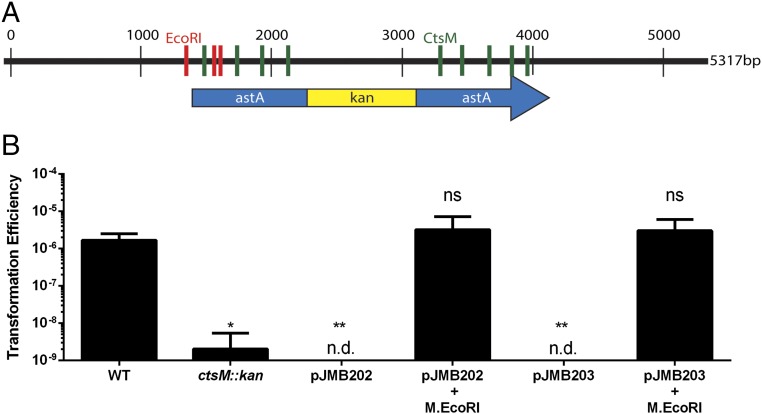
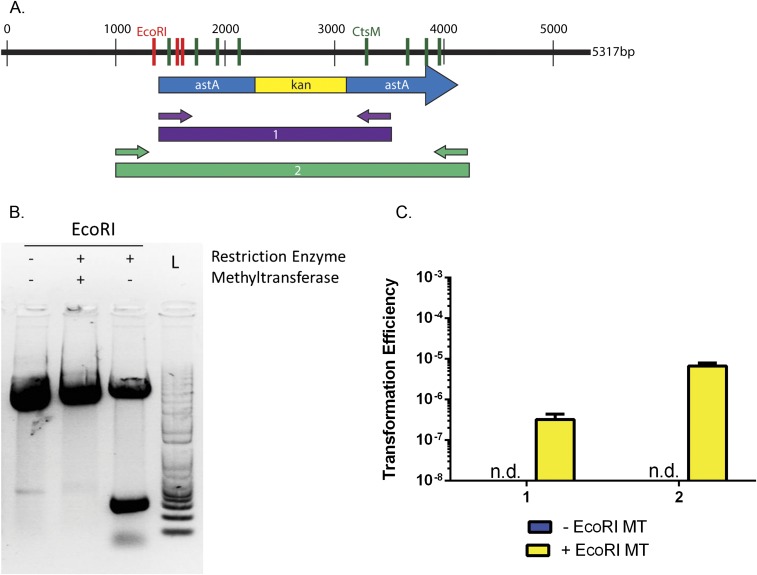
We constructed antibiotic cassettes flanked with the C. jejuni astA sequence to allow for homologous recombination, and cloned them into pUC19, which does not replicate in C. jejuni. The resulting plasmids have three EcoRI sites, two in astA and one in the pUC19 vector (Fig. 6A). We purified these plasmids from E. coli, methylated them with M.EcoRI, confirmed their methylation using restriction digest (Fig. S3B), and assessed their ability to transform DRH212 by selecting for the antibiotic cassette (Fig. 6B). Plasmids treated with M.EcoRI transformed C. jejuni at frequencies similar to WT gDNA, and at 1,000-fold greater frequencies than unmethylated DNA of the same origin.
Fig. 6.
In vitro methylation by M.EcoRI allows for transformation of plasmid DNA. (A) Plasmid map of pJMB202 with pertinent features denoted. KanR refers to a kanamycin resistance cassette. It is inserted in astA. Red lines indicate EcoRI sites (GAATTC) that are methylated by M.EcoRI. Green lines indicate CtsM sites (RAATTY) that cannot be methylated by M.EcoRI. (B) WT cells were transformed with various types of DNA as indicated. These DNA fragments were then used to transform WT cells. Transformation efficiency was calculated as described previously. The data represent three biological replicates, each of which contained three technical replicates. n.d. indicates that colonies were not detected. Error bars indicate SD. *P < 0.001; **P < 0.001, one-way ANOVA with Tukey’s posttest. ns, not significant.
Fig. S3.
Digests of plasmids treated with M. EcoRI and PCR transformation data. (A) Diagram of pJMB202 and PCR products generated from this plasmid. GAATTC (M.EcoRI sites) are shown in red, and other RAATTY sites are shown in green. Small arrows indicate primers. (B) pJMB202 was treated as indicated. DNA was then electrophoresed and visualized using Gel Red. (C) PCR fragments derived from pJMB202 as indicated in A were methylated in vitro using M.EcoRI. ctsM+ cells were then transformed with DNA that either had not been methylated (blue) or had been methylated (yellow). Transformation efficiency was calculated as described in Materials and Methods. The dashed line indicates the limit of detection. n.d. indicates that colonies were not detected. Error bars indicate SD.
We also tested the ability of methylated PCR products to transform C. jejuni. We designed primers to amplify two PCR products with increasing amounts of flanking DNA from the plasmid described above. These fragments were amplified and methylated with M.EcoRI. Both products transformed DRH212 at frequencies much higher than their nonmethylated cognate fragments (Fig. S3C).
One Methylated RAATTY Site Is Sufficient for Transformation.
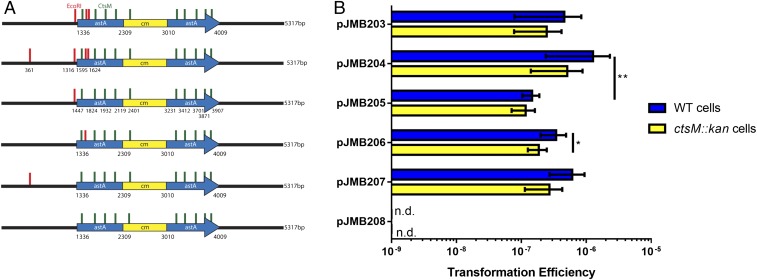
To determine whether the location and number of methylated sites affect transformation, we performed site-directed mutagenesis on pJMB203 to generate plasmids pJMB204-208, either adding (JMB204) or eliminating (JMB205-208) EcoRI sites (Fig. 7A). This allowed us to determine the number of methylated RAATTY sites required for transformation, as well as the location of the sites relative to the recombining DNA. A plasmid, JMB207, which has only one methylated site located 1,300 bases away from the recombining DNA, transformed to similar efficiencies as plasmids with one or more sites located within the recombining DNA. Based on our data, we determined that one site is sufficient to allow for transformation, and that the site does not need to be located within the DNA segment that will ultimately recombine (Fig. 7B). The site can be at least 1 kb away without significantly impacting transformation efficiency.
Fig. 7.
One methylated EcoRI site is sufficient for transformation. (A) Diagram of the plasmids used. EcoRI sites (GAATTC) are in red, and CtsM sites (RAATTY) that are not methylated by M.EcoRI are shown in green. The exact sites of the EcoRI sites are indicated for pJMB204. The plasmids are identical except for the location and number of EcoRI sites. The plasmid backbone is indicated in black, and regions homologous to C. jejuni are indicated in blue. The antibiotic cassette is in yellow. (B) WT cells (blue) or ctsM::kan cells (yellow) were transformed with different plasmids as indicated, and transformation efficiency was measured as before. n.d. indicates that colonies were not detected. Error bars indicate SD. *P < 0.01; **P < 0.01, Kruskal–Wallis test with Dunn’s multiple-comparison posttest.
Partially Methylated DNA Transforms Cells Lacking CtsM, as well as Cells Containing CtsM.
As discussed earlier, in the context of DNA discrimination, MTases generally function as part of a RM system in which DNA that lacks the appropriate methylation is degraded by REases. However, CtsM is an orphan MTase and does not have a cognate REase. This is based on (i) the absence of an adjacent REase gene [although in rare instances cognate MTase and REase genes are widely separated on the chromosome (28)] and (ii) the ability to inactivate CtsM without also inactivating a corresponding REase, which would otherwise degrade the chromosome. Therefore, the basis for DNA discrimination against unmethylated DNA is unclear. Our data demonstrate that partially methylated DNA transforms WT cells.
It is possible that the partial methylation allows the DNA to enter the cell, where endogenous CtsM will methylate the remaining sites and protect the DNA from degradation by an unidentified restrictive factor. To further rule out such a restrictive factor, we asked whether cells lacking CtsM could be transformed with DNA that is partially methylated at the RAATTY site. Because these recipient cells lack CtsM, they cannot further methylate the DNA, leaving unmethylated sites susceptible to degradation by a restrictive factor. We again used DNA methylated in vitro by M.EcoRI. As mentioned earlier, this enzyme can methylate only a subset (∼25%) of all the RAATTY sites. Using gDNA purified from ctsM− cells and methylated in vitro with M.EcoRI, M.HpaII, or M.TaqI, we determined how well that DNA transformed cells lacking CtsM (Fig. 5, yellow bars). If a restrictive factor were present, it would be able to degrade the partially methylated (i.e., M.EcoRI-modified) DNA, as well as the control methylated DNAs. As shown in Fig. S2, the number of unmethylated sites (green) greatly outnumbered the number of EcoRI methylated sites (red); however, the partially methylated DNA transformed CtsM− cells to almost an equivalent degree as CtsM+ cells. While we did observe a small difference (<0.5 log), this difference is much smaller than that in cases where the DNA lacks any RAATTY methylation and thus is nontransformable.
We also tested the same hypothesis using E. coli-derived plasmid DNA. The plasmids used to determine the number of methylated sites required for successful transformation were methylated in vitro, after which we measured how well they transformed cells lacking CtsM. In this case, we know that there are eight RAATTY sites located within the homologous DNA and one site located within the antibiotic cassette that will not be methylated by M.EcoRI, aas well as between zero and four sites that will be methylated. Both CtsM+ and CtsM− cells were transformed equivalently by these plasmids (Fig. 7B).
Methylated RAATTY Promotes DNA Uptake.
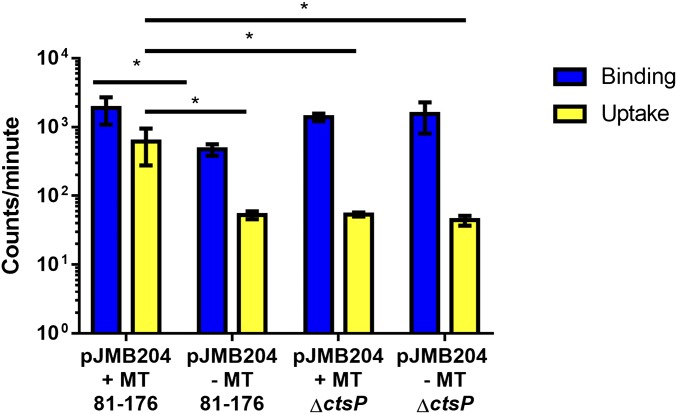
To determine mechanistically how the cell discriminates between methylated and unmethlyated DNA and where in the transformation process that discrimination takes place, we performed a DNA binding and uptake assay (29) as described in Materials and Methods. This assay can determine whether the DNA efficiently binds to the outside of the cell and is transported into the cells, where it becomes resistant to DNase degradation. We observed that DNA methylated by M.EcoRI was taken up by WT cells with significantly more efficiency compared with unmethylated DNA (Fig. 8). The low uptake of nonmethylated DNA is similar to that observed with methylated DNA uptake by a mutant lacking ctsP, a previously described putative NTPase essential for transformation in C. jejuni (29, 30). Nonmethylated DNA consistently bound less well to WT cells compared with M.EcoRI-methylated DNA, while previous methylation did not influence DNA binding to ctsP mutant bacteria.
Fig. 8.
Methylated RAATTY promotes DNA uptake. DRH212 cells were incubated with radiolabled pJMB204 that had been methylated with M.EcoRI (+MT) or mock-methylated (−MT) for 1 h. The methyltransferase experiments were initiated with 5 ng/µL of DNA. Cells were centrifuged, washed, split, and either mock-treated or treated with DNase. Mock-treated cells contain the bound DNA (blue), and DNase-treated cells contain DNA that has been transported into the cells (yellow); 100 ng of DNA was used in each experiment. After treatment, cells were washed and resuspended in scintillation fluid. Total cpm was measured using a scintillation counter. Data are representative of the experiment repeated three times. Error bars indicate SD of three technical replicates. *P < 0.05, two-way ANOVA and Tukey’s posttest.
Discussion
The goal of this study was to determine how C. jejuni discriminates extracellular DNA for incorporation during natural transformation. We identified a methylation motif, RAm6ATTY, that plays a critical role in the discrimination mechanism. DNA with this site methylated transforms cells at a frequency several orders of magnitude greater than DNA lacking this methylation mark. The 26,609 methylated RAATTY sites are distributed fairly equally around the genome, with the exception of three regions with significantly fewer sites (Fig. 1). These regions harbor the three copies of the 81–176 ribosomal gene cluster, where there are only five RAATTY sites over ∼5 kb. In contrast, the 5 kb flanking these clusters average 40 RAATTY sites. This dearth of RAATTY sites might affect the ability of sites to be exchanged during natural transformation. Analyses of uptake sequences in several species suggests that at a minimum, such sequences are found with above-average frequencies in genome maintenance genes (including ribosomal gene clusters) compared with other coding sequences (31, 32), although the significance of this is unclear (32).
A DNA fragment does not need to have all of its RAATTY sites methylated to be a substrate for transformation. This was demonstrated using EcoRI MTase, which methylates the more specific, and thus less frequent, GAATTC. There are ∼550 GAATTC sites in 81–176 gDNA, as opposed to 26,609 methylated RAATTY sites. Furthermore, E. coli-derived or PCR-generated DNA, which is typically a poor substrate for transformation, became a much better substrate after methylation by the EcoRI MTase. Furthermore, we demonstrated, using plasmids containing different numbers of EcoRI sites, that one methylated EcoRI site on the plasmid is sufficient for transformation. This argues strongly against a REase-based system, because the plasmids and the PCR products contain unmethylated RAATTY sites within DNA that will ultimately recombine onto the chromosome that would be targeted by a restriction enzyme. That a single methylated RAATTY site on a relatively long piece of DNA confers transformability calls into question the need for so many methylated RAATTY sites within the genome, and suggests that these sites may play other roles in the biology of the cell. For example, they could be epigenetic marks that influence patterns of gene expression.
To our knowledge, there are no previous reports of a methylation-dependent DNA discrimination process for natural transformation that does not involve a restriction enzyme. The discrimination process is distinct from what has been observed in bacteria such as H. pylori, N. meningitidis, and P. stutzeri, all of which use methylation-associated RM systems that degrade DNA lacking the correct methylation motif (12–16). CtsM is an orphan MTase lacking an obvious cognate REase, analogous to the Dam methylase of many γ-proteobacteria. Furthermore, inactivation of ctsM did not alter the fitness of C. jejuni during laboratory growth, indicating an absence of DNA damage caused by an unidentified REase. In addition, we were able to use plasmid DNA containing nine unmethylated RAATTY sites to transform the cells. If a restriction enzyme existed, it would cleave the plasmid at these sites, eliminating transformation. For our plasmid experiments, while most of the unmethylated RAATTY sites are located within astA, one site is located 282 bases into the chloramphenicol gene. If a restriction enzyme existed, it would render this gene nonfunctional, and we would not be able to detect chloramphenicol-resistant transformants. Linearizing DNA can increase its transformability by recruiting RecBCD (33); however, the plasmid the we used has nine cleavable RAATTY sites, including one site in the middle of the chloramphenicol gene. Therefore, even if there were an increase in transformation due to the linearity of the DNA, the chloramphenicol gene would be disrupted, and both pieces of this gene would have to independently recombine onto the chromosome perfectly to restore the double-stranded break and allow for chloramphenicol-resistant transformants.
We have also demonstrated that methylated RAATTY functions effectively as an uptake sequence for C. jejuni. RAATTY-methylated DNA is transported into the cell, whereas DNA lacking methylation at RAATTY is not. This is distinct from other uptake sequences, which are longer and not methylation-dependent. If this system functions similarly to those in Neisseria and Haemophilus, then we predict that there is an outer membrane protein that will recognize methylated RAATTY and promote translocation of the DNA into the cell. In Neisseria, this protein is ComP, a minor pilin (34). C. jejuni has two minor pilin genes, ctsG and ctsT, which have been found to be defective in DNA uptake, but their role in transformation has yet to be determined (29). It is possible that these pilins may play a similar role as ComP in Neisseria.
Although the difference is not great, unmethylated DNA binds less well to WT C. jejuni than to ctsP mutant bacteria. This finding warrants further exploration to uncover the underlying mechanism; however, speculation about it is reasonable. The ctsP gene encodes a putative NTPase that presumably is required for some energy-dependent step in the process of binding and/or uptake. CtsP is one of two NTPases (along with CtsE) required for transformation (29, 30). A translocation ratchet mechanism has been proposed for DNA uptake in Neisseria gonorrhoeae, although not directly through pilus retraction per se (35). If CtsP somehow controls a similar ratchet mechanism by regulating proteins that selectively bind CtsM-methylated DNA better than nonmethylated DNA, then in a CtsP mutant, that protein (and the corresponding bound DNA) might accumulate on the surface of the cell, because the cell would no longer be capable of ratcheting it in. This hypothesis is not perfectly consistent with the proposed ratchet mechanism, but provides a basis for further investigation.
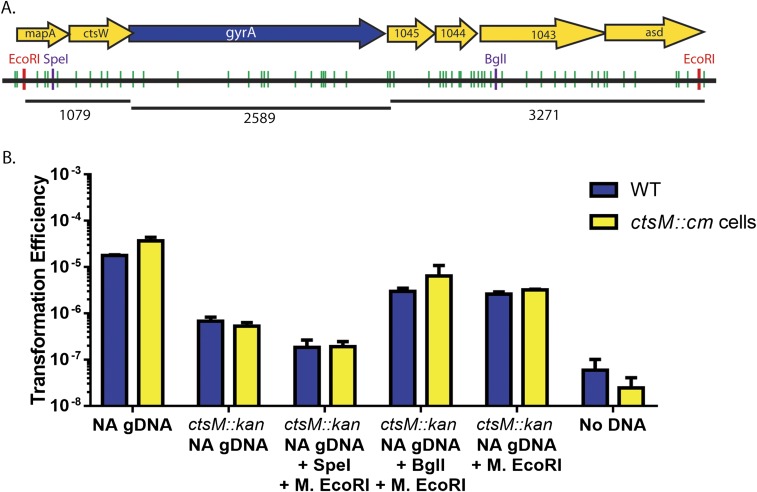
In addition to identifying a DNA discrimination motif, we have introduced a potentially powerful tool for genome editing in C. jejuni. We have shown that to successfully transform C. jejuni, it is only necessary to methylate DNA in vitro with M.EcoRI. This may enable transformation of E. coli plasmids or PCR products to make mutations on the chromosome. Furthermore, only a single nearby methylation event is required, and that methylation event does not have to be on the DNA segment that will ultimately recombine into the chromosome. Both our plasmid data (Fig. 7) and experiments using in vitro methylation of the chromosome (Fig. 5 and Fig. S4) indicate that the methylated site can be up to 1.6 kb away from the antibiotic cassette; however, there does seem to be an upper limit to this as a single methylated site located 3.2 kb away from the antibiotic cassette does not allow for efficient transformation (Fig. S4). This upper limit could be related to two different mechanisms. First, large pieces of single-stranded DNA eventually become degraded inside the cell, decreasing the transformation efficiency. Second, recombination might be inefficient over large stretches of DNA, and so the upper limit is based on recombination efficiency.
Fig. S4.
Sites 3.2 kb away from an antibiotic cassette are not sufficient for transformation. (A) Chromosome locations of GAATTC and RAATTY sites at gyrA. WT chromosome with RAATTY sites (green) and EcoRI sites (red) shown for DNA surrounding ctsM. ORFs are indicated by yellow arrows; those smaller than 200 aa are not shown. gyrA is shown in blue. Numbers under chromosomes indicate bases. SpeI and BglI sites are indicated in purple. A point mutation in gyrA confers naladixic acid (NA) resistance. (B) gDNA was purified from either WT NAR cells or ctsM::kan NAR cells. The DNA was then digested with either SpeI or BglI and in vitro methylated with M.EcoRI. The treated DNA was used to transform WT (blue) or ctsM::kan (yellow) cells. Transformation efficiency was measured as described in Materials and Methods. Error bars indicate SD. Data are the results of three biological replicates.
There is evidence of a restriction barrier in C. jejuni. This barrier is partially explained by identification of the RM system encoded in C. jejuni strain 11168. Inactivation of this RM system significantly increased the transformation efficiency of plasmids derived from Campylobacter; however, it did not allow for transformation of E. coli-derived plasmids (17). Furthermore, this REase is not found in all strains of C. jejuni and is in fact absent in 81–176. Because WT 81–176 cannot be transformed by E. coli-derived plasmids, this RM system cannot fully explain the demonstrated restriction barrier. CtsM appears to explain more of the observed restriction barrier, as it is present in all sequenced Campylobacter strains examined to date (60 of 60 as of December 2016). Moreover, we were able to transform WT C. jejuni with both plasmid- and PCR-derived DNA after methylation. In this case, we simply had to modify the substrate, and not the cell, to allow transformation. Finally, we found that RAATTY methylation allows transformation of E. coli-derived plasmids into strain 11168 (Fig. S5). This strain transformed less well than 81–176, presumably due to the Cj1051c RM system.
Fig. S5.
Transformation efficiencies of 11168. (A) 11168 and 81–176 were transformed with gDNA derived from WT 81–176 or 81–176 ctsM::kan. (B) 11168 was transformed by the indicated DNA. WT and ctsM::kan are from 81–176. Transformation efficiency was calculated as before. Error bars indicate SD.
CtsM and its cognate methylation motif are conserved among the PacBIo-sequenced Campylobacter strains. These strains are diverse and range from laboratory C. jejuni isolates to the more distantly related Campylobacter subantarcticus and Campylobacter peloridis isolates. Of more than 35 identified methylation motifs, this is the only motif conserved among all SMRT-sequenced strains evaluated thus far. If this is a conserved requirement for DNA in Campylobacter competence, then we would predict that this motif will be found in all naturally competent strains of Campylobacter to allow the strain to share its DNA with related cells. Our ability to insertionally inactivate the DNA MTase demonstrated that it is not required for growth in vitro, although its strict conservation throughout the genera indicates that it might have other roles. To this point, while remaining quite viable and able to form single colonies, the ctsM mutant strain does grow less well than WT and reaches a lower optical density in standard Campylobacter media compared with the WT strain. Loss of CtsM would not limit a strain’s ability to import DNA from other cells, but its own genes would lose the capacity for horizontal transfer, such that any alleles unique to CtsM− strains would possibly tend to decrease in frequency relative to alleles in WT strains.
Orphan MTases are a growing area of study, and are now recognized to play diverse roles in gene regulation. For example, Dam MTases in Gammaproteobacteria regulate the timing of DNA replication, and CcrM MTases in Alphaproteobacteria regulate cell cycle progression (36–42). Because orphan MTases are more conserved at the genus, family, and class levels than MTases in RM systems, it is hypothesized that they have a functional role distinct from host genome protection (43). Among the more than 230 diverse bacteria, the largest class of orphan MTases comprises those that methylate RAATTY (43). These orphan MTases are found in Campylobacter, Akkermanisia muciniphilia, three Spirochaeta species, and three Treponema species (43). In Spirochaeta smaragdinae, RAATTY sites in the promoters of transcriptional regulators were not methylated, leading to the suggestion that that these orphan MTases play a role in gene regulation (43). Hypomethylation of RAATTY also has been identified in C. jejuni in front of rRNAs, as well as the flagellar genes FlgK and a flagellar basal-body rod protein (20). This implies a role for CtsM in regulation of flagella.
In conclusion, we have identified a methylation-dependent DNA uptake sequence in C. jejuni. RAATTY methylation is required for efficient transformation, and when this site is methylated, previously nontransformable substrates become capable of transforming C. jejuni. Our data show that RAATTY methylation is required for transformation, and that DNA lacking RAATTY methylation is not transported into the cell. One hypothesis is that, analogous to Neisseria (34), there is a protein on the outer membrane that binds and recognizes methylated RAATTY DNA such that only methylated DNA is brought into the cell.
Materials and Methods
Bacterial Strains and Media.
The bacterial strains used in this work are listed in Table S2. C. jejuni was routinely cultured on Muller Hinton (MH) agar with 10% sheep’s blood in microaerophilic conditions (5% CO2, 10% O2, balanced with N2) in a trigas incubator at 42 °C. When necessary, media were supplemented with antibiotics at the following concentrations: trimethoprim, 10 µg mL−1; kanamycin, 150 µg mL−1; naladixic acid, 30 µg mL−1; and chloramphenicol, 15 µg mL−1. All C. jejuni strains were stored at −80 °C in MH broth with 20% glycerol.
Table S2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source |
| Bacteria | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (Ф80lacZΔM15) hsDr17 recA1 endA1gyrA96 thi-1 relA1 | Lab strain |
| GM2163 | dam-, dcm-, camR | New England BioLabs |
| DH5α pRK212.1 | Contains conjugative plasmid for transferring DNA into Campylobacter | (46) |
| C. jejuni | ||
| 81–176 | Clinical isolate | (47) |
| 11168 | Clinical isolate | |
| DRH153 | 81–176 astA::aphA3; Kanr | (48) |
| DRH154 | 81–176 astA::cat, CmR | (48) |
| DRH212 | 81–176 rpsLSm (this strain is referred to as “wild type” throughout) | (48) |
| JMB200 | DRH212 ctsM::kan, KanR | This study |
| JMB202 | DRH212 NAR | This study |
| JMB204 | DRH212 ctsM::cm, CmR | This study |
| Plasmids | ||
| pUC19 | AmpR | New England BioLabs |
| pEC0102 | pRY112 derivative with cat promoter | (29) |
| pJMB200 | pUC19 ctsM::kan, KanR | This study |
| pJMB201 | pUC19 ctsM::cm, CmR | This study |
| pJMB202 | pUC19 astAkan, KanR | This study |
| pJMB203 | pUC19 astAcm, CmR | This study |
| pJMB204 | pJMB203 + new EcoRI site at 4053, CmR | This study |
| pJMB205 | pJMB203 only 1 EcoRI site in MCS, CmR | This study |
| pJMB206 | pJMB203 only 1 EcoRI, in astA, CmR | This study |
| pJMB207 | pJMB203 only 1 EcoRI site at 4053 (away from insert), CmR | This study |
| pJMB208 | pJMB203 no EcoRI sites, CmR | This study |
E. coli strains were routinely cultured at 37 °C in LB broth or agar. When necessary, antibiotics were used at the following concentrations: ampicillin, 100 µg mL−1; chloramphenicol, 30 µg mL−1; kanamycin, 50 µg mL−1; and tetracycline, 12.5 µg mL−1. All E. coli strains were stored at −80 °C in LB broth with 20% glycerol.
Construction of ctsM::kan and ctsM::cm Mutants.
Insertion mutants were created using Gibson assembly (44). We designed primers using the NEBuilder Assembly Tool (New England BioLabs). The primers used in this study are listed in Table S3. We amplified 500 bp upstream and downstream of ctsM, as well as either the kanamycin cassette or a chloramphenicol cassette from DRH153 or DRH154, respectively. These fragments were purified using the QIAgen PCR Purification Kit (Qiagen) and quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific). pUC19 was also digested with BamHI and SalI. The digested plasmid was purified using the QIAgen PCR Purification Kit and quantified using a Qubit 2.0 fluorometer. The PCR fragments and the digested plasmid were then mixed in equimolar ratios using Gibson Assembly Master Mix (New England BioLabs) following the manufacturer’s instructions, followed by a 60-min incubation at 50 °C. They were then transformed into NEB 5-alpha Competent E. coli (High Efficiency) cells (New England BioLabs) according to the manufacturer’s instructions, and then plated on LB kanamycin or LB chloramphenicol. All subsequent clones were verified by sequence determination. Plasmids were transformed into C. jejuni using electroporation and plated on MH agar with appropriate antibiotics. gDNA was purified, and generation of the mutant was confirmed using PCR.
Table S3.
Primers used in this study
| Primer no. | Primer name | Sequence (5′-3′) | Description |
| 145 | 0240 kan/CM GA 1 | ATT CGA CGT CGG TAC CCG GGT GTG TTG GCG ATG ATG GC | Used to make 0240::kan and 0240::cm insertional mutants |
| 146 | 0240 Kan GA 2 | TCA TTT TAG CCA TAA TTT AAA ACC TTT AAA AGC CTT ATC CAC | Used to make 0240::kan insertional mutant |
| 147 | 0240 Kan GA 3 | GGT TTT AAA TTA TGG CTA AAA TCA CAA TAT CAC | Used to make 0240::kan insertional mutant |
| 148 | 0240 Kan GA 4 | AGC ACA AAA TCT AAA ACA ATT CAT CCA GTA AAA TAT AAT ATT TTA TTT TTC | Used to make 0240::kan insertional mutant |
| 149 | 0240 Kan GA 5 | ATT GTT TTA GAT TTT GTG CTA AAA TTA TAT TTA TAA AAA GG | Used to make 0240::kan insertional mutant |
| 150 | 0240 Kan/CM GA 6 | CAA GCT TGC ATG CCT GCA GGC TTT TAT TGA GAC TAA TCT TAC TCA TTT TTA AAA C | Used to make 0240::kan and 0240::cm insertional mutants |
| 151 | 0240 CM GA 2 | TTA ACT TGG AAA TTT AAA ACC TTT AAA AGC CTT ATC CAC | Used to make 0240::cm insertional mutant |
| 152 | 0240 CM GA 3 | GGT TTT AAA TTT CCA AGT TAA TTG CGT GAT ATA G | Used to make 0240::cm insertional mutant |
| 153 | 0240 CM GA 4 | AGC ACA AAA TTG CGC CCT TTA CTT CCT AAA G | Used to make 0240::cm insertional mutant |
| 154 | 0240 CM GA 5 | AAA GGG CGC AAT TTT GTG CTA AAA TTA TAT TTA TAA AAA GG | Used to make 0240::cm insertional mutant |
| 183 | pUC19astA F | GGC ACT TTT GGC GGG TTC TAC C | Used to create astAkan PCR fragment |
| 184 | pUC19astA R | GGA TCA TAA TCA ACG CTA TTT ACA TGA GCC C | Used to create astAkan PCR fragment |
| 185 | M13 F | TGT AAA ACG ACG GCC AGT | Used to create M13 PCR product |
| 186 | M13 R | CAG GAA ACA GCT ATG ACC | Used to create M13 PCR product |
| 187 | astA1 F GA | AGT GAA TTC GAG CTC GGT ACC CGG GAT GAG ACT TAG CAA AAC TCT TTG | Used to make pUC19astAkan and pUC19astAcm |
| 188 | astA1 cm R GA | CAA TTA ACT TGG AAA CAA CAT TAC CAT TCT CAT C | Used to make pUC19astAcm |
| 189 | CM F GA | TGG TAA TGT TTC CAA GTT AAT TGC GTG ATA TAG | Used to make pUC19astAcm |
| 190 | CM R GA | ATC TCC AAT CAT CAA TGC GCC CTT TAG TTC C | Used to make pUC19astAcm |
| 191 | astA 2 CM F GA | TAA AGG GCG CAT TGA TGA TTG GAG ATT GTA TGA G | Used to make pUC19astAcm |
| 192 | astA 2 R GA | TAC GCC AAG CTT GCA TGC CTG CAG GTT ATT TTT TAG GAT TGA ATG CTT GAT C | Used to make pUC19astAkan and pUC19astAcm |
| 193 | astA 1 Kan R GA | TCA TTT TAG CCA TTC ATT TTA GCC ATA ACA ACA TTA C | Used to make pUC19astAkan |
| 194 | Kan F GA | ATG GCT AAA ATG AAT GGC TAA AAT GAG AAT ATC AC | Used to make pUC19astAkan |
| 195 | Kan R GA | ATC TCC AAT CAT CCT AAA ACA ATT CAT CCA GTA AAA TAT AAT ATT TTA TTT TC | Used to make pUC19astAkan |
| 196 | astA2 Kan F GA | TGA ATT GTT TTA GGA TGA TTG GAG ATT GTA TGA G | Used to make pUC19astAkan |
| 197 | Used for binding and uptake assay to amplify off kan fragment | ||
| 198 | Used for binding and uptake assay to amplify off kan fragment |
Construction of pJMB202 and pJMB203.
These plasmids were created using Gibson assembly, as described above. We amplified the first 975 nucleotides of astA from 81 to 176, either the kanamycin or chloramphenicol cassette from DRH153 and DRH154, respectively, and then the remainder of astA. All subsequent clones were verified by sequence determination.
Construction of pJMB204-209.
These plasmids were created using site-directed mutagenesis of pJMB203, which has three EcoRI sites. We used the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent), and followed the manufacturer’s instructions to design primers to either add a new EcoRI site on the plasmid backbone, >1 kb from the insert, or eliminate EcoRI sites so there was only one site left on the plasmid. We made single nucleotide changes in all cases (indicated in the primers), and the resulting sites cannot be recognized by either EcoRI or CtsM.
gDNA Purification.
Campylobacter cells were grown on MH agar plates with appropriate antibiotics as described previously. For transformation assays, a small number of cells (one or two passes with an inoculating loop) were scraped off the plate and resuspended in 300-µL filter-sterilized genomic lysis buffer (50 mM Tris base pH 7.5, 50 mM EDTA, 1% SDS, and 10 mM NaCl). The cells were heated at 95 °C for 10 min, after which 150 µL of Epicenter MPC Protein Precipitation Reagent was added. The cells were vortexed for 20 s, incubated on ice for 5 min, and then spun at 14,000 rpm for 15 min. After centrifugation, the supernatant was transferred to a clean 1.5-mL tube, and 750 µL of isopropanol was added, with mixing by inversion. After another centrifugation at 14,000 rpm for 5 min, the pellet was then washed twice with 70% ethanol. After washing, all ethanol was removed, and the pellet was dried in a SpeedVac for 10 min and then resuspended in 100 µL of dH2O. Concentration was determined using the Qubit 2.0 fluorometer.
Transformation Assays.
Transformation assays were performed as described previously with minor changes (29, 45). In brief, C. jejuni was grown for 16–18 h on MH agar plates supplemented with appropriate antibiotics. Cells were resuspended in MH broth to an OD600 of 0.5, and 500-μL aliquots were added to 13-mm test tubes. Unless ndicated otherwise, 1 µg of DNA (genomic, plasmid, or PCR product) was added to the tubes, and the cultures were incubated for 4 h at 37 °C in 5% CO2. The number of transformants and the total number of bacteria were determined by dilution plating on MH agar with appropriate antibiotics. Transformation efficiency represents the number of transformants per total number of bacteria per microgram of DNA. Transformations were conducted in triplicate and repeated at least three times. The reported transformation efficiency represents the average of three experiments.
In Vitro Methylation.
DNA MTases (M.EcoRI, M.HpaII, and M.TaqI) were purchased from New England BioLabs. DNA was methylated according to the manufacturer’s instructions, using the appropriate MTase concentration, time, buffer, and AdoMet concentration. After incubation, the DNA was purified. Plasmid and PCR fragments were purified using the QIAgen PCR Purification Kit, and genomic DNA was purified using ethanol precipitation. After purification, the DNA was quantified using the Qubit 2.0 fluorometer, and the methylation status was tested using appropriate restriction enzyme digestion. Methylation was deemed complete if the DNA was protected from cleavage after a 1-h digest as determined by gel electrophoresis.
DNA Binding and Uptake Assays.
Here 0.1 µg of purified plasmid DNA (with or without M.EcoRI methylation) per reaction was radiolabeled with 20 µCi [α-32P] dCTP using a Nick Translation Kit (Roche) in accordance with the manufacturer’s instructions. The DNA was purified using Sephadex G-25 Quick Spin Columns for radiolabeled DNA purification (Roche), following the manufacturer’s instructions. The purified DNA was mixed with 500 µL of 0.5 OD600 C. jejuni and then incubated at 37 °C for 30 min. To evaluate binding, the cell/DNA mixture was centrifuged, washed twice with MH broth, and then resuspended in 100 µL of dH2O. The mixture was then added to 5 mL of Scintillation Saf-Solve (VWR), and counts were measured with a scintillation counter. To evaluate uptake, the cell/DNA mixture was centrifuged, washed twice with MH broth, and then resuspended in 500 µL of MH broth. Then 1 µL of Turbo DNase (Life Technologies) was added, followed by a 45-min incubation at 37C. The DNase step was repeated once, and then cells were centrifuged, washed twice with MH broth, and resuspended in 100 µL of dH2O. The cells were then added to 5 mL of Scintillation Saf-Solve, and counts were measured with a scintillation counter.
Acknowledgments
We thank Robert Blumenthal for helpful comments on the manuscript. This work was supported in part by the National Institute of Allergy and Infectious Diseases (Grant R01 AI069383) and the Rudolph Hugh Endowment at Michigan State University (to V.J.D.). J.M.B. was supported by the Michigan Predoctoral Training Program in Genetics (Grant T32GM007544).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703331114/-/DCSupplemental.
References
- 1.Johnsborg O, Eldholm V, Håvarstein LS. Natural genetic transformation: Prevalence, mechanisms and function. Res Microbiol. 2007;158:767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Guschinskaya N, et al. Random mutagenesis of the hyperthermophilic archaeon Pyrococcus furiosus using in vitro mariner transposition and natural transformation. Sci Rep. 2016;6:36711. doi: 10.1038/srep36711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: Molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 4.de Boer P, et al. Generation of Campylobacter jejuni genetic diversity in vivo. Mol Microbiol. 2002;44:351–359. doi: 10.1046/j.1365-2958.2002.02930.x. [DOI] [PubMed] [Google Scholar]
- 5.Wassenaar TM, Fry BN, van der Zeijst BA. Genetic manipulation of Campylobacter: Evaluation of natural transformation and electro-transformation. Gene. 1993;132:131–135. doi: 10.1016/0378-1119(93)90525-8. [DOI] [PubMed] [Google Scholar]
- 6.Elkins C, Thomas CE, Seifert HS, Sparling PF. Species-specific uptake of DNA by gonococci is mediated by a 10-base pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman SD, Scocca JJ. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mell JC, Hall IM, Redfield RJ. Defining the DNA uptake specificity of naturally competent Haemophilus influenzae cells. Nucleic Acids Res. 2012;40:8536–8549. doi: 10.1093/nar/gks640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiesner RS. 2006. Physiological, genetic, and biochemical studies of natural competence in Campylobacter jejuni. Doctoral dissertation (University of Michigan, East Lansing, MI)
- 10.Loenen WA, Raleigh EA. The other face of restriction: Modification-dependent enzymes. Nucleic Acids Res. 2014;42:56–69. doi: 10.1093/nar/gkt747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin LF, Posfai J, Roberts RJ, Kong H. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc Natl Acad Sci USA. 2001;98:2740–2745. doi: 10.1073/pnas.051612298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humbert O, Salama NR. The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008;36:6893–6906. doi: 10.1093/nar/gkn718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humbert O, Dorer MS, Salama NR. Characterization of Helicobacter pylori factors that control transformation frequency and integration length during inter-strain DNA recombination. Mol Microbiol. 2011;79:387–401. doi: 10.1111/j.1365-2958.2010.07456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aras RA, Small AJ, Ando T, Blaser MJ. Helicobacter pylori interstrain restriction-modification diversity prevents genome subversion by chromosomal DNA from competing strains. Nucleic Acids Res. 2002;30:5391–5397. doi: 10.1093/nar/gkf686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambur OH, Frye SA, Nilsen M, Hovland E, Tønjum T. Restriction and sequence alterations affect DNA uptake sequence-dependent transformation in Neisseria meningitidis. PLoS One. 2012;7:e39742. doi: 10.1371/journal.pone.0039742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berndt C, Meier P, Wackernagel W. DNA restriction is a barrier to natural transformation in Pseudomonas stutzeri JM300. Microbiology. 2003;149:895–901. doi: 10.1099/mic.0.26033-0. [DOI] [PubMed] [Google Scholar]
- 17.Holt JP, Grant AJ, Coward C, Maskell DJ, Quinlan JJ. Identification of Cj1051c as a major determinant for the restriction barrier of Campylobacter jejuni strain NCTC11168. Appl Environ Microbiol. 2012;78:7841–7848. doi: 10.1128/AEM.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray IA, et al. The methylomes of six bacteria. Nucleic Acids Res. 2012;40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Loughlin JL, et al. Analysis of the Campylobacter jejuni genome by SMRT DNA sequencing identifies restriction-modification motifs. PLoS One. 2015;10:e0118533. doi: 10.1371/journal.pone.0118533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mou KT, et al. A comparative analysis of methylome profiles of Campylobacter jejuni sheep abortion isolate and gastroenteric strains using PacBio data. Front Microbiol. 2015;5:782. doi: 10.3389/fmicb.2014.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zautner AE, et al. SMRT sequencing of the Campylobacter coli BfR-CA-9557 genome sequence reveals unique methylation motifs. BMC Genomics. 2015;16:1088. doi: 10.1186/s12864-015-2317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller WG, et al. Comparative genomics of the Campylobacter lari group. Genome Biol Evol. 2014;6:3252–3266. doi: 10.1093/gbe/evu249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 24.Gardner SP, Olson JW. Barriers to horizontal gene transfer in Campylobacter jejuni. Adv Appl Microbiol. 2012;79:19–42. doi: 10.1016/B978-0-12-394318-7.00002-4. [DOI] [PubMed] [Google Scholar]
- 25.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson M, Christ C, Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984;12:5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allan BW, et al. DNA bending by EcoRI DNA methyltransferase accelerates base flipping but compromises specificity. J Biol Chem. 1999;274:19269–19275. doi: 10.1074/jbc.274.27.19269. [DOI] [PubMed] [Google Scholar]
- 28.Ershova AS, et al. Solitary restriction endonucleases in prokaryotic genomes. Nucleic Acids Res. 2012;40:10107–10115. doi: 10.1093/nar/gks853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiesner RS, Hendrixson DR, DiRita VJ. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J Bacteriol. 2003;185:5408–5418. doi: 10.1128/JB.185.18.5408-5418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beauchamp JM, Erfurt RS, DiRita VJ. Characterization and localization of the Campylobacter jejuni transformation system proteins CtsE, CtsP, and CtsX. J Bacteriol. 2015;197:636–645. doi: 10.1128/JB.02434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidsen T, et al. Biased distribution of DNA uptake sequences towards genome maintenance genes. Nucleic Acids Res. 2004;32:1050–1058. doi: 10.1093/nar/gkh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Findlay WA, Redfield RJ. Coevolution of DNA uptake sequences and bacterial proteomes. Genome Biol Evol. 2009;1:45–55. doi: 10.1093/gbe/evp005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conley EC, Saunders JR. Recombination-dependent recircularization of linearized pBR322 plasmid DNA following transformation of Escherichia coli. Mol Gen Genet. 1984;194:211–218. doi: 10.1007/BF00383519. [DOI] [PubMed] [Google Scholar]
- 34.Cehovin A, et al. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci USA. 2013;110:3065–3070. doi: 10.1073/pnas.1218832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hepp C, Maier B. Kinetics of DNA uptake during transformation provide evidence for a translocation ratchet mechanism. Proc Natl Acad Sci USA. 2016;113:12467–12472. doi: 10.1073/pnas.1608110113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehling JS, Lavender H, Clegg S. A Dam methylation mutant of Klebsiella pneumoniae is partially attenuated. FEMS Microbiol Lett. 2007;268:187–193. doi: 10.1111/j.1574-6968.2006.00581.x. [DOI] [PubMed] [Google Scholar]
- 37.Oshima T, et al. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol Microbiol. 2002;45:673–695. doi: 10.1046/j.1365-2958.2002.03037.x. [DOI] [PubMed] [Google Scholar]
- 38.Løbner-Olesen A, Marinus MG, Hansen FG. Role of SeqA and Dam in Escherichia coli gene expression: A global/microarray analysis. Proc Natl Acad Sci USA. 2003;100:4672–4677. doi: 10.1073/pnas.0538053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robbins-Manke JL, Zdraveski ZZ, Marinus M, Essigmann JM. Analysis of global gene expression and double-strand break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J Bacteriol. 2005;187:7027–7037. doi: 10.1128/JB.187.20.7027-7037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 41.Berdis AJ, et al. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proc Natl Acad Sci USA. 1998;95:2874–2879. doi: 10.1073/pnas.95.6.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahng LS, Shapiro L. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J Bacteriol. 2001;183:3065–3075. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blow MJ, et al. The epigenomic landscape of prokaryotes. PLoS Genet. 2016;12:e1005854. doi: 10.1371/journal.pgen.1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Taylor DE. Natural transformation in Campylobacter species. J Bacteriol. 1990;172:949–955. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- 48.Hendrixson DR, Akerley BJ, DiRita VJ. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol. 2001;40:214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]