Significance
Caspases are central players in programmed cell death. Among caspases, caspase-6 is unique for its association with neurological disorders, including Alzheimer’s, Huntington’s, and Parkinson’s diseases. The structural details underlying caspase-6 activation are still limited but are requisites in understanding caspase-6 function. The prodomain and linker play essential roles in caspase function and regulation; however, while the long prodomains in initiator caspases are known, the structures of the short prodomains in executioner caspases remain elusive, despite efforts using crystallography and NMR. Here, we used hydrogen/deuterium exchange MS and revealed two important findings: the prodomain and intersubunit linker are intrinsically disordered, and the presence or absence of these regions results in distinct structural dynamics as procaspase-6 progresses through its proteolytic activation.
Keywords: hydrogen exchange MS, apoptosis, neurodegeneration, cysteine protease, conformational dynamics
Abstract
Caspase-6 is critical to the neurodegenerative pathways of Alzheimer’s, Huntington’s, and Parkinson’s diseases and has been identified as a potential molecular target for treatment of neurodegeneration. Thus, understanding the global and regional changes in dynamics and conformation provides insights into the unique properties of caspase-6 that may contribute to achieving control of its function. In this work, hydrogen/deuterium exchange MS (H/DX–MS) was used to map the local changes in the conformational flexibility of procaspase-6 at the discrete states that reflect the series of cleavage events that ultimately lead to the fully active, substrate-bound state. Intramolecular self-cleavage at Asp-193 evoked higher solvent exposure in the regions of the substrate-binding loops L1, L3, and L4 and in the 130s region, the intersubunit linker region, the 26–32 region as well as in the stabilized loop 2. Additional removal of the linker allowed caspase-6 to gain more flexibility in the 130s region and in the L2 region converting caspase-6 to a competent substrate-binding state. The prodomain region was found to be intrinsically disordered independent of the activation state of caspase-6; however, its complete removal resulted in the protection of the adjacent 26–32 region, suggesting that this region may play a regulatory role. The molecular details of caspase-6 dynamics in solution provide a comprehensive scaffold for strategic design of therapeutic approaches for neurodegenerative disorders.
Caspases are a family of cysteine aspartyl-specific peptidases that are central players in apoptosis and inflammation. Based on their cellular function and domain organization, caspases are classified as initiator (caspase-2, -8, and -9) or executioner (caspase-3, -6, and -7). In the canonical caspase activation route, initiator caspases activate executioner caspases through proteolytic processing, which promotes apoptosis. The executioner caspases are expressed as dimeric yet inactive zymogens (procaspases) and activated via proteolytic cleavage at conserved aspartate residues to generate the large and small subunits.
Caspase-6 is unique among caspases for its role in neurological disorders, including Alzheimer’s (1–3), Huntington’s (4–7), and Parkinson’s (8) diseases. Neuronal proteins that are known substrates of caspase-6 include microtubule-associated protein Tau (9), amyloid precursor protein (10), presenilin I and II (10), polyglutamine-expanded and native huntingtin protein (7), and Parkinson disease protein 7, also known as protein deglycase DJ-1 (8). Cleavage of these neuronal substrates by caspase-6 is recognized to impact the physiological outcomes in these neurological disorders, thus making caspase-6 an attractive molecular target for treatment of neurodegeneration.
Although it is categorized as an executioner caspase, caspase-6 sometimes plays roles as a noncanonical executioner or even as an inflammatory caspase (11), making its classification complex. Although caspase-6 is a weak executioner of apoptosis, its overexpression in mammalian cells results in apoptosis (12), where it is the only known caspase to cleave the nuclear lamellar protein, lamin A/C (13–15). Caspase-6 is also associated with the inflammatory pathway by being activated by caspase-1 (11). Moreover, a noncanonical route for activation has been reported for caspase-6. Caspase-6 is often activated by caspase-3 rather than by initiator caspases (16, 17); however, it can also be activated in the absence of caspase-3 (18–20) and is reported to self-activate in vitro and in vivo (21, 22). The crystal structure of procaspase-6 provides clear insight into the molecular details of self-activation (22). In particular, the long caspase-6 intersubunit linker is seen occupying the substrate-binding groove in preparation for autoactivation in cis. Nevertheless, the key regulatory signals that control the caspase-6 self-activation in a neurodegenerative context still are not understood.
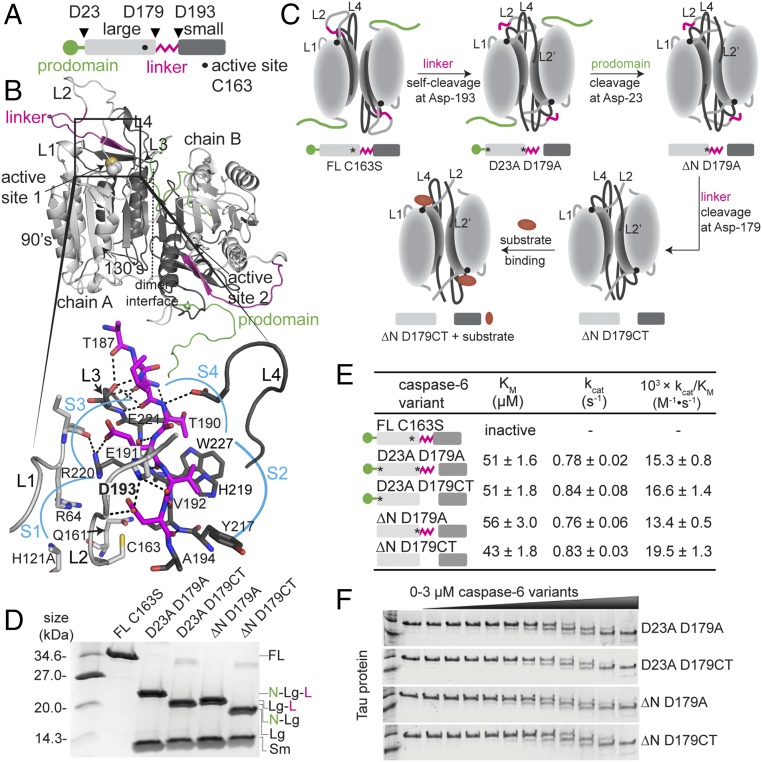
Caspase-6 cleavage sites are located at the prodomain (Asp-23) and at both sides of the intersubunit linker (Asp-179 and Asp-193) (Fig. 1A). The intersubunit linker joins the large and the small subunits of procaspase-6 with the active site Cys-163 located within the large subunit. The active site cavity of caspase-6 is composed of four flexible loops (L1, L2, L3, L4) that undergo structural rearrangement to accommodate the substrate in the substrate-binding pockets (S1, S2, S3, S4) (Fig. 1B). Current evidence of a caspase-6 activation pathway (Fig. 1C) proposed that the initial intramolecular self-cleavage at Asp-193 in the intersubunit linker is required for procaspase-6 self-activation (21, 22), which we surmise is the relevant pathway in a neurodegenerative context, whereas caspase-3 activates caspase-6 by cleavage at Asp-179 during apoptosis. Subsequent proteolytic cleavage by caspase-3, caspase-1, or caspase-6 itself (intermolecularly) at Asp-23 in the prodomain and at Asp-179 in the intersubunit linker (11, 21–23) leads to full maturation of caspase-6. All substrate-binding loops are then available to accommodate the substrate in the substrate-binding pocket to enable proteolysis.
Fig. 1.
Caspase-6 maturation-state variants. (A) Linear cartoon of procaspase-6 illustrating the prodomain (green), large subunit (light gray), intersubunit linker (magenta), and small subunit (dark gray). The active site C163 is denoted by a dot. Triangles indicate proteolytic cleavage sites. (B) Model of full-length (FL) procaspase-6 zymogen colored as in A was generated based on zymogen structures 4IYR and 3NR2 with missing regions modeled de novo. Inset shows details of binding interactions of the intersubunit linker residues (magenta) binding into the S1–S4 subsites. (C) Cartoon of expected conformational changes in caspase-6 constructs representing various points along the zymogen maturation (cleavage) pathway colored as in A, with substrate as an ellipsoid (red). FL indicates FL uncleaved procaspase-6 zymogen. The C163S catalytic site substitution renders caspase-6 inactive and incapable of self-activation. ΔN indicates removal of the N-terminal prodomain (1–23). D23A substitution renders caspase-6 uncleavable after the prodomain. D179A prevents cleavage of the intersubunit linker. D179CT constructs have a stop codon inserted after residue 179 to allow expression of a constitutively two-chain caspase-6, mimicking native cleavage at D179. The asterisk designates D to A mutation in the proteolytic cleavage site in caspase-6. (D) Maturation-state caspase-6 variants purified from overexpression in E. coli. (E) Catalytic properties of caspase-6 maturation-state variants. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. (F) Cleavage of Tau-383 protein (3 μM) is not impacted by the maturation state of caspase-6, tested at increasing concentrations (0–3 μM).
The prodomain is the most distinctive region of caspases in terms of its sequence conservation (24, 25) and is, therefore, perhaps the most promising region for achieving selective regulation. As a result, the function of the prodomain varies among caspases. The prodomain of initiator caspases comprises caspase recruitment domain or death effector domains, which are typically longer than executioner caspase prodomains. The long prodomain facilitates recruitment to the activating scaffold, which is important for initiator caspase activation (26, 27). In executioner caspases, the short prodomain has a variety of functions. In caspase-3, it facilitates folding by acting as a chaperone (28), suppresses zymogen activation in vivo (29), and binds Hsp27 for inhibition of its proteolytic activation (30). In caspase-7, the prodomain-adjacent region acts as an exosite for substrate recognition (31), while in caspase-6, the prodomain inhibits its self-activation in vivo (21) and in vitro (32). Taken together, these findings clearly show that the prodomain is an integral part of caspase function and regulation.
Despite its importance in caspases, structural and functional details of the prodomain as well as the intersubunit linker are inadequately understood because the electron density in these regions has been missing in all caspase crystal structures to date. In particular, these regions in procaspase-6 (32) or in cleaved caspase-6 (25) have never been visualized. In addition to the properties of the prodomain and intersubunit linker being ethereal, the impact of the prodomain and linker on the conformational flexibility and dynamics in other regions of caspase-6 also remains elusive. Thus, probing the fine details of the influence of the cleavage state (e.g., the presence or absence of the prodomain and linker) on the individual conformational flexibility in all regions of caspase-6 is expected to provide a more complete picture detailing the underlying mechanism of caspase-6 proteolytic activation. Furthermore, insight into the functional and structural consequences of caspase-6 activation is likely to add to our understanding of other caspases as well.
Since the prodomain and the intact linker have eluded crystallization and our NMR structure determination has also not been successful, solution phase techniques, including the use of hydrogen/deuterium exchange MS (H/DX-MS), are anticipated to be valuable to study the structural behavior over the entirety of the caspase-6 structure. H/DX-MS has emerged as a powerful tool to study changes in protein structure and dynamics resulting from protein–protein or protein–ligand interactions in solution (33–35). Previously in the realm of caspases, H/DX-MS has been used to probe the unique helix-strand interconversion in caspase-6 (36) and the conformational changes associated with the activation of Pak2 after caspase-3 cleavage and autophosphorylation (37). Nevertheless, a detailed understanding of the changes in the dynamics of a caspase as it progresses through the stages of proteolytic activation has not been undertaken. In this work, we report the use of H/DX-MS to trace the discrete conformational flexibility profile at discrete cleavage states along the pathway of proteolytic activation of procaspase-6 as it matures from initial intramolecular self-cleavage to subsequent intermolecular cleavage of the prodomain and intersubunit linker.
Results
H/DX-MS Profile of Caspase-6 Maturation-State Variants.
It is clear that caspase-6 can be self-activated by cleavage at Asp-193 or activated by caspase-3 cleavage at Asp-179. Our model is that Asp-179 cleavage is the most relevant activation pathway during apoptosis but that self-activation at Asp-193 is likely to be the most relevant in a neurodegenerative context. To map the changes in the conformational dynamics accompanied by caspase-6 proteolytic self-activation, a panel of caspase-6 variants that represent each maturation state of caspase-6 (Fig. 1 C and D) was prepared. The full-length (FL) procaspase-6 C163S, which is the catalytic site-substituted, inactivated variant, represents the zymogen form of procaspase-6. Caspase-6 D23A D179A represents the form of caspase-6 after initial activation by intramolecular self-cleavage at Asp-193. This variant maintains the prodomain but is self-cleaved at Asp-193 in the intersubunit linker. Caspase-6 D23A D179CT has the prodomain intact but lacks the entire intersubunit linker (residues 180–193), representing the form of caspase-6 that has been initially activated by caspase-3. Caspase-6 ΔN D179CT represents the fully cleaved, mature form of caspase-6, lacking both the prodomain and intersubunit linker. All caspase-6 variants used in this study (except FL caspase-6 C163S) were active for cleaving both tetrapeptide substrate Ac-Val-Glu-Ile-Asp-7-amino-4-methylcoumarin (VEID-AMC) (Fig. 1E) and protein substrate Tau (Fig. 1F).
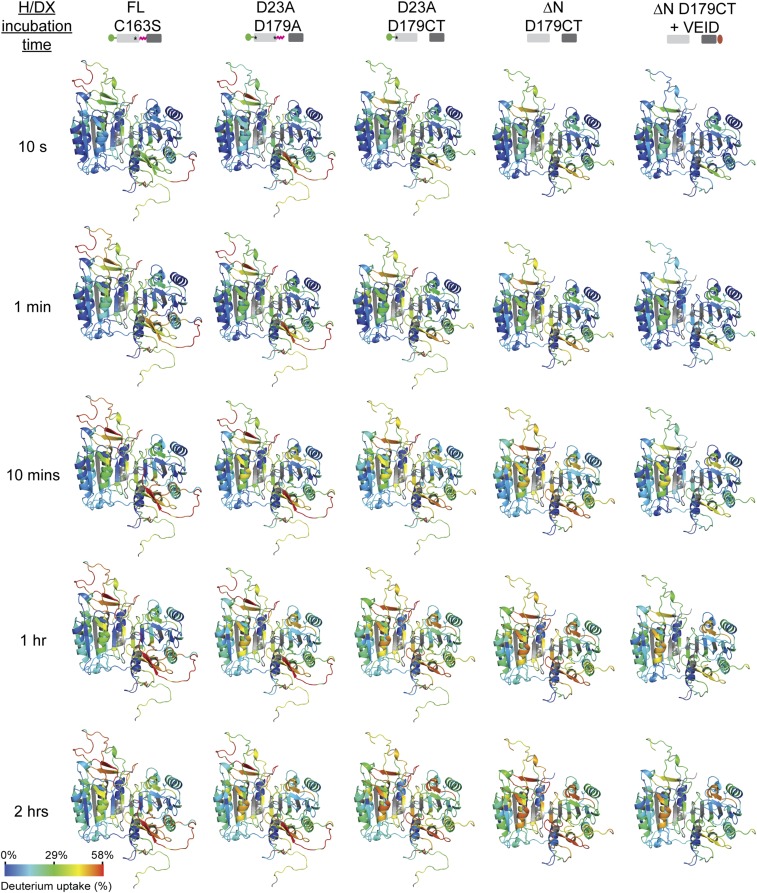
H/DX-MS of the panel of caspase-6 maturation-state variants was performed using a protocol identical to that previously reported (36). After incubation of proteins in deuterium for between 10 s and 2 h to allow hydrogen/deuterium (H/D) exchange on intact caspase-6, samples were subjected to on-column pepsin digestion followed by MS for peptide mass analysis. A heat map of the relative differential deuteration profiles of various regions in caspase-6 over the course of H/D exchange incubation time is depicted for each of the maturation-state variants from self-activation to full maturation and eventual binding to a substrate (Fig. 2). The corresponding relative deuterium uptake profiles were also mapped onto a model of FL procaspase-6 zymogen (Fig. S1), where the crystal structures of the zymogen [Protein Data Bank (PDB) ID codes 4IYR and 3NR2] were used as templates, and the missing residues were modeled using Chimera/Modeller (38, 39). The deuterium uptake plots of peptic peptides are shown in Figs. S2 and S3. The use of this hybrid model structure, which has complete sequence coverage of caspase-6, ensures that all observed peptides could be represented during visualization of the results of the H/DX-MS. As anticipated, the ordered and buried regions in the core of caspase-6 had lower H/D exchange levels than flexible or exposed regions of caspase-6. These most highly exchangeable regions comprised mainly the substrate-binding loops, the intersubunit linker, and the prodomain. Notably, the H/D exchange at the proteolytic cleavage sites (TETD23, DVVD179, and TEVD193) are among the highest in caspase-6, including in the zymogen form, suggesting that the higher conformational flexibility of this region is required for greater access to proteolytic cleavage by caspase-6 and other proteases. Approximately 75% of the backbone amide hydrogens underwent less than 30% of H/D exchange within 10 s, suggesting that caspase-6 is a properly folded and dynamically stable protein overall. The peptic peptide coverage ranges from 90.8 to 92.1% across the linear amino acid sequences of the caspase-6 maturation state variants, with an average redundancy of 2.46 (Fig. S4). The representative MS spectra of key peptic peptides are shown in Figs. S5 and S6. Together, these results suggest that all caspase-6 variants used were amenable to H/DX-MS experiments, which enable precise detection of the diverse conformational flexibility of caspase-6 at the discrete cleavage states attained during proteolytic activation.
Fig. 2.
H/D exchange heat map of the relative deuterium incorporation for caspase-6 maturation-state variants. For each peptic peptide from the caspase-6 maturation-state variants, the percentage relative deuterium level for each H/D exchange incubation time (minutes: 0.17, 1, 10, 60, and 120) is mapped onto the corresponding linear sequence of caspase-6. The percentage relative deuterium incorporation is calculated by dividing the observed deuterium uptake by the theoretical maximum deuterium uptake for each peptide. The H/DX-MS experiments followed all observed peptides and cover 90.8–92.1% of the combined linear sequences. Peptic peptides with no H/D exchange data at a given incubation time or regions absent in various constructs are colored white. Secondary structural elements are depicted above the primary sequence. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. The percentage relative deuterium level of each peptic peptide represents the average values of duplicate experiments performed on 2 separate days. §This H/DX-MS dataset has been reported in ref. 36.
Fig. S1.
The percentage relative deuterium incorporations of caspase-6 mutation variants over the course of the H/D exchange incubation period are mapped onto the model structure of caspase-6. The percentage relative deuterium incorporation is calculated by dividing the observed deuterium uptake by the theoretical maximum deuterium uptake for each peptide. The intensities of the red and blue colors indicate increase and decrease in H/D exchange, respectively. The regions of the protein for which H/D exchange data are not available are shown in gray. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable.
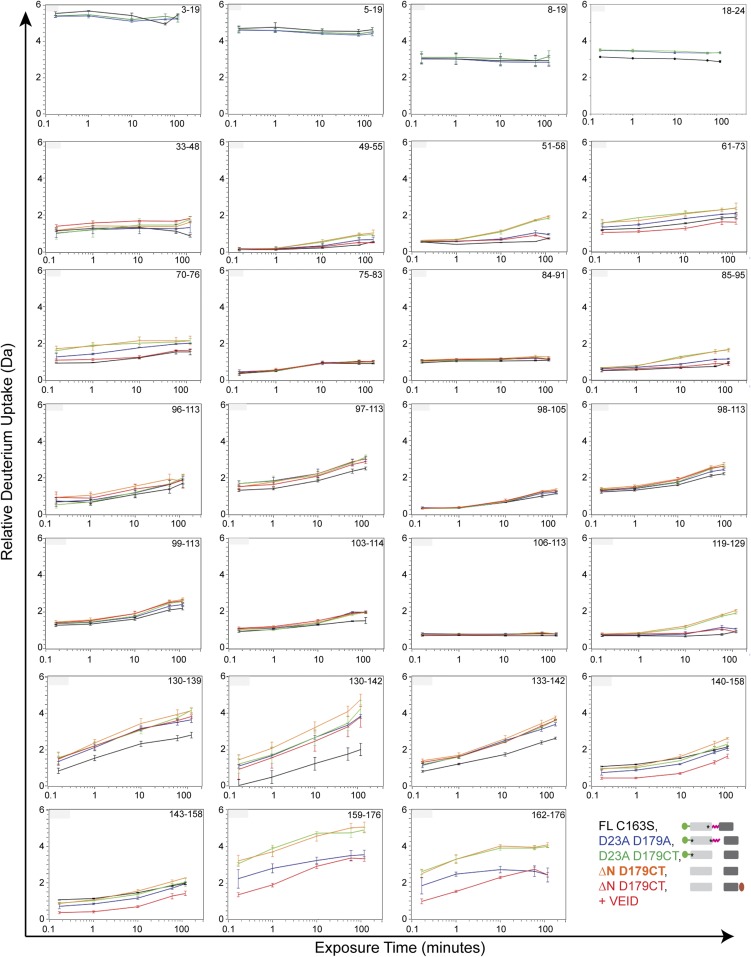
Fig. S2.
Relative deuterium uptake plots of caspase-6 maturation variants over the course of the H/D exchange experiment. The different colors represent the deuterium uptake profiles of caspase-6 mutation variants FL C163S (black), D23A D179A (blue), D23A D179CT (green), ΔN D179CT (orange), and ΔN D179CT + VEID (red). The covered residues for each peptic peptide of caspase-6 mutation variants are indicated. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. Error bars, SD of duplicate H/DX-MS measurements done on 2 separate days.
Fig. S3.
Relative deuterium uptake plots of caspase-6 maturation variants over the course of the H/D exchange experiment. The different colors represent the deuterium uptake profile of caspase-6 mutation variants FL C163S (black), D23A D179A (blue), D23A D179CT (green), ΔN D179CT (orange), and ΔN D179CT + VEID (red). The covered residues for each peptic peptide of caspase-6 mutation variants are indicated. The asterisk designates the D to A mutation in the proteolytic cleavage site in caspase-6. Error bars, SD of duplicate H/DX-MS measurements done on 2 separate days. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable.
Fig. S4.
Coverage map of the peptic peptides of caspase-6 maturation variants identified from H/DX-MS. The teal color represents the set of peptic peptides found in all caspase-6 maturation variants covering the core regions of caspase-6. Peptic peptides highlighted in green were found in FL C163S, D23A D179A, and D23A D179CT covering the prodomain region. Peptic peptides highlighted in gray represent the linker region found only in FL C163S and D23A D179A. Peptic peptides highlighted in black and blue colors were found in FL C163S and D23A D179A, respectively. Peptic peptides exclusively found in one or more caspase-6 maturation variants were the result from the variation in the caspase-6 constructs, including the presence of the prodomain or linker and mutations in the active site cysteine and proteolytic cleavage sites of caspase-6. The peptic peptide coverage ranges from 90.8 to 92.1% across the linear amino acid sequences of the caspase-6 maturation variants, with an average redundancy of 2.46.
Fig. S5.
Representative MS spectra of key peptic peptides after H/D exchange experiments of caspase-6 maturation variants. The relative locations of the highlighted peptic peptides for the following regions are mapped onto the hybrid model of procaspase-6: purple blue, peptide including the 26–32 region; green, top of 130s region; orange, 130s region; cyan, part of L1; red, L2. The amino acid sequence, the MS charge-state distribution, and the residue numbers covered by the representative peptic peptide are also indicated. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable.
Fig. S6.
Representative MS spectra of key peptic peptides after H/D exchange experiments of caspase-6 maturation variants. The relative location and charge-state distribution of peptides from additional highlighted regions: yellow, L3; light pink, L4; blue, N-terminal part of linker; magenta, C-terminal part of linker. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable.
Intramolecular Self-Cleavage at Asp-193 Influences Local Conformation and Dynamics.
In the crystal structure of the procaspase-6 zymogen (Fig. 1B), the active site pocket is occupied with the 190TEVDA194 region of the intersubunit linker, which forms an antiparallel β-sheet with 217YSHRE221 residues in L3 through six main-chain hydrogen bonds (22). Consequently, the scissile peptide bond between the Asp-193 and Ala-194 sits atop the active site Cys-163, and is primed for hydrolysis by intramolecular self-cleavage (21, 22). The H/D exchange of peptide 186–193, which derive from the 190TEVDA194 region, significantly increased on self-cleavage at Asp-193 (Fig. 3 A and E and Fig. S6). This increase in H/D exchanges suggests that the 190TEVDA194 site is more exposed in the Asp-193-cleaved form (D23A D179A) compared with the zymogen (FL C163S).
Fig. 3.
Caspase-6 displays variable conformational flexibility across the states of proteolytic activation. Differences in deuterium uptake (daltons) of the corresponding peptic peptides identified in caspase-6 maturation variants [(A) D23A D179A, (B) D23A D179CT, (C) ΔN D179CT, and (D) ΔN D179CT + VEID)] compared with the zymogen (FL C163S) at the indicated time points of exposure to deuterated solvent. The residue numbers for each peptic peptide are listed with the corresponding secondary structural elements. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. These statistically significant differences in the H/D exchange between the caspase-6 maturation variants [(E) D23A D179A, (F) D23A D179CT, (G) ΔN D179CT, and (H) ΔN D179CT + VEID)] and the zymogen (FL C163S) after 2 h of incubation were mapped onto the model structure of caspase-6 shown in both ribbon and surface representations. The asterisk designates D to A mutation in the proteolytic cleavage site in caspase-6. For these data, a deuterium uptake difference greater than 0.6 Da is considered significant at the 98% confidence interval. The intensities of the blue and red colors indicate peptides that undergo either a statistically significant decrease (less exchangeable/flexible) or increase (more exchangeable/flexible), respectively, during H/D exchange along the path of caspase-6 proteolytic activation. (I) Representative deuterium incorporation plots of key peptic peptides identified in caspase-6 maturation variants covering the regions of 26–32, 90s, and 130s; the linker; and the substrate-binding loops L1–L4. The representative MS spectra of the highlighted peptic peptides are shown in Figs. S5 and S6. Error, SD of duplicate H/DX-MS measurements done on 2 separate days.
Our data show that initial cleavage at Asp-193 impacts the overall conformational flexibility of caspase-6 substrate-binding loops (L1, L2, L4), the 130s region, and the 26–32 region (Fig. 3A). In the crystal structure of procaspase-6 zymogen, the side chains of 190TEVDA194 occupy the substrate-binding pockets S1–S4 (Fig. 1B). It was, therefore, not surprising to observe higher H/D exchange in the L1 region (peptide 56–74), where Arg-64 is expected to be engaged as part of the S1 pocket for substrate binding. Likewise, the part of 130s region (peptide 130–142) that sits below the activity site cavity also had higher H/D exchange in the Asp-193–cleaved form compared with the zymogen. The increase in the H/D exchange levels in the L1 and 130s region suggests that these regions are relatively protected in the zymogen state and become more exposed after initial cleavage at Asp-193 because of an overall increase in the mobility of this region.
L2 in caspase-6 is relatively long compared with L2 in caspase-3 and caspase-7. Substitution of residues in L2 of caspase-6 with L2 residues from caspase-3 and caspase-7 as well as truncation of at least three amino acid residues (170–173) from the N terminus of L2 prevented self-activation of procaspase-6 (22), suggesting that a long L2 is required for binding the sequence 190TEVDA194 of the intersubunit linker into the substrate-binding groove. This suggests a critical role of L2 in regulating the intramolecular self-cleavage at Asp-193. In terms of conformational dynamics, the L2 region (peptide 159–176) was found to have a significant decrease in H/D exchange on cleavage at Asp-193 (Fig. 3 A and I). Interestingly, in the crystal structure of the procaspase-6 zymogen, part of L4 (residues 261–271) is flexible and not engaged in binding with 190TEVDA194. This same region of L4 is covered by peptide 264–278, where the H/D exchange profile was slightly perturbed (protected at early time points but exposed after 2 h) on Asp-193 cleavage. The L4 and the L2—which are located on the same face of the protein—were the only regions of caspase-6 that experienced significant protection on Asp-193 cleavage. The L2 peptide 159–176 and the L4 peptide 264–278 show charge complementarity: part of L2 is negatively charged, and part of L4 is positively charged (Fig. S7). The result that both the L2 and L4 regions experienced protection from H/D exchange and exhibited charged complementarity to each other suggests that L2 and L4 are interacting after self-activation of caspase-6 at Asp-193, potentially initiating loop reorganization for proper substrate binding.
Fig. S7.
Map of the electrostatic potential of the model structure of procaspase-6 built from the zymogen crystal structures (PDB ID codes 4iyr and 3NR2). The missing residues of each PDB template structure were built by de novo modeling using Chimera/Modeler platform. The electrostatic potential map (red, acidic regions; blue, basic regions; white, hydrophobic/neutral regions) was generated using PyMOL Molecular Graphics System (Schrödinger, LLC). Highlighted are key regions of caspase-6 that undergo statically significant changes in the H/D exchange along caspase-6 proteolytic activation.
Finally, it is interesting that self-activation at Asp-193 resulted in a statistically significant increase in H/D exchange of the 26–32 region after subtractive analysis of overlapping peptides 26–48 and 33–48 (Fig. 3 A, E, and I). This suggests that the 26–32 region is relatively protected in the zymogen and is exposed after initial self-cleavage at Asp-193. This region immediately follows the prodomain in sequence. Intriguingly, the 26–32 region is situated in the same loop as one of the residues (K36) in the allosteric and regulatory site for zinc binding in caspase-6 (40) and the identified exosite in capase-7 (38KKKK41) that promotes efficient cleavage of the poly(ADP ribose) polymerase 1 substrate (31). These data collectively suggest that the initial intramolecular cleavage at Asp-193 impacts the overall flexibility of caspase-6 in the context of the proper assembly of structural elements that enables caspase-6 activation and regulation. In particular, we hypothesize that cleavage at the intersubunit linker at Asp-193 may expose a substrate-binding exosite present in or adjacent to the 26–32 region.
Complete Removal of the Linker Is Important to Attain the Fully Exposed Substrate-Binding Cavity.
The linker region contains two proteolytic cleavage sites within the regulatory L2 loop in caspase-6. In the zymogen structure of procaspase-6, part of this region (residues 186–193) is ordered and accommodated in the substrate-binding groove, and the rest of the linker residues are disordered (22). After initial intramolecular self-cleavage at Asp-193 (D23A D179A), caspase-6 can exist in a state where the linker is still attached to the large subunit with the intact 176DVVD179 site. This Asp-179 is not a good substrate for intramolecular cleavage by caspase-6, because the intersubunit linker is not long enough to position Asp-179 for self-cleavage (22). However, Asp-179 is the first site to be intermolecularly cleaved by caspase-3 and other caspases (22, 23). A crystal structure of cleaved caspase-6 with intact linker was solved, but the electron density of the entire linker region (180–193) was missing. Thus, the structural behavior of the linker and its impact on the dynamics during caspase-6 activation are still unexplored. In this H/DX-MS study, 100% of the linker region was covered by peptides 178–185 and 186–193 (Fig. 3 A, B, and I). The significant increase in the H/D exchange of the C-terminal part of the linker region (186–193) between the zymogen and the Asp-193-cleaved form suggests that, before cleavage at Asp-193 in the zymogen form, the region of the 186–193 is bound to the substrate-binding groove—consistent with the crystal structure of procaspase-6 zymogen. However, the exchange profile of the 178–185 region between the zymogen and Asp-cleaved form was unaffected, suggesting that this region, a contiguous part of L2, has the same conformational flexibility in both the zymogen and Asp-193 cleaved form. The shape of the curve for deuterium incorporation over the time course of H/D exchange often reveals interesting structural features for particular regions of proteins and provides meaningful interpretation of the H/D exchange data as has been described elsewhere (41). The H/D exchange profile of peptide 178–185 in both the zymogen and the Asp-193–cleaved form showed a nearly maximal uptake of deuterium, even at the earliest time point (10 s), and remained at that same high level throughout the H/D exchange experiment (Fig. 3I). This deuterium uptake profile suggests that the 178–185 region has high solvent exposure and the absence of H-bond interactions, such as is characteristic for unstructured regions. Conversely, peptide 186–193 exchange parameters suggest a partially protected, moderately dynamic region. Peptide 186–193 showed a one-deuterium difference between the zymogen and Asp-193 self-cleaved forms at the earliest time point (10 s), suggesting that this region is more exposed after Asp-193 cleavage. This one-deuterium difference persisted throughout the H/D exchange time course (Fig. 3I). In the crystal structure of the zymogen, the 190TEVDA194 cleavage site is present as a β-strand, which rests in the substrate-binding groove and is expected to be highly protected from H/D exchange. The fact that the 186–193 peptide in the Asp-193 cleaved form of caspase-6 shows less protection than in the zymogen suggests that only before zymogen activation does the intersubunit linker remain stably bound to the active site.
Farther along the activation pathway of caspase-6, the complete removal of the intersubunit linker occurs when Asp-179 is cleaved (D23A D179CT or ΔND179CT). Removal of the linker resulted in a global H/D exchange profile that is significantly altered compared with the zymogen (Fig. 3 B, C, F, and G). Removal of the intersubunit linker resulted in significant exposure—relative to the zymogen—of the regions in the substrate-binding loops: L1 (peptide 56–74), L2 (peptide 159–176), L3 (peptides 210–216 and 217–227), L4 (peptides 264–278 and 269–278), and the 130s region (peptides 119–130, 130–142, and 133–142). Moreover, the differences in the H/D exchange between cleaved forms of capase-6 in the absence or presence of the linker showed a similar trend where removal of the linker resulted in a statistically significant increase in the H/D exchange in the substrate-binding loops and the top of the 130s region (Fig. 4 A and B). Collectively, these data suggest that the removal of the linker renders the substrate-binding groove fully exposed and competent to bind substrate.
Fig. 4.
Intersubunit linker cleavage increases conformational flexibility of all substrate-binding loops. Comparative differences in the H/D exchange profiles among cleaved caspase-6 maturation variants [(A) D23A D179A vs. D23A D179CT, (B) D23A D179A vs. ΔN D179CT, and (C) D23A D179CT vs. ΔN D179CT] at the indicated time points of exposure to deuterated solvent. These statistically significant differences in the H/D exchange among cleaved caspase-6 maturation variants [(D) D23A D179A vs. D23A D179CT, (E) D23A D179A vs. ΔN D179CT, and (F) D23A D179CT vs. ΔN D179CT]) after 2 h of incubation were mapped onto the model structure of caspase-6 shown in both ribbon and surface representations. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. For these data, a deuterium uptake difference greater than 0.6 Da is considered significant at the 98% confidence interval. The intensities of the blue and red colors represent the peptides that undergo either a statistically significant decrease (less exchangeable/flexible) or increase (more exchangeable/flexible), respectively, in the H/D exchange along the path of caspase-6 proteolytic activation.
After the substrate-binding groove is exposed, substrate can bind to the fully cleaved, mature caspase-6 (ΔND179CT + VEID substrate mimic). The H/D exchange profiles of the loop bundles (L1–L4) and the top of the 130s region (peptide 119–130) were significantly decreased relative to the zymogen on substrate binding (Fig. 3 D, H, and I). This suggests that these regions undergo significant structural reorganization and protection on engaging substrate, resulting in a lower conformational flexibility of these regions.
Substrate-Binding Groove Is Accessible in Cleaved Caspase-6, Independent of the Presence of the Prodomain or Linker.
The H/D exchange data show that the overall accessibility of the substrate-binding groove increases even further as caspase-6 matures into its substrate-binding competent state. To further explore this observation, we sought a complementary technique to interrogate the conformation of the substrate-binding groove before and after substrate binding. Caspase-6 has only two tryptophan residues, which are found in L1 (Trp-57) and L3 (Trp-227) (Fig. 5A). Trp-57 is completely exposed in crystal structures of both the zymogen and mature forms of caspase-6. However, in the zymogen, Trp-227 sits immediately adjacent to the substrate-binding groove as part of the S4 specificity pocket within L3 (Fig. 1B), which forms an antiparallel β-strand with the linker in the zymogen state. Intrinsic tryptophan fluorescence is influenced by the polarity of the environment, where shifts in the emission maxima toward longer wavelengths (red shift) indicate higher solvent exposure. All maturation variants of caspase-6 were subjected to intrinsic fluorescence measurements (Fig. 5B and Fig. S8). Notably, all caspase-6 cleavage variants clustered in two classes of emission spectra, where all linker-cleaved (singly cleaved or complete removal of the linker) forms of caspase-6 showed a 9-nm red shift toward a more solvent-exposed state relative to the zymogen, irrespective of the presence of the prodomain (Fig. 5B). This suggests that initial Asp-193 cleavage in the linker leads to the relative exposure of the bound intersubunit linker. Thus, Asp-193 cleavage activates caspase-6 by generating an open and more dynamic substrate-binding competent active site.
Fig. 5.
Intersubunit linker cleavage leads to increased exposure of the substrate-binding groove. (A) Comparison of the crystal structures of procaspase-6 zymogen (PDB ID code 3NR2; Left) and mature, unliganded caspase-6 (PDB ID code 2WDP; Right) highlighting the only two tryptophan residues, Trp-57 and Trp-227, as well as the relative location of the intersubunit linker (magenta) and the active site Cys-163. (B) The intrinsic tryptophan fluorescence profiles of caspase-6 mutation variants. Fluorescence emission scans of 305–400 nm were collected after excitation at 295 nm of 3 μM proteins in 10 mM phosphate buffer, pH 7.5, 120 mM NaCl, and 2 mM DTT. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. Data presented here are a representative from three independent experiments.
Fig. S8.
Intrinsic fluorescence profile of caspase-6 mutation variants excited at 280 nm. Fluorescence emission scans from 305 to 400 nm were collected after excitation at 280 nm of proteins (3 μM) in 10 mM phosphate buffer, pH 7.5, 120 mM NaCl, and 2 mM DTT. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. Data presented here are representative of three independent experiments.
The Prodomain Is Intrinsically Disordered.
An attempt to solve the crystal structure of procaspase-6 zymogen with intact prodomain has been reported (32), but no electron density for the prodomain was visible, suggesting that the prodomain may be a mobile region. Although both biological (21) and biochemical studies (32) revealed that the prodomain impacts caspase-6 function, the existing structural data on the prodomain are extremely limited. In this work, the structural properties of the prodomain were explored using H/DX-MS as well as its impact on the dynamics of caspase-6 during proteolytic activation. The electrostatic potential map of the 23-aa prodomain of caspase-6 showed an obvious charged–polarized region (Fig. 6A). The N and C termini were packed with positively and negatively charged residues, respectively. H/DX-MS studies covered 22 of 23 amino acids of the prodomain represented by five overlapping peptic peptides (2–19, 3–19, 5–19, 8–19, 18–24) (Figs. 2–4 and Fig. S2). The H/D exchange profile of the prodomain region showed no statistically significant changes between the zymogen and the cleaved forms of caspase-6, irrespective of the presence or absence of the linker (Figs. 3 A and B and 6B). Likewise, the H/D exchange profile of the prodomain region showed no statistically significant changes between cleaved forms of caspase-6 with or without the linker (Figs. 4A and 6B). The H/D exchange profiles of the prodomain peptides in both the zymogen and cleaved forms of caspase-6 with intact prodomain (Fig. 6B) showed nearly maximal uptake of deuterium by the earliest time point, which persisted at that same level over the course of the H/D exchange experiment. The corresponding MS spectra of prodomain peptides in all forms of caspase-6 with intact prodomains showed a well-defined isotopic distribution (Fig. 6C). Collectively, these data suggest that, independent of the activation state of caspase-6, the prodomain is highly solvent exposed, consistent with this region being unstructured or intrinsically disordered.
Fig. 6.
The prodomain is intrinsically disordered. (A) An electrostatic potential map of the procaspase-6 prodomain with its corresponding amino acid sequence. The blue and red regions represent the relative localization of the positive and negative potentials, respectively. (B) The deuterium uptake profile of a representative peptic peptide (2–19) in the prodomain identified in the prodomain-containing caspase-6 maturation variants. The asterisk designates the D to A substitution at the indicated proteolytic cleavage site, rendering that site uncleavable. Error, SD of duplicate H/DX-MS measurements done on 2 separate days. (C) The corresponding MS spectra of peptide 2–19 presented in B. (D) Prediction of the disordered (blue) and protein-binding (orange) regions of caspase-6 using DISOPRED server. (E) CD spectra of cleaved caspase-6 with (D23A D179CT) and without (ΔN D179CT) the prodomain.
Subtractive analysis of the H/D exchange profiles of the cleaved caspase-6 variants (with intact prodomain but with or without the linker) and the fully cleaved, mature caspase-6 (Fig. 4 B and C) revealed a statistically significant decrease in the deuterium uptake at positions 26–32 after cleavage at Asp-23. In mammalian caspases, the 26–32 region of has never been visualized crystallographically. However, in the Spodoptera frugiperda caspase-1 structure (PDB ID code 1M72), the region equivalent to caspase-6 residues 25–27 forms a β-strand that makes backbone H bonds with L2′ residues Asp-193 and Ala-195. Unfortunately, the peptide containing Asp-193 and Ala-195 was among the 8% of caspase-6 that was not observed in the H/D exchange analysis. Nevertheless, given the decrease in the H/D exchange in the 26–32 region upon removal of the prodomain and the absence of any significant changes in the H/D exchange to the rest of the region of caspase-6 (Fig. 4C), we hypothesize that the interaction between the 26–32 region and the L2′ seen in S. frugiperda caspase-1 is also present in caspase-6 but only occurs in the procaspase form or after prodomain cleavage.
The full sequence of caspase-6 was analyzed for protein disorder using DISOPRED (39) (Fig. 6D). Regions of the prodomain and the linker were predicted to be in a highly disordered state and further categorized these regions as being involved in protein binding. In addition, CD spectra of caspase-6 with or without the prodomain (Fig. 6E) showed no dramatic changes in the secondary structures of caspase-6, consistent with the prodomain being in a disordered state. In addition, the intrinsic tryptophan fluorescence profiles of both procaspase-6 zymogen and cleaved forms with or without the prodomain (Fig. 5B) were superimposable, suggesting that the prodomain does not impact the Trp-227 environment near the active site. Altogether, the H/DX-MS and the biophysical measurements imply that the prodomain is an unstructured region of caspase-6, which does not form any stable contacts with either the active site or the caspase-6 core at any point during the course of caspase-6 proteolytic maturation.
Discussion
The H/D exchange reported here has allowed us to follow the diverse conformational changes that occur in caspase-6 at discrete cleavage states as it progresses through the zymogen self-maturation pathway (Fig. 7). These data provide insight on the prodomain and intersubunit linker of caspase-6, as these regions have been absent in all crystal structures of the enzyme. The initial intramolecular cleavage of the zymogen at Asp-193 in the linker resulted in changes in the conformational dynamics of the substrate-binding loops L1–L4, the 130s, and the 26–32 regions. Moreover, the cleavage at Asp-193 also allowed the L2 region to engage with L4. Farther along the proteolytic activation pathway, removal of the linker increases the conformational flexibility of all substrate-binding loops and the 130s region. On substrate binding, all substrate-binding loops are engaged to accommodate the substrate, including the top of the 130s region. Importantly, the prodomain is found to be intrinsically disordered independent of the activation state of caspase-6 (i.e., zymogen or Asp-193 self-cleaved form). However, the presence of the prodomain impacts the conformational flexibility of the 26–32 region dependent on the proteolytic activation state of caspase-6.
Fig. 7.
A schematic model highlighting the changes in the conformational flexibility of caspase-6 along its path to proteolytic maturation. Initial intramolecular self-cleavage of the procaspase-6 zymogen at Asp-193 in the intersubunit linker results in significant exposure of the linker, the 130s region, the 26–32 region, and the substrate-binding loop L1, whereas L2 and L4 are engaged in a stabilizing interaction. The subsequent intermolecular cleavage of the prodomain at Asp-23 results in the protection of the 26–32 region. The complete removal of the linker on intermolecular cleavage at the Asp-179 site results in more exposure of regions in the 130s and all of the substrate-binding loops L1–L4. Also, on substrate binding (red ellipsoids), the 130s and the substrate-binding loops L1–L4 become protected to engage the substrate in the active site cavity. The intensities of the red and blue colors represent the exposure or protection, respectively, relative to the conformational flexibility of the procaspase-6 zymogen.
The changes in the dynamics of caspase-6 reported here square with previously reported changes in the thermal stability profile of maturation variants of caspase-6 (25). The zymogen was found to be the least stable form of caspase-6, with at least an 8 °C decrease in the apparent melting temperature compared with its cleaved forms. The Asp-193 self-cleaved form (with intact prodomain and linker) is the most stable form of caspase-6. The dramatic gain in the apparent thermal stability of the zymogen after the initial intramolecular self-cleavage at Asp-193 can likely be accounted for as the outcome of the changes in the conformational dynamics of caspase-6. In particular, we observed a stabilizing interaction of L2 and L4 promoted by the presence of Asp-193-cleaved linker. Previous work on proteome-wide thermal stability across various cell types from Escherichia coli to mammalian cells showed a direct association between protein stability and intracellular abundance and an inverse relationship between protein stability and aggregation (42). Thus, the gain of stability as a consequence of altered conformational dynamics during proteolytic activation in caspase-6 may be relevant to the lifetime of the protein in the cell and its interaction with the proteasome for degradation.
Crystal structures of the mature unliganded caspase-6 showed that, before substrate binding, the 130s region can exist in either the canonical (strand) (43) or noncanonical (helical) (25, 44) conformations. Mature caspase-6 exists exclusively in the canonical (strand) conformation in all substrate-bound structures—a unique structural feature not observed in any other caspases. We have recently reported H/DX-MS coupled with molecular dynamics simulations, which shows that the 130s region of the mature unliganded caspase-6 is in constant interconversion between the helical and strand conformations before substrate binding and converts completely to the strand conformation after substrate binding (36). H/DX-MS results presented here show that the presence of the linker affects slow H/D exchange in the 130s region in caspase-6 (Fig. 3I). At long time points, the H/D exchange profile of the Asp-193 cleaved caspase-6 with intact linker was similar to the canonical (strand) caspase-6 forms (the zymogen and substrate-bound caspase-6); however, it is distinct from the H/D exchange profile of the mature unliganded caspase-6, irrespective of the presence of the prodomain. Thus, these data suggest that the presence of the linker promotes a higher fractional residence in the canonical (strand) conformation of caspase-6. This dynamic conformational state of caspase-6 may represent one of the conformational ensembles that enables fine-tuning of pathways in the cell leading to caspase-6 activation.
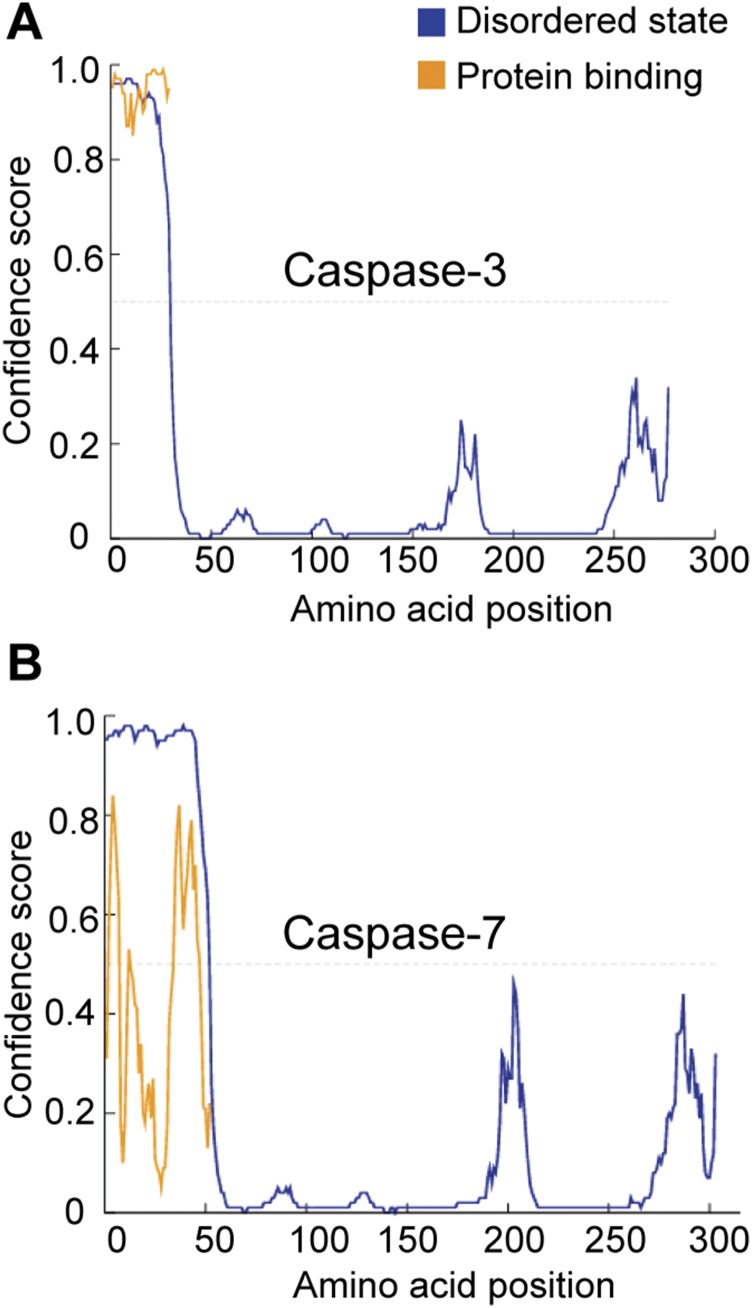
Although the biological consequences of the dynamic nature of the 130s region are still unknown, there have been previous cell-based studies that reported the relative activity of the maturation-state variants of caspase-6. Each maturation-state variant showed unique in cellulo activity in HEK293T cells (21), suggesting that each cleavage state of caspase-6 may play unique functional roles in cells. This is not unprecedented in caspases. Caspase-8 and -10 show altered substrate repertoire based on their maturation state and binding partners (45–47). These differences in caspase-6 cellular activity are certainly related to the dynamics changes observed in this study. In addition, the caspase-6 intersubunit linker is long relative to the other executioners, caspase-3 and -7, and is the only linker for which DISOPRED (48) analysis suggests the presence of a protein-binding motif (Fig. 6D and Fig. S9). This insinuates the linker as potentially playing a role in substrate selection and specificity as well as in the several protein–protein interactions, in which caspase-6 has been identified to participate (49, 50).
Fig. S9.
Prediction of disorder and protein binding in caspases. Amino sequences of caspase-3 (A) and caspase-7 (B) were submitted to the DISOPRED for the prediction of regions of the protein that are disordered (blue) as well as possible motifs for protein binding (orange).
In this work, we present several lines of evidence that the prodomain in caspase-6 is intrinsically disordered in all activation states. In addition, the crystal structure of the procaspase-6 zymogen with an intact prodomain showed no visible electron density for the prodomain region (32). In addition, the prodomain of caspase-6 was computationally predicted to be both disordered and a protein-binding region. Consistent with these observations, the H/D exchange profile indicated that the prodomain is unstructured or intrinsically disordered. In general, disordered regions of proteins are often involved in molecular recognition (reviewed in ref. 51). It is thought that several metastable conformations of proteins with disordered binding sites enable recognition of molecular targets with remarkable specificity and low affinity. Intriguingly, the presence of the prodomain in cleaved caspase-6 is dispensable for caspase-6 activity in vitro against peptide substrates but has been reported to impact caspase-6 activity intracellularly (21). This may indicate that the prodomain binds proteins, potential substrates, or regulatory partners. The caspase-6 prodomain has also been implicated in limiting self-activation through intermolecular cleavage at Asp-193 at low protein concentrations in vitro (32). Despite growing evidence of the roles of the prodomain in regulating caspase-6 activity, its biological role remains poorly understood; however, our characterization as an intrinsically disordered protein might provide the insight needed to further the elucidation of prodomain roles.
The prodomains of caspase-3 and -7 are also predicted to be intrinsically disordered (Fig. S9), and the prodomains of several caspases have been reported to participate in protein–protein interactions (30, 31, 52) impacting enzyme activity and apoptotic signal transduction as well as mediating binding to molecular chaperones, cellular localization, and substrate selection. Caspase prodomains also appear to be “hot spots” for regulatory posttranslational modifications (53–55) modulating enzyme activity and binding to initiator caspases, resulting in decreased apoptosis as well as impacting proteasomal degradation. The prodomain also aids folding of procaspase-3 (28). The prodomain is one of the least conserved regions of the caspase family (24, 25), and therefore, it is possible or even likely that each prodomain plays unique roles that have yet to be fully uncovered.
The exchangeability of the 26–32 region, which is immediately adjacent to the prodomain, changes in unforeseen ways as caspase-6 progresses along the proteolytic activation pathway. Initial self-cleavage at Asp-193 of the zymogen results in increased exposure of the 26–32 region, suggesting that an interaction of the 26–32 region, which is present in the zymogen, is lost on initial cleavage of the intersubunit linker. However, after caspase-6 fully matures by proteolytic cleavage of the prodomain and linker, the 26–32 region exhibits a less flexible conformation similar to the zymogen state. This finding may be relevant to fine-tuning caspase-6 activity that stems from the diverse conformational flexibility profile of caspase-6 along its pathway to full maturation. Notably, the 26–32 region is located in close proximity to Lys-36, one of the ligands in the allosteric site for zinc binding identified in caspase-6 (40). Moreover, this region is also positioned in the same loop as two regulatory sites in caspase-7. PAK2-mediated phosphorylation of S30 in this region prevents caspase-7 activation by caspase-9 (55). The 38KKKK site within the caspase-7 N-terminal domain is an exosite used for substrate selection and is the only identified exosite in any caspase. The differential conformational flexibility of the 26–32 region could potentially expose or protect key regulatory sites in caspases. For example, the differential exposure or protection of the 26–32 region in various activation states may explain differential substrate recruitment that has been observed intracellularly (21). While it is exciting to observe enzymatic differences in the cellular context, we recognize that insights into the molecular mechanisms can only be accomplished by in vitro investigations, such as those included in this work.
In summary, H/DX-MS revealed the distinct conformational dynamics in critical regions of the caspase-6 structure, including regions like the prodomain and intersubunit linker, which have not been observable by any other techniques. At each stage along the proteolytic activation pathway, changes in distinct regions of caspase-6 have been observed. These changes are confined to the substrate-binding loops, the 130s, and the 26–32 region. We anticipate that many of these changes would also be relevant when procaspase-6 is activated by caspase-3 via cleavage at Asp-179 and to the activation pathways of other caspases. In addition, we have observed that the prodomain is intrinsically disordered. The structural dynamic changes provided in this study afforded insights into the underlying molecular mechanism of caspase-6 activation and regulation. Caspase-6 is implicated in neurological disorders, including Alzheimer’s, Huntington’s, and Parkinson’s diseases. The structural features and conformational changes described here may inspire approaches for manipulating caspase-6 in the context of neurodegeneration.
Materials and Methods
Generation of Caspase Variants.
The caspase-6 variants FL C163S, D23A D179A, D23A D179CT, ΔN D179A, and ΔN D179CT were derived from the synthetic, E. coli codon-optimized (His)6 C-terminally tagged caspase-6 gene (Celtek Bioscience) ligated into the NdeI/BamHI sites of a pET11a vector (25). ΔN C163S with C-terminal (His)6 tag was generated using FL C136S as s template through Phusion mutagenesis (Thermo Scientific).
Caspase Expression, Purification, and Activity Assays.
Caspase-6 variants were expressed, purified, and assayed as previously described (36).
Tau Protein Expression and Purification.
The human Tau-383 (0N4R) variant in pMXB10 vector (gift from Bing Zhou, Tsinghua University, Beijing) was expressed in BL21(DE3) T7 express E. coli strain and purified as described (56, 57). Briefly, cells were grown in 2x yeast extract tryptone media until A600 of 0.6. Cells were harvested, lysed, and centrifuged at 30,600 × g for 1 h at 4 °C. Supernatant was loaded into chitin beads (New England Biolabs) and washed with binding buffer (20 mM Tris, pH 8.5, 500 mM NaCl, 1 mM EDTA, 0.1% Tween-20) until absorbance reached baseline levels. The column was flushed with three-column volumes of binding buffer with 50 mM DTT and incubated for 16 h at 4 °C to allow removal of the intein/maltose-binding protein tag. After incubation, the protein was eluted with binding buffer. The eluted protein was loaded into HiLoad 26/600 Superdex 200 Column and eluted using 200 mM Hepes, pH 7.5, and 150 mM NaCl. The purified protein was analyzed by SDS/PAGE to confirm identity and purity.
Proteolysis of Tau Protein by Caspase-6 Variants.
Human Tau protein (3 μM) was incubated with varying concentrations of active caspase-6 activation-state variants (0–3 μM; twofold dilution) in caspase-6 assay buffer (100 mM Hepes, pH 7.5, 10% sucrose, 0.1% CHAPS, 120 mM NaCl, 5 mM DTT) at 37 °C for 6 h. SDS loading buffer was added to the samples and boiled for 10 min before analysis by 16% SDS/PAGE. The gels were imaged using ChemiDoc MP imaging system (Bio-Rad).
H/DE-MS.
H/D exchange experiments on caspase-6 were performed as described previously (36) and as described in SI Materials and Methods.
CD Spectroscopy.
Caspase-6 activation-state variants (8 μM) were concentrated and diluted three times in 10 mM phosphate buffer, pH 7.5, with 120 mM NaCl in an Amicon Ultra 0.5-mL centrifugal filter (MWCO 10K; Millipore). After buffer exchange, caspase-6 concentrations were determined by 280-nm absorbance. CD spectra (250–190 nm) were measured on a J-1500 CD spectrometer (Jasco) with Peltier temperature controller. All data were collected at triplicate on different days.
Intrinsic Fluorescence Spectroscopy.
Caspase-6 activation-state variants (3 μM) in 20 mM phosphate buffer, pH 7.5, 120 mM NaCl, and 2 mM DTT were prepared. Fluorescence emission scans (305–400 nm) were collected after excitation at 280 or 295 nm using J-1500 spectrometer (Jasco) equipped with fluorescence emission monochromator (FMO-522) and detector (FDT-538). Signal was acquired by setting the emission detector at high-tension voltage value of 700 V, the data integration time to 1 s, and the data pitch to 1 nm. The bandwidths used for excitation and emission were 2 and 10 nm, respectively.
Caspase-6 Model Building and Disorder Prediction.
The FL caspase-6 zymogen model (residues 1–293) was built from crystal structures of caspase-6 zymogen. In this model, chain A was derived from PDB ID code 4iyr (chain A); chain B of this model was derived from PDB ID code 3NR2 (chain A) as templates. The missing residues [1–30, 174–186, and 292–293 in PDB ID code 4iyr (chain A); 1–30, 167–186, 261–271, and 292–293 in PDB ID code 3NR2 (chain A)] were built by de novo modeling using Chimera/Modeller platforms (38, 39). All illustrations with molecular visualization were generated using the PyMOL Molecular Graphics System (Schrödinger, LLC). Amino acid sequences of caspase-3, -6, and -7 were submitted to the DISOPRED (48) server for the prediction of protein disorder.
SI Materials and Methods
H/D exchange experiments on caspase-6 were performed as described previously (36). Briefly, an initial stock of 15 μM proteins in 20 mM Tris, pH 8.5, 200 mM NaCl, and 2 mM DTT in H2O was prepared. The protein samples were then introduced into the nanoACQUITY system equipped with H/D exchange technology for ultra-performance liquid chromatography (UPLC) separation (58) (Waters Corp.), which performed all subsequent manipulations for the H/D exchange. Accurate mass and collision-induced dissociation in data-independent acquisition mode (MSE) (59) and ProteinLynx Global Server (PLGS) 3.0 software (Waters Corp.) were used to determine the peptic peptides in the undeuterated protein samples analyzed on the same UPLC-electrospray ionization quadrupole time-of-flight (ESI-Q-ToF) system used for H/DX-MS experiments. Data from peptic peptides generated from PLGS were imported into DynamX 3.0 (Waters Corp.), with peptide quality thresholds of MS1 signal intensity ≥ 5,000, maximum sequence length of 25 aa, maximum mass error of 1 ppm, and minimum products per amino acid of ≥0.3. Automated results were manually inspected to ensure the corresponding m/z and isotopic distributions at various charge states were properly assigned to the appropriate peptic peptide. DynamX 3.0 was then used to generate the relative deuterium incorporation plot and H/D exchange heat map for each peptic peptide. The relative deuterium incorporation of each peptide was determined by subtracting the weight-averaged centroid mass of the isotopic distribution of undeuterated control sample from that of the weight-averaged centroid mass of the isotopic distribution of deuterium-labeled samples at each labeling time point. All comparisons were performed under identical experimental conditions, negating the need for back exchange correction in the determination of the deuterium incorporation. Thus, H/D exchange levels are reported as relative (60). The fractional relative deuterium uptake was calculated by dividing the relative deuterium uptake of each peptic peptide by its theoretical maximum uptake. All H/DX-MS experiments were performed in duplicate on 2 separate days, and a 98% confidence limit for the uncertainty of the mean relative deuterium uptake of ±0.6 Da was calculated as described (61). Differences in deuterium uptake between two states that exceed 0.6 Da were considered significant.
Acknowledgments
We thank Stephen J. Eyles (director of the UMass Institute of Applied Life Sciences Mass Spectrometry Core Facility) for abundant assistance with H/DX-MS data collection and processing. This work was supported by NIH Grant GM080532.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704640114/-/DCSupplemental.
References
- 1.Albrecht S, et al. Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol. 2007;170:1200–1209. doi: 10.2353/ajpath.2007.060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H, et al. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer’s disease. Am J Pathol. 2004;165:523–531. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeBlanc AC. Caspase-6 as a novel early target in the treatment of Alzheimer’s disease. Eur J Neurosci. 2013;37:2005–2018. doi: 10.1111/ejn.12250. [DOI] [PubMed] [Google Scholar]
- 4.Aharony I, et al. A Huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant Huntingtin-induced motor and behavioral deficits. Hum Mol Genet. 2015;24:2604–2614. doi: 10.1093/hmg/ddv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham RK, et al. Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci. 2010;30:15019–15029. doi: 10.1523/JNEUROSCI.2071-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong BK, et al. Partial rescue of some features of Huntington disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol Dis. 2015;76:24–36. doi: 10.1016/j.nbd.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Giaime E, et al. Loss of function of DJ-1 triggered by Parkinson’s disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death Differ. 2010;17:158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horowitz PM, et al. Early N-terminal changes and caspase-6 cleavage of tau in Alzheimer’s disease. J Neurosci. 2004;24:7895–7902. doi: 10.1523/JNEUROSCI.1988-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albrecht S, Bogdanovic N, Ghetti B, Winblad B, LeBlanc AC. Caspase-6 activation in familial Alzheimer disease brains carrying amyloid precursor protein or presenilin I or presenilin II mutations. J Neuropathol Exp Neurol. 2009;68:1282–1293. doi: 10.1097/NEN.0b013e3181c1da10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H, et al. Caspase-1 activation of caspase-6 in human apoptotic neurons. Cell Death Differ. 2006;13:285–292. doi: 10.1038/sj.cdd.4401753. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki A, et al. Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene. 2004;23:7067–7075. doi: 10.1038/sj.onc.1207963. [DOI] [PubMed] [Google Scholar]
- 13.Orth K, Chinnaiyan AM, Garg M, Froelich CJ, Dixit VM. The CED-3/ICE-like protease Mch2 is activated during apoptosis and cleaves the death substrate lamin A. J Biol Chem. 1996;271:16443–16446. [PubMed] [Google Scholar]
- 14.Srinivasula SM, et al. The Ced-3/interleukin 1beta converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2alpha are substrates for the apoptotic mediator CPP32. J Biol Chem. 1996;271:27099–27106. doi: 10.1074/jbc.271.43.27099. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi A, et al. Cleavage of lamin A by Mch2 alpha but not CPP32: Multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci USA. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon DJ, et al. A caspase cascade regulating developmental axon degeneration. J Neurosci. 2012;32:17540–17553. doi: 10.1523/JNEUROSCI.3012-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slee EA, et al. Ordering the cytochrome c-initiated caspase cascade: Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allsopp TE, et al. Caspase 6 activity initiates caspase 3 activation in cerebellar granule cell apoptosis. Cell Death Differ. 2000;7:984–993. doi: 10.1038/sj.cdd.4400733. [DOI] [PubMed] [Google Scholar]
- 19.Doostzadeh-Cizeron J, Yin S, Goodrich DW. Apoptosis induced by the nuclear death domain protein p84N5 is associated with caspase-6 and NF-kappa B activation. J Biol Chem. 2000;275:25336–25341. doi: 10.1074/jbc.M000793200. [DOI] [PubMed] [Google Scholar]
- 20.LeBlanc A, Liu H, Goodyer C, Bergeron C, Hammond J. Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer’s disease. J Biol Chem. 1999;274:23426–23436. doi: 10.1074/jbc.274.33.23426. [DOI] [PubMed] [Google Scholar]
- 21.Klaiman G, Champagne N, LeBlanc AC. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta. 2009;1793:592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Wang XJ, et al. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010;11:841–847. doi: 10.1038/embor.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasula SM, et al. Generation of constitutively active recombinant caspases-3 and -6 by rearrangement of their subunits. J Biol Chem. 1998;273:10107–10111. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- 24.Clark AC. Caspase allostery and conformational selection. Chem Rev. 2016;116:6666–6706. doi: 10.1021/acs.chemrev.5b00540. [DOI] [PubMed] [Google Scholar]
- 25.Vaidya S, Velázquez-Delgado EM, Abbruzzese G, Hardy JA. Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J Mol Biol. 2011;406:75–91. doi: 10.1016/j.jmb.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin H, et al. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 27.Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 28.Feeney B, Clark AC. Reassembly of active caspase-3 is facilitated by the propeptide. J Biol Chem. 2005;280:39772–39785. doi: 10.1074/jbc.M505834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meergans T, Hildebrandt AK, Horak D, Haenisch C, Wendel A. The short prodomain influences caspase-3 activation in HeLa cells. Biochem J. 2000;349:135–140. doi: 10.1042/0264-6021:3490135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voss OH, et al. Binding of caspase-3 prodomain to heat shock protein 27 regulates monocyte apoptosis by inhibiting caspase-3 proteolytic activation. J Biol Chem. 2007;282:25088–25099. doi: 10.1074/jbc.M701740200. [DOI] [PubMed] [Google Scholar]
- 31.Boucher D, Blais V, Denault JB. Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc Natl Acad Sci USA. 2012;109:5669–5674. doi: 10.1073/pnas.1200934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Q, Wang XJ, Li LF, Su XD. The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays. Acta Crystallogr D Biol Crystallogr. 2014;70:58–67. doi: 10.1107/S1399004713024218. [DOI] [PubMed] [Google Scholar]
- 33.Chalmers MJ, et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 34.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 36.Dagbay KB, Bolik-Coulon N, Savinov SN, Hardy JA. Caspase-6 undergoes a distinct helix-strand interconversion upon substrate binding. J Biol Chem. 2017;292:4885–4897. doi: 10.1074/jbc.M116.773499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu YH, Johnson DA, Traugh JA. Analysis of conformational changes during activation of protein kinase Pak2 by amide hydrogen/deuterium exchange. J Biol Chem. 2008;283:36397–36405. doi: 10.1074/jbc.M805581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 40.Velázquez-Delgado EM, Hardy JA. Zinc-mediated allosteric inhibition of caspase-6. J Biol Chem. 2012;287:36000–36011. doi: 10.1074/jbc.M112.397752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan CR, Engen JR. Investigating solution-phase protein structure and dynamics by hydrogen exchange mass spectrometry. Curr Protoc Protein Sci. 2009;58:17.6.1–17.6.17. doi: 10.1002/0471140864.ps1706s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leuenberger P, et al. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355:eaai7825. doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- 43.Müller I, et al. A new apo-caspase-6 crystal form reveals the active conformation of the apoenzyme. J Mol Biol. 2011;410:307–315. doi: 10.1016/j.jmb.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Baumgartner R, et al. The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J. 2009;423:429–439. doi: 10.1042/BJ20090540. [DOI] [PubMed] [Google Scholar]
- 45.Hughes MA, et al. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell. 2009;35:265–279. doi: 10.1016/j.molcel.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 46.Pop C, et al. FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem J. 2011;433:447–457. doi: 10.1042/BJ20101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu JW, Jeffrey PD, Shi Y. Mechanism of procaspase-8 activation by c-FLIPL. Proc Natl Acad Sci USA. 2009;106:8169–8174. doi: 10.1073/pnas.0812453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 49.Jung JY, et al. Identification of novel binding partners for caspase-6 using a proteomic approach. J Microbiol Biotechnol. 2014;24:714–718. doi: 10.4014/jmb.1312.12068. [DOI] [PubMed] [Google Scholar]
- 50.Riechers SP, et al. Interactome network analysis identifies multiple caspase-6 interactors involved in the pathogenesis of HD. Hum Mol Genet. 2016;25:1600–1618. doi: 10.1093/hmg/ddw036. [DOI] [PubMed] [Google Scholar]
- 51.Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 52.Choi YE, et al. The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase-3 and -7 via unique mechanisms at distinct steps in their processing. J Biol Chem. 2009;284:12772–12782. doi: 10.1074/jbc.M807550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin MC, Allan LA, Mancini EJ, Clarke PR. The docking interaction of caspase-9 with ERK2 provides a mechanism for the selective inhibitory phosphorylation of caspase-9 at threonine 125. J Biol Chem. 2008;283:3854–3865. doi: 10.1074/jbc.M705647200. [DOI] [PubMed] [Google Scholar]
- 54.Nutt LK, et al. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell. 2005;123:89–103. doi: 10.1016/j.cell.2005.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eron SJ, Raghupathi K, Hardy JA. Dual site phosphorylation of caspase-7 by PAK2 blocks apoptotic activity by two distinct mechanisms. Structure. 2017;25:27–39. doi: 10.1016/j.str.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venter PA, et al. Multivalent display of proteins on viral nanoparticles using molecular recognition and chemical ligation strategies. Biomacromolecules. 2011;12:2293–2301. doi: 10.1021/bm200369e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, Kang JH, Kim KH, Lee EK. Expression of intein-tagged fusion protein and its applications in downstream processing. J Chem Technol Biotechnol. 2010;85:11–18. [Google Scholar]
- 58.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geromanos SJ, et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9:1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 60.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 61.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]