Significance
There is increasing literature that links the overexpression of protein arginine methyltransferase (PRMT) 5 and PRMT7 to cancer metastasis and tumorigenesis and that suggests these enzymes may be good therapeutic targets. An important question remaining is how PRMT7 may control PRMT5 activity in mammalian cells. In this work, we demonstrate that PRMT7-dependent monomethylation at one site in histone H4 can activate another site for methylation by PRMT5. Such allosteric regulation has not been previously seen in this class of protein modification enzymes.
Keywords: epigenetics, allosteric regulation, PRMT5, PRMT7, histone methylation
Abstract
Arginine methylation on histones is a central player in epigenetics and in gene activation and repression. Protein arginine methyltransferase (PRMT) activity has been implicated in stem cell pluripotency, cancer metastasis, and tumorigenesis. The expression of one of the nine mammalian PRMTs, PRMT5, affects the levels of symmetric dimethylarginine (SDMA) at Arg-3 on histone H4, leading to the repression of genes which are related to disease progression in lymphoma and leukemia. Another PRMT, PRMT7, also affects SDMA levels at the same site despite its unique monomethylating activity and the lack of any evidence for PRMT7-catalyzed histone H4 Arg-3 methylation. We present evidence that PRMT7-mediated monomethylation of histone H4 Arg-17 regulates PRMT5 activity at Arg-3 in the same protein. We analyzed the kinetics of PRMT5 over a wide range of substrate concentrations. Significantly, we discovered that PRMT5 displays positive cooperativity in vitro, suggesting that this enzyme may be allosterically regulated in vivo as well. Most interestingly, monomethylation at Arg-17 in histone H4 not only raised the general activity of PRMT5 with this substrate, but also ameliorated the low activity of PRMT5 at low substrate concentrations. These kinetic studies suggest a biochemical explanation for the interplay between PRMT5- and PRMT7-mediated methylation of the same substrate at different residues and also suggest a general model for regulation of PRMTs. Elucidating the exact relationship between these two enzymes when they methylate two distinct sites of the same substrate may aid in developing therapeutics aimed at reducing PRMT5/7 activity in cancer and other diseases.
Posttranslational modifications (PTMs) of proteins such as histones and transcription factors have been shown to regulate gene expression and contribute to epigenetic control (1–4). PTMs that occur on histones commonly include methylation marks on lysine and arginine residues. Histone arginine methylation has been recently linked to stem cell pluripotency (5), DNA damage repair (6), and cancer metastasis and tumorigenesis (7–10). As such, the enzymes that catalyze these modifications have become popular targets for therapeutic treatments (11–14).
In mammals, there are nine enzymes in the seven–β-strand family of protein arginine methyltransferases, designated PRMT1–9 (3, 4). These PRMTs are further divided into three types based on the different methylarginine derivatives they produce: type I PRMTs (PRMT1–4, 6, and 8) catalyze the production of ω-monomethylarginine (MMA) and asymmetric dimethylarginine (ADMA); type II PRMTs (PRMT5 and 9) catalyze MMA and symmetric dimethylarginine (SDMA) production; and type III enzymes (PRMT7) catalyze only the production of MMA residues (3, 4).
PRMT5, often in complex with methylosome protein 50 (MEP50), is the most prolific type II mammalian PRMT and is responsible for almost all of the SDMA marks in the cell (4, 15–18). Studies of the symmetric dimethylation of arginine-3 in histone H4 indicate that this modification is affected by the expression of PRMT5 (15, 16, 19–21). Since R3 SDMA is a repressive mark, changes in this modification can lead to the loss of tumor suppressor proteins and contribute to proliferative diseases such as lymphoma (8), mixed-lineage leukemia (MLL) (22), and acute myeloid leukemia (AML) (20).
Recently, similar observations of a link between protein arginine symmetric dimethylation of histone H4 at position 3 and a distinct enzyme, PRMT7, have been made (4, 6, 22–26). Initially, PRMT7 was incorrectly identified as an SDMA-catalyzing enzyme due to contamination with PRMT5 (3, 10, 27, 28). As such, PRMT7 was reported to symmetrically dimethylate histone H4 at R3 (29, 30). However, since those initial studies, it has been clearly shown that PRMT7 does not catalyze dimethylarginine production and that it is only able to produce MMA (27, 31, 32). The corrected characterization of PRMT7 as a type III PRMT does not explain, however, why PRMT7 expression levels also seem to affect SDMA levels at R3 on histone H4 (4, 6, 22, 24, 26) and R2 on histone H3 (23). One hypothesis suggests that PRMT7 monomethylates substrates for PRMT5 to subsequently symmetrically dimethylate; thus, a depletion of PRMT7 may result in fewer “primed” arginine residues, causing lower PRMT5-mediated SDMA (10). This hypothesis, however, has not been experimentally supported (4, 26). In fact, it has been shown that PRMT7 has distinctly different substrate recognition from PRMT5 and specifically does not methylate R3 on histone H4 in vitro (27). Nevertheless, PRMT7 is able to monomethylate arginine-17 and 19 on the same histone H4 N-terminal tail (27). A recent proteomic study documented the presence of monomethylation at R17 on histone H4 in mice testes (33). These observations lend themselves to another hypothesis: Monomethylation of R17 and/or R19 by PRMT7 may direct PRMT5 activity on R3 of histone H4.
Kinetic studies on PRMT5 and different forms of histone H4, including peptides with variable numbers of residues and amino acid substitution and modifications, have demonstrated the sensitivity of the enzyme activity to the residues downstream (distal site) of the methylatable R3 site (15). Similar studies have been done with PRMT1, and they generally also demonstrate the importance of residues, and their modifications, downstream from the primary methylation site for PRMT-mediated activity (34–36). For example, lysine acetylation at K5, K8, K12, and K16 on histone H4 significantly effects PRMT1 activity at residue R3 (36). Likewise, a study of Caenorhabditis elegans PRMT5 revealed the importance of the chemical properties of residues downstream from R3 for catalytic activity (15), indicating that a second or “distal” site on substrates and enzymes alike might exist as a means for regulation of methylation activity. Interestingly, with one recent exception (37), none of these studies reported effects of distal substrate recognition sites on the type of kinetics displayed by PRMT5 and PRMT1 such as allostery (cooperativity). The experiments in our current study, however, demonstrate not only the allosteric nature of PRMT5 and PRMT1, key regulators of gene transcription, but also that downstream methylation of histone H4 (R17MMA) can significantly affect the methylation by PRMT5 of R3, an important gene repression marker, on the same protein.
Results
Histone H4 Monomethylation at R17 Affects Methylation of R3 by HsPRMT5/MEP50.
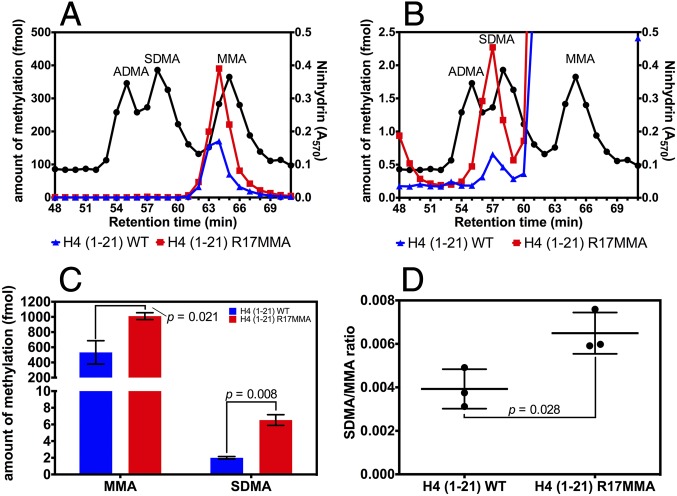
Symmetric dimethylation of histone H4 residue R3 in mammals has been reported to be affected by the expression levels of PRMT7 (4, 6, 22–26). However, in vitro assays of PRMT7 show that it monomethylates residues R17 and R19 in histone H4 and does not modify R3 (27, 31). Methylation of R17 and R19 in histone H4 from intact cells has not been observed with the exception of monomethylation at R17 in one proteomic study of mouse testes (33). To resolve this apparent paradox, we investigated the effect of the methylation state of histone H4 R17 in an N-terminal peptide [H4 (1-21)] on the activity of HsPRMT5/MEP50. Cation exchange chromatography on the acid hydrolysates of the methylation products of reactions with HsPRMT5/MEP50 and the H4 (1-21) peptide, tested either with unmodified sequence (WT) or with R17 monomethylated (R17MMA) (Fig. 1), was performed. When the R17MMA peptide was used as a substrate, we found significantly higher MMA (∼twofold increase) and SDMA production (∼threefold increase) than with the unmodified peptide (Fig. 1 A–C). Additionally, the ratio of SDMA to MMA was also significantly higher with the R17MMA peptide (Fig. 1D). These results suggest that the methylation state of a distal residue can markedly affect the activity of PRMT5 on this peptide.
Fig. 1.
Analysis of methylarginine production by HsPRMT5/MEP50 on histone H4 (1-21) peptide. (A) A representative cation exchange chromatogram (n = 3) for hydrolysates of reactions with H4 (1-21) WT (blue) and H4 (1-21) R17MMA (red) as substrates with [methyl-3H]-AdoMet. Fractions (1 min) were collected and analyzed for the presence of nonradioactive methylarginine standards and radioactivity. The black line indicates the retention profile of the standards as determined by ninhydrin assays (47, 48). The colored lines represent radioactive methylarginine derivatives which elute about 1 min before the nonradioactive standards due to the isotope effect (50). For details of the reaction and chromatography conditions, see Methods. (B) An expanded view of A to emphasize the differences in SDMA levels. (C) Data from three replicate experiments were used to show changes in 3H-MMA and 3H-SDMA produced with H4 (1-21) WT (blue bars) or its R17MMA derivative (red bars); the P values were determined through two-tailed t tests. The error bars represent SDs. (D) The SDMA/MMA ratio was calculated from the data in C. The P value was determined as for C, and the error bars represent SDs.
HsPRMT5/MEP50 Methylation Displays Positive Cooperativity.
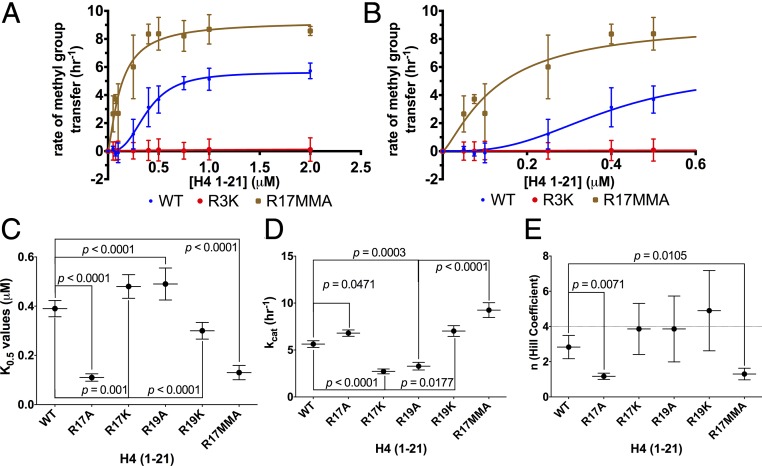
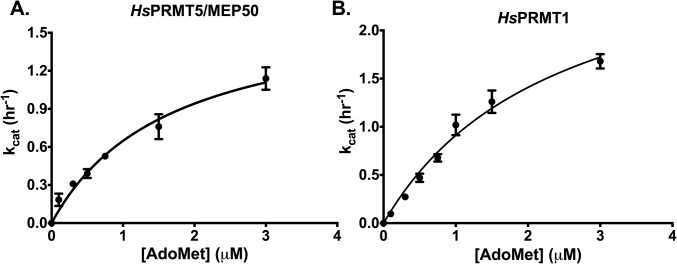
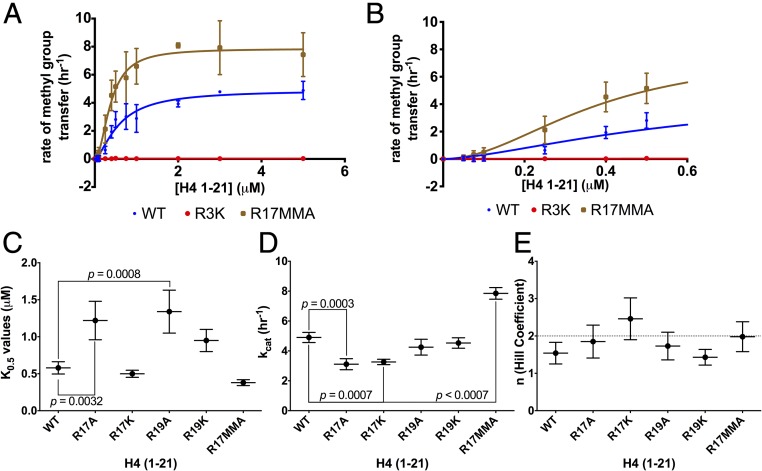
To confirm that R3 and not R17/R19 is the site of methylation carried out by HsPRMT5/MEP50, we demonstrated that no methylation by this enzyme was observed with the R3K-modified H4 (1-21) peptide (Fig. 2A). We then determined the kinetic parameters of HsPRMT5/MEP50 with the cofactor AdoMet and the substrate H4 (1-21), finding that this enzyme showed typical Michaelis–Menten kinetics as a function of AdoMet concentration with a Km of 1.66 ± 0.37 μM (Fig. S1A), a value that is consistent with previous measurements (15, 21). However, a different result was found when the peptide substrate concentration was varied. Importantly, these kinetic data revealed that HsPRMT5/MEP50 does not follow simple Michaelis–Menten kinetics but appears to show positive cooperativity with the H4 (1-21) WT substrate and a Hill coefficient (n) > 1 (Fig. 2 A and B). Cooperativity had not been observed for PRMT5 in previous studies. These findings suggest a possible allosteric mechanism for the regulation of PRMT5 via binding of second-site arginine residues. The kinetic data best fits the Hill equation (38) for positive cooperativity, with kcat, K0.5, kcat/K0.5, and Hill coefficients shown in Table 1. We note that the kcat of the human enzyme measured here is about 10-fold lower than that of the C. elegans enzyme (15), although its kcat/K0.5 is about threefold greater.
Fig. 2.
Monomethylation of H4 R17 affects the positive cooperativity exhibited by HsPRMT5/MEP50. Initial kinetic measurements were made, and the data were fit to the Hill equation (38). (A) Enzyme activity of HsPRMT5/MEP50 with H4 (1-21) WT (blue), H4 (1-21) R17MMA (brown), and H4 (1-21) R3K (red) is shown for triplicate assays (error bars represent SD). (B) An expanded view of A at the low substrate concentrations. Best fit curves are shown for K0.5, kcat, and Hill coefficient values for the H4 (1-21) WT substrate of 0.39 μM, 5.63 h−1, and 2.83, respectively. For H4 (1-21) R17MMA, the parameters were 0.13 μM, 9.25 h−1, and 1.3, respectively. For details about reaction conditions and concentrations, see Methods. Statistical analysis of K0.5 values (C), statistical analysis of kcat values (D), and statistical analysis of the Hill coefficient values (E). The dashed line represents a Hill coefficient of 4. Data were taken from the triplicate assays shown in A and B; error bars represent SD. The P values were calculated using a one-way ANOVA test with a Dunnett test for multiple comparisons using the GraphPad Prism 6.0 software.
Fig. S1.
Binding affinity of AdoMet with HsPRMT5/MEP50 and HsPRMT1. Initial kinetic measurements were made, and the data were fit to the Michaelis–Menten equation. (A) Enzyme activity of HsPRMT5/MEP50 with varying [AdoMet] and 10 μM H4 (1-21) WT peptide in triplicate (error bars represent SD). The solid line was best fit to the Michaelis–Menten equation with the following parameters: a Km of 1.66 ± 0.37 μM and a kcat of 1.72 ± 0.14 h−1. (B) Enzyme activity of HsPRMT1 with varying [AdoMet] and 10 μM H4 (1-21) WT peptide in triplicate (error bars represent SD). The solid line was best fit to the Michaelis–Menten equation with the following parameters: a Km of 2.41 ± 0.35 μM and a kcat of 3.11 ± 0.27 h−1. For details about reaction conditions and concentrations, see Methods.
Table 1.
Initial kinetic parameters
| HsPRMT5/MEP50 | HsPRMT1 | |||||||
| Substrate | K0.5, μM | kcat, h−1 | kcat/K0.5, h−1·μM−1 | n (Hill coefficient) | K0.5, μM | kcat, h−1 | kcat/K0.5, h−1·μM−1 | n (Hill coefficient) |
| H4 (1-21) WT* | 0.39 ± 0.033 | 5.63 ± 0.36 | 14.44 | 2.83 ± 0.66 | 0.58 ± 0.083 | 4.90 ± 0.34 | 8.45 | 1.54 ± 0.29 |
| H4 (1-21) R17A* | 0.11 ± 0.015 | 6.80 ± 0.34 | 61.82 | 1.17 ± 0.18 | 1.22 ± 0.26 | 3.11 ± 0.37 | 2.55 | 1.85 ± 0.44 |
| H4 (1-21) R17K* | 0.48 ± 0.048 | 2.72 ± 0.26 | 5.67 | 3.87 ± 1.45 | 0.50 ± 0.048 | 3.26 ± 0.18 | 6.52 | 2.46 ± 0.56 |
| H4 (1-21) R17MMA* | 0.13 ± 0.029 | 9.25 ± 0.79 | 71.15 | 1.3 ± 0.33 | 0.38 ± 0.040 | 7.85 ± 0.39 | 20.66 | 1.98 ± 0.40 |
| H4 (1-21) R19A* | 0.49 ± 0.065 | 3.28 ± 0.41 | 6.69 | 3.86 ± 1.87 | 1.34 ± 0.29 | 4.25 ± 0.53 | 3.17 | 1.73 ± 0.37 |
| H4 (1-21) R19K* | 0.30 ± 0.034 | 7.02 ± 0.57 | 23.4 | 4.90 ± 2.28 | 0.95 ± 0.15 | 4.53 ± 0.35 | 4.77 | 1.43 ± 0.21 |
| H4 (1-21) R3K* | nd† | nd† | nd† | nd† | nd† | nd† | nd† | nd† |
Twenty micromolar AdoMet used (20:1 molar ratio of AdoMet: [methyl-3H]-AdoMet).
Amount of product formation was too low to accurately measure kinetic parameters.
Monomethylation of Histone H4 R17 Has Significant Effects on the Kinetics of HsPRMT5 and Its Allosteric Enzymatic Activity.
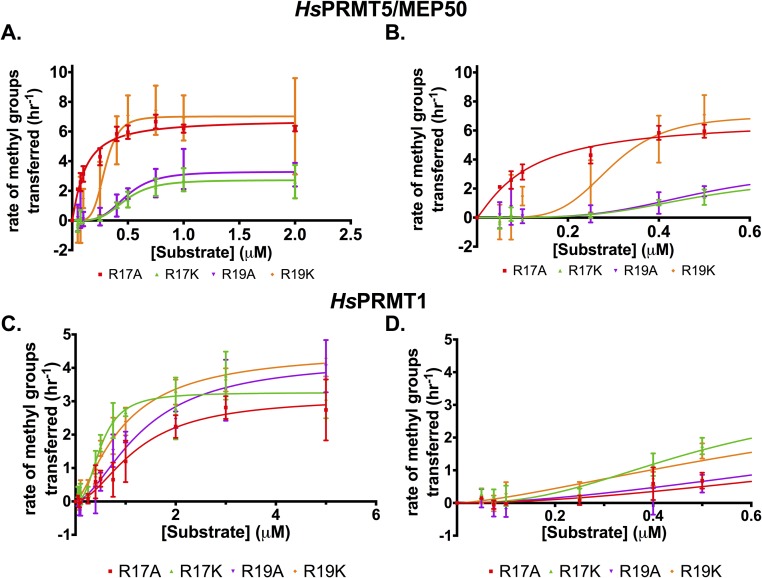
Having observed the increase in methylation of H4 (1-21) in the presence of MMA at position R17 (Fig. 1) and the cooperative nature of PRMT5 with the unmodified H4 (1-21) peptide (Fig. 2), we then tested the effect of monomethylation at position R17 on the activity of HsPRMT5/MEP50. We were able to determine kinetic parameters and characterize the activity of HsPRMT5/MEP50 as a function of R17 methylation (Fig. 2 and Table 1). When presented with the H4 (1-21) peptide synthetically monomethylated at position 17 [H4 (1-21) R17MMA], HsPRMT5/MEP50 exhibits about a twofold increase in maximal activity relative to the WT peptide and a much larger—fivefold or more—increase in activity at substrate concentrations below 0.5 μM (Fig. 2, brown vs. blue and Table 1), consistent with the data from the cation exchange experiments (Fig. 1). This methyltransferase also shows positive cooperativity with the modified H4 (1-21) peptides described below (Fig. S2 A and B and Table 1).
Fig. S2.
HsPRMT5/MEP50 and HsPRMT1 exhibit positive cooperativity as a function of modifications on the substrate H4 peptide. Initial kinetic measurements were made, and the data were fit to the Hill equation (37). (A) Enzyme activity of HsPRMT5/MEP50 with H4 (1-21) R17A (red), H4 (1-21) R17K (green), H4 (1-21) R19A (purple), and H4 (1-21) R19K (orange) in triplicate assays (error bars represent SD). (B) A closeup of the graph from A to make differences at low [substrate] clearer. (C) Enzyme activity of HsPRMT1 with H4 (1-21) R17A (red), H4 (1-21) R17K (green), H4 (1-21) R19A (purple), and H4 (1-21) R19K (orange) in triplicate assays (error bars represent SD). (D) A close-up of the graph from C to make differences at low [substrate] clearer. For details about reaction conditions and concentrations, see Methods.
Given the positive cooperativity of PRMT5 with H4 (1-21) (Fig. 2), we characterized the kinetics of this enzyme with the R17MMA modification and either alanine or lysine substitutions at positions R17 and R19 on H4 (1-21) (Fig. S2 A and B). To determine significant differences in kinetic parameters with the H4 (1-21) peptides used, we compared the value of each parameter relative to the WT peptide value (Fig. 2 C–E). There was a significant decrease in K0.5 relative to WT when H4 (1-21) R17MMA was used, indicating an increase in binding affinity (Fig. 2 A–C and Table 1); a similar effect was seen for the R17A derivative (Fig. 2C). The R19A peptide exhibited a significant decrease in binding affinity while the other peptides did not have a significant effect on the K0.5. Enzymatic activity, kcat, appeared to vary significantly for most of the H4 (1-21) derivatives relative to WT; notably, the R17MMA peptide had the highest activity at 2.31 ± 0.20 h−1 and a P value of 0.0001 (Fig. 2D and Table 1). The statistical analysis of Hill coefficients revealed that PRMT5 had maximal cooperativity with H4 (1-21) WT with an n value of nearly 3. However, the Hill coefficient for the R17MMA peptide was significantly lower (about 1.3), indicating that the PRMT5 exhibited mostly noncooperative (Michaelis–Menten) kinetics with this peptide (Table 1).
HsPRMT1 also Exhibits Positive Cooperativity.
To see if the results observed with PRMT5 were unique for that enzyme, kinetics experiments were conducted with HsPRMT1, an enzyme that also targets R3 on histone H4 for ADMA formation (4, 39, 40), and the various H4 (1-21) peptides. Significantly, we also observed positive cooperativity with this enzyme (Fig. 3). A comparison of PRMT1 kinetics with the WT H4 peptide and the R3K derivative shows similar results as with PRMT5; PRMT1 only methylates residue R3 on the H4 (1-21) peptide (Fig. 3). HsPRMT1 also exhibits about a twofold increase in overall activity relative to the WT peptide with H4 (1-21) R17MMA as a substrate (Fig. 3 and Table 1), although there is no apparent increase in activity at the low substrate concentrations, unlike PRMT5. Again, as with PRMT5, PRMT1 showed positive cooperativity with H4 (1-21) WT (Fig. 3E and Table 1). While PRMT1 did display similar kinetics to PRMT5 with histone H4 (1-21) WT, the difference between the degree of cooperativity for the R17MMA and WT peptide was greater for PRMT5 than for PRMT1 (Figs. 2E and 3E and Table 1). In fact, none of the modified H4 peptides had a significant effect on PRMT1 cooperativity; this indicates that the R17MMA modification may more selectively affect PRMT5 kinetics than those for PRMT1. Experiments to assess kinetic parameters of PRMT1 with AdoMet as the varying substrate gave similar results as seen with PRMT5; no cooperativity was observed (Fig. S1B). The results of these kinetic studies indicate PRMT1 to be an allosteric enzyme as well, whose activity can be modulated by binding downstream residues, albeit to a more minor degree than PRMT5.
Fig. 3.
HsPRMT1 exhibits positive cooperativity. Initial kinetic measurements were made, and the data were fit to the Hill equation (38). (A) Enzyme activity of HsPRMT1 with H4 (1-21) WT (blue), H4 (1-21) R17MMA (brown), and H4 (1-21) R3K (red) is shown for triplicate assays (error bars represent SD). (B) An expanded view of A at the low substrate concentrations. Best fit curves are shown for K0.5, kcat, and Hill coefficient values for the H4 (1-21) WT substrate of 0.58 μM, 4.90 h−1, and 1.54, respectively. For H4 (1-21) R17MMA, the parameters were 0.38 μM, 7.85 h−1, and 1.98, respectively. For details about reaction conditions and concentrations, see Methods. Statistical analysis of K0.5 values (C), statistical analysis of kcat values (D), and statistical analysis of the Hill coefficient values (E). The dashed line represents a Hill coefficient of 2. Data were taken from the triplicate assays shown in A and B; error bars represent SD. The P values were calculated using a one-way ANOVA test with a Dunnett test for multiple comparisons using the GraphPad Prism 6.0 software.
Discussion
As the molecular mechanisms of PRMT5 and PRMT7 have become clearer and their impact on the biological landscape of disease more pronounced, the need to understand how these enzymes engage in cross-talk has become more important. Several studies have already demonstrated that PRMT7 expression levels influence the amount of PRMT5-catalyzed methylation of histone H4 R3 (4, 6, 22–26). Both of these enzymes are involved in major biological functions such as DNA damage repair and cellular proliferation as well as being dysregulated in diseases such as cancer (7–9). With the knowledge that PRMT7 prefers to methylate histone H4 downstream from position R3 (27) and that chemical changes in such regions—distal sites—can, in general, affect PRMT activity (15, 34–36), we set out to determine if there was a link between PRMT5- and PRMT7-mediated methylation of different residues on the same protein.
With the exception of one recent study of PRMT1 (37), previous studies have not reported PRMT5 and PRMT1 to behave in a cooperative fashion (15, 16, 21, 34–36). Our experiments, however, not only show that PRMT5 and PRMT1 each demonstrate positive cooperativity when methylating one of their endogenous substrates, but that the allosteric activity of PRMT5—very low enzyme activity at lower substrate concentrations—is alleviated when R17 on the same peptide substrate is monomethylated (Fig. 2 B and E and Table 1). For both enzymes, we found that the degree of cooperativity, as well as the level of activity, is affected by the chemical makeup of residues R17 and 19 on the histone H4 peptide. Intriguingly, monomethylation of R17 in histone H4 had the largest effect on the activity of PRMT5 at low substrate concentrations (Fig. 2). Given the fact that both PRMT1 and PRMT5 are highly active and promiscuous enzymes, there has been surprisingly little uncovered about how their activity is regulated. Allosteric dependence on “distal sites” of methylation substrates may help highlight a mode of regulation through which the activity of PRMT5 and PRMT1 activity is modulated.
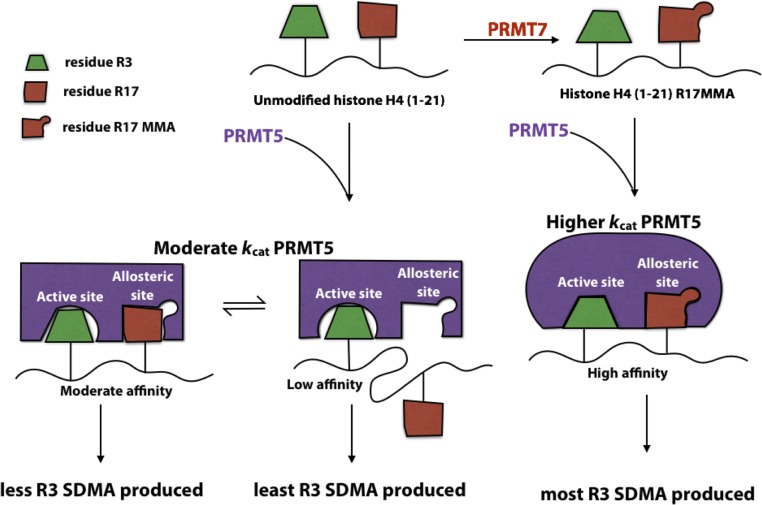
We know from our previous work that PRMT7 does not methylate R3 on histone H4 and instead catalyzes MMA production on R17 and/or R19 (27). Recent literature, however, links changes in SDMA levels at R3 on histone H4 with expression of PRMT7 (4, 6, 22–26), suggesting that this enzyme may be responsible for aiding PRMT5-mediated methylation at this residue. We thus propose that methylation of R17 by PRMT7 may be responsible for the indirect activation of PRMT5-mediated methylation of R3 on histone H4 in mammals (Fig. 4). Because R17MMA appears to be an allosteric regulator of PRMT5 activity, it is possible that this methylated residue binds to PRMT5 at an allosteric site, causing conformational changes in the enzyme which increase its activity toward its native substrate, residue R3. In fact, similar binding phenomena have previously been suggested (35) for PRMT1, although not in the context of allosteric regulation.
Fig. 4.
Model for allosteric regulation of PRMT5/MEP50 activity by PRMT7. The green blocks represent residue R3 on histone H4 (1-21), while the red blocks represent residue R17. PRMT5/MEP50 is shown in purple.
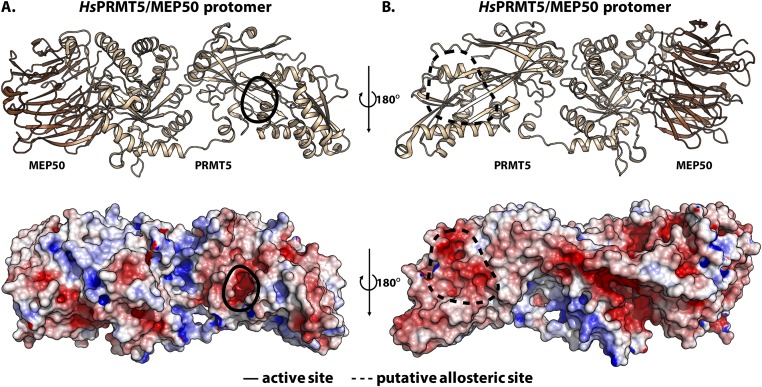
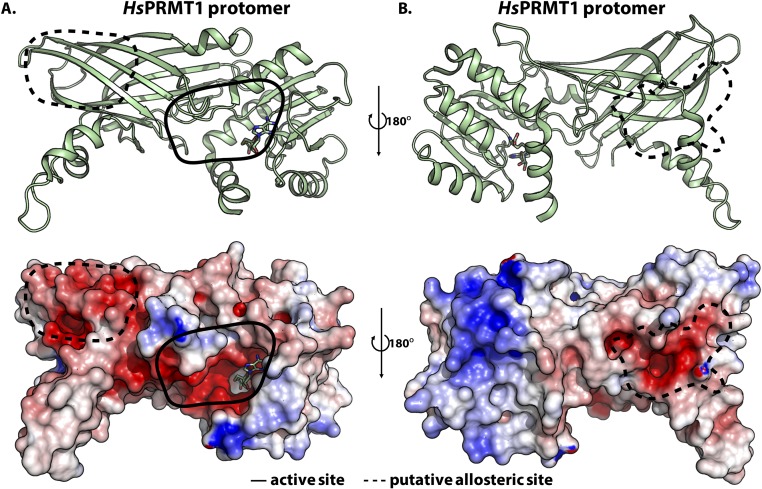
By looking at the electrostatic potentials for PRMT5 and PRMT1 structures, it may be possible to identify potential allosteric binding regions (Figs. S3 and S4, respectively). The sequence around histone H4 R17 contains basic residues, so allosteric binding sites on the enzyme would ideally be negatively charged. It is interesting, therefore, that PRMT5 appears to have a negatively charged cavity on the face opposite to its active site (solid black enclosure in Fig. S3A) and on part of the postmethyltransferase domain β-barrel (dashed black enclosure in Fig. S3B); no such regions appear on the same face as the active site (Fig. S3). The large negatively charged furrow illustrated in Fig. S3B is unlikely to be a potential allosteric binding site because the ∼70 Å distance from the active site is greater that the ∼50 Å distance from R3 to R17 in the most extended conformation. PRMT1’s structure also reveals similar allosteric sites at negatively charged regions after its methyltransferase domain (dashed black enclosures in Fig. S4A). However, due to its simpler oligomeric structure, PRMT1 may have more surface area accessible, making other allosteric sites possible as well (Fig. S4B).
Fig. S3.
Electrostatic potential map of HsPRMT5/MEP50 protomer reveals potential allosteric sites. (A) The protomer of HsPRMT5/MEP50 (PDB ID code: 4GQB) is shown; the beige subunit represents PRMT5, while the brown subunit represents MEP50. The active site is highlighted by a solid black enclosure. (B) Dashed black enclosures on the opposite face of the structure in A indicate negatively charged cavities as potential allosteric binding sites on the electrostatic potential map, generated using APBS in PyMOL (red to blue corresponds to −5 kT/e to 5 kT/e).
Fig. S4.
Electrostatic potential map of HsPRMT1 homodimer reveals potential allosteric sites. The protomer of HsPRMT1 (PDB ID code: 1ORI) is shown. (A) The cartoon representation and electrostatic map for the PRMT1 dimer with the active site (solid black enclosure) facing forward; the dashed black enclosure indicates a negatively charged cavity as potential allosteric binding site on the electrostatic potential map. (B) A 180° rotation of the molecules in A about the y axis shows an additional putative allosteric site. Electrostatic potentials were generated using APBS in PyMOL (red to blue corresponds to −5 kT/e to 5 kT/e).
Although both PRMT5 and PRMT1 exhibit positive cooperativity in the presence of the unmodified histone H4 (1-21) peptide, it is unclear whether this occurs in a symmetrical/concerted fashion, as theorized by the Monod–Wyman–Changeux model (41), or in a sequential manner, as theorized by the Koshland–Nemethy–Filmer model (42). However, our data suggests that when residue R17 is monomethylated, there is likely to be an altered conformation of PRMT5 which results not only in higher affinity binding of the substrate, but also higher maximal velocity. Further structural studies must be undertaken to determine where in the methyltransferase such allosteric sites are and the nature of the different conformational states.
Until recently, PRMTs were thought to behave like canonical Michaelis–Menten enzymes (15, 21, 34–36), but our work has shown that at least PRMT1 and PRMT5 can catalyze methylation via positive cooperativity. As these enzymes are key players in controlling gene transcription, it is logical to assume that there are mechanisms to regulate their activity. Furthermore, the PRMTs’ role in disease-related processes such as cancer metastasis and tumorigenesis has been established. Recently, there has been considerable work in the development of small molecular inhibitors of these enzymes (11, 43). Regulation of methyltransferase activity by cross-talk with other modifications has been seen in a number of other systems (22, 44–46), such as in the interactions between methylation on histone H3 K27 and K36 by PRC2 and Setd2/Ash1, respectively. Specifically, our studies show how positive cooperativity can play an integral part in cross-talk between PRMTs. With this understanding of PRMT behavior and regulation, it may be possible to generate new and more selective types of drugs which target a previously unexplored facet of arginine methyltransferases—their allosteric kinetics.
Methods
Peptide Substrates.
H4 (1-21) WT and R17MMA peptides were purchased as trifluoroacetic acid (TFA) salts from GenScript Inc. at >95% purity by HPLC. Histone H4 (1-21) R17/A/K and R19A/K peptides were generous gifts from Paul Thompson, University of Massachusetts Medical School, Worcester, MA. All of the peptide masses were confirmed by MALDI-TOF mass spectrometry (Table S1).
Table S1.
Histone H4 (1-21) peptides
| Peptide | Sequence | Expected mass, Da | Observed mass, Da* |
| H4 (1-21) WT | Ac-SGRGKGGKGLGKGGAKRHRKV | 2,133 | 2,133 |
| H4 (1-21) R17A | Ac-SGRGKGGKGLGKGGAKAHRKV | 2,048 | 2,049 |
| H4 (1-21) R17K | Ac-SGRGKGGKGLGKGGAKKHRKV | 2,105 | 2,106 |
| H4 (1-21) R17MMA | Ac-SGRGKGGKGLGKGGAKR(me)HRKV | 2,147 | 2,148 |
| H4 (1-21) R19A | Ac-SGRGKGGKGLGKGGAKRHAKV | 2,048 | 2,049 |
| H4 (1-21) R19K | Ac-SGRGKGGKGLGKGGAKRHKKV | 2,105 | 2,106 |
| H4 (1-21) R3K | Ac-SGKGKGGKGLGKGGAKRHRKV | 2,105 | 2,106 |
Data from MALDI-TOF analysis show monoisotopic M+1 values; bold residues indicate the modified positions in each peptide.
Protein Expression and Purification.
His-tagged human PRMT1 (HsPRMT1) was obtained in a pET28b plasmid from Paul Thompson and was expressed and purified as previously described (47). Human PRMT5/MEP50 (HsPRMT5/MEP50) complex protein was purchased from BioSciences as recombinantly coexpressed and purified proteins in HEK293T cells (0.65 mg/mL, 51045, Lot 150126; BPS Biosciences).
Phosphocellulose Paper Kinetics Assay.
Methylation reactions were performed with 2.45 nM HsPRMT5/MEP50 (calculated as the tetramer complex) or 10 nM PRMT1 (calculated as the dimer) buffered with 50 mM Hepes (pH 8.0), 10 mM NaCl, and 1 mM DTT containing 20 μM of a 20:1 molar ratio of S-adenosyl-l-methionine p-toluenesulfonate salt (AdoMet) (Sigma A2408; ≥80% purity) to S-adenosyl-l-[methyl-3H]-methionine ([methyl-3H]-AdoMet) [stock solution of 7 μM (78.2 Ci/mmol) in 10 mM H2SO4/EtOH (9:1, vol/vol); PerkinElmer Life Sciences] as a methyl donor. A H4 (1-21) peptide substrate concentration range of 0.05–2 μM was used in each reaction with PRMT5 and a concentration range of 0.05–5 μM for PRMT1 reactions. When determining the kinetic parameters for the enzymes with AdoMet as the varying substrate, a range of 0–3 μM AdoMet (20:1 molar ratio of AdoMet to [methyl-3H]-AdoMet) and 10 μM H4 (1-21) WT were used. The reaction volume was brought up to 30 μL with deionized water. Reactions were incubated for 1 h for HsPRMT5/MEP50 and 1.5 h for HsPRMT1 at 37 °C, and then quenched with 0.5 μL of 100% (vol/vol) TFA. Each reaction was run in triplicate.
Twenty-five microliters of the reaction products were spotted onto 1.5 cm × 1.5 cm P81 phosphocellulose ion exchange filter paper (IEP-01; Reaction Biology Corp.) and air dried for 30 min. The papers were subsequently washed in 1 L of 50 mM NaHCO3 at pH 9.0 for 45 min. The papers were placed in scintillation vials and allowed to further air dry for 45 min. Radioactivity was counted with 5 mL of Safety-Solve scintillation mixture (111177; Research Products International) for three cycles of 5 min using a Beckman LS6500 instrument.
Amino Acid Analysis of Protein and Peptide Substrates.
In vitro methylation assays with 12.3 nM HsPRMT5/MEP50 tetramer, 10 μM H4 (1-21) peptide (WT and R17MMA), and 0.7 μM [methyl-3H]-AdoMet were carried out in 50 mM K-Hepes (pH 8.0), 10 mM NaCl, 1 mM DTT, and 5% glycerol at 37 °C for 2 h. Reactions were quenched with 0.5 μL of 100% TFA, and peptides were purified via RP-HPLC and acid hydrolyzed as described previously (47, 48). Acid hydrolysates were also analyzed through cation exchange chromatography as previously described (47, 48).
Statistical Analysis.
All error bars indicate the SD of triplicate measurements. Data were analyzed using the GraphPad Prism 6.0 software. The one-way ANOVA test was used to compare kinetic parameters with a Dunnett test for multiple comparisons (49). A two-tailed t test was used to compare MMA and SDMA levels from cation exchange chromatography.
Acknowledgments
We thank Paul Thompson (University of Massachusetts Medical School) for providing the H41-21 R17A/K, R19A/K, and R17/19A/K peptides and the human His-PRMT1 plasmid; Jon Lowenson for thoughtful discussions and critique of the manuscript; and Duilio Cascio for aiding in generating electrostatic potential maps [University of California, Los Angeles (UCLA)]. This work was supported by grants from the NIH, including Grant GM026020 (to S.G.C.), Ruth L. Kirschstein National Service Award GM007185 (to K.J.), a Faculty Research Grant from the UCLA Academic Senate (to S.G.C.), a Zymo Summer Research Scholarship (to C.Y.J.), and an Edith and Lew Wasserman Fellowship (to C.Y.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706978114/-/DCSupplemental.
References
- 1.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 2.Fuhrmann J, Clancy KW, Thompson PR. Chemical biology of protein arginine modifications in epigenetic regulation. Chem Rev. 2015;115:5413–5461. doi: 10.1021/acs.chemrev.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc RS, Richard S. Arginine methylation: The coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y-C, Peterson SE, Loring JF. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014;24:143–160. doi: 10.1038/cr.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karkhanis V, et al. Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1. J Biol Chem. 2012;287:29801–29814. doi: 10.1074/jbc.M112.378281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldwin RM, et al. Protein arginine methyltransferase 7 promotes breast cancer cell invasion through the induction of MMP9 expression. Oncotarget. 2015;6:3013–3032. doi: 10.18632/oncotarget.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koh CM, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 9.Hu D, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun. 2015;6:8419. doi: 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira de Freitas R, et al. Discovery of a potent class I protein arginine methyltransferase fragment inhibitor. J Med Chem. 2016;59:1176–1183. doi: 10.1021/acs.jmedchem.5b01772. [DOI] [PubMed] [Google Scholar]
- 12.Eram MS, et al. A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem Biol. 2016;11:772–781. doi: 10.1021/acschembio.5b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaniskan HÜ, et al. A potent, selective and cell-active allosteric inhibitor of protein arginine methyltransferase 3 (PRMT3) Angew Chem Int Ed Engl. 2015;54:5166–5170. doi: 10.1002/anie.201412154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan-Penebre E, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11:432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Xu R-M, Thompson PR. Substrate specificity, processivity, and kinetic mechanism of protein arginine methyltransferase 5. Biochemistry. 2013;52:5430–5440. doi: 10.1021/bi4005123. [DOI] [PubMed] [Google Scholar]
- 16.Antonysamy S, et al. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci USA. 2012;109:17960–17965. doi: 10.1073/pnas.1209814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonysamy S. The structure and function of the PRMT5:MEP50 complex. Subcell Biochem. 2017;83:185–194. doi: 10.1007/978-3-319-46503-6_7. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun. 2015;6:6428. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho MC, et al. Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One. 2013;8:e57008. doi: 10.1371/journal.pone.0057008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarighat SS, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: Gene activation and repression via histone arginine methylation. Leukemia. 2016;30:789–799. doi: 10.1038/leu.2015.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L, et al. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc Natl Acad Sci USA. 2011;108:20538–20543. doi: 10.1073/pnas.1106946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhar SS, et al. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012;26:2749–2762. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migliori V, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 24.Blanc RS, Vogel G, Chen T, Crist C, Richard S. PRMT7 preserves satellite cell regenerative capacity. Cell Rep. 2016;14:1528–1539. doi: 10.1016/j.celrep.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Ying Z, et al. Histone arginine methylation by PRMT7 controls germinal center formation via regulating Bcl6 transcription. J Immunol. 2015;195:1538–1547. doi: 10.4049/jimmunol.1500224. [DOI] [PubMed] [Google Scholar]
- 26.Blanc RS, Richard S. Regenerating muscle with arginine methylation. Transcription. 2017;8:175–178. doi: 10.1080/21541264.2017.1291083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem. 2013;288:37010–37025. doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishioka K, Reinberg D. Methods and tips for the purification of human histone methyltransferases. Methods. 2003;31:49–58. doi: 10.1016/s1046-2023(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 29.Lee J-H, et al. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem. 2005;280:3656–3664. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- 30.Gonsalvez GB, et al. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol. 2007;178:733–740. doi: 10.1083/jcb.200702147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Y, Hadjikyriacou A, Clarke SG. Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): The importance of acidic residues in the double E loop. J Biol Chem. 2014;289:32604–32616. doi: 10.1074/jbc.M114.609271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J Biol Chem. 2012;287:7859–7870. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luense LJ, et al. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin. 2016;9:24. doi: 10.1186/s13072-016-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obianyo O, Osborne TC, Thompson PR. Kinetic mechanism of protein arginine methyltransferase 1. Biochemistry. 2008;47:10420–10427. doi: 10.1021/bi800904m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: Positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry. 2007;46:13370–13381. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Y, et al. Histone H4 acetylation differentially modulates arginine methylation by an in cis mechanism. J Biol Chem. 2011;286:20323–20334. doi: 10.1074/jbc.M110.207258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fulton MD, Zhang J, He M, Ho M-C, Zheng YG. The intricate effects of alpha-amino and lysine modifications on arginine methylation on the N-terminal tail of histone H4. Biochemistry. 2017;56:3539–3548. doi: 10.1021/acs.biochem.7b00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss JN. The Hill equation revisited: Uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- 39.Feng Y, et al. A transient kinetic analysis of PRMT1 catalysis. Biochemistry. 2011;50:7033–7044. doi: 10.1021/bi200456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 42.Koshland DE, Jr, Némethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 43.Ye Y, et al. Discovery and optimization of selective inhibitors of protein arginine methyltransferase 5 by docking-based virtual screening. Org Biomol Chem. 2017;15:3648–3661. doi: 10.1039/c7ob00070g. [DOI] [PubMed] [Google Scholar]
- 44.Lu C, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molina-Serrano D, Schiza V, Kirmizis A. Cross-talk among epigenetic modifications: Lessons from histone arginine methylation. Biochem Soc Trans. 2013;41:751–759. doi: 10.1042/BST20130003. [DOI] [PubMed] [Google Scholar]
- 46.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain K, et al. Protein arginine methyltransferase product specificity is mediated by distinct active-site architectures. J Biol Chem. 2016;291:18299–18308. doi: 10.1074/jbc.M116.740399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Debler EW, et al. A glutamate/aspartate switch controls product specificity in a protein arginine methyltransferase. Proc Natl Acad Sci USA. 2016;113:2068–2073. doi: 10.1073/pnas.1525783113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunnett CW. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50:1096. [Google Scholar]
- 50.Gottschling H, Freese E. A tritium isotope effect on ion exchange chromatography. Nature. 1962;196:829–831. doi: 10.1038/196829a0. [DOI] [PubMed] [Google Scholar]