Significance
Current models of virus entry by type 1 viral envelope glycoprotein-mediated membrane fusion place the fusion domains (fusion peptides or fusion loops) and transmembrane (TM) domains of these proteins in close proximity, but a direct molecular interaction and functional cooperation of these domains have not been previously demonstrated for any viral envelope glycoprotein. In the present work, we determined the structure of the only missing pieces of the Ebolavirus glycoprotein 2 [namely, its membrane proximal external region (MPER) and TM domains], demonstrate MPER’s direct molecular interaction with the fusion loop of the same protein, and provide evidence for the functional significance of this interaction.
Keywords: Ebola virus, envelope protein, membrane fusion, structure, membrane protein
Abstract
Ebolavirus (EBOV), an enveloped filamentous RNA virus causing severe hemorrhagic fever, enters cells by macropinocytosis and membrane fusion in a late endosomal compartment. Fusion is mediated by the EBOV envelope glycoprotein GP, which consists of subunits GP1 and GP2. GP1 binds to cellular receptors, including Niemann-Pick C1 (NPC1) protein, and GP2 is responsible for low pH-induced membrane fusion. Proteolytic cleavage and NPC1 binding at endosomal pH lead to conformational rearrangements of GP2 that include exposing the hydrophobic fusion loop (FL) for insertion into the cellular target membrane and forming a six-helix bundle structure. Although major portions of the GP2 structure have been solved in pre- and postfusion states and although current models place the transmembrane (TM) and FL domains of GP2 in close proximity at critical steps of membrane fusion, their structures in membrane environments, and especially interactions between them, have not yet been characterized. Here, we present the structure of the membrane proximal external region (MPER) connected to the TM domain: i.e., the missing parts of the EBOV GP2 structure. The structure, solved by solution NMR and EPR spectroscopy in membrane-mimetic environments, consists of a helix-turn-helix architecture that is independent of pH. Moreover, the MPER region is shown to interact in the membrane interface with the previously determined structure of the EBOV FL through several critical aromatic residues. Mutation of aromatic and neighboring residues in both binding partners decreases fusion and viral entry, highlighting the functional importance of the MPER/TM–FL interaction in EBOV entry and fusion.
Ebolavirus (EBOV) is an enveloped, negative strand RNA virus that causes severe hemorrhagic fever associated with a 25 to 90% mortality rate; for example, the mortality rate in the recent outbreak in Western Africa was ∼40% (1–4). EBOV enters cells by macropinocytosis and releases its genetic material into the cytoplasm by fusion of the viral membrane envelope with the membrane of a late endosomal compartment (5–8). The entry and membrane fusion of EBOV is governed by the viral surface glycoprotein (GP), which is enzymatically cleaved by cathepsins L and B in endosomes, followed by binding to the late endosomal/lysosomal receptor Niemann-Pick C1 (NPC1) (9–15). The enzymatic processing and binding to NPC1 are thought to lead to conformational changes in the GP subunit 2 (GP2) that are responsible for the fusion process. It is proposed that the EBOV fusion loop (EBOV FL), a hydrophobic disulfide-linked sequence within GP2, is released and inserts into the target membrane in the course of this multistep activation process (16, 17). At this stage, GP2 is hypothesized to form an intermediate prehairpin conformation, in which the N- and C-terminal heptad repeats of GP2 are extended, thereby bridging the viral and endosomal membranes. The FL inserts into the host membrane in the form of a hydrophobic fist, a conformation that appears to initiate the fusion process (18). In a further conformational change, the N- and C-terminal heptad repeats of GP2 form a six-helix bundle that provides the energy to bring the two membranes into fusion contact (19, 20). At this point, the transmembrane (TM) domain of GP2 is speculated to be in close physical proximity to the FL, and the TM domain and FL are proposed to cooperate to open a fusion pore and complete the fusion process. There are also numerous studies, primarily from other viral envelope proteins, that a polypeptide sequence close to the membrane surface and adjacent to the TM domain [i.e., the membrane proximal external region (MPER)] may be critically important for membrane fusion (21–23). However, despite the attractive working model of FL, TM, and perhaps MPER domain cooperation in viral fusion, currently, only very limited structural information is available on EBOV MPER (24), and the structure of the EBOV TM domain is completely unknown. Moreover, it is unclear how the MPER/TM domains would cooperate with the FL to promote membrane fusion.
To investigate the structural role of the MPER/TM domain of EBOV GP2 in the fusion process of EBOV cell entry, we first determined by solution NMR the structure of this domain in dodecylphosphocholine (DPC) micelles at pH 5.5, the pH that prevails in endosomes where the viral envelope fuses. We also performed NMR relaxation, paramagnetic relaxation enhancement (PRE), and electron paramagnetic resonance (EPR) experiments to determine protein dynamics and the disposition of the protein in lipid bilayers. We found that, independent of pH, EBOV MPER/TM consists of an unstructured N-terminal region and a short helix in the MPER region that is followed by a turn and the TM helix: i.e., MPER/TM forms a helix-turn-helix motif. Measuring chemical shift changes upon titration of the FL to the MPER/TM domain indicates that the FL interacts with the sequence WTGW in the MPER region, but not with the TM region. This interaction was further found to be functionally important in GP2-mediated lipid mixing assays in vitro and in virus-like particle (VLP) entry assays in cells as, compared with WT, the WTGW mutant GP constructs were strongly impaired in fusion. We conclude that two tryptophans in the MPER region are critical for the interaction of the FL with MPER and its promotion of GP-mediated membrane fusion in EBOV cell entry.
Results
Solution NMR Structure of EBOV MPER/TM in DPC Micelles at Neutral and Mildly Acidic pH.
A key feature of the conformational rearrangement of EBOV GP2 (see Fig. 1A for its domain structure and Fig. 1B for its amino acid sequence) during fusion activation is its strong dependence on pH, which drops by about two units when the virus moves from the cell surface into the endosome. Previous structural studies on the soluble ectodomain and the FL of EBOV GP have shown very different structures at neutral and low pH (18, 25). The pre- and postfusion structures of EBOV GP suggest that GP2 undergoes a dramatic conformational change, from a complex trimeric helical arrangement at neural pH to a highly ordered six-helix bundle at mildly acidic pH. A similar helical packing has been reported at low pH for the related Marburg virus (26). The EBOV FL also exhibits very different conformations when bound to DPC micelles at neutral and low pH. While the structure is only peripherally associated with the membrane/micelle surface at neutral pH, it forms a hydrophobic fist that penetrates more deeply into the membrane/micelles at low pH (18). In contrast to these well-studied fragments of GP2, there is no structure available for the MPER/TM domain of GP2. It is also not known how it might change its conformation in response to the lower endosomal pH. To address questions that require knowledge of the structure of the MPER/TM domain of GP2, we successfully expressed and purified a construct encompassing both of these regions of GP2. Our strategy involved linking the protein to a Trp leader protein system that targets the toxic TM domain to inclusion bodies rather than the cell surface (Methods). Using Ni affinity and size exclusion chromatography and thrombin cleavage in the presence of detergent ultimately produced high yields of pure MPER/TM protein (Fig. 1 C and D and SI Appendix, Fig. S1).
Fig. 1.
Expression, purification, and solution NMR structure of the EBOV MPER/TM domain. (A) EBOV GP2 domain structure. CHR, C-heptad repeat region (orange); FL, fusion loop (blue); MPER, membrane proximal external region (red); NHR, N-heptad repeat region (green); TM, transmembrane region (red). (B) Primary structure of EBOV GP2 (Zaire strain). Color codes are matched with domain structure. (C) SDS/PAGE gel showing the isolation of the MPER/TM domain from the expression fusion protein construct with the Trp leader protein. Lane 1, markers; lane 2, expressed fusion protein after solubilization from inclusion bodies; lane 3, cleaved MPER/TM domain; lane 4, Trp leader protein. (D) Size exclusion chromatography of the MPER/TM domain in DPC micelles (20 mM phosphate, 100 mM NaCl, 0.2% DPC) at pH 7.0 showing a single homogeneous symmetric peak. (E) HSQC spectra of the MPER/TM domain in DPC micelles at pH 7.0 (blue) and pH 5.5 (red). All backbone atoms were assigned as shown. (F) Twenty lowest energy structures of the MPER/TM domain in DPC micelles at pH 5.5.
The proteins were concentrated in DPC micelles, and 1H-15N heteronuclear quantum coherence (HSQC) spectra were recorded at pH 7.0 and 5.5 (Fig. 1E). Interestingly, the two spectra were nearly superimposable, indicating that the EBOV MPER/TM domain does not undergo a major conformational change in response to low pH. No major chemical shift changes were observed between the two different values of pH. This indicates that the EBOV MPER/TM domain assumes very similar structures in its pre- and postfusion forms. Therefore, and since EBOV fusion happens only in late endosomal compartments under mildly acidic conditions, we proceeded with solving the solution NMR structure of the EBOV MPER/TM domain only at pH 5.5.
We recorded sets of heteronuclear triple-resonance spectra of 13C,15N-labeled samples in fully deuterated DPC as indicated in Methods and ultimately were able to fully assign the backbone and side chain resonances of all 45 residues in the construct. The assignments of the HSQC cross-peaks are shown in Fig. 1E. Dihedral angles were calculated from the chemical shifts using TALOS+, and nuclear Overhauser effect (NOE) distance constraints were obtained from 15N- and 13C-edited NOESY spectra. The 20 lowest energy structures calculated using CYANA and CNS representing the pH 5.5 conformational ensemble are depicted in Fig. 1F. The structural statistics are reported in SI Appendix, Table S1. The ensemble is characterized by an unstructured N-terminal region (residues D632 to N641), a helical MPER region (D642 to Q650), a turn (W651 to I656), and a TM helical region (G657 to F676).
Comparison of Secondary Structure in DPC Micelles and DMPC/DHPC Bicelles.
To check if the DPC micelle is an adequate environment for maintaining the structure of MPER/TM in lipid bilayers, we prepared samples in 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) bicelles at a molar ratio of q = 0.5. In these experiments, MPER/TM was reconstituted in 15% (wt/vol) DMPC/DHPC bicelles by reconstituting MPER/TM into DMPC vesicles followed by addition of an appropriate amount of DHPC that dispersed the vesicles into bicelles (Methods). NMR spectra of the bicelle samples were collected at 45 °C instead of 30 °C to obtain sharper resonance lines. Excellent HSQC spectra were obtained under these conditions that were fully assigned de novo by obtaining appropriate heteronuclear triple resonance spectra (SI Appendix, Fig. S2A). Based on chemical shift index (CSI) analysis, the secondary structure of MPER/TM was very similar and essentially indistinguishable with a few minor exceptions in the DPC and DMPC/DHPC environments (SI Appendix, Fig. S2C). Likewise, when MPER/TM was labeled with a methane-thiosulfonate spin-label (MTSL) at its C terminus, completely superimposable continuous wave EPR data were collected in micelle and bicelle environments at room temperature (SI Appendix, Fig. S2B), which suggests that the internal molecular mobility and conformation of MPER/TM is very similar in the two systems. Despite this close similarity of secondary structure, we did not attempt to calculate the complete 3D structure in bicelles because the resonances of the aliphatic side chains overlapped in the bicelle system and therefore could not be easily assigned. However, the main conclusion from this and nuclear and paramagnetic relaxation data presented below is that the structures of MPER/TM in DPC micelles and DMPC/DHPC bicelles are very similar to one another based on the obtained NMR data.
We also performed NMR experiments on full-length EBOV GP2 and attempted to solve its structure in DPC micelles. Although we could not observe all expected cross-peaks of the 175-residue protein, the HSQC cross-peaks were dispersed well enough (SI Appendix, Fig. S3A) to pursue the assignment of large parts of the MPER/TM segment and to show that, again, a very similar secondary structure, as observed in the micellar or bicellar system, is preserved in this structure. The problem with the full-length construct is that it contains a well-known trimerization motif in the N-heptad repeat region (NHR) of the ectodomain. This could be demonstrated in native Bis-Tris gels, which showed that the protein was monomeric in DPC, but trimeric in DMPC/DHPC or in DMPC vesicles (SI Appendix, Fig. S4). Interestingly, DPC seems to be capable of disrupting the NHR trimerization sites of GP2. Therefore, we also investigated full-length GP2 in lipid bicelles by NMR (SI Appendix, Fig. S3B). Although reasonable quality HSQC spectra could be obtained with this system, they were still not good enough to pursue assignments or a full structure determination. Even when specifically labeled with 15N-Leu, only about half of the 16 expected Leu cross-peaks could be observed in an HSQC spectrum of GP2 at pH 7 and 5.5, perhaps reflecting conformational exchange or heterogeneity of the sample.
Membrane Depth and Backbone Dynamics of MPER/TM in Lipid Micelles and Bicelles.
To better understand the disposition of the MPER/TM domain in the membrane, the depth of the protein in the bicelle was investigated by PRE experiments in the presence of soluble gadolinium(III) diethylenetriaminepentaacetic acid [Gd(DTPA)] or the bicelle-incorporated spin-labeled fatty acids 5-DSA or 16-DSA (Fig. 2). The paramagnetic quenching profiles in Fig. 2 A–C show that the N-terminal and front of the MPER region were accessible to Gd(DTPA) but not to 5- and 16-DSA. In marked contrast, the TM region was highly quenched by 16-DSA, but less by the shallower membrane probe 5-DSA and the soluble probe Gd(DTPA). These data demonstrate that the MPER domain is interfacially located and that the TM domain is deeply immersed into the bicelle. Similar and even more clear-cut data were obtained in DPC micelles (SI Appendix, Fig. S5 A–C). Spin-lattice and spin-spin relaxation, as well as heteronuclear NOE data, indicated that the membrane-embedded regions of MPER/TM were well-ordered and that the solvent-exposed regions were significantly more dynamic in lipid bicelles (Fig. 2 D–F) and micelles (SI Appendix, Fig. S5 D–F).
Fig. 2.
Paramagnetic relaxation enhancement (PRE) and dynamics measurements of the MPER/TM domain in q = 0.5 DMPC/DHPC bicelles at pH 5.5 and 45 °C. The PRE experiments were performed in the presence of (A) soluble Gd(DTPA), (B) bicelle-bound 5-doxyl stearic acid, and (C) bicelle-bound 16-doxyl stearic acid. (D) Heteronuclear {1H}15N-NOEs, (E) 15N T1 spin-lattice, and (F) T2 spin–spin relaxation times recorded in q = 0.5 DMPC/DHPC bicelles. Helical regions in MPER and TM domains are shown at the top of the figure.
To further investigate the depth of the protein in lipid bilayers, we performed EPR power saturation experiments with several single cysteine mutants (N643C, W644C, T659C, I663C, I666C, and A670C) that were each labeled with the nitroxide spin label MTSL. The mutation sites were chosen based on the NMR structure of the MPER/TM domain: Somewhat bulky residues facing away from MPER helix were selected in the TM region to match the bulkiness and avoid protein clashes of the MTSL spin probes. N643 and W644 were selected because they are positioned just before the critical MPER region, and T569 was chosen to mark the turn in the bilayer. The proteins were reconstituted into 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) (85:15) vesicles, and power saturation EPR experiments were performed in the presence of water-soluble Ni-ethylenediamine-N,N′-diacetic acid (NiEDDA) and membrane-permeant O2 and N2 (Fig. 3A), which yielded NiEDDA and O2 accessibilities (Fig. 3B). Depth parameters were calculated from these data (Fig. 3C). Comparisons of these depth parameters with values from reference proteins of known structure indicate that the TM domain transverses the lipid bilayer with Ile663 situated close to the center of the bilayer and that the MPER region resides in the membrane interface.
Fig. 3.
Structural model of the MPER/TM domain in a lipid bilayer based on structural restraints obtained by NMR in DPC micelles and by EPR in lipid bilayers. (A) Diagram explaining the EPR power saturation experiment performed in the presence of O2, which favorably partitions into the hydrophobic core of the bilayer, and NiEDDA, which is highly soluble in water and does not favor the core of the bilayer. The chemical structure of the MTSL spin label that was attached to various surface-located and membrane interior-located residues is shown. (B) Accessibilities of spin labels attached to N643C, W644C, T659C, I663C, I666C, and A670C, respectively, to NiEDDA (green) and O2 (red) are shown. (C) Depth parameters of the six labeled residues calculated from the experimental NiEDDA and O2 accessibilities. Higher numbers indicate deeper positions within the lipid bilayer, with 0 corresponding to the average level of the lipid headgroup phosphates and 4 corresponding to approximately the center of the bilayer. (D) The five lowest energy structures of the MPER/TM domain calculated using XPLOR-NIH within an implicit membrane model using power saturation EPR data as additional restraints.
To determine if MPER/TM forms a monomer or trimer in micellar and bilayered membrane mimetics, we performed double electron resonance (DEER) experiments of the Cys677-nitroxide–labeled protein in DPC micelles, DMPC/DHPC bicelles, and DMPC/membrane scaffolding protein 1D1 (MSP1D1) nanodiscs (SI Appendix, Fig. S6). No spin–spin interactions were detected in either situation, indicating that this construct is monomeric in lipid micelles, bicelles, and nanodiscs.
Next, we examined if the angle between the MPER and TM domain that was determined by solution NMR in DPC micelles was preserved in lipid bilayers. To this end, we double-labeled with nitroxide spin labels the MPER/TM construct at cysteine residues introduced in positions 643 and 666 or 643 and 670 in the MPER and TM portions of the molecule, respectively. These sites were chosen because they did not interfere with each other or clash with other residues based on the NMR structure. Intramolecular DEER experiments were performed and compared in DPC micelles and DMPC/MSP1D1 nanodiscs (SI Appendix, Fig. S7 A and C). The derived distance distributions centered at about 3.5 and 4.0 nm for the two spin label pairs, respectively, (SI Appendix, Fig. S7 B and D) and were similar in micelles and nanodiscs although they were somewhat more broadly distributed in nanodiscs. Importantly, the calculated distance distributions between these two pairs of residues matched the distributions calculated from the NMR structures quite well, lending support to the validity of these structures in lipid micelles and bilayers.
To quantitatively position the MPER/TM structure in the lipid bilayer, the NMR structure was recalculated using XPLOR-NIH, but within an implicit membrane model using the PRE and EPR power saturation data as additional restraints. The five lowest energy structures calculated by this procedure are shown in Fig. 3D. This calculation suggests that the MPER helix resides slightly beneath the phosphate level of the lipid headgroups and that the TM helix transverses the lipid bilayer at an angle. Although this calculation makes perhaps overly optimistic assumptions about the transferability of the NMR structural data into a lipid bilayer and although the number of membrane restraints are almost certainly not sufficient to determine the membrane-embedded structure with high precision, the calculations, which are all based on experimental measurements, are still instructive and indicate how this structure is most likely embedded in the lipid bilayer. Regardless, it can be concluded from our data with certainty that MPER/TM consists of a soluble and dynamic N-terminal region, a structured interfacial α-helical MPER region, a more flexible turn, and a well-ordered TM region that transverses the lipid bilayer. We also obtained highly resolved spectra of a construct comprising the C-terminal heptad repeat (CHR) followed by MPER and TM (SI Appendix, Fig. S8). Comparison with the shorter MPER/TM construct shows virtually all corresponding peaks in the CHR/MPER/TM construct, indicating that the CHR domain unlikely influences the structure of the MPER/TM domain.
Interaction of MPER/TM with FL of EBOV GP2.
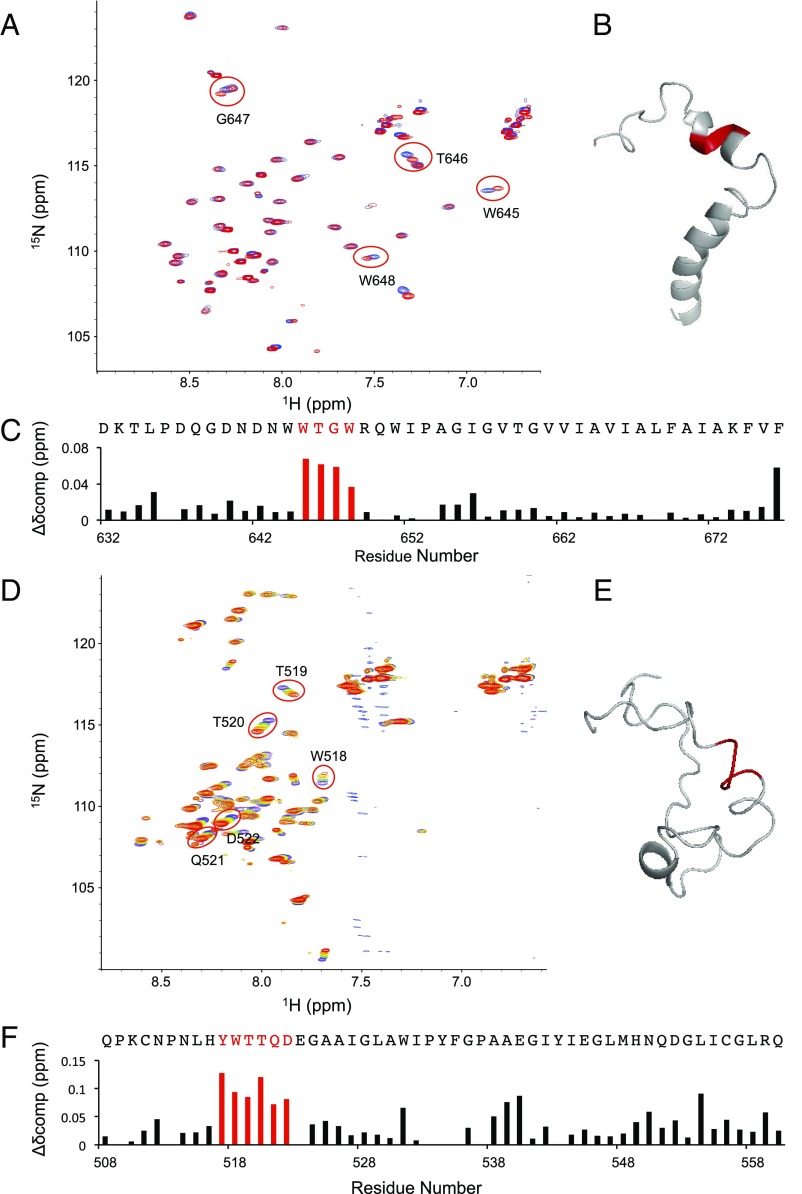
Structural and functional results suggest that the TM and fusion domains (fusion peptides or fusion loops) of class 1 viral fusion proteins might interact during a late stage in membrane fusion (27). However, there is relatively little direct experimental evidence that such interactions in fact occur between the relevant protein segments. An exception to this is some evidence for an interaction between the TM and fusion domains of HIV gp41 (28), but, even in this case, the interaction has not been demonstrated structurally. Like for fusion proteins of other class 1 viruses, the TM and fusion domains of EBOV consist of highly conserved sequences within each family of viruses. The results of this work, together with prior work from our laboratories, indicate that the MPER and N-terminal part of the TM regions of the MPER/TM domain and the FL (29) reside at a similar depth in lipid bilayers, allowing for a potential physical interaction between them in the plane of the membrane. To test this hypothesis, we collected HSQC spectra of the MPER/TM domain in DPC after adding increasing amounts of unlabeled FL and monitored the chemical shifts of affected residues (Fig. 4 A–C). The chemical shifts of most residues were unaffected by this titration, but significant chemical shift changes were observed for the MPER/TM residues Trp645, Thr646, Gly647, and Trp648. These four consecutive residues all reside in the short interfacial MPER helix. Perhaps surprisingly, the TM region of the domain was not affected by this titration. We also carried out the reciprocal titration, in which chemical shifts in the FL were monitored when increasing amounts of unlabeled MPER/TM were added (Fig. 4 D–F). In this case, significant chemical shift changes were observed for FL residues Tyr517, Trp518, Thr519, Thr520, Gln521, and Asp522. These sites of interaction are also located in a helical region of the FL, the helix that precedes the most hydrophobic front end of the pH 5.5 FL fist structure (18). The identified sites of interaction are consistent with the depths of membrane insertion of the MPER/TM and FL domains. They also support the notion that the two domains indeed interact with each other in membrane environments, consistent with current models of late steps of type 1 viral fusion protein-mediated membrane fusion.
Fig. 4.
NMR chemical shift perturbations upon the mutual interactions of the EBOV MPER/TM and FL domains. (A) HSQC spectra of 15N-labeled MPER/TM titrated with increasing amounts (blue to red) of unlabeled FL. (B) Structure of the MPER/TM domain with residues shown in red whose chemical shifts are most perturbed by the addition of the FL. (C) Chemical shift perturbations calculated as Δδcomp = [ΔδHN2 + (ΔδN/6.5)2]1/2 with the most significantly perturbed residues highlighted in red. (D) HSQC spectra of 15N-labeled FL titrated with increasing amounts (blue to red) of unlabeled MPER/TM domain. (E) Structure of the FL, with residues shown in red whose chemical shifts are most perturbed by the addition of the MPER/TM domain. (F) Chemical shift perturbations calculated as Δδcomp = [ΔδHN2 + (ΔδN/6.5)2]1/2 with the most significantly perturbed residues highlighted in red.
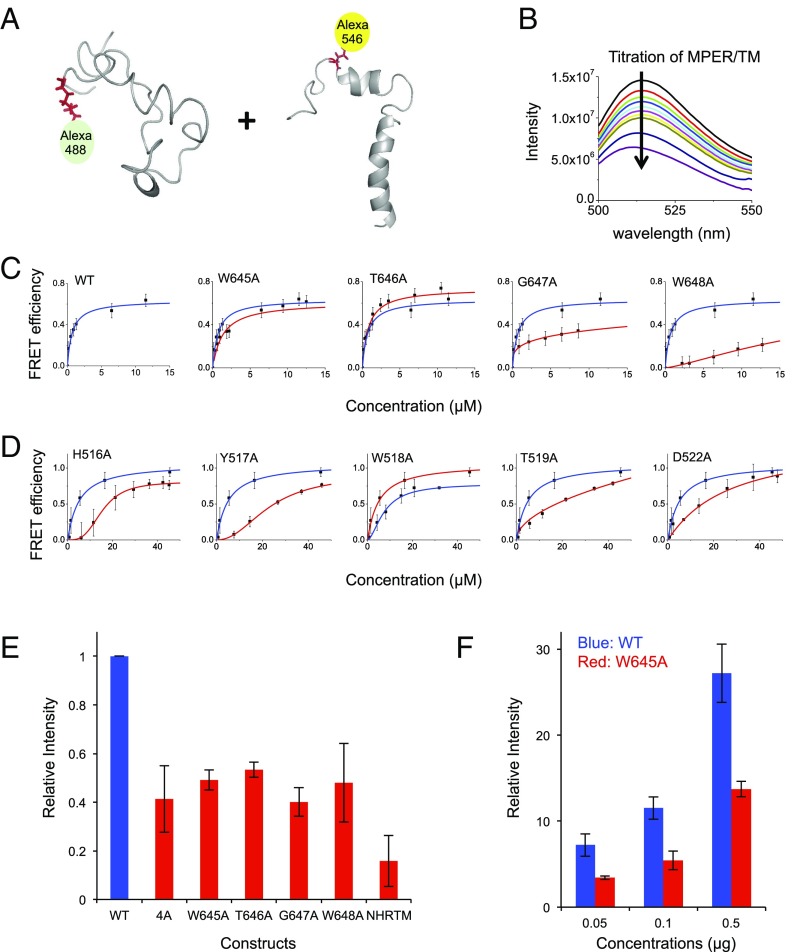
To further confirm the interaction between the membrane-active domains of EBOV GP2 and to assess the importance of the specific residues mediating this interaction, we labeled the FL with Alexa 488 and the MPER/TM domain with Alexa 546 fluorophors and performed Förster resonance energy transfer (FRET) experiments (Fig. 5 A and B). Strong binding of the two WT sequences to each other was observed. This binding was reduced when the four interacting residues of the MPER were changed to alanines in single site mutations (Fig. 5C). Similarly, five residues were replaced with alanines in the critical region of the FL structure (namely, His516, Tyr517, Trp518, Thr519, and Asp522), and all of these single site mutants exhibited reduced binding to the MPER/TM domain (Fig. 5D). All mutants expressed well and could be easily purified and refolded as described in Methods for the WT protein. The single site mutations unlikely introduced significant conformational changes to the WT structure, as shown by relatively unperturbed chemical shifts of the mutant molecules outside of the site that was mutated (SI Appendix, Fig. S9).
Fig. 5.
Functional consequences of EBOV FL-MPER/TM domain interaction site mutations. (A–D) Förster resonance energy transfer (FRET) showing the binding of Alexa 488-labeled FL to Alexa 546-labeled MPER/TM domain and their single site mutants in DPC micelles at pH 5.5. (A) Molecular models of the FL and MPER/TM domain, with sites where respective labels were attached. (B) Representative dynamic range of FRET experiment upon EBOV MPER/TM titration to EBOV FL. (C) Effect of various single site mutations in the MPER/TM domain upon titration of MPER/TM domain to the FL. (D) Effect of various single site mutations in the FL upon titration of the MPER/TM domain to the FL. In C and D, the data for binding with the mutated residues are shown in red, and the WT data are reproduced in blue as references in each plot. (E) Lipid mixing fusion assay with reconstituted proteoliposomes containing WT and several mutant full-length GP2 proteins. Most are single-site mutations, but the 4A construct contains four consecutive alanine mutations on the MPER region residues Trp645 to Trp648. The NHRTM construct, which was used as a control, comprises the GP2 sequence starting with the NHR region: i.e., lacking the entire FL. (F) Entry of WT and mutant virus-like particles (VLPs) into HEK 293AD cells. The critical FL-interactive residue Trp645 in the MPER region promotes cell entry at various concentrations compared with an alanine in this position.
Functional Confirmation of EBOV MPER/TM and FL Interaction Sites.
To investigate if the residues that are critical for the interaction between MPER/TM and the FL are also critical for EBOV GP2-mediated membrane fusion, we examined the effects of these same mutations on lipid mixing in fusion experiments in vitro (Fig. 5E). To this end, full-length GP2 was reconstituted into proteoliposomes (virosomes) that were then reacted with fluorescent lipid-labeled liposomes to measure fusion between the virosomes and liposomes at various pH values (SI Appendix, Fig. S10). This system gave much better control of the fusion reaction and ensured a precise 1:1 stoichiometry of the FL and MPER/TM domains, compared with a less well-defined fusion reaction that could have been performed with the two unconnected GP2 fragments. When all four critical residues on MPER were simultaneously replaced by alanines, fusion decreased by 60%, compared with WT GP2-mediated fusion (Fig. 5E). Interestingly, all single alanine mutants in this region produced similar deficiencies in fusion activity. When, in a control experiment, the entire FL was deleted from GP2 (NHR-TM construct), the residual fusion activity was less than 20% of the full-length WT GP2 fusion activity. These results prove that the FL is needed for observing appreciable fusion in this system and that each of the four interacting residues of the MPER is critical to support this activity.
To confirm that the MPER region and its interaction with the FL is also important for virus entry into cells, we prepared virus-like particles (VLPs) expressing GP1/GP2 on their surface with the W645A mutation built into the GP2 protein. W645A VLPs exhibited a cell entry efficiency of only about 40% of WT VLPs (Fig. 5F). This result demonstrates that the MPER helix is critical for virus entry into cells and, in combination with the presented domain interaction studies, that the critical function of MPER is to directly interact with the FL of EBOV GP2. We also attempted to perform the same experiment with the single MPER mutant W648A. In contrast to W645A, this mutant either did not express well or did not traffic correctly to the cell surface in VLP-producing cells so that we could not obtain VLPs with sufficient copy numbers of GP1/GP2 on their surface (SI Appendix, Fig. S11). Similarly, a previous study showed that the W518A mutation in the FL did not express well on the surface of VLPs (29). Therefore, membrane-interfacial tryptophans of EBOV GP2 seem to be important not only for fusion, but also for the biogenesis of the viral particles.
Discussion
Class 1 viral fusion glycoproteins, including those of influenza virus, HIV, and EBOV, are characterized by similar domain structures consisting of N-terminal or near N-terminal fusion domains, NHR and CHR regions, followed by MPER and TM domain regions (30–32). Depending on the particular virus, these domains are connected by shorter or longer loop regions. In the resting state on the viral surface, the NHR regions typically form a three-helix bundle, but the CHR regions are embedded elsewhere in the structures of these proteins. Upon activation, the fusion domains are exposed to the lipid bilayer of the target membrane, and the CHR regions fold back onto the NHR trimers to form six-helix bundle structures (Fig. 6 B–D). Whether or not the NHR trimers temporarily dissociate in the course of these transitions is currently debated (33). Recent evidence suggests that, at least in the case of HIV gp41, connected NHR and CHR constructs “melt” into the lipid bilayer as monomeric amphipathic helices (34, 35). Whether such a temporary dissociation of the NHR and CHR domains also occurs for EBOV GP2 in the presence of lipids is currently not known. However, structures of the EBOV FL have been determined at pH 7 and pH 5.5, and residues supporting a structural transition from an extended to a more compact “fist”-like loop have been identified (18). The pH trigger cannot be localized to a single or even a few isolated residues on the FL but seems to be distributed over many residues (36). Models, such as those depicted in Fig. 6 B–D, put the fusion domains in close contact with the TM and MPER domains, and there is some biophysical evidence for an interaction between these domains in the literature for a few class 1 fusion proteins (21, 28, 37, 38). However, structural evidence for this interaction has not yet been presented for any of these fusion proteins. In fact, there are almost no TM domain structures of viral fusion proteins that have been solved to date. Exceptions are the TM domains of influenza hemagglutinin (HA) and HIV gp41, whose structures have been solved in the organic solvent hexafluoroisopropanol (39, 40) and in DMPC/DHPC bicelles for the case of the HIV gp41 (41). The secondary structure, oligomerization, and orientation of the HA TM domain were also determined in lipid bilayers (42). As a first step toward solving this problem for EBOV GP2, we developed an expression and purification system that allowed us to obtain large quantities of a construct that comprised the MPER and TM domain regions of EBOV GP2. The construct behaved well in DPC micelles and DMPC/DHPC lipid bicelles so that a structure could be solved by NMR and its disposition in the membrane could be determined by PRE and EPR experiments.
Fig. 6.
Structural model of the proposed FL-MPER/TM domain interaction. (A) Proposed structural model of the two membrane-interacting domains of EBOV GP2 in a bilayer, with the most significant residues contributing to the binding interface highlighted in green. (B–D) Standard model of the three major steps thought to be involved in the membrane fusion process of type 1 viral fusion glycoproteins including EBOV GP2. (B) Extended prehairpin structure of GP2, with the N-terminal and C-terminal heptad repeats (NHR and CHR) labeled in addition to the FL and MPER/TM inserted into the target and viral membrane, respectively. (C) Six-helix bundle structure of GP2 and formation of a lipid-connected hemifusion intermediate. (D) Completed membrane fusion pore, with postfusion six-helix bundle structures of GP2 with the interacting FL and MPER/TM domains as determined in this work.
The structure of the EBOV MPER/TM domain consists of a helix-turn-helix motif, and the short MPER helix lies in the membrane interface, well-embedded in the lipid headgroup region. Two critical tryptophans, Trp645 and Trp648, which are part of this helix, seem to be responsible for this location. This structure is different from a model of the HIV MPER/TM domain that was proposed to be a straight helix for the MPER/TM domain transition region and to have a kink in the TM region based on partial NMR structures obtained in organic solvent of fragments of the entire sequence (40). Other authors found the HIV TM domain (without the MPER domain) to be a well-ordered trimer in DMPC/DHPC bicelles (41). Although the structural ensemble presented in Fig. 1F shows a well-defined turn between the two helices, we acknowledge that the definition of this turn depends to some extent on the stringency of the experimental distance and angle information applied to the structure calculation. Despite the relatively limited number of experimental NMR restraints in the turn region, we feel that the models are good representations of the structures. Importantly, our DEER experiments confirmed that the MPER and TM helices are roughly perpendicular to one another and that the angle between them is approximately preserved between the structures in micelles, bicelles, and bilayers in nanodiscs. Moreover, the refined structure that was obtained with the EPR restraints in lipid bilayers (Fig. 3D) did not indicate major contradictions between the micelle and bilayer structures. The main conclusion of all of these results combined from multiple magnetic resonance techniques is that the MPER domain is helical and well-positioned in the membrane interface to interact with the EBOV FL.
Trp645 and Trp648 and the intervening residues Thr646 and Gly647 of the MPER region were also critical for interaction with the EBOV FL, based on our chemical shift perturbation data (Fig. 4). Beyond more indirect evidence for an interaction of the HIV gp41 fusion peptide with its TM domain (28), this study offers direct structural evidence for a fusion domain–MPER/TM domain interaction in EBOV GP2, i.e., an interaction that may occur in many other or all class I viral fusion proteins. Interestingly, the EBOV FL also features a tryptophan, Trp518, which is central to its interaction with the MPER/TM domain. The predominant interaction region of the FL is characterized by a distorted helical turn comprised of the sequence Tyr517-Trp-Thr-Thr-Gln-Asp521 (Fig. 4). Interestingly, we found earlier that full-length GP1/GP2 with the W518A mutation could not be expressed well to form VLPs (29). It thus appears that this residue of the FL is also critical for proper folding of the FL in the full-length protein. We found the same folding problem with the W648A MPER region mutation in the current work. The interacting surfaces of the two protein domains fit together approximately as shown in the model of Fig. 6A. Although this model (and similar models built using the docking programs ZDOCK and HADDOCK) is useful to conceptually visualize the interaction, this is not an atomic structural model. The model is based on bringing the residues with significant chemical shift perturbations into close contact and does not rely on an actual structure determination of the complex. It is possible that both structures shift upon interaction and that the fluid-ordered structure of the lipid bilayer might influence details of the interaction.
Despite these caveats, we are confident that the interaction does take place via the highlighted residues on both the FL and MPER/TM domain sides. This notion is corroborated by our interaction and functional studies using mutations in the critical regions of both interaction partners. Replacements of MPER residues Gly647 and Trp648 with alanines were more severe in inhibiting association with the FL than replacements of the MPER region residues Trp645 and Thr646 (Fig. 5C). The reason for this behavior may be that Gly647 and Trp648 in the MPER structure are better exposed to interact with the FL than the other two residues. On the FL side, replacements of all residues in the identified interaction region had quite significant effects on binding to the MPER/TM domain. Some of these mutations led to different shapes of the FRET-based binding curves, which we do not completely understand and did not attempt to further analyze. The main point is that mutations of all of the critical residues identified by NMR in the FL altered binding to the MPER/TM domain. The functional fusion data of Fig. 5 E and F further confirm that disruption of the FL-MPER/TM domain interaction by multiple alanine mutations on the MPER side compromised fusion of reconstituted GP2 proteoliposomes with protein-free liposomes and virus entry in a cell-based VLP entry assay.
It is interesting that the FL apparently does not interact with the TM portion of the MPER/TM domain. This is consistent with the shallow location of the FL in the membrane that we determined previously (29). On the other hand, it has been shown that the TM domain of influenza HA is critical for late steps in fusion because HA constructs with short TM domains were arrested at a hemifusion stage (43), even though the influenza HA fusion peptide also resides in an interfacial location in the membrane (44, 45). The fusion peptide of HIV gp41 is also interfacially located (46). These structural and functional results are not contradictory. It is likely that the fusion domains first locate in the membrane interface to interact with the MPER regions of their own proteins and penetrate the membrane more deeply only in a later step: i.e., when the bilayers get distorted and form a hemifusion intermediate. It could well be that they would interact with the TM domain deep in the membrane during such late fusion transitions. Such a mechanism has been proposed for the fusion peptide of parainfluenza virus 5 (38). Several critical aromatic residues (Tyr517 and Trp518 on the FL and Trp645 and Trp648 on the MPER domain) are likely important for the initial location of these domains in the membrane interface. GXXXG/A sequence motifs may become important for later protein–protein interactions deeper in the membrane. Such motifs are well known to promote helix–helix interactions in membranes (47). The EBOV GP2 has a conserved GXXGXXXA (residues 657 to 664) motif in its TM domain, which may interact with the GXXXG motif in residues 524 to 528 or the GPXXXG motif in residues 536 to 541 of the FL. Previous studies, in which these motifs in the TM domains of other viruses were changed by mutation, have demonstrated their importance in membrane fusion (48–50). Alternatively, such motifs have also been shown to bind cholesterol (51), which is important for fusion of some enveloped viruses and SNARE-mediated intracellular membrane fusion (52).
In summary, we found that the structure of the EBOV MPER/TM domain is independent of pH and consists of a membrane interfacial-membrane spanning helix-turn-helix motif. We also provide physical evidence for the interaction of the MPER helix with the FL in the membrane interface and show that this interaction is important for fusion and cell entry of EBOV. Crucial tryptophans and a tyrosine in the MPER and FL domains localize both domains to the membrane interface where they mediate their mutual interaction and membrane fusion.
Methods
Design and Cloning of the MPER/TM Domain and GP2 Constructs.
To express the EBOV MPER/TM domain and full-length GP2, fusion proteins were designed with the Trp leader protein in front of a thrombin cleavage site and the MPER/TM or GP2 sequence. The two palmitylation sites C670 and C672 in the MPER/TM domain were mutated to alanines to avoid the formation of nonnative disulfides. Previous studies have shown that this modification does not result in loss of the virus’s capability to enter host cells (53, 54). The Ebola MPER/TM or GP2 genes were cloned into the pET-24a vector containing the Trp leader protein, which was a kind gift from K. J. Glover, Lehigh University, Bethlehem, PA. The vector contains a kanamycin resistance site and an N-terminal His-tag for purification by Ni affinity chromatography. Details of the cloning are described in SI Appendix, SI Methods.
Expression and Purification of the MPER/TM Domain, GP2, and FL Domain.
The expression and purification of the EBOV MPER/TM domain and the entire GP2 subunit were carried out using a method described previously for caveolin TM domains (55) with some modifications that are described in SI Appendix, SI Methods. The FL of EBOV was prepared as previously described (18).
Bicelle and Nanodisc Sample Preparation.
The bicelle and nanodisc samples containing EBOV constructs were prepared by the following protocol. For EBOV MPER/TM incorporation into bicelles or nanodisc, DMPC liposomes were prepared in 20 mM Na phosphate, 100 mM NaCl, pH 7.4, buffer by extrusion through 100-nm polycarbonate filters. Then, 20 mM Na phosphate, 100 mM NaCl, pH 7.4, buffer containing 1% n-octyl-β-d-glucopyranoside (β-OG) was added to the extruded liposomes to a final concentration of 0.125% β-OG and then incubated at room temperature for at least 1 h. Then, EBOV MPER/TM in DPC was added to give an estimated protein:lipid ratio of 1:200 and incubated for at least 1 h before dialysis. Extensive dialysis against 20 mM Na phosphate, 100 mM NaCl, pH 7.4, buffer was performed to remove all detergent and to incorporate EBOV MPER/TM into the liposomes. After formation of the proteoliposomes, samples were concentrated, and buffers were exchanged to the desired final buffers. Appropriate amounts of DHPC or scaffolding proteins were added to form bicelles or nanodiscs, respectively. Even if proteoliposome solutions were somewhat turbid because of the high concentration of lipids, they became fully transparent when DHPC or scaffolding proteins were added. The longer EBOV constructs were prepared in the same manner.
NMR Experiments and Structure Calculation.
All NMR spectra of the MPER/TM domain were acquired at 30 °C on Bruker Avance III 600- or 800-MHz spectrometers, except for the experiments in bicelles and full-length GP2, which were recorded at 45 °C. Pulse programs from the Bruker Topspin suite were used to obtain heteronuclear 2D and 3D spectra and to measure T1 and T2 relaxation times. HSQC, HNCA, HNCOCA, HNCO, HNCACO, and HNCACB spectra were recorded to obtain backbone information. For side-chain assignments, HCCONH- and HCCH-total correlated spectroscopy (TOCSY) experiments were performed. 15N-edited NOESY and 13C-edited NOESY were performed with 120-ms mixing times to obtain NOE-based distance information. An aromatic 13C-edited NOESY experiment was also performed with 80-ms mixing time to obtain additional distance restraints. All NMR data were processed using NMRPipe and SPARKY (56, 57). Dihedral angle restraints were calculated using TALOS+, and CYANA was used to obtain initial structures (58, 59). Two hundred structures were next calculated using CNS (60), and the final 20 lowest energy structures were chosen to represent the overall structure of the MPER/TM domain. PROCHECK-NMR (61) was used to validate the structural ensemble, and MOLMOL (62) and PYMOL (63) were used to visualize the structures.
Spin-Label EPR Spectroscopy.
Site-directed nitroxide-labeled samples of MPER/TM for EPR spectroscopy were prepared by introducing single cysteine mutations in the MPER/TM domain as described previously (64). All samples were treated with DTT before they were reacted with the methane-thiosulfonate spin-label (MTSL) overnight at room temperature. All buffers were degassed before use, and DTT was removed on a PD-10 column before the MTSL coupling reaction. Excess of MTSL was removed on a PD-10 column when MTSL coupling reaction was complete. The spin-labeled proteins were incorporated into POPC:POPG (85:15) liposomes at a protein:lipid molar ratio of 1:400 by combining appropriate amounts of each component in 20 mM Tris, 100 mM NaCl, 1% n-octyl-β-d-glucopyranoside, pH 8, followed by extensive dialysis against 20 mM Na phosphate, 100 mM NaCl buffer, pH 7.0. Continuous-wave power saturation EPR measurements were performed on a Bruker X-band EMX EPR spectrometer. Power saturation curves were obtained from the peak-to-peak amplitudes of the central (M = 0) lines of the first derivative EPR spectra under three conditions: equilibrated with O2, equilibrated with N2, and equilibrated with N2 in the presence of 20 mM Ni-ethylenediamine-N,N′-diacetic acid (NiEDDA). The depths of the spin labels in the lipid bilayer were derived from the depth parameter Φ = ln{[P1/2(O2) − P1/2(N2)]/[P1/2(NiEDDA) − P1/2(N2)]} (65).
For DEER experiments, ∼15 μL of sample with 15% deuterated glycerol were loaded into quartz capillary tubes with an inner diameter of 1.1 mm and outer diameter of 1.6 mm (Vitrocom). Samples were frozen in liquid nitrogen and loaded into a Bruker E580 spectrometer with an EN5107D2 resonator. DEER data were collected at Q-band and 80 K using a dead-time free four-pulse sequence with 16-step phase cycling (66). Pump and observe pulses were separated by 75 MHz. DeerAnalysis2015 was used for the removal of background functions from initial V(t)/V(0) data, and Tikhonov regularization was used to extract distance distributions from the resulting F(t)/F(0) form factors (67). Error ranges shown are within 15% root-mean-square deviation of the best fit. The distances obtained from DEER experiments were compared with predicted distance distributions. The three lowest energy structures were selected from the family of structures generated from structure calculation using XPLOR-NIH and the implicit membrane potential. For each structure, the program MMM was then used to generate predicted distances for each of the spin label pairs used in the DEER experiments (68). In this program, we used a rotamer library for the R1 side chains that was based on the use of a density functional theory (69).
Structure Calculation Within the Implicit Membrane Model.
The NMR structure of the MPER/TM domains was recalculated using XPLOR-NIH, but with an implementation of the EPR-derived membrane depth information as additional structural restraints (70, 71). The initial folding and refinement were undertaken using only NMR restraints and an implicit membrane potential (eefx) with scripts provided by Charles Schwieters, NIH, Bethesda. Briefly, the extended structure of the MPER/TM domain was generated, and NOE and dihedral angle restraints were initiated. Then, calculations in the implicit membrane with a hydrophobic thickness of 27 Å and with 5 ∼ 6 Å for the lipid headgroups on both sides of a standard POPC bilayer were initiated. The first minimization was performed in an effective vacuum, and the protein was translated to the membrane center. After an annealing cycle was performed, the annealed structure was subjected to Cartesian minimization. The lowest energy structure generated was subjected to a further refinement cycle, which was similar to that applied to the extended structure described above. After refinement, the lowest energy structure was loaded into the program MMM (68), and the site-scan feature was used to compute rotamers for the MTSL in positions 643, 644, 666, and 670 using the Warsh rotamer library. Next, another annealing step was performed on the obtained structure. In this step, the segment comprising residues 645 to 676 was treated as a single rigid body based on PRE data, and the backbone atoms, beta carbons, and the proximal sulfur atom were grouped together for residues 643 and 644. This resulted in a fixed spin label conformation for residues 666 and 670 while residues 643 and 644 were effectively free with regard to rotations of the χ2–χ5 angles.
Förster Resonance Energy Transfer Experiments.
For FRET experiments, a single cysteine mutation was introduced in position 643 of WT and various mutant EBOV MPER/TM domains. We figured that Asn643, which is right before the MPER helix, would not affect its structure and interaction, yet would be in close enough proximity to the FL interaction site to sense the interaction by FRET. Expression and purification of the mutant MPER/TM domains and EBOV FLs were performed as described in SI Appendix, SI Methods, and all buffers were degassed before use. Protein samples in DPC were treated with a 20-fold molar excess of DTT for 2 h at room temperature and passed over a PD-10 column, and labels were attached per the manufacturer’s (Life Technologies) instruction. Alexa 546 C5 maleimide was conjugated to the MPER/TM domain using cysteine chemistry, and Alexa 488 succinimidyl ester was conjugated to the single lysine 510 of the FL using primary amine chemistry because the two cysteines of the sequence are required for functional disulfide bond formation within the FL (18). The amine labeling of the FL was done before the N-terminal His-tag was cleaved with factor X (18), which ensured that any label on the N-terminal amine was removed by factor X cleavage and that a single label was present on Lys510. To measure FRET, 0.1 μM FL-Alexa 488 in DPC was placed in a quartz cuvette of a Jobin–Yvon Fluorolog 3 spectrometer, and increasing amounts MPER/TM-Alexa 546 in DPC were titrated to this solution. The emission spectra were recorded from 500 to 600 nm at each titration point with the excitation wavelength set at 488 nm. Excitation and emission slit widths were set to 1 nm. Spectra without FL-Alexa 488 were collected and used to subtract background.
Fusion Assay of GP2 Proteoliposomes by Lipid Mixing.
WT and mutant full-length GP2 proteins were reconstituted into unlabeled POPC:POPG (85:15) liposomes at a protein:lipid ratio of 1:100 by detergent dialysis. Briefly, unlabeled liposomes were prepared in 10 mM Hepes/Mes/Tris, 100 mM NaCl, pH 7.4 (HMA) buffer by extrusion through 100-nm polycarbonate filters. Then, 20 mM Tris, 100 mM NaCl, pH 8.0, buffer containing 1% n-octyl-β-d-glucopyranoside (β-OG) was added to the extruded liposomes to a final concentration of 0.125% β-OG and then incubated at room temperature for at least 1 h. Then, GP2 in DPC was added to give an estimated protein:lipid ratio of 1:100 (based on absorbance at 280 nm; typically around 20 μL of GP2 was added to an ∼0.5-mL lipid/detergent solution) and incubated for at least 1 h before dialysis. Extensive dialysis against HMA buffer was performed to remove all detergent and to incorporate GP2 into the liposomes. Bicinchoninic acid (BCA) and phosphate assays were performed to confirm that the actual protein:lipid ratios of all constructs were ∼1:100: i.e., close to the input ratio.
Lipid mixing fusion assays were performed using a SpectraMaxM5 plate reader as described previously with slight modification (18). Liposomes composed of POPC:POPG (85:15) with 1.5 mol% rhodamine(Rh)-1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and N-(7-nitro-2-1,3-benzoxadiazol-4-yl) [NBD]-DOPE each were prepared by extrusion of appropriately mixed lipid dispersions in HMA buffer (10 mM Hepes, 10 mM Mes, 10 mM Na acetate, and 100 mM NaCl, pH 7.4) through 100-nm-pore-size polycarbonate filters. To measure fusion, fluorescent liposomes and unlabeled proteoliposomes were mixed at a ratio of 1:9 in HMA buffer. Relief of NBD-Rh FRET (i.e., an increase in NBD fluorescence) was recorded at room temperature as a function of time with mixing between each reading. Excitation and emission wavelengths were set at 460 nm and 538 nm, respectively. Percent lipid mixing (fusion) was determined as the fraction of the maximal FRET relief observed after addition of 2% Triton X-100 at the end of each reaction. All mutant lipid mixing data were normalized to the extent of lipid mixing observed with the WT protein.
Virus-Like Particle Production and Entry Assay.
The production of EBOV VLPs and the assay to measure their entry into cells were performed as described previously (72). In brief, HEK293T/17 cells were cotransfected with plasmids encoding for VP40, β-lactamase-VP40, mCherry-VP40, and WT, W645A, or W648A GP with a C-terminal V5 tag using the polyethylenimine method. The ratio of respective plasmids was 1:2.25:2.25:1.5. After 48 h of incubation at 37 °C, the cell medium was collected and cleared of cellular debris by centrifugation, and the cleared media supernatant containing VLPs was ultracentrifuged through a 20% sucrose cushion. The VLP pellets were resuspended in HM buffer (20 mM Hepes, 20 mM Mes, 130 mM NaCl, pH7.4) overnight and subsequently repelleted. The final VLP pellets were resuspended in 10% sucrose-HM buffer, aliquotted, and stored at −80 °C. All VLPs were analyzed by Western blotting for the presence of GP and VP40, and total protein concentrations were determined by BCA assay. To measure cell entry, VLPs were spinfected onto 293AD cells (250 × g) for 1 h at 4 °C and then allowed to enter for 3 h at 37 °C in a 5% CO2 incubator. The fluorescent CCF2-AM β-lactamase substrate was then fed to the cells for 1 h at room temperature in the dark. Cells were then washed and incubated overnight in the dark at room temperature. Cells were fixed and analyzed by flow cytometry on a FACSCalibur flow cytometer. The degree of the shift in fluorescence from green to blue was used to measure entry (73). All data were analyzed using FlowJo software.
Supplementary Material
Acknowledgments
We thank Dr. Kerney J. Glover (Lehigh University) for the pET-24a expression plasmid and Dr. Charles Schwieters (NIH) for providing scripts for the implicit membrane model used in the membrane-based structure refinements. We also thank Sarah B. Nyenhuis for help with the EPR power saturation experiments and Alyson Barnes for help with VLP entry assays. This work was supported by NIH Grants R01 AI030557 (to L.K.T.), R01 AI114776 (to J.M.W.), and P01 GM072693 (to L.K.T. and D.S.C.). D.A.N. is supported by NIH Training Grant T32 GM080186.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5T42), and the NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 30162).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708052114/-/DCSupplemental.
References
- 1.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoenen T, Groseth A, Falzarano D, Feldmann H. Ebola virus: Unravelling pathogenesis to combat a deadly disease. Trends Mol Med. 2006;12:206–215. doi: 10.1016/j.molmed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 3.de La Vega MA, Stein D, Kobinger GP. Ebolavirus evolution: Past and present. PLoS Pathog. 2015;11:e1005221. doi: 10.1371/journal.ppat.1005221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization 2016 Ebola situation reports. Available at apps.who.int/ebola/ebola-situation-reports. Accessed June 2017.
- 5.Hunt CL, Lennemann NJ, Maury W. Filovirus entry: A novelty in the viral fusion world. Viruses. 2012;4:258–275. doi: 10.3390/v4020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nanbo A, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mingo RM, et al. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: Evidence that transport to NPC1+ endolysosomes is a rate-defining step. J Virol. 2015;89:2931–2943. doi: 10.1128/JVI.03398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JM, Schornberg KL. A new player in the puzzle of filovirus entry. Nat Rev Microbiol. 2012;10:317–322. doi: 10.1038/nrmicro2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller EH, Chandran K. Filovirus entry into cells – New insights. Curr Opin Virol. 2012;2:206–214. doi: 10.1016/j.coviro.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schornberg K, et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Côté M, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dube D, et al. Cell adhesion promotes Ebola virus envelope glycoprotein-mediated binding and infection. J Virol. 2008;82:7238–7242. doi: 10.1128/JVI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory SM, et al. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc Natl Acad Sci USA. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malashkevich VN, et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 21.Liao Y, Zhang SM, Neo TL, Tam JP. Tryptophan-dependent membrane interaction and heteromerization with the internal fusion peptide by the membrane proximal external region of SARS-CoV spike protein. Biochemistry. 2015;54:1819–1830. doi: 10.1021/bi501352u. [DOI] [PubMed] [Google Scholar]
- 22.Salzwedel K, West JT, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sáez-Cirión A, Gómara MJ, Agirre A, Nieva JL. Pre-transmembrane sequence of Ebola glycoprotein. Interfacial hydrophobicity distribution and interaction with membranes. FEBS Lett. 2003;533:47–53. doi: 10.1016/s0014-5793(02)03747-x. [DOI] [PubMed] [Google Scholar]
- 24.Regula LK, et al. Conformational properties of peptides corresponding to the ebolavirus GP2 membrane-proximal external region in the presence of micelle-forming surfactants and lipids. Biochemistry. 2013;52:3393–3404. doi: 10.1021/bi400040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison JS, Higgins CD, Chandran K, Lai JR. Designed protein mimics of the Ebola virus glycoprotein GP2 alpha-helical bundle: Stability and pH effects. Protein Sci. 2011;20:1587–1596. doi: 10.1002/pro.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison JS, Koellhoffer JF, Chandran K, Lai JR. Marburg virus glycoprotein GP2: pH-dependent stability of the ectodomain α-helical bundle. Biochemistry. 2012;51:2515–2525. doi: 10.1021/bi3000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamm LK, Han X, Li Y, Lai AL. Structure and function of membrane fusion peptides. Biopolymers. 2002;66:249–260. doi: 10.1002/bip.10261. [DOI] [PubMed] [Google Scholar]
- 28.Reuven EM, et al. HIV-1 gp41 transmembrane domain interacts with the fusion peptide: Implication in lipid mixing and inhibition of virus-cell fusion. Biochemistry. 2012;51:2867–2878. doi: 10.1021/bi201721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory SM, et al. Ebolavirus entry requires a compact hydrophobic fist at the tip of the fusion loop. J Virol. 2014;88:6636–6649. doi: 10.1128/JVI.00396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilen CB, Tilton JC, Doms RW. HIV: Cell binding and entry. Cold Spring Harb Perspect Med. 2012;2:a006866. doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison SC. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White JM, Whittaker GR. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamm LK, Lee J, Liang B. Capturing glimpses of an elusive HIV gp41 prehairpin fusion intermediate. Structure. 2014;22:1225–1226. doi: 10.1016/j.str.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roche J, Louis JM, Grishaev A, Ying J, Bax A. Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion. Proc Natl Acad Sci USA. 2014;111:3425–3430. doi: 10.1073/pnas.1401397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis JM, et al. Insights into the conformation of the membrane proximal regions critical to the trimerization of the HIV-1 gp41 ectodomain bound to dodecyl phosphocholine micelles. PLoS One. 2016;11:e0160597. doi: 10.1371/journal.pone.0160597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Gregory SM, Nelson EA, White JM, Tamm LK. The roles of histidines and charged residues as potential triggers of a conformational change in the fusion loop of Ebola virus glycoprotein. PLoS One. 2016;11:e0152527. doi: 10.1371/journal.pone.0152527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang DK, Cheng SF, Kantchev EA, Lin CH, Liu YT. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008;6:2. doi: 10.1186/1741-7007-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donald JE, et al. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc Natl Acad Sci USA. 2011;108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mineev KS, et al. Structural investigation of influenza virus hemagglutinin membrane-anchoring peptide. Protein Eng Des Sel. 2013;26:547–552. doi: 10.1093/protein/gzt034. [DOI] [PubMed] [Google Scholar]
- 40.Apellániz B, et al. The atomic structure of the HIV-1 gp41 transmembrane domain and its connection to the immunogenic membrane-proximal external region. J Biol Chem. 2015;290:12999–13015. doi: 10.1074/jbc.M115.644351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dev J, et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science. 2016;353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatulian SA, Tamm LK. Secondary structure, orientation, oligomerization, and lipid interactions of the transmembrane domain of influenza hemagglutinin. Biochemistry. 2000;39:496–507. doi: 10.1021/bi991594p. [DOI] [PubMed] [Google Scholar]
- 43.Armstrong RT, Kushnir AS, White JM. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat Struct Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 45.Lai AL, Tamm LK. Shallow boomerang-shaped influenza hemagglutinin G13A mutant structure promotes leaky membrane fusion. J Biol Chem. 2010;285:37467–37475. doi: 10.1074/jbc.M110.153700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai AL, Moorthy AE, Li Y, Tamm LK. Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. J Mol Biol. 2012;418:3–15. doi: 10.1016/j.jmb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 48.Langosch D, Brosig B, Pipkorn R. Peptide mimics of the vesicular stomatitis virus G-protein transmembrane segment drive membrane fusion in vitro. J Biol Chem. 2001;276:32016–32021. doi: 10.1074/jbc.M102579200. [DOI] [PubMed] [Google Scholar]
- 49.Miyauchi K, et al. Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions. Jpn J Infect Dis. 2006;59:77–84. [PubMed] [Google Scholar]
- 50.Melikyan GB, Markosyan RM, Roth MG, Cohen FS. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol Biol Cell. 2000;11:3765–3775. doi: 10.1091/mbc.11.11.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett PJ, et al. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang ST, Kreutzberger AJ, Lee J, Kiessling V, Tamm LK. The role of cholesterol in membrane fusion. Chem Phys Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeffers SA, Sanders DA, Sanchez A. Covalent modifications of the ebola virus glycoprotein. J Virol. 2002;76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito H, Watanabe S, Takada A, Kawaoka Y. Ebola virus glycoprotein: Proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J Virol. 2001;75:1576–1580. doi: 10.1128/JVI.75.3.1576-1580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J, Glover KJ. The transmembrane domain of caveolin-1 exhibits a helix-break-helix structure. Biochim Biophys Acta. 2012;1818:1158–1164. doi: 10.1016/j.bbamem.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 57.Goddard TD, Kneller DG. 2008. SPARKY: NMR Assignment and Integration Software (University of California, San Francisco), Version 3.114.
- 58.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 59.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 60.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 61.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 62.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 63.Schrödinger LLC. 2015. The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.8.
- 64.Liang B, Kiessling V, Tamm LK. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proc Natl Acad Sci USA. 2013;110:19384–19389. doi: 10.1073/pnas.1314699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: Application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 67.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 68.Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys Chem Chem Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 69.Warshaviak DT, Serbulea L, Houk KN, Hubbell WL. Conformational analysis of a nitroxide side chain in an α-helix with density functional theory. J Phys Chem B. 2011;115:397–405. doi: 10.1021/jp108871m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marassi FM, Ding Y, Schwieters CD, Tian Y, Yao Y. Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation. J Biomol NMR. 2015;63:59–65. doi: 10.1007/s10858-015-9963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian Y, Schwieters CD, Opella SJ, Marassi FM. A practical implicit membrane potential for NMR structure calculations of membrane proteins. Biophys J. 2015;109:574–585. doi: 10.1016/j.bpj.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shoemaker CJ, et al. Multiple cationic amphiphiles induce a Niemann-Pick C phenotype and inhibit Ebola virus entry and infection. PLoS One. 2013;8:e56265. doi: 10.1371/journal.pone.0056265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yonezawa A, Cavrois M, Greene WC. Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: Involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J Virol. 2005;79:918–926. doi: 10.1128/JVI.79.2.918-926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.