Significance
This paper describes the finding that mitochondria synthesize and release melatonin and have their selective G protein-coupled receptor (GPCR) in the outer membrane. We further demonstrate that mitochondrial melatonin type 1 receptors respond to melatonin by activating heterotrimeric G proteins located in the intermembrane space and inhibit stress-mediated cytochrome c release. This remarkable insight changes our classical understanding of biological GPCR function by showing that a cellular organelle both synthesizes and has a signaling receptor for a specific ligand. Implicit with our original work is the existence of an automitocrine signaling pathway by which melatonin prevents neurodegeneration associated with mitochondrial cytochrome c release and downstream caspase activation.
Keywords: mitochondria, G protein-coupled receptor, melatonin, ischemia, neuroprotection
Abstract
G protein-coupled receptors (GPCRs) are classically characterized as cell-surface receptors transmitting extracellular signals into cells. Here we show that central components of a GPCR signaling system comprised of the melatonin type 1 receptor (MT1), its associated G protein, and β-arrestins are on and within neuronal mitochondria. We discovered that the ligand melatonin is exclusively synthesized in the mitochondrial matrix and released by the organelle activating the mitochondrial MT1 signal-transduction pathway inhibiting stress-mediated cytochrome c release and caspase activation. These findings coupled with our observation that mitochondrial MT1 overexpression reduces ischemic brain injury in mice delineate a mitochondrial GPCR mechanism contributing to the neuroprotective action of melatonin. We propose a new term, “automitocrine,” analogous to “autocrine” when a similar phenomenon occurs at the cellular level, to describe this unexpected intracellular organelle ligand–receptor pathway that opens a new research avenue investigating mitochondrial GPCR biology.
Melatonin has strong neuroprotective properties (1–8) including its ability to inhibit mitochondrial cytochrome c release and ensuing caspase activation (9–14) as well as decrease reactive oxygen species (ROS) levels in vivo after ischemia (15–17). However, the mechanism by which melatonin mediates neuroprotection is unknown. We recently reported the presence of the melatonin type 1 (MT1) receptor in mitochondria isolated from brain lysates (4). However, it is unclear how melatonin is accumulated in the mitochondria, whether mitochondrial MT1 is found in neurons, and if mitochondrial MT1 can transduce the classic G protein-coupled receptor (GPCR) signal after melatonin binding. These questions limit our understanding of the fundamental neurobiological processes of the melatonin signal transduction system. Here we show that melatonin is produced in the mitochondrial matrix, is released by the organelle, and is bound to high-affinity MT1 located in the outer mitochondrial membrane (OMM) with its ligand-binding domain facing the cytosol and that its signal transduction apparatus is located in the intermembrane space. We further demonstrate that melatonin activates the mitochondrial MT1/G protein signal system and inhibits the release of cytochrome c, thus blocking caspase activation and the neurodegenerative process.
Results
Melatonin Synthesis in Brain Mitochondrial Matrix.
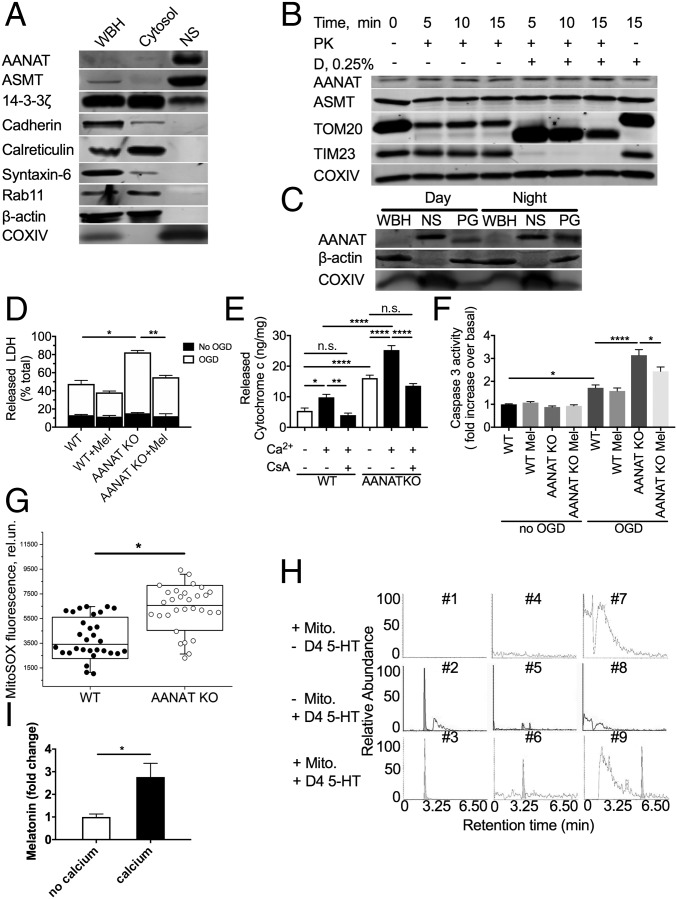
Because mitochondria have high concentrations of melatonin (18), we reasoned that melatonin might be produced in the mitochondria. Aralkylamine N-acetyltransferase (AANAT) is the penultimate and rate-limiting enzyme in melatonin biosynthesis and generates N-acetylserotonin from serotonin (19). Melatonin is thereafter produced by acetylserotonin O-methyltransferase (ASMT) (20). We thus assessed whether these key enzymes responsible for melatonin biosynthesis are located in the mitochondria. In mouse whole-brain lysate collected during daylight hours, we fractionated the mouse brain into cytosol and nonsynaptosomal mitochondria. The cytosolic fraction was free of mitochondria, given the lack of the mitochondrial marker cytochrome c oxidase subunit IV (COX IV) (Fig. 1A). ASMT was detected as a weak band in whole-brain lysate. Purified nonsynaptosomal brain mitochondria, as noted by transmission electron microscopy (SI Appendix, Fig. S1), and the absence of actin, E-cadherin, calreticulin, syntaxin-6, and Rab11 (Fig. 1A) showed the presence of the two key melatonin biosynthetic enzymes, AANAT and ASMT (Fig. 1A and SI Appendix, Fig. S2 A and B). Therefore, the two terminal enzymes, including the rate-limiting enzyme AANAT, are located in brain mitochondria. The chaperone 14-3-3ζ plays an important role, inhibiting AANAT degradation and increasing affinity for its substrate serotonin (21). Since we identified localization of AANAT in the mitochondria, we next evaluated whether 14-3-3ζ also colocalizes to mitochondria. We found that 14-3-3ζ was present in brain cytosolic and mitochondrial fractions (Fig. 1A).
Fig. 1.
Key enzymes in melatonin synthesis are located in neuronal mitochondria matrix, and melatonin is synthesized in mitochondria. (A) Immunoblotting of fractionated 6-wk-old B6CBA mouse brain; n = 3. (B) Immunoblotting of lysates following proteinase K (PK) digestion with/without digitonin (D). Nonsynaptosomal mitochondria from 6-wk-old B6CBA mice were incubated with proteinase K or proteinase K plus digitonin for the times indicated. Following digestion, protease inhibitors were added, and lysates were subjected to immunoblotting; n = 3. (C) Whole-brain homogenate (WBH), nonsynaptosomal mitochondria (NS), and pineal gland (PG) lysates were isolated during the daytime (2:00 PM) or night (2:00 AM) for immunoblotting with anti-AANAT antibody; n = 3. (D) LDH release from N2a and N2a-AANAT–KO cells under optimal conditions demonstrating background LDH release for each cell line (black portion of bars) compared with LDH release after OGD-induced stress (white portion of bars); n = 3. (E) Calcium-induced cytochrome c release from crude mitochondria isolated from N2a and N2a-AANAT–KO cells with or without cyclosporine A (10 μM) treatment; n = 3. After stress, AANAT-KO cells are more vulnerable than parental cells. (F) Caspase 3 activity from N2a wild-type and N2a-AANAT–KO cells was evaluated following OGD-induced stress with or without melatonin presence; n = 3. (G) N2a cells (WT and AANAT-KO) plated in the same multiwell plates were incubated with MitoSOX reagent, and fluorescence was measured using λex/em = 510/580 nm. P < 0.001 (n = 30). (H) Representative chromatograms showing metabolites extracted from mitochondria incubated in control buffer with mitochondria but no 4d-serotonin (#1, 4, and 7), with d4-serotonin in the absence of mitochondria (#2, 5, and 8), or with d4-serotonin and mitochondria (#3, 6, and 9). Samples incubated with buffer alone (#1, 3, and 5) show no presence of d4-labeled metabolites. Samples incubated with d4-serotonin show detected levels of unprocessed d4-serotonin (#2 and 3, retention time 1.65 min), d4-N-acetyl-serotonin (#6, retention time 2.28 min), and d4-melatonin (#9, retention time 4.45 min). This experiment was repeated three times; a representative result is shown. (I) Melatonin release from purified mouse brain mitochondria upon calcium-induced stress. The value expressed is after normalization to no calcium control, n = 4. For all panels, *P < 0.05, **P < 0.01, ****P < 0.0001, n.s., not significant. Error bars represent SEM.
To determine the intramitochondrial localization of AANAT and ASMT, we treated brain mitochondria with proteinase K and digitonin. The addition of digitonin, which permeabilizes the outer membrane and leads to ultrastructure changes but leaves the inner membrane intact, renders intermembrane proteins and proteins on the inner membrane facing the intermembrane space accessible to proteolysis. Controls demonstrated that TOM20, an OMM protein, was partly degraded by proteinase K alone and was fully degraded following digitonin addition (Fig. 1B and SI Appendix, Fig. S2 B and C). TIM23, an inner mitochondrial membrane protein, was more resistant to proteinase K digestion and was fully degraded after digitonin addition, and COX IV, a matrix protein, was resistant to proteinase K/digitonin proteolysis. AANAT and ASMT were also fully resistant to proteinase K/digitonin digestion (Fig. 1B and SI Appendix, Fig. S2 B and C), indicating that they are located in the mitochondrial matrix.
Because pineal AANAT levels change with circadian cycles (22), we evaluated whether nonpineal AANAT levels vary during the day. We isolated brain nonsynaptosomal mitochondria at 2:00 AM and 2:00 PM and determined AANAT levels, using pineal gland lysates as controls. In contrast with circadian pineal AANAT fluctuations, there was no difference in mitochondrial AANAT content between day and night times, indicating that extrapineal brain AANAT is not regulated in a circadian manner (Fig. 1C and SI Appendix, Fig. S2D).
We determined the importance of AANAT expression for cellular survival using AANAT-KO cells. Mouse neuroblastoma (N2a) cells had AANAT expression knocked out using a CRISPR-Cas9 vector system with guide RNAs specific to AANAT (SI Appendix, Fig. S1C). After knockout, we found that AANAT-KO cells were more vulnerable to oxygen–glucose deprivation (OGD)-induced stress than were parental N2a cells, as measured by lactate dehydrogenase (LDH) release, caspase-3 activation, and calcium-induced cytochrome c release of isolated mitochondria (Fig. 1 D–F). Melatonin treatment inhibited OGD-induced LDH release and caspase activation, suggesting that exogenous melatonin treatment can functionally replace endogenously synthesized melatonin (Fig. 1 D and F). Furthermore, knockout of AANAT in cells increases the levels of mitochondrial-generated superoxide (Fig. 1G) but does not alter mitochondrial membrane potential (SI Appendix, Fig. S2E).
We next examined whether melatonin can be synthesized in mitochondria. To this end, isolated brain mitochondria were incubated with deuterated (d4)-serotonin, the precursor of melatonin and AANAT substrate, in the presence of respiratory substrate and ATP to determine if mitochondrial melatonin synthesis enzymes are active. Qualitative mass spectrometry was then used to detect the conversion of d4-serotonin into intermediate (d4-n-acetylserotonin) and final (d4-melatonin) products, thus demonstrating AANAT and ASMT activity, respectively. These data demonstrated that d4-melatonin is indeed produced by the mitochondria and is not detectable in the absence of either substrate or mitochondria (Fig. 1H). Furthermore, we demonstrate that calcium triggers melatonin release from purified nonsynaptosomal mitochondria (Fig. 1I).
Mitochondrial Melatonin Binding and Downstream Effects.
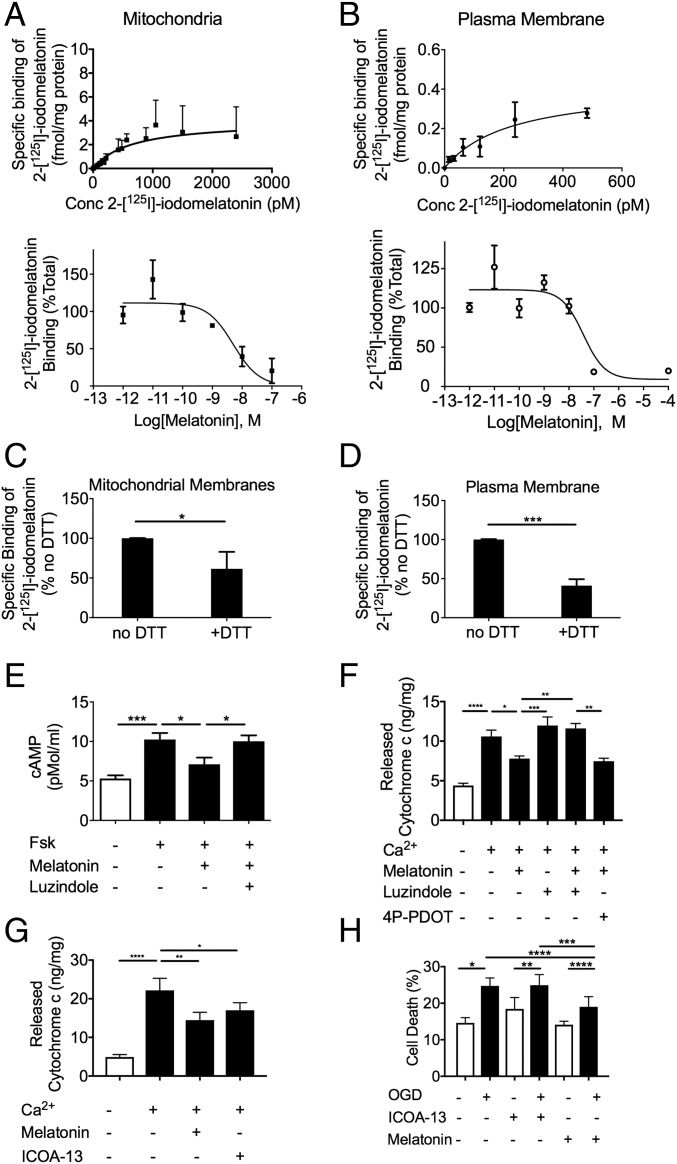
We then tested the ability of melatonin to bind receptors on the OMM using purified brain mitochondria from mice or cultured N2a cells. We first measured the affinity of melatonin for binding to melatonin receptors located in brain plasma membrane (PM) and mitochondria. As established by competition-binding isotherms and saturation curves, the affinity of melatonin (Ki = 1.0 ± 0.6 nM) (n = 3) and 2-[125I]-iodomelatonin (Kd = 2.2 ± 0.6 nM) (n = 7) for binding to melatonin receptors located in mitochondria are in a similar range as those determined for melatonin receptors located in the PM [Ki = 2.2 ± 0.9 nM (n = 6) and Kd = 342 ± 232 pM (n = 3)] and thus are consistent with high-affinity melatonin receptors in the OMM (Fig. 2 A and B). Saturation binding data were best fit by nonlinear regression analysis (SI Appendix, Fig. S3A). Total levels of melatonin receptor expression (Bmax) in mouse brain mitochondria were calculated to be 8 ± 2 fmol/mg protein, which is consistent with published studies (20) but is ≈16 times higher than observed in the PM (Bmax = 0.5 ± 0.1 fmol/mg protein). This difference in melatonin receptor expression levels was likely due to the relative enrichment of the mitochondrial fraction compared with PM. Because the cytosolic environment is presumed to be a reducing environment, we also measured the binding of [125I]-iodomelatonin in the presence of DTT (Fig. 2 C and D). For both PM (Fig. 2D) and mitochondria (Fig. 2C), binding was reduced by ∼50% but not eliminated, consistent with past studies (23, 24) and also consistent with the hypothesis that MT1 is functional in the reducing environment of the cytoplasm. Consistent with the capacity of melatonin to activate inhibitory G proteins (Gi) for adenylate cyclases through MT1, melatonin inhibited forskolin-mediated cAMP generation in purified mouse brain mitochondria with an EC50 value of 44.5 ± 0.5 nM (Fig. 2E and SI Appendix, Fig. S3B). The melatonin antagonist luzindole blocked the inhibitory effect produced by melatonin, in accordance with its competitive and antagonistic binding nature (Fig. 2E). These results demonstrate that melatonin activates a fully functional GPCR signaling system at the OMM.
Fig. 2.
MT1 signaling in isolated mitochondria from mice brains. (A and B) Comparison between the binding properties (Ki, Kd, Bmax) of melatonin receptors located in mitochondria (A) or PM (B) isolated from mouse brains. Composite graphs were fit by nonlinear regression yielding Kd = 641 pM, Bmax = 4.0 fmol/mg protein, Ki = 2.4 nM (A) and Kd = 225 pM, Bmax = 0.42 fmol/mg protein, Ki = 20 nM (B). (C and D) Quantification of 2-[125I]-iodomelatonin binding in the absence or presence of 1 mM DTT on mitochondrial membrane extracts (C) or PM (D) isolated from mouse brains. Nonspecific binding was determined in the presence of 10 μM melatonin. (E) cAMP levels of isolated synaptosomal mitochondria from wild-type mice treated with or without 10 μM forskolin (Fsk), 10 μM melatonin, and/or 100 μM luzindole. (F) Calcium-induced cytochrome c release from isolated mitochondria. Melatonin (10 μM) was incubated alone or in combination with luzindole (100 μM) or 4P-PDOT (100 μM). (G) Calcium-induced cytochrome c release from isolated mitochondria incubated with either melatonin (10 μM) or ICOA-13 (100 μM). (H) N2a cells exposed to OGD for 6 h and then grown in regular medium at normoxia in the presence of melatonin (40 μM) or ICOA-13 (40 μM). LDH was used to evaluate cell death. Bars represent the mean value ± SEM of n = 3 in A–C; n = 5 in E; and n = 4 in F. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 indicate a significant difference.
Since melatonin-binding properties are consistent with the presence of MT1 on the mitochondrial membrane, we then questioned whether the action of melatonin on mitochondria MT1 could prevent cytochrome c release as a possible mechanism of melatonin neuroprotection. Mitochondrial release of cytochrome c is a key event resulting in neuronal cell death (1). Cytochrome c release results in apoptosome assembly that mediates the sequential activation of caspase-9 and caspase-3, ultimately leading to cell death. The regulation of mitochondrial cytochrome c release is thus critical to neuronal survival. We found that Ca2+-mediated cytochrome c release from purified mitochondria was blocked by melatonin. Consistent with its antagonistic action, luzindole prevented the inhibitory effect mediated by melatonin. To differentiate whether the inhibitory effect of melatonin is mediated by the MT1 or MT2 receptor, we used a selective MT2 receptor antagonist (4P-PDOT). Unlike luzindole, 4P-PDOT was unable to inhibit melatonin inhibition of cytochrome c release, providing further evidence that MT1 mediates this activity (Fig. 2F). To confirm that this was an effect of mitochondrial-localized MT1, we used a cell-impermeable MT1 agonist, ICOA-13 (25) (SI Appendix, Fig. S4). ICOA-13 does not inhibit cell death (Fig. 2H) but prevents calcium-induced cytochrome c release from purified mitochondria (Fig. 2G), providing evidence that the melatonin-mediated neuroprotection results from its binding to the mitochondrial and not to the PM receptor.
Mitochondrial MT1 Expression and Signaling.
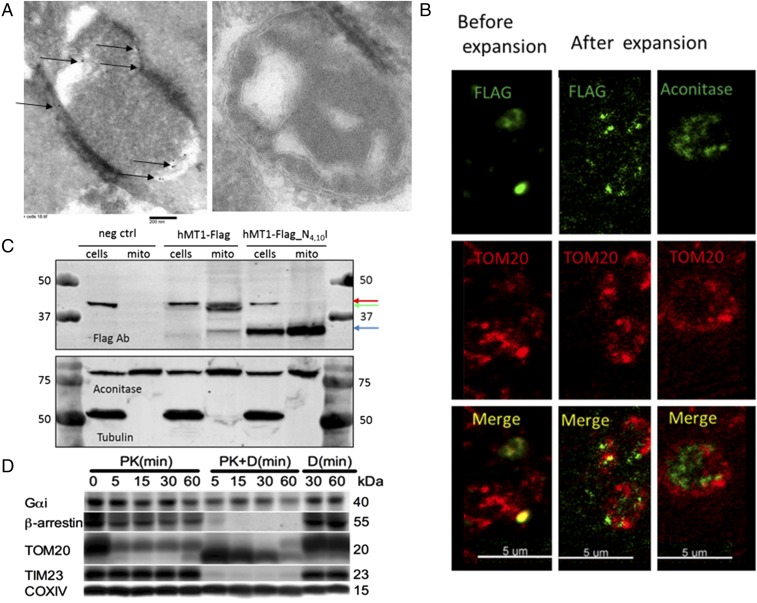
The binding and signaling results shown in Fig. 2 are consistent with expression of MT1 in the mitochondria, but the unavailability of suitable antibodies does not permit direct visualization of endogenous MT1. We thus used constructs to express human or mouse MT1 C-terminally tagged with the FLAG tag epitope (hMT1-FLAG and mMT1-FLAG, respectively) in N2a cells to determine MT1 compartmentalization in mitochondria along a series of experiments. First, we assessed mitochondria from hMT1-FLAG– and mMT1-FLAG–transfected N2a cells by electron microscopy. We detected mMT1-FLAG immunogold staining in crude mitochondria pellets extracted from cells expressing mMT1-FLAG but not from control cells (Fig. 3A). Second, we used expansion microscopy that enables nanoscale microscopic fluorescent imaging (26). By staining nanobead-purified mitochondria followed by expansion microscopy, we identified colocalization of the OMM marker TOM20 and hMT1-FLAG but not with the mitochondrial matrix protein aconitase (Fig. 3B and SI Appendix, Fig. S2F). Third, immunoblotting experiments in mitochondria isolated from cells expressing hMT1-FLAG revealed the presence of two selective protein bands specific to hMT1-FLAG at ≈40 and 33 kDa. To better characterize these two receptor bands, we mutated the glycosylation sites in MT1, asparagine 4 and 10, to isoleucine. When the glycosylation sites were mutated, these two bands resolved into one band at 33 kDa, demonstrating that the higher band is the glycosylated form of the receptor, which is present in both whole-cell lysates and isolated mitochondria (Fig. 3C). Taken together, these results indicate the presence of MT1 in the mitochondrial membrane.
Fig. 3.
MT1-FLAG in neuronal mitochondria. (A) Transmission electron microscopy of an isolated mitochondrion from a MT1-FLAG–transfected (Left) or untransfected (Right) N2a cell. Sections were incubated with FLAG antibody and immunogold secondary antibody before imaging. Arrows indicate positive staining. (B) Expansion microscopy of mitochondria isolated from MT1-FLAG–transfected N2a cells and then fixed with 4% PFA for 5 min and permeabilized with Triton X-100 for 10 min. (Left) Mitochondria before expansion. (Center) Mitochondrial immunostaining of MT1-FLAG and TOM20 after expansion. (Right) Controls demonstrate aconitase staining in the mitochondrial matrix and TOM20 staining on the outer membrane. (C) Immunoblot of control N2a cell lysates and MT1-FLAG– or MT1-FLAG glycosylation mutant vector–transfected N2a cell lysates. The red arrow indicates a background band, the green arrow indicates glycosylated MT1, and the blue arrow indicates nonglycosylated MT1. (D) Orientation of MT1 signaling partners on the mitochondrial membrane. Purified mitochondria were treated with proteinase K (PK) alone or in combination with digitonin (PK+D) for the times indicated followed by immunoblot analysis of Gαi, β-arrestins, TOM20, TIM23, and COX IV. Representative images are shown; all experiments were performed at least in triplicate.
To further support the hypothesis that melatonin signals through MT1 in mitochondria, we tested whether central components of the MT1 signaling system were also present in mitochondria. Using immunoblotted lysates obtained from purified mouse brain mitochondria, we found Gαi and β-arrestins. They were resistant to proteinase K digestion but degraded following the addition of digitonin, thus indicating that Gi and β-arrestin are located in the intermembrane mitochondrial space (Fig. 3D). Although Gαi was not fully degraded, its levels were reduced following the addition of digitonin. This may be due to its secondary structure or due to its relationship with the outer membrane. These findings are consistent with the hypothesis that MT1 signaling occurs in the mitochondrial intermembrane space.
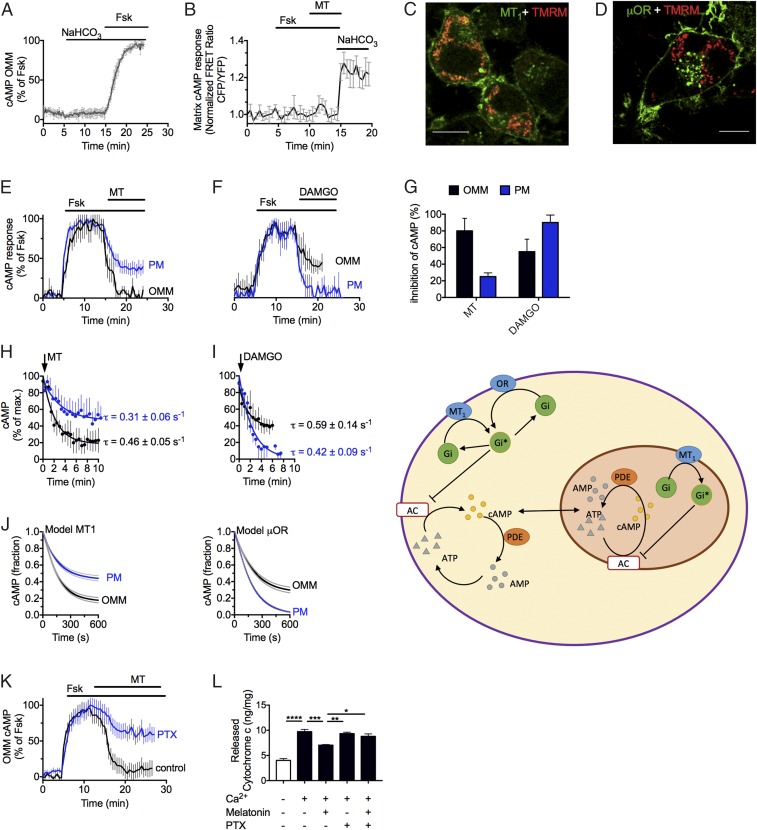
We next confirmed the mitochondrial MT1 functionality in N2a cells. We verified the presence of mitochondrial MT1 (Fig. 4C), Gαi, and AC5 (SI Appendix, Fig. S5) by imaging their strong colocalization with the fluorescent live mitochondrial marker TMRM. Cells transiently expressing MT1 were then used to measure the capacity of melatonin to inhibit cAMP production mediated by forskolin. To this end, we used FRET-based cAMP sensors localized either in the PM (27, 28) or in the OMM (SI Appendix, Fig. S6) (29). Control experiments ascertained that cAMP elevation recorded at the OMM originated only from forskolin-activated membrane adenylyl cyclases (mAC) and not from bicarbonate (NaHCO3)-sensitive soluble adenylyl cyclases (sAC) (Fig. 4A). We also found that forskolin-mediated cAMP cannot accumulate in the mitochondrial matrix (Fig. 4B) as previously reported (29). We compared receptor activity between MT1-GFP, which is localized at the cell surface and in the OMM (Fig. 4C), and the µ-opioid receptor (µOR), a Gi-coupled receptor that localized exclusively at the PM (Fig. 4D). Melatonin more efficiently inhibited the production of cAMP mediated by forskolin in the OMM than in the PM (≈40%) (Fig. 4E), with an average inhibition of 80 ± 15% and 28 ± 5%, respectively (Fig. 4G). Conversely, with the µOR-selective agonist DAMGO, cAMP inhibition was greater at the PM than at the OMM (Fig. 4 F and G). This opposite change in efficacy of reducing cAMP levels was not due to a difference in kinetics between melatonin and DAMGO (Fig. 4 H and I) but more likely was due to the expression of MT1 in both the plasma and OMM and the expression of µOR only at the PM. We next quantitatively evaluated our experimental results by constructing a computational model (SI Appendix, Fig. S7) comparing cAMP dynamics of a GPCR localized only in the PM (μOR–model 1) with one that is localized in the plasma and OMM (MT1–model 2). We used mass action kinetics to model the biochemical reactions, resulting in a set of coupled ordinary differential equations (ODEs) that described the changes of cAMP levels as a function of time upon receptor ligand activation (Fig. 4J). The model agrees with our observations: The μOR pathway inhibited cAMP production more efficiently in the PM environment than in mitochondria, whereas the MT1 pathway showed the opposite effect. Our computational analysis of cAMP dynamics thus highlights MT1 location as a key determinant of melatonin efficacy in terms of forskolin-mediated cAMP inhibition. It also indicates that model 2 better describes the action of melatonin in neurons. Consistent with this model is the observation that the cell-nonpermeant melatonin analog ICOA-13 exhibited a markedly reduced efficacy in inhibiting cAMP accumulation in mitochondria compared with melatonin (SI Appendix, Fig. S4C). To further examine the involvement of Gi in mitochondrial MT1 signaling, we pretreated N2a cells expressing MT1 with pertussis toxin (PTX) to inactivate Gi. We found that PTX markedly prevented the inhibitory action of melatonin in forskolin-mediated cAMP production in the mitochondrial intermembrane space (Fig. 4K). We confirmed this result by treating isolated mitochondria with PTX and measuring cytochrome c release. Consistent with a model in which Gi is determinant for transmitting the action of melatonin in mitochondria, we found that PTX blocked the capacity of melatonin to reduce the release of cytochrome c from isolated brain mitochondria (Fig. 4L). Collectively these results demonstrated that MT1 localizes to the OMM, that melatonin engages a fully active mitochondrial MT1/Gαi signaling system, and that the inhibitory effect of melatonin on cytochrome c release is mediated by mitochondrial MT1.
Fig. 4.
MT1 signaling in living N2a cells. (A and B) Averaged time courses of mitochondrial cAMP production in response to forskolin (Fsk) and bicarbonate (NaHCO3) (A) or to melatonin (MT) (B) in N2a cells when the FRET-based cAMP biosensor is localized in either the outer membrane (A) or the matrix (B). Cells were continuously perfused with buffer or stimuli (10 µM) for the time indicated by the horizontal bar. Data represent the mean value ± SEM of n = 16 cells. (C and D) Confocal imaging of N2a cells expressing MT1-GFP (C, green) or μOR-GFP (D, green) together with the mitochondrial marker TMRM (red). µOR-GFP localizes exclusively in the cell surface, whereas MT1 is also detected at mitochondria. (Scale bars, 10 μm.) (E and F) Examples of cAMP recording in single cells expressing FRET-based cAMP sensors at either the PM or the OMM and MT1 (E) or µOR (F). (G–I) Averaged maximal inhibition of cAMP (G) and averaged kinetics (H and I). Data represent the mean value ± SEM of four independent experiments (n = 15–40 cells). (J, Left) Comparison of simulated time courses of cAMP inhibition for model 1 (µOR–GPCR at the PM only) and model 2 (MT1–GPCR at the PM and OMM). (Right) A schematic of these models. (K) Averaged time courses of the inhibitory action of melatonin on forskolin-stimulated cAMP in the OMM of N2a cells without (control) or with PTX. Data shown are the mean value ± SEM of three independent experiments (n = 25–35 cells). (L) Effect of PTX (1 ng/mL) on calcium-induced cytochrome c release from isolated mitochondria with or without melatonin (10 μM). Bars represent the mean value ± SEM of five independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 indicate a significant difference.
Biological Consequence of MT1 Localization in Brain Mitochondria.
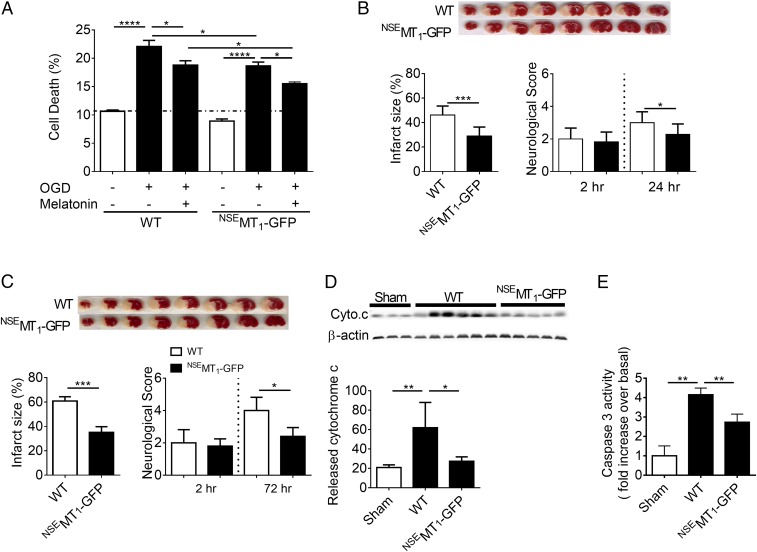
We then evaluated the biological role of mitochondrial MT1 in vivo by developing a transgenic mouse specifically expressing MT1 N-terminally tagged with GFP under the control of the neuron-specific enolase promoter (NSE) (NSEMT1-GFP or NSE-MT1). The NSE promoter targets transgene expression specifically to neurons (30). Transgenic mice had normal development, including normal brain weight and the same number of cortical neurons as wild-type littermates (SI Appendix, Fig. S8). As observed in cultured cells (Fig. 4C) and similarly to native MT1 (Fig. 2A), NSEMT1-GFP was located in mitochondria of primary cerebrocortical neurons (PCN) after transfection with the NSEMT1-GFP plasmid as well as in PCN isolated from transgenic mice (SI Appendix, Fig. S8 A and B). Since melatonin inhibits ischemia/hypoxia-mediated neuronal death in vivo and in vitro (1), we evaluated whether mitochondrial MT1 overexpression could enhance the neuroprotective action of melatonin by preventing neuronal death caused by OGD. We confirmed that exogenous melatonin inhibits OGD-induced death of PCN isolated from wild-type mice (Fig. 5A). PCN overexpressing NSEMT1-GFP significantly reduced cell death induced by OGD, and the addition of melatonin resulted in additive neuroprotection compared with wild-type PCN, thus implying a direct role of the receptor in reducing cell death (Fig. 5A). Taken together these results indicate that expression of the mitochondrial MT1 system mediates neuroprotection in vitro.
Fig. 5.
MT1 overexpression ameliorates brain injury after permanent cerebral ischemia. (A) OGD-induced cell death of PCN from NSEMT1-GFP mice. Melatonin (MT, 10 μM) treatment of PCN from NSEMT1-GFP embryos further reduced OGD-induced cell death. Bars represent the mean value ± SEM of three to six experiments. (B and C) Representative TTC-stained images of wild-type and NSEMT1-GFP murine brain sections at 24 h (B, n = 10 WT and n = 11 NSEMT1-GFP sections) and 72 h (C, n = 4 WT and n = 5 NSEMT1-GFP sections). Infarct size and neurological scores were measured at 24 h (B) and 72 h (C) after induction of cerebral ischemia. (D and E) Release of cytochrome c from mitochondria into cytosol (D) and activation of caspase-3 (E) in wild-type or NSEMT1-GFP mouse brains following 24 h permanent cerebral ischemia compared with sham surgical (anesthesia and carotid dissection only) controls in wild-type mice. Data represent the mean value ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 indicate a significant difference.
We next evaluated the effect of MT1 overexpression on cerebral ischemic injury in vivo, since studies have demonstrated that it may be possible to decrease infarct size by preventing caspase-mediated cell death in the penumbra using melatonin (1). For this study, we compared neurological scores and infarct size between wild-type and NSEMT1-GFP transgenic mice at different time points following permanent middle cerebral artery occlusion (pMCAO). Similar to previous reports (30, 31), 2 h after pMCAO there was no difference in neurological score between wild-type and NSEMT1-GFP mice (line 1). However, at 24 and 72 h of pMCAO, neurological deficits and infarct size were reduced in NSEMT1-GFP mice compared with wild-type littermates (Fig. 5 B and C). To confirm that the observed protection was not related to the transgene insertion site but rather to the physiologic effect of neuronal MT1 overexpression, we performed pMCAO in a second MT1-GFP transgenic line (line 2). Similar to line 1, the second line demonstrated milder ischemia-mediated neurologic deficits and smaller lesion size compared with littermate controls (SI Appendix, Fig. S9). We then evaluated the MT1 overexpression effect on ischemia-induced cytochrome c release and caspase-3 activation. Following pMCAO, we found that NSEMT1-GFP mice had markedly reduced cytochrome c release and caspase-3–activation levels (Fig. 5 D and E). The PCN and the in vivo stroke data provide evidence that mitochondrial MT1 contributes to inhibit neuronal death resulting from hypoxic/ischemic injury.
Discussion
We demonstrate that in the brain melatonin is exclusively synthesized and is functional at the level of neuronal mitochondria. Since melatonin is a potent free radical scavenger, it is of interest that melatonin is synthesized in the mitochondria, given that it is a major site of free radical generation. Therefore, melatonin serves a dual role in the mitochondria, as a free radical scavenger and as an intrinsic modulator of cytochrome c release.
Our findings also identify the presence of a fully functional mitochondrial GPCR signaling pathway activated by melatonin as a key determinant blocking cytochrome c release. This was unexpected, since GPCRs are thought to be primarily localized on the PM. However, recent identification of GPCRs on intracellular membranes including endosomal, endoplasmic reticulum, nuclear, and mitochondrial membranes point to previously unrecognized cellular functions for intracellular GPCR signaling systems (32–39). Furthermore, although melatonin synthesis enzymes, and GPCR signaling proteins do not have known mitochondrial targeting sequences, many known mitochondrial proteins do not have the canonical targeting sequences, and numerous types of noncleavable targeting sequence have been described. OMM proteins, the majority of intermembrane space proteins, numerous multispanning inner membrane proteins, and a few matrix proteins have an internal targeting region in the mature protein instead of the canonical N-terminal targeting sequence (40, 41). The mechanism responsible for translocating MT1 and other members of the GPCR signaling pathway to the mitochondria is unknown. Because we were unable to find an antibody that specifically labels MT1, a limitation of this study is that we cannot detect MT1 endogenously. However, several results demonstrate that melatonin can signal through a mitochondrial G protein-coupled MT1 pathway. These include the following: (i) characterization of a high-affinity melatonin-binding site in brain mitochondria membranes, (ii) location of Gi in the intermembrane space of brain mitochondria, (iii) inhibition of forskolin-mediated cAMP in isolated brain mitochondria by melatonin, (iv) antagonist actions of luzindole on the inhibitory effects of melatonin on cAMP production and cytochrome c release from isolated mitochondria, and (v) visualization and signaling characteristics of the recombinant receptor and adenylate cyclase in N2a cells’ mitochondria. In light of these data, we conclude that mitochondria harbor a functional high-affinity melatonin receptor on the outer membrane that signals through Gi to regulate cytochrome c release.
This study shows that melatonin acting on mitochondrial MT1 activates Gαi, blocks adenylate cyclase activity, and inhibits stress-induced cytochrome c release, thus damping caspase activation. The mechanism by which activation of the mitochondrial MT1/G protein system by melatonin inhibits cytochrome c release is unknown. Further understanding of this process will require studies with multiple tools, including individual knockdown not only of AC5 and Gαi but also of Gαq, Gβγ, and β-arrestins. Additionally, we demonstrate that melatonin is synthesized in and released from the mitochondrial matrix. An intriguing question is thus whether the release of melatonin from the matrix acts directly in the mitochondrial MT1 as a negative feedback mechanism that prevents cytochrome c release (SI Appendix, Fig. S10). Although a limitation of our model is the inability to directly demonstrate in vivo protection by melatonin synthesized in the mitochondria, our data provide the necessary foundation for understanding the protective action of melatonin on brain injury caused by ischemia via a mitochondrial GPCR signaling pathway regulating cytochrome c release, likely by protecting the penumbra, and opens a research avenue for mitochondrial GPCR pharmacology and biology. For example, PTX activity is often associated with ADP ribosylation factors, which have not been investigated in mitochondria.
In summary, our findings demonstrate an unexpected intracellular GPCR signaling pathway centered upon the mitochondria with its binding domain in the cytoplasm and its ligand, melatonin, being produced in the mitochondrial matrix. Given the lipophilic nature of melatonin, and the fact that we did not detect AANAT or ASMT in the cytosolic fraction, it is likely that melatonin is synthesized exclusively in the mitochondrial matrix and thereafter is released into the cytosol where it binds to the mitochondrial MT1. Analogous to a cellular autocrine pathway, we delineate an organelle-based intracellular receptor–ligand (i.e., automitocrine) pathway.
Materials and Methods
Additional detailed materials and methods are included in the SI Appendix.
Animals.
The University of Pittsburgh Institutional Animal Care and Use Committee approved all experimental procedures (protocol no. 13112568). All procedures conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (42). In all experiments, we used male mice that were 8–10 wk of age with 23–28 g body weight except as otherwise indicated. Experimentally naive wild-type (B6CBA) MT1 male littermate mice maintained on a 12-h light/dark cycle were used as described (43). For the MT1 mouse colony, MT1 male mice were mated with F1 B6CBA females.
Mitochondria Isolation.
Synaptosomal and nonsynaptosomal mitochondria were isolated from wild-type and NSEMT1-GFP transgenic mice brains as described (43–45). The purified mitochondria were then diluted to a concentration of 1 mg/mL in mitochondrial incubation buffer and treated with 10 µM melatonin or DMSO at 30 °C for 30 min, followed by pelleting the mitochondria at 6,900 × g. To ensure the purity of mitochondria, we verified PM, cytosol, and endoplasmic reticulum contaminations with antibodies specific for these components of cells. For each experiment, five forebrains were pooled for mitochondria isolation. All experiments using isolated mouse brain mitochondria were done at least three times, for a total of 15 mice (each n, as indicated in the legend, is the result of five pooled forebrains).
Immunoblot Analysis.
Protein was immunoblotted and quantified by fluorescence intensity on a LiCOR Odyssey imaging system.
Proteinase K Digestion.
Purified mitochondria diluted in isolation buffer were incubated with proteinase K in the presence or absence of digitonin. The reaction was stopped by adding protease inhibitor mixture (Roche) and was centrifuged. Each pellet was suspended in sample buffer and analyzed by immunoblot.
Pineal Gland Removal.
Animals housed under a 12-h light/12-h dark lighting regimen (lights on at 0700 h) were killed at 1400 h for daytime pineal gland isolation (n = 5–10 mice per experiment) and at 0200 h for nighttime pineal gland isolation (n = 5–10 mice per experiment). For nighttime pineal gland removal, all procedures were carried out in the dark under a dim red safe light. The pineal glands were immediately homogenized in lysis buffer with protease inhibitors and frozen at −80 °C for later use. Brains were kept in mitochondrial isolation medium, and mitochondria were isolated immediately after pineal gland removal. All homogenates were centrifuged, and supernatants were used for immunoblot analysis.
Melatonin Synthesis.
Isolated nonsynaptosomal and synaptosomal mitochondria (1 mg/mL) were incubated with 50 μM d4-serotonin (58264-95-2; Alsachim) or buffer alone (5 mM Hepes⋅Tris, 125 mM KCl, 2 mM Pi, 3 μM EDTA, 1 mM MgCl2, 1 mM ATP, 5 mM glutamate/malate, 5 mM succinate, 10 μM deprenyl) for 30 min at room temperature, with a total incubation volume of 100 μL. Sodium hydroxide (0.5 N) (SS255; Fisher Chemical) was added to a total volume of 1 mL melatonin, and its intermediates were extracted using either chloroform (C2432; Sigma Aldrich) or acetonitrile. Then samples were dried down under nitrogen and were reconstituted in 100 µL double-distilled H2O. Reconstituted samples were injected onto an Acquity UPLC BEH C18 column (2.1 × 100 mm) and eluted with 0.1% formic acid in water and acetonitrile using a gradient from 97:3 (initial) to 60:40 over 5 min and maintained 60:40 for 1 min before reequilibration at 97:3. Deuterated melatonin (d4) was detected in positive mode with a Thermo Fisher TSQ Quantum Ultra mass spectrometer interfaced via an electrospray ionization probe with the Waters UPLC Acquity solvent delivery system. Transitions used for analysis were 237→178 for d4 melatonin and 181→164 for d4 serotonin. Neat solutions (20 μM) of d4-serotonin, d4-melatonin (66521-38-8; AlsaChim), and N-acetyl-serotonin (1210-83-9; Sigma) were run to verify retention times and optimize detection.
AANAT Knockout in N2a Cells.
Guide RNAs against mouse AANAT were designed and cloned into pLenti CRISPR vectors by the Magee-Womens Research Institute Transgenic Core Facility. N2a cells were cotransfected with pLenti AANAT-CRISPR A-eGFP and pLenti AANAT-CRISPR B-mCherry, and clonal colonies were cultured. AANAT-KO was verified by PCR.
Culture and OGD of PCN and N2a Cells.
Culture and OGD treatment of PCN were performed as previously described (4). PCN or N2a cells were preincubated with or without melatonin at 10 μM or 40 μM concentration for 2 h (PCN) or 30 min (N2a cells) and were subjected to OGD (1). OGD was terminated by a return to normal culture conditions. Cell death was determined by LDH assay.
LDH Assay.
The extent of cell death was determined by the LDH assay according to the manufacturer’s instructions (Roche Products).
Cytochrome c Release from N2a Cells.
N2a or N2a-AANAT–KO cells were collected, and mitochondria were isolated. Mitochondria were diluted to a concentration of 1 mg/mL in mitochondrial incubation buffer and used for the cytochrome c release experiment. The concentration of cyclosporine A was 10 μM.
Reactive Oxygen Quantification.
N2a cells (wild type and AANAT-KO) plated in the same multiwell plates were incubated with 1 μM of MitoSOX reagent (Thermo Fisher Scientific) for 15 min. Cells were washed with DMEM culture medium, and MitoSOX fluorescence was measured using λex/em = 510/580 nm. The fluorescence intensity measured from cells without MitoSOX treatment was counted as background signal and was subtracted before analysis.
MT1-FLAG Vectors Construction and Transfection.
The mMT1-FLAG vector was purchased from OriGene (MR224599). The hMT1-FLAG vector was created by modifying the pcDNA3.1 DNA vector to hMT1-FLAG-mCherry in our laboratory to insert a stop codon in frame with the hMT1 starting codon. The mutation was confirmed by sequencing. The hMT1-FLAG_N4,10I vector was created by site-directed mutagenesis of the hMT1-FLAG vector. The vectors were purified with the QIAGEN Plasmid Maxi Kit, and the plasmid DNA was used to transfect N2a using Lipofectamine 2000 as recommended by the manufacturer.
Confocal Microscopy.
Cells plated on coverslips were mounted in Attofluor cell chambers (Life Technologies) and were viewed using the Nikon Ti-E microscope (Nikon) equipped with a Z-driven piezo motor. Imaging were acquired using a Nikon A1 confocal unit through a 60× NA = 1.45 objective (Nikon) and using 440-, 488-, 514-, and 560-nm lasers (Melles Griot), respectively. Emission fluorescence was acquired using a Spectral Detection mode and collected by a 32-channel photomultiplier tube. Typically, a Z-stack of four to eight images (Z step = 500 nm) was acquired every 5 min during 45–60 min. Data acquisitions were done using Nikon Element Software (Nikon Corporation). After acquisition, raw data were spectrally deconvoluted using Nikon Element Software (Nikon). Every different analysis was done at the single-cell level.
Transmission Electron Microscopy.
The specimens were fixed in glutaraldehyde. The specimens were rinsed in PBS, postfixed in 1% osmium tetroxide (osmium tetroxide crystals; Electron Microscopy Sciences) with 1% potassium ferricyanide (Fisher), dehydrated through a graded series of ethanol, and embedded in Epon (dodecenyl succinic anhydride, Nadic methyl anhydride, Scipoxy 812 resin, and dimethylaminomethyl) (Energy Beam Sciences). Semithin (300-nm) sections were cut on a Reichert UltraCut ultramicrotome (Leica), stained with 0.5% Toluidine Blue (Toluidine Blue O and sodium borate) (Fisher), and examined under the light microscope. Ultrathin sections (65 nm) were stained with 2% uranyl acetate (uranyl acetate dihydrate) (Electron Microscopy Sciences) and methanol (Fisher) and with Reynold’s lead citrate (lead nitrate, sodium citrate, and sodium hydroxide) (Fisher) and were examined on Jeol 1011 transmission electron microscope with a side-mount AMT 2k digital camera (Advanced Microscopy Techniques).
Immuno-Electron Microscopy.
The specimens were fixed in paraformaldehyde and glutaraldehyde. The supernatant was removed and pelleted, and specimens were resuspended in soluble 10–15 µL of 3% gelatin in PBS. The gelatin was heated to 36 °C before resuspension. Specimens were pelleted in this solution to concentrate them at the tip of the gelatin plug before gelling. Once solidified, the gelatin pellets were fixed further in the paraformaldehyde with glutaraldehyde fixative. The pellets were infused with 20% polyvinylpyrrolidone (PVP)/1.6 M sucrose buffered with 0.055 M sodium carbonate overnight and then were frozen to small stubs in liquid nitrogen. Semithin (300-nm) sections were cut on a Leica UltraCut 7 ultramicrotome with a Cryokit (Leica) at −90 °C, stained with 0.5% Toluidine Blue (Toluidine Blue O and sodium borate) (Fisher), and examined under the light microscope. Ultrathin sections (65 nm) were labeled with antibody and then visualized with a colloidal gold-labeled secondary. After rinsing in PBS, sections were fixed in 2.5% glutaraldehyde and then rinsed in PBS and distilled H2O. The rinsed sections were counterstained with 2% neutral uranyl acetate (uranyl acetate dihydrate) (Electron Microscopy Sciences) and 4% uranyl acetate, coated with methyl cellulose and were examined on a Jeol 1400 transmission electron microscope (Grant no.1S10RR016236-01) with a side-mount AMT 2k digital camera (Advanced Microscopy Techniques).
Expansion Microscopy.
Isolated mitochondria were placed on the coverslip, fixed with 4% paraformaldehyde (PFA), and permeabilized with 0.1% Triton X-100 for 5 min. Then mitochondria were incubated with a combination of either anti-TOM20 and anti-FLAG or anti-TOM20 and anti-aconitase antibodies at 4 °C overnight. Then the mitochondria were incubated with secondary antibodies for 1 h at room temperature and subjected to the expansion procedure described in ref. 26. Imaging was done using an Olympus IX-81 fluorescent microscope equipped with an UPLSAPO 100XO objective.
Immunofluorescence Analysis of N2a Cells, PCN, and Isolated Mitochondria.
Primary cortical neurons were fixed, solubilized, and immunostained using standard methods. Images were captured using a confocal laser-scanning microscope system (FluoView FV1000; Olympus, Inc.). Isolated mitochondria were incubated with MitoTracker deep red at 37 °C for 45 min. MitoTracker-incubated mitochondria were then loaded onto the slide and fixed, followed by MT1 immunostaining. Images were taken using Olympus confocal laser-scanning microscope (FluoView FV1000; Olympus, Inc.) equipped with a 100× objective.
Assay for Melatonin Receptor-Binding Affinity and Expression in Mitochondria.
The affinity (Ki) of melatonin for melatonin receptors located in B6CBA wild-type mouse brain mitochondria (0.06–0.09 mg protein per tube) and PM (0.24–0.26 mg protein per tube) was assessed by competition binding analysis using 100 pM 2-[125I]-iodomelatonin and 1 pM–100 nM melatonin as described (46). Total melatonin receptor expression (Bmax) and affinity (Kd) of 2-[125I]-iodomelatonin binding to melatonin receptors were assessed in mitochondria (0.06–0.22 mg protein per tube) and whole-brain homogenates (0.25–0.46 mg protein per tube) by saturation isotherms using 0–500 pM 2-[125I]-iodomelatonin for PM and 0–2.4 nM for mitochondria. Nonspecific binding was determined using 10 μM melatonin. Parallel binding assays were performed in the presence of 1 mM DTT using 250–500 pM 2-[II25]-iodomelatonin. Data were best-fit analyzed by nonlinear regression analysis least-squares fit (GraphPad Prism, Inc.) (SI Appendix, Fig. S4).
Time-Course Measurements of cAMP Production in Live N2a Cells.
cAMP was assessed using FRET-based assays. Cells were transiently transfected with FRET-based biosensors for measuring cAMP at the PM, the OMM (27), or the mitochondrial matrix (29). Measurements were performed and analyzed as previously described (47). The FRET ratio for single cells was calculated and corrected as previously described (48). Individual cells were continuously perfused with buffer or with the ligand for the time indicated by the horizontal bar in Fig. 4 A, B, E, F, and K.
Assay for cAMP Measurement in Isolated Mitochondria.
The cAMP assay was performed on isolated mitochondria using the Direct cAMP ELISA kit (Enzo Life Sciences) according to the manufacturer’s instructions in the absence or presence of melatonin or melatonin plus luzindole at 30 °C for 30 min.
In Vitro Cytochrome c Release.
Mouse brain nonsynaptosomal mitochondria were preincubated with melatonin (10 µM) or with melatonin plus luzindole (100 µM) for 5 min in mitochondria incubation buffer (13, 45) followed by incubation with 40 µM CaCl2 at 30 °C for 10 min. Aliquots of 25 µL were centrifuged, and the supernatant and pellet were evaluated by immunoblot. Cytochrome c in the supernatant was also measured by ELISA (Life Technologies).
Melatonin Release from Isolated Mitochondria.
The mitochondria pellet was resuspended in mitochondria incubation buffer (45) to a final concentration of 1 mg/mL immediately after isolation. An aliquot of 500 µg was used as control or was treated with 100 µM CaCl2 at 30 °C for 1 min followed by centrifugation at 7,000 × g for 2 min. Melatonin in the supernatant was extracted with chloroform and vacuum dried. The dried melatonin extracts were then resuspended in the diluted wash buffer, and the quantification of melatonin was performed using the melatonin ELISA kit according to the manufacturer’s protocol (IBL International GMBH).
Caspase Activity Assay.
Caspase-3–like activity was assayed by the fluorometric protease assay kit according to the manufacturer’s instructions (Abcam). Briefly, tissue from ipsilateral brain of wild-type and NSEMT1-GFP mice at 24 h after pMCAO was homogenized. The corresponding brain region of sham-operated wild-type mice was used as control. The fluorescence intensity of the cytosolic fraction was then quantified as caspase-3 activity by a plate reader (49).
MT1 Transgenic Mice.
The Magee-Womens Institute and Foundation Transgenic and Molecular Core Facility generated two mouse lines overexpressing human MT1 selectively in neurons. We constructed and used a vector encoding human MT1 C-terminally fused to the Flag-tag epitope and to eGFP, which is under the control of NSE promoter, permitting selective expression of the neurons. Vector construction was confirmed by sequencing. Transgenic mice were generated on the hybrid B6CBA background.
pMCAO.
To minimize variation in endogenous melatonin levels, all experiments were performed between 9 AM and 1 PM. For these experiments, we used permanent MCAO models to reflect the type of ischemic strokes seen in patients (30). pMCAO methods followed published protocols (1). Furthermore, we followed neurologic outcomes of mice for up to 3 d to determine the effect of MT1 on the survival of the neurons in the penumbra, which continue to be vulnerable after the first 24 h of recovery from ischemia. Animals were allowed to recover and were neurologically evaluated at 2, 24, and 72 h following MCAO.
Infarction Volume and Neurological Deficits.
After mouse brain was sliced into 1 mm sections, the sections were stained with 2,3,5-triphenyltetrazolium chloride (TTC) (50). Areas that were not stained red with TTC were identified as the infarct area. After staining, the brain slices were fixed and scanned. The TTC-stained areas of the ipsi- and contralateral hemispheres were quantified separately with ImageJ. The infarct area was calculated indirectly by subtracting the area of intact tissue in the ipsilateral hemisphere from that in the contralateral hemisphere. These values were then used to calculate the infarct volume expressed as a percentage of the contralateral hemisphere. Each animal was assessed for neurological deficits at 2 and 24 or 72 h after the onset of focal ischemia. The following scoring was used: 0, no observable deficits; 1, forelimb flexion when lifted by the tail; 2, forelimb flexion and consistently reduced resistance to lateral push; 3, forelimb flexion, reduced resistance to lateral push, and unilateral circling toward the paretic side; 4, forelimb flexion and ambulation inability or difficulty; 5, dead (30, 31).
Analysis of Cytochrome c Release from Tissue.
Wild-type and transgenic mouse brains were removed, and the ipsilateral hemisphere was dissected at 24 h after occlusion. The corresponding regions of wild-type brains from sham-operated mice were separated as well. Dissected tissues were immediately placed in mitochondria isolation buffer (45). Tissues were then homogenized and centrifuged at 1,300 × g. Supernatants were collected and centrifuged again at 7,000 × g for 10 min. The resulting supernatants were used for the quantification of released cytochrome c. The amount of cytochrome c release was measured by cytochrome c ELISA (Life Technologies).
qPCR.
Wild-type or MT1 mouse brain was isolated and lysed for total RNA using an RNAeasy (Qiagen) kit. RT-PCR was done using a high-capacity RNA-to-cDNA kit (Applied Biosystems). The cDNA products of RT-PCR were then amplified (CFX97 Touch; Bio-Rad) using the RT2 qPCR primer assay from QIAGEN (catalog nos. PPH02532A and PPM02946E), and mRNA expression was quantified using the ΔΔCt method (51). In addition, to amplify both mouse and human MT1, we used the following primers: forward, 5′-ACCATCGTGGTGGACAT-3′, reverse, 5′-GTTCCTGAGCTTCTTGTT-3′.
Model Construction and Analysis.
We constructed a simple mathematical model of the proposed mechanism (SI Appendix, Fig. S7). Forskolin-treated cells have active adenylyl cyclases (AC) on both the PM and OMM. Active AC localized to the PM converts cytosolic ATP to cytosolic cAMP; active AC on the OMM converts ATP in the mitochondrial lumen to luminal cAMP. Nucleotides were permitted to diffuse through the OMM, but proteins were constrained to their cellular compartments. cAMP in the cytosol and mitochondrial lumen was converted via phosphodiesterases (PDE) to AMP, which was converted to ATP. In forskolin-treated cells under steady-state conditions, relative ATP and cAMP levels were therefore regulated by AC and PDE activity.
MT1 and μOR were overexpressed and localized to the PM. MT1 also localizes to the OMM, but μOR did not. Upon stimulation with a saturating concentration of DAMGO, μOR became active and activates cytosolic Gαi proteins. Active Gαi bound to membrane AC, where it inhibited the conversion of ATP to cAMP. Similarly, stimulation with saturating quantities of melatonin initiated activation of MT1 at both the PM and the OMM. MT1-activated Gαi inhibited cAMP production in both the cytosol and the mitochondrial lumen.
The model was constructed in compartmental BioNetGen (52) and assumes mass action kinetics. The final model contains 30 molecular species, the time evolution of which was governed by 30 ODEs containing 12 kinetic parameters (SI Appendix, Table S1). Initial quantities of Gαi, AC, PDE, ATP, and cAMP were assigned in the cytosol or PM as shown in SI Appendix, Table S2. Additional assumptions and parameters are described in the SI Appendix.
Statistical Analysis.
Quantitative data were expressed as the mean value ± SD or SEM as indicated. Statistical comparisons were conducted using ANOVA for multiple comparisons or Student’s t test for difference between two conditions. Differences with P < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Ed Boyden of Massachusetts Institute of Technology for his help implementing expansion microscopy, Drs. Kyle Orwig and Yi Sheng at the Magee-Women’s Research Institute and Foundation Transgenic and Molecular Research Core for generating the MT1 transgenic mouse lines, and Dr. David Klein of the Eunice Kennedy Shriver National Institute of Child Health and Human Development for sharing his AANAT antibody. This work was supported by NIH Grants R01NS039324 (to R.M.F.), R01NS077748 (to R.M.F.), R01 DK087688 (to J.-P.V.), and R01 DK102495 (to J.-P.V.); The David Scaife Family Charitable Foundation (R.M.F.); the Cotswold Foundation Fellowship Award (to F.G.J.-A.); Agence Nationale de la Recherche (ANR) Grant ANR-12-RPIB-0016 “MED-HET-REC-2” (to R.J.); Fondation de la Recherche Médicale Equipe Grant FRM DEQ20130326503 (to R.J.); INSERM (R.J.); CNRS and the ANR Who am I? Laboratory of Excellence Grant ANR-11-LABX-0071 (to R.J.); ANR Grant ANR-11-IDEX-0005-01 (to R.J.); La Région Centre Grants APR2009-LOIREMEL (to G.G.) and APR2012-LIFERMEL (to F.S.); Labex SynOrg Grant ANR-11-LABX-0029 (to F.S.); Marie-Clement Rodier CSSp Endowed Chair Funds (PAW-E); and University of Pittsburgh Center for Biologic Imaging NIH Grants 1S10RR019003 and 1S10RR025488.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705768114/-/DCSupplemental.
References
- 1.Wang X, et al. Methazolamide and melatonin inhibit mitochondrial cytochrome C release and are neuroprotective in experimental models of ischemic injury. Stroke. 2009;40:1877–1885. doi: 10.1161/STROKEAHA.108.540765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poeggeler B, et al. Melatonin reverses the profibrillogenic activity of apolipoprotein E4 on the Alzheimer amyloid Abeta peptide. Biochemistry. 2001;40:14995–15001. doi: 10.1021/bi0114269. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, et al. Melatonin inhibits the caspase-1/cytochrome c/caspase-3 cell death pathway, inhibits MT1 receptor loss and delays disease progression in a mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2013;55:26–35. doi: 10.1016/j.nbd.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. The melatonin MT1 receptor axis modulates mutant Huntingtin-mediated toxicity. J Neurosci. 2011;31:14496–14507. doi: 10.1523/JNEUROSCI.3059-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiter RJ, et al. Melatonin ameliorates neurologic damage and neurophysiologic deficits in experimental models of stroke. Ann N Y Acad Sci. 2003;993:35–47, discussion 48–53. doi: 10.1111/j.1749-6632.2003.tb07509.x. [DOI] [PubMed] [Google Scholar]
- 6.Kilic E, Ozdemir YG, Bolay H, Keleştimur H, Dalkara T. Pinealectomy aggravates and melatonin administration attenuates brain damage in focal ischemia. J Cereb Blood Flow Metab. 1999;19:511–516. doi: 10.1097/00004647-199905000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Wakatsuki A, Okatani Y, Shinohara K, Ikenoue N, Fukaya T. Melatonin protects against ischemia/reperfusion-induced oxidative damage to mitochondria in fetal rat brain. J Pineal Res. 2001;31:167–172. doi: 10.1034/j.1600-079x.2001.310211.x. [DOI] [PubMed] [Google Scholar]
- 8.Farez MF, et al. Melatonin contributes to the seasonality of multiple sclerosis relapses. Cell. 2015;162:1338–1352. doi: 10.1016/j.cell.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HM, Zhang Y. Melatonin: A well-documented antioxidant with conditional pro-oxidant actions. J Pineal Res. 2014;57:131–146. doi: 10.1111/jpi.12162. [DOI] [PubMed] [Google Scholar]
- 10.Reiter RJ, Tan DX, Manchester LC, El-Sawi MR. Melatonin reduces oxidant damage and promotes mitochondrial respiration: Implications for aging. Ann N Y Acad Sci. 2002;959:238–250. doi: 10.1111/j.1749-6632.2002.tb02096.x. [DOI] [PubMed] [Google Scholar]
- 11.Acuña-Castroviejo D, et al. Melatonin, mitochondria, and cellular bioenergetics. J Pineal Res. 2001;30:65–74. doi: 10.1034/j.1600-079x.2001.300201.x. [DOI] [PubMed] [Google Scholar]
- 12.Genade S, Genis A, Ytrehus K, Huisamen B, Lochner A. Melatonin receptor-mediated protection against myocardial ischaemia/reperfusion injury: Role of its anti-adrenergic actions. J Pineal Res. 2008;45:449–458. doi: 10.1111/j.1600-079X.2008.00615.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, et al. Inhibitors of cytochrome c release with therapeutic potential for Huntington’s disease. J Neurosci. 2008;28:9473–9485. doi: 10.1523/JNEUROSCI.1867-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 15.Li XJ, Zhang LM, Gu J, Zhang AZ, Sun FY. Melatonin decreases production of hydroxyl radical during cerebral ischemia-reperfusion. Zhongguo Yao Li Xue Bao. 1997;18:394–396. [PubMed] [Google Scholar]
- 16.Sinha K, Degaonkar MN, Jagannathan NR, Gupta YK. Effect of melatonin on ischemia reperfusion injury induced by middle cerebral artery occlusion in rats. Eur J Pharmacol. 2001;428:185–192. doi: 10.1016/s0014-2999(01)01253-5. [DOI] [PubMed] [Google Scholar]
- 17.Lee EJ, et al. Melatonin attenuates gray and white matter damage in a mouse model of transient focal cerebral ischemia. J Pineal Res. 2005;38:42–52. doi: 10.1111/j.1600-079X.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 18.Venegas C, et al. Extrapineal melatonin: Analysis of its subcellular distribution and daily fluctuations. J Pineal Res. 2012;52:217–227. doi: 10.1111/j.1600-079X.2011.00931.x. [DOI] [PubMed] [Google Scholar]
- 19.Falcón J, et al. Structural and functional evolution of the pineal melatonin system in vertebrates. Ann N Y Acad Sci. 2009;1163:101–111. doi: 10.1111/j.1749-6632.2009.04435.x. [DOI] [PubMed] [Google Scholar]
- 20.Stefulj J, et al. Gene expression of the key enzymes of melatonin synthesis in extrapineal tissues of the rat. J Pineal Res. 2001;30:243–247. doi: 10.1034/j.1600-079x.2001.300408.x. [DOI] [PubMed] [Google Scholar]
- 21.Ganguly S, et al. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci USA. 2005;102:1222–1227. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasahara T, Abe K, Mekada K, Yoshiki A, Kato T. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc Natl Acad Sci USA. 2010;107:6412–6417. doi: 10.1073/pnas.0914399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mseeh F, Gerdin MJ, Dubocovich MI. Identification of cysteines involved in ligand binding to the human melatonin MT(2) receptor. Eur J Pharmacol. 2002;449:29–38. doi: 10.1016/s0014-2999(02)01903-9. [DOI] [PubMed] [Google Scholar]
- 24.Dubocovich ML, Markowska M. Functional MT1 and MT2 melatonin receptors in mammals. Endocrine. 2005;27:101–110. doi: 10.1385/ENDO:27:2:101. [DOI] [PubMed] [Google Scholar]
- 25.Gbahou F, et al. Design and validation of the first cell-impermeant melatonin receptor agonist. Br J Pharmacol. 2017;174:2409–2421. doi: 10.1111/bph.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tillberg PW, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiPilato LM, Cheng X, Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc Natl Acad Sci USA. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feinstein TN, et al. Noncanonical control of vasopressin receptor type 2 signaling by retromer and arrestin. J Biol Chem. 2013;288:27849–27860. doi: 10.1074/jbc.M112.445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Benedetto G, Scalzotto E, Mongillo M, Pozzan T. Mitochondrial Ca2+ uptake induces cyclic AMP generation in the matrix and modulates organelle ATP levels. Cell Metab. 2013;17:965–975. doi: 10.1016/j.cmet.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Friedlander RM, et al. Expression of a dominant negative mutant of interleukin-1 beta converting enzyme in transgenic mice prevents neuronal cell death induced by trophic factor withdrawal and ischemic brain injury. J Exp Med. 1997;185:933–940. doi: 10.1084/jem.185.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara H, et al. Inhibition of interleukin 1beta converting enzyme family proteases reduces ischemic and excitotoxic neuronal damage. Proc Natl Acad Sci USA. 1997;94:2007–2012. doi: 10.1073/pnas.94.5.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates D. Learning and memory: The cannabinoid connection. Nat Rev Neurosci. 2016;18:4. doi: 10.1038/nrn.2016.171. [DOI] [PubMed] [Google Scholar]
- 33.Vilardaga JP, Jean-Alphonse FG, Gardella TJ. Endosomal generation of cAMP in GPCR signaling. Nat Chem Biol. 2014;10:700–706. doi: 10.1038/nchembio.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang X, Benovic JL, Wedegaertner PB. Plasma membrane and nuclear localization of G protein coupled receptor kinase 6A. Mol Biol Cell. 2007;18:2960–2969. doi: 10.1091/mbc.E07-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irannejad R, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto C, et al. G protein-coupled receptor 30 localizes to the endoplasmic reticulum and is not activated by estradiol. Endocrinology. 2008;149:4846–4856. doi: 10.1210/en.2008-0269. [DOI] [PubMed] [Google Scholar]
- 37.Belous A, et al. Mitochondrial P2Y-Like receptors link cytosolic adenosine nucleotides to mitochondrial calcium uptake. J Cell Biochem. 2004;92:1062–1073. doi: 10.1002/jcb.20144. [DOI] [PubMed] [Google Scholar]
- 38.Bénard G, et al. Mitochondrial CB1 receptors regulate neuronal energy metabolism. Nat Neurosci. 2012;15:558–564. doi: 10.1038/nn.3053. [DOI] [PubMed] [Google Scholar]
- 39.Abadir PM, et al. Identification and characterization of a functional mitochondrial angiotensin system. Proc Natl Acad Sci USA. 2011;108:14849–14854. doi: 10.1073/pnas.1101507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stojanovski D, Bohnert M, Pfanner N, van der Laan M. Mechanisms of protein sorting in mitochondria. Cold Spring Harb Perspect Biol. 2012;4:a011320. doi: 10.1101/cshperspect.a011320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies; Washington, DC: 2011. [Google Scholar]
- 43.Yano H, et al. Inhibition of mitochondrial protein import by mutant huntingtin. Nat Neurosci. 2014;17:822–831. doi: 10.1038/nn.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kristian T. Isolation of mitochondria from the CNS. Curr Protoc Neurosci. 2010;Chapter 7:Unit 7.22. doi: 10.1002/0471142301.ns0722s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khattar NK, et al. Isolation of functionally active and highly purified neuronal mitochondria from human cortex. J Neurosci Methods. 2016;263:1–6. doi: 10.1016/j.jneumeth.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Witt-Enderby PA, Dubocovich ML. Characterization and regulation of the human ML1A melatonin receptor stably expressed in Chinese hamster ovary cells. Mol Pharmacol. 1996;50:166–174. [PubMed] [Google Scholar]
- 47.Ferrandon S, et al. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat Chem Biol. 2009;5:734–742. doi: 10.1038/nchembio.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilardaga JP. Studying ligand efficacy at G protein-coupled receptors using FRET. Methods Mol Biol. 2011;756:133–148. doi: 10.1007/978-1-61779-160-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, et al. Huntingtin inhibits caspase-3 activation. EMBO J. 2006;25:5896–5906. doi: 10.1038/sj.emboj.7601445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suofu Y, et al. Matrix metalloproteinase-2 or -9 deletions protect against hemorrhagic transformation during early stage of cerebral ischemia and reperfusion. Neuroscience. 2012;212:180–189. doi: 10.1016/j.neuroscience.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeatts K. Quantitative polymerase chain reaction using the comparative C q method. Methods Mol Biol. 2011;700:171–184. doi: 10.1007/978-1-61737-954-3_12. [DOI] [PubMed] [Google Scholar]
- 52.Faeder JR, Blinov ML, Hlavacek WS. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol Biol. 2009;500:113–167. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.