Significance
Neurons develop diverse dendrite morphologies to achieve specialized functions. Certain neurons completely fill their receptive fields with evenly spaced dendrites, while others only partially occupy available space with sparse dendrites. How a similar extracellular environment regulates dendrite patterning of different neurons has been elusive. Here we show that Drosophila space-filling neurons require epidermis-derived heparan sulfate proteoglycans (HSPGs) to cover the body wall, while other sensory neurons sharing the same receptive fields are insensitive to extracellular HSPGs. HSPGs promote dendritic growth and maintenance of space-filling neurons by stabilizing microtubules in dynamic high-order branches. HSPGs do not function through the only known HSPG receptor leukocyte antigen-related (Lar) nor by transporting extracellular diffusible ligands. Our results reveal important mechanisms by which HSPGs regulate neuronal morphogenesis.
Keywords: space-filling neurons, dendritic arborization neurons, dendrite, receptor protein tyrosine phosphatase, heparan sulfate proteoglycan
Abstract
Neurons sometimes completely fill available space in their receptive fields with evenly spaced dendrites to uniformly sample sensory or synaptic information. The mechanisms that enable neurons to sense and innervate all space in their target tissues are poorly understood. Using Drosophila somatosensory neurons as a model, we show that heparan sulfate proteoglycans (HSPGs) Dally and Syndecan on the surface of epidermal cells act as local permissive signals for the dendritic growth and maintenance of space-filling nociceptive C4da neurons, allowing them to innervate the entire skin. Using long-term time-lapse imaging with intact Drosophila larvae, we found that dendrites grow into HSPG-deficient areas but fail to stay there. HSPGs are necessary to stabilize microtubules in newly formed high-order dendrites. In contrast to C4da neurons, non–space-filling sensory neurons that develop in the same microenvironment do not rely on HSPGs for their dendritic growth. Furthermore, HSPGs do not act by transporting extracellular diffusible ligands or require leukocyte antigen-related (Lar), a receptor protein tyrosine phosphatase (RPTP) and the only known Drosophila HSPG receptor, for promoting dendritic growth of space-filling neurons. Interestingly, another RPTP, Ptp69D, promotes dendritic growth of C4da neurons in parallel to HSPGs. Together, our data reveal an HSPG-dependent pathway that specifically allows dendrites of space-filling neurons to innervate all target tissues in Drosophila.
The sensory or synaptic input of a neuron is greatly influenced by the morphological characteristics of its dendritic arbor. Different neurons in the same tissue and extracellular microenvironment can develop drastically different dendrite morphologies. While some neurons grow simple dendritic trees with sparse and stereotypic branches, some others are capable of completely and adaptively occupying large receptive fields with a high density of dendrites. The phenomenon of a neuron filling its receptive field with dendrites, sometimes referred to as dendritic space-filling, depends on two intrinsic properties of the neuron: (i) a capacity to grow copious high-order dendrites, and (ii) repulsions among homotypic dendrites to keep neighboring branches at an optimal distance. Notable examples of space-filling neurons include cerebellar Purkinje cells (1), retinal ganglion cells (RGCs) and amacrine cells that innervate the inner plexiform layer of the vertebrate retina (2, 3), zebrafish somatosensory neurons that innervate the skin (4), and class IV dendritic arborization (C4da) neurons on the Drosophila larval body wall (5). By completely and evenly covering their 2D target domains, these neurons are able to uniformly sample sensory or synaptic information across the receptive fields (6).
The developmental regulation of space-filling likely relies on both intrinsic properties of a neuron and the extrinsic environment in which it grows. Indeed, a number of neuronal intrinsic factors have been identified that specifically regulate space-filling of neurons. These factors include transcription factors that collectively determine the neuronal identity and bestow upon neurons the capacity to elaborate exuberant neurites (7, 8), motor proteins and components of the secretary pathway that control the number and position of high-order dendrites (9–12), and an amino acid transporter that allows neurons to grow total dendritic length beyond a certain threshold (13). In contrast to intrinsic control of dendritic growth, extracellular regulation of space-filling is more mysterious. Although guidance cues are known to target neurites of some space-filling neurons to the correct spatial domains (14, 15), how neurites interact with the extracellular microenvironment to fill the receptive field once there is more elusive. In particular, whether dendritic space-filling requires unique extracellular permissive signals is currently unknown.
Heparan sulfate proteoglycans (HSPGs) are good candidates for extracellular regulation of space-filling, because they are required for the functions of a large number of extracellular signaling molecules and because HSPGs are ligands for neuronal receptors. HSPGs are membrane and extracellular glycoproteins that contain heparan sulfate (HS) glycosaminoglycan (GAG) chains attached to the protein core (16). With the negatively charged HS chains serving as binding sites for many secreted ligands such as growth factors, morphogens, and axonal guiding molecules, HSPGs are required for the extracellular transport and full biological activities of these ligands (17). In addition, HSPGs are ligands for leukocyte antigen-related (LAR) members of the receptor protein tyrosine phosphatase (RPTP) family. HS chains can bind the Ig ectodomains of LAR and induce LAR clustering (18). In the nervous system, HSPGs and LAR together regulate axon guidance, synaptogenesis, and axonal regeneration after injury (19, 20). In contrast to the known involvement of HSPGs in axon guidance and outgrowth (15, 18), the roles of HSPGs in dendrite morphogenesis are only beginning to be understood. In Caenorhabditis elegans, UNC-52/Perlecan, a secreted HSPG in the basement membrane, indirectly regulates dendritic growth and branching of PVD sensory neurons through patterning hypodermal-derived adhesion signal SAX-7/L1CAM (21). However, whether target tissue-derived HSPGs directly regulate dendritic patterns of specific neurons and how HSPGs affect the cytoskeletal and membrane trafficking machinery in neurite growth and guidance remain to be answered.
Drosophila C4da neurons are a good model system for investigating mechanisms of space-filling. C4da neurons extend dendrites mostly between the basal surface of epidermal cells and the ECM (22, 23). Distinct from other non–space-filling da neurons, larval C4da neurons have a high capacity for growing dynamic, high-order dendritic branches (5, 24). This capacity is essential for C4da neurons to fill the space on the rapidly expanding larval body wall during development (25) or to invade empty receptive fields caused by the loss of neighboring neurons (5, 26). Here we identify epidermis-derived HSPGs as a permissive signal uniquely required for dendritic growth and maintenance of space-filling C4da neurons. This neuronal type-specific permissive role is carried out redundantly by two distinct classes of HSPGs, the Syndecan and the glypican Dally. Using a long-term time-lapse live imaging method, we found that HSPGs are not required for extension or branching of high-order dendrites but rather stabilize dynamic dendritic branches by promoting microtubule stabilization. Furthermore, we present evidence that this HSPG-dependent dendritic growth is not mediated by extracellular ligands diffusing along the epidermal sheet or by potential HSPG receptors in the RPTP family. Our results therefore reveal critical mechanisms by which the extracellular microenvironment specifically regulates dendritic growth of space-filling neurons.
Results
C4da Neurons Require Epidermis-Derived HSPGs for Local Dendritic Growth.
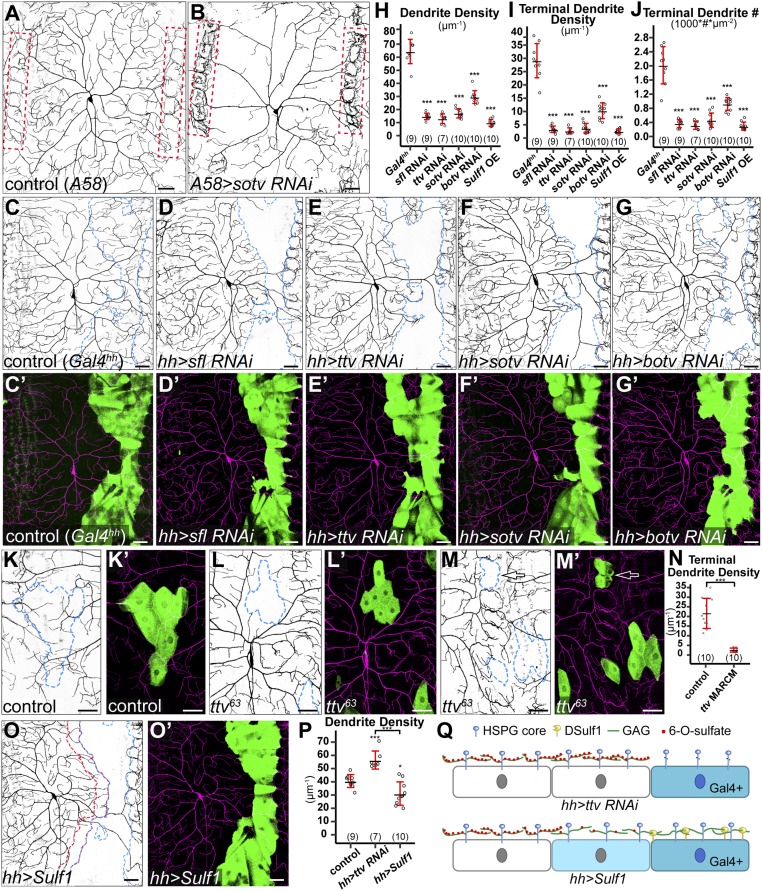
To determine whether the space-filling property of C4da neurons is regulated by extracellular signals, we first asked whether target tissue-derived HSPGs contribute to the dendritic growth of C4da neurons by disrupting enzymes that are specifically required for the biosynthesis of HS GAG chains (16). Among these enzymes are HS-glucosamine N-sulfotransferase Sulfateless (Sfl), HS polymerase Brother of tout-velu (Botv), and two HS copolymerases, Tout-velu (Ttv) and Sister of tout-velu (Sotv or Ext2). Because C4da dendrites innervate epidermal cells, we knocked down sotv (Fig. 1B), ttv (Fig. S1B), and sfl (Fig. S1C) by RNA interference (RNAi) using a pan-epidermal driver Gal4A58. Compared with the control (Fig. 1A), all of these manipulations resulted in drastic reductions of C4da dendrites within the intrasegmental area at late third instar [96 h after egg laying (AEL)] as measured by the dendrite density (defined as dendritic length per unit area) (Fig. S1D). The reductions are most evident in high-order dendrites, as reflected by the significant decreases of the terminal dendrite density (Fig. S1E) and the terminal dendrite number per unit area (referred to as terminal dendrite number hereafter) (Fig. S1F). Also associated with these manipulations are more dendrites around tendon cells at the anterior and posterior borders of each segment (outlined by red dots in Fig. 1B). Tendon cells are muscle attachment sites (MASs) and express very low levels of Gal4 in Gal4A58. The dendrite phenotypes were consistent for all three C4da neurons in each hemisegment, but for simplicity, we focused on the dorsal ddaC neuron in this study. These data demonstrate that the dendritic growth of C4da neurons requires substrate-derived HSPGs.
Fig. 1.
Epidermis-derived HSPGs are required for local growth of C4da dendrites. (A and B) DdaC neurons in the Gal4A58 control (A) and an animal expressing Gal4A58-driven sotv RNAi (B). Red dots outline the MASs. (C–G′) DdaC neurons in the Gal4hh control (C and C′) and animals expressing sfl RNAi (D and D′), ttv RNAi (E and E′), sotv RNAi (F and F′), and botv RNAi (G and G′) in the hh domain. Blue dots outline hh domains (Upper). Lower panels show hh domains (green) and C4da dendrites (magenta). (H–J) Quantification of total dendrite density (H), terminal dendrite density (I), and terminal dendrite number (J) in hh domains. ***P ≤ 0.001; ANOVA and Tukey’s HSD test. (K–M′) Epidermal MARCM clones of wild type (K and K′) and ttv63 (L–M′). Arrows point to a ttv63 epidermal clone at the MAS (M and M′). Merged panels show epidermal clones in green and C4da dendrites in magenta. (N) Quantification of terminal dendrite density in epidermal MARCM clones. ***P ≤ 0.001; Student’s t test. (O and O′) A ddaC neuron in an animal expressing membrane-tethered DSulf1 in the hh domain. Blue dots outline the hh domain, and red dots outline a one cell diameter-wide area anterior to the hh domain (O). Gal4hh-expressing cells are in green, and C4da dendrites are in magenta (O′). (P) Quantification of total dendrite density in the one cell diameter-wide area anterior to the hh domain. *P ≤ 0.05; ***P ≤ 0.001; ANOVA and Tukey’s HSD test. (Q) A diagram showing the effects of expressing ttv RNAi and membrane-tethered DSulf1 on epidermal HSPGs. Knockdown of ttv leads to the loss of HSPG GAG chains only on RNAi-expressing cells (dark blue), while membrane-tethered DSulf1 removes 6-O-sulfates of HSPG GAG chains on both DSulf1-expressing cells and cells directly contacting them (light blue). For all quantifications, each circle represents a neuron. The numbers of neurons are indicated. Black bar, mean; red bars, SD. (Scale bars, 50 µm.)
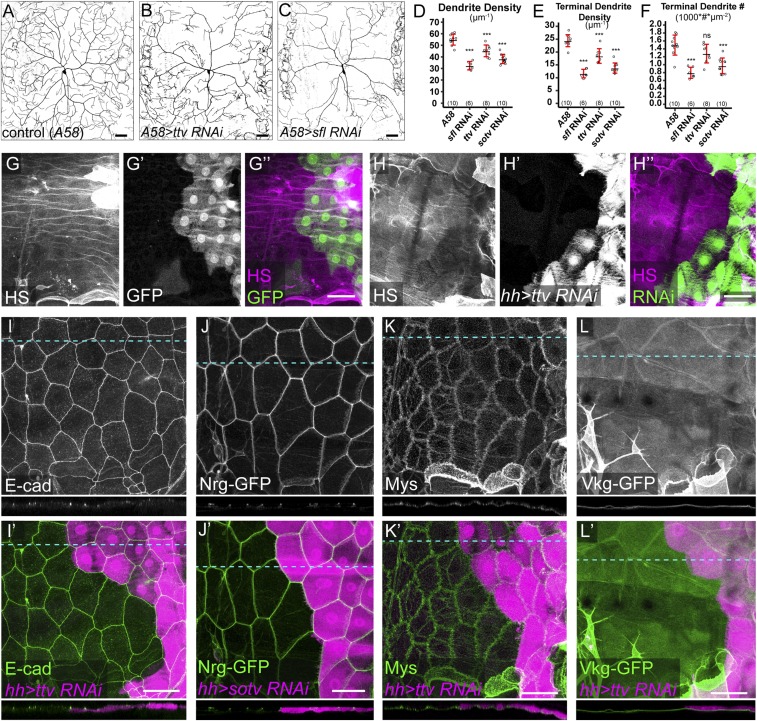
Fig. S1.
Effects of knockdown of HS synthesis genes on dendrite morphology, HS level, and epithelial morphology. (A–C) DdaC dendritic fields of a Gal4A58 control animal (A) and animals expressing Gal4A58 > ttv RNAi (B) and Gal4A58 > sfl RNAi (C). (D–F) Quantification of total dendrite density (D), terminal dendrite density (E), and terminal dendrite number (F) of control and RNAi animals. ***P ≤ 0.001; ns, not significant; one-way analysis of variance and Tukey’s HSD test. For all quantifications, each circle represents an individual neuron. The number of neurons for each genotype is indicated. The black bars represent the mean, and the red bars represent the SD. (G–H′′) Immunostaining of HS in a Gal4hh > GFP control animal (G–G′′) and an animal expressing Gal4hh > ttv RNAi (H–H′′). HS was stained with the antibody 3G10, which recognizes heparinase III-digested HS. (I–L′) Expression of E-Cad (I and I′), Nrg-GFP (J and J′), Mys (K and K′), and Vkg-GFP (L and L′) in animals expressing Gal4hh > ttv RNAi (I, I′, K, K′, L, and L′) and Gal4hh > sotv RNAi (J and J′). E-Cad and Mys expressions were detected by antibody staining, and Nrg-GFP and Vkg-GFP expressions were detected using GFP trap lines. A cross-section for each marker is shown below each 2D projection image. Blue dotted lines indicate locations of cross-sections. (Scale bars, 50 µm.)
HSPGs could promote dendritic growth by modulating global gene transcription in neurons or by regulating local dendritic sprouting. To distinguish between these possibilities, we knocked down sfl, ttv, sotv, and botv using Gal4hh, which is expressed in a stripe of epidermal cells located at the posterior side of each segment (Fig. 1 D–G). We predict that if HSPGs regulate transcription in neurons, knockdown in the hh-expressing cells should have no or global effects on dendritic growth in the entire dendritic field. However, if HSPGs promote dendritic growth locally, the effect on dendrite growth should be restricted to the hh domain. Strikingly, knockdown of each of the four genes in hh epidermal cells led to a more dramatic reduction of dendrites (Fig. 1H) compared with pan-epidermal knockdowns (Fig. S1D). In particular, sfl RNAi, ttv RNAi, and sotv RNAi almost completely eliminated high-order dendrites from the hh domain (Fig. 1 I and J). In these domains, only “naked” primary dendritic branches persisted (Fig. 1 D–F). Importantly, the dendrite reductions were only observed in the hh domain. These results strongly suggest that epidermis-derived HSPGs locally and posttranscriptionally promote the innervation of receptive fields by C4da dendrites.
Two additional lines of evidence further suggest that the dendrite reductions were due to defects of HS rather than RNAi off-target effects. First, ttv RNAi in the hh domain caused a reduction of HS staining (Fig. S1 G–H′′). Second, using mosaic analysis with a repressible cell marker (MARCM) (27) in which ttv63 homozygous mutant epidermal cells were generated in an otherwise ttv63 heterozygous background, we observed a cell-autonomous loss of terminal dendrites on ttv63 mutant epidermal cells (Fig. 1 L–N) compared with wild-type epidermal clones (Fig. 1 K, K′, and N). ttv63 clones in tendon cells also inhibited dendritic growth locally (arrow in Fig. 1 M and M′), suggesting that dendritic innervation at the MASs requires HSPGs too.
As HSPGs play important roles in epidermal cell patterning and differentiation during embryogenesis, one concern of our experiments is that the lack of dendrites on HSPG-deficient cells may be due to defects of epidermal cells. We examined the morphological characteristics of epidermal cells including adherens junctions, septate junctions, focal adhesions, and the ECM. Knockdown of ttv or sotv by Gal4hh did not alter the expression levels or subcellular localizations of the adherens junction marker E-cad (28) (Fig. S1 I and I′), the septate junction marker Nrg (29) (Fig. S1 J and J′), the focal adhesion maker Mys (30) (Fig. S1 K and K′), and an ECM component Vkg (23) (Fig. S1 L and L′), suggesting that the above genetic manipulations did not cause obvious morphological defects of epidermal cells.
We further asked whether C4da dendrite innervation depends on the modification state of HS chains. HS activity requires sulfation of its disaccharide units at 2-O and 6-O positions (31). A secreted endosulfatase Sulf1 can effectively remove 6-O sulfate from extracellular HSPGs (32). We overexpressed a membrane-tethered Sulf1 (33) in hh cells and observed a nearly complete loss of high-order dendrites on the expressing cells (Fig. 1 H–J, O, and O′). This dendrite reduction is as strong as in the knockdown of HS synthesis genes, suggesting that HS mediates most, if not all, of the activity of HSPGs. However, we noticed a distinction between Sulf1 overexpression and knockdown of HS synthesis genes in a one cell diameter-wide area anterior and adjacent to the hh domain (outlined in red dots in Fig. 1O). In these areas, the total dendrite density was increased in knockdown of HS synthesis genes such as ttv but reduced in Sulf1-overexpressing animals (Fig. 1 O and P). The cell-autonomous loss of dendritic coverage on ttv RNAi-expressing epidermal cells is consistent with the idea that HS synthesis is only affected in the RNAi-expressing cells (Fig. 1Q). In contrast, plasma membrane-located Sulf1 on hh-expressing cells can potentially act on the wild-type cells directly contacting Sulf1-expressing cells (Fig. 1Q) and cause nonautonomous dendrite reduction. Together, our results indicate that HS on the epidermal surface is required for local innervation of epidermal cells by space-filling C4da dendrites.
HSPGs Are Specifically Required for the Dendritic Growth and Maintenance of Space-Filling Sensory Neurons.
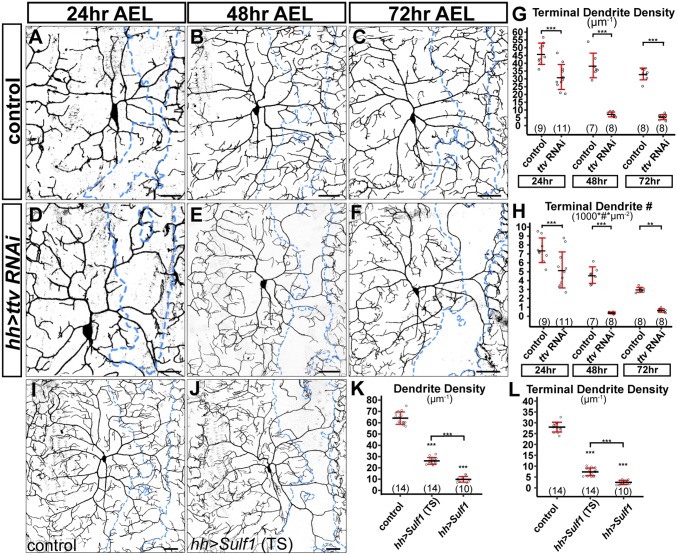
The dendrite reduction associated with HSPG deficiency observed at the late third instar larval stage could be due to a lack of dendritic growth or a failure to maintain existing dendrites. To distinguish between these two possibilities, we examined control and Gal4hh > ttv RNAi animals at younger larval stages, including the beginning of the first instar (24 h AEL), the beginning of the second instar (48 h AEL), and early third instar (72 h AEL). Compared with the control (Fig. 2 A–C), ttv RNAi (Fig. 2 D–F) caused dendritic reduction as early as 24 h AEL. The reduction became more severe at 48 h AEL and 72 h AEL (Fig. 2 G and H). These data demonstrate that HSPGs are required for dendrite growth of C4da neurons.
Fig. 2.
HSPGs are specifically required for dendrite development of C4da neurons. (A–F) DdaC in the Gal4hh control (A–C) and Gal4hh-driven ttv RNAi (D–F) at three developmental stages. (G and H) Quantification of terminal dendrite density (G) and terminal dendrite number (H) in hh domains. **P ≤ 0.01; ***P ≤ 0.001; Student’s t test. (I and J) DdaC in the control (tubP-Gal80ts; Gal4hh) (I) and an Sulf1-expressing animal (tubP-Gal80ts; hh > Sulf1) in which Sulf1 expression was induced by temperature shift (TS) (J). Both genotypes were subjected to TS. (K and L) Quantification of dendrite density (K) and terminal dendrite density (L) in hh domains of Gal4hh control, hh > Sulf1 (TS), and hh > Sulf1 (constant expression). ***P ≤ 0.001; ns, not significant; ANOVA and Tukey’s HSD test. For all quantifications, each circle represents a neuron. The numbers of neurons are indicated. Black bar, mean; red bars, SD. (Scale bars, 10 µm in A and D, 35 µm in B and E, and 50 µm in the rest of panels.)
To determine if HSPGs are continuously required to maintain dendrites, we investigated the effects of late removal of HSPGs by controlling Sulf1 overexpression using temperature-sensitive Gal80 (Gal80ts) (34). Animals were allowed to develop at 18 °C to suppress Gal4 activity until early third instar and then shifted to 28 °C to induce Sulf1 expression in the hh domain for 24 h. Although dendritic patterns were indistinguishable between the control and hh > Sulf1 before the temperature shift, Sulf1 induction caused dramatic dendrite reduction (Fig. 2 I–L). The Sulf1-expressing regions contained few high-order dendrites, suggesting that HSPG removal caused regression of earlier high-order dendrites. However, late induction of Sulf1 led to a relatively weaker dendrite reduction (i.e., more low-order dendrites) compared with that caused by early and persistent Sulf1 expression (Fig. 2 K and L). This result suggests that some dendritic branches had been stabilized before Sulf1 induction and they persisted with late HSPG removal. Together, these data suggest that HSPGs are required to maintain high-order dendrite branches but not those that are already stabilized.
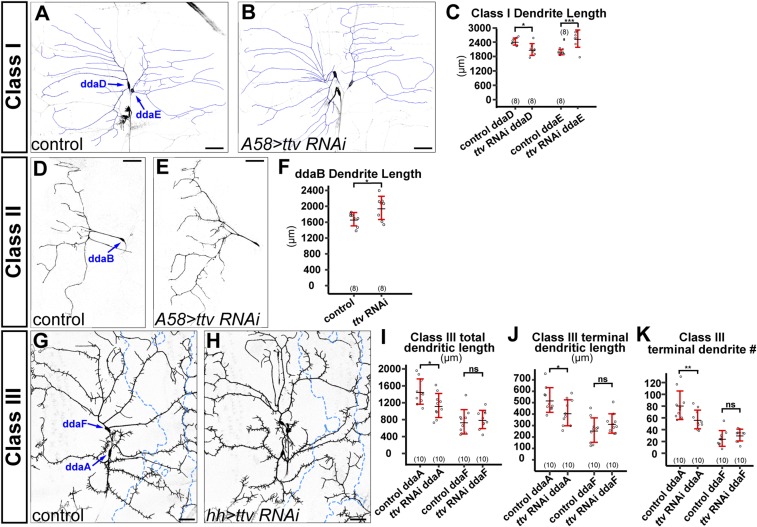
The Drosophila peripheral nervous system (PNS) has four classes of da sensory neurons with overlapping dendritic fields (35), with C4da as the only space-filling neurons. We wondered whether HSPGs are required for the arborization of the other three classes. We examined the dendritic patterns of dorsal class I da neurons ddaD and ddaE (Fig. S2 A–C), the ventral class II da neuron ddaB (Fig. S2 D–F), and dorsal class III da neurons ddaA and ddaF (Fig. S2 G–K) in epidermal knockdown of ttv. Pan-epidermal knockdown of ttv caused a slight reduction of class I ddaD dendrites and a slight increase of class I ddaE dendrites (Fig. S2C). Pan-epidermal knockdown of ttv also caused a mild increase of class II ddaB dendrites (Fig. S2F). For C3da neurons, knockdown of ttv in the hh domain did not affect the dendrites of ddaF but mildly reduced the total dendritic length, terminal dendritic length, and total terminal dendrite number of ddaA within the hh domain (Fig. S2 I–K). These data demonstrate that although HSPGs can mildly influence the dendrites of class I–III, the dendritic growth of these neurons during larval development does not rely on HSPGs. Therefore, HSPGs are specifically required for the dendritic development of space-filling C4da neurons.
Fig. S2.
HSPGs are not required for dendrite growth of other classes of da neurons. (A and B) Dendritic fields of class I ddaD and ddaE neurons in a Gal4A58 control animal (A) and an animal expressing Gal4A58-driven ttv RNAi (B). The dendrites are overlaid with blue lines. (C) Quantification of class I total dendrite length of control and ttv RNAi animals. *P ≤ 0.05; ***P ≤ 0.001; Student’s t test. (D and E) Dendritic fields of class II ddaB neurons in a Gal4A58 control animal (D) and an animal expressing Gal4A58-driven ttv RNAi (E). (F) Quantification of ddaB total dendrite length of control and ttv RNAi animals. *P ≤ 0.05; Student’s t test. (G and H) Dendritic fields of class III ddaF and ddaA neurons in a Gal4hh control animal (G) and an animal expressing Gal4hh-driven ttv RNAi (H). Blue dots outline hh domains. (I–K) Quantification of total dendrite density (I), terminal dendrite density (J), and terminal dendrite numbers (K) in hh domains of control and ttv RNAi animals. *P ≤ 0.05; **P ≤ 0.01; ns, not significant; Student’s t test. For all quantifications, each circle represents an individual neuron. The number of neurons for each genotype is indicated. The black bars represent the mean, and the red bars represent the SD. (Scale bars, 50 µm.)
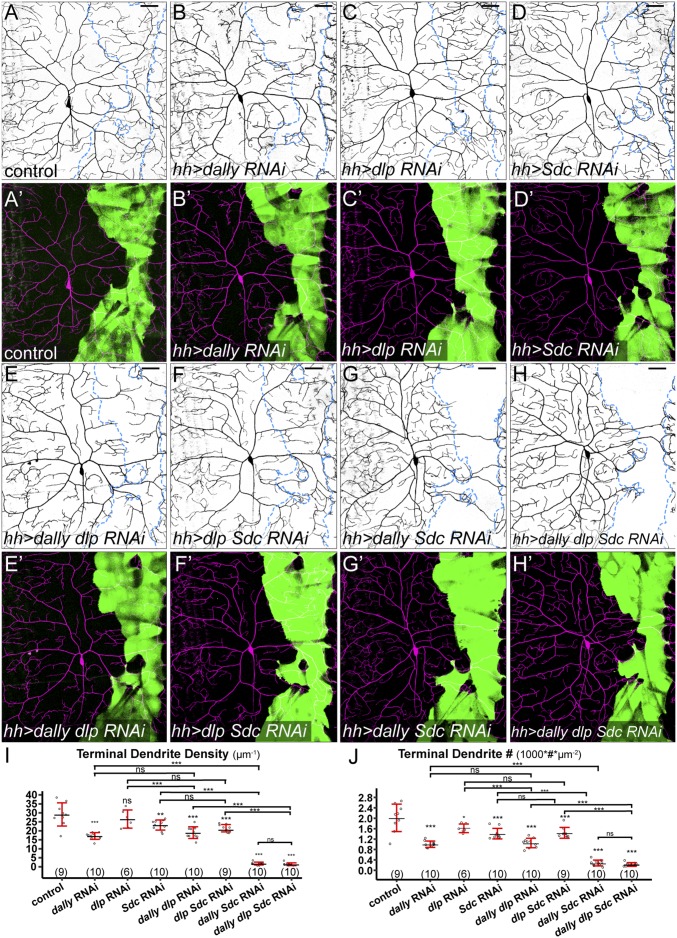
Sdc and Dally Play Partially Redundant Roles in Promoting C4da Dendritic Growth.
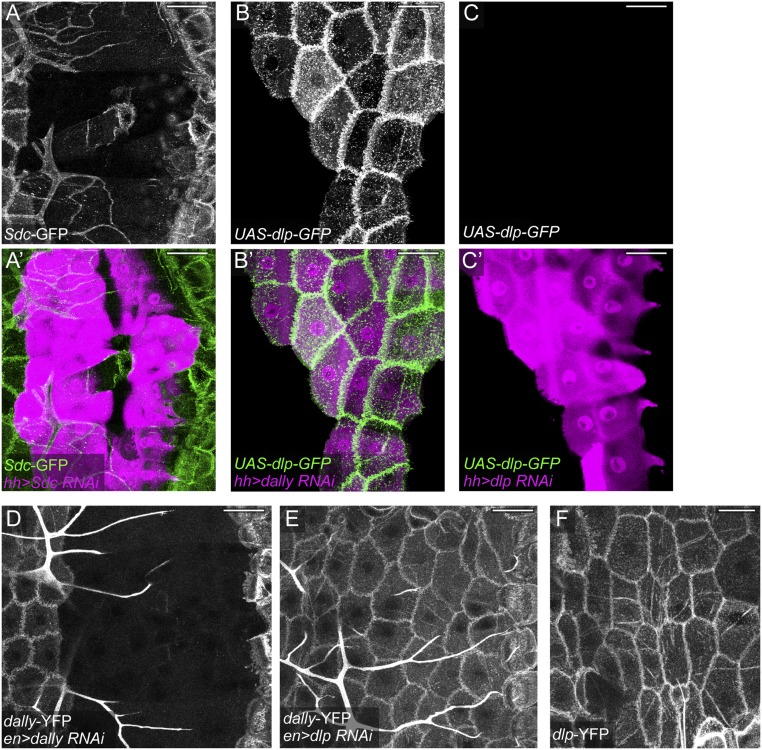
Drosophila has four HSPG core proteins, including two GPI-anchored glypicans [Division abnormally delayed (Dally) and Dally-like (Dlp)], one transmembrane syndecan (Sdc), and one secreted perlecan (Trol) (16). We next asked which HSPG core proteins are necessary for C4da dendritic growth. We focused on glypicans and the syndecan, as they are more commonly involved in signaling than perlecans (17). Knockdown of either dally (Fig. 3 B and B′) or Sdc (Fig. 3 D and D′) caused a mild dendrite reduction (Fig. 3 I and J), while dlp knockdown had little effect (Fig. 3 C, C′, I, and J). Knockdown of trol did not produce obvious dendritic defects either. The lack of strong phenotypes in these experiments may be due to poor knockdown efficiency or functional redundancy among HSPGs. To evaluate these possibilities, we first tested the efficiency of RNAi in knocking down endogenous or overexpressed HSPGs in epidermal cells. Imaging of protein trap lines Sdc-GFP (Fig. S3 A and A′), dally-YFP (Fig. S3 D and E), and dlp-YFP (Fig. S3F) showed that larval epidermal cells express all three HSPGs with the expected membrane localization. Sdc-GFP (Fig. S3 A and A′) and Dally-YFP (Fig. S3D) were efficiently knocked down in cells expressing their corresponding RNAi constructs. For dlp, we coexpressed UAS-dlp-GFP with either UAS-dally-RNAi (as a negative control) (Fig. S3 B and B′) or UAS-dlp-RNAi (Fig. S3 C and C′) in the hh domain. Dlp-GFP was strongly expressed in the presence of dally RNAi but is absent when coexpressed with dlp RNAi. While dlp RNAi is effective in suppressing Dlp-GFP expression, it had no detectable effect on Dally-YFP expression (Fig. S3E). These data demonstrate that HSPG knockdowns are effective and RNAi constructs for dally and dlp are specific.
Fig. 3.
Sdc and dally play partially redundant roles in promoting dendritic growth of C4da neurons. (A–H′) DdaC in the Gal4hh control (A and A′) and animals expressing dally RNAi (B and B′), dlp RNAi (C and C′), Sdc RNAi (D and D′), dally dlp RNAi (E and E′), dlp Sdc RNAi (F and F′), dally Sdc RNAi (G and G′), and dally dlp Sdc RNAi (H and H′) in the hh domain. Blue dots outline hh domains (Upper). Lower panels show Gal4hh-expressing cells in green and C4da dendrites in magenta. (I and J) Quantification of terminal dendrite density (I) and terminal dendrite number (J) in hh domains. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns, not significant; ANOVA and Tukey’s HSD test. For all quantifications, each circle represents a neuron. The numbers of neurons are indicated. Black bar, mean; red bars, SD. (Scale bars, 50 µm.)
Fig. S3.
Effectiveness and specificity of HSPG RNAi. (A and A′) Sdc-GFP protein trap expression in an animal expressing Gal4hh > Sdc RNAi. A′ shows RNAi-expressing cells in magenta and Sdc-GFP in green. (B–C′) An animal expressing Gal4hh UAS-dlp-GFP UAS-dally RNAi (B and B′) and an animal expressing Gal4hh UAS-dlp-GFP UAS-dlp RNAi (C and C′). B′ and C′ show RNAi-expressing cells in magenta and Dlp-GFP expression in green. (D and E) dally-YFP protein trap expression in an animal expressing Gal4en > dally RNAi (D) and an animal expressing Gal4en > dlp RNAi (E). (F) dlp-YFP protein trap expression in the epidermis. All animals are 96 h AEL larvae. (Scale bars, 50 µm.)
We next tested whether HSPGs act redundantly to promote C4da dendritic growth by knocking down dally, dlp, and Sdc in all possible combinations. Knockdown of both dally and Sdc in epidermal cells (Fig. 3 G and G′) almost completely blocked high-order dendritic growth (Fig. 3 I and J), mirroring the phenotypes of sfl, ttv, and sotv knockdown (Fig. 1). In contrast, the additional knockdown of dlp did not enhance the dendritic reduction in dally RNAi (Fig. 3 E and E′), Sdc RNAi (Fig. 3 F and F′), or dally Sdc RNAi (Fig. 3 H and H′), indicating that Dlp does not play a significant role in C4da dendritic growth. Together, these results demonstrate that Dally and Sdc play partially redundant roles in promoting C4da dendrite development.
HSPGs Stabilize High-Order C4da Dendrites at Least Partially by Promoting Microtubule Stabilization.
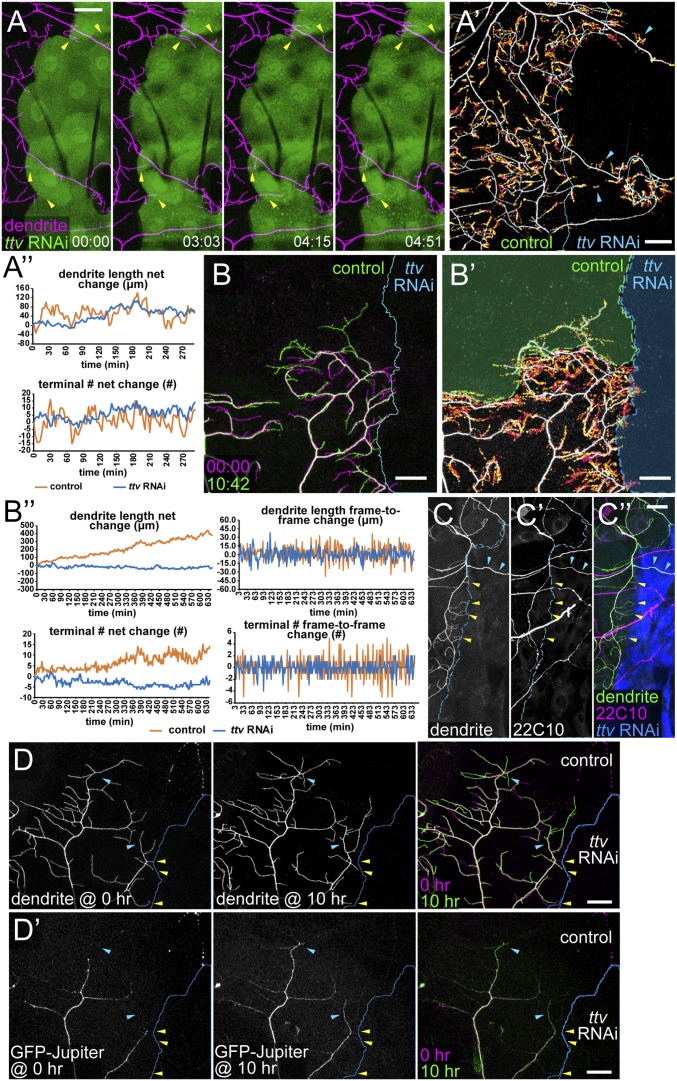
C4da neurons are unique among da neurons in that they continuously sprout new branches to innervate unoccupied epidermal space during development. To understand the molecular mechanism and neuronal specificity of HSPG-dependent dendritic growth, we first investigated how growing C4da dendrites behave when encountering HS-deficient epidermal cells. Addressing this question required long-term live imaging of C4da dendrites. However, common methods for immobilizing and mounting Drosophila larvae generally cause animal lethality within 30 min and thus do not permit live imaging long enough for capturing both short-term dendrite dynamics and long-term changes of dendritic patterns. Therefore, we developed a method of long-term time-lapse imaging for Drosophila larvae (SI Methods). This method allows for continuous live imaging of da neurons in partially immobilized larvae for longer than 10 h. Using this method, we examined the dendrite dynamics of C4da neurons in animals expressing ttv RNAi in hh epidermal cells between 72 h AEL and 84 h AEL. Our time-lapse analyses show that the high-order C4da dendrites are highly dynamic at the border of HS-deficient zones (Fig. 4A and Movie S1). Instead of halting at the border, growing dendritic tips could extend tens of microns into the HS-deficient zone. In addition, the primary dendrites that persisted in the HS-deficient zone also sprouted many new branches during the course of the time lapse. The presence of high-order dendrites in the HS-deficient zone during the time-lapse imaging is also demonstrated by plotting the locations of dendrite endings in all frames (Fig. 4A′). However, high-order dendrites in the HS-deficient zone were transient, and there was no obvious net increase of the total dendrite length or the terminal dendrite number during the imaging (Fig. 4A′′). We imaged 55 neurons and observed consistent results.
Fig. 4.
HSPGs stabilize high-order C4da dendrites by promoting microtubule bundling. (A) Selected frames from a time-lapse series showing ddaC dendrites (magenta) at the border of ttv RNAi-expressing epidermal cells (green). Yellow arrows indicate terminal branches temporarily invading the HS-deficient zone. Times (HH:MM) are relative to the first frame. (A′) Projection of dendrite endings over time with ending locations in each frame temporally color-coded (early, magenta; late, yellow). The blue line indicates borders of HS-deficient regions. Arrowheads point to dendrite endings in the HS-deficient region. (A′′) Net changes in dendrite length and terminal dendrite numbers (relative to the first frame) in HS-positive (“control,” orange lines) and HS-deficient (“ttv RNAi,” blue lines) regions at each time point. (B–B′′) Time-lapse imaging of ddaC dendrites next to an HS-deficient region (Gal4hh > ttv RNAi) on the right and an empty wild-type epidermal region on the top. (B) Dendritic patterns of the first frame (magenta) and the last frame (green). Times (HH:MM) are relative to the first frame. (B′) Projection of dendritic endings over time. The dendritic pattern and the border are from the last frame. The green overlay indicates a wild-type epidermal region (control), which was empty in the first frame, and the blue overlay indicates the HS-deficient region (ttv RNAi). (B′′) Dendrite dynamics in control (orange lines) and ttv RNAi (blue lines) regions at each time point. Graphs on Left show net changes in dendrite length and terminal dendrite numbers relative to the first frame. Graphs on Right show frame-to-frame changes in dendrite length and terminal dendrite numbers. The control and ttv RNAi regions show similar sizes of frame-to-frame changes in the early part of the time lapse, but later the control region displays greater frame-to-frame changes due to the more dendrites accumulated in the control region. (C–C′′) Immunostaining of 22C10 in Gal4hh > ttv RNAi. Yellow arrowheads point to 22C10-negative terminal dendrites at the border of the HS-deficient zone. Blue arrowheads point to 22C10-positive primary branches within the HS-deficient zone. The merge (C′′) shows hh-expressing cells in blue, 22C10 staining in magenta, and C4da dendrites in green. (D and D′) Time-lapse snapshots of ddaC dendrites that are next to an HS-deficient region (Gal4hh > ttv RNAi) on the right and an empty wild-type region on the top. Times 0 h and 10 h are shown in separate and merged panels. D shows dendritic patterns and D′ shows GFP-Jupiter that labels bundled microtubules in the dendrites. Yellow arrowheads point to GFP-Jupiter–negative terminal dendrites at the border of the HS-deficient zone. Blue arrowheads point to newly stabilized high-order dendrites that are invaded by GFP-Jupiter at 10 h. (Scale bars, 25 µm.)
C4da dendrites are known to be dynamic during early third instar (24). Indeed, our time-lapse data revealed that high-order C4da dendrites in the control region were active in exploratory behaviors including extension, retraction, turning, and branching (Movie S1). Dendrite-dense control regions often did not exhibit robust net dendrite growth within a few hours (Fig. 4A′′), likely due to the inhibitory effect of homotypic dendritic repulsion (26, 35). To rule out the possibility that high-order C4da dendrites are intrinsically unstable and therefore cannot permanently invade empty spaces at this stage, we did similar time-lapse experiments, except that we additionally laser-ablated the ddaC neurons on the right side of the larva between 48 and 60 AEL. The ablation presented the ddaC neurons on the left side with an empty, but otherwise wild-type, territory at the dorsal midline. Such HS-positive empty territory does not impose homotypic repulsion onto dendrites and therefore is a better internal control for the HS-deficient zone. In these experiments, although high-order dendrites at the dorsal midline were still very dynamic, dendritic arbors gradually expanded into the empty wild-type spaces (Fig. 4 B–B′′ and Movie S2). In contrast, there was no net growth of dendrites into HS-deficient zones in the same experiments, even though high-order dendrites in wild-type empty zones and HS-deficient zones were similarly dynamic from frame to frame (Fig. 4B′′). We imaged 16 neurons and observed consistent results. These data strongly suggest that HSPGs are not required for short-term dynamics of C4da dendrites but are necessary to stabilize growing dendrites.
An important mechanism of dendrite stabilization is microtubule stabilization and bundling by microtubule-associated proteins (MAPs) (36). A hallmark of stabilized microtubules in da dendrites is the presence of Futsch, the Drosophila MAP1, which is restricted to stable neuronal microtubule bundles (37). To further investigate how HSPGs stabilize growing dendrites, we examined the distribution of Futsch in C4da dendrites using the antibody 22C10 (37). Futsch was detected in the primary dendrites that persisted in the HS-deficient zone (cyan arrowheads in Fig. 4 C–C′′), consistent with these dendrites being stabilized. In contrast, the high-order C4da dendrites at the border of the HS-deficient zone lacked detectable Futsch staining (yellow arrowheads in Fig. 4 C–C′′). The correlation between epidermal HS deficiency and the lack of bundled microtubules in high-order C4da dendrites supports the idea that HS may stabilize growing dendrites by promoting microtubule stabilization and bundling. To further investigate the dynamics of bundled microtubules in C4da neurons, we expressed GFP-Jupiter in C4da neurons and simultaneously knocked down ttv in hh-expressing epidermal cells. Jupiter is a Drosophila MAP (38), and its distribution upon exogenous expression in da neurons matches that of Futsch. In this experiment, we ablated ddaC neurons on one side of the larva so as to induce long-term dendritic growth of the remaining ddaC neurons. Over the course of 10 h, we indeed observed gradual appearance of GFP-Jupiter in newly stabilized dendritic branches at the border of the empty, wild-type space (cyan arrowheads in Fig. 4 D and D′) (100%, n = 10). In contrast, GFP-Jupiter was never detected in the high-order dendrites that straddled the border of the HS-deficient zone (yellow arrowheads in Fig. 4 D and D′). Together, these data suggest that HSPGs promote C4da dendritic growth at least partially by stabilizing newly formed dendrites through microtubule stabilization and bundling.
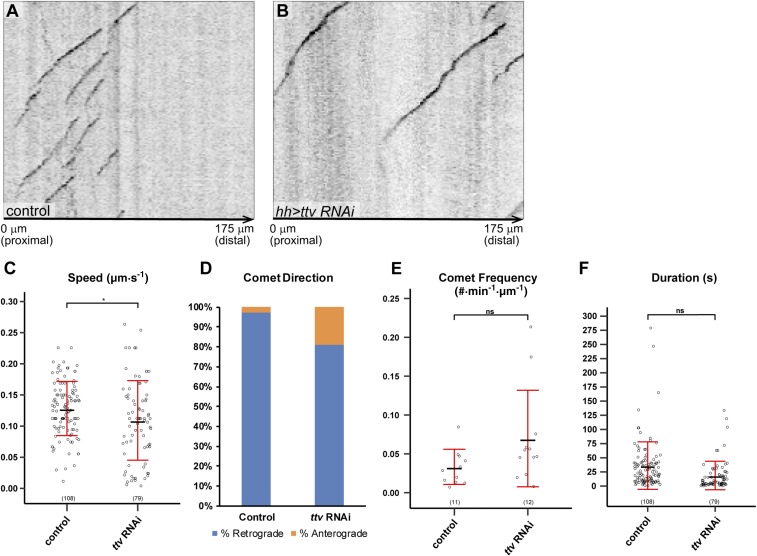
To further understand the impact of HS deficiency on microtubule dynamics in C4da neurons, we also examined microtubule growth behaviors using EB1-GFP (39), which binds growing microtubule plus ends. As EB1-GFP is difficult to detect in terminal dendrites, we mainly focused on the primary dendritic branches in the HS-deficient zone (Fig. S4 A and B). We noticed a slightly more variable and decreased speed of EB1-GFP comets (Fig. S4C) and a mild increase of the fraction of anterograde comets (Fig. S4D) in HS deficiency but no difference in comet frequency (Fig. S4E) or duration (Fig. S4F). These weak effects suggest that changes in microtubule growth dynamics probably are not a major cause of the dendritic reduction in HS deficiency.
Fig. S4.
Dynamics of EB1-GFP in ttv knockdown. (A and B) Example kymographs of EB1-GFP comets in primary branches in the hh domain in a wild-type (A) and a Gal4hh > ttv RNAi (B) animal. The left edge of each kymograph marks the left border of the hh domain. The proximal-distal orientation of the dendrite and the distance from the border of the hh domain are labeled at the bottom of each panel. (C–F) Quantification of EB1-GFP comet speed (μm/s) (C), comet direction (D), comet frequency (#⋅min−1·µm−1) (E), and comet duration (s) (F) in wild-type and Gal4hh > ttv RNAi animals. For comet direction, the percentage of retrograde comets is shown in blue, and the percentage of anterograde comets is shown in orange. *P ≤ 0.05; ns, not significant; Student’s t test. For all quantifications, each circle represents a single EB1-GFP comet. The number of comets for each genotype is indicated. The black bars represent the mean, and the red bars represent the SD.
Dendritic Growth of C4da Neurons Does Not Involve HSPG-Mediated Transport of Diffusible Signaling Molecules.
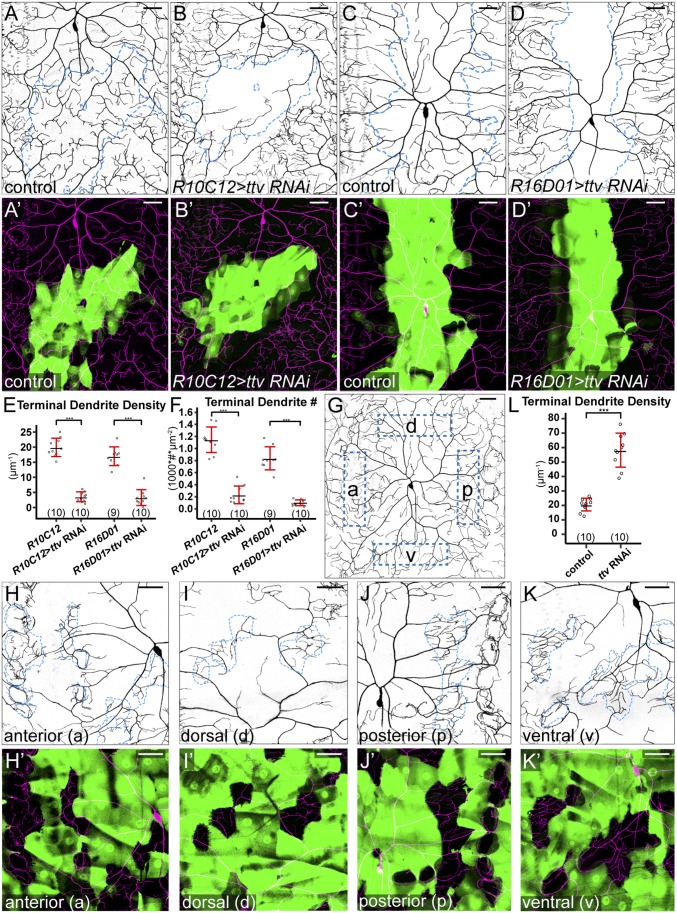
HSPGs regulate activities of many growth factors and formation of morphogen gradients (16, 17). In particular, HSPGs are necessary for the restricted diffusion of extracellular signaling molecules from signal-producing cells to signal-receiving cells (40–42). Therefore, a plausible model for the role of HSPGs in dendritic growth is that they mediate the action of a signaling molecule that is secreted by a subset of epidermal cells and diffuses along the epithelial sheet. This model has at least two predictions. First, the location of HSPG knockdown relative to the source of the signal should influence the severity of the dendritic growth defect. For example, HSPG knockdown in signal-producing cells should block spreading of the signal and reduce dendritic growth in surrounding wild-type regions. Second, a patch of wild-type epidermal cells completely surrounded by HS-deficient epidermal cells should not receive the signal and hence show reduction of high-order dendrites. To test the first prediction, we knocked down ttv in two additional domains in the dendritic field (Fig. 5 A–D′). R10C12-Gal4 drives expression in a patch of epidermal cells on the lateral body wall that overlaps with the border of ddaC and lateral C4da v’ada (Fig. 5 A and A′). R16D01-Gal4 drives expression in a stripe of epidermal cells in the middle of the segment (Fig. 5 C and C′). Knockdown with either driver caused a strong reduction of terminal dendrites in the RNAi-expressing domains (Fig. 5 E and F) but no apparent dendritic loss in neighboring wild-type regions. These results suggest that the action of the presumptive HSPG-mediated signal is not location-dependent.
Fig. 5.
HSPGs do not promote dendritic growth by transporting diffusible signals. (A–B′) DdaC in an R10C12-Gal4 control (A and A′) and an animal expressing R10C12-Gal4–driven ttv RNAi (B and B′). (C–D′) DdaC in an R16D01-Gal4 control animal (C and C′) and an animal expressing R16D01-Gal4–driven ttv RNAi (D and D′). Blue dots in A–D outline Gal4-expressing cells, and A′–D′ show Gal4-expressing cells in green and C4da dendrites in magenta. (E and F) Quantification of terminal dendrite density (E) and terminal dendrite number (F). ***P ≤ 0.001; Student’s t test. (G) DdaC in a control animal with anterior (a), dorsal (d), posterior (p), and ventral (v) locations in the dendritic field indicated. (H–K′) DdaC dendrites in animals expressing ttv RNAi in random epidermal patches, showing wild-type epidermal cells surrounded by RNAi-expressing epidermal clones in anterior (H and H′), dorsal (I and I′), posterior (J and J′), and ventral (K and K′) regions of the dendritic field. Blue dots in H–K outline wild-type epidermal cells. H′–K′ show RNAi-expressing cells in green and C4da dendrites in magenta. (L) Quantification of terminal dendrite density in wild-type epidermal cells surrounded by control and ttv-RNAi–expressing epidermal clones. ***P ≤ 0.001; Student’s t test. For all quantifications, each circle represents a neuron. The numbers of neurons are indicated. Black bar, mean; red bars, SD. (Scale bars, 50 µm.)
To test the second prediction, we generated random epidermal clones that expressed ttv RNAi using the “Flp-out” technique (43). We made a highly efficient epidermal Flipase (Flp) that turns on expression in most epidermal cells and leaves islands of wild-type epidermal cells completely surrounded by RNAi-expressing cells (Fig. 5 H–K′) at various locations in the dendritic field (Fig. 5G). Surprisingly, terminal dendritic densities significantly increased, rather than decreased, in all wild-type clones surrounded by ttv RNAi-expressing cells compared with similar clones surrounded by wild-type cells (Fig. 5L). The lack of dendritic reduction in clones surrounded by HS deficiency strongly suggests that the dendrites do not receive the growth-promoting signal through HSPG-mediated extracellular diffusion. Instead, similar to membrane HSPGs, this signal is likely produced by and associated with every epidermal cell.
Ptp69D but Not Lar Is Required for Dendritic Growth of C4da Neurons.
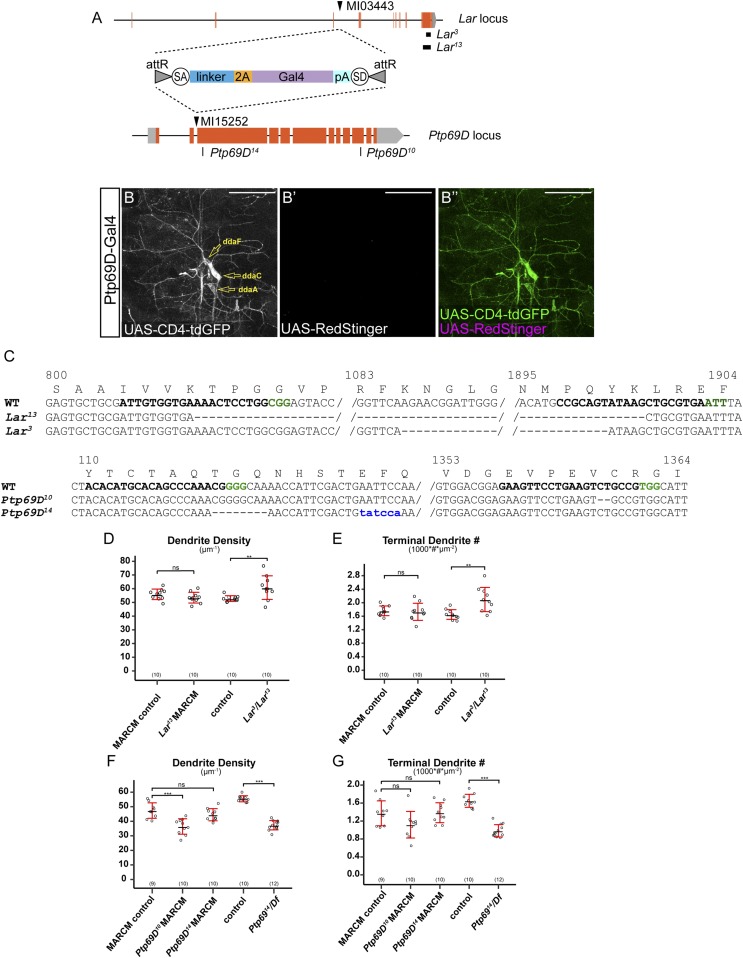
The effects of HSPGs on axonal outgrowth and guidance are mediated by LAR members of the RPTP family in mice, zebrafish, and Drosophila (15, 18, 44). Both Sdc and Dlp are known ligands for Drosophila Lar (44, 45). We therefore wondered if HSPGs promote C4da dendritic growth by acting together with Lar on the neuronal membrane. Besides Lar, Drosophila has another Ig-containing RPTP, Ptp69D. Although Ptp69D belongs to a different class of RPTP from Lar (46), we also tested its role in C4da dendritic growth under the assumption that Ptp69D Ig domains may interact with HS in a similar way to those of Lar. We first examined Lar and Ptp69D expression by generating transcriptional reporters using the Trojan-exon Gal4 strategy (47). We converted MiMIC cassettes inserted between coding exons of Lar and Ptp69D into 2A-Gal4 insertions (Fig. S5A), which are predicted to be spliced into all isoforms and produce in-frame Gal4 proteins. In this way, we generated Gal4 drivers under the endogenous regulatory control of Lar and Ptp69D loci. Using a membrane marker UAS-CD4-tdGFP and a nuclear marker UAS-RedStinger, we found that Lar is expressed in a subset of epidermal cells and in all classes of da neurons (Fig. 6 A–A′′). At the late third instar stage, Ptp69D is primarily expressed in C3da and C4da neurons but is also weakly and variably expressed in C1da and C2da neurons (Fig. 6 B–B′′). At earlier larval stages, we only observed CD4-tdGFP labeling of C3da and C4da neurons (Fig. S5 B–B′′). The expression patterns of Lar and Ptp69D suggest that they could play a role in dendrite development of C4da neurons.
Fig. S5.
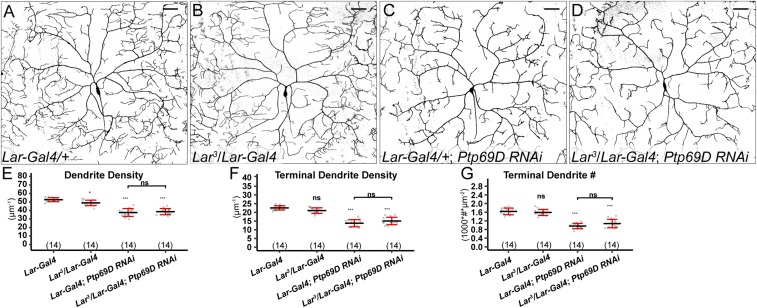
Lar and Ptp69D reporters, mutations, expressions, and LOF phenotypes. (A) A diagram showing the Trojan Exon structure and MiMIC insertion sites in Lar and Ptp69D loci. Coding exons are in orange, and untranslated regions are in light gray. The splice acceptor (SA) and splice donor (SD) sites allow incorporation of the transgene into the gene’s mRNA. The T2A peptide allows two protein products to be made from a single mRNA transcript (63). The black boxes in the Lar locus show the locations of CRISPR-induced deletions, and the black lines in the Ptp69D locus show the sites of the CRISPR-induced indels. (B–B′′) Expression of CD4-tdGFP (B) and RedStinger (B′) in a Ptp69D-Gal4 UAS-CD4-tdGFP UAS-RedStinger larva at 48 h AEL. B′′ shows the merge of both markers. RedStinger was not expressed in CD4-tdGFP–labeled neurons most likely because the Gal4 expression is very low at this stage and UAS-CD4-tdGFP is much more responsive to Gal4 than UAS-RedStinger (59). (Scale bars, 50 µm.) (C) Sequences of mutations induced by CRISPR/Cas9 in Lar and Ptp69D loci. The wild-type sequence is shown at Top with the gRNA target site in bold and the PAM sequence in green. The amino acid sequence and position are shown above the wild-type DNA sequence. Deleted residues are shown as dashes, and inserted residues are shown in blue lowercase letters. (D and E) Quantification of total dendrite density (D) and terminal dendrite numbers (E) in wild-type, Lar13 mutant neurons, and Lar3/Lar13 mutant animals. **P ≤ 0.01; ns, not significant; one-way analysis of variance and Tukey’s HSD test. (F and G) Quantification of total dendrite density (F) and terminal dendrite numbers (G) in wild-type, Ptp69D10, Ptp69D14 mutant neurons, and Ptp69D14/Df(3L)8ex34 mutant animals. ***P ≤ 0.001; ns, not significant; one-way analysis of variance and Tukey’s HSD test. For all quantifications, each circle represents an individual neuron. The number of neurons for each genotype is indicated. The black bars represent the mean, and the red bars represent the SD.
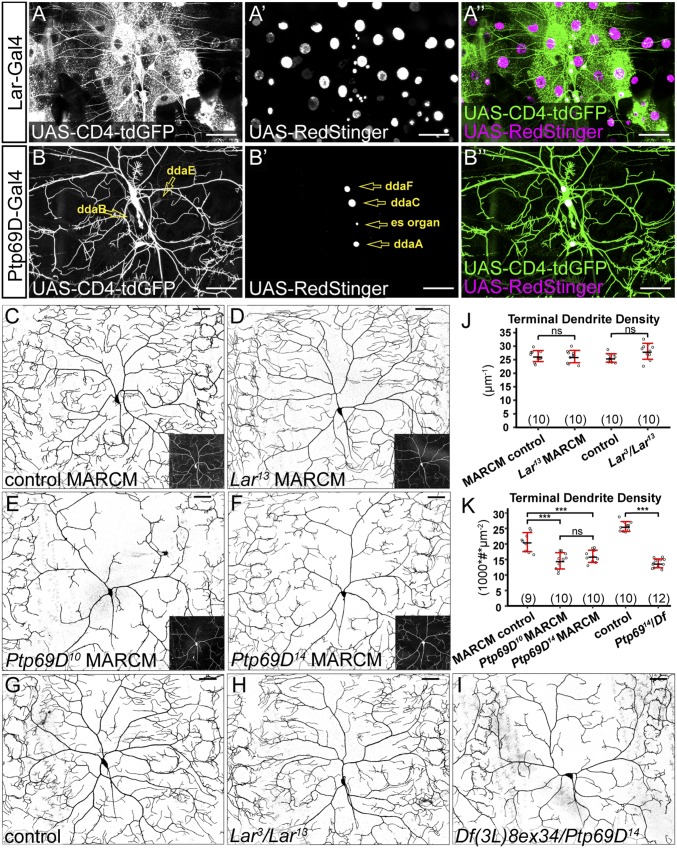
Fig. 6.
Ptp69D but not Lar is necessary for C4da dendritic growth. (A–A′′) Lar-expressing cells at 96 h AEL labeled by Lar-Gal4–driven membrane marker CD4-tdGFP and nuclear marker RedStinger. (B–B′′) Ptp69D-expressing cells at the wandering stage (120 h AEL) labeled by Ptp69D-Gal4–driven CD4-tdGFP and RedStinger. Ptp69D-expressing neurons are labeled in B and B′. (C–F) DdaC MARCM clones of wild type (C) and Lar13 (D), Ptp69D10 (E), and Ptp69D14 (F). Insets show MARCM clones labeled by tdTom (C and D) and mCD8-GFP (E and F). (G–I) DdaC in a control animal (G), a Lar3/Lar13 transheterozygote animal (H), and a Ptp69D14/Df(3L)8ex34 animal (I). (J and K) Quantification of terminal dendrite density in genotypes indicated. ns, not significant; ***P ≤ 0.001; ANOVA and Tukey’s HSD test. For all quantifications, each circle represents a neuron. The numbers of neurons are indicated. Black bar, mean; red bars, SD. (Scale bars, 50 µm.)
To determine if Lar and Ptp69D are important for dendritic growth of C4da neurons, we generated mutations for each of them using CRISPR/Cas9 (48) (Fig. S5 A and C). Lar3and Lar13 are large deletions that remove parts of the extracellular domain, the transmembrane domain, the first intracellular phosphatase domain, and parts of the second phosphatase domain and are thus predicted to be null alleles. Ptp69D10 carries an indel that shifts the reading frame in the second phosphatase domain and therefore may behave as a hypomorph. Ptp69D14 carries a reading frame-shifting indel in the first extracellular Ig domain and thus is predicted to be null. We generated MARCM clones for Lar13 in C4da neurons (Fig. 6D) but did not detect dendrite defects (Fig. 6J and Fig. S5 D and E) compared with the control (Fig. 6C). Lar3/Lar13 transheterozygotes (Fig. 6H) did not show defects in terminal dendrite density (Fig. 6J) but exhibited slight increases in total dendrite density (Fig. S5D) and terminal dendrite numbers (Fig. S5E). In contrast, Ptp69D10 and Ptp69D14 C4da mutant neurons generated by MARCM (Fig. 6 E and F) showed strong reductions of the terminal dendrite density (Fig. 6K). Transheterozygotes of Ptp69D14 and Df(3L)8ex34, a deficiency that deletes the entire Ptp69D locus (49), showed more consistent dendrite reduction (Fig. 6 I and K and Fig. S5 F and G). We did not detect statistically significant differences in the terminal dendrite number between wild-type and Ptp69D mutant neurons generated by the MARCM technique (Fig. S5G), possibly due to the more variable phenotypes of MARCM clones than neurons in the whole-animal mutant. Together, these data demonstrate that Ptp69D, but not Lar, is required for dendrite development of C4da neurons.
Ptp69D and HSPGs Function in Separate Genetic Pathways.
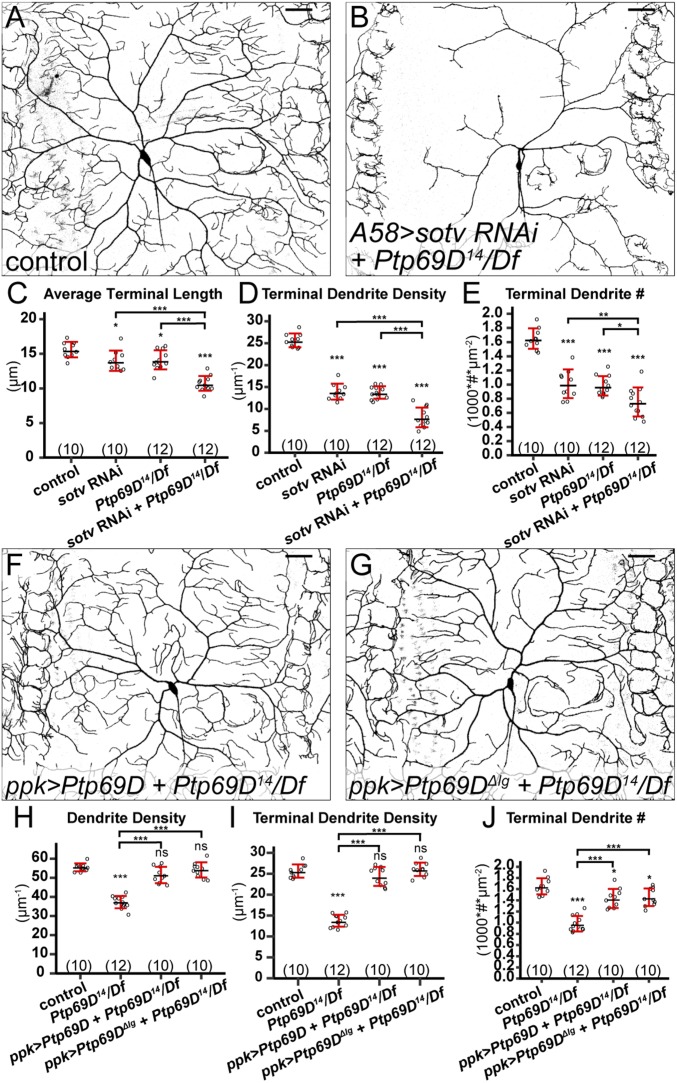
Because both Ptp69D and HSPGs are involved in dendrite development of C4da neurons, we next wanted to know whether they function in the same genetic pathway and, in particular, whether Ptp69D is the neuronal receptor for epidermal HSPGs in promoting dendritic growth. Because of the lack of a dominant active Ptp69D allele that can enhance dendritic growth when overexpressed and the difficulty to simultaneously knock down HSPGs in epidermal cells and express transgenes in C4da neurons, we investigated the epistatic relationship of HSPGs and Ptp69D by knocking down epidermal HSPGs in the Ptp69D mutant background. If Ptp69D is indeed the receptor for HSPGs, the combined loss of HSPGs and Ptp69D should be phenotypically similar to the loss of Ptp69D alone; but if HSPGs and Ptp69D function in separate pathways, loss of both of them should have combined effects. Pan-epidermal knockdown of sotv in Df(3L)8ex34/Ptp69D14 transheterozygotes (Fig. 7B) exhibited stronger dendrite reductions than either sotv RNAi (Fig. 1B) or Df(3L)8ex34/Ptp69D14 (Fig. 6J) alone, as shown by further reduced terminal dendrite density (Fig. 7D) and terminal dendrite number (Fig. 7E). In addition, the average length of terminal dendrites further decreased in the loss of both Ptp69D and epidermal sotv (Fig. 7C). Phenotypically, Ptp69D mutant neurons in epidermal knockdown of sotv exhibited characteristics of wild-type neurons in sotv knockdown, such as more crowded dendrites at the MAS (Fig. 7B), which is not predicted to occur if the HSPG function depends on Ptp69D. Therefore, these data show that the loss of both HSPGs and Ptp69D has combined effects on dendrite development.
Fig. 7.
HSPGs and Ptp69D act in different genetic pathways. (A and B) DdaC in a Gal4A58 control (A) and a Ptp69D14/Df(3L)8ex34 animal expressing Gal4A58 > sotv RNAi (B). (C–E) Quantification of average terminal length (C), terminal dendrite density (D), and terminal dendrite number (E) in genotypes indicated. (F and G) DdaC in animals expressing ppk > Ptp69D (F) and ppk > Ptp69D-ΔIg (G) in the Ptp69D14/Df(3L)8ex34 background. (H–J) Quantification of total dendrite density (H), terminal dendrite density (I), and terminal dendrite number (J) in genotypes indicated. For all quantifications: ns, not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ANOVA and Tukey’s HSD test. Each circle represents a neuron. The numbers of neurons are indicated. Black bar, mean; red bars, SD. (Scale bars, 50 µm.)
We then tested whether Ptp69D requires Ig domains for its function. As RPTPs are known to rely on their Ig domains to interact with HS (18), the Ig domains should be important for Ptp69D function if it is the receptor for HSPGs. We compared the ability of a full-length Ptp69D and a version of Ptp69D lacking both Ig domains (50) (Ptp69DΔIg) to rescue C4da dendrite defects in the Df(3L)8ex34/Ptp69D14 background (Fig. 7 F and G). Neuronal overexpression of Ptp69D completely rescued the total dendrite density (Fig. 7H) and terminal dendrite density (Fig. 7I) and largely rescued the terminal dendrite number (Fig. 7J). Surprisingly, Ptp69DΔIg rescued Ptp69D mutant neurons as well as did the full-length version (Fig. 7 H–J). These data demonstrate that Ig domains are dispensable for the function of Ptp69D in regulating dendritic growth and are inconsistent with the idea that Ptp69D is a receptor for HSPGs.
Finally, we investigated the possibility that Lar and Ptp69D may be redundant in mediating HSPG function. Such a possibility predicts a stronger phenotype in the loss of both Lar and Ptp69D than that of Lar or Ptp69D mutant alone. To examine C4da neurons lacking both Ptp69D and Lar, we knocked down Ptp69D in the Lar3/Lar-Gal4 background. The Trojan-exon Lar-Gal4 is predicted to truncate the Lar transcript (Fig. S5A) and therefore should behave as a null allele. Lar3/Lar-Gal4 did not differ from the control, except showing a slight reduction in dendrite density (Fig. S6 A, B, and E–G). Ptp69D knockdown by Lar-Gal4 (Fig. S6 C and E–G) also mirrored the dendrite reduction phenotype of Df(3L)8ex34/Ptp69D14 (Fig. 7 H–J). Importantly, removing Lar in addition to Ptp69D knockdown did not enhance the dendrite reduction (Fig. S6 D–G). Combined with the HSPG–Ptp69D epistatic analysis and Ptp69D rescue experiments, these results demonstrate that neither Lar nor Ptp69D is a receptor for HSPGs and Ptp69D regulates C4da dendrite development in a pathway independent of HSPGs.
Fig. S6.
Lar and Ptp69D do not play redundant roles in dendrite development of C4da neurons. (A–D) DdaC dendritic fields of an animal expressing Lar-Gal4 (A), a Lar3/Lar-Gal4 transheterozygote animal (B), an animal expressing Lar-Gal4 > Ptp69D RNAi (C), and a Lar3/Lar-Gal4 transheterozygote expressing Ptp69D RNAi (D). (E–G) Quantification of total dendrite density (E), terminal dendrite density (F), and terminal dendrite number (G) in Lar-Gal4, Lar mutant animals, Ptp69D RNAi animals, and Lar mutant animals expressing Ptp69D RNAi. ns, not significant; one-way analysis of variance and Tukey’s HSD test. For all quantifications, each circle represents an individual neuron. The number of neurons for each genotype is indicated. The black bars represent the mean, and the red bars represent the SD. (Scale bars, 50 µm.)
SI Methods
Fly Stocks.
Gal4hh (41), Gal4A58 (56), Gal4en (41), ttv63 (57), Act5C > y+ > Gal4 (42), tubP-Gal80ts (34), UAS-DSulf1-Golgi (33), UAS-dlp-GFP (58), UAS-CD4-tdGFP (59), UAS-dcr2 (23), vkg-GFP (23), UAS-mCherry-Jupiter (60), and ppk-EB1-GFP (39) have been described previously. UAS-Sdc-RNAi was a gift from Xinhua Lin (Cincinnati Children's Hospital Medical Center, Cincinnati). UAS-mIFP-2A-HO1 was a gift from Yuh Nung Jan, University of California, San Francisco. NompC-LexA::p65 (#52240), Df(3L)8ex34 (50), UAS-Ptp69D-myc (50), UAS-Ptp69D-ΔIg (50), UAS-RedStinger (#8547), Nrg-GFP (#6844), R10C12-Gal4 (#47841), R16D01-Gal4 (#48722), nos-Cas9 (#54591), and UAS-Ptp69D-RNAi (#29462) were obtained from Bloomington Stock Center (BDSC). UAS-ttv-RNAi (v4871), UAS-sotv-RNAi (v4902), UAS-botv-RNAi (v37186), UAS-sfl-RNAi (v5070), UAS-dally-RNAi (v14136), and UAS-dlp-RNAi (v10298) were obtained from Vienna Drosophila Resource Center (VDRC). Sdc-GFP (#109775), dally-YFP (#115064), and Dlp-YFP (#115031) were obtained from Drosophila Genomics and Genetic Resources Center (DGRC).
We used the following neuronal markers to label specific classes of da neurons: ppk-CD4-tdGFP (59) and ppk-CD4-tdTom (23) for C4da; R10D05-CD4-tdTom (this study) for C1da; R20C11-CD4-tdGFP (this study) for C2da; and NompC-LexA::p65 LexAop-CD4-tdTom (this study) for C3da. To generate MARCM and Flp-out clones in the epidermis, we created zk-Flp, which is active in epidermal progenitor cells. To monitor microtubule bundling in da neuron dendrites, we made LexAop-GFP-Jupiter.
Molecular Cloning.
R10D05-CD4-tdTom.
The R10D05 enhancer was PCR-amplified from R10D05-Gal4 (#48438; BDSC) genomic DNA using primers ggggACAAGTTTGTACAAAAAAGCAGGCTGCAACGGACCTCTTGTATATGGAC and ggggACCACTTTGTACAAGAAAGCTGGGTGGTTTCGGATTGAAAGTAACCCTGG. The resulting DNA fragment was used to create an entry vector through a Gateway BP reaction (Thermo Fisher Scientific). The entry vector was then combined with pDEST-HemmarR (59) to generate pR10D05-CD4-tdTom expression vector through a Gateway LR reaction.
R20C11-CD4-tdGFP.
An entry vector containing R20C11 enhancer (a gift from Gerald Rubin, Howard Hughes Medical Institute, Janelia Research Campus, Ashburn, VA) was combined with pDEST-HemmarG (59) to generate pR20C11-CD4-tdGFP expression vector through a Gateway LR reaction.
zk-Flp.
An entry vector was first constructed to contain zen VR element (61) and two copies of kr AD1 element (62) in tandem. The zen VR element was amplified from w1118 genomic DNA using primers atatGGTACCCATACTTAGCCCATATTTCCGTC and aataGCTAGCGGATTCAGAGCTTGGTTTTCC; the kr AD1 element was amplified from w1118 genomic DNA using primers tttttctagaGTTTTCCTATTGCACCACACTC and ttttGCTAGCTCCACGCCTTTTTGTTTCTG. A destination vector pDEST-APIC-Flp1 was then constructed by replacing the CD4-tdTom (XhoI/XbaI) fragment in pDEST-HemmarR with the Flp1 coding sequence. Flp1 was amplified by PCR from plasmid pJFRC150-20XUAS-IVS-Flp1::PEST (#32132; Addgene) using primers TTATCCTTTACTTCAGGCGGC and aattTCTAGATTAAATACGGCGATTGATGTAGGAGC and then digested by XhoI and XbaI. The entry vector was then combined with pDEST-APIC-Flp1 to make the pzk-Flp1 expression vector through a Gateway LR reaction.
LexAOP-CD4-tdTom.
A 13xLexAop2 fragment was released from pJFRC19-13XLexAop2-IVS-myr::GFP (#26224; Addgene) by digesting with HindIII (blunted by T4 polymerase) and SpeI. It was then used to replace the KpnI (blunted by T4 polymerase)/NheI fragment of pDEST-APPHIH (59). The resulting plasmid was then digested by XhoI/PacI and religated with the XhoI/PacI fragment (containing CD4-tdTom coding sequence) released from pDEST-HemmarR to make the final construct pAPLO-CD4-tdTom.
LexAOP-GFP-jupiter.
pAPLO-CD4-tdTom was digested with BglII and AscI and then religated after blunting to remove an XbaI site 5′ to 13xLexAop. The resulting plasmid was digested by XhoI and XbaI and assembled with a superfolder GFP (sfGFP) PCR fragment and a Jupiter PCR fragment through three-fragment Gibson Assembly (NEBuilder HiFi DNA Assembly kit). The sfGFP fragment was amplified from pBS-sfGFP (a gift from Thomas Kornberg, University of California, San Francisco) using primers cagttcaattacagctctcgagaaatcaaaATGTCCAAGGGCGAGGAGC and agagatcatCGAGCCACCACCGCCACT. The Jupiter fragment was amplified from UAS-mCherry-Jupiter (a gift from Chi-Hon Lee, National Institute of Child Health and Human Development, Bethesda) genomic DNA using primers gtggctcgATGATCTCTAACTTTGATTGCACCG and ctggcttagttaattaattctagaTTACCACAGGCCCGACGA.
The above constructs were injected in-house or by Rainbow Transgenic Flies to transform flies through either P-element–mediated transformation or φC31 integrase-mediated transformation.
Generation of Lar and Ptp69D Transcription Reporters Using Trojan Exons.
Gal4 transcriptional reporters were generated using the previously described in vivo Trojan-MiMIC system (47). Briefly, MiMIC lines for Lar (MI03443) and Ptp69D (MI15252) were crossed to flies with the triplet Trojan donor construct. The progeny of this cross were then crossed to females expressing Cre recombinase and ΦC31 integrase in the germ line, which allow the Trojan exons to replace the MiMIC attP cassettes. Progeny were then crossed to UAS-GFP for screening Gal4 expression by fluorescence microscopy. Adults positive for GFP expression were used to establish the line. The 2A-Gal4 insertions from established lines were sequenced to confirm the accuracy of the sites and reading frames.
Generation of Lar and Ptp69D Mutations Through CRISPR/Cas9.
We first generated two transgenic fly strains each expressing two gRNAs driven by U6 promoters for targeting either Lar or Ptp69D. The transgenic gRNA constructs were similar to pCFD4-U6.1_U6.3 (48) except that the selection marker is miniwhite. The targeting sequences are AATTCACGCAGCTTATACTGCGG and ATTGTGGTGAAAACTCCTGGCGG for Lar and ACACATGCACAGCCCAAACGGGG and GAAGTTCCTGAAGTCTGCCGTGG for Ptp69D (the PAM sequences are underlined). The constructs were injected by Rainbow Transgenic Flies to transform attP docker lines using φC31 integrase-mediated transformation, with gRNA-Lar inserted to attPVK19 and gRNA-Ptp69D inserted to attPVK27.
CRISPR-induced mutations were generated by crossing gRNA-Lar and gRNA-Ptp69D to y nos-Cas9 w; FRTG13 FRT42D; Tm2/Tm6B and y nos-Cas9 w; Sp/CyOweep; FRT2A FRT80B females, respectively. Male progeny containing nos-Cas9, gRNA, and FRT sites were crossed to balancers to separate FRT chromosomes potentially carrying indel mutations. The progeny containing balanced FRT chromosomes were individually crossed to deficiencies uncovering Lar or Ptp69D to screen for noncomplements. The mutations recovered from the screens were then determined by sequencing. The sequences of both the Lar and Ptp69D mutant alleles are shown in Fig. S5C. Lar3 carries a deletion of amino acids 1085–1899, and Lar13 carries a deletion of amino acids 806–1901; Ptp69D10 has a deletion of two nucleotides at amino acids 1360 and 1361, causing a frameshift mutation, and Ptp69D14 has a deletion of amino acids 116–118 and insertion of TATCCA at amino acids 123 and 124.
Generation of MARCM and Flp-Out Clones.
MARCM analysis of ttv63 was performed by crossing ttv63 FRTG13; zk-Flp ppk-CD4-tdTom with tub-Gal80 FRTG13; tub-Gal4 UAS-CD8-GFP to generate marked ttv63 epidermal clones. MARCM analysis of Lar13 was performed by crossing Lar13 FRTG13; ppk-CD4-tdGFP males with SOP-Flp; tub-Gal80 FRTG13; n-syb-Gal4 UAS-tdTom (a gift from Yuh Nung Jan) females to generate marked neurons mutant for Lar. MARCM analysis of Ptp69D mutations was performed by crossing ppk-CD4-tdTom; Ptp69D10 (or Ptp69D14) FRT2A flies with Gal4elav UAS-mCD8-GFP SOP-Flp; tubP-Gal80 FRT2A (a gift from Yuh Nung Jan) flies to generate marked neurons mutant for Ptp69D. Wild-type MARCM clones in the epidermis or in da neurons were generated using wild-type FRT chromosomes.
Flp-out clones in the epidermis were generated by crossing ppk-spGFP11-CD4-tdTom Act5c > y+ > Gal4 UAS-EGFP; zk-Flp UAS-Dcr-2 flies with UAS-ttv-RNAi or w1118 (as the control).
RNAi.
RNAi knockdown of ttv, sotv, botv, and sfl in epidermal cells was carried out with ppk-CD4-tdTom UAS-dcr2; Gal4hh UAS-EGFP and ppk-CD4-tdTom UAS-dcr2; Gal4A58. RNAi knockdown of ttv in different epidermal domains was carried out with ppk-CD4-tdTom UAS-dcr2; R10C12-Gal4 UAS-mIFP-2A-HO1 and ppk-CD4-tdTom, UAS-dcr2; R16D01-Gal4 UAS-mIFP-2A-HO1. UAS-mIFP-2A-HO1 and UAS-GFP were used to label RNAi-expressing epidermal cells. ppk-CD4-tdTom was used to label C4da neurons. RNAi knockdown of dally, dlp, and Sdc (individually and in combination) was carried out with ppk-CD4-tdTom UAS-dcr2; Gal4hh UAS-EGFP. The effectiveness of dlp RNAi was determined by crossing UAS-dlp-GFP; UAS-dlp-RNAi flies with ppk-CD4-tdTom UAS-dcr2; Gal4hh UAS-mIFP-2A-HO1 flies. The effectiveness of dally RNAi was determined by crossing Gal4en; dally-YFP flies with UAS-dally-RNAi flies. The effectiveness of Sdc RNAi was determined by crossing Sdc-GFP; UAS-Sdc-RNAi flies with ppk-CD4-tdTom UAS-dcr2; Gal4hh UAS-mIFP-2A-HO1 flies. For RNAi knockdown of Ptp69D, either UAS-Ptp69D-RNAi or Lar3/CyOweep; UAS-Ptp69D-RNAi/Tm6B was crossed to Lar-Gal4; ppk-CD4-tdTom UAS-dcr2.
For C1da and C2da neurons, we use UAS-dcr2; Gal4A58 for pan-epidermal expression of ttv RNAi and use R10D05-CD4-tdTom and R20C11-CD4-tdGFP to label C1da and C2da neurons, respectively. For C3da neurons, we used NompC-LexA::p65 LexAOP-CD4-tdTom to label C3da neurons and Gal4hh UAS-mIFP-2A-HO1 to express ttv RNAi.
Immunohistochemistry.
Antibodies used in this study are mouse anti-Mys (DSHB CF.6G11, 1:100), mouse anti-Futsch (DSHB 22C10, 1:100), and mouse anti-HS mAb F69-3G10 (1:100; Amsbio). Secondary antibodies conjugated to DyLight dyes (Jackson ImmunoResearch) were used at 1:400 dilution. Immunostaining of Drosophila larvae was performed as previously described (23). Briefly, third instar larvae were dissected in cold PBS, fixed in 4% formaldehyde/PBS for 30 min at room temperature, and stained with the proper primary antibodies and subsequent secondary antibodies, each for 2 h at room temperature. HS staining was performed as described previously (57). Briefly, larval fillets were incubated with 500 mU/mL heparinase III (Sigma) in 50 mM Tris·HCl (pH 7.2), 100 mM NaCl, 1 mM CaCl2, 0.1% Triton-X100, 5 mg/mL BSA for 6 h at 37 °C. Then, larval fillets were stained with primary (F69-3G10) and secondary antibodies.
Long-Term Time-Lapse Live Imaging.
Larvae at the appropriate age were anesthetized by isoflurane for 2 min. Each larva was then placed on a polydimethylsiloxane (PDMS) block. The PDMS block is 2 mm wide, 8 mm long, 2 mm thick, with a groove in the center (2 mm wide, 0.1–0.3 mm thick, depending on the size of larvae). Six larvae/PDMS were mounted per coverslip. A small amount of UV glue (glue NOA61; Norland) was then added to the two sides of PDMS (about 0.2 µL) and the dorsal side of larvae (0.05–0.1 µL). After that, a coverslip was placed on top of the PDMS and larvae. The coverslip was pressed slightly, so that the PDMS provides a weak pressure to keep larvae in place. Then, glue was cured by UV light (a modified version of WorkStar 2003 NDT 365 nm UV-A Inspection Light) at Hi setting for 4–6 min when the larvae were 5 cm away from the UV light. Next, the coverslip with PDMS and larvae was placed on and glued to a home-made aluminum chamber, which was used for providing ventilation and maintaining moisture. The chamber has two holes drilled on the side and contained a small piece of Kimwipe soaked with water during the imaging. Time-lapse imaging was performed on a Leica SP8 confocal at different time intervals. For ablation experiments, da neurons in the A2, A4, and A6 segments on the right dorsal side of larvae at 48–60 h AEL with appropriate genotypes were ablated using a two-photon laser on a Zeiss 880 confocal microscope. The larvae were then recovered on grape agar plates for 8 h before mounting and imaging on the chamber. For imaging short-term dynamics of dendrites, 3-min time intervals were used. For neurons expressing GFP-Jupiter, 5-h time intervals were used to minimize phototoxicity.
For time-lapse imaging of EB1-GFP, primary dendrites in the hh domain were imaged with a 40× N.A. 1.3 oil objective at a digital zoom of 2.5. Each time-lapse series lasted 5 min with a 2-s time interval. Ten and 12 positions from eight larvae were imaged for control and RNAi-expressing animals, respectively.
Image Analysis and Quantification.
For tracing and measuring C4da and C3da dendrites in Fiji/ImageJ, images of dendrites (1,024 × 1,024 pixels) taken with a 20× objective were first processed by Gaussian Blur (0.8; Sigma) and then Auto Local Threshold (Phansalkar method; radius, 50). Isolated particles below the size of 120 pixels were removed by the Particles4 plugin (www.mecourse.com/landinig/software/software.html). The dendrites were then converted to single pixel-width skeletons using the Skeletonize (2D/3D) plugin and processed using Analyze Skeleton (2D/3D) plugin. The length of skeletons was calculated based on pixel distance; terminal dendrites in the dendritic field or regions of interest were counted based on the endings of terminal dendrites. Skeletons of terminal dendrites in regions of interest were isolated using the Strahler Analysis plugin (imagej.net/Strahler_Analysis). Dendrite density was calculated using the following formula: 1,000 × dendritic length (µm)/dendritic area (µm2); normalized terminal dendrite number was calculated using the following formula: 1,000 × terminal dendrite number/dendritic area (µm2). For C1da and C2da neurons, dendrite tracing and measuring were done using the Simple Neurite Tracer plugin, and the total dendritic lengths were presented. The dynamics of EB1-GFP comets were analyzed using the KymographBuilder plugin. Comet speed was defined as distance the comet moved (µm) per second. Comet speed was calculated using the following formula: total number of comets/[length of the dendrite (µm) × imaging time (5 min)]. R was used to conduct statistical analyses and generate graphs.
Dendrite endings in time-lapse images were identified using the OpenCV library and a custom-written Python script. Each still frame was first processed by Gaussian Blur (ksize, 5) and then Binary Threshold (thresh, 30). Feature points were subsequently identified using Shi-Tomasi Corner-Detection (maxCorners, 500; qualityLevel, 0.1; minDistance, 4; useHarrisDetector, False), and the endings were determined by examining the local topology in the vicinity of each feature point. To follow the movements of the endings, Lucas-Kanade Optical Flow analysis (winSize, 5 × 5; maxLevel, 0; maxCount, 10; epsilon, 0.03) was conducted, and a distance threshold of five pixels was used to merge the displaced endings in each frame with the endings in the subsequent frame.
Discussion
How neurons interact with substrates to fill a receptive field is an unsolved problem. Previous studies have demonstrated the importance of dendrite spatial restriction and homotypic repulsion in spreading dendrites and preventing branch overlaps of the same neuron (self-avoidance) and neighboring like-neurons (tiling) (6, 23, 51). Although much has recently been learned about molecular mechanisms of dendritic spatial restriction and homotypic repulsion (52–54), the roles of the extracellular microenvironment in space-filling remain poorly understood. In particular, it is unknown whether dendritic innervation of target tissues by space-filling neurons in vivo requires neuronal type-specific extrinsic signals, even though earlier work suggested that RGCs have an intrinsic capacity for establishing space-filling patterns in vitro (3). Here we demonstrate that Drosophila space-filling C4da neurons rely on two HSPGs of distinct types, the Syndecan and the glypican Dally, as redundant local permissive signals for their innervation of the skin, while other somatosensory neurons do not require HSPGs for their dendritic growth. Consistent with the neuronal-type specificity, HSPGs stabilize dendritic microtubules and thus dynamic high-order dendritic branches of C4da neurons. Surprisingly, HSPGs promote C4da dendritic growth neither through transporting extracellular diffusible signals nor by interacting with Lar. Interestingly, another RPTP, Ptp69D, regulates the growth of C4da high-order dendrites through an HSPG-independent pathway. Our study, therefore, reveals an HSPG-dependent pathway specific for the dendritic growth of space-filling neurons.
A Model for HSPG-Dependent Dendritic Growth of Spacing-Filling Neurons.
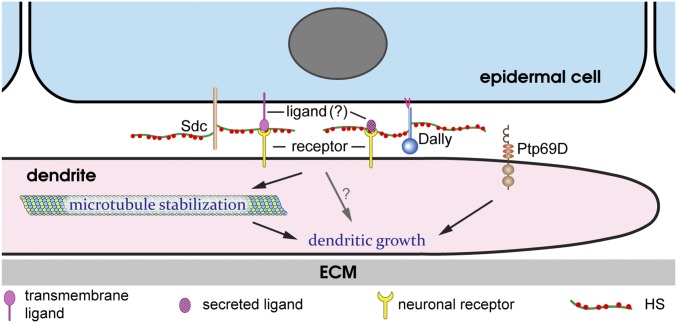
Our results demonstrate that the HS chains of Dally and Sdc on the surface of epidermal cells serve as extracellular permissive signals for dendritic growth of C4da neurons. However, unlike many other contexts in which HSPGs regulate neural development, Lar is not required for the HSPG-dependent dendritic growth. This observation raises the question of whether HS is the sole permissive signal or if it functions together with other extracellular ligands. Our results strongly suggest that for the HSPG-dependent growth, dendrites do not receive morphogen-like molecules that diffuse from specific locations of the epidermis. Consistent with this conclusion, we have failed to rescue the loss of high-order dendrites in HSPG deficiency by epidermal overexpression of many secreted and HS-dependent ligands, including Slit and members of Wnt, BMP, and EGF families. Instead, the cell-autonomous loss of dendrite coverage on HS-deficient epidermal cells indicates that the dendritic growth-promoting signal is present and attached to each individual epidermal cell. Therefore, we propose two possible scenarios (Fig. 8): (i) The HS chains of Dally and Sdc interact with and activate a novel receptor located on the dendritic membrane, or (ii) HS activates a neuronal receptor by acting as a coligand for a membrane protein expressed by all epidermal cells or for a secreted molecule that tightly adheres to the epidermal surface by binding HS. In both scenarios, the activation of the neuronal receptor leads to downstream signaling events that stabilize microtubules. Ptp69D regulates dendritic growth in a separate pathway. This model does not exclude the possibility that there may be other diffusible growth factors that can influence the global pattern of the C4da dendritic arbor.
Fig. 8.
A model for the roles of HSPGs and Ptp69D in dendritic growth. A diagram showing Dally and Sdc and their potential interactions with an unknown ligand and an unknown neuronal receptor. Ptp69D functions in a parallel pathway. See Discussion for details.
The Mechanism of Neuronal Type-Specific Roles of HSPGs in Regulating Dendritic Growth.
An important finding of our study is that HSPGs are specifically required for dendritic growth of C4da neurons. As space-filling neurons, C4da neurons possess highly dynamic high-order dendrites that undergo constant branch turnover. By developing a long-term live-imaging method, we were able to continuously monitor the dynamics of larval da dendrites for more than 10 h. This method revealed that HSPGs are not required for transient dendritic extension or branching. Instead, HSPGs stabilize newly formed and existing high-order branches and enhance long-term dendritic growth at least partially by promoting microtubule stabilization and bundling. The effects of HSPGs on microtubule bundling may be direct or indirect, and HSPGs may have additional effects on other molecular processes that regulate long-term growth of dendrites, such as actin cytoskeleton stability and vesicular trafficking. Nevertheless, our data suggest that for C4da neurons to expand the dendritic arbor or to fill a receptive field, dendrite stabilization is an essential step.
The Drosophila larva has four classes of multidendritic da neurons. Unlike C4da neurons, other da classes lack highly branched high-order dendrites and do not fill their receptive fields. During larval development, once the initial dendritic territory is established, the size of dendritic arbors of da neurons expands in proportion with the overall larval body size (25). For non–space-filling da neurons, the dendritic arbor maintains its shape and expands mostly by elongating existing dendrites. However, space-filling neurons have to generate new branches to invade the space created during body expansion. Therefore, HSPG-dependent stabilization of newly formed dendrites not only serves as a mechanism for C4da neurons to sense and elaborate dendrites into the correct spatial domain but may also underlie the neuronal type-specificity of HSPGs in regulating dendritic growth.
Coexistence of space-filling and non–space-filling neurons in the same tissue is also common in vertebrates. For example, basket cells grow sparse dendrites in the molecular layer of the cerebellum, the same tissue where Purkinje cells extend expansive and numerously branched dendritic arbors (55). It will be interesting to find out whether Purkinje cells also require neuronal type-specific permissive signals for arbor growth.
Cell-Nonautonomous Increase of Dendrites Induced by HSPG Deficiency.
Our experiments also revealed an unexpected cell-nonautonomous increase of dendrites induced by HSPG deficiency. Knockdown of ttv in the hh domain caused an increase of dendrites on epidermal cells anterior to the hh domain (Fig. 1P). Similarly, wild-type epidermal clones completely surrounded by ttv RNAi-expressing epidermal cells showed higher terminal dendrite density than similar clones surrounded by wild-type epidermal cells (Fig. 5L). In addition, pan-epidermal knockdowns of sotv, ttv, and sfl by Gal4A58 all led to increases of dendrites at MASs (Fig. 1B and Fig. S1 B and C). These results demonstrate that when innervation in one spatial domain is blocked by HS loss, the dendrite density is increased in nearby permissive domains. A tantalizing interpretation is that C4da neurons may have an intrinsic “drive” to elaborate a certain length of dendrites and the lack of dendrites in HS-deficient zones is compensated by a dendrite increase in other areas. We found that neurons lacking dendrite coverage in the hh domain, or in the middle of the segment (driven by R16D01-Gal4), as a result of HS deficiency are similar to Gal4-only control neurons in total dendrite lengths, supporting this hypothesis. Two other possible mechanisms may also explain the results. In the first one, HS loss in one epidermal region may somehow increase the concentration of the dendrite growth-promoting signal on neighboring wild-type epidermal cells. In the second, HS deficiency may trap intracellular dendrite growth-promoting organelles, such as Golgi outposts (9), to dendrite segments covering nearby wild-type epidermal cells. It will be interesting to distinguish these possibilities in the future.
Conserved and Diverse Roles of HSPGs in Insect and Vertebrate Space-Filling Neurons.
C4da neurons are analogous to the trigeminal sensory neurons and Rohon–Beard (RB) neurons of zebrafish in that they are all space-filling somatosensory sensory neurons that innervate the skin (4, 14). Consistent with the conclusion that HSPGs play a permissive, but not instructive, role, C4da dendrites do not misroute to other areas in the absence of HSPGs. Instead, they can extend and branch into HS-deficient zones but fail to be stabilized. In contrast, HSPGs seem to play a different role in axonal morphogenesis of zebrafish space-filling neurons: They function as attractants to direct growing sensory axons of RB neurons to the skin, and loss of HS in the skin leads to misrouting of axons to internal tissues (15). This apparent difference may be related to zebrafish neurons having alternative but disfavored substrates (i.e., internal tissues), whereas Drosophila C4da neurons cannot innervate tissues other than the epidermis. To some extent, the increase of C4da dendrites outside the HS-deficient zone may be seen as resembling the misrouting phenotype of zebrafish RB neurons.
On the other hand, the pathways through which HSPGs regulate neurite outgrowth of Drosophila C4da neurons and zebrafish RB neurons are clearly distinct. Zebrafish RB neurons express two LAR homologs, PTPRFa and PTPRFb, which act as redundant receptors for HS to control the guidance of sensory axons (15). The interaction between these LAR proteins and extracellular HS is essential as mutations of the HS-interaction motif in the Ig domains of the LARs abolish their ability to respond to HS (15). In contrast, the Drosophila Lar does not play a detectable role in dendritic growth of C4da neurons. Although the Ig-containing RPTP Ptp69D is expressed in da neurons and is required for high-order dendritic growth of C4da neurons, it does not function in the same pathway as HSPGs. Therefore, it appears that HSPGs can regulate neurite growth of different types of space-filling neurons through different downstream signaling pathways. These similarities and distinctions raise several interesting questions. First, does the HSPG–PTPRFa/PTPRFb pathway control axon guidance of RB neurons through stabilizing microtubules in selective branches? Second, do HSPGs regulate space-filling independent of LAR proteins in other neuronal systems? Lastly, do HSPGs play any role in neurite growth of vertebrate non–space-filling neurons that exhibit dynamic branch turnover? Answering these questions will broaden our understanding of the roles the extracellular microenvironment plays in neuronal morphogenesis.
Methods
Snapshot live imaging of dendritic morphology of da neurons was performed as described previously (23). For long-term time-lapse imaging, larvae at the right stage were glued to a coverslip using a UV-sensitive glue (NOA 61; Norland) and then mounted on a chamber constructed with aluminum. See SI Methods for details of fly stocks, molecular cloning, generation of CRISPR mutants and transcription reporters, MARCM, RNAi, immunohistochemistry, live imaging, image analysis, and quantification.
Supplementary Material
Acknowledgments
We thank Xinhua Lin, Yuh Nung Jan, Hiroshi Nakato, Chi-Hon Lee, Kai Zinn, Benjamin White, Jill Wildonger, Bloomington Stock Center, Kyoto Stock Center, and Vienna Drosophila Resource Center (VDRC) for fly stocks; Scott Emr, Mariana Wolfner, and Joe Fetcho for critical reading and suggestions on the manuscript; Gerald Rubin, Thomas Kornberg, and Addgene for plasmids; Developmental Studies Hybridoma Bank (DSHB) for antibodies; and Halocarbon Products Corporation for Halocarbon oils. This work was supported by a start-up fund from Cornell University and NIH Grant R01 NS099125 (to C.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707467114/-/DCSupplemental.
References
- 1.Fujishima K, Horie R, Mochizuki A, Kengaku M. Principles of branch dynamics governing shape characteristics of cerebellar Purkinje cell dendrites. Development. 2012;139:3442–3455. doi: 10.1242/dev.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panico J, Sterling P. Retinal neurons and vessels are not fractal but space-filling. J Comp Neurol. 1995;361:479–490. doi: 10.1002/cne.903610311. [DOI] [PubMed] [Google Scholar]
- 3.Montague PR, Friedlander MJ. Expression of an intrinsic growth strategy by mammalian retinal neurons. Proc Natl Acad Sci USA. 1989;86:7223–7227. doi: 10.1073/pnas.86.18.7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sagasti A, Guido MR, Raible DW, Schier AF. Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr Biol. 2005;15:804–814. doi: 10.1016/j.cub.2005.03.048. [DOI] [PubMed] [Google Scholar]
- 5.Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 6.Grueber WB, Sagasti A. Self-avoidance and tiling: Mechanisms of dendrite and axon spacing. Cold Spring Harb Perspect Biol. 2010;2:a001750. doi: 10.1101/cshperspect.a001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 8.Jinushi-Nakao S, et al. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 9.Ye B, et al. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoh D, et al. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- 11.Zheng Y, et al. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol. 2008;10:1172–1180. doi: 10.1038/ncb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng Y, et al. Regulation of dendrite growth and maintenance by exocytosis. J Cell Sci. 2015;128:4279–4292. doi: 10.1242/jcs.174771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin WY, et al. The SLC36 transporter pathetic is required for extreme dendrite growth in Drosophila sensory neurons. Genes Dev. 2015;29:1120–1135. doi: 10.1101/gad.259119.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X, Shen K, Bülow HE. Intrinsic and extrinsic mechanisms of dendritic morphogenesis. Annu Rev Physiol. 2015;77:271–300. doi: 10.1146/annurev-physiol-021014-071746. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Wolfson SN, Gharib A, Sagasti A. LAR receptor tyrosine phosphatases and HSPGs guide peripheral sensory axons to the skin. Curr Biol. 2012;22:373–382. doi: 10.1016/j.cub.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 17.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coles CH, et al. Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science. 2011;332:484–488. doi: 10.1126/science.1200840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoker AW. RPTPs in axons, synapses and neurology. Semin Cell Dev Biol. 2015;37:90–97. doi: 10.1016/j.semcdb.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Van Vactor D, Wall DP, Johnson KG. Heparan sulfate proteoglycans and the emergence of neuronal connectivity. Curr Opin Neurobiol. 2006;16:40–51. doi: 10.1016/j.conb.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Liang X, Dong X, Moerman DG, Shen K, Wang X. Sarcomeres pattern proprioceptive sensory dendritic endings through UNC-52/perlecan in C. elegans. Dev Cell. 2015;33:388–400. doi: 10.1016/j.devcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim ME, Shrestha BR, Blazeski R, Mason CA, Grueber WB. Integrins establish dendrite-substrate relationships that promote dendritic self-avoidance and patterning in Drosophila sensory neurons. Neuron. 2012;73:79–91. doi: 10.1016/j.neuron.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han C, et al. Integrins regulate repulsion-mediated dendritic patterning of Drosophila sensory neurons by restricting dendrites in a 2D space. Neuron. 2012;73:64–78. doi: 10.1016/j.neuron.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrish JZ, Emoto K, Jan LY, Jan YN. Polycomb genes interact with the tumor suppressor genes hippo and warts in the maintenance of Drosophila sensory neuron dendrites. Genes Dev. 2007;21:956–972. doi: 10.1101/gad.1514507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in Drosophila sensory neurons. Neuron. 2009;63:788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugimura K, et al. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci. 2003;23:3752–3760. doi: 10.1523/JNEUROSCI.23-09-03752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 28.Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- 29.Genova JL, Fehon RG. Neuroglian, gliotactin, and the Na+/K+ ATPase are essential for septate junction function in Drosophila. J Cell Biol. 2003;161:979–989. doi: 10.1083/jcb.200212054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delon I, Brown NH. The integrin adhesion complex changes its composition and function during morphogenesis of an epithelium. J Cell Sci. 2009;122:4363–4374. doi: 10.1242/jcs.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamimura K, et al. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J Cell Biol. 2006;174:773–778. doi: 10.1083/jcb.200603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhoot GK, et al. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 33.Kleinschmit A, et al. Drosophila heparan sulfate 6-O endosulfatase regulates wingless morphogen gradient formation. Dev Biol. 2010;345:204–214. doi: 10.1016/j.ydbio.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 35.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 36.Farah CA, Leclerc N. HMWMAP2: New perspectives on a pathway to dendritic identity. Cell Motil Cytoskeleton. 2008;65:515–527. doi: 10.1002/cm.20284. [DOI] [PubMed] [Google Scholar]
- 37.Hummel T, Krukkert K, Roos J, Davis G, Klämbt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- 38.Karpova N, Bobinnec Y, Fouix S, Huitorel P, Debec A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil Cytoskeleton. 2006;63:301–312. doi: 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- 39.Arthur AL, Yang SZ, Abellaneda AM, Wildonger J. Dendrite arborization requires the dynein cofactor NudE. J Cell Sci. 2015;128:2191–2201. doi: 10.1242/jcs.170316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belenkaya TY, et al. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 41.Han C, Belenkaya TY, Wang B, Lin X. Drosophila glypicans control the cell-to-cell movement of hedgehog by a dynamin-independent process. Development. 2004;131:601–611. doi: 10.1242/dev.00958. [DOI] [PubMed] [Google Scholar]
- 42.Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular wingless morphogen gradient in the wing disc. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- 43.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 44.Fox AN, Zinn K. The heparan sulfate proteoglycan syndecan is an in vivo ligand for the Drosophila LAR receptor tyrosine phosphatase. Curr Biol. 2005;15:1701–1711. doi: 10.1016/j.cub.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 45.Johnson KG, et al. The HSPGs syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 2006;49:517–531. doi: 10.1016/j.neuron.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 46.Andersen JN, et al. Computational analysis of protein tyrosine phosphatases: Practical guide to bioinformatics and data resources. Methods. 2005;35:90–114. doi: 10.1016/j.ymeth.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Diao F, et al. Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desai CJ, Gindhart JG, Jr, Goldstein LS, Zinn K. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell. 1996;84:599–609. doi: 10.1016/s0092-8674(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 50.Garrity PA, et al. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron. 1999;22:707–717. doi: 10.1016/s0896-6273(00)80730-8. [DOI] [PubMed] [Google Scholar]
- 51.Matthews BJ, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 52.Zipursky SL, Grueber WB. The molecular basis of self-avoidance. Annu Rev Neurosci. 2013;36:547–568. doi: 10.1146/annurev-neuro-062111-150414. [DOI] [PubMed] [Google Scholar]
- 53.Soba P, et al. The Ret receptor regulates sensory neuron dendrite growth and integrin mediated adhesion. Elife. 2015;4:e05491. doi: 10.7554/eLife.05491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meltzer S, et al. Epidermis-derived semaphorin promotes dendrite self-avoidance by regulating dendrite-substrate adhesion in Drosophila sensory neurons. Neuron. 2016;89:741–755. doi: 10.1016/j.neuron.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Markram H, et al. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 56.Han C, et al. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron. 2014;81:544–560. doi: 10.1016/j.neuron.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han C, et al. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- 58.Yan D, Wu Y, Feng Y, Lin SC, Lin X. The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev Cell. 2009;17:470–481. doi: 10.1016/j.devcel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han C, Jan LY, Jan YN. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc Natl Acad Sci USA. 2011;108:9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDonald JA, Khodyakova A, Aranjuez G, Dudley C, Montell DJ. PAR-1 kinase regulates epithelial detachment and directional protrusion of migrating border cells. Curr Biol. 2008;18:1659–1667. doi: 10.1016/j.cub.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doyle HJ, Kraut R, Levine M. Spatial regulation of zerknüllt: A dorsal-ventral patterning gene in Drosophila. Genes Dev. 1989;3:1518–1533. doi: 10.1101/gad.3.10.1518. [DOI] [PubMed] [Google Scholar]
- 62.Hoch M, Schröder C, Seifert E, Jäckle H. cis-acting control elements for Krüppel expression in the Drosophila embryo. EMBO J. 1990;9:2587–2595. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.