Preface
The 4D Nucleome Network aims to develop and apply approaches to map the structure and dynamics of the human and mouse genomes in space and time with the goal of gaining deeper mechanistic understanding of how the nucleus is organized and functions. The project will develop and benchmark experimental and computational approaches for measuring genome conformation and nuclear organization, and investigate how these contribute to gene regulation and other genome functions. Validated experimental approaches will be combined with biophysical modeling to generate quantitative models of spatial genome organization in different biological states, both in cell populations and in single cells.
The human genome contains over 20,000 of genes, and a larger number of regulatory elements. Large-scale studies over the last decade have cataloged these components of our genome and the cell types in which they are active. The ENCODE, Roadmap Epigenome, International Human Epigenome Project, EpiGeneSys (http://www.epigenesys.eu/en/) and FANTOM projects1–4 annotated thousands of genes and millions of candidate regulatory elements. However, our understanding of the mechanisms by which they exert regulatory effects on specific target genes across distances of kilobases to in some cases megabases, is decidedly incomplete.
The spatial folding of chromosomes and their organization in the nucleus have profound impact on gene expression. For example, spatial proximity is necessary for enhancers to modulate transcription of target genes (e.g.5–7), and clustering of chromatin near the nuclear lamina is correlated with gene silencing and replication timing8,9. Meanwhile, genome-wide association studies have identified large numbers of disease-associated loci, and the majority of them are located in distal, potentially regulatory, noncoding regions (e.g.10). In cancer cells genomic rearrangements frequently occur and these are at least in part guided by the 3D organization of the nucleus11,12. These results emphasize the importance of distal elements for gene regulation and suggest an exciting opportunity to uncover fundamental mechanisms of disease through the mapping of long-range chromatin interactions and 3D genome organization. Therefore, to determine how the genome operates, we need to understand not only the linear encoding of information along chromosomes, but also its 3-dimensional organization and its dynamics across time, i.e. the “4D nucleome”. Concomitantly, we must pursue deeper knowledge of the biophysical and molecular factors that determine genome organization, and how this organization contributes to gene regulation and other nuclear activities. Here we outline the goals and strategies of the 4D Nucleome (4DN) Network. This Network builds on other consortia and efforts focusing on (epi-) genome analysis outlined above and adds the spatial and time dimensions to explore how the genome is organized inside cells and how this relates to genome function.
The nucleus is not a homogeneous organelle, but consists of distinct nuclear structures and non-chromatin bodies as well as defined chromosomal regions, such as centromeres, telomeres and insulator bodies, recognized to cluster with each other and other genomic regions to define distinct nuclear compartments13,14. Examples of nuclear structures include the nuclear periphery and the heterochromatic compartment, while examples of nuclear bodies include nucleoli, nuclear speckles, paraspeckles, and Cajal and PML bodies. Chromosome conformation capture (3C) approaches15,16 have yielded additional insights by characterizing chromatin folding genome-wide at kilobase-resolution6,17,18. These studies show that the genome is compartmentalized in active and inactive spatial compartments at the scale of the nucleus, and within each compartment folding of chromatin fibers brings together loci and regulatory elements that are otherwise separated by large genomic distances. CTCF, the cohesin complex, and other DNA binding proteins as well as RNAs play roles in organizing chromatin domains and long-range interactions between DNA loci18–24. These studies point to the genome being intricately organized within the nucleus, with this organization playing a critical role in gene regulation and activity.
The past decade witnessed significant innovation in chromosome and nuclear structure analysis. Genomic approaches for mapping chromatin interactions, such as 3C, 4C, 5C, Hi-C, and ChIA-PET16, are yielding genome-wide chromatin interaction maps at unprecedented resolution. Live cell and super-resolution microscopic approaches, combined with application of new ways (e.g. CRISPR/Cas9 –based systems) to visualize loci and sub-nuclear structures are beginning to provide detailed views of the organization and dynamics of chromatin inside (living) cells25–31. There has also been tremendous progress in analyzing chromosome structural data, producing structural models for chromosome folding32,33. However, despite this progress, a comprehensive understanding of the 4D nucleome is still lacking. This is partly due to the fact that different experimental cell systems and approaches are used, which together with the absence of shared benchmarks for assay performance have led to observations that cannot be directly compared. Additionally, we currently have limited ability to integrate different datatypes (e.g. chromatin interaction data and imaging-based distance measurements) and lack approaches that can measure and account for cell-to-cell variability in chromosome and nuclear organization. Finally, we lack mechanistic insights into the relationships between chromosome conformation and nuclear processes including transcription, DNA replication, and chromosome segregation. These major gaps can be addressed by a highly synergistic, multidisciplinary and integrated approach in which groups with different expertise and knowledge, ranging from imaging and genomics to computer science and physics, work closely together to study common cell systems using complementary methods.
Goals and strategy of the project
The 4DN Network aims to develop a set of approaches to map the structures and dynamics of the genome and to relate these features to its biological activities. The Network aims to generate quantitative models of nuclear organization in diverse cell types and conditions, including in single cells. Overall, we anticipate that these efforts will lead to new mechanistic insights into how the genome is organized, maintained, expressed, and replicated, in both normal and disease states.
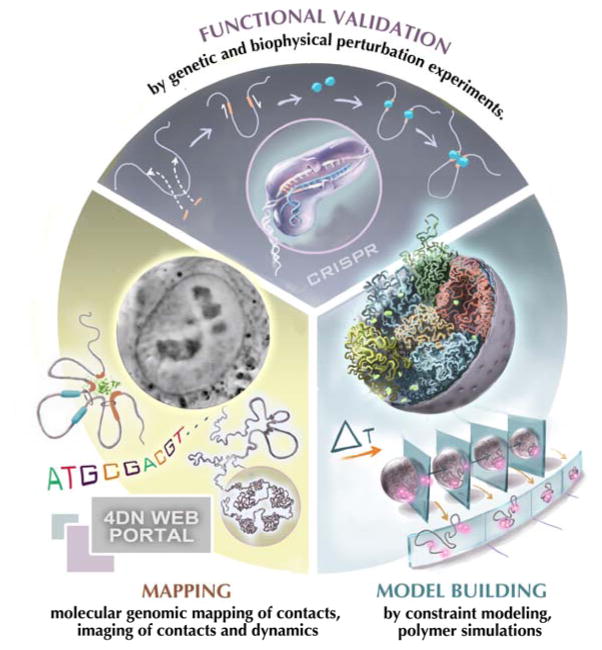
The 4DN Network will 1) develop, benchmark, validate, and standardize a wide array of technologies to probe the 4D nucleome; 2) integrate, analyze, and model datasets obtained with these technologies to obtain a comprehensive view of the 4DN; and 3) investigate the functional role of various structural features of chromosome organization in transcription, DNA replication and other nuclear processes. These three main components are illustrated in Fig. 1.
Figure 1. The 4D Nucleome project.
The project encompasses three components: First, experimental mapping approaches are employed to measure a range of aspects of the spatial organization of the genome including chromatin loops, domains, nuclear bodies etc. Second, computational and modeling approaches are used to interpret experimental observations and build (dynamic) spatial models of the nucleus. Third, perturbation experiments, e.g. using CRISPR/cas9-mediated genome engineering, are used for functional validation. In these studies chromatin structures are altered, e.g. removing chromatin loops, creating novel loops at defined positions or tethering regulatory components in selected regions in order to test their architectural function. These perturbation studies can be complemented with functional studies, e.g. analysis of gene expression to assess the functional implications of chromatin folding. (Picture of cell nucleus was provided by Hanhui Ma and Thoru Pederson).
To achieve these objectives, we defined following key steps. First, a set of common cell lines will be studied to enable direct cross-validation of data obtained with different methods (Table 1). Important criteria include a stable, haplotype-phased and normal karyotype, ease of growth, ease of genome editing, and suitability for (live-cell) imaging. Further, given that cell populations are characterized by cell-to-cell variation in their biological state (e.g. cell cycle stage), it will be important to employ clonal cell populations that can be synchronized, activated, induced or differentiated in a controlled manner.
Table 1.
Common cell lines used by the 4DN network. Tier 1 cells are used for all studies, while Tier 2 cell lines are used for specific projects.
| Tier 1 | Description | Availability |
|---|---|---|
| H1-ESC (WA01) | Human embryonic stem cells (male) | WiCell |
| hTert-HFF | hTert-immortalized human foreskin fibroblasts (male) | Dekker lab |
| IMR90 | Primary lung fibroblasts (female) | ATCC |
| GM12878 | EBV-immortalized B-lymphocytes (female) | Coriell |
|
| ||
| Tier 2 | Description | Availability |
|
| ||
| F121-9 | Mouse Cast-129 F1 hybrid embryonic stem cells (female) | Gilbert lab, FSU |
| F123 | Mouse Cast-129 F1 hybrid embryonic stem cells (male) | Jaenisch lab, MIT |
| JM8.N4 | Mouse C57BL/6N inbred embryonic stem cells (male) | KOMP/UCDavis |
| H9-ESC (WA09) | Human embryonic stem cells (female) | WiCell |
| hTert-RPE | hTert-immortalized retina pigmented epithelium (female) | ATCC |
| K562 | Chronic myelogenous leukemia (female) | ATCC |
| HEK293 | Human embryonic kidney (likely female) | ATCC |
| U-2 OS | Osteosarcoma cells (female) | ATCC |
| HAP1 | Haploid derivative of chronic myelogenous leukemia (CML) cell line KBM-7 (male) | van Steensel lab, NKI |
| WTC-11 | iPSC (male) | Conklin lab, Gladstone/UCSF |
Second, standards for data formats and quality will be established so that data can be shared broadly. This includes defining metrics for reproducibility and assessment of the sensitivity, specificity, resolution and precision with which aspects of the 4D nucleome can be measured.
Third, computational and analytical tools will be developed to analyze individual datasets and to integrate, compare and cross-validate data obtained with different technologies. Importantly, they will allow for the integration of the diverse datasets necessary to build comprehensive models of the 4D nucleome.
Fourth, genetic, biochemical and biophysical approaches will be developed to measure and perturb the roles of DNA sequences and trans-acting factors (proteins, RNA) in the formation of local and global aspects of the 4D nucleome and their impact on transcription and other nuclear functions.
Fifth, a common vocabulary will be developed to describe nuclear features and biophysically derived principles guiding chromosome folding. This is important because currently different structural descriptions and interpretations have been put forward to describe features detected by different technologies, or even by the same methods. We need better and more precise descriptions of the underlying reality of structural features that make up the 4D nucleome, e.g. loops and domains, and develop a consistent terminology as these features are detected by different technologies. This can be achieved by integrated analysis of data obtained with the wide range of technologies employed and under development by the Network.
A major goal is to compare and integrate the wealth of information that is anticipated to be generated by the Network. This will allow both benchmarking of experimental and computational approaches and better interpretation of what each data type (e.g. chromosome conformation capture data on the one hand and imaging on the other hand) reveals about the structure, dynamics and cell-to-cell variation in folding of chromosomes. The Network will analyze a small set of common cell lines (Table 1), and select a set of loci, to be studied by all participating groups. A joint analysis group with members from across the Network will integrate and analyze this diverse set data to produce benchmarks for each methodology and provide an integrated view of the 4D nucleome. Such a first modeling of the 4D nucleome can produce models that represents the folded state of chromosome and how this is dynamic in real time, variable between cells, and how it relates to gene regulation.
Finally, to facilitate rapid dissemination of data to the larger scientific community, a shared database and a public 4DN data browser will be established which includes all data, detailed protocols, engineered cell lines, and reagents used across the Network.
Structure of the 4DN Network
The 4DN Network encompasses several related efforts (http://www.4dnucleome.org/). First, six Centers make up the Nuclear Organization and Function Interdisciplinary Consortium (NOFIC). These Centers will develop genomic and imaging technologies, and implement computational models for understanding the 4D nucleome. NOFIC centers will work together with other components of the Network to benchmark experimental and computational tools, and to identify the most appropriate repertoire of methods to study the 4D nucleome. These studies will be combined with structural and functional validation of observations and models. Ultimately, the NOFIC aims to deliver integrated approaches that can be used towards generating a first draft of a model of the 4D nucleome.
Second, ongoing technology development is addressed by the 4DN Network in three ways. (i) New genomic interaction technologies will be developed to study the 4D nucleome at the single cell level, to probe the roles of RNA in chromatin architecture, and to engineer new chromatin interactions. (ii) New imaging and labeling methods are developed to visualize the genome at high resolution, in live cells as well as in tissues, and in relation to genome activity. Chromatin dynamics will be assayed at high resolution over time scales of seconds (e.g. mitotic compaction) to minutes (transcription) to hours (cell cycle) to days (differentiation). (iii) New methods will be developed to probe the DNA, RNA and protein composition of subnuclear structures such as the nuclear envelope and the nucleolus.
Third, a Data Coordination and Integration Center (DCIC; http://dcic.4dnucleome.org/) stores all data generated by the Network, and coordinates data analysis. The DCIC will maintain a web portal to share data and models with the Network and the larger scientific community. A separate Organizational Hub coordinates communication across the Network and organizes Network meetings. Finally, a 4DN Network Outreach/Education Working Group works to increase the visibility of the 4DN Network and its associated resources, and foster interactions and collaborations with the larger biomedical community.
Research Plans
Genomic technologies to reveal the 4D nucleome
3C technologies have been developed to examine long-range interactions across the genome15,16. Genome-wide 3C technologies, e.g. Hi-C, have revealed patterns of interactions that define genome structures at various resolutions, including loops and topologically associating domains (TADs)17,18,34,35. TADs can be hundreds of kilobases in size, often containing several genes and multiple enhancers, at least some of which appear to interact by looping mechanisms. The ChIA-PET method provides finer resolution to detect structures defined by architectural proteins such as CTCF and cohesin, as well as enhancer-promoter interactions associated with RNAPII and other TFs6. Furthermore, genome-wide mapping at base pair resolution to detect haplotype specific interactions are in progress, which will enable the connection of chromatin topology to the vast genetic information regarding complex traits and diseases. The Network will continue to develop 3C-based technologies, including genome-wide methods that enable exploration of higher-order (beyond pairwise) DNA contacts, detection of chromatin interactions in (thousands of) individual cells36, and mapping of RNA-DNA interactions (Table 2).
Table 2.
Genomic technologies currently in use or in development in the 4DN network. UD = under development.
| Assay Name | Assay Abbreviation. | Key features |
|---|---|---|
| Chromosome Conformation Capture | All methods use proximity ligation to measure interacting genomic loci | |
| 3C 15,60 | Interactions between specific genomic loci, including genome-wide studies | |
| 4C 61 | Genome-wide interactions of a specific genomic locus | |
| 5C 62 | Many loci against many loci | |
| Hi-C 17 | Genome wide map of all interactions in the nucleus | |
| Single Cell Hi-C 36 | Hi-C variant that enables mapping contacts within single cells | |
| Combinatorial single-cell Hi-C 63 | Single cell Hi-C variant using split-pool barcoding to map single cells | |
| In situ Hi-C 63 | Hi-C variant that performs digestion and ligation in intact nuclei | |
| DNase Hi-C 64 | Hi-C variant that digests chromatin using DNase | |
| Micro-C 65, 66 | Hi-C variant that digests chromatin using micococcal nuclease | |
| Capture Hi-C 67,68 | Hi-C variant that incorporates selection of targeted genomic loci | |
| TCC 69 | A variant of Hi-C using bead-coupling of complexes | |
| Distance Hi-C UD | Variant of Hi-C using photo-activated crosslinkers to measure the distance between interacting DNA regions. | |
| COLA 70 | A variant of Hi-C using frequent restriction cutters to map >2 simultaneous DNA interactions | |
| Chromatin Interaction Analysis by paired-end-tag sequencing | ChIA-PET 6,71 | Genome-wide map of interactions bound to a specific protein. |
| Genome Architecture Mapping | GAM 37 | A cryosectioning method to map colocalized DNA regions in a ligation-independent manner. |
| Split-pool barcoding of RNA and DNA | SPRITE | A ligation-independent method to barcode interacting RNA and DNA. Enables mapping of higher-order contacts. |
| RNA interaction with chromatin by paired-end-tag sequencing | RICh-PET UD | Genome-wide mapping for all ncRNA-chromatin interactions. |
| Chromatin IP aided RICh-PET | ChIP RICh-PETUD | Genome-wide mapping for 3-way interactions involving ncRNA, DNA loci, and protein factors. |
| DNA Adenine Methyltrans-ferase Identification | DamID and Single-Cell DamID 72,8 | Genome-wide mapping of molecular contact frequency of DNA locus to a Dam methylase fusion protein: will be used to measure DNA proximity to different nuclear compartments |
| Tyramide Signal Amplification-Seq | TSA-Seq UD | Genome-wide mapping of estimated mean cytological distance (in microns) of DNA locus to a nuclear compartment. |
| Replication Labeling-Seq | Repli-Seq 73,74 | Genome-wide mapping of timing of DNA replication |
| Thousands of Reporters Integrated in Parallel | TRIP 50 | Genome-wide mapping of chromosome position effects on transcription and post-transcriptional processing |
| RNA Antisense Purification | RAP 48,49, 75,76 | Mapping of DNA regions, RNA species, and proteins that are in proximity to a specific noncoding RNA in the nucleus. |
A limitation of current 3C methods is that they depend on a single cross-linker, formaldehyde, which has known biases in the type of residues it can cross-link. Because of the nature of formaldehyde, which is known to polymerize and crosslink molecules across a large range of distances, this approach lacks precise distance information. The Network will explore bivalent photo-activated cross-linkers that are separated by linkers of defined length and flexibility. Other strategies entirely eliminate the use of cross-linkers, e.g. GAM37.
Imaging the 4D genome
4DN investigators will develop and integrate imaging platforms that enable visualizing dynamics, interactions, and structural organization of the nucleus at unprecedented temporal and spatial resolutions (Table 3). Each of these approaches has unique and complementary abilities for probing different aspects of genome organization. In particular, platforms enabling live cell imaging allow the dynamics of select chromatin regions and nuclear features to be studied in real time (seconds to hours).
Table 3.
Imaging technologies currently in use or in development in the 4DN network
| Application | Method (PMID) | Label/Stain/Dye | Feature |
|---|---|---|---|
|
| |||
| Visualizing DNA and RNA sequences | 3D DNA and RNA FISH 77, seqFISH 78, MERFISH 79,80 | Fl-labeled oligo and BAC probes | Fixed cells, single and multiple target DNA and RNA detection |
| HIPMap 81 | Fl-labeled probes | High throughput FISH and automated microscopy in fixed cells | |
| Cas-FISH 31,82), CRISPRainbow (29), Cas live cell imaging 30,38 | Fl-labeled dCas9 and guide RNAs (gRNAs) to label genomic loci; gRNAs can contain stem loop bound by fl-labeled coat proteins | Live and fixed cells-gRNAs against repetitive targets and/or collections of gRNAs that cover unique sequences | |
| Track first, identify later 83 | Bar coded fl-labeled oligos and fl-dCas9 | Combination of SeqFISH and Cas9 live cell imaging | |
| MS2, PP7, mSpinach, aptamers 84,85 | Fl-fusions that bind tagged RNA sequences | Live and fixed cells: transcription | |
| ParS, LacO or TetO labeled genomic loci 86,87 | Visualized by Fl-labeled ParB, lac or tet repressor proteins | Live and fixed cells: genomic tags | |
|
| |||
| High Resolution Ultrastructur e and 3D organization | Transmission EM, SBEM, multiple-tilt 88 and serial section EM tomography 89 | Heavy metals, colloidal nano gold, inorganic and organic probes and tags, ChromEM, ALEXA 633, Fluoronanogold, Time-STAMP-YFP-MiniSOG | High resolution ultrastructure of macromolecules in situ ranging from 2D transverse projections through 70–250 nm sections to reconstruction of large 3D volumes (250 to >500nm thick) |
| Multi-Color EM 90, ChromEMT (under development) | Orthogonal correlated light and EM probes and DAB conjugates | Local ultrastructure and global 3D organization of chromatin as a continuum at nucleosome resolutions and megabase scales | |
| X-Ray Tomography 41 | Quantitative linear absorption coefficient (LAC) generated contrast that reflects the bioorganic composition of unlabeled DNA, RNA and proteins | Mesoscale resolution (20–50 nm) in intact, unprocessed cells | |
| Correlated SIM and X-Ray Tomography 42 | Fl-labeled probes | Visualize ultrastructure and interactions of selectively labeled chromatin and protein simultaneously | |
|
| |||
| Visualizing spatial and dynamic 3D nuclear organization | Wide-field, Confocal/Multiphoton | Fl probes: Fl protein, SNAP and Halo genomic tags, Alexa and Cy dyes, atto488, Suntag, SH2, Quantum dots, Janelia fluor dyes 100 | Live and fixed cells: Diffraction limited (>250 nm) |
| Super resolution: 3D SIM, PALM, STORM 91, STED, adaptive optics 92, tcPALM, 28, SAIM 93 | Fl probes, photoactivatable and photoswitchable proteins; caged organic fluorophores 100 and organic fluorophores in thiol buffer coupled to SNAP, Halo, nanobodies or FISH oligos. | Super-resolution imaging of nuclear organization in live and fixed cells (ca. 10–20 nm). | |
| Lattice Light Sheet (LLS), LLS-PAINT 94,95 | Fl-labeled probes | Live cell: 3D dynamics and nuclear organization | |
| Single molecule imaging and tracking 96–99 | Fl-labeled and photoactivatable probes, ArrayG | Live cell: Binding and search dynamics: residence time, search time, search mechanism | |
Abbreviations: Fl, fluorescent, BAC, bacterial artificial chromosome, FISH, Fluorescent in situ hybridization; seqFISH, sequential live and fixed FISH; MERFISH, multiplexed error robust fluorescence in situ hybridization; HIPMap, High-throughput imaging position mapping; CasFISH, CRISPR associated protein 9 FISH, CRISPRainbow, multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs; MS2 and PP7, bacteriophage RNA stem loop motifs; SBEM, serial block face scanning EM; SIM, Structured Illumination Microscopy, PALM, Photoactivated Localization Microscopy; STORM, stochastic optical reconstruction microscopy; STED, Stimulated emission depletion; tcPALM, time-correlated PALM; SAIM, Scanning Angle Interference Microscopy
Standard and high-throughput fluorescent in situ hybridization (FISH) using oligonucleotide probes or guide RNA-mediated recruitment of fluorescently labeled dCas9 (“CASFISH”31) in fixed cells will be exploited to image genomic interactions over different spatial distances in different cell types and states. These imaging tools will play an important role in benchmarking, validating and complementing data obtained with genomic and proteomic mapping technologies. CRISPR/dCas9 FISH in live cells29,30,38 and other live cell imaging approaches (Table 3) will be employed to assay the dynamic behavior of particular chromatin regions and/or nuclear structures in real time.
New technologies will be developed to label DNA, RNA, and protein that occur in proximity of specific nuclear bodies. These technologies include HRP-labeled antibodies in TSA-SEQ (Box 1), APEX (ascorbate peroxidase39) and Killer Red40. Genome-editing technologies will also be applied to tag a subset of key genomic loci, loops, TADs and potentially newly discovered structures to help visualize these moieties and document their interactions with other nuclear regions in live cells.
Super resolution microscopy, single-molecule tracking techniques and multiplex fluorescent/chemical tags will be applied in living cells to determine the dynamic interactions, diffusion and motion of fluorescently labeled proteins, ncRNAs and genomic loci (Table 3). These live cell imaging approaches are expected to provide information regarding search mechanism, binding and residence time of DNA and protein interactions and will also be used to validate and complement genomic methods used by the Network.
Soft X-ray Tomography (SXT41) will be used to visualize the 3D organization of chromatin in nuclei of cells in the native state (cryo-immobilized). SXT will be used to directly measure chromatin compaction, e.g. in relation to sub-nuclear position, at different stages of cell differentiation and in different cell types. Correlated microscopy approaches will be used to augment ultrastructural data with molecular localization information. Cryogenic Fluorescence Tomography (CFT42) will be used to precisely locate molecules in 3D reconstructions of intact, native state cells imaged using Soft X-ray Tomography.
Members of the Network will develop new Electron Microscopy (EM) technologies that enable the local and global structural organization of chromatin to be visualized as a continuum from nucleosome to Mbp scale in both interphase and mitotic cells. One such method, ChromEM, will be combined with new genetic tags and nanoparticle labeling technologies to develop the EM equivalent of ‘multi-color’ fluorescence.
The development of automated imaging analysis pipelines and data standards will be important to extract the maximum structural information possible from these datasets. Further development of software for analyzing, annotating, and archiving imaging data, together with implementation of new approaches for correlating imaging and genomics datasets are major goals of the 4DN Network (see below).
Nuclear bodies and non-chromatin structures
The nucleus consists of distinct nuclear structures, such as the nuclear lamina and nuclear pores, chromatin-associated bodies such nucleoli that are initiated at specific genomic loci, as well as non-chromatin bodies such as nuclear speckles, and PML bodies43,44. Increasing evidence indicates that specific genomic regions associate with these structures, suggesting that these chromosomal associations may have a functional role in regulating genome function8,45,46.
Goals of the 4DN Network further include development of new mapping methodologies to measure the genome-wide molecular interaction frequency and cytological distance of chromosome loci to major nuclear compartments, including the nuclear lamina, nuclear pores, nuclear speckles, nucleoli, and pericentric heterochromatin (Table 2). Concurrently, new and improved technologies will be developed, including localized APEX (ascorbate peroxidase)-mediated protein biotinylation47, fractionation by cryomilling, and RNA Antisense Purification (RAP48,49), to catalog and measure the protein and RNA components of these nuclear compartments, as well as both optogenetic and degron-based approaches to alter or disrupt sub-nuclear bodies and compartments. Functional mapping approaches based on Repli-Seq9 and TRIP (thousands of reporters integrated in parallel50) will provide genome-wide correlations of DNA replication timing and chromosome position effects on transcription and RNA processing that can be correlated with these structural maps (Table 2). New imaging approaches will be developed to correlate chromosome and nuclear compartment dynamics with changes in DNA replication timing, transcriptional activation, and other functional states (Table 3). Computational analyses of these genome mapping data from several cell types will be aimed at identifying possible cis and trans determinants of nuclear compartmentalization.
Modeling the 4D Nucleome
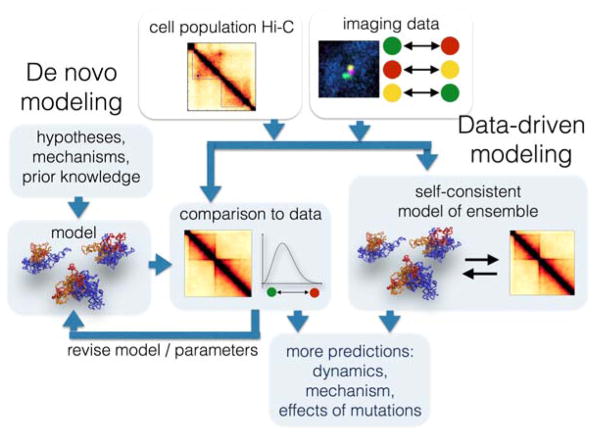
In parallel with the emergence of increasingly powerful experimental methods has been the development of computational approaches for modeling the spatial organization of the genome. There are at least two major computational approaches for modeling genome architecture on the basis of experimental data: data-driven and de novo approaches33 (Figure 2). Data-driven approaches directly use experimental data (Hi-C, imaging etc.) to produce an ensemble of conformations that best match an experimentally observed set of contact probabilities and distances51,52. De novo modeling, on the other hand, produces ensembles of conformations that result from known or hypothesized physical or biological processes, and tests whether these ensembles are consistent with features of experimental contact frequency maps and imaging data (e.g. 19,53). Such de novo models can suggest specific molecular mechanisms and principles of chromosome organization, and can be predictive of chromosome dynamics thus going far beyond the experimental data33.
Figure 2. Modeling the 4D genome.
Data obtained with imaging and chromosome conformation capture based assays can be used for building spatial and dynamic models of chromosomes using two main approaches. In the data-driven approach, experimental data are used directly to generate ensembles of conformations that reproduce the experimental observations. In the de novo approach, ensembles of conformations are built according to known or hypothesized physical or biological processes. Models are then selected based on their agreement with experimental data.
There are several challenges and promises of current modeling approaches. The first is in a wide diversity of technologies that capture complementary aspects of genome organization: contact frequencies, distances, proximities to various nuclear bodies, etc. Relationships between these data can be complicated: e.g. contact frequency is distinct from an average spatial distance, possibly creating a seemingly paradoxical relationship between Hi-C and FISH 54). Current modeling approaches, however, can systematically integrate a variety of data to generate comprehensive structural and dynamic models of the 4D nucleome. Such models can be validated against data not used for model selection, e.g. predicting dynamics from static data and testing using live imaging.
Second, most current genomic methods yield data from ensembles of thousands to millions of cells, obscuring structural heterogeneity that exists among single cells. A number of groups within the Network are developing methods for generating data from large numbers of single cells, which will present new computational challenges for integrating with current modeling approaches36. It is possible that some of these methods will yield functional data on these same single cells (i.e. Hi-C and RNA-seq, from each of many single cells), which would represent a significant opportunity for directly relating structure to function.
Third, most current models ignore the fact that mammalian cells are diploid, i.e. they do not distinguish or separately model homologous chromosomes, which will be particularly important for modeling based on single-cell data. The haplotype-resolution of the genomes of the common cell types chosen by the consortium will aid in this goal.
Fourth, contemporary approaches for modeling genome architecture are typically static rather than dynamic, reflecting the static nature of available Hi-C data and the majority of imaging data. As we are increasingly able to visualize (via direct imaging) or infer (via single cell or bulk Hi-C analyses of time series) chromatin dynamics, e.g. in differentiation, and cell cycle progression, it will be essential that these observations can be integrated into computational models. Two classes of modeling approaches can tackle dynamics differently. Data-driven modeling can use Hi-C data obtained for different time points (e.g. stages of differentiation or cell cycles) to build conformational ensembles for each point and then hypothesize about possible mechanisms that led to observed reorganizations. De-novo modeling, in turn, can test whether a particular mechanism that needs to be stipulated first, could lead to observed temporal changes in Hi-C data. Polymer models can further show whether observed temporal reorganizations (through differentiation or cell cycle) can reflect slow equilibration process of generally non-equilibrium chromosomes. Moreover, mechanistic de novo models can be further validated by dynamic data from live-cell imaging experiments, e.g. by examining mean-squared displacements of different chromosomal loci vs, time in experiment and simulations 55.
Fifth, new data and future models should help to connect genome architecture and other aspects of genome function, e.g. by suggesting molecular mechanisms of how transcription factor binding or epigenetic modifications can lead to formation of active/inactive chromatin compartments56. The inferred mechanisms could generate testable predictions of sequence-structure-function relationships, i.e. how nuclear architecture relates to nuclear function. Availability of temporal Hi-C, functional data, and models of chromatin organization and dynamics can allow identifying causality if certain functional characteristics at earlier time points are predictive of later chromosomal states, or vice versa. Such associations and inferred causations can then be further tested experimentally.
Perturbation and manipulation of the 4D nucleome to relate structure to function
A critical and overarching goal is to determine how genome structure and chromatin conformation modulate genome function in health and disease. To this end the 4DN Network will explore experimental approaches to manipulate and perturb different features of the 4D nucleome. First, using CRISPR/Cas9 technologies DNA elements involved in specific chromatin structures, e.g. domain boundaries or chromatin loops, can be altered, re-located or deleted57,58. Second, defined chromatin structures such as chromatin loops will be engineered de novo by targeting proteins that can (inducibly) dimerize with their partner looping proteins (e.g.7). Third, other CRISPR/Cas9 approaches will be used to target enzymes (e.g. histone modifying enzymes, structural proteins) or ncRNAs to specific sites in the genome. Fourth, several groups will perturb nuclear compartmentalization by developing methods for “rewiring” chromosome regions to different nuclear compartments, either by integrating specific DNA sequences that are capable of autonomous targeting the locus to different nuclear compartments or by tethering certain proteins to these loci to accomplish similar re-positioning. Fifth, cell lines will be generated for conditional/temporal ablation of nuclear bodies or candidate chromosome architectural proteins (such as CTCF and cohesin) or RNAs. Sixth, additional methods will be developed to nucleate nuclear bodies at specific chromosomal loci. Finally, biophysical approaches will be developed to micro-mechanically perturb cell nuclei and chromosomes followed by direct imaging of specific loci59. Although it remains challenging to establish direct cause-and-effect relationships, analysis of the effects of any of these perturbations on processes such as gene expression and DNA replication can provide deeper mechanistic insights into the roles of chromosome structure and nuclear organization in regulating the genome.
Data sharing and standards
The Network will develop guidelines for data formats, metadata (descriptions of how the data was acquired), standards, quality control measures, and other key data-related issues. Another goal is to make this data rapidly accessible, both within the Network and the entire scientific community. These efforts will be of particular importance for new technologies for which standards for sharing data and assessing data quality have not yet been established. Such standards will greatly enhance the usefulness of the datasets for the broader scientific community beyond those who generate the data.
For sequencing-based technologies, data format standards to represent sequences and alignments have long been present (e.g. fastq, bam/sam). However, common formats to represent three-dimensional interactions are yet to be developed. These formats need to account for large data sizes and the constraints imposed by different computer architectures. For Hi-C data, for example, the genome-wide contact probability map is an N^2 matrix, where N is the spatial resolution (for 10kb resolution, N=300,000), with most of the entries being empty. There are multiple ways to represent such sparse matrices, appropriate for different analysis and storage approaches. For imaging technologies, the situation is even more challenging, as the types of microscopes employed are highly variable, and the data formats and analysis tools are often manufacturer-dependent. Standards to unify data and metadata from different manufacturers, such as the Open Microscopy Environment (https://www.openmicroscopy.org/site), are under development. These standards also need to accommodate the rapid developments in super-resolution microscopy.
A related issue is to define a set of appropriate metadata fields and minimum metadata requirements such that sufficient and useful details are available to other investigators outside the Network. While not all information can be captured about an experiment, collecting pertinent information will increase the reproducibility of experiments and the likelihood that the data will be re-used by other investigators. The 4DN Network has established formal working groups, including the 4DN Data Analysis Group, Omics Data Standards Group and Imaging Data Standards Group, which will define these 4DN standards and data analysis protocols.
Developing a set of measures for assessing data quality and determining appropriate thresholds will be important for ensuring high-quality 4DN data. An important measure of data quality and reliability of new technologies is the reproducibility of results between repeated experiments. Reproducibility can be assessed at multiple levels: broadly speaking, technical reproducibility measures how well a technique performs for the same starting material, whereas biological reproducibility should also capture all other variations including heterogeneity among samples. The 4DN Analysis Group will compute and make available quality control measures and provide recommendations on expected quality standard thresholds so that investigators can make decisions regarding the utility of specific datasets for addressing their specific questions.
Finally, to ensure rapid dissemination of findings made by the 4DN Network with the larger scientific community the Network has adopted a transparent and open publication policy where all work supported by the Network is submitted to a public preprint server such as BioRxiv before submission to a peer reviewed journal.
Outlook
After determining the complete DNA sequence of the human genome and subsequent mapping of most genes and potential regulatory elements, we are now in a position that can be considered the third phase of the human genome project. In this phase, which builds upon and extends other epigenome mapping efforts mentioned above, the spatial organization of the genome is elucidated and its functional implications revealed. This requires a wide array of technologies from the fields of imaging, genomics, genetic engineering, biophysics, computational biology and mathematical modeling. The 4DN Network, as presented here, provides a mechanism to address this uniquely interdisciplinary challenge. Further, the policy of openness and transparency both within the Network and with the broader scientific community, and the public sharing of all methods, data and models will ensure rapid dissemination of new knowledge, further enhancing the potential impact of the work. This will also require fostering collaborations and establishing connections to other related efforts around the world, e.g. the initiative to start a European 4DN project (https://www.4dnucleome.eu), that are currently under development. Together these integrated studies promise to allow moving from a one-dimensional representation of the genome as a long DNA sequence to a spatially and dynamically organized three dimensional structure of the living and functional genome inside cells.
Supplementary Material
References
- 1.ENCODE-Project-Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stunnenberg H, Hirst M International Human Epigenome Consortium. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167:1145–1149. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 5.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and Interaction between Hypersensitive Sites in the Active beta-globin Locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 6.Tang Z, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng W, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 9.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurano MT, et al. Sytematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 14.Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14 doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- 15.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 16.Denker A, de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30:1357–1382. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fudenberg G, et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wit E, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Vietri Rudan M, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dekker J, Mirny LA. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 25.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophysical journal. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 27.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cisse II, et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 29.Ma H, et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nature biotechnology. 2016;34:528–530. doi: 10.1038/nbt.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng W, Shi X, Tjian R, Lionnet T, Singer RH. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci U S A. 2015;112:11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marti-Renom MA, Mirny LA. Bridging the Resolution Gap in Structural Modeling of 3D Genome Organization. PLoS Comput Biol. 2011;7:e1002125. doi: 10.1371/journal.pcbi.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imakaev MV, Fudenberg G, Mirny LA. Modeling chromosomes: Beyond pretty pictures. FEBS letters. 2015;589:3031–3036. doi: 10.1016/j.febslet.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beagrie RA, et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017;543:519–524. doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma H, et al. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nature biotechnology. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulina ME, et al. A genetically encoded photosensitizer. Nature biotechnology. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 41.Le Gros MA, et al. Soft X-Ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis In Vivo. Cell reports. 2016;17:2125–2136. doi: 10.1016/j.celrep.2016.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith EA, et al. Quantitatively imaging chromosomes by correlated cryo-fluorescence and soft x-ray tomographies. Biophysical journal. 2014;107:1988–1996. doi: 10.1016/j.bpj.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dundr M. Nuclear bodies: multifunctional companions of the genome. Current opinion in cell biology. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in genetics : TIG. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Németh A, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Koningsbruggen S, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SY, et al. APEX Fingerprinting Reveals the Subcellular Localization of Proteins of Interest. Cell reports. 2016;15:1837–1847. doi: 10.1016/j.celrep.2016.04.064. [DOI] [PubMed] [Google Scholar]
- 48.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akhtar W, et al. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 51.Serra F, et al. Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS letters. 2015;589:2987–2995. doi: 10.1016/j.febslet.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Zhu Y, et al. Comprehensive characterization of neutrophil genome topology. Genes & development. 2017;31:141–153. doi: 10.1101/gad.293910.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fudenberg G, Imakaev M. FISH-ing for captured contacts: towards reconciling FISH and 3C. bioRxiv. 2016 doi: 10.1101/081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lucas JS, Zhang Y, Dudko OK, Murre C. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158:339–352. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jost D, Carrivain P, Cavalli G, Vaillant C. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 2014;42:9553–9561. doi: 10.1093/nar/gku698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong X, Chen M, Lim WA, Zhao D, Qi LS. CRISPR/Cas9 for Human Genome Engineering and Disease Research. Annual review of genomics and human genetics. 2016;17:131–154. doi: 10.1146/annurev-genom-083115-022258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 59.Poirier MG, Marko JF. Micromechanical studies of mitotic chromosomes. Current topics in developmental biology. 2003;55:75–141. doi: 10.1016/s0070-2153(03)01002-0. [DOI] [PubMed] [Google Scholar]
- 60.Rodley CD, Bertels F, Jones B, O’Sullivan JM. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet Biol. 2009;46:879–886. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 62.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): A Massively Parallel Solution for Mapping Interactions between Genomic Elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramani V, et al. Massively multiplex single-cell Hi-C. Nat Methods. 2017;14:263–266. doi: 10.1038/nmeth.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma W, et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2014 doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsieh TS, et al. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsieh TS, Fudenberg G, Goloborodko A, Rando OJ. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat Methods. 2016;13:1009–1011. doi: 10.1038/nmeth.4025. [DOI] [PubMed] [Google Scholar]
- 67.Dryden NH, et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 2014;24:1854–1868. doi: 10.1101/gr.175034.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hughes JR, et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 69.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2011;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darrow EM, et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A. 2016;113:E4504–4512. doi: 10.1073/pnas.1609643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 73.Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strongin DE, Groudine M, Politz JC. Nucleolar tethering mediates pairing between the IgH and Myc loci. Nucleus. 2014;5:474–481. doi: 10.4161/nucl.36233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma H, Reyes-Gutierrez P, Pederson T. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc Natl Acad Sci U S A. 2013;110:21048–21053. doi: 10.1073/pnas.1319097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nature structural & molecular biology. 2013;20:1321–1324. doi: 10.1038/nsmb.2680. [DOI] [PubMed] [Google Scholar]
- 81.Shachar S, Voss TC, Pegoraro G, Sciascia N, Misteli T. Identification of Gene Positioning Factors Using High-Throughput Imaging Mapping. Cell. 2015;162:911–923. doi: 10.1016/j.cell.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma H, et al. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. The Journal of cell biology. 2016;214:529–537. doi: 10.1083/jcb.201604115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takei Y, Shah S, Harvey S, Qi LS, Cai L. Multiplexed Dynamic Imaging of Genomic Loci by Combined CRISPR Imaging and DNA Sequential FISH. Biophysical journal. 2017 doi: 10.1016/j.bpj.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods. 2013;10:119–121. doi: 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fukaya T, Lim B, Levine M. Enhancer Control of Transcriptional Bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robinett CC, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen H, Fujioka M, Gregor T. Direct visualization of transcriptional activation by physical enhancer-promoter proximity. BioRxiv. 2017 doi: https://doi.org/10.1101/099523.
- 88.Phan S, et al. 3D reconstruction of biological structures: automated procedures for alignment and reconstruction of multiple tilt series in electron tomography. Advanced structural and chemical imaging. 2017;2:8. doi: 10.1186/s40679-016-0021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soto GE, et al. Serial section electron tomography: a method for three-dimensional reconstruction of large structures. NeuroImage. 1994;1:230–243. doi: 10.1006/nimg.1994.1008. [DOI] [PubMed] [Google Scholar]
- 90.Adams SR, et al. Multicolor Electron Microscopy for Simultaneous Visualization of Multiple Molecular Species. Cell chemical biology. 2016;23:1417–1427. doi: 10.1016/j.chembiol.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szymborska A, et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science. 2013;341:655–658. doi: 10.1126/science.1240672. [DOI] [PubMed] [Google Scholar]
- 92.Izeddin I, et al. PSF shaping using adaptive optics for three-dimensional single-molecule super-resolution imaging and tracking. Optics express. 2012;20:4957–4967. doi: 10.1364/OE.20.004957. [DOI] [PubMed] [Google Scholar]
- 93.Paszek MJ, et al. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nat Methods. 2012;9:825–827. doi: 10.1038/nmeth.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen BC, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Legant WR, et al. High-density three-dimensional localization microscopy across large volumes. Nat Methods. 2016;13:359–365. doi: 10.1038/nmeth.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen J, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Izeddin I, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 2014;3 doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. CTCF abd Cohesin regulate chromatin loop stability with distinct dynamics. bioRxiv. 2016 doi: 10.7554/eLife.25776. doi: https://doi.org/10.1101/093476. [DOI] [PMC free article] [PubMed]
- 99.Ghosh R, Draper W, Franklin JM, Shi Q, Liphardt JT. A fluorogenic nanobody array tag for prolonged single molecule imaging in live cells. bioRxiv. 2017 doi: https://doi.org/10.1101/111690.
- 100.Grimm JB, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12:244–250. 243–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.