Significance
The phytohormone abscisic acid (ABA) is the key signal for regulation of a plant’s water status. ABA signaling mediates drought resistance and can ameliorate water-use efficiency in plants. Arabidopsis has different ABA receptors and coreceptors, which form heterodimers that regulate the various ABA responses. This study reveals that the three receptor subfamilies have different sensitivities to ABA in Arabidopsis cells and the vast majority of receptor–coreceptor combinations are functional. The insights gained here will support the development of targeted approaches for harnessing ABA receptor components to improve crop performance under a limited water supply.
Keywords: RCAR/PYL, PP2C, abiotic stress, drought, ABA
Abstract
The phytohormone abscisic acid (ABA) is induced in response to abiotic stress to mediate plant acclimation to environmental challenge. Key players of the ABA-signaling pathway are the ABA-binding receptors (RCAR/PYR1/PYL), which, together with a plant-specific subclade of protein phosphatase 2C (PP2C), form functional holoreceptors. The Arabidopsis genome encodes nine PP2C coreceptors and 14 different RCARs, which can be divided into three subfamilies. The presence of these gene families in higher plants points to the existence of an intriguing regulatory network and poses questions as to the functional compatibility and specificity of receptor–coreceptor interactions. Here, we analyzed all RCAR–PP2C combinations for their capacity to regulate ABA signaling by transient expression in Arabidopsis protoplasts. Of 126 possible RCAR–PP2C pairings, 113 were found to be functional. The three subfamilies within the RCAR family showed different sensitivities to regulating the ABA response at basal ABA levels when efficiently expressed. At exogenous high ABA levels, the RCARs regulated most PP2Cs and activated the ABA response to a similar extent. The PP2C AHG1 was regulated only by RCAR1/PYL9, RCAR2/PYL7, and RCAR3/PYL8, which are characterized by a unique tyrosine residue. Site-directed mutagenesis of RCAR1 showed that its tyrosine residue is critical for AHG1 interaction and regulation. Furthermore, the PP2Cs HAI1 to HAI3 were regulated by all RCARs, and the ABA receptor RCAR4/PYL10 showed ABA-dependent PP2C regulation. The findings unravel the interaction network of possible RCAR–PP2C pairings and their different potentials to serve a rheostat function for integrating fluctuating hormone levels into the ABA-response pathway.
ABA regulates a plethora of responses associated with plant growth and the homeostatic control of water relations. ABA controls root extension and branching, stomatal opening and density, and tolerance to water deficit during seed maturation and drought (1, 2). Core ABA signaling can be considered as a three-step regulatory process involving the receptor complex, protein kinases as intermediate signaling components, and downstream targets such as ion channels and transcription factors (1). The heteromeric receptor complex consists of the ABA-binding RCAR/PYR1/PYL receptor and the PP2C coreceptor. The protein phosphatase activity is regulated by ABA that stabilizes the PP2C–RCAR interaction, which blocks substrate access and thereby inhibits the catalytic activity of the coreceptor (3, 4).
The clade A of Arabidopsis PP2Cs comprises nine members and forms a plant-specific subgroup among the large family of PP2Cs, which are Mg2+- and Mn2+-dependent serine–threonine protein phosphatases (5). The ABA-mediated inactivation of PP2C activity releases SNF1-related kinase 2 (SnRK2) from inhibition, and it subsequently targets downstream components such as transcription factors and ion channels (6–12). The 14 different RCARs of Arabidopsis can be divided into three subfamilies according to their sequence homology (13–15). RCARs show overlapping gene expression patterns (16–18). Genetic analyses revealed additive effects of ABA receptor deficiency in the control of ABA sensitivity during seed germination, root growth, and in guard cells (15, 17–20). Likewise, multiple deficiency mutants in the PP2Cs ABI1, ABI2, PP2CA, and HAB1 or HAI1–HAI3 exhibited additive phenotypes (20–24). The incremental contributions of RCARs and PP2Cs in establishing ABA sensitivity of plant organs and single cells indicate that several different receptors and coreceptors are simultaneously expressed and regulate ABA responsiveness.
The Arabidopsis genome encodes 14 RCARs and 9 clade A PP2Cs. This translates into 126 possible holo-receptor combinations. How many combinations exist and how many of these function as true ABA receptor complexes in vivo remains to be determined. Thus far, biochemical and protein interaction studies have analyzed a fraction of the possible RCAR–PP2C combinations, frequently without addressing ABA-response regulation (13–15, 22, 25–33).
This study elucidates the functionality of all possible RCAR–PP2C pairings of Arabidopsis in regulating ABA-responsive gene expression. Three major features of ABA receptors emerged from the analysis: the remarkable capability of PP2Cs to form functional holo-receptor complexes with many different RCARs, the overall similar PP2C regulation by individual RCARs, and the different potency of RCAR subfamilies in regulating the ABA response at endogenous basal ABA levels. The in vivo analysis also shows the uniqueness of the PP2C AHG1 in being regulated by only three different RCARs and the effective regulation of HAI1–HAI3 by ABA receptors.
Results
ABA Dependencies of RCAR-Mediated Signaling.
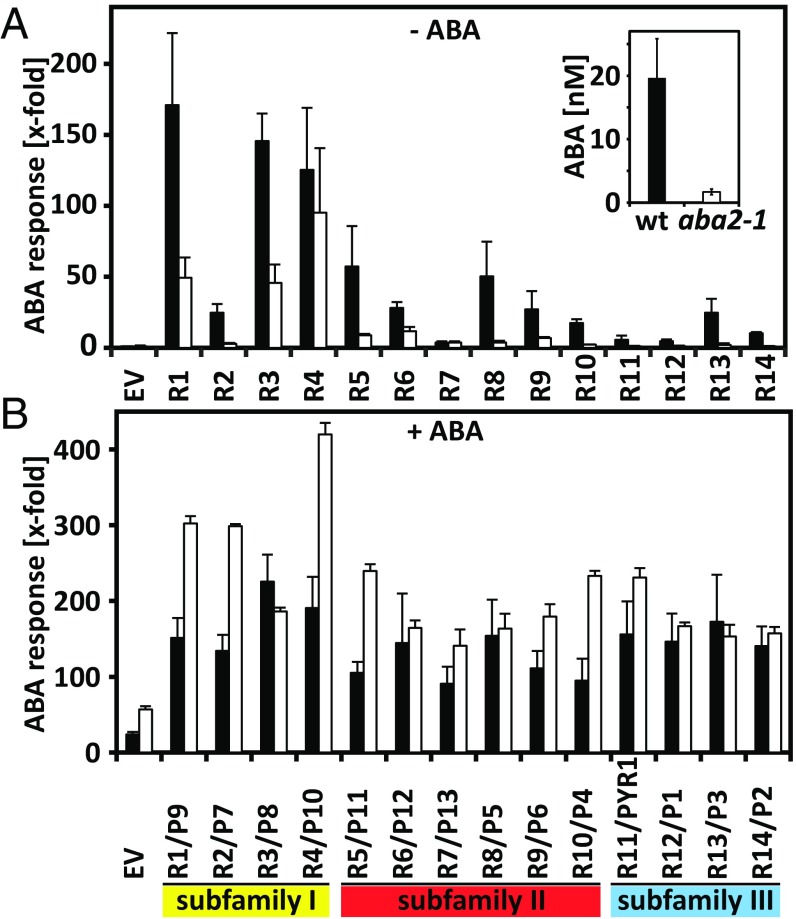
In the presence of ABA, ABA signaling is stimulated by ectopic expression of RCARs; however, the response was found to be quite variable (13, 27, 32, 34, 35). The variations might be caused by different ABA affinities and ABA-independent action of RCARs (28) or by the abundance of target PP2Cs and RCARs. To address the first two possibilities, RCAR- and ABA-dependent signaling was analyzed in Arabidopsis protoplasts of both WT and the ABA-deficient mutant aba2-1 (36). In the absence of exogenous ABA, ectopic expression of the 14 ABA receptors revealed that members of each of the three RCAR subfamilies had similar tendencies with respect to their ability to regulate ABA responses. Immunodetection of ectopically expressed RCARs indicated a comparable expression level with the exception of a lower RCAR3 abundance (Fig. S1). In WT protoplasts, subfamily I members, comprising RCAR1–RCAR4, induced the ABA response on average more than 110-fold with RCAR2 being the exception (25-fold, Fig. 1A). Subfamily II receptors, RCAR5–RCAR10, showed a moderate ABA response (31-fold mean). Expression of subfamily III RCARs had little effect on ABA signaling for RCAR11, -12, and -14 (sevenfold mean), while RCAR13 expression activated ABA signaling 25-fold. In aba2-1 protoplasts, the RCAR-mediated ABA response was clearly reduced or even abrogated compared with the WT, with the exception of RCAR4 (P > 0.45; Fig. 1A). Protoplasts of aba2-1 contained an ∼11-fold lower level of ABA: 1.7 nM compared with 19.6 nM ABA in WT protoplasts (Fig. 1A, Inset), which corresponded to 18 pmol and 209 pmol ABA per gram dry weight of leaf tissue, respectively. The ABA content found in WT protoplasts is in the range reported for Arabidopsis rosettes grown in soil under well-watered conditions (153 pmol and 395 pmol ABA per gram dry weight, ref. 37). Administration of exogenous ABA to WT cells enhanced the ABA response primarily for subfamily II and III member-transfected cells, leading to a similar activation level among all RCARs that deviated from the average (∼150-fold, Fig. 1B) by a factor of 1.6. Taken together, the findings support the notion that all RCARs regulate the ABA response in an ABA-dependent manner with the possible exception of RCAR4/PYL10, which is a matter of debate in the field (28, 33). Basal ABA levels as found in WT and aba2-1 protoplasts are sufficient to mediate ABA responses by subfamily I and, to a lesser extent, by subfamily II members.
Fig. S1.
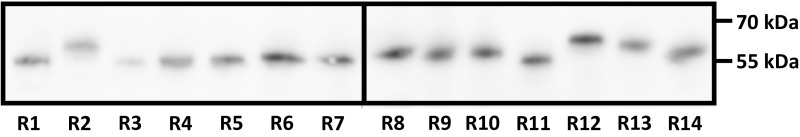
Immunodetection of RCAR expression in Arabidopsis protoplasts. Protoplasts of WT Col-0 (105 cells) were transfected with DNA for expression of the ABA receptors (RCAR1–RCAR14, R1–R14) tagged with GFP (5 µg) and a β-glucuronidase reporter construct (4 µg). Cell-free protein extracts of transfected protoplasts were normalized to the β-glucuronidase activity and analyzed for protein expression using GFP-specific antibodies. The β-glucuronidase activity among the samples prior to normalization was 1.11 ± 0.25 RFUs (mean ± SD).
Fig. 1.
Regulation of ABA signaling by the RCAR family of Arabidopsis. Protoplasts of WT Col-0 (filled bars) and the ABA-deficient mutant aba2-1 (open bars) were transfected with 3 µg effector DNA for expression of the different RCARs (R1–R14)/PYR1/PYLs (P1–P13) or the empty vector (EV) in absence (−ABA; A) and presence (+ABA; B) of 5 µM exogenous ABA. (Inset) The endogenous ABA level of protoplasts of WT and aba2-1 plants (mean of triplicates ± SD). In the presence of exogenous ABA, the RCAR-mediated inductions of ABA signaling in aba2-1 protoplasts were significantly higher compared with the ABA-treated cells transfected with the empty vector (P < 0.02). The three subfamilies of ABA receptors are color-coded as follows: subfamily I, yellow; subfamily II, red; subfamily III, blue. ABA-responsive luciferase expression driven by the RD29B promoter is expressed relative to the control without ABA and was normalized to a constitutively expressed control gene. The reporter expression in protoplasts was 3.1 kRLU (relative light units)/RFU (relative fluorescence units) for Col-0 cells and 25.7 kRLU/RFU for aba2-1 cells without both RCAR effector and ABA. Both values were set to 1. The analyses were conducted in triplicates (mean ± SD).
ABA-Response Regulation by PP2Cs.
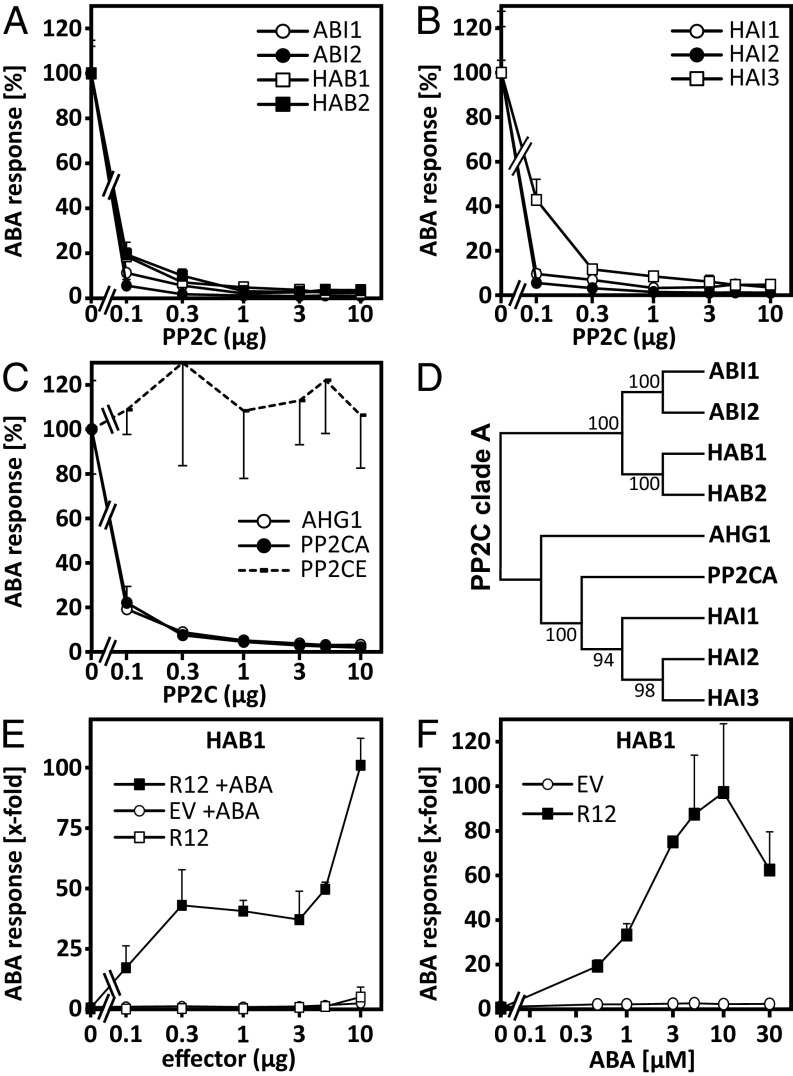
Considering that several PP2Cs are expressed in plant cells, the different extents of ABA-response activation observed might be caused by the presence and abundance of suitable target PP2Cs for RCAR regulation. To examine this possible limitation, all nine coreceptors of Arabidopsis were ectopically expressed and assessed for their ability to down-regulate ABA signaling (Fig. 2 A–C). Clade A PP2Cs can be subdivided into three subgroups: an ABA-INSENSITIVE 1 (ABI1)-homologous subgroup; a PP2CA-homologous subgroup; and the unique PP2C HYPERSENSITIVE GERMINATION 1 (AHG1) (Fig. 2D), which lacks a Trp residue important for RCAR interaction (5, 38). All clade A PP2Cs blocked the ABA response more than 90%; this was not true for a clade E member (Fig. 2 A–C). The regulation of the ABA signal pathway by a PP2C-RCAR pair can be evaluated by comparing the ABA response of cells inhibited by ectopic expression of a single PP2C with cells in which a specific RCAR was coexpressed in the presence and absence of exogenous ABA (32). To exemplify the potential of the assay system, ABA signaling was reduced to ∼10% by ectopic expression of HAB1 and activated by coexpression of RCAR12 (Fig. 2E). The induction triggered by RCAR12 is higher with ectopic PP2C coexpression than without and increased from sixfold (Fig. 1B) to more than 30-fold (Fig. 2E) compared with the empty vector control. ABA signaling was strictly RCAR12- and ABA-dependent and reached saturation at 10 µM exogenous ABA (Fig. 2F). The analysis unravels the potential of RCAR12 to control ABA signaling and the strict ABA dependence on the regulation and provides evidence for a functional HAB1–RCAR12 interaction.
Fig. 2.
Inhibition of ABA signaling by clade A PP2Cs. (A–C) Inhibition of ABA-induced reporter expression by different amounts of PP2C effector constructs in the presence of 10 µM exogenous ABA. The ABA response without PP2C effector was set to 100%, and the analysis was performed as outlined in Fig. 1. The clade E PP2C (PP2CE, At5g27930) served as a negative control (C, interrupted line). (D) Phylogenetic tree of the nine clade A PP2Cs of Arabidopsis. The percentage of conserved clusters in the bootstrap test with 1,000 replicates is shown. (E and F) Activation of the HAB1-inhibited ABA response (0.25 µg effector, 90% inhibition) by (E) various levels of RCAR12 (R12) in the absence (open squares) and presence of 5 µM exogenous ABA (filled squares) compared with the empty vector control (EV, open circles). (F) ABA dependence of reporter expression in protoplasts transfected with 5 µg DNA of RCAR12 effector or EV. Each data point represents the mean value of at least three independent transfections (mean ± SD).
Interaction Matrix of ABA Receptor Components.
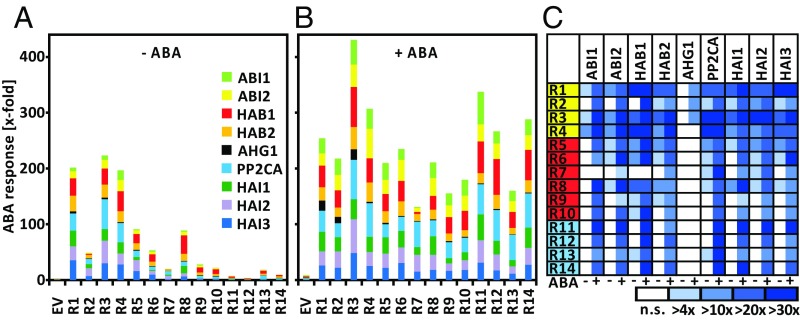
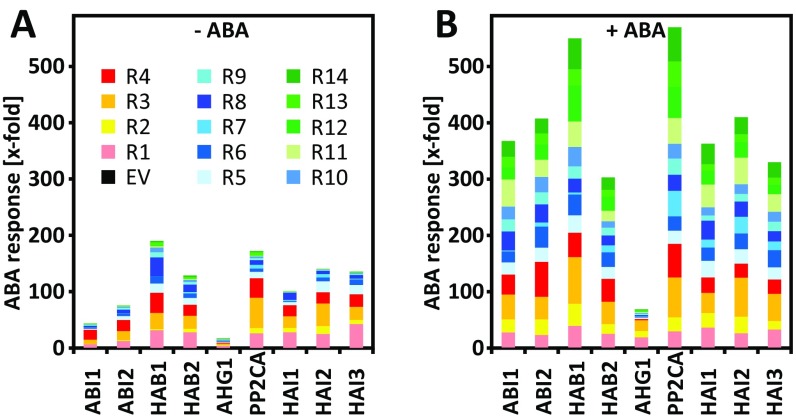
This assay system permits the analysis of all possible PP2C-RCAR interactions for their capacity to regulate ABA signaling. There are 126 potential combinations between the 14 RCARs and the 9 clade A PP2Cs from Arabidopsis. For 113 combinations of all 126 combinations tested, we found evidence for a regulatory interaction, 56 were active in the presence of endogenous ABA, and 57 required elevated ABA levels provided by exogenous hormone administration (Fig. 3). At endogenous ABA levels, ectopic expression of single RCARs, particularly of subfamily I, restored the ABA response as observed before (Fig. 1A). RCAR1, RCAR3, and RCAR4 efficiently activated ABA signaling in cells where HAB1, HAB2, PP2CA, HAI1, HAI2, and HAI3 were coexpressed. RCAR2 was somewhat less effective in recovering the ABA response, similar in extent to the most active subfamily II members RCAR5, RCAR6, and RCAR8. RCAR8 strongly regulated HAB1. In agreement with the previous analysis (Fig. 1A), expression of subfamily II and subfamily III receptors generally showed moderate and low activity, respectively.
Fig. 3.
ABA-response regulation by the combinatorics of ABA receptors and coreceptors. (A and B) RCAR (R1–R14)-mediated activation of ABA signaling attenuated by clade A PP2C expression in the absence and presence of exogenous ABA (±ABA) in WT protoplasts. The analysis was performed as described in Fig. 2F with 10 µM ABA. PP2C effector levels were chosen to inhibit ABA signaling by ∼90% (Fig. 2 A–C and Supporting Information). Note that the ABA response of cells transfected with various PP2Cs and the empty RCAR expression cassette (EV) in the presence of 10 µM ABA was set to 1. The values (in kRLU/RFUs) were the following: ABI1, 22; ABI2, 41; HAB1, 10; HAB2, 56; AHG1, 51; PP2CA, 34; HAI1, 38; HAI2, 71; and HAI3, 58. Each data point represents the mean value of three independent transfections (SD ± 27%). (C) Matrix structure of RCAR–PP2C combinations and their potential to regulate ABA signaling at resting ABA levels (−ABA, left half of box) and at high exogenous ABA (+ABA, right half of box). The data are compiled from results presented in A and B. The three subfamilies of ABA receptors are color-coded as in Fig. 1. Combinations of receptor components with no significant RCAR regulation are depicted as a white box. Regulation above the threshold is categorized from light blue to dark blue as indicated.
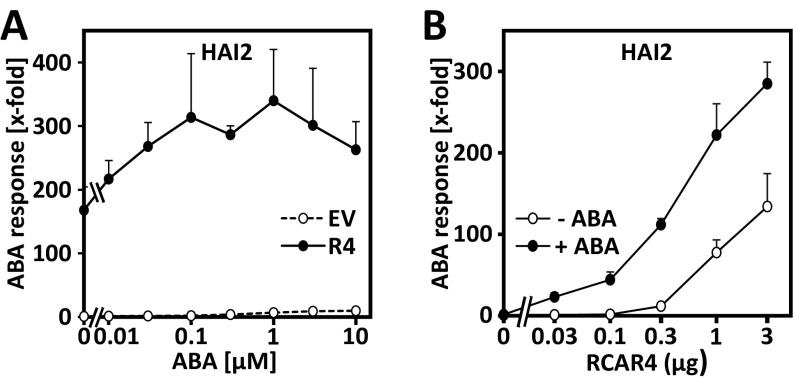
In the presence of 10 µM exogenous ABA, however, all RCARs stimulated ABA signaling, and the PP2C regulation by single ABA receptors was remarkably similar (Fig. 3B). Six of nine PP2Cs (ABI2, HAB2, PP2CA, and HAI1–HAI3) were regulated by all RCARs (Fig. 3C and Fig. S2), and ABI1 and HAB1 by all except RCAR7. AHG1, however, was regulated only by RCAR1–RCAR3 in the transient ABA-signaling assay. The overall efficiency of RCARs to regulate PP2Cs in the presence of exogenous ABA differed from the mean 240-fold (sum of induction values by single RCARs) by a maximum twofold with RCAR7 and RCAR3 showing the lowest and highest combined regulation of 130- and 430-fold, respectively (Fig. 3B). The three HIGHLY ABA-INDUCED PP2C GENE (HAI) PP2Cs emerged as good targets for RCAR regulation and were efficiently regulated by subfamily I receptors at resting ABA levels. Interestingly, RCAR4 showed ABA responsiveness in a number of PP2C combinations such as ABI1, ABI2, HAB2, HAI1, and HAI2 (Fig. 3). The ABA dependency of the RCAR4-HAI2 pairing was corroborated by titration of the exogenous ABA level showing an approximately twofold increase in ABA signaling at saturating exogenous ABA levels (100 nM, Fig. S3A). Lowering the amount of RCAR effector revealed a strong ABA-dependent action of RCAR4 (Fig. S3B). Generally, ABA signaling attenuated by ABI1, ABI2, and AHG1 was less efficiently regulated by RCARs at basal ABA levels, whereas in the presence of high exogenous ABA only AHG1 was poorly regulated (Fig. S2).
Fig. S2.
Compilation of PP2C regulation by ABA receptors from Arabidopsis. The figure depicts combined RCAR (R1–R14) regulation of single PP2Cs in the abscence (A) and presence (B) of 10-µM exogenous ABA. The data presented is taken from Fig. 3. Members of the RCAR subfamily I are shown in yellow to red colors, subfamily II members in blue, and subfamily III RCARs in different green shades as indicated. Note that AHG1 is poorly regulated by RCARs. In the presence of exogenous ABA, only three subfamily I members substantially regulate AHG1.
Fig. S3.
ABA-dependent activation of ABA signaling by RCAR4. (A) Protoplasts of the aba2-1 mutant were transfected with HAI2 effector (0.1 µg) to achieve ∼90% inhibition of ABA signaling at 10 µM exogenous ABA. Cotransfection of RCAR4 effector (5 µg, filled circle) restored ABA signaling, expressed as a fold increase of reporter activity. Cells transfected with the empty vector construct (EV, open circle) served as the control. Protoplasts were exposed to exogenous ABA levels as indicated for ∼16 h before analysis. The ABA response is given as fold induction relative to the control without ABA and RCAR4 expression and was set to 1. (B) Titration of the RCAR4 effector. The analysis was performed as described in A except that the effector DNA varied as indicated in the presence of 100 nM exogenous ABA (+ABA) or without (−ABA). The reporter expression in protoplasts without effector and exogenous ABA was set to 1. Each data point represents the mean of three independent transfections (mean ± SD).
Specificity of AHG1 Regulation.
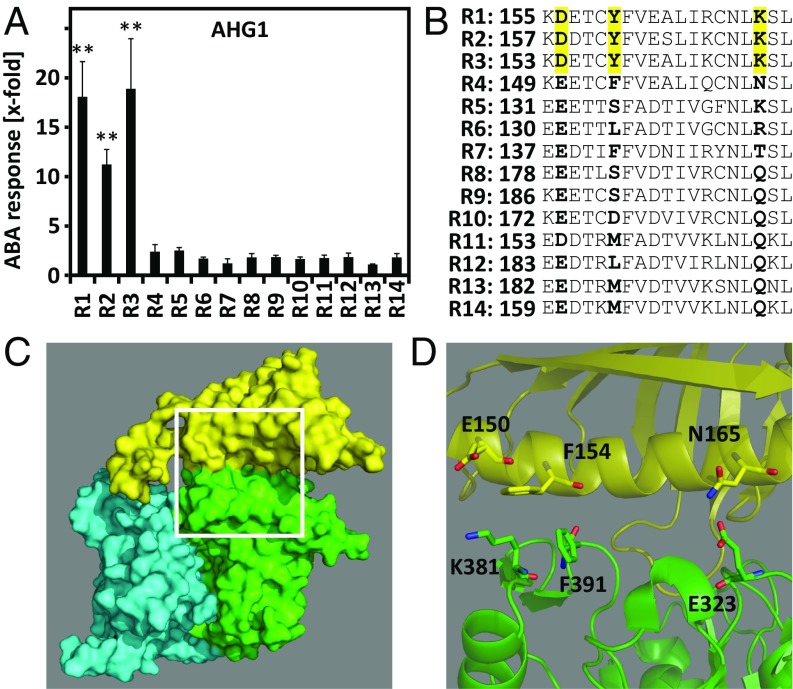
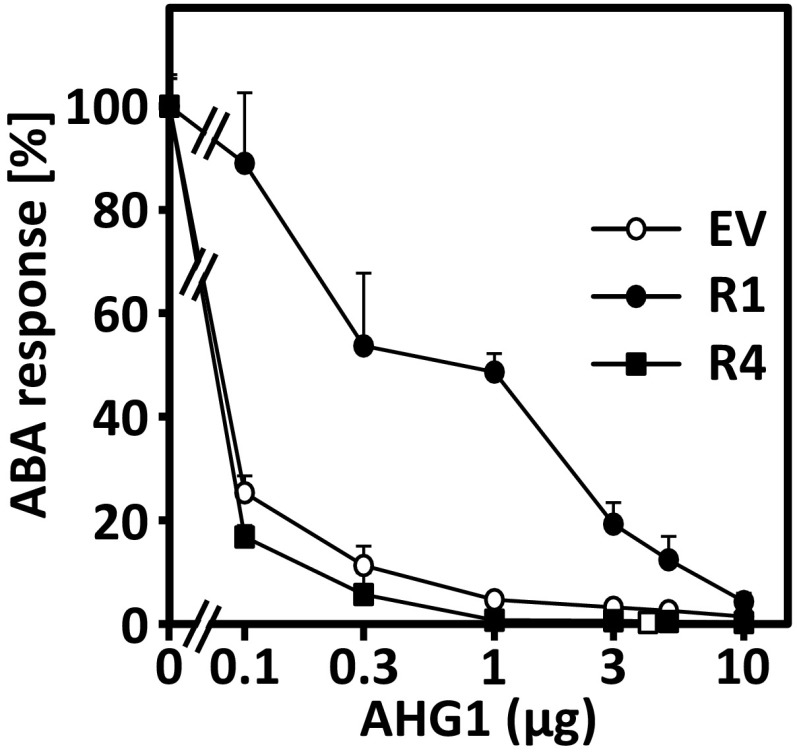
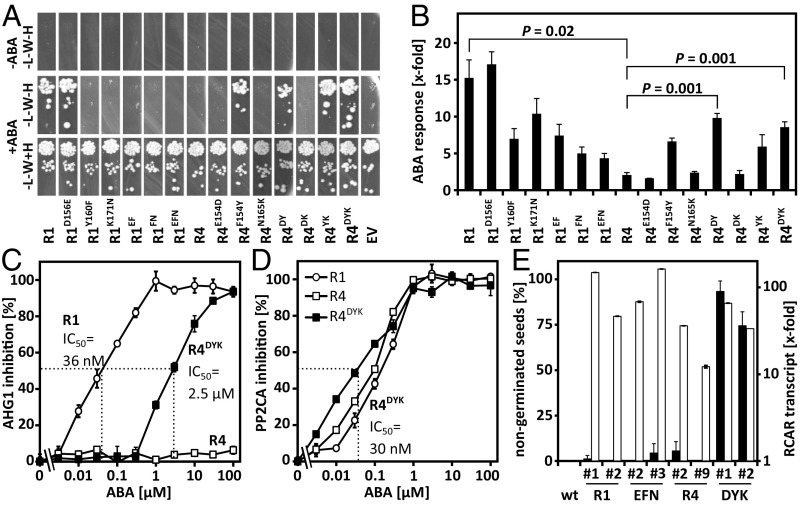
AHG1-inhibited ABA signaling was clearly activated by coexpression of RCAR1, RCAR2, or RCAR3 while expression of RCAR4, the fourth member of subfamily I, had no effect (Fig. 4A). The comparison of RCAR1- and RCAR4-expressing cells at varying AHG1 effector levels confirmed this observation (Fig. S4).
Fig. 4.
Inhibition of the ABA response by the ABA coreceptor AHG1 is overcome by RCAR1, RCAR2, and RCAR3 containing unique amino acid residues at the PP2C interaction surface. (A) Activation of AHG1-attenuated ABA response by ectopic RCAR expression as shown in Fig. 3B in the presence of 10 µM ABA. The double asterisks (**) mark P values below 0.01 compared with the EV transfection. Induction values below fourfold were considered not significant. (B) Alignment of the primary structure segment of RCARs encompassing the alpha helix 2 at the interaction surface of PP2Cs. The amino acid residues conserved in RCAR1, RCAR2, and RCAR3 and different from RCAR4 are Asp (D), Tyr (Y), and Lys (K), which are highlighted in yellow. The numbers indicate the position of the first amino acid shown in the RCAR protein. (C) Three-dimensional structure of RCAR4 (yellow) in complex with HAB1 (green) from Protein Data Bank 3RT0 using PyMOL (www.pymol.org) (28). A second HAB1 molecule is shown in cyan. (D) Close-up of alpha helix 2 of RCAR4 and its contact sites with HAB1. Amino acid residues equivalent in position to the three highlighted amino acid residues of the subfamily I receptors shown in B are presented as yellow sticks (E150, F154, N165) and juxtapositioned amino acid residues of HAB1 as green sticks (K381, F391, E323).
Fig. S4.
Comparison of RCAR1 and RCAR4 efficiency in rescuing the ABA response curbed by AHG1 expression in the presence of 10 µM ABA. Arabidopsis protoplasts were cotransfected with different levels of AHG1 effector DNA and 5 µg DNA of the RCAR effector cassette or the empty vector control (EV). The values of ABA reporter expression without ectopic AHG1 expression were set to 100% as reference points; this corresponds to 229; 1,161; and 1,294 kRLU/RFU for cells transfected with EV, RCAR1, and RCAR4 effector, respectively. Each data point represents the mean of three independent transfections (mean ± SD).
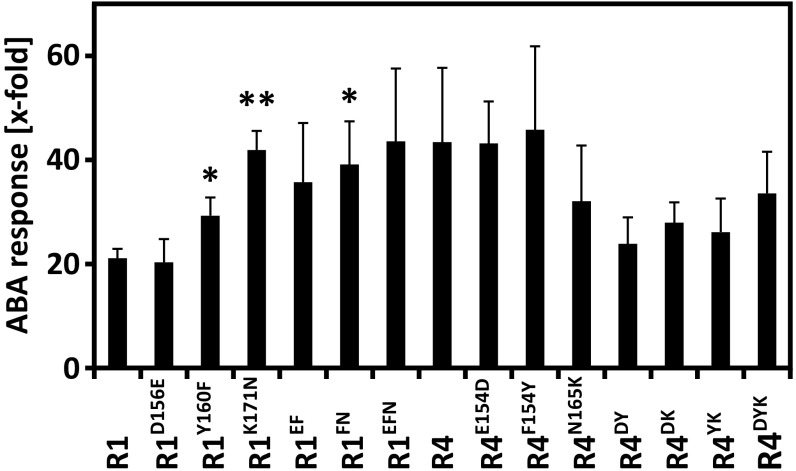
Analysis of the PP2C interaction surface of RCARs identified three amino acid residues in RCAR4 that differed from the other subfamily I members (Fig. 4 B–D). To analyze the contribution of these amino acid residues to AHG1-RCAR specificity, the three amino acid residues were reciprocally exchanged between RCAR1 and RCAR4. Protein interaction analysis in yeast confirmed interaction of AHG1 with RCAR1 but not with RCAR4 (Fig. 5A). Replacing the Tyr (Y160) and Lys (K171) residues of RCAR1 with the RCAR4 residues Phe and Asn, respectively, disrupted the RCAR1–AHG1 interaction. Reciprocal amino acid exchanges in RCAR4 revealed that RCAR4F154Y, carrying a Tyr residue instead of Phe, was able to bind to AHG1, but no other single variant (Fig. 5A). Double- and triple-mutated versions of RCAR1 and RCAR4 corroborated the critical role of this Tyr residue for AHG1 interaction. Subsequent analysis of the RCAR variants for AHG1 regulation in the ABA response of Arabidopsis cells showed that the RCAR4 mutant forms, which contained the F154Y exchange, conferred ABA-dependent regulation of AHG1, albeit less effectively than RCAR1 (Fig. 5B). All variants activated ABA signaling, which was inhibited by PP2CA expression (Fig. S5). The RCAR1 variants without the critical Tyr residue were impaired but still active in stimulating the AHG1-inhibited response, even RCAR1EFN, with all three RCAR4-specific residues (Fig. 5B).
Fig. 5.
Specificity determinants of RCAR1 and RCAR4 for AHG1 regulation and interaction. (A) Protein interaction analysis of AHG1 and variants of RCAR1 and RCAR4 in yeast shows the critical role of a Tyr residue in RCAR1 and ABA for AHG1 binding. Histidine autotrophic growth of yeast (−H) spotted as a 10-fold dilution series on media deficient in Leu (−L) and Trp (−W) indicates RCAR–AHG1 interaction. The RCAR variants were modified at the amino acid residues highlighted in yellow in Fig. 4B. (B) Rescue of AHG1-imposed inhibition of ABA signaling by different RCAR variants in the presence of 10 µM exogenous ABA. The analysis was performed in protoplasts with the RCAR variants as mentioned in Fig. 4A (triplicates, mean ± SD). (C and D) Biochemical analysis of PP2C regulation by ABA in the presence of RCAR1 (open circle), RCAR4 (open squares), and the modified RCAR4DYK (black squares). (C) The phosphatase activity of AHG1 (2 µM) and (D) PP2CA (0.4 µM) was assayed in the presence of twofold molar excess of RCAR protein and different ABA concentrations as indicated. The IC50 values for ABA of the RCAR-AHG1 regulation are depicted. AHG1 is not regulated by RCAR4 (open squares) even at 0.1 mM ABA (mean ± SD). (E) Analysis of RCAR-overexpressing Arabidopsis lines for seed germination and RCAR overexpression. The percentage of nongerminated seeds was assessed in independently generated lines overexpressing RCAR1/RCAR1-EFN variant and RCAR4/RCAR4-DYK variant in comparison with the parental line Col-0 after 2 d of stratification at 4 °C and 3 d of incubation at room temperature (filled bars, n > 90 per data point, mean ± SD). The RCAR1 and RCAR4 transcript levels were determined in seedlings and expressed relative to WT (open bars).
Fig. S5.
Activation of ABA signaling by different RCAR1 and RCAR4 variants in PP2CA-inhibited cells. The analysis was performed in the presence of 10 µM exogenous ABA as mentioned in Fig. 5B, except that PP2CA replaced AHG1 (triplicates, mean ± SD, *P < 0.05, **P < 0.01, compared with their RCAR WT forms). The ABA response without RCAR effector expression was set to 1.
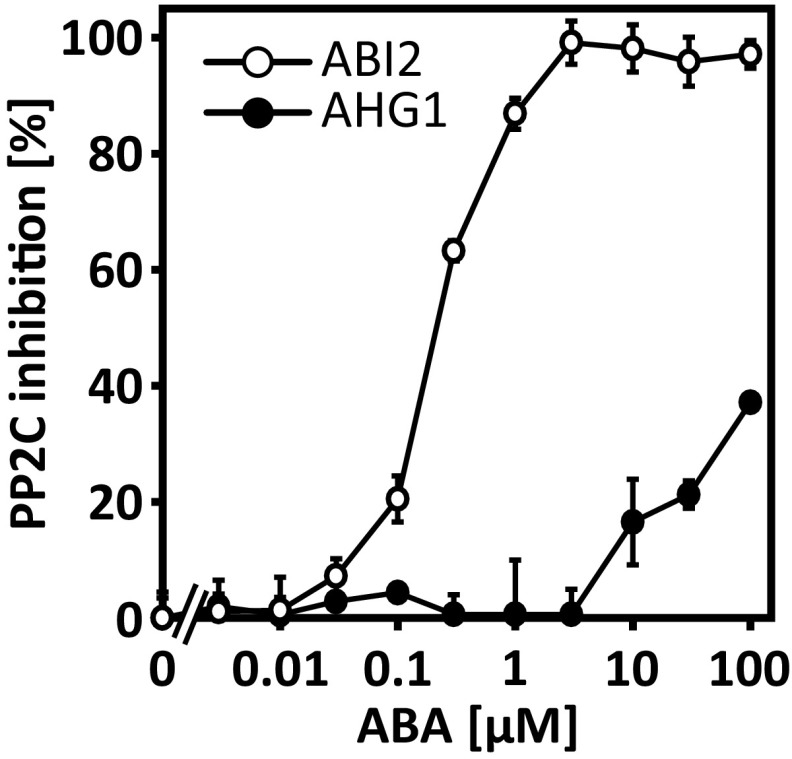
Heterologous expression and protein purification of RCAR1, RCAR4, and the triple-modified RCAR4DYK allowed a biochemical assay for coreceptor regulation. The analysis confirmed the ABA-mediated inhibition of AHG1 phosphatase activity by RCAR1 but not by RCAR4. Interestingly, RCAR3 was a poor inhibitor of AHG1 in the presence of micromolar ABA concentrations pointing to a prominent role of RCAR1, and possibly RCAR2, in regulating AHG1 (Fig. S6). RCAR4 was not able to inhibit the AHG1 activity even in the presence of 0.1 mM ABA, while half-maximum inhibition occurred at 36 ± 5 nM ABA in the presence of RCAR1 (Fig. 5C). The triple mutated RCAR4 inhibited AHG1 with an IC50 value of ∼2.5 ± 0.3 µM ABA. Both RCAR4 and RCAR4DYK sensitively regulated PP2CA with IC50 values of ∼75 ± 9 and 30 ± 6 nM ABA, respectively (Fig. 5D). AHG1 is a central regulator of seed dormancy and strongly expressed in dry seeds (16, 39, 40). To examine the effect of modified RCAR1 and RCAR4 variants on seed germination, the triple variants and their RCAR WT forms were ectopically expressed in Arabidopsis. Analysis of seed material of the same age revealed that expression of RCAR4DYK reduced germination to 7% and 25% in two independent lines compared with ∼97% in RCAR4-overexpressing lines (Fig. 5E). Extended stratification and incubation for 4 and 7 d, respectively, resulted in an ∼60% germination rate of both RCAR4DYK lines. The RCAR1EFN and RCAR1 lines had germination frequencies varying between 96 and 100%, similar to the control.
Fig. S6.
Biochemical analysis of ABA-dependent AHG1 (closed circle) and ABI2 (open circle) regulation in the presence of RCAR3. The phosphatase activity of AHG1 (2 µM) and ABI2 (0.1 µM) set to 100% was assayed in the presence of twofold molar excess of RCAR3 protein and different ABA concentrations as indicated. AHG1 is poorly regulated by RCAR3 even at 10 µM ABA (n = 3; mean ± SD).
Discussion
The diversity of responses controlled by ABA and the dynamics in stress development associated with different ABA levels might require a large number of different ABA receptors and coreceptors for fine-tuned adjustments in gas exchange and metabolism. Our study uncovers three major properties of the ABA receptor components in Arabidopsis, the remarkable capability of PP2Cs to form functional holo-receptor complexes with many different RCARs, the overall similar regulation of single RCARs across the family of PP2Cs with only a few exceptions, and the different potency of RCAR subfamilies to regulate the ABA response at endogenous, basal ABA levels.
At resting ABA levels, subfamily I receptors efficiently regulated PP2Cs, while those of subfamily II were moderately active, and those from subfamily III were overall only weakly active when ectopically expressed. Exceptions to this general trend were RCAR2/PYL7 and RCAR13/PYL3. In the presence of high concentrations of exogenous ABA, subfamily III members activated PP2C-inhibited ABA signaling comparatively to subfamily I or II, arguing against major differences in expression levels, which is also supported by immunodetection analysis. The different ABA sensitivities provided by the RCAR subfamilies were corroborated in ABA-deficient protoplasts of the aba2-1 mutant, which revealed markedly reduced or abolished ABA responses for all RCARs.
The observations are in agreement with the concept of a generally higher ABA affinity of holo-receptor complexes containing subfamily I RCARs in comparison with complexes of members of subfamily III (13, 25, 27, 28). Subfamily III receptors with the prototype PYR1/RCAR11 are characterized as homodimers while other RCARs are monomeric proteins in the absence of bound ABA (26, 28, 41–43). This might explain the strict ABA requirement of RCAR11, RCAR12/PYL1, and RCAR14/PYL2 for regulating ABA signaling (Fig. 3B). RCAR13 has been characterized as a transdimeric ABA receptor that readily interacts with PP2Cs (42), which is consistent with the moderate activity of RCAR13 found at endogenous ABA levels.
The functionality of RCAR–PP2C combinations was assessed by enforced expression of ABA receptor components. While the analysis as such is robust, the question remains as to whether those RCAR–PP2C combinations exist in planta and whether the cellular concentrations of the expressed proteins achieved by the transient analysis are relevant. Genome-wide expression analyses (16–18, 27, 39, 44) and analyses of multiple RCAR as well as PP2C mutants (14, 15, 17–24) support the notion that different RCARs and PP2Cs are present simultaneously within a single cell or plant tissue. However, tissue-specific, developmental stage-specific, and environmental cue-specific expression of ABA receptor components probably restrict the complexity of RCAR–PP2C interactions. Hence, our analysis provides an assessment of the maximum possible number of functional holo-receptor complexes based on transcriptional regulation as output. ABA receptors can exert their action not only in the nucleus but also at cell membranes and in the cytosol (1), and PP2Cs can target different subcellular sites (7, 8, 23). Those intracellular sites may contribute to the specificity of PP2C-RCAR pairings as well as differential binding to downstream-acting SnRK2s. In addition, endogenous ABA levels and protein levels of ABA receptor components affect which receptor–coreceptor combinations are preferentially realized. At very low endogenous ABA levels, RCAR4/PYL10 and other subfamily I members seem to be the most efficient regulators of PP2Cs.
The RCAR-mediated regulation of the ABA coreceptors was generally quite uniform with two major exceptions, AHG1 and RCAR7. The selective discrimination of certain ABA receptors by PP2Cs such as AHG1 implies that PP2C-specific features contribute to selectivity. In the same context, selective discrimination of PP2Cs by specific RCARs such as RCAR7/PYL13 point to RCAR-specific features. In fact, analysis-of-specificity determinants for RCARs (30, 33, 45, 46) and PP2C HAB1 (32) and AHG1 (this study) showed that both receptor components have evolved key-lock features. RCAR7 did not rescue ABI1-, AHG1-, and HAB1-inhibited ABA signaling in this study in agreement with previous analyses (32, 35). Characterization of the RCAR7-specific repulsion by HAB1 revealed that changes in two amino acid residues of HAB1 located at the RCAR interaction surface confer RCAR7 binding and regulation (32). Similarly, analysis of the specificity determinants for the AHG1–RCAR interaction identified a Tyr residue of RCARs located at the PP2C interaction surface as being critical for regulation.
Subfamily I RCARs possess the above-mentioned Tyr residue with the exception of RCAR4, in which a Phe residue replaces the Tyr residue. Substituting the Phe with Tyr conferred RCAR4 interaction and regulation of AHG1. In the crystal structure of the RCAR4–HAB1 complex (28), the Phe residue of RCAR4 establishes a hydrophobic interaction with a Phe residue at position 391 of the HAB1 primary structure that is embedded in the conserved sequence GARVF391G. AHG1 has a Glu residue in this motif, GARVE296G, which provides a negatively charged side chain that is predicted to repulse the interaction with the hydrophobic moiety of the juxtaposed Phe in RCAR4. The Glu residue of AHG1, however, supports a hydrophilic interaction with the Tyr residue conserved in the AHG1-regulating RCARs. Replacing that Tyr residue in RCAR1 with Phe severely impaired AHG1 regulation but did not fully abolish it, suggesting the presence of additional structural features contributing to AHG1 interaction.
AHG1 is a negative regulator of ABA-sensitive seed germination and dormancy (39). Seed dormancy is regulated by ABA and a protein network in which AHG1, PP2CA, and DELAY OF GERMINATION1 (DOG1) are central players (47, 48). Gene expression analysis revealed low RCAR3 expression and high transcript abundances of RCAR1, RCAR2, and AHG1 during seed development and in dormant seeds (16). Our study shows that AHG1 is specifically regulated in vivo by RCAR1, RCAR2, and RCAR3. In vitro experiments corroborate an efficient regulation of AHG1 by RCAR1, not by RCAR3 as shown before (23). Overexpression of the RCAR4 triple variant, in which the critical Tyr residue for AHG1 interaction was introduced, efficiently reduced seed germination compared with the RCAR4-overexpressing Arabidopsis line. The findings provide evidence for a prominent role of RCAR1 and possibly RCAR2 in regulating AHG1-controlled seed development and germination.
Understanding ABA signaling is critical for the development of targeted approaches to improve crop performance under limiting water supply. Analysis of the RCAR–PP2C combinatorics unravels the general suitability of ABA receptors to regulate PP2Cs. The moderate effects of subfamily II members on the ABA response at basal ABA levels make these RCARs prime candidates to reduce transpiration under nonstress conditions without curbing photosynthesis. Recent analysis of all Arabidopsis RCARs showed that overexpression of two subfamily II members, RCAR6/PYL12 and RCAR10/PYL4, increased water-use efficiency and resulted in water-productive plants, which are characterized by reduced transpiration with little or no trade-offs in CO2 uptake and biomass accumulation (49). Important for the trait is the altered ABA response at resting ABA levels. RCAR6 and RCAR10 share the capacity to regulate HAB1 at basal ABA levels while they strongly regulate HAB1 at high ABA levels. The observation points to a critical function of HAB1 in the control of plant’s water status.
The interaction network of the ABA receptors with their cognate coreceptors provides a major challenge in elucidating the molecular details of ABA action. This study provides insights into the combinatorics of RCAR–PP2C interactions and their differing ABA sensitivity, supporting a rheostat function for integrating different hormone levels in the ABA-response pathway.
Methods
Analysis of ABA signaling in protoplasts was performed by transfection of effector DNA constructs together with an ABA-responsive luciferase reporter and normalized to a constitutively expressed control gene (50). All analyses were performed in triplicate (mean ± SD). The in vitro ABA receptor assay was carried out at twofold molar excess of RCAR over the PP2C (32). Further details on methods, including the plant material and chemicals used, are provided in Supporting Information.
SI Methods
Chemicals.
Chemicals were obtained from Sigma-Aldrich (www.sigmaaldrich.com), J. T. Baker (www.fishersci.de), (S)-ABA from CHEMOS (www.chemos-group.com) and d6-ABA (2-cis,4-transabscisic acid-[2H6], catalog no. 034 272x) from OlChemim (www.olchemim.cz).
Plant Material.
The Arabidopsis lines were provided by the Nottingham Arabidopsis Stock Center. Col-0 and aba2-1 plants were grown in a perlite-soil mixture at 23 °C under long-day conditions with 16 h of light (250 µE m−2⋅s−1). Lines overexpressing RCAR1 or RCAR4 were established by transferring an expression cassette for RCARs under control of the 35S promoter into Arabidopsis and selecting for homozygous lines, as carried out for RCAR1 (13). RCAR transcript abundance was determined by RT-PCR analysis (CFX96; Bio-Rad) using ubiquitin 10 as a reference and target-specific primers (32). RNA extraction and subsequent cDNA biosynthesis from 12-d-old seedlings were performed as described (32).
Seed Germination Assay.
Arabidopsis seeds (3 × 30) were plated on Murashige and Skoog agar medium under sterile conditions and stratified at 4 °C for 2 d in the dark, unless otherwise stated. The plates were then transferred to a culture room with continuous light (∼100 µE m−2⋅s−1) at 22 °C, and the germination rate was determined after 3 d (13).
RCAR and PP2C Analyses in Protoplasts.
Preparation and analysis of Arabidopsis protoplasts was performed as described (50). For ABA-signaling analysis, Arabidopsis protoplasts of the ABA-deficient aba2-1 mutant and Col-0 (105 protoplasts in 0.1 mL) were transfected with 5 µg DNA of reporter construct (pRD29B::LUC), 3 µg of p35S::GUS plasmid as a control for internal normalization of expression, and various levels of effector expression cassettes as indicated. The effector cassettes drive expression of RCAR- and PP2C-coding sequences (CDSs) under control of the viral 35S promoter (13). RCARs were additionally expressed under the 35S promoter with a carboxyterminal GFP tag (13) for immunodetection analysis. All constructs used were verified by DNA sequence analysis. The protoplast suspensions were incubated in the presence or absence of exogenous ABA, and reporter expression was determined after 16–18 h of incubation at 25 °C. Assays were carried out in three replicates per data point. To examine the RCAR-mediated release of ABA signaling from PP2C inhibition, PP2C effector levels were chosen to inhibit ABA signaling by ∼90%: ABI1 (0.2 µg), ABI2 (0.1 µg), HAB1 (0.25 µg), HAB2 (0.3 µg), AHG1 (0.3 µg), PP2CA (0.25 µg), HAI1 (0.1 µg), HAI2 (0.1 µg), and HAI3 (0.35 µg). For immunodetection analysis, proteins were isolated by lysis of protoplasts transfected with effector and reporter constructs (5 µg DNA effector construct, 4 µg of p35S::GUS plasmid, 5 µM ABA). Aliquots of protein fractions corresponding to ∼1/6 of the transfected protoplasts were normalized by the β-glucuronidase activity, subjected to SDS/PAGE, and subsequently transferred onto a polyvinylidene fluoride membrane by electroblotting. The membrane was blocked with 2.5% dry milk in the presence of 0.05% Tween 20 and incubated with horseradish peroxidase-conjugated anti-GFP (https://vectorlabs.com/?SID=ac191456df451325188092816d7f2444) according to the provider’s instructions. The immunoblot was visualized using the SuperSignal West Femto Maximum Sensitivity Substrate obtained from Thermo Fisher Scientific and the ImageQuant LAS 4000 (GE Healthcare Life Sciences) imaging system.
Interaction Analysis of ABA Receptors in Vitro and in Yeast.
Heterologous expression of RCARs and PP2Cs and subsequent purification were performed as described (32). RCAR and PP2Cs were expressed in Escherichia coli strains Rosetta pLys and BL21 and purified to homogeneity. Phosphatase activity of purified AHG1 was measured using 4-methyl-umbelliferyl-phosphate as a substrate. The receptor assay was performed at 2 µM concentration of AHG1, 0.4 µM of PP2CA, or 0.1 µM of ABI2 protein and a twofold molar excess of RCAR. Values are means ± SDs of four replicates. Yeast two-hybrid assays were carried out as described (32). AHG1 (AT5G51760), RCAR1 (AT1G01360), and RCAR4 (AT4G27920) were amplified from a cDNA library as mentioned (13). The AHG1 and RCAR CDSs were cloned into the pGAD vector and the pBridge vector (Clontech), respectively. Site-directed mutagenesis was performed according to the manufacturer’s instructions (QuikChange, www.genomics.agilent.com), and all constructs were DNA sequence-verified.
Quantification of ABA Content.
ABA was analyzed essentially as reported (51). Briefly, ABA was extracted from 0.2 mL sedimented protoplasts (60 × g, 2 min) by adding 1 mL methanol and mixing and incubating for 5 min on ice. After mixing and centrifugation (14,000 × g, 4 min), an aliquot per sample (10 µl) was removed for chlorophyll measurement. The chlorophyll level was used for normalizing the extracts and to express ABA levels of protoplasts relative to plant leaf material on a fresh and dry weight basis. Deuterated ABA (final concentration 20 nM) was added as an internal standard to the supernatant, which was subsequently acidified by administration of acetic acid (10 µL), mixed, and again centrifuged (13,000 × g, 10 min, 4 °C). The supernatant was passed through a 0.2-µM syringe filter (Minisart; Sartorius AG), and the eluate was dried by vacuum centrifugation. The sample residues were dissolved in 0.1 mL methanol, centrifuged (3,000 × g, 10 min), and subjected to HPLC-MS/MS analysis by using a Prominence HPLC system (Shimadzu) connected to a 4000 QTRAP (Applied Biosystems). The separation was performed using a XB-C18 100 A column (100 × 2.1 mm, 5 µm Kinetex; Phenomenex) at a flowrate of 400 µL/min and the solvents A and B [A: water with 0.1% (vol/vol) formic acid; B: methanol with 0.1% formic acid]. The conditions were 70% A/30% B for 1 min, a linear gradient to 100% B in 6.6 min, and 3 min 100% B. Ions were analyzed by MS in the negative mode using a Turbo V Ion Source (Applied Biosystems). The ion spray voltage was set to −4,500 V at a source temperature of 500 °C with nitrogen as the collision gas. The parameters for the collision-activated dissociation were −2; curtain gas: 35 psi; ion source gas 1: 55 psi; ion source gas 2: 65 psi. The multiple reaction monitoring parameters for the declustering potential (DP), entrance potential (EP), collision energy (CE), and collision cell exit potential (CXP) were as follows: ABA quantifier m/z 262.991 → 153.0 (DP: −76; EP: −10; CE: −16; CXP: −1); ABA qualifier m/z 262.991 → 203.9 (DP: −76; EP: −10; CE: −30; CXP: −1), d6-ABA m/z 269.100 → 158.8 (DP: −76; EP: −10; CE: −16; CXP: −1) at a dwell time of 0.150 s.
Statistical Analysis.
Data were analyzed using OriginPro 2016 for determining IC50 values, Analyst (Version 1.6.2) for LC-MS/MS data, and other data by an unpaired, two-sided t test (Microsoft Excel, version 2016). For the phylogenetic analysis, the neighbor-joining tree with genetic distance matrices of protein sequences was constructed in MEGA v7.0 using the Poisson model, pairwise deletion, and the bootstrap test with 1,000 replications (52).
Acknowledgments
We thank Christoph Heidersberger and Christian Kornbauer for technical assistance and Farhah Assaad for comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grants EG938 and SFB924 “Molecular mechanisms regulating yield and yield stability in plants”.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706593114/-/DCSupplemental.
References
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein R. Abscisic acid synthesis and response. Arabidopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs S, Grill E, Meskiene I, Schweighofer A. Type 2C protein phosphatases in plants. FEBS J. 2013;280:681–693. doi: 10.1111/j.1742-4658.2012.08670.x. [DOI] [PubMed] [Google Scholar]
- 6.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiger D, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato A, et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- 10.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandt B, et al. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA. 2012;109:10593–10598. doi: 10.1073/pnas.1116590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imes D, et al. Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 2013;74:372–382. doi: 10.1111/tpj.12133. [DOI] [PubMed] [Google Scholar]
- 13.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura N, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park S-Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winter D, et al. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Guzman M, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoni R, et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013;161:931–941. doi: 10.1104/pp.112.208678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin Y, et al. Difference in abscisic acid perception mechanisms between closure induction and opening inhibition of stomata. Plant Physiol. 2013;163:600–610. doi: 10.1104/pp.113.223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merilo E, et al. PYR/RCAR receptors contribute to ozone-, reduced air humidity-, darkness-, and CO2-induced stomatal regulation. Plant Physiol. 2013;162:1652–1668. doi: 10.1104/pp.113.220608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubio S, et al. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhaskara GB, Nguyen TT, Verslues PE. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 2012;160:379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antoni R, et al. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 2012;158:970–980. doi: 10.1104/pp.111.188623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saez A, et al. Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 2006;141:1389–1399. doi: 10.1104/pp.106.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szostkiewicz I, et al. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 28.Hao Q, et al. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell. 2011;42:662–672. doi: 10.1016/j.molcel.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Lim CW, Kim JH, Baek W, Kim BS, Lee SC. Functional roles of the protein phosphatase 2C, AtAIP1, in abscisic acid signaling and sugar tolerance in Arabidopsis. Plant Sci. 2012;187:83–88. doi: 10.1016/j.plantsci.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Pizzio GA, et al. The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 2013;163:441–455. doi: 10.1104/pp.113.224162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SC, Lim CW, Lan W, He K, Luan S. ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol Plant. 2013;6:528–538. doi: 10.1093/mp/sss078. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs S, Tischer SV, Wunschel C, Christmann A, Grill E. Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc Natl Acad Sci USA. 2014;111:5741–5746. doi: 10.1073/pnas.1322085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, et al. The HAB1 PP2C is inhibited by ABA-dependent PYL10 interaction. Sci Rep. 2015;5:10890. doi: 10.1038/srep10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, et al. The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 2013;23:1380–1395. doi: 10.1038/cr.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.González-Guzmán M, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Priest DM, et al. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006;46:492–502. doi: 10.1111/j.1365-313X.2006.02701.x. [DOI] [PubMed] [Google Scholar]
- 38.Dupeux F, et al. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol. 2011;156:106–116. doi: 10.1104/pp.110.170894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura N, et al. ABA-hypersensitive germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 40.Lynch T, Erickson BJ, Finkelstein RR. Direct interactions of ABA-insensitive(ABI)-clade protein phosphatase(PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol Biol. 2012;80:647–658. doi: 10.1007/s11103-012-9973-3. [DOI] [PubMed] [Google Scholar]
- 41.Dupeux F, et al. A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J. 2011;30:4171–4184. doi: 10.1038/emboj.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, et al. Complex structures of the abscisic acid receptor PYL3/RCAR13 reveal a unique regulatory mechanism. Structure. 2012;20:780–790. doi: 10.1016/j.str.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto M, et al. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA. 2013;110:12132–12137. doi: 10.1073/pnas.1305919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, et al. Molecular character of a phosphatase 2C (PP2C) gene relation to stress tolerance in Arabidopsis thaliana. Mol Biol Rep. 2013;40:2633–2644. doi: 10.1007/s11033-012-2350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosquna A, et al. Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc Natl Acad Sci USA. 2011;108:20838–20843. doi: 10.1073/pnas.1112838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagawa M, Kagiyama M, Shibata N, Hirano Y, Hakoshima T. Mechanism of high-affinity abscisic acid binding to PYL9/RCAR1. Genes Cells. 2014;19:386–404. doi: 10.1111/gtc.12140. [DOI] [PubMed] [Google Scholar]
- 47.Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 48.Nakabayashi K, et al. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell. 2012;24:2826–2838. doi: 10.1105/tpc.112.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, et al. Leveraging abscisic acid receptors for efficient water use in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:6791–6796. doi: 10.1073/pnas.1601954113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moes D, Himmelbach A, Korte A, Haberer G, Grill E. Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 2008;54:806–819. doi: 10.1111/j.1365-313X.2008.03454.x. [DOI] [PubMed] [Google Scholar]
- 51.Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc. 2010;5:986–992. doi: 10.1038/nprot.2010.37. [DOI] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]