Significance
TGF-β and related growth factors critically regulate cell potency and functions. Smad7 is induced by TGF-βs and inhibits the physiological functions of TGF-β signaling. This study describes an unexpected finding that Smad7 promotes self-renewal of embryonic stem cells (ESCs) in a manner independent of its inhibition on TGF-β signaling. Instead, Smad7 acts to induce activation of transcription factor signal transducers and activators of transcription 3 (STAT3) in ESCs. Smad7 activates STAT3 through its direct binding to the cytokine receptor upstream of STAT3 activation. In agreement with the role of STAT3 in maintaining ESC pluripotency, Smad7 promotes ESC self-renewal and induced pluripotent stem cell reprogramming. This finding illustrates a regulatory mechanism for Smad7 in maintaining pluripotency, and likely in cancer and inflammation.
Keywords: Smad7, gp130, STAT3, TGF-β, pluripotency, differentiation
Abstract
Smad7 is a negative feedback product of TGF-β superfamily signaling and fine tunes a plethora of pleiotropic responses induced by TGF-β ligands. However, its noncanonical functions independent of TGF-β signaling remain to be elucidated. Here, we show that Smad7 activates signal transducers and activators of transcription 3 (STAT3) signaling in maintaining mouse embryonic stem cell pluripotency in a manner independent of the TGF-β receptors, yet dependent on the leukemia inhibitory factor (LIF) coreceptor glycoprotein 130 (gp130). Smad7 directly binds to the intracellular domain of gp130 and disrupts the SHP2–gp130 or SOCS3–gp130 complex, thereby amplifying STAT3 activation. Consequently, Smad7 facilitates LIF-mediated self-renewal of mouse ESCs and is also critical for induced pluripotent stem cell reprogramming. This finding illustrates an uncovered role of the Smad7–STAT3 interplay in maintaining cell pluripotency and also implicates a mechanism involving Smad7 underlying cytokine-dependent regulation of cancer and inflammation.
Members of the TGF-β superfamily, including TGF-β, Activin, Nodal, and BMP, play a major role in maintaining pluripotency in stem cells and controlling cell fate determination during development (1–4). In cell culture, BMP4 and leukemia inhibitory factor (LIF) are required to maintain pluripotency (5). BMP induces Id proteins to suppress differentiation and sustain embryonic stem cell (ESC) self-renewal (6). Although being indispensable for ESC propagation, TGF-β/Activin/Nodal induce differentiation of ESCs in the absence of LIF (7). During mouse fibroblast reprogramming into ES-like cells by Oct4, KLF4, c-Myc, and Sox2 or alternatives (8), BMP enhances reprogramming into induced pluripotent stem cells (iPSCs) (9, 10), while TGF-β signaling exerts an inhibitory effect on iPSC induction (11, 12). Thus, in response to morphogen gradients of the TGF-β superfamily ligands, the cellular outcome is determined by integration of their balanced signaling activities.
Signals of the TGF-β superfamily are transduced by intracellular R-Smads, i.e., BMP-activated Smad1, Smad5, and Smad8, and TGF-β/Activin/Nodal-activated Smad2 and Smad3. Smad7, induced by all ligands of the TGF-β superfamily, can act as a negative feedback product to inhibit TGF-β signaling (13, 14). Smad7 can compete with R-Smads for binding to the type I receptor (e.g., TβRI) (15–17), recruit the HECT E3 ubiquitin ligases to promote proteasomal degradation of the receptor proteins (18), or recruit protein phosphatase 1 to inactivate the type I receptor (19, 20). In addition, Smad7 also disrupts the association of functional R-Smad–Smad4 complexes as well as binding of R-Smads complex to DNA in the nucleus (21). Certain cytokines such as IFN-γ induce expression of Smad7 to suppress TGF-β/BMP signaling (22). Although it modulates NF-κB, c-Jun N-terminal kinase (JNK)/p38, and Wnt signaling (23–25), Smad7 has been thought to function primarily through its inhibitin on both TGF-β and BMP signaling. It has been reported that Smad7 directly converts human ESCs to telencephalic fate (26) and promotes self-renewal of mouse hematopoietic stem cells (27). In mouse ESCs, an increased level of Smad7 due to loss of its E3 ligase RNF12 impairs both activin-induced anterior mesoderm formation and BMP-mediated repression of neural induction (28). Despite all these studies on Smad7, it remains elusive whether Smad7 acts through a non–TGF-β pathway to impact ESC pluripotency.
LIF and related cytokines signal through the glycoprotein 130 (gp130) and signal transducers and activators of transcription 3 (STAT3) (29–34). Following the engagement of LIF to its receptor complex containing gp130, Janus kinases (JAKs) become catalytically activated. Activated JAKs phosphorylate tyrosine residues in the intracellular domain of gp130, which serves as docking sites for the Src homology 2 (SH2) domains of STATs (35, 36). When bound to gp130 and JAK, STATs are phosphorylated, translocate to the nucleus, and then bind to the STAT-binding elements in the target genes. Termination of the gp130–STAT3 signaling is effectuated by several mechanisms, including dephosphorylation of gp130 by the SH2-containing phosphatase (SHP2) (37) and negative feedback inhibition by the suppressor of cytokine signaling (SOCS) proteins (38).
In regulating ESC pluripotency, LIF-activated STAT3 functions through its cooperation with the core pluripotency transcription factors such as Oct4, Nanog, and Sox2 (39). Smad1/5/8 also cooperate with Oct4, Nanog, and Sox2 as well as STAT3 to maintain pluripotency (39). In addition, STAT3 can promote expression of TGF-β1 (40) and Smad7 (41, 42). STAT3 also selectively interacts with Smad3 to antagonize TGF-β signaling (43), whereas Smads attenuate the STAT3 signaling through an inhibition of STAT3 binding to DNA and cooperation with p300 (44). However, it remains unknown whether Smad7, induced by both TGF-β and STAT3 signaling, directly cross-talks with STAT3 signaling to influence ESC pluripotency. In this study, we identified and characterized a direct interaction between Smad7 and gp130 that leads to Smad7-mediated amplification of the gp130–STAT3 signaling and maintenance of embryonic pluripotency. This unexpected action of Smad7 is independent of its inhibitory effect on TGF-β/BMP signaling. Our findings elucidate a mechanism underlying the modulation of the gp130–STAT3 axis by Smad7 and identify the essential role of Smad7 as a critical cell fate regulator.
Results
Smad7 Attenuates Mouse ESC Differentiation.
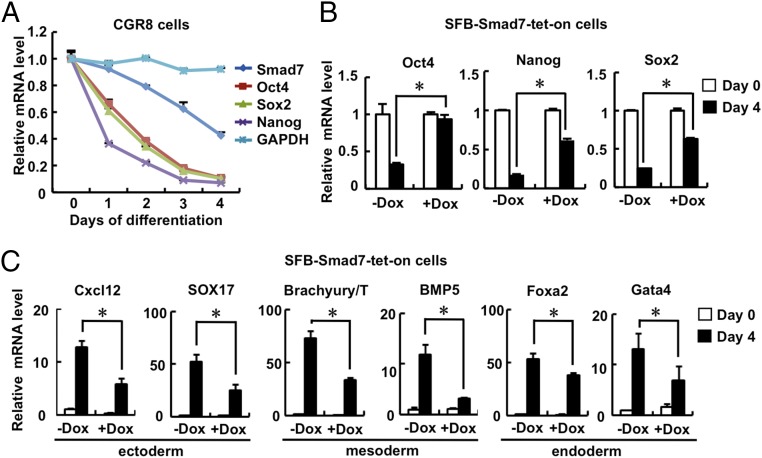
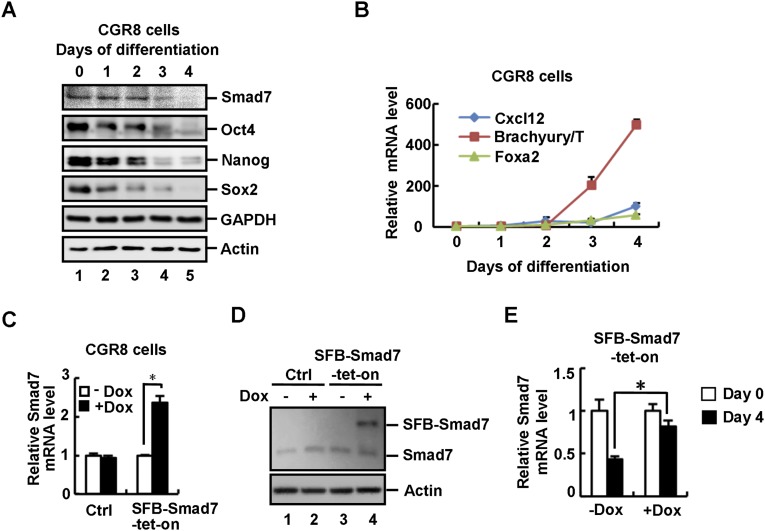
Smad7 has been identified as a STAT3 target gene, and it is abundantly expressed in mouse ESCs (41, 42). We first investigated whether the expression of Smad7 changes during embryoid body (EB) differentiation derived from mouse ESCs. During cell differentiation, we observed an apparently gradual decrease in expression of Smad7, which was accompanied by the decrease of pluripotency markers including Oct4, Nanog, and Sox2 (Fig. 1A and Fig. S1A), whereas expression of differentiation markers such as Brachyury/T, Foxa2, and Cxcl12 profoundly increased (Fig. S1B). These results indicate a high expression level of Smad7 may be required for the undifferentiated state of ESCs.
Fig. 1.
Smad7 promotes self-renewal and inhibits differentiation of ESCs. (A) Smad7 is down-regulated during EB formation in CGR8 cells. qRT-PCR was used to analyze mRNA levels of Smad7, Oct4, Sox2, Nanog, and GAPDH. Data are shown as mean ± SEM; n = 3. (B) Smad7 maintains a high expression level of pluripotency markers during EB formation. SFB–Smad7–tet-on CGR8 underwent EB differentiation for 4 d in the absence or presence of 1 μg/mL Dox. Total RNAs were subjected to qRT-PCR to examine expression levels of Oct4, Nanog, Sox2, and Smad7. Data are shown as mean ± SEM; n = 3. *P < 0.05. (C) Smad7 inhibits ESC differentiation during EB formation. The experiment was essentially performed as described in Fig. 1B, and qRT-PCR was used to examine mRNA levels of ectoderm, mesoderm, and endoderm markers. Data are shown as mean ± SEM; n = 3. *P < 0.05.
Fig. S1.
Smad7 promotes self-renewal and inhibits differentiation of ESCs. (A) Smad7 is down-regulated during EB formation in CGR8 cells. Cell lysates were subjected to Western blot analysis to examine protein levels of Smad7, Oct4, Sox2, Nanog, and GAPDH (as a negative control) with appropriate antibodies. (B) Differentiation markers are up-regulated during EB formation in CGR8 cells. Total RNAs were subjected to qRT-PCR to examine mRNA levels of Cxcl12, Brachyury/T, and Foxa2. Data are shown as mean ± SEM; n = 3. (C) Dox induces an approximately two-fold increase in Smad7 mRNA in SFB–Smad7–tet-on CGR8 cells. Wild-type and SFB–Smad7–tet-on CGR8 cells were cultured in the absence or presence of 1 μg/mL Dox for 72 h. Total RNAs were extracted from cells and subjected to qRT-PCR to examine expression levels of Smad7. Data are shown as mean ± SEM; n = 3. *P < 0.05. (D) Dox induction of Smad7 gives a moderate increase in total Smad7 protein in SFB–Smad7–tet-on CGR8 cells. Wild-type and SFB–Smad7–tet-on CGR8 cells were cultured in the absence or presence of 1 μg/mL Dox for 72 h. Cells were subjected to Western blot analysis to examine the protein level of Smad7. (E) Dox treatment maintains the mRNA level of Smad7 during EB formation. SFB–Smad7–tet-on CGR8 underwent EB differentiation for 4 d in the absence or presence of 1 μg/mL Dox. Total RNAs were extracted from cells and subjected to qRT-PCR analysis to examine the mRNA level of Smad7. Data are shown as mean ± SEM; n = 3. *P < 0.05.
To further investigate whether Smad7 regulates ES cell fate determination, stable and inducible expression of Smad7 was established in the mouse ES cell line CGR8 using the tetracycline-inducible (tet-on) system, designated as SFB–Smad7–tet-on cells. Doxycycline (Dox) treatment induced a moderate expression of Smad7 in SFB–Smad7–tet-on cells (Fig. S1 C and D). During EB differentiation, Oct4, Nanog, and Sox2 as well as endogenous Smad7 were reduced at day 4 of differentiation, yet Dox-induced expression of Smad7 maintained high expression levels of these pluripotency markers (Fig. 1B and Fig. S1E). In contrast, induced expression of Smad7 markedly decreased expression of differentiation markers of all three germ layers, including ectodermal markers (i.e., Cxcl12 and SOX17), mesodermal markers (i.e., Brachyury/T and BMP5), and endodermal markers (i.e., Foxa2 and Gata4) at day 4 of EB differentiation (Fig. 1C). These results imply that Smad7 promotes self-renewal of mouse ESCs.
Smad7 Is Essential in Promoting ESC Self-Renewal and iPSC Reprogramming.
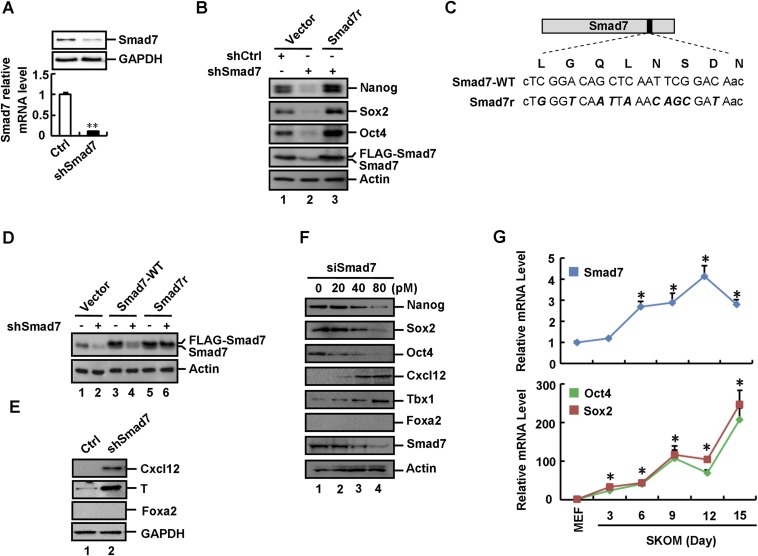
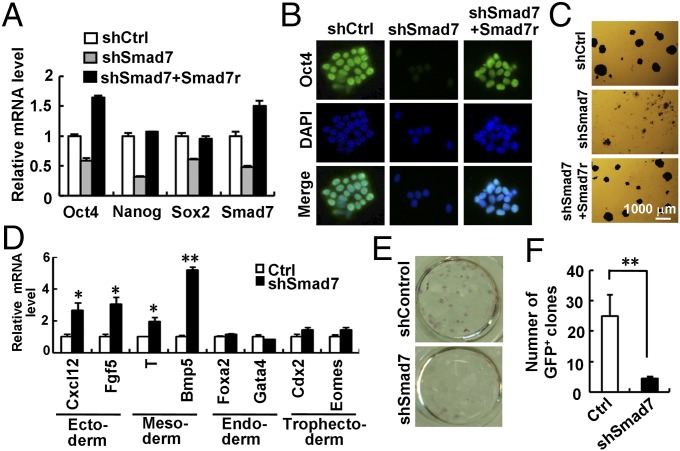
We next determined whether loss of Smad7 expression could enhance ESC differentiation. In mESCs, stably expressed shSmad7 (Fig. S2A) reduced the mRNA and protein levels of Oct4, Nanog, and Sox2 (Fig. 2A and Fig. S2B). By using immunofluorescence, a profound loss of Oct4 expression was observed in shSmad7-expressing cells (Fig. 2B). Consistently, shSmad7 resulted in low alkaline phosphatase (AP) activity (Fig. 2C). Notably, an shRNA-resistant variant of Smad7 (FLAG-tagged Smad7r, Fig. S2 C and D) completely rescued the effect of shSmad7 on ESC differentiation (Fig. 2 A–C and Fig. S2B), demonstrating the specific on-target effect of shSmad7. In addition, shSmad7 induced the mRNA and protein levels of ectodermal markers (i.e., Cxcl12 and Fgf5) and mesodermal markers (i.e., Brachyury/T and BMP5), but not endodermal or trophectodermal markers (Fig. 2D and Fig. S2E). Furthermore, transient knockdown of Smad7 exhibited the same effect on ESC self-renewal and differentiation (Fig. S2F). Thus, our results strongly support a direct role of Smad7 in maintaining ESC self-renewal.
Fig. S2.
Smad7 is essential in maintenance of pluripotency. (A) Stable knockdown of Smad7 in CGR8 cells. Cell lysates from pLKO.1–control and pLKO.1–shSmad7 CGR8 stable cells were subjected to qRT-PCR and Western blot analysis. qRT-PCR data are shown as mean ± SEM; n = 3. **P < 0.01. (B) shSmad7 down-regulates expression of pluripotency marker proteins in ESCs. pLKO.1–control and pLKO.1–shSmad7 CGR8 stable cells were transfected with the indicated plasmids for 48 h. Cells were subjected to Western blot analysis to examine protein levels of Nanog, Sox2, Oct4, Smad7, and β-actin. (C) Schematic representation of the FLAG-tagged Smad7r expression vector. The shSmad7 target sequence (encoding amino acids 893–913, above the nucleotide sequence) is indicated by capital letters. Mutations introduced are indicated by bold/italic letters. (D) pLKO.1–control and pLKO.1–shSmad7 CGR8 stable cells were transfected with the empty vector or the indicated Smad7 expression vectors. Cell lysates were subjected to Western blot analysis to examine Smad7 protein levels. Smad7-WT is the FLAG-tagged wild-type Smad7, whereas Smad7r is a FLAG-tagged Smad7 variant resistant to shSmad7. (E) Depletion of Smad7 enhances expression of Cxcl12 (ectodermal marker) and Brachyury/T (mesodermal marker), but not Foxa2 (endodermal marker). Cell lysates of shSmad7 and control ESCs were subjected to Western blot analysis to examine protein levels of Cxcl12, T, and Foxa2. (F) Knockdown of Smad7 inhibits stemness and enhances ES cell differentiation into ectoderm and mesoderm. CGR8 cells were transfected with different concentrations of Smad7 siRNA as indicated and cultured for 48 h. Cell lysates were subjected to Western blot analysis to examine protein levels of pluripotency and differentiation markers. (G) Smad7 increases during the MEF reprogramming process. Cells were collected every 3 d from day 0 to day 15 after infection with pMXs-based OSKM. Total RNAs were extracted from cells and subjected to qRT-PCR to examine expression levels of Smad7, Oct4, and Sox2. Data are shown as mean ± SEM; n = 3. *P < 0.05.
Fig. 2.
Smad7 is essential in maintenance of pluripotency. (A–C) Depletion of Smad7 down-regulates expression of pluripotency markers and induces differentiation in ESCs. CGR8 cells stably expressing shSmad7 were established as described in Supporting Information. Smad7r is a RNAi-resistant variant of Smad7. (A) qRT-PCR analysis of Oct4, Nanog, Sox2, and Smad7. Data are shown as mean ± SEM; n = 3. (B) Immunofluorescence analysis of Oct4. DNA was stained with DAPI. (C) AP staining of CGR8 cells stably expressing shSmad7 and/or Smad7r. (D) Depletion of Smad7 enhances ESC differentiation into ectoderm and mesoderm, but not endoderm or trophectoderm. qRT-PCR was used to examine mRNA levels of indicated differentiation markers. Data are shown as mean ± SEM; n = 3. *P < 0.05, **P < 0.01. (E and F) Smad7 depletion inhibits the reprogramming efficiency in reprogrammable MEFs. Reprogrammed iPSC colonies were identified by AP staining (E) and quantitation of GFP-positive clones (F) at day 14. Data are shown as mean ± SEM; n = 3. **P < 0.01.
Given the positive role of Smad7 in promoting ESC self-renewal, we were interested in determining whether Smad7 has a critical role in iPSC reprogramming. We used four conventional reprogramming factors, i.e., Oct4, Sox2, KLF4, and c-Myc (OSKM), to induce pluripotency in mouse embryonic fibroblasts (MEFs). Accompanied by the increased expression of OSKM, we observed an increase in the expression of Smad7 (Fig. S2G). We then further determined the role of Smad7 in iPSC reprogramming. While OSKM could produce as high as 25% reprogramming efficiency in Oct4–GFP MEFs, as indicated by AP- and GFP-positive iPSC colonies, shSmad7 markedly reduced the number of OSKM-induced iPSC colonies (Fig. 2 E and F). Our results indicate that depletion of Smad7 profoundly blocks OSKM-mediated reprogramming into iPSCs.
Smad7 Regulates Pluripotency Independent of TGF-β/BMP Signaling.
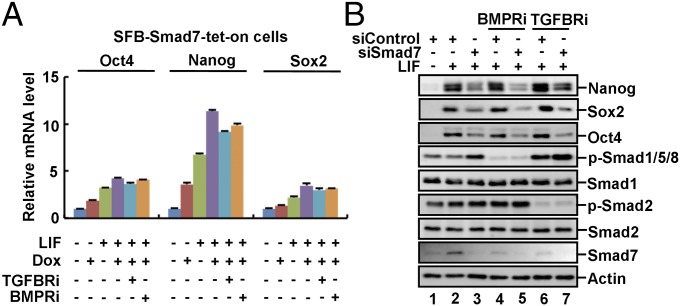
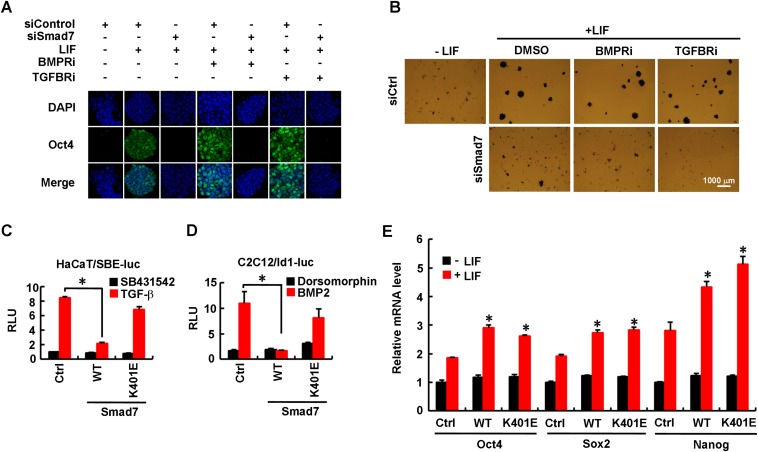
We next attempted to determine whether Smad7 regulates pluripotency through canonical inhibition of TGF-β/BMP signaling. We first examined the effects of small molecule inhibitors SB431542 against TGF-β type I receptor or Dorsomorphin against BMP type I receptor, named TGFBRi and BMPRi, respectively. Dox-induced expression of Smad7 could suffice to moderately increase expression of pluripotency markers, especially Oct4 and Nanog, which was further increased by addition of exogenous LIF (Fig. 3A). It is not surprising that TGFBRi or BMPRi had no effects on Smad7-induced up-regulation of Nanog, Oct4, and Sox2 (Fig. 3A) as Smad7 itself is a potent inhibitor of TGF-β/BMP signaling. Remarkably, the effect of Smad7 depletion on expression of pluripotency markers, as either measured by Western blotting analysis (Fig. 3B) or immunofluorescence (Fig. S3A) or AP activity (Fig. S3B), remained unchanged after treatment with TGFBRi or BMPRi. Furthermore, the Smad7 mutant K401E, which was defective in binding to the type I receptor (45), failed to effectively inhibit TGF-β or BMP signaling (Fig. S3 C and D) and notably retained its ability to promote expression of ESC pluripotency markers (Fig. S3E). These findings clearly demonstrated that the function of Smad7 in controlling ESC pluripotency may not rely on its negative regulation of TGF-β–Smad signaling.
Fig. 3.
Smad7 promotes pluripotency independent of canonical TGF-β/Smad signaling. (A) Smad7 promotes ESC self-renewal independent of TGF-β/BMP signaling. SFB–Smad7–tet-on cells were pretreated with 5 μM SB431542 (TGFBRi) or 10 μM Dorsomorphin (BMPRi) for 12 h and then cultured in indicated medium for another 3 d. qRT-PCR was used to analyze expression of indicated pluripotency markers. Data are shown as mean ± SEM; n = 3. (B) Inhibition of TGF-β/BMP signaling does not reverse the effect of siSmad7 in ESC pluripotency. CGR8 cells were transfected with 40 pM Smad7 siRNA or control siRNA and cultured with TGFBRi or BMPRi for 2 d. Cell lysates were subjected to Western blot analysis.
Fig. S3.
Smad7 promotes pluripotency independent of canonical TGF-β/Smad signaling. (A) siSmad7 inhibits Oct4 expression independent of TGF-β/BMP signaling. CGR8 cells were transfected with 40 pM Smad7 siRNA or control siRNA and then split into six-well plates at a low density. Transfected cells were treated with TGFBRi or BMPRi for 3 d. Cells were fixed and immune stained with anti-Oct4 antibody. DNA was stained with DAPI. (B) siSmad7 inhibits ES cell colony formation independent of TGF-β/BMP signaling. CGR8 cells were transfected with siRNAs as described in Fig. S3A. Transfected cells were treated with TGFBRi or BMPRi for 5 d, and then fixed and subjected to AP staining. (C) Smad7 mutant K401E does not inhibit the TGF-β–induced SBE–luc reporter activity. HaCaT cells were cotransfected with wild-type Smad7 (WT) or Smad7–K401E together with the Smad-binding element (SBE)–luc reporter (a synthetic TGF-β–responsive reporter), followed by stimulation with 5 μM SB431542 (TGF-β type I receptor inhibitor) or 2 ng/mL TGF-β for 12 h. Cells were harvested for luciferase assay. Data are shown as mean ± SEM; n = 3. *P < 0.05. (D) Smad7 mutant K401E does not inhibit the BMP-induced Id–luc reporter activity. C2C12 cells were cotransfected with wild-type Smad7 (WT) or Smad7–K401E together with the expression plasmid for Id1–luc reporter (a luciferase reporter driven by the Id1 promoter containing the BMP-responsive element), followed by stimulation with 10 μM Dorsomorphin (BMP type I receptor inhibitor) or 50 ng/mL BMP2 for 12 h. Cells were harvested for luciferase assay. Data are shown as mean ± SEM; n = 3. *P < 0.05. (E) Smad7 promotes ESC stemness independent of its ability to bind to TβRI or BMPRI in ESCs. CGR8 cells were transfected with FLAG–Smad7 (WT), the K401E mutant or control plasmid and then treated with or without 0.1 ng/mL LIF for 48 h. Total RNAs were extracted from cells and subjected to qRT-PCR to examine expression level of indicated pluripotency markers. Data are shown as mean ± SEM; n = 3. *P < 0.05.
Smad7 Activates STAT3 Independent of TGF-β Receptor Signaling.
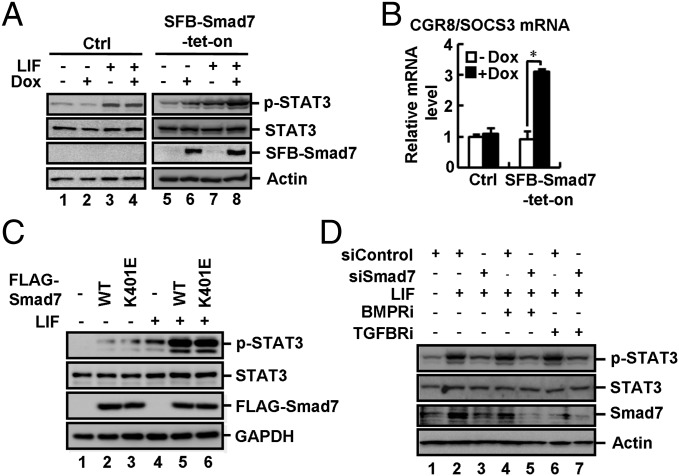
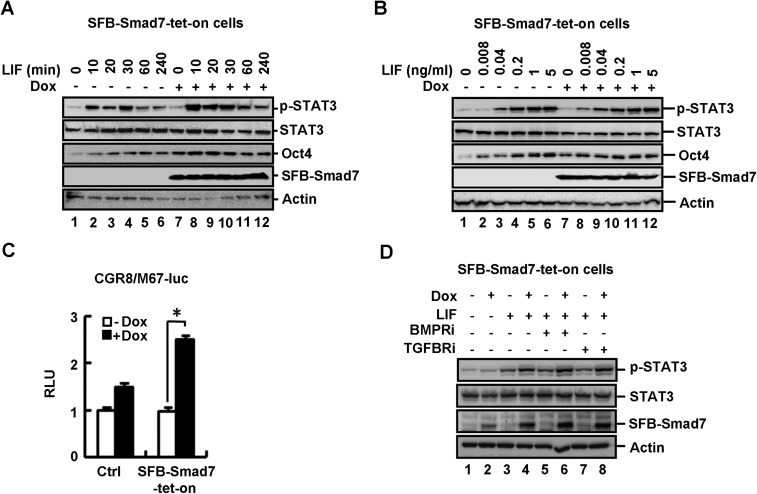
Smad7 is not only induced by TGF-β signaling, but also by JAK–STAT signaling (41, 42, 46). We sought to determine whether increased expression of Smad7 could affect STAT3 activation. In CGR8 SFB–Smad7–tet-on cells, Dox induced expression of SFB-tagged Smad7 (Fig. 4A, lanes 6 and 8) and promoted LIF-induced STAT3 phosphorylation at Y705 (p-STAT3), indicative of STAT3 activation (Fig. 4A, lane 8). Smad7 apparently enabled ESCs to be more sensitive to LIF (short time or low dosage) in STAT3 activation and Oct4 expression (Fig. S4 A and B). As a consequence, Dox induced a higher level of endogenous SOCS3 mRNA (a STAT3 target gene) (Fig. 4B) and also a significantly higher level of LIF-induced M67-luc reporter activity (Fig. S4C). In sharp contrast, Dox did not alter the levels of p-STAT3, SOCS3 mRNA, and M67-luc activity in control (Ctrl) cells (Fig. 4 A and B and Fig. S4C).
Fig. 4.
Smad7 potently activates gp130-mediated STAT3 signaling. (A) Smad7 stimulates LIF-induced STAT3 activation in ESCs. CGR8 control (Ctrl) and SFB–Smad7–tet-on cells were cultured ± Dox (1 μg/mL, 72 h) and then treated with LIF (0.1 ng/mL, 20 min). Cell lysates were subjected to Western blot analysis. (B) Smad7 enhances SOCS3 expression in ESCs. SFB–Smad7–tet-on or control cells were treated with or without 1 μg/mL Dox for 72 h. qRT-PCR was used to examine the SOCS3 mRNA level. Data are shown as mean ± SEM; n = 3. *P < 0.05. (C) Smad7 activates LIF-induced STAT3 signaling independent of TβRI or BMPRI in ESCs. WT and the K401E mutant of Smad7 are indicated. CGR8 cell transfection, LIF treatment, and Western blot analysis were done as described in Supporting Information. (D) Inhibition of TβRI/BMPRI does not influence the effect of siSmad7 in LIF-induced STAT3 signaling in ESCs. CGR8 cell transfection, LIF treatment, and Western blot analysis were done as described in Supporting Information.
Fig. S4.
Smad7 potentiates gp130-mediated STAT3 signaling. (A) Smad7 enhances LIF-induced STAT3 activation and Oct4 expression in ES cells. SFB–Smad7–tet-on cells were cultured with or without 1 μg/mL Dox for 72 h and then treated with 0.1 ng/mL LIF for indicated time. Cell lysates were subjected to Western blot analysis. (B) Smad7 increases the sensitivity of ES cells to LIF-induced STAT3 activation. SFB–Smad7–tet-on cells were cultured with or without 1 μg/mL Dox for 72 h and then treated with indicated concentrations of LIF for 20 min. Cell lysates were subjected to Western blot analysis. (C) Smad7 up-regulates LIF-activated reporter activity in ESCs. SFB–Smad7–tet-on or control cells were transfected with M67–luc reporter (a synthetic luciferase reporter with STAT3-responsive elements) and Renilla luciferase plasmids, and then treated with or without 1 μg/mL Dox for 72 h. Cell were harvested for luciferase assay. Data are shown as mean ± SEM; n = 3. *P < 0.05. (D) Inhibition of TβRI/BMPRI does not affect the ability of Smad7 in enhancing LIF-induced STAT3 signaling in ESCs. SFB–Smad7–tet-on cells were pretreated with TGFBRi or BMPRi for 12 h and then cultured with or without 1 μg/mL Dox for 72 h and then treated with 0.1 ng/mL LIF for another 20 min. Cell lysates were subjected to Western blot analysis.
We then determined whether Smad7-enabled STAT3 activation requires TGF-β receptor signaling. We found that the Smad7 mutant K401E, defective in binding to the type I receptor, was as potent as wild-type Smad7 in activating STAT3 (Fig. 4C). TGFBRi or BMPRi had no effects on Smad7-induced STAT3 activation (Fig. S4D). Notably, TGFBRi or BMPRi could not reverse the effect of shSmad7 on LIF-mediated STAT3 activation (Fig. 4D). Thus, our results suggest that Smad7 potentiates STAT3 signaling independent of its inhibitory effects on TGF-β signaling.
Smad7 Directly Binds to gp130 and Disrupts the SHP2/SOCS3 Binding to gp130.
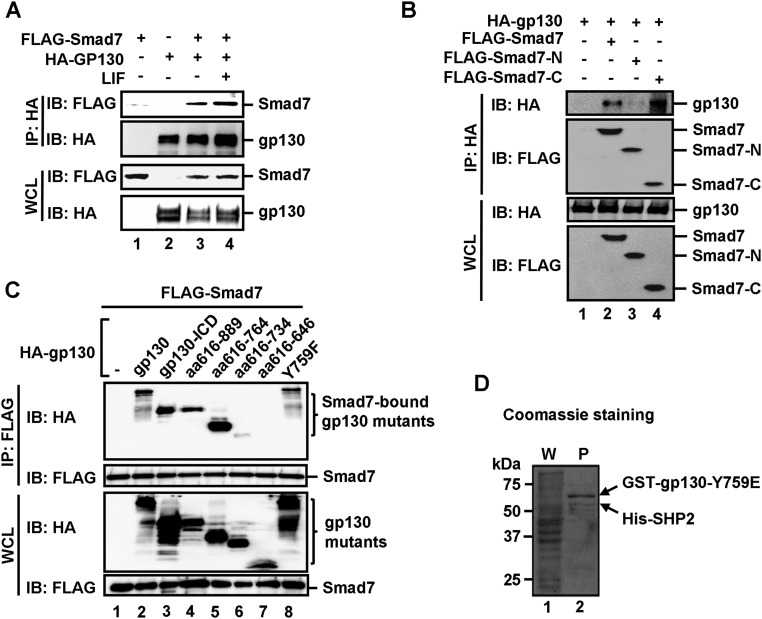
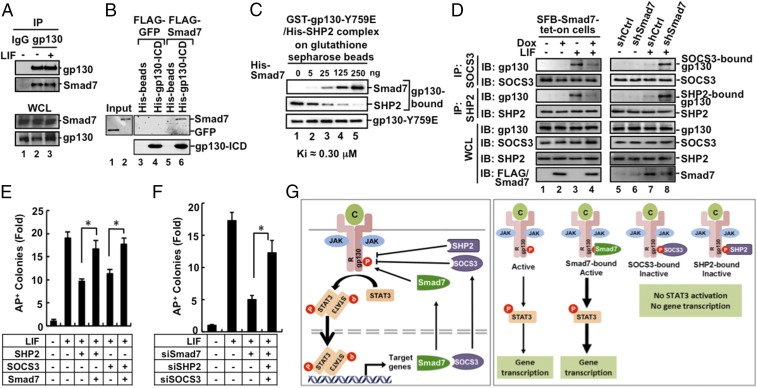
Because Smad7 promotes LIF-induced STAT3 activation, we speculated that Smad7 might interact with the LIF–gp130–STAT3 pathway. Indeed, we found that Smad7 bound to gp130, a coreceptor for LIF in transfected cells in coimmunoprecipitation (co-IP) assays (Fig. S5A). Further, Smad7 could interact with gp130 at the endogenous levels (Fig. 5A). We also conducted an in vitro binding assay using purified recombinant proteins. As shown in Fig. 5B, His-tagged gp130–ICD protein (the cytoplasmic domain of gp130) could bind to Smad7, but not GFP protein, indicating that Smad7 directly binds to the cytoplasmic domain of gp130.
Fig. S5.
Smad7 directly binds to the cytoplasmic domain of gp130 and disrupts the binding of SHP2/SOCS3 to gp130. (A) Smad7 interacts with gp130 in vivo. HEK293T cells were cotransfected with FLAG–Smad7 and HA–gp130 and then treated with 1 ng/mL LIF for 2 h before harvest. The cell lysates were harvested and immunoprecipitated with anti-HA antibody. Smad7 and gp130 were detected from the immunoprecipitates by Western blot analysis. (B) The MH2 domain (or C domain) of Smad7 binds to gp130. HEK293T cells were transfected with indicated plasmids. Levels of these proteins in IP products and whole cell lysates were analyzed by Western blot analysis. (C) The 734–764 aa region in the cytoplasmic domain of gp130 is essential in Smad7 binding. HEK293T cells were cotransfected with FLAG–Smad7 and a HA-tagged gp130 wild-type or a deletion mutant, and then treated with 1 ng/mL LIF for 2 h before harvest. Cell lysates were harvested and immunoprecipitated with anti-FLAG antibody. Smad7 and gp130 variants were detected from the immunoprecipitates by Western blot analysis. (D) Formation of the Y759E–SHP2 complex in E. coli. Fusion proteins of GST–gp130–Y759E and His–SHP2 were coexpressed in E. coli. The Y759E–SHP2 complex was retrieved by glutathione Sepharose beads, as indicated by Coomassie staining. P, purified proteins on glutathione Sepharose beads; W, whole cellular protein lysates.
Fig. 5.
Smad7 directly competes with SHP2/SOCS3 for gp130 binding and enables STAT3 signaling in maintaining pluripotency. (A) Smad7 interacts with gp130 under physiological conditions. Immunoprecipitation and Western blot analysis were done as described in Supporting Information. (B) Smad7 directly interacts with gp130. In vitro binding was carried out with purified His–gp130–ICD and in vitro translated Smad7. Experimental details are described in Supporting Information. (C) Smad7 displaces SHP2 on gp130. Increasing concentrations of purified His–Smad7 proteins were added to the gp130Y759E–SHP2 complex, and followed by Western blot analysis with indicated antibodies. (D) Smad7 competes with endogenous SHP2 and SOCS3 for gp130 binding in CGR8 cells. Cell lysates were immunoprecipitated with anti-SOCS3 antibody or anti-SHP2 antibody. Cell culture, LIF treatment, immunoprecipitation, and Western blot analysis were done as indicated and described in Supporting Information. (Left) SFB–Smad7–tet-on cells treated with or without Dox; (Right) CGR8 cells with shCtrl and shSmad7. In the bottom blots, FLAG/Smad7 means the use of anti-FLAG in lanes 1–4 and anti-Smad7 in lanes 5–8. (E) Smad7 overcomes SHP2- or SOCS3-mediated suppression of ESC colony formation. CGR8 cell transfection and AP staining were performed as described in Supporting Information. The bar graph represents the fold change of numbers of uniform AP+ colonies in Fig. S6F. Data are shown as mean ± SEM; n = 3. *P < 0.05. (F) SiSmad7 inhibition of ESC self-renewal is reversed by simultaneous knockdown of SHP2 and SOCS3. Experiments and data analysis were done as described in Fig. 5E and Supporting Information. The bar graph represents the fold change of numbers of uniform AP+ colonies in Fig. S6H. Data are shown as mean ± SEM; n = 3. *P < 0.05. (G) A working model for Smad7 potentiating STAT3 activation. (Left) LIF and related cytokines (C) bind to the gp130 receptor complex. Receptor-associated JAK kinases phosphorylate STAT3 leading to STAT3 accumulation in the nucleus, where STAT3 controls expression of target genes, including Smad7 and SOCS3. SOCS3 and SHP2 bind to gp130 to inhibit STAT3 activation. Smad7 can compete for the gp130 binding, maintaining STAT3 activation. (Right) Active and inactive forms of the cytokine-receptor–gp130 complex are shown.
To determine the structural features for the Smad7–gp130 interaction, we first mapped the domain of Smad7 for gp130 binding. Our co-IP assay revealed that both wild-type and the MH2 or C domain (aa 228–426) of Smad7, but not the N domain (amino acids 1–228), bound to gp130 (Fig. S5B). On the gp130 side, a series of HA-tagged deletions in gp130–ICD were tested for their interactions with FLAG–Smad7 in HEK293T cells (Fig. S5C). The mutant containing amino acids 616–918, 616–889, or 616–764 retained the ability to interact with Smad7, whereas the amino acids 616–734 and 616–646 mutants did not bind to Smad7 (Fig. S5C), indicating that residues 734–764 could be potentially critical for the Smad7 binding.
The 734–764 aa region of gp130 has a critical phosphotyrosine-759 (pY759). SHP2 and SOCS3 are recruited to the pY759 residue to block STAT3 activation (37, 38). We then asked whether Smad7 blocks the binding of SHP2/SOCS3 to gp130. GST–gp130–Y759E (mimicking Y759 phosphorylation) and His–SHP2 were coexpressed in Escherichia coli and the preformed complex between gp130–Y759E and SHP2 was retrieved using glutathione beads (Fig. S5D). Interestingly, when added to the purified gp130–Y759E/SHP2 complex in vitro, increasing amounts of recombinant His–Smad7 protein competitively replaced His–SHP2 for binding to GST–gp130–Y759E with an approximate Ki of 0.30 μM (Fig. 5C). These results demonstrate that Smad7 directly competes with SHP2 for gp130 binding.
We further assessed the effect of Smad7 on endogenous gp130–SHP2 or gp130–SOCS3 interactions in CGR8 cells. Lentiviral expression of exogenous Smad7 profoundly blocked binding of endogenous SHP2 or SOCS3 to gp130 (Fig. 5D, lane 4). Furthermore, shSmad7 markedly increased the physiological interaction of either SHP2 or SOCS3 with gp130 in CGR8 cells (Fig. 5D, lane 8). Collectively, our data suggest that Smad7 promotes gp130-mediated STAT3 signaling by overriding SHP2/SOCS3-mediated inhibition.
Smad7 Promotes Pluripotency Through Blocking SHP2 and SOCS3.
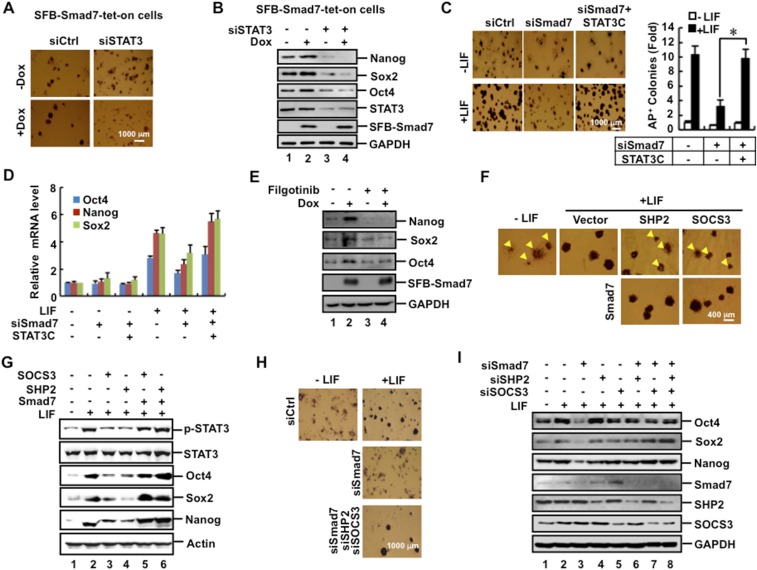
Having established the molecular antagonism between Smad7 and SHP2 or SOCS3 during STAT3 activation, we further assessed the relationship among Smad7, STAT3, and SHP2/SOCS3 in maintaining ESC pluripotency. While Dox-induced expression of Smad7 could induce formation of AP-positive colonies and enhanced expression of pluripotency markers, knockdown of STAT3 markedly attenuated the effect of Smad7 (Fig. S6 A and B). Conversely, whereas Smad7 depletion reduced the mRNA levels of pluripotency markers and abolished production of LIF-induced AP-positive colonies, overexpression of STAT3C (a constitutively active mutant of STAT3) completely rescued pluripotency in Smad7-depleted cells (Fig. S6 C and D). Moreover, JAK inhibitor Filgotinib also strongly attenuated the effect of Smad7 in ESC pluripotency (Fig. S6E). These results suggest that Smad7 stimulates stemness through JAK-dependent STAT3 activation.
Fig. S6.
Smad7 stimulates stemness through STAT3 activation. (A) STAT3 depletion blocks the positive effect of Smad7 in ESC colony formation. SFB–Smad7–tet-on cells were transfected with 40 pM STAT3 siRNA or control siRNA and then split into six-well plates at a low density. Transfected cells were cultured with or without 1 μg/mL Dox for 5 d and then fixed and subjected to AP staining. (B) STAT3 depletion blocks the effect of Smad7 in ESC pluripotency. SFB–Smad7–tet-on cells were transfected with 40 pM STAT3 siRNA or control siRNA and then cultured with or without 1μg/mL Dox for 3 d. Cell lysates were subjected to Western blot analysis. (C) siSmad7 inhibition of ESC self-renewal is rescued by constitutive activation of STAT3 signaling. CGR8 cells were cotransfected with Smad7 siRNA (40 pM) and FLAG–STAT3C and then split into six-well plates at a low density. Transfected cells were cultured with or without 1 ng/mL LIF for 5 d and then fixed and subjected to AP staining. The bar graph displays the fold change of numbers of colonies with uniform AP staining. Data are shown as mean ± SEM; n = 3. *P < 0.05. (D) Constitutive active STAT3C reverses the effect of siSmad7 inhibition on pluripotency marker expression. CGR8 cells were cotransfected with Smad7 siRNA (40 pM) and FLAG–STAT3C and then cultured with or without 1 ng/mL LIF for 2 d. Total RNAs were extracted from cells and subjected to qRT-PCR to examine mRNA levels of indicated pluripotency markers. (E) Inhibition of JAK kinase activity attenuates the effect of Smad7 in ES cell pluripotency. SFB–Smad7–tet-on cells were treated with 20 nM JAK inhibitor Filgotinib for 12 h and then cultured with or without 1 μg/mL Dox and 0.1 ng/mL LIF for 72 h. Cell lysates were subjected to Western blot analysis. (F) Smad7 overcomes the SHP2/SOCS3-mediated suppression of ESC colony formation. CGR8 cells were transfected with FLAG–Smad7, FLAG–SHP2, and/or MYC–SOCS3 as indicated. Experiments were performed as described in Fig. S6A. Arrowheads point to ESC colonies that are not uniformly ALP+. Statistical analysis of numbers of colonies with uniform ALP staining is presented in Fig. 5E. (G) Smad7 antagonizes the suppressive effects of SHP2 and SOCS3 on STAT3 activation and ESC pluripotency. CGR8 cells were transfected with FLAG–Smad7, FLAG–SHP2, and/or MYC–SOCS3 as indicated. Cell lysates were subjected to Western blot analysis. (H) siSmad7 inhibition of ESC self-renewal is reversed by simultaneous knockdown of SHP2 and SOCS3. CGR8 cells were transfected with siRNA (40 pM) specific to Smad7, SHP2, and/or SOCS3 as indicated. Experiments were performed as described in Fig. S6A. Statistical analysis of numbers of colonies with uniform ALP staining is presented in Fig. 5F. (I) siSmad7 inhibition of ES cell self-renewal is rescued by simultaneous knockdown of SHP2 and SOCS3. CGR8 cells were transfected with siRNA (40 pM) specific to Smad7, SHP2, or SOCS3 as indicated for 2 d. Cell lysates were subjected to Western blot analysis.
Although overexpression of SHP2 or SOCS3 reduced ESC colony formation, ectopic expression of Smad7 could reverse the action of SOCS3 or SHP2 to rescue ESC colony formation (Fig. 5E and Fig. S6F), STAT3 activation, and expression of Oct4, Sox2, and Nanog (Fig. S6G). Conversely, knockdown of Smad7 alone attenuated STAT3 activation and ESC pluripotency (Figs. 4D and 5F and Fig. S6 C and H). Notably, simultaneous double knockdown of SHP2 and SOCS3 in Smad7-depleted ESCs could restore AP-positive colony formation (Fig. 5F and Fig. S6H) and expression of pluripotency markers (Fig. S6I). Collectively, our findings illustrate that Smad7 antagonizes the negative role of SHP2 and SOCS3 in LIF/STAT3 signaling in pluripotency maintenance (Fig. 5G).
Discussion
Numerous investigations have elucidated the function of Smad7 in differentiated cells and adult stem cells. It is generally thought that the primary function of Smad7 is to negatively impact TGF-β/BMP signaling. However, Smad7 actions outside of the TGF-β/BMP signaling have rarely been explored. A previous study reported that Smad7 is highly expressed in undifferentiated ESCs (41). Consistently, we found that expression of Smad7 decreases during ESC differentiation (Fig. 1A and Fig. S1A), implying a possible function of Smad7 in maintaining ESC pluripotency. Here we report that Smad7 promotes ESC self-renewal and attenuates ESC differentiation and identify the direct role of Smad7 in maintaining pluripotency through a gp130–STAT3-dependent yet TGF-β/BMP-independent signaling pathway. In the current study, we not only reveal a function of Smad7 in controlling pluripotency, but also offer an underlying mechanism for previously unexplained signaling interplays.
A few lines of experimental evidence convincingly demonstrate the role of Smad7 in controlling pluripotency. First, induced expression of Smad7 up-regulates expression of the core pluripotency markers, whereas it down-regulates expression of differentiation makers of all three germ-layer lineages (Fig. 1). As a result, Smad7 promotes ESC colony formation. Second, depletion of Smad7 severely attenuates expression of the core pluripotency markers and colony formation and markedly enhances expression of differentiation makers in the ectoderm and mesoderm lineages (Fig. 2 A–D and Fig. S2 B, E, and F). In addition, we failed to generate complete knockout of the Smad7 gene in mouse ESCs using the CRISPR/Cas9 technology, implying the critical function of Smad7 in maintaining pluripotency. Third, depletion of Smad7 in MEFs drastically reduces the reprogramming efficiency (Fig. 2 E and F). Together with the molecular actions and interactions of Smad7 with the gp130–STAT3 pathway, these findings support the important function of Smad7 in maintaining pluripotency.
Although Smad7 is a well-established negative regulator in TGF-β/BMP signaling, our study has clearly revealed that Smad7 does not require TGF-β/BMP signaling to enable STAT3 activation and maintenance of pluripotency (Fig. 3 and Fig. S3). Small molecule inhibitors against TβRI or BMPRI fail to reverse the effect of Smad7 depletion on attenuating STAT3 activation and pluripotency. Moreover, Smad7 mutants deficient in binding to TβRI and BMPRI retain the ability to activate STAT3 signaling and expression of pluripotency markers. These results support the notion that Smad7 promotes STAT3 activation independently of TGF-β/BMP signaling.
Instead, our work has revealed that Smad7 specifically promotes pluripotency through the LIF–gp130–JAK–STAT3 pathway. STAT3 depletion or JAK1 inhibitor blocks Smad7-mediated promotion of ESC self-renewal. Moreover, constitutively activated STAT3 completely reverses the effect of shSmad7 on ESC differentiation. These results strongly suggest that Smad7 promotes LIF-induced STAT3 activation to stimulate ESC pluripotency. Our work has further revealed the molecular mechanism underlying Smad7-induced STAT3 signaling in pluripotency. Smad7 directly interacts with the cytoplasmic domain of gp130 (Fig. 5 A and B and Fig. S5A) and blocks the binding of SHP2 or SOCS to gp130, thereby ensuring the maintenance or amplification of STAT3 activation (Fig. 5 C and D). Indeed, ectopic expression of Smad7 can override the negative action of SOCS3 or SHP2 to rescue ESC pluripotency (Fig. 5E and Fig. S6 F and G), whereas the destructive effect of Smad7 depletion on STAT3 activation and ESC pluripotency can be counterbalanced by simultaneous knockdown of SHP2 and SOCS3 (Fig. 5F and Fig. S6 H and I). Collectively, our findings illustrate that Smad7-mediated disruption of the SHP2/SOCS3-dependent negative impact in LIF/STAT3 signaling is an essential regulatory means in pluripotency maintenance (Fig. 5G).
Our study also implicates that TGF-β/BMP signaling may regulate pluripotency through various mechanisms. Previous reports mostly attribute the actions of TGF-β/BMP signaling in controlling pluripotency to their direct role in regulating cell proliferation and differentiation. For example, Activin/Nodal/TGF-β is indispensable for ESC propagation (7), while BMP induces Id proteins to suppress differentiation and sustain ESC self-renewal (6). Providing the fact that Smad7 is induced by TGF-β/BMP signaling, it is plausible that Smad7 can act as an effector in mediating TGF-β/BMP signaling likely in promoting STAT3 activation. In addition, the BMP–Smad signaling and LIF–STAT3 pathways collaboratively control the maintenance of mouse ESC self-renewal (5). Smad1/5/8 can cooperate with the core pluripotency factors to maintain pluripotency (39). BMP increases LIF responsiveness in epiblast stem cells through a p300-bridged complex between Smad1 and STAT3 (47). Our study adds another layer of signaling cross-talk that BMP4-induced Smad7 may act to sensitize ESCs to respond to LIF in activating STAT3. Thus, as a transcriptional product in response to TGF-β/BMP ligands, Smad7 may positively effectuate certain TGF-β/BMP-induced responses such as pluripotency control.
Therefore, in addition to its well-established role in blocking canonical TGF-β–Smad signaling via binding to the TGF-β/BMP type I receptor, Smad7 can exert its cellular function through direct binding to a cytokine receptor and enhancement of downstream STAT3 signaling. Because the interplay between TGF-β and gp130–STAT3 signaling exists in various physiological contexts, it is conceivable that through its interaction with the gp130–STAT3 axis, Smad7 may have a broader role in bridging the collaborative functions of the TGF-β–Smad and gp130–STAT3 signaling pathways in other pathophysiological processes such as inflammation and tumorigenesis.
Materials and Methods
Cell Culture, Transfection, Immunoprecipitation, Immunofluorescence, qRT-PCR, and Western Blotting.
Culture and transfection of CGR8, HEK293T, C2C12, and HaCaT cell lines, and subsequent molecular analysis were done as described in Supporting Information.
Secondary Colony Formation Assay and Alkaline Phosphatase Staining.
Establishment of CGR8 and its stable lines with Dox-induced expression or knockdown of Smad7, cell transfection, LIF treatment, colony formation, and alkaline phosphatase staining were carried out as described in Supporting Information.
Full materials and methods are outlined in Supporting Information.
SI Materials and Methods
Cell Culture and Transfection.
CGR8 mouse embryonic stem cells (mESCs) were maintained in knockout DMEM (Gibco) supplemented with 15% FBS (Gibco), 2 mM l-glutamine (Gibco), 100 μM nonessential amino acids (Gibco), 0.1 mM β-mercaptoethanol, 1,000 units ml−1 LIF (Millipore) and penicillin/streptomycin on gelatin-coated plates. SFB–Smad7–tet-on cells, a stable CGR8 cell line with doxycycline (Dox)-inducible expression of SFB-tagged Smad7, were established and selected by G418 (500 μg/mL). HEK293T and C2C12 cells were cultured in DMEM (Corning) supplemented with 10% FBS (Gibco). HaCaT cells were cultured in MEM (Corning) supplemented with 10% FBS (Gibco).
Plasmids.
The following mammalian expression plasmids have been previously described: FLAG–Smad7 (48), Smad7–K401E (45), Smad7-N and Smad7-C (21). FLAG–Smad7r is a RNAi-resistant variant of Smad7 (designated Smad7r) that contains 10 silent mutations in the 21 bp Smad7–shRNA target sequence, which was generated by using PCR. The mutations were confirmed by sequence analysis. DNA fragments encoding HA-tagged gp130 variants, including gp130 (amino acids 1–918), gp130–ICD (616–918 aa, 616–889 aa, 616–764 aa, 616–734 aa, 616–646 aa), or Y759F were generated by PCR and subcloned into pRK3HA using BamHI and SalI sites. FLAG-tagged SHP2 and MYC-tagged SOCS3 were generated by PCR and subcloned into pRK5F (C-terminal FLAG tag) and pXF3HM, respectively. N-terminally FLAG-tagged STAT3C were generated by PCR and subcloned into pXF6F (N-terminal FLAG tag). All mammalian vectors are derived from pRK5 (Genentech). His-tagged gp130–ICD (intracellular domain, 642–918 aa) and Smad7 (FL) were generated by PCR and subcloned into pET28a (His tag). Bacterial expression plasmids coexpressing His–SHP2:GST–gp130–Y759E were generated according to Scheich et al. (49).
Antibodies and Reagents.
Antibodies and their commercial sources are as follows: Nanog (D73G4), Sox2 (L1D6A2), Oct-4A (C30A3), p-Smad1/5/8 (9511), Smad1 (D59D7), p-Smad2 (138D4), Smad2 (D43B4), p-STAT3 (D3A7), STAT3 (9139), HA (C29F4), SHP-2 (3397), SOCS3 (2923), and Cxcl12 (3740) from Cell Signaling Technology; Smad7 (sc-7004), MYC (sc-40), and gp130 (sc-656) from Santa Cruz Biotechnology; IgG from rabbit serum (I5006), FLAG (F3165), GAPDH (G8795), and β-actin (A5441) from Sigma-Aldrich; Tbx1 (ab109313) and Foxa2 (ab108422) from Abcam; and His (04905318001) from Roche.
The following chemical compounds and recombinant proteins were commercially obtained: SB431542 (S4317) and Dorsomorphin (P5499) from Sigma-Aldrich; Filgotinib (GLPG0634) (S7605) from Selleck; BMP2 (335-BM-050) from R&D Systems; and TGF-β (TGFB1-100) from StemRD.
If not specified, the following concentrations of the reagents or chemicals were generally used in cell culture: TGF-β at 2 ng/mL, BMP2 at 50 ng/mL, LIF at 0.1 ng/mL, Dox at 1 μg/mL, SB431542 at 5 μM, Dorsomorphin at 10 μM, and JAK inhibitor Filgotinib at 20 nM.
Immunoprecipitation and Western Blot Analysis.
Coimmunoprecipitation (co-IP) was carried out using antibodies and protein A Sepharose (GE Healthcare). After several washes, precipitated proteins were eluted in SDS loading buffer and separated by SDS/PAGE, transferred onto PVDF membranes (Millipore), and detected in Western blotting with appropriate antibodies.
Immunofluorescence.
Cells grown on coverslips were fixed with 4% formaldehyde for 20 min, and then incubated with 0.3% Triton X-100 and 5% BSA for 1 h. Cells were subsequently probed with primary antibodies, and then Alexa Fluor 546- or Alexa Fluor 488-conjugated secondary antibodies (Invitrogen). Fluorescence images were acquired by Zeiss LSM710 confocal microscope (Carl Zeiss).
In Vitro Protein Binding and Coexpression Assays.
His fusion protein of gp130 intracellular domain (gp130–ICD) was prepared from Escherichia coli strain DE3. In vitro translation of Smad7 and GFP were carried out using Quick Coupled Transcription/Translation System (Promega). In vitro binding was carried out using His–gp130–ICD on nickel Sepharose beads incubated with in vitro translated Smad7 and GFP for 2 h in the binding buffer (0.5% Nonidet P-40, 150 mM NaCl, 50 mM Tris⋅HCl, 5 mM EDTA), and followed by Western blot analysis.
To examine the preformed complex between gp130–Y759E and SHP2 in E. coli, GST–gp130–Y759E and His–SHP2 were coexpressed in E. coli growing in LB medium (2 L) with 0.5 mM IPTG for 16 h at 16 °C. Cells were harvested, resuspended in a bacterial lysis buffer (20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 2 mM β-mercaptoethanol, 1 mM PMSF, and one tablet/50 mL lysate of Roche complete EDTA-free protease inhibitor mixture) and lysed via incubation with 1 mg/mL lysozyme for 20 min on ice, followed by addition of 50 μg/mL DNaseI for 10 min on ice. Insoluble materials were removed by centrifugation (60 min, 75,000 × g) and the lysate was incubated with glutathione Sepharose beads for 2 h in the binding buffer (0.5% Nonidet P-40, 150 mM NaCl, 50 mM Tris⋅HCl, 5 mM EDTA), and followed by SDS/PAGE and Coomassie Blue staining.
RNA Interference and qRT-PCR.
siRNAs were synthesized by RIOBIO Co. and transfected at 40 pM into cells using Lipofectamine RNAiMAX reagent (Invitrogen). siRNA sequences targeting mouse genes were as follows: siSmad7, GAGGCTGTGTTGCTGTGAA; siSHP2, GAACCTTCATTGTGATTGA; and siSOCS3, GGAGTTCCTGGATCAGTAT. Total RNA (1 μg) isolated from cells using TRIzol Reagent (Sigma) was reverse transcribed to cDNA using Transcriptor Reverse Transcriptase (Roche). cDNA was then diluted and used for quantification by real-time PCR, which was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and the 7300 Real-Time PCR System (Applied Biosystems). qRT-PCR primers are listed as follows: m-Oct4 (For) 5′-TGAGAACCTTCAGGAGATATGCAA-3′ and m-Oct4 (Rev) 5′-CTCAATGCTAGTTCGCTTTCTCTTC-3′; m-Nanog (For) 5′-CAGAAAAACCAGTGGTTGAAGACTAG-3′ and m-Nanog (Rev) 5′-GCAATGGATGCTGGGATACTC-3′; m-Sox2 (For) 5′-CAGGAGAACCCCAAGATGCACAA-3′ and m-Sox2 (Rev) 5′-AATCCGGGTGCTCCTTCATGTG-3′; m-Cxcl12 (For) 5′-CTTCCTCCCAGAAGTCAGTCATCC-3′ and m-Cxcl12 (Rev) 5′-ACACAACACTGAACCCATCGCTG-3′; m-Brachyury/T (For) 5′-CATTACACACCACTGACGCACA and m-Brachyury/T (Rev) 5′-AGAAGACGAGGACGTGGCAG-3′; m-Foxa2 (For) 5′-CATCTCGCTCATCACCATGG-3′ and m-Foxa2 (Rev) 5′-CAGCGTCAGCATCTTGTTGG-3′; m-Cdx2 (For) 5′-AGGCTGAGCCATGAGGAGTA-3′ and m-Cdx2 (Rev) 5′-CGAGGTCCATAATTCCACTCA-3′; m-Eomes (For) 5′-CCTGGTGGTGTTTTGTTGTG-3′ and m-Eomes-(Rev) 5′-TTTAATAGCACCGGGCACTC-3′; m-Sox17 (For) 5′-AGCTCAGCGGTCTACTATTGCA-3′ and m-Sox17 (Rev) 5′-GGTCGGCAACCGTCAAAT-3′; m-BMP5 (For) 5′-GCTGCGCTCCAACCAAACTAAATG-3′ and m-BMP5 (Rev) 5′-ATTCCCCATTCCCCTGCTTTTTAT-3′; m-Gata4 (For) 5′-CTGTCATCTCACTATGGGCA-3′ and m-Gata4 (Rev) 5′-CCAAGTCCGAGCAGGAATTT-3′; m-Fgf5 (For) 5′-CTGTATGGACCCACAGGGAGTAAC-3′ and m-Fgf5 (Rev) 5′-ATTAAGCTCCTGGGTCGCAAG-3′; m-Smad7 (For) 5′-CCAACTGCAGACTGTCCAGA-3′ and m-Smad7 (Rev) 5′-CAGGCTCCAGAAGAAGTTGG-3′; m-SOCS3 (For) 5′-ATGGTCACCCACAGCAAGTTT-3′ and m-SOCS3 (Rev) 5′-TCCAGTAGAATCCGCTCTCCT-3′; m-GAPDH (For) 5′-CTCGCTCCTGGAAGATGGTG-3′ and m-GAPDH (Rev) 5′-GGTGAAGGTCGGTGTGAACG-3′; and m-β-actin (For) 5′-TGAGCGCAAGTACTCTGTGTGGAT-3′ and m-β-actin (Rev) 5′-ACTCATCGTACTCCTGCTTGCTGA-3′.
To generate stable knockdown of gene expression, a lentiviral shRNA expression system was used for knocking down expression of endogenous Smad7. A DNA fragment containing the target sequence of mouse Smad7 ORF (nt 893–913, TCGGACAGCTCAATTCGGACA) (23) or nonspecific control was cloned into the pLKO.1 lentiviral vector (Addgene plasmid no. 24150). The pLKO.1–shRNA plasmid was then transfected into 293T cells together with the packaging system. After 48 h, viral supernatants were prepared by filtration through a 0.45-μm filter and added to the target cells for infection for 6 h in the presence of polybrene (10 μg/mL). After incubation for 6 h, the medium was exchanged with a complete fresh medium. Cells were pooled and subjected to puromycin (1 μg/mL) selection for 4 d.
Luciferase Reporter Assays.
Reporter plasmids SBE–luc (50), Id1–luc (51), and M67–luc (52) were used to measure transcriptional responses induced by TGF-β, BMP, and STAT3, respectively. Cells were transfected with reporter plasmids together with a Renilla luciferase expression plasmid to normalize transfection efficiency. The luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). All assays were carried out in triplicates and normalized against Renilla luciferase activity.
Embryoid Body Formation.
The hanging drop method was used for embryoid body (EB) formation. ESCs were dissociated with 0.05% trypsin-EDTA solution to form a single cell suspension. A total of 1,000 cells in 30 μL of differentiation medium (ES culturing medium without LIF) were pipetted on the inside surface of a bacterial Petri dish lid. To prevent drying of the cell droplets, the Petri dish bottom was filled with PBS. The cells were forced to aggregate with the help of gravity by reversing the Petri dish lid. Two days later, EBs were transferred onto Ultra-Low attachment culture six-well plates (Corning).
Secondary Colony Formation Assay and Alkaline Phosphatase Staining.
ES cells were transfected with indicated plasmids and cultured for 24 h. Cells were then seeded at very low density to form colonies in six-well plates containing culture media in the presence or absence of LIF. Five days later, cells were stained with alkaline phosphatase (AP) staining kit (SCR004; Millipore), according to the manufacturer’s instructions. AP-stained and differentiated colonies were counted to score ES cell self-renewal ability.
Somatic Cell Programming.
Mouse embryonic fibroblasts (MEFs) from Oct4–GFP transgenic mice were seeded at a density of 30,000 cells in 12-well plates and infected with either Smad7 knockdown or control lentivirus for 24 h, and then infected with pMXs-based OSKM (Oct4, Sox2, Klf4, and c-Myc) for another 24 h. The virus supernatants were removed and fresh iPSC culture media were added to the plates. Media were changed every day. After 14 d, iPSC clones were examined by both AP staining and GFP fluorescence. Expression of Smad7 was determined by using real-time PCR 4 d postinfection.
Acknowledgments
This research was partly supported by various current and past grants, including National Natural Science Foundation of China (NSFC) Grants NSFC 91540205, NSFC 31571447, and NSFC 31090360; Ministry of Sciences and Technology of China (MOST) Grants 2015CB553800 and 2013CB966600; Department of Defense Grant 1W81XWH-15-1-0650; NIH Grants R21CA209007, R01AR053591, and R01DK073932; and the Fundamental Research Funds for the Central Universities.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705755114/-/DCSupplemental.
References
- 1.Derynck R, Miyazono K. The TGF-β Family. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer TA, Narimatsu M, Weiss A, David L, Wrana JL. The TGFβ superfamily in stem cell biology and early mammalian embryonic development. Biochim Biophys Acta. 2013;1830:2268–2279. doi: 10.1016/j.bbagen.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 3.Sakaki-Yumoto M, Katsuno Y, Derynck R. TGF-β family signaling in stem cells. Biochim Biophys Acta. 2013;1830:2280–2296. doi: 10.1016/j.bbagen.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watabe T, Miyazono K. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–115. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- 5.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 6.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa K, et al. Activin-Nodal signaling is involved in propagation of mouse embryonic stem cells. J Cell Sci. 2007;120:55–65. doi: 10.1242/jcs.03296. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Ichida JK, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan X, Chen YG. Smad7: Not only a regulator, but also a cross-talk mediator of TGF-β signalling. Biochem J. 2011;434:1–10. doi: 10.1042/BJ20101827. [DOI] [PubMed] [Google Scholar]
- 14.Briones-Orta MA, Tecalco-Cruz AC, Sosa-Garrocho M, Caligaris C, Macías-Silva M. Inhibitory Smad7: Emerging roles in health and disease. Curr Mol Pharmacol. 2011;4:141–153. [PubMed] [Google Scholar]
- 15.Hayashi H, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 16.Nakao A, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 17.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 18.Ebisawa T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 19.Shi W, et al. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164:291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valdimarsdottir G, et al. Smad7 and protein phosphatase 1alpha are critical determinants in the duration of TGF-beta/ALK1 signaling in endothelial cells. BMC Cell Biol. 2006;7:16. doi: 10.1186/1471-2121-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, et al. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol. 2007;27:4488–4499. doi: 10.1128/MCB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SH. Fine tuning and cross-talking of TGF-beta signal by inhibitory Smads. J Biochem Mol Biol. 2005;38:9–16. doi: 10.5483/bmbrep.2005.38.1.009. [DOI] [PubMed] [Google Scholar]
- 23.Hong S, et al. Smad7 binds to the adaptors TAB2 and TAB3 to block recruitment of the kinase TAK1 to the adaptor TRAF2. Nat Immunol. 2007;8:504–513. doi: 10.1038/ni1451. [DOI] [PubMed] [Google Scholar]
- 24.Edlund S, et al. Transforming growth factor-beta1 (TGF-beta)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-beta-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell. 2003;14:529–544. doi: 10.1091/mbc.02-03-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozair MZ, Noggle S, Warmflash A, Krzyspiak JE, Brivanlou AH. SMAD7 directly converts human embryonic stem cells to telencephalic fate by a default mechanism. Stem Cells. 2013;31:35–47. doi: 10.1002/stem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blank U, et al. Smad7 promotes self-renewal of hematopoietic stem cells. Blood. 2006;108:4246–4254. doi: 10.1182/blood-2006-02-005611. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, et al. RNF12 controls embryonic stem cell fate and morphogenesis in zebrafish embryos by targeting Smad7 for degradation. Mol Cell. 2012;46:650–661. doi: 10.1016/j.molcel.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Kishimoto T. Interleukin-6: From basic science to medicine–40 years in immunology. Annu Rev Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. Differentiation and growth arrest signals are generated through the cytoplasmic region of gp130 that is essential for Stat3 activation. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 31.Wu YY, Bradshaw RA. Induction of neurite outgrowth by interleukin-6 is accompanied by activation of Stat3 signaling pathway in a variant PC12 cell (E2) line. J Biol Chem. 1996;271:13023–13032. doi: 10.1074/jbc.271.22.13023. [DOI] [PubMed] [Google Scholar]
- 32.Minami M, et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukada T, et al. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: Involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 34.Takeda K, et al. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: Generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 35.Murakami M, et al. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci USA. 1991;88:11349–11353. doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami M, et al. IL-6-induced homodimerization of gp130 and associated activation of a tyrosine kinase. Science. 1993;260:1808–1810. doi: 10.1126/science.8511589. [DOI] [PubMed] [Google Scholar]
- 37.Anhuf D, et al. Signal transduction of IL-6, leukemia-inhibitory factor, and oncostatin M: Structural receptor requirements for signal attenuation. J Immunol. 2000;165:2535–2543. doi: 10.4049/jimmunol.165.5.2535. [DOI] [PubMed] [Google Scholar]
- 38.Babon JJ, et al. Suppression of cytokine signaling by SOCS3: Characterization of the mode of inhibition and the basis of its specificity. Immunity. 2012;36:239–250. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 40.Ogata H, et al. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene. 2006;25:2520–2530. doi: 10.1038/sj.onc.1209281. [DOI] [PubMed] [Google Scholar]
- 41.Bourillot PY, et al. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27:1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- 42.Luwor RB, et al. Targeting Stat3 and Smad7 to restore TGF-β cytostatic regulation of tumor cells in vitro and in vivo. Oncogene. 2013;32:2433–2441. doi: 10.1038/onc.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G, et al. STAT3 selectively interacts with Smad3 to antagonize TGF-beta. Oncogene. 2016;35:4388–4398. doi: 10.1038/onc.2015.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zauberman A, Lapter S, Zipori D. Smad proteins suppress CCAAT/enhancer-binding protein (C/EBP) beta- and STAT3-mediated transcriptional activation of the haptoglobin promoter. J Biol Chem. 2001;276:24719–24725. doi: 10.1074/jbc.M005813200. [DOI] [PubMed] [Google Scholar]
- 45.Mochizuki T, et al. Roles for the MH2 domain of Smad7 in the specific inhibition of transforming growth factor-beta superfamily signaling. J Biol Chem. 2004;279:31568–31574. doi: 10.1074/jbc.M313977200. [DOI] [PubMed] [Google Scholar]
- 46.Bitzer M, et al. A mechanism of suppression of TGF-β/SMAD signaling by NF-κ B/RelA. Genes Dev. 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]
- 47.Onishi K, Tonge PD, Nagy A, Zandstra PW. Local BMP-SMAD1 signaling increases LIF receptor-dependent STAT3 responsiveness and primed-to-naive mouse pluripotent stem cell conversion frequency. Stem Cell Rep. 2014;3:156–168. doi: 10.1016/j.stemcr.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin X, et al. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol Cell Biol. 2003;23:9081–9093. doi: 10.1128/MCB.23.24.9081-9093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheich C, Kümmel D, Soumailakakis D, Heinemann U, Büssow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zawel L, et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 51.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 52.Besser D, Bromberg JF, Darnell JE, Jr, Hanafusa H. A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation. Mol Cell Biol. 1999;19:1401–1409. doi: 10.1128/mcb.19.2.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]