Eukaryotic genomes encode thousands of long noncoding RNAs, and many of these function in transcriptional regulation.
Abstract
It has recently become apparent that RNA, itself the product of transcription, is a major regulator of the transcriptional process. In particular, long noncoding RNAs (lncRNAs), which are so numerous in eukaryotes, function in many cases as transcriptional regulators. These RNAs function through binding to histone-modifying complexes, to DNA binding proteins (including transcription factors), and even to RNA polymerase II. In other cases, it is the act of lncRNA transcription rather than the lncRNA product that appears to be regulatory. We review recent progress in elucidating the molecular mechanisms by which lncRNAs modulate gene expression and future opportunities in this research field.
INTRODUCTION
Deep sequencing of mammalian transcriptomes has revealed the remarkable fact that there are plausibly more than 100,000 different RNAs produced in the organism, far exceeding the ~20,000 protein-coding genes. Most of these RNA sequences are noncoding. It has been useful in the field to define the long noncoding RNAs (lncRNAs) as those >200 nucleotides (nt), thereby separating them from the distinct classes of microRNAs (miRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) that function through distinct mechanisms. However, this is admittedly an arbitrary cutoff, and in this Review, we will include B2 RNA (~180 nt) as an “honorary lncRNA” because it reveals an important mechanism about how noncoding RNAs can inhibit transcription.
We are deliberately vague about the number of lncRNAs, because deep-sequencing experiments always require a cutoff of some minimum number of reads. It is more difficult to conceive a function for a lncRNA that is present at less than one copy per cell than for lncRNAs that are more abundant. Very rare or unstable lncRNAs could be transcriptional noise. Furthermore, many researchers study one cell type, but lncRNAs are often specific to particular cell types (1). Thus, their total numbers may be underestimated. Enhancer RNAs (eRNAs) alone constitute an enormous class [reviewed by Li et al. (2)]; if eRNAs were transcribed from every active enhancer, then this class alone would number in the hundreds of thousands when all cell types are considered.
The lncRNAs can be nuclear, nucleolar, or cytoplasmic or occupy several cellular compartments. The cellular localization of a lncRNA is informative regarding its function. For example, nuclear lncRNAs could plausibly have functions in histone modification or direct transcriptional regulation. If a lncRNA has a substantial cytoplasmic component, then the possibility that it contains a short open reading frame that is translated should be considered.
Some lncRNAs may function at the RNA level, for example, as ribozymes or riboswitches (3). Most commonly, however, lncRNAs perform their functions as ribonucleoprotein particles (RNPs). In terms of the protein partners of the lncRNAs, it is useful to know whether they bind RNAs highly specifically or more promiscuously. For example, U1A protein appears to have predominantly two RNA partners (U1 snRNA and its own mRNA), and TERT protein has one established RNA partner [telomerase RNA (TR)]. Admittedly, the number of known RNA partners tends to increase as more research is done. For example, yeast Pop1/Pop6/Pop7 was initially found as a component of two RNPs, ribonuclease (RNase) P and RNase MRP, and then more recently as a functional component of yeast telomerase (4), and additional functional partners could exist. At the other extreme, heterogeneous nuclear RNP (hnRNP) proteins, PRC2 (Polycomb repressive complex 2) and FUS proteins have very large transcriptomes, binding many thousands of RNAs in cells (5); correspondingly, they bind many RNAs in vitro with similar binding constants (6, 7).
Many proteins known to be involved in eukaryotic transcription, most of which are therefore DNA binding proteins, also bind lncRNAs in vitro and in vivo. The proteins may have separate sites for binding RNA and DNA, or the two nucleic acids may bind mutually exclusively in overlapping binding modes [reviewed by Hudson and Ortlund (8)]. In some of the latter cases, it is possible that the dual binding is fortuitous: The protein has a DNA binding site, which therefore has more or less ability to bind RNA, and the RNA binding may have little consequence. In many cases, however, there is evidence that the RNA binding is regulatory. In a very general sense, we can think of several ways in which lncRNAs could regulate transcription, as follows:
(1) Recruitment
RNA can recruit a regulatory protein complex to a gene or an entire chromosome in cis (when the nascent RNA still occupies its site of transcription) or in trans (for example, by base pairing with another RNA, by RNA binding directly to DNA, or by RNA-protein interactions).
(2) Inhibition
RNA can inhibit the binding of a transcriptional regulatory factor by acting as a “decoy” or inhibit its activity by direct active-site occlusion, by allosteric effects, or more indirectly.
(3) Indirectly, through the act of transcription
Transcription of a lncRNA may regulate the transcription of nearby mRNA genes, either positively (maintaining active chromatin structure) or negatively (for example, colliding polymerases). In these cases, the RNA product may have no importance at all, or it could have an additional function.
(4) Indirectly, through genome organization and the architecture of the nucleus
Organizing hetero- or euchromatic regions into close proximity may stabilize these domains and/or control the spreading of posttranslational modifications (PTMs) to nearby chromatin.
As we traverse the landscape of recent research in this area, we will encounter examples where each of these features appears to be used. Because this is a very new field, the mechanistic understanding is still a work in progress. Therefore, at the end of the review, we will present our ideas about the type of experiments that should be considered if one wants to really nail down the mechanisms of RNA-mediated transcriptional regulation.
B2 RNA REGULATES RNA POLYMERASE II BY DIRECT BINDING
Transcription is broadly dysregulated in response to heat shock (9). Chaperones and heat shock proteins, such as HSP70, are transcriptionally up-regulated, whereas housekeeping genes, such as actin and hexokinase II, are down-regulated (10–12). The mouse B2 noncoding RNA is both up-regulated in response to heat shock and coordinates proper gene expression during organogenesis (13–16). The ~180-nt B2 RNA is synthesized by polymerase III (Pol III) and belongs to the short interspersed nuclear elements (SINEs) family, transcribed from retrotransposons that are interspersed throughout the mouse genome (17–20). It was not until this past decade that B2 RNA became appreciated as one of the master regulators of gene expression during heat shock.
To understand the function of B2 up-regulation, Allen and colleagues (18) inhibited total Pol III transcription and performed antisense oligonucleotide knockdowns of B2 RNA during heat shock. Decreased accumulation of B2 RNA corresponded to an up-regulation of coding genes that were previously down-regulated (18). This result was further corroborated by the finding that RNA Pol II transcription in nuclear lysates was significantly inhibited in the presence of B2 RNA, but not by the control B1 RNA (18, 20). These data prompted the inquiry of how the mature B2 noncoding transcript inhibits RNA Pol II transcription.
RNA immunoprecipitation (RIP) studies demonstrated that B2 RNA is enriched in RNA Pol II purifications (18). Although RIP is useful for identifying candidate RNA-protein interactions, it is unable to define direct binding partners and can suffer from RNA-protein exchange in extracts (21). However, in the current case, follow-up electrophoretic mobility shift studies and functional in vitro transcriptional assays showed that B2 RNA inhibits transcription by associating directly with the preinitiation complex (Fig. 1) (20). This mechanism of inhibition paralleled findings in Escherichia coli, where the 184-nt 6S RNA was found to inhibit transcription by binding to the RNA polymerase-σ70 holoenzyme (22).
Fig. 1. B2 RNA directly inhibits transcription.
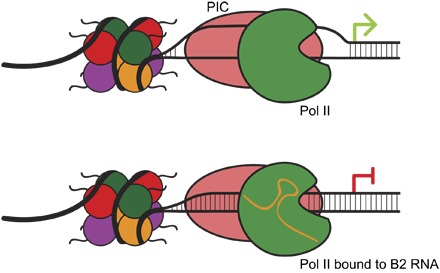
In the absence of B2 RNA, a functional closed preinitiation complex (PIC) can assemble. (Top) This complex can melt the dsDNA duplex, forming an open preinitiation complex to promote transcription initiation. In contrast, B2 RNA directly binds to RNA Pol II in a nonfunctional closed complex. (Bottom) B2 RNA precludes Pol II from making functional DNA contacts and thereby prevents dsDNA melting and open complex formation.
Deletion analysis on B2 RNA coupled with RNA Pol II RNase protection assays determined that the 3′ region of the transcript is responsible for binding to RNA Pol II (20). Additionally, experimental determination of the B2 RNA secondary structure showed that the 3′ end of the noncoding transcript has several stem-loop and single-stranded regions (20). Deletion of a single-stranded region affects the ability of B2 RNA to repress transcription, whereas it has no effect on binding to RNA Pol II (23). This contrasts with how disruption of the RNA stem loop prevents binding to RNA Pol II and transcriptional repression (20).
Further work identified additional small RNAs, such as Alu and Fc, that compete with B2 RNA for Pol II binding and inhibition (24, 25). A recent cocrystal structure of RNA Pol II and the Fc RNA revealed that the Fc RNA inhibitor binding site is unique but has significant overlap with the canonical nucleic acid docking site in the elongation complex (25). This cocrystal structure is entirely consistent with the previously reported mechanism of RNA-mediated inhibition.
Multiple independent research groups conducting both in vitro and in vivo assays have accumulated a conclusive body of evidence defining the mechanism of B2 RNA–mediated transcriptional repression. It is now clear that B2 RNA inhibits transcription through a direct interaction with RNA Pol II and prevents the formation of a functional closed preinitiation complex. However, the downstream function of B2-mediated repression and determining why certain RNA Pol II transcripts are up-regulated in response to heat shock are still areas of active research. For example, a recent model suggested that whereas there is global up-regulation of B2 RNA in heat shock, there is local degradation of B2 RNA at sites of active transcription (26). This local degradation of B2 RNA at stress response genes then induces an up-regulation of these RNA Pol II transcripts relative to the level of global RNA Pol II transcription (26).
roX RNAs ARE ESSENTIAL FOR THE HYPERACTIVE X CHROMOSOME IN DROSOPHILA MALES
Drosophila melanogaster and Homo sapiens both require X chromosome dosage compensation, because the female genome comprises two X chromosomes and the male genome carries one. H. sapiens resolve the problem by compacting one X chromosome in females through epigenetic modifications (27). The result is a silent X chromosome, referred to as the Barr body. In contrast, D. melanogaster acetylate histones on the male X chromosome, resulting in hyperactive transcription (28, 29). Although these processes seem entirely divergent, they are linked by the common feature of lncRNAs directing epigenetic modifications (30).
D. melanogaster express two lncRNAs on the male X chromosome, referred to as RNA on the X 1 and 2 (roX1 and roX2) (31, 32). roX1 (3.7 kb) and roX2 (1.1 kb) display overlapping functions; either one can be deleted, but deletion of both results in male lethality (31, 33). Additionally, transgenic expression of either roX RNA can rescue the lethal phenotype of roX RNA deletion (32, 34).
Elegant immunofluorescence and RNA fluorescence in situ hybridization studies identified that roX RNAs normally coat one X chromosome, and they spread in cis across autosomal chromosomes when they are ectopically expressed (31–35). Additional imaging studies showed that roX RNAs colocalize perfectly with a male-specific protein complex that includes five proteins: maleless (MLE), male-specific lethal 1 to 3 (MSL1 to MSL3), and males absent on the first (MOF) (31–35). It was quickly suggested, and later confirmed, that together these proteins and RNAs form the dosage compensation complex (DCC), which generates the hyperactive X chromosome (36).
In males, the DCC ribonucleoprotein complex coats one X chromosome and acetylates Lys16 on histone 4 (H4K16Ac) (Fig. 2) (36–39). H4K16Ac is deposited across much of the X chromosome, and there is a consensus that this PTM is essential for creating hyperactive transcription (36–40).
Fig. 2. roX1 and roX2 RNAs are essential for the hyperactive Drosophila X chromosome.
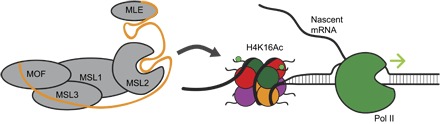
The DCC consists of five proteins (gray) and roX1 or roX2 RNA (orange line). The MLE helicase remodels roX RNAs into a tandem stem-loop structure that is incorporated into a functional DCC. When fully assembled, this ribonucleoprotein complex is recruited to the X chromosome and acetylates Lys16 on histone 4. This PTM results in chromosome decompression and hyperactive transcription. H4K16 acetylation of the X chromosome does not occur in the absence of roX RNA transcripts.
The roX RNAs naturally adopt a repetitive tandem stem-loop conformation (41, 42). This was predicted by a chemical probing method [SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension)], as well as experimentally corroborated (41, 42). Cross-linking studies coupled to RNA mutagenesis identified MLE and MSL2 as the major RNA binding partners of the roX RNAs (41). Additionally, the MLE helicase remodels the tandem stem-loop structure of roX RNAs into a different stem-loop structure that attains a higher binding affinity toward MLE and MSL2 (42–44). This remodeled structure was also experimentally observed as the active chromatin-bound RNA in vivo and is dependent on the adenosine triphosphatase (ATPase) activity of MLE (42–44).
Although MLE and MSL2 appear to be the major RNA binding proteins within the DCC (41), the chromodomains of MOF and MSL3 have also been suggested to play a role in roX RNA recognition (37). Furthermore, protein mutagenesis and immunoprecipitation studies suggest that the affinity of MSL3 toward roX RNAs is affected by acetylation at a single lysine residue (45).
It is quite clear that the DCC is a ribonucleoprotein complex and that roX RNAs are essential for targeting the DCC to the X chromosome. However, we still do not fully understand how roX RNAs contribute to DCC recruitment. The DCC is known to have high affinity toward certain genomic sites (46–48). One model is that incorporation of roX RNAs into the DCC increases the affinity toward these high-affinity DNA binding sites and decreases the affinity toward nonspecific regions (49). Another model is that additional protein partners could be driving the specificity of the DCC toward DNA recognition elements (30, 50). Thus, after 20 years of active research, the precise role of roX RNAs in X-chromosome hyperactivation remains an active question in the field.
PRC2, Spen/SHARP, AND Xist: INACTIVATION OF A WHOLE CHROMOSOME
PRC2 is an essential histone methyltransferase required for gene silencing during development and cancer. PRC2 binds both lncRNAs and pre-mRNAs (Fig. 3), and it can be said to interact with RNAs promiscuously in vitro and in vivo because of the broad spectrum of transcripts that it binds (7, 51, 52). Recently, it was revealed that PRC2 reads RNA motifs consisting of short repeats of consecutive guanines, which are ubiquitous in the transcriptome (53); this clarifies the promiscuous nature of PRC2 targeting to RNA.
Fig. 3. lncRNAs regulate transcription through histone modifiers.
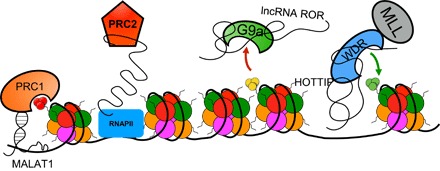
PRC1 interacts with lncRNA, either TUG1 or MALAT1. These interactions regulate methylation status and localization of PRC1. PRC2 is inhibited by binding lncRNA or nascent pre-mRNA. lncRNAs Kcnq1ot1, Air, and ROR (regulator of reprogramming) regulate the activity of G9a, an enzyme that methylates H3K9. HOTTIP interacts with the WDR5-MLL complex and localizes the complex to the 5′HOXA locus. RNAPII, RNA polymerase II; MALAT1, metastasis-associated lung adenocarcinoma transcript 1.
One important biological system involving PRC2 and lncRNAs is X-chromosome inactivation (XCI). As described in the previous section, female mammalian cells inactivate one of their two X chromosomes to equalize X-linked gene expression with that of males. One chief mediator of XCI is the 17-kb lncRNA Xist (X-inactive specific transcript), which is transcribed from the Xist gene on one X chromosome (54). Although the Xist RNA is capped, spliced, and polyadenylated, it remains exclusively in the nucleus and coats one X chromosome in cis. This RNA coating initiates XCI, coinciding with the exclusion of RNA Pol II and the eventual silencing of most gene expression on that X chromosome. One of the first histone modifications detected on the inactive X chromosome is H3K27me3, which is dependent on PRC2. The possibility that Xist RNA might directly recruit PRC2 has been much discussed (55, 56). A knockout of EED (embryonic ectoderm development) does not affect random XCI in the mouse embryo, suggesting that PRC2 may be dispensable for the initiation and maintenance of random XCI (57).
Recent studies have provided intriguing evidence for indirect recruitment of PRC2. McHugh and colleagues (58) identified RNA binding proteins associated with Xist RNA by ultraviolet (UV) cross-linking followed by purification and quantitative mass spectrometry (RAP-MS). Strikingly, the top hit was not PRC2, but instead three factors: SHARP/Spen, SAF-A, and LBR. In brief, subsequent work discovered that SHARP/Spen binds directly to Xist RNA and recruits the SMRT (silencing mediator of retinoid and thyroid receptor)–histone deacetylase 3 (HDAC3) complex. This leads to deacetylation of histone H3 and facilitates the enrichment of PRC2. As validation, the knockdown of SHARP and HDAC3 reactivates gene expression on the silenced X chromosome and leads to the depletion of PRC2. Another study used a similar approach but instead relied on formaldehyde cross-linking under nondenaturing conditions [chromatin isolation by RNA purification–mass spectrometry (ChIRP-MS)] (59). This study identified 81 proteins that directly or indirectly interact with Xist. Furthermore, the authors proposed that heterogeneous nuclear ribonucleoprotein K (hnRNPK) might have a direct influence on H3K27me3 and Polycomb recruitment.
What is consistent between the two studies is that neither found components of PRC2 in the Xist RNA interactome. However, it is worthwhile to emphasize that another proteomics paper used an approach similar to that of McHugh et al. but came to a different conclusion (60). Specifically, they identified RBBP4/7 as possibly interacting with Xist. Although RBBP4 is a known subunit of the PRC2 complex, it is also a component of other chromatin-modifying complexes, including the NuRD and SIN3-HDAC complexes (61, 62). The study did not identify any of the core PRC2 subunits Ezh2, Suz12, and Eed.
Finally, two independent groups (63, 64) used elegant genetic screens to determine which genes are required for Xist-mediated XCI. Both groups identified the RNA binding protein Spen as the top hit. In total, these studies discovered dozens of new factors, each of which warrants further exploration in relation to Xist RNA function.
Beyond XCI, PRC2 has genome-wide roles. Another system that has received much attention in the field is HOTAIR lncRNA’s interaction with PRC2. However, it has now been shown that HOTAIR is dispensable for the regulation of Hoxd transcription (65). Artificially tethering HOTAIR to chromatin does lead to an increase of H3K27me3, but in a PRC2-independent manner (66). Although it remains possible that lncRNAs recruit PRC2 to specific loci in some cases, currently, the best-characterized function of RNAs is to inhibit PRC2 methyltransferase activity by inhibiting PRC2 binding to chromatin (67–69).
OTHER HISTONE MODIFIERS
In recent years, the theme of lncRNAs controlling gene activity by mediating epigenetic mechanisms has become much more pervasive. Broadly, lncRNAs exercise the roles of recruiters, decoys, stimuli, and scaffolds, or some combinations thereof. This section will provide examples of lncRNAs driving gene regulation through the activity of chromatin modifiers. These recent studies establish that lncRNAs contribute to many biological systems. Understanding how these lncRNAs function, on the other hand, is challenging, and therefore, the mechanisms proposed require more validation and may be modified by future studies.
G9a
G9a is a histone methyltransferase that deposits repressive methyl marks on H3K9. Its essential role is highlighted by lethality and severe growth defects observed in G9a-deficient embryos (70). It has been suggested that G9a targets transcriptionally active euchromatin regions, as opposed to repressive pericentric heterochromatin. G9a targeting has been associated with the regulation of genes important to development (71).
Three lncRNAs—Kcnq1ot1, Air, and ROR—have been suggested to interact with G9a via either the recruitment model or decoy model. Kcnq1ot1 is a 91-kb transcript, transcribed from the antisense strand of the Kcnq1 gene by RNA Pol II (72). It is exclusively localized in the nuclear compartment with moderate stability. In a ChIRP study, Kcnq1ot1 was shown to interact with chromatin. Additional RIP experiments used antibodies raised against G9a to pull out Kcnq1ot1 in a lineage-specific manner. This is consistent with a lineage-specific difference in the H3K9me3 modification in the Kcnq1 gene. The G9a-Kcnq1ot1 interaction also contributes to imprinting in the mouse placenta. A similar recruiting mechanism has been observed with Air lncRNAs, which are largely unspliced and retained in the nucleus. It has been shown that Air is involved in silencing clusters of multiple imprinted genes in cis on chromosome 17 in mice (73). This silencing mechanism involves Air recruiting G9a to the paternal Slc22a3.
In the case of lncRNA ROR, it has been demonstrated that ROR evicts G9a. Human ROR is 2.6 kb in length, and true to its name, it functions by reprogramming human induced pluripotent stem cells (iPSCs) and shares regulatory miRNAs with the transcription factors OCT4, SOX2, and NANOG. Fan et al. (74) show that ROR occupies and activates the TESC promoter by repelling the histone G9a methyltransferase and promoting the release of histone H3K9 methylation. This decoying mechanism leads to a reduction in tumor growth and metastasis.
Mixed lineage leukemia
MLL (mixed lineage leukemia) is a protein first identified as a functional ortholog of the trithorax (trx) complex in Drosophila (75). The canonical role of the MLL protein is to methylate H3K4, a trigger of gene activation. MLL has been shown to be required for the maintenance of activated genes during normal embryogenesis, hematopoiesis, and neurogenesis (76–78). The essentiality of MLL is underscored by the embryonic lethality of MLL-knockout mice.
How MLL might be recruited to specific genomic loci still remains to be fully elucidated. However, one study has shed light on a possible mechanism involving a lncRNA called HOTTIP (79, 80), which appears to play a role in the trafficking of MLL to specific HOXA genes. Specifically, Wang and colleagues show that HOTTIP interacts with the WDR5-MLL complex and localizes the complex to the 5′HOXA locus. Quite remarkably, a follow-up investigation identified a single residue (F266) on WDR5 that is necessary for RNA binding and indispensable for gene activation. A similar mechanism has been proposed with the lncRNA HoxBlinc (81), which recruits the Set1/MLL complex. This recruitment is followed by the transcriptional activation of the Hoxb gene, thus regulating cardiac/hematopoietic differentiation. These studies together highlight the profound role of lncRNA binding in the regulation of active chromatin states.
Another compelling aspect of the scaffold-like property of some lncRNAs is the ability of a single transcript to bind multiple chromatin-modifier complexes. For instance, the Fendrr lncRNA is specifically expressed in the nascent lateral mesoderm in a developing embryo and has been reported to interact with both MLL and PRC2 complexes (82). Fendrr targets complexes to specific promoters to alter the epigenetic landscape. These epigenetic changes lead to attenuation of the expression of transcription factors, which are important in lateral mesoderm development. Therefore, as knowledge about the factors binding lncRNAs increases, it will become important to begin investigating how different factors engage in functional cross-talk.
Heterochromatin protein 1
Heterochromatin protein 1 (HP1) was first characterized in Drosophila as localizing to heterochromatin and being involved in position-effect variegation (83, 84). HP1 binds methyl marks on H3K9 and elicits chromatin packaging and gene silencing. Early embryonic lethality in Drosophila is caused by the loss of HP1. In humans, the loss of HP1 has been shown to correlate with metastatic breast cancer (85).
The HP1 protein contains a conserved N-terminal chromodomain, followed by a variable hinge region, and finally a conserved chromoshadow domain at the C terminus. The chromodomain has been suggested to be an RNA binding module; for example, the MOF histone acetyltransferase (HAT) in Drosophila (described above) specifically interacts with roX RNA via its chromodomain (39), and these protein-RNA interactions may contribute to the recruitment of MOF to the X chromosome in male Drosophila. Evolutionarily, HP1’s chromodomain shares homology to the MOF variant; therefore, it has been speculated that RNA acts similarly to recruit HP1 to pericentromeric loci. One early study found that the RNase treatment of cells induces dispersion of HP1 from the pericentromeric foci (86). Furthermore, replenishing RNase-treated cells with purified nuclear RNA rescues the pericentric structures. Later, it was shown that HP1 directly binds nuclear RNA using the electrophoretic mobility shift assay (EMSA). These early studies collectively suggested that the mechanism of HP1-mediated chromatin compaction in cells involves RNA actors. However, they did not offer a functional connection between specific RNAs and HP1.
In 2011, Maison and colleagues (87) reported that strand-specific centromeric RNAs (transcribed in the forward direction) colocalize with HP1 in mouse cells. Using HP1 chromatin immunoprecipitation (ChIP) experiments, it was observed that HP1 is enriched at the genomic regions encoding the centromeric RNAs. This study helped confirm a particular link between the subnuclear localization of RNA transcripts and HP1 recruitment. Maison et al. also revealed that the posttranslational SUMOylation of HP1 actively promotes binding of the protein to the purine-rich sense RNA transcripts, and the combinatorial effects of SUMO and RNA binding together initiate targeting of HP1 to pericentric heterochromatin. This work reveals how PTMs of chromatin readers and remodelers might regulate their intrinsic binding properties and, subsequently, recruitment.
In addition to RNA having a recruitment role that captures free HP1 to pericentric heterochromatin, an antagonizing “eviction” role of lncRNA has also been proposed. One such study has identified a lncRNA called BORDERLINE in Schizosaccharomyces pombe (88), which, when processed into short RNAs, evicts HP1 and prevents the spreading of HP1 and histone H3K9 methylation beyond the pericentromeric repeat region.
Suv4-20h
At pericentric and telomeric regions of chromosomes, heterochromatin formation is orchestrated by a series of interactions involving Suv39h, HP1, and Suv4-20h. Current literature shows that Suv39h methylates H3K9, which serves as a precursor to binding of HP1. Upon binding of methylated H3K9, HP1 recruits Suv4-20h by direct protein-protein interaction. Then, Suv4-20h proceeds to establish H4K20me3 marks. Alternative ways of targeting Suv4-20h to H4K20 have been proposed. One study provides evidence implicating lncRNA in this mechanism. Specifically, pre–ribosomal RNA (rRNA) antisense transcripts (PAPAS) bind pre-rRNA coding regions and recruit Suv4-20h2 in quiescent cells (89). This recruitment promotes H4K20me3-mediated transcriptional silencing of ribosomal DNA (rDNA). In addition, the authors observed a similar scheme at retrotransposon elements, where lncRNA triggers H4K20me3 and transcriptional repression.
Polycomb repressive complex 1
PRC1 has a core that consists of four proteins: Bmi1, HPH, Ring1, and CBX. The chromodomain of CBX binds to trimethylated histone H3 Lys27 and initiates the direct catalysis of H2A119 ubiquitination. These ubiquitination marks have been thought for years to recruit PRC2 in hierarchical fashion and subsequently enforce gene silencing and chromatin compaction. However, recent studies have provided alternative models that reveal emerging roles for PRC1. Details can be found in the review by Gil and O’Loghlen (90).
Regarding PRC1, an early study by Bernstein and colleagues (91) suggested that CBX proteins bind RNAs in vitro. Around 2010, two more studies provided mechanistic insights into the functional connections between lncRNA and CBX proteins. In particular, the antisense lncRNA ANRIL, which is transcribed from the INK4b/ARF/INK4a tumor suppressor locus, recruits PRC1 to that specific locus for transcriptional repression via a direct interaction with the CBX7 subunit. This repression regulates senescence and proliferation of prostate cancer cells. The authors observed a possible ternary complex consisting of H3K27me3-ANRIL-CBX7 (92).
Another CBX subunit of PRC1 that binds lncRNA is CBX4. Some lncRNA transcripts known to interact with CBX4 include TUG1 and MALAT1/NEAT2. These CBX4-RNA interactions stimulate the SUMOylation of the E2F1 growth factor (93), a PTM that results in increased cellular proliferation. Intriguingly, the methylation status of CBX4 appears to dictate whether CBX4 interacts with TUG1 or MALAT1/NEAT2, with the unmethylated variant binding the latter. Given that TUG1 and MALAT1/NEAT1 exhibit differential subnuclear localization, with TUG1 being localized to Polycomb bodies (PcGs) and MALAT1/NEAT2 located in interchromatin granules (ICGs), the methylation status of CBX4 can therefore dictate where PRC1 traffics in the subnuclear environment. This work provides a clear example of how lncRNAs can act as scaffolds to organize nuclear architecture and influence recruitment of chromatin-modifier complexes.
p300/CBP
p300 and CBP [cyclic adenosine monophosphate response element–binding protein (CREB)–binding protein] are two highly homologous and conserved proteins that have intrinsic HAT activity, which plays a critical role in regulating gene expression through lysine acetylation of histone H3 (94). These proteins act as transcriptional coactivators for a number of nuclear genes. Unlike other HATs, p300 and CBP are able to acetylate all four histones both in vitro and in vivo (95). Therefore, they are capable of coupling with a variety of transcription factors during chromatin remodeling. Not surprisingly, they are involved in a wide array of basic cellular processes, such as DNA damage repair and cell proliferation, and are inherently crucial for embryonic development and cancer.
For the first time in 2015 (96), p300 and CBP were implicated to interact with lncRNA. The study presents a fascinating model where the antisense lncRNA Khps1 forms a DNA/RNA triplex with the SPHK1 promoter, and these triplexes recruit CBP/p300. This recruitment triggers an open chromatin structure, followed by binding of transcription factors, and eventually leads to the activation of SPHK1 transcription.
Recently, a genome-wide analysis of p300/CBP binding to RNA using PAR-CLIP (photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation) was published (97). Bose and colleagues suggest that RNA transcribed locally directly interacts with CBP and stimulates catalytic HAT activity, thereby promoting gene expression. They also suggest that eRNAs at enhancers may interact with p300/CBP and control transcription activation.
Lysine-specific demethylase 1
Lysine-specific demethylase 1 (LSD1) is a protein responsible for removing mono- and dimethyl modifications from H3K4 and H3K9 of histones, and it plays a pivotal role during embryonic development and cancer (98). This is highlighted by a variety of tumors that display LSD1 overexpression (99). LSD1 is the first identified histone demethylase and has been found to associate with a number of transcriptional corepressor complexes (including CoREST and CtBP) and a subset of HDAC complexes (99). The lncRNA HOTAIR, transcribed from the HoxC locus, has been reported to interact with LSD1 and also with PRC2 (100). This illustrates the scaffold-like function of some lncRNAs, which has been studied in detail in the case of yeast TR (101).
Curiously, the TERRA (telomeric repeat–containing RNA) RNA transcribed from telomeres has been shown to interact with LSD1. TERRA is bound by LSD1 at TRF2-depleted telomeres, and the RNA promotes the physical interaction between LSD1 and MRE11 (102). This physical interaction stimulates MRE11 nuclease activity and consequently stimulates removal of 3′G-strand overhangs at uncapped telomeres.
DNA METHYLTRANSFERASES
DNA methylation is widely involved in transcriptional repression in mammals, with DNA methyltransferases (DNMTs) methylating cytosines (m5C) in CpG-rich sequences. Growing evidence has demonstrated that all three major DNMTs (DNMT1, DNMT3A, and DNMT3B) in mammals bind to and can be regulated by noncoding RNAs. This regulation by noncoding RNAs usually results in alteration of DNA methylation and expression of target genes (Fig. 4). DNMT2, the enigmatic DNMT homolog, has little DNA methylation activity but instead methylates transfer RNAs (tRNAs) (103, 104), and this could imply that the RNA binding properties of DNMTs have a deep evolutionary origin.
Fig. 4. ncRNAs regulate transcription through DNA binding proteins.
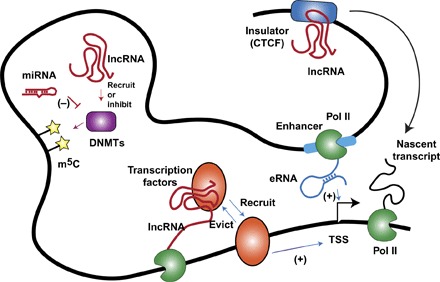
lncRNAs (and sometimes miRNAs) interact with DNMTs, resulting in recruitment or inhibition of DNMTs at chromatin loci. Alteration of DNA methylation (m5C) level generally affects local transcription. lncRNA–transcription factor interactions can either recruit or evict transcription factors from chromatin, and this action can be either in cis (demonstrated in figure) or in trans. eRNA transcribed from an enhancer region can contribute to chromatin looping and gene activation. lncRNAs interact with the chromatin insulator CCCTC-binding factor (CTCF) and regulate transcription. The mechanism may involve CTCF’s action in chromatin looping and nuclear architecture. TSS, transcription start site.
DNA methyltransferase 1
DNMT1 is the key maintenance DNMT and is ubiquitously expressed in proliferating cells. A few RNAs have been identified to regulate the activity of DNMT1. Kcnq1ot1, a lncRNA that regulates the Kcnq1 imprinting control region (105), interacts with and recruits DNMT1 to differentially methylated regions (106). However, whether the interaction between DNMT1 and Kcnq1ot1 lncRNA is direct or indirect is unclear because of the limitation of RIP experiments, as discussed earlier in this review.
Later, a noncoding RNA named ecCEBPA arising from the CEBPA (CCAAT/enhancer-binding protein α) gene locus was found to interact with DNMT1, resulting in decreased methylation at the CEBPA gene (107). Surprisingly, in this study, DNMT1 bound to RNA with a greater affinity than to its own substrate DNA in vitro, and DNMT1 seemed to prefer binding with stem loop–structured RNAs. This study also provided mutagenesis scanning of the DNMT1 protein and suggested that the catalytic domain is the minimal RNA binding motif. DNMT1-RNA interaction is not limited to the CEBPA locus, and RNA species associated with DNMT1 and their regulation of DNA methylation and gene expression have been globally identified, indicating that RNA could block unwanted DNA methylation at many sites of active transcription.
Moreover, two later studies supported the inhibition model. A lncRNA named DBCCR1-003 binds to DNMT1 and prevents DNMT1-mediated methylation of DBCCR1 in bladder cancer (108), and lncRNA RBMY2FP interacts with DNMT1 and hampers its binding to promoters of the RBMY gene family (109). Besides lncRNAs, miRNAs including miR-155-5p also bind to and inhibit DNMT1 in vitro and in vivo (110), suggesting a widespread role of RNA interaction in regulating the activity of DNMT1.
DNA methyltransferase 3A
DNMT3A and DNMT3B are the de novo DNMTs and show no preference between hemimethylated and unmethylated DNA substrates (111, 112). Tsix RNA (the antisense of Xist) can form a complex with DNMT3A, as demonstrated by an RNA-ChIP analysis, but not with DNMT1, DNMT2, or DNMT3B (113). Tsix RNA might activate DNMT3A at the Xist promoter and repress the Xist gene. The molecular details of the DNMT3A-RNA interaction were later demonstrated in vitro as two modes: allosteric regulation (no change in catalysis) and catalytic domain binding (potent inhibition) (114), supporting a model that the molecular basis of the DNMT3A-RNA interaction determines the effects on DNA methylation activity of DNMT3A.
DNA methytransferase 3B
DNMT3B can be recruited by a DNA/RNA triplex, formed by a noncoding RNA [promoter-associated RNA (pRNA)] and the rDNA promoter region (115). DNMT3B, but not DNMT1 or DNMT3A, shows a preference for DNA/RNA triplexes in vitro, suggesting a potentially widespread RNA-guided DNMT3B targeting mechanism in epigenetic regulation.
TRANSCRIPTION FACTORS
Transcription factors are sequence-specific DNA binding proteins that can activate or repress transcription. Growing evidence suggests that a large number of transcription factors interact with RNA, and these interactions could play an important role in their regulation [Fig. 4; also reviewed by Hudson and Ortlund (8)]. A classic example is the feedback inhibition of zinc finger protein transcription factor IIIA (TFIIIA) by 5S rRNA. TFIIIA activates 5S rRNA transcription Xenopus oocytes, and increasing levels of the 5S rRNA products strip TFIIIA off chromatin (116–120). Similar to this inhibition mechanism, GAS5 lncRNA acts as a decoy RNA by binding to the DNA binding domain of glucocorticoid receptor (GR), inhibiting GR’s DNA binding and transcriptional activation activity (121).
In contrast to the inhibition and decoy mechanism, RNA can also activate or recruit transcription factors. YY1 is a ubiquitous transcription factor that can activate or repress individual promoters. In mammals, X-chromosome inactivation requires the interaction between YY1 and Xist lncRNA. YY1 interacts with the Repeat C region of Xist RNA and contributes to docking Xist RNA onto the X chromosome (122). YY1 binds to both gene regulatory elements and their associated RNA species (transcribed from the same regulatory elements) across the entire genome, and artificial tethering of RNA enhances YY1 occupancy at these elements, potentially providing a positive feedback for robust transcription (123). A lncRNA transcribed from the YY1 gene promoter (linc-YY1) interacts with YY1 through its middle domain to evict YY1-PRC2 from target promoters, thus activating gene expression in trans (124). A recent SELEX (systematic evolution of ligands by exponential enrichment) study found that YY1 interacts with RNA with low sequence specificity (125), and the lack of RNA sequence specificity has been commonly observed among chromatin binders more generally. The biochemical and structure nature of the protein-RNA interaction may determine the inhibition or activation mechanism. If the DNA and RNA binding are shared as in the cases of TFIIIA and GR, then an inhibition mechanism is more likely to be used. Otherwise, the inhibition mechanism may apply when the DNA and RNA binding are mutually exclusive as for YY1 (123).
Besides these well-studied interactions, an increasing number of lncRNAs have been demonstrated to regulate transcription through transcription factors. A lncRNA transcribed from the CDKN1A promoter, PANDA, differentially interacts with the transcription factor NF-YA (nuclear transcription factor Y subunit alpha) or PRCs (PRC1 and PRC2) to either promote or suppress senescence (126). The lncRNA rhabdomyosarcoma 2–associated transcript (RMST) interacts directly with SOX2 protein to activate gene expression, and chromatin occupancy of SOX2 was reduced following RMST depletion (127, 128). A few other examples are listed in Table 1. Most of the lncRNA–transcription factor interactions have been demonstrated by RIP experiments, so indirect versus direct interactions could not be differentiated. Identification of the RNA binding motif in the protein and the corresponding RNA identity element would be helpful to confirm these interactions.
Table 1. Transcription factors and their interacting RNAs.
N/A, not applicable; PVT1, plasmacytoma variant translocation 1; HSF1, heat shock factor 1; HSR1, heat shock RNA 1; DHFR, dihydrofolate reductase; ICR1, interfering Crick RNA 1; PWR1, promoting Watson RNA 1.
|
Transcription factor |
RNA binder | Evidence of interaction | Function of interaction | References |
| YY1 | Xist, linc-YY1, and gene regulatory element–associated RNAs |
RIP, EMSA, and in vivo tethering | YY1 helps to tether Xist RNA to the inactive X nucleation center; YY1 trapping by lncRNA for robust activation |
(122) (123) (124) (125) |
| NFAT | NRON noncoding RNA | N/A | Represses NFAT | (129) |
| NF-YA | PANDA lncRNA | RNA pull down | Attenuates NF-YA’s occupancy at target gene promoters | (126) |
| SOX2 | RMST lncRNA | RIP | Activates gene transcription | (127, 128) |
| MYC | PVT1 lncRNA | Genetic disruption | Prevents phosphorylation and degradation of MYC | (160) |
| TFIIIA | 5S rRNA | Mutagenesis, binding assays |
Regulates TFIIIA activity | (116–120) |
| TFIIB | DHFR upstream transcripts | EMSA and RIP | Dissociates the preinitiation complex from the major promoter | (161) |
| HSF1 | HSR1 lncRNA | In vitro activation assay and in vivo RIP |
Activates HSF1 | (162) |
| Flo8 and Sfl1 | ICR1 and PWR1 lncRNAs | Genetic disruption and ChIP analysis |
Dictates the variegated expression of FLO11 mRNA in yeast | (149, 163) |
| PURB | linc-HOXA1 RNA | RIP | Represses Hoxa1 by recruiting PURB as a transcriptional cofactor |
(164) |
A few other studies infer roles of lncRNAs in regulating transcription factor activities, although no direct physical interaction was reported. For example, the NRON lncRNA forms a complex with a few other proteins to repress the transcription factor NFAT (nuclear factor of activated T cell), potentially by regulating NFAT’s subcellular localization (129). Future in vitro EMSA or in vivo pull-down experiments would be helpful to identify these interactions.
OTHER DNA BINDING TRANSCRIPTION REGULATORS
CCCTC-binding factor
CTCF is a ubiquitous zinc finger protein that binds DNA and serves as a chromatin insulator, activator, or repressor, depending on the epigenetic context [reviewed by Ong and Corces (130) and Phillips and Corces (131)]. CTCF was first demonstrated to interact with RNA in the initiation of XCI (132). CTCF represses Xist expression by binding to its promoter and is later titrated away from the Xist promoter by Jpx RNA when XCI is initiated. The interaction between CTCF and Xist is supported by in vivo UV-crosslinked RIP (UV-RIP) and in vitro gel shift assays.
Later, a number of lncRNAs were found to regulate CTCF and affect transcription. CTCF forms a complex with DEAD-box RNA helicase p68 (DDX5) and an associated noncoding RNA, steroid receptor RNA activator (SRA), and this complex may be essential for CTCF’s insulator function (133). In another study, CTCF was found to regulate p53 expression through its physical interaction with Wrap53 RNA, and an RNA binding region (residues 576 to 614) of CTCF was identified by in vitro scanning mutagenesis (134). This study also discovered that CTCF interacts with a variety of RNAs in vivo using a PAR-CLIP experiment. A later study further confirmed the observation that CTCF binds thousands of transcripts in vivo in mouse embryonic stem cells, including Tsix, Xite, and Xist RNAs, many in close proximity to CTCF’s genomic binding sites (135). The function of the RNP formed by CTCF in XCI was further expanded by a study demonstrating that lncRNA Firre (functional intergenic repeating RNA element) colocalizes with CTCF on the X chromosome, although no evidence for direct interaction was presented (136). A mutagenesis study of CTCF and one of its RNA binders—MYCNOS lncRNA—implicated zinc fingers 9 to 11 (500 to 576 amino acids) of CTCF and exons 1 and 3 of MYCNOS lncRNA as the crucial interacting regions (137).
α-Thalassemia/mental retardation X-linked
α-Thalassemia/mental retardation X-linked (ATRX) is a transcriptional regulator that belongs to the SWI/SNF (switch/sucrose nonfermentable) family of chromatin remodeling proteins. It has ATPase activity that is stimulated by naked DNA and mononucleosomes (138, 139) and also has a DNA translocase activity (139, 140). In a more recent study, ATRX was unexpectedly found to function as a high-affinity RNA binding protein, which directly interacts with RepA/Xist RNA to promote loading of PRC2 in vivo (141). Interaction between ATRX and Xist RNA is supported by UV-RIP analysis and in vitro gel shift assays, and a minimal RepA region on Xist has been identified as the specific identity element (141). Although ATRX seems to be able to bind to both double-stranded DNA (dsDNA) and RNA with similar affinities and probably using different regions of the protein, an RNA-ATRX-dsDNA ternary complex was not detected using a pull-down experiment. Furthermore, a separation-of-function mutant of ATRX that still binds DNA but no longer binds RNA would be useful to confirm the importance of RNA binding in XCI. In addition, ATRX is also related to the function of TERRA lncRNA [reviewed by Rippe and Luke (142)], but a direct interaction has not been demonstrated.
Distal-less homeobox 2
Homeobox protein DLX2 (distal-less homeobox 2) is a DNA binding transcription regulator. EVF2 lncRNA is a transcriptional coactivator of DLX2 and recruits methyl-CpG–binding protein 2 (MECP2) to intergenic enhancers. A direct interaction between DLX2 and EVF2 has been demonstrated by the immunoprecipitation of DLX2, followed by reverse transcription polymerase chain reaction of the EVF2 lncRNA, and is supported by their intranuclear colocalization (143, 144). A conserved region of EVF2 has been shown to be important for the interaction with DLX2, and the interaction is important in maintaining the transcriptional activity of the DLX5/6 enhancer.
THE IMPACT OF TRANSCRIPTION ITSELF
It has also been shown that the act of transcription, rather than the RNA product of transcription, can have regulatory effects on neighboring loci in the mammalian nucleus. Numerous groups have recently reported on this phenomenon. One group found that transcription of the mouse noncoding Airn gene leads to silencing of the overlapping Igf2r gene in mice, but the Air lncRNA product is not required (145). Another study found that genetically disrupting transcription at both lncRNA and protein-coding loci was sufficient to dysregulate the expression of neighboring genes (146). An additional report discovered an essential interplay between the heart development gene HAND2 and active transcription of an upstream lncRNA termed upperhand (Uph) (147). The researchers found that disrupting Uph transcription abolished Hand2 expression, whereas knockdown of the mature Uph transcript had no effect (147).
These observations were presaged by studies in budding yeast. For example, transcriptional repression of the SER3 gene depends on active transcription of nearby noncoding sequences, which interferes with the binding of activators to the SER3 promoter (148). In another case, two yeast lncRNAs transcribed in opposite directions provide a “toggle switch” that can either repress or activate the transcription of an adjacent protein-coding gene (149). Thus, in general, many lncRNAs may function not at the RNA level, but rather simply because they are transcribed, and this transcription can lead to activation or repression of nearby genes.
lncRNAs ORGANIZING NUCLEAR ARCHITECTURE
There are several prominent examples of lncRNAs acting as platforms for nuclear organization [reviewed by Melé and Rinn (150)]. One well-studied example is NEAT1, a lncRNA that is sufficient to generate nuclear paraspeckles (150, 151). Recently, a lncRNA, termed Firre, was identified as essential for adipocyte differentiation, and it displays a unique function in nuclear organization (136, 152, 153). Genetic deletion studies of Firre in embryonic stem cells showed that the lncRNA interacts with hnRNPU and mediates cross-chromosomal contacts between five chromosomes (153). Knockout of the transcript had no effect on the expression of the neighboring genomic loci (153). Additionally, oligonucleotide-mediated knockdown of this transcript was shown to dysregulate a myriad of RNA processing genes (154).
LOOKING INTO THE FUTURE, WHAT KINDS OF EXPERIMENTS ARE NEEDED TO ESTABLISH MECHANISM?
Many published studies show an association between a lncRNA and transcriptional repression or activation. A reasonable next question to ask is whether the association is causative; establishing causation requires perturbing the production, stability, nucleotide sequence, or cellular location of the lncRNA and observing a concomitant change in expression of specific gene(s). This set of experiments provides the starting point for understanding mechanism; that is, how does the lncRNA perform its function?
In considering mechanisms, we start with cases in which a lncRNA represses transcription. One possibility is that the lncRNA acts as a “sponge” or “decoy,” binding a transcription-activating protein and preventing it from associating with its DNA or RNA target. Deciding whether this mechanism is tenable requires measuring the stoichiometry of the lncRNA relative to the protein and the relative affinity of the protein for the lncRNA versus the target nucleic acid. For example, a lncRNA present at 100 copies per cell is unlikely to provide an efficient sponge for a transcriptional protein present at 10,000 copies per cell, unless there is some special compartmentalization. A related possibility is that RNA binding functionally inactivates the protein. For example, if a transcriptional regulatory protein binds its DNA target and a lncRNA at the same site (or mutually exclusively), then the mechanism of lncRNA inhibition is clear. Mutually exclusive binding can be tested, for example, by competition binding experiments (155) or by determining cocrystal structures of the protein-DNA and protein-lncRNA complexes (8).
We next consider cases in which the lncRNA activates transcription. One possible mechanism involves the lncRNA recruiting an activating protein to its site of action. To establish this mechanism, one must first find the sequences and/or structures on the lncRNA that bind the protein, for example, using motif searching, mutagenesis, and protein-RNA footprinting. If transcriptional activation occurs in cis, then binding to nascent transcripts may, by itself, accomplish recruitment. However, if activation is occurring in trans, then establishing the protein-lncRNA binding is only half the story, and one must next ask how the lncRNA facilitates recruitment. The three most obvious possibilities are (i) the lncRNA binds to a protein bound at the target locus, (ii) the lncRNA base pairs to nascent transcripts at the target locus, or (iii) the lncRNA binds directly to the DNA at the target locus by triplex formation or potentially R-loop formation. In cases where intermolecular RNA-RNA base pairing is implicated, the observation of sequence complementarity is insufficient to indicate pairing. Convincing evidence can be obtained by introducing mutations that disrupt pairing and compensatory second-site mutations and by psoralen photocrosslinking (156).
Another potential function of lncRNAs is to act as a scaffold, bringing proteins together in an RNP complex. In the simplest cases, the RNA will have individual sequence and structure motifs, each of which binds a single protein or protein complex. If the only function of the lncRNA is to act as a scaffold, then deletion of a protein binding motif will result in loss of function, but reinsertion of that motif at an unnatural location within the RNA will restore function (157).
Finally, how does one test the hypothesis that active transcription of noncoding sequences, rather than the lncRNA product of this transcription, controls the expression of another gene(s)? As has been discussed by Bassett et al. (158), this is particularly challenging. Deletion of the promoter for the lncRNA [for example, by CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 genome editing] is often too crude of an approach, because the deletion may remove DNA elements that regulate the transcription of a proximal mRNA gene (146). Inserting multiple polyadenylation sequences downstream from a lncRNA promoter and interfering with lncRNA transcription using CRISPR interference (159) are more surgically incisive, but negative results must be interpreted with caution because these techniques truncate rather than eliminate lncRNA transcription. Replacing a lncRNA sequence with a different sequence without functional consequence does not necessarily indicate that the lncRNA is unimportant, given the fact that many histone-modifying complexes bind RNA promiscuously and thus might still bind the substituted sequence. Moreover, knockdown of the lncRNA product with antisense nucleic acids (Gapmers) that function in the nucleus may suggest the unimportance of the lncRNA product, but unlike genome editing, these knockdowns are incomplete. Thus, it is best to use multiple approaches when assessing whether it is the act of transcription or the lncRNA product that is functional in any particular instance.
In conclusion, the era of cataloging mammalian and other eukaryotic lncRNAs is well under way. Researchers are determining their tissue distribution, subcellular localization, expression patterns, abundance, and splicing patterns, which requires enormous effort but is reasonably straightforward. The era of determining the function of these lncRNAs, on the other hand, is just beginning.
Acknowledgments
We appreciate helpful discussions and suggestions from J. L. Rinn, J. A. Goodrich, and J. F. Kugel (all from University of Colorado Boulder). Funding: T.R.C. is an investigator of the Howard Hughes Medical Institute, which provided funding in support of this work. This work was also supported by a grant from the NIH (GM099705 to T.R.C.). Author contributions: All authors contributed to the writing of this article. Competing interests: T.R.C. is on the board of directors of Merck Inc., which provides no funding for his research. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the articles cited herein.
REFERENCES AND NOTES
- 1.Liu S. J., Horlbeck M. A., Cho S. W., Birk H. S., Malatesta M., He D., Attenello F. J., Villalta J. E., Cho M. Y., Chen Y., Mandegar M. A., Olvera M. P., Gilbert L. A., Conklin B. R., Chang H. Y., Weissman J. S., Lim D. A., CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, eaah7111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., Notani D., Rosenfeld M. G., Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 17, 207–223 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Cech T. R., Steitz J. A., The noncoding RNA revolution—Trashing old rules to forge new ones. Cell 157, 77–94 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Lemieux B., Laterreur N., Perederina A., Noël J.-F., Dubois M.-L., Krasilnikov A. S., Wellinger R. J., Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell 165, 1171–1181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrickson D. G., Kelley D. R., Tenen D., Bernstein B., Rinn J. L., Widespread RNA binding by chromatin-associated proteins. Genome Biol. 17, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Schwartz J. C., Cech T. R., Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 43, 7535–7543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidovich C., Zheng L., Goodrich K. J., Cech T. R., Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 20, 1250–1257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson W. H., Ortlund E. A., The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 15, 749–760 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiDomenico B. J., Bugaisky G. E., Lindquist S., The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell 31, 593–603 (1982). [DOI] [PubMed] [Google Scholar]
- 10.Findly R. C., Pederson T., Regulated transcription of the genes for actin and heat-shock proteins in cultured Drosophila cells. J. Cell Biol. 88, 323–328 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmour D. S., Lis J. T., In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol. Cell. Biol. 5, 2009–2018 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien T., Lis J. T., Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 13, 3456–3463 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W. M., Chu W. M., Choudary P. V., Schmid C. W., Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 23, 1758–1765 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornace A. J. Jr, Mitchell J. B., Induction of B2 RNA polymerase III transcription by heat shock: Enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res. 14, 5793–5811 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lunyak V. V., Prefontaine G. G., Núñez E., Cramer T., Ju B.-G., Ohgi K. A., Hutt K., Roy R., García-Díaz A., Zhu X., Yung Y., Montoliu L., Glass C. K., Rosenfeld M. G., Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317, 248–251 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Li T.-H., Spearow J., Rubin C. M., Schmid C. W., Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene 239, 367–372 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Kramerov D. A., Vassetzky N. S., SINEs. Wiley Interdiscip. Rev. RNA 2, 772–786 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Allen T. A., Von Kaenel S., Goodrich J. A., Kugel J. F., The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 11, 816–821 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Kramerov D. A., Vassetzky N. S., Short retroposons in eukaryotic genomes. Int. Rev. Cytol. 247, 165–221 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Espinoza C. A., Allen T. A., Hieb A. R., Kugel J. F., Goodrich J. A., B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 11, 822–829 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Mili S., Steitz J. A., Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wassarman K. M., Storz G., 6S RNA regulates E. coli RNA polymerase activity. Cell 101, 613–623 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Espinoza C. A., Goodrich J. A., Kugel J. F., Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA 13, 583–596 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yakovchuk P., Goodrich J. A., Kugel J. F., B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. U.S.A. 106, 5569–5574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettenberger H., Eisenführ A., Brueckner F., Theis M., Famulok M., Cramer P., Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat. Struct. Mol. Biol. 13, 44–48 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Zovoilis A., Cifuentes-Rojas C., Chu H.-P., Hernandez A. J., Lee J. T., Destabilization of B2 RNA by EZH2 activates the stress response. Cell 167, 1788–1802.e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rastan S., X chromosome inactivation and the Xist gene. Curr. Opin. Genet. Dev. 4, 292–297 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Kelley R. L., Kuroda M. I., Equality for X chromosomes. Science 270, 1607–1610 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Lucchesi J. C., Dosage compensation in Drosophila and the ‘complex’ world of transcriptional regulation. BioEssays 18, 541–547 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Rutenberg-Schoenberg M., Sexton A. N., Simon M. D., The properties of long noncoding RNAs that regulate chromatin. Annu. Rev. Genomics Hum. Genet. 17, 69–94 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Amrein H., Axel R., Genes expressed in neurons of adult male Drosophila. Cell 88, 459–469 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Meller V. H., Rattner B. P., The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21, 1084–1091 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franke A., Baker B. S., The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell 4, 117–122 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Kelley R. L., Meller V. H., Gordadze P. R., Roman G., Davis R. L., Kuroda M. I., Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98, 513–522 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Meller V. H., Wu K. H., Roman G., Kuroda M. I., Davis R. L., roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88, 445–457 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Smith E. R., Pannuti A., Gu W., Steurnagel A., Cook R. G., Allis C. D., Lucchesi J. C., The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20, 312–318 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akhtar A., Zink D., Becker P. B., Chromodomains are protein-RNA interaction modules. Nature 407, 405–409 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Hallacli E., Akhtar A., X chromosomal regulation in flies: When less is more. Chromosome Res. 17, 603–619 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akhtar A., Becker P. B., Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Smith E. R., Allis C. D., Lucchesi J. C., Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 276, 31483–31486 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Ilik I. A., Quinn J. J., Georgiev P., Tavares-Cadete F., Maticzka D., Toscano S., Wan Y., Spitale R. C., Luscombe N., Backofen R., Chang H. Y., Akhtar A., Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell 51, 156–173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maenner S., Müller M., Fröhlich J., Langer D., Becker P. B., ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol. Cell 51, 174–184 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Izzo A., Regnard C., Morales V., Kremmer E., Becker P. B., Structure-function analysis of the RNA helicase maleless. Nucleic Acids Res. 36, 950–962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prabu J. R., Muller M., Thomae A. W., Schüssler S., Bonneau F., Becker P. B., Conti E., Structure of the RNA helicase MLE reveals the molecular mechanisms for uridine specificity and RNA-ATP coupling. Mol. Cell 60, 487–499 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Buscaino A., Köcher T., Kind J. H., Holz H., Taipale M., Wagner K., Wilm M., Akhtar A., MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol. Cell 11, 1265–1277 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Kind J., Vaquerizas J. M., Gebhardt P., Gentzel M., Luscombe N. M., Bertone P., Akhtar A., Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133, 813–828 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Alekseyenko A. A., Peng S., Larschan E., Gorchakov A. A., Lee O.-K., Kharchenko P., McGrath S. D., Wang C. I., Mardis E. R., Park P. J., Kuroda M. I., A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134, 599–609 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Straub T., Grimaud C., Gilfillan G. D., Mitterweger A., Becker P. B., The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLOS Genet. 4, e1000302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li F., Schiemann A. H., Scott M. J., Incorporation of the noncoding roX RNAs alters the chromatin-binding specificity of the Drosophila MSL1/MSL2 complex. Mol. Cell. Biol. 28, 1252–1264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soruco M. M. L., Chery J., Bishop E. P., Siggers T., Tolstorukov M. Y., Leydon A. R., Sugden A. U., Goebel K., Feng J., Xia P., Vedenko A., Bulyk M. L., Park P. J., Larschan E., The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 27, 1551–1556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davidovich C., Wang X., Cifuentes-Rojas C., Goodrich K. J., Gooding A. R., Lee J. T., Cech T. R., Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell 57, 552–558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaneko S., Son J., Shen S. S., Reinberg D., Bonasio R., PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1258–1264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Goodrich K. J., Gooding A. R., Naeem H., Archer S., Paucek R. D., Youmans D. T., Cech T. R., Davidovich C., Targeting of Polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell 65, 1056–1067.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Augui S., Nora E. P., Heard E., Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429–442 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Brockdorff N., Noncoding RNA and Polycomb recruitment. RNA 19, 429–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidovich C., Cech T. R., The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 21, 2007–2022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalantry S., Magnuson T., The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLOS Genet. 2, e66 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McHugh C. A., Chen C.-K., Chow A., Surka C. F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A., Sweredoski M. J., Shishkin A. A., Su J., Lander E. S., Hess S., Plath K., Guttman M., The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chu C., Zhang Q. C., da Rocha S. T., Flynn R. A., Bharadwaj M., Calabrese J. M., Magnuson T., Heard E., Chang H. Y., Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minajigi A., Froberg J. E., Wei C., Sunwoo H., Kesner B., Colognori D., Lessing D., Payer B., Boukhali M., Haas W., Lee J. T., A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark M. D., Marcum R., Graveline R., Chan C. W., Xie T., Chen Z., Ding Y., Zhang Y., Mondragón A., David G., Radhakrishnan I., Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex. Proc. Natl. Acad. Sci. U.S.A. 112, E3669–E3678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Millard C. J., Varma N., Saleh A., Morris K., Watson P. J., Bottrill A. R., Fairall L., Smith C. J., Schwabe J. W. R., The structure of the core NuRD repression complex provides insights into its interaction with chromatin. eLife 5, e13941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monfort A., Di Minin G., Postlmayr A., Freimann R., Arieti F., Thore S., Wutz A., Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moindrot B., Cerase A., Coker H., Masui O., Grijzenhout A., Pintacuda G., Schermelleh L., Nesterova T. B., Brockdorff N., A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amândio A. R., Necsulea A., Joye E., Mascrez B., Duboule D., Hotair is dispensible for mouse development. PLOS Genet. 12, e1006232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Portoso M., Ragazzini R., Brenčič Ž., Moiani A., Michaud A., Vassilev I., Wassef M., Servant N., Sargueil B., Margueron R., PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 36, 981–994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaneko S., Son J., Bonasio R., Shen S. S., Reinberg D., Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 28, 1983–1988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cifuentes-Rojas C., Hernandez A. J., Sarma K., Lee J. T., Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell 55, 171–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beltran M., Yates C. M., Skalska L., Dawson M., Reis F. P., Viiri K., Fisher C. L., Sibley C. R., Foster B. M., Bartke T., Ule J., Jenner R. G., The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 26, 896–907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y., G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zylicz J. J., Dietmann S., Günesdogan U., Hackett J. A., Cougot D., Lee C., Surani M. A., Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. eLife 4, e09571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pandey R. R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-DiNardo D., Kanduri C., Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Wagschal A., Sutherland H. G., Woodfine K., Henckel A., Chebli K., Schulz R., Oakey R. J., Bickmore W. A., Feil R., G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell. Biol. 28, 1104–1113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fan J., Xing Y., Wen X., Jia R., Ni H., He J., Ding X., Pan H., Qian G., Ge S., Hoffman A. R., Zhang H., Fan X., Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 16, 139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schuettengruber B., Martinez A.-M., Iovino N., Cavalli G., Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Lim D. A., Huang Y.-C., Swigut T., Mirick A. L., Garcia-Verdugo J. M., Wysocka J., Ernst P., Alvarez-Buylla A., Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morey L., Santanach A., Di Croce L., Pluripotency and epigenetic factors in mouse embryonic stem cell fate regulation. Mol. Cell. Biol. 35, 2716–2728 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McMahon K. A., Hiew S. Y.-L., Hadjur S., Veiga-Fernandes H., Menzel U., Price A. J., Kioussis D., Williams O., Brady H. J. M., Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1, 338–345 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Wang K. C., Yang Y. W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B. R., Protacio A., Flynn R. A., Gupta R. A., Wysocka J., Lei M., Dekker J., Helms J. A., Chang H. Y., A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Y. W., Flynn R. A., Chen Y., Qu K., Wan B., Wang K. C., Lei M., Chang H. Y., Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 3, e02046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deng C., Li Y., Zhou L., Cho J., Patel B., Terada N., Li Y., Bungert J., Qiu Y., Huang S., HoxBlinc RNA recruits Set1/MLL complexes to activate Hox gene expression patterns and mesoderm lineage development. Cell Rep. 14, 103–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M., Herrmann B. G., The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Delattre M., Spierer A., Tonka C. H., Spierer P., The genomic silencing of position-effect variegation in Drosophila melanogaster: Interaction between the heterochromatin-associated proteins Su(var)3-7 and HP1. J. Cell Sci. 113, 4253–4261 (2000). [DOI] [PubMed] [Google Scholar]
- 84.James T. C., Elgin S. C. R., Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6, 3862–3872 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Norwood L. E., Moss T. J., Margaryan N. V., Cook S. L., Wright L., Seftor E. A., Hendrix M. J. C., Kirschmann D. A., Wallrath L. L., A requirement for dimerization of HP1Hsα in suppression of breast cancer invasion. J. Biol. Chem. 281, 18668–18676 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Muchardt C., Guillemé M., Seeler J.-S., Trouche D., Dejean A., Yaniv M., Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 3, 975–981 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maison C., Bailly D., Peters A. H. F. M., Quivy J.-P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Keller C., Kulasegaran-Shylini R., Shimada Y., Hotz H.-R., Bühler M., Noncoding RNAs prevent spreading of a repressive histone mark. Nat. Struct. Mol. Biol. 20, 1340 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Bierhoff H., Dammert M. A., Brocks D., Dambacher S., Schotta G., Grummt I., Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell 54, 675–682 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Gil J., O’Loghlen A., PRC1 complex diversity: Where is it taking us? Trends Cell Biol. 24, 632–641 (2014). [DOI] [PubMed] [Google Scholar]
- 91.Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., Allis C. D., Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 26, 2560–2569 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yap K. L., Li S., Muñoz-Cabello A. M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M. J., Zhou M.-M., Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang L., Lin C., Liu W., Zhang J., Ohgi K. A., Grinstein J. D., Dorrestein P. C., Rosenfeld M. G., ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vo N., Goodman R. H., CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276, 13505–13508 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Bannister A. J., Kouzarides T., The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643 (1996). [DOI] [PubMed] [Google Scholar]
- 96.Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., Grummt I., LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell 60, 626–636 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Bose D. A., Donahue G., Reinberg D., Shiekhattar R., Bonasio R., Berger S. L., RNA binding to CBP stimulates histone acetylation and transcription. Cell 168, 135–149.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Foster C. T., Dovey O. M., Lezina L., Luo J. L., Gant T. W., Barlev N., Bradley A., Cowley S. M., Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol. Cell. Biol. 30, 4851–4863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hino S., Kohrogi K., Nakao M., Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells. Cancer Sci. 107, 1187–1192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsai M.-C., Manor O., Wan Y., Mosammaparast N., Wang J. K., Lan F., Shi Y., Segal E., Chang H. Y., Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zappulla D. C., Cech T. R., Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. U.S.A. 101, 10024–10029 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Porro A., Feuerhahn S., Lingner J., TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Rep. 6, 765–776 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Hermann A., Schmitt S., Jeltsch A., The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem. 278, 31717–31721 (2003). [DOI] [PubMed] [Google Scholar]
- 104.Goll M. G., Kirpekar F., Maggert K. A., Yoder J. A., Hsieh C.-L., Zhang X., Golic K. G., Jacobsen S. E., Bestor T. H., Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Thakur N., Tiwari V. K., Thomassin H., Pandey R. R., Kanduri M., Göndör A., Grange T., Ohlsson R., Kanduri C., An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 24, 7855–7862 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mohammad F., Mondal T., Guseva N., Pandey G. K., Kanduri C., Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137, 2493–2499 (2010). [DOI] [PubMed] [Google Scholar]
- 107.Di Ruscio A., Ebralidze A. K., Benoukraf T., Amabile G., Goff L. A., Terragni J., Figueroa M. E., De Figueiredo Pontes L. L., Alberich-Jorda M., Zhang P., Wu M., D’Alò F., Melnick A., Leone G., Ebralidze K. K., Pradhan S., Rinn J. L., Tenen D. G., DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503, 371–376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qi D., Li J., Que B., Su J., Li M., Zhang C., Yang M., Zhou G., Ji W., Long non-coding RNA DBCCR1-003 regulate the expression of DBCCR1 via DNMT1 in bladder cancer. Cancer Cell Int. 16, 81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu F., Yang F., Wu X., Huang J.-F., Yuan J.-H., Tao Q.-f., Jiang G.-h., Zheng J.-s., Sun S.-H., Long non-coding RNA RBMY2FP promotes proliferation of male hepatocellular carcinoma by directing DNA methylation and activating RBMY1A1 via DNMT1. Oncotarget (2016). [Google Scholar]
- 110.Zhang G., Estève P.-O., Chin H. G., Terragni J., Dai N., Corrêa I. R. Jr, Pradhan S., Small RNA-mediated DNA (cytosine-5) methyltransferase 1 inhibition leads to aberrant DNA methylation. Nucleic Acids Res. 43, 6112–6124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Okano M., Xie S., Li E., Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220 (1998). [DOI] [PubMed] [Google Scholar]
- 112.Gowher H., Jeltsch A., Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: The enzyme modifies DNA in a non-processive manner and also methylates non-CpG sites. J. Mol. Biol. 309, 1201–1208 (2001). [DOI] [PubMed] [Google Scholar]
- 113.Sun B. K., Deaton A. M., Lee J. T., A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21, 617–628 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Holz-Schietinger C., Reich N. O., RNA modulation of the human DNA methyltransferase 3A. Nucleic Acids Res. 40, 8550–8557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schmitz K.-M., Mayer C., Postepska A., Grummt I., Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Picard B., Wegnez M., Isolation of a 7S particle from Xenopus laevis oocytes: A 5S RNA–protein complex. Proc. Natl. Acad. Sci. U.S.A. 76, 241–245 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pelham H. R., Brown D. D., A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl. Acad. Sci. U.S.A. 77, 4170–4174 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Engelke D. R., Ng S.-Y., Shastry B. S., Roeder R. G., Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell 19, 717–728 (1980). [DOI] [PubMed] [Google Scholar]
- 119.Guddat U., Bakken A. H., Pieler T., Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell 60, 619–628 (1990). [DOI] [PubMed] [Google Scholar]
- 120.Setzer D. R., Menezes S. R., Del Rio S., Hung V. S., Subramanyan G., Functional interactions between the zinc fingers of Xenopus transcription factor IIIA during 5S rRNA binding. RNA 2, 1254–1269 (1996). [PMC free article] [PubMed] [Google Scholar]
- 121.Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P., Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jeon Y., Lee J. T., YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sigova A. A., Abraham B. J., Ji X., Molinie B., Hannett N. M., Guo Y. E., Jangi M., Giallourakis C. C., Sharp P. A., Young R. A., Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou L., Sun K., Zhao Y., Zhang S., Wang X., Li Y., Lu L., Chen X., Chen F., Bao X., Zhu X., Wang L., Tang L.-Y., Esteban M. A., Wang C.-C., Jauch R., Sun H., Wang H., Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 6, 10026 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Wai D. C. C., Shihab M., Low J. K. K., Mackay J. P., The zinc fingers of YY1 bind single-stranded RNA with low sequence specificity. Nucleic Acids Res. 44, 9153–9165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hung T., Wang Y., Lin M. F., Koegel A. K., Kotake Y., Grant G. D., Horlings H. M., Shah N., Umbricht C., Wang P., Wang Y., Kong B., Langerød A., Børresen-Dale A.-L., Kim S. K., van de Vijver M., Sukumar S., Whitfield M. L., Kellis M., Xiong Y., Wong D. J., Chang H. Y., Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ng S.-Y., Johnson R., Stanton L. W., Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ng S.-Y., Bogu G. K., Soh B. S., Stanton L. W., The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359 (2013). [DOI] [PubMed] [Google Scholar]
- 129.Willingham A. T., Orth A. P., Batalov S., Peters E. C., Wen B. G., Aza-Blanc P., Hogenesch J. B., Schultz P. G., A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573 (2005). [DOI] [PubMed] [Google Scholar]
- 130.Ong C.-T., Corces V. G., CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Phillips J. E., Corces V. G., CTCF: Master weaver of the genome. Cell 137, 1194–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun S., Del Rosario B. C., Szanto A., Ogawa Y., Jeon Y., Lee J. T., Jpx RNA activates Xist by evicting CTCF. Cell 153, 1537–1551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yao H., Brick K., Evrard Y., Xiao T., Camerini-Otero R. D., Felsenfeld G., Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saldaña-Meyer R., González-Buendía E., Guerrero G., Narendra V., Bonasio R., Recillas-Targa F., Reinberg D., CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 28, 723–734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kung J. T., Kesner B., An J. Y., Ahn J. Y., Cifuentes-Rojas C., Colognori D., Jeon Y., Szanto A., del Rosario B. C., Pinter S. F., Erwin J. A., Lee J. T., Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell 57, 361–375 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang F., Deng X., Ma W., Berletch J. B., Rabaia N., Wei G., Moore J. M., Filippova G. N., Xu J., Liu Y., Noble W. S., Shendure J., Disteche C. M., The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16, 52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao X., Li D., Pu J., Mei H., Yang D., Xiang X., Qu H., Huang K., Zheng L., Tong Q., CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene 35, 3565–3576 (2016). [DOI] [PubMed] [Google Scholar]
- 138.Tang J., Wu S., Liu H., Stratt R., Barak O. G., Shiekhattar R., Picketts D. J., Yang X., A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 279, 20369–20377 (2004). [DOI] [PubMed] [Google Scholar]
- 139.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T. L., Sechi S., Qin J., Zhou S., Higgs D., Wang W., The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. U.S.A. 100, 10635–10640 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mitson M., Kelley L. A., Sternberg M. J. E., Higgs D. R., Gibbons R. J., Functional significance of mutations in the Snf2 domain of ATRX. Hum. Mol. Genet. 20, 2603–2610 (2011). [DOI] [PubMed] [Google Scholar]
- 141.Sarma K., Cifuentes-Rojas C., Ergun A., Del Rosario A., Jeon Y., White F., Sadreyev R., Lee J. T., ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 159, 869–883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rippe K., Luke B., TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 22, 853–858 (2015). [DOI] [PubMed] [Google Scholar]
- 143.Feng J., Bi C., Clark B. S., Mady R., Shah P., Kohtz J. D., The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bond A. M., Vangompel M. J. W., Sametsky E. A., Clark M. F., Savage J. C., Disterhoft J. F., Kohtz J. D., Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Latos P. A., Pauler F. M., Koerner M. V., Senergin H. B., Hudson Q. J., Stocsits R. R., Allhoff W., Stricker S. H., Klement R. M., Warczok K. E., Aumayr K., Pasierbek P., Barlow D. P., Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 (2012). [DOI] [PubMed] [Google Scholar]
- 146.Engreitz J. M., Haines J. E., Perez E. M., Munson G., Chen J., Kane M., McDonel P. E., Guttman M., Lander E. S., Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Anderson K. M., Anderson D. M., McAnally J. R., Shelton J. M., Bassel-Duby R., Olson E. N., Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martens J. A., Laprade L., Winston F., Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429, 571–574 (2004). [DOI] [PubMed] [Google Scholar]
- 149.Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R., Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 106, 18321–18326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Melé M., Rinn J. L., “Cat’s Cradling” the 3D genome by the act of LncRNA transcription. Mol. Cell 62, 657–664 (2016). [DOI] [PubMed] [Google Scholar]
- 151.Clemson C. M., Hutchinson J. N., Sara S. A., Ensminger A. W., Fox A. H., Chess A., Lawrence J. B., An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sun L., Goff L. A., Trapnell C., Alexander R., Lo K. A., Hacisuleyman E., Sauvageau M., Tazon-Vega B., Kelley D. R., Hendrickson D. G., Yuan B., Kellis M., Lodish H. F., Rinn J. L., Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 3387–3392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hacisuleyman E., Goff L. A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D. G., Sauvageau M., Kelley D. R., Morse M., Engreitz J., Lander E. S., Guttman M., Lodish H. F., Flavell R., Raj A., Rinn J. L., Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bergmann J. H., Li J., Eckersley-Maslin M. A., Rigo F., Freier S. M., Spector D. L., Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 25, 1336–1346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pfingsten J. S., Goodrich K. J., Taabazuing C., Ouenzar F., Chartrand P., Cech T. R., Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell 148, 922–932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lu Z., Zhang Q. C., Lee B., Flynn R. A., Smith M. A., Robinson J. T., Davidovich C., Gooding A. R., Goodrich K. J., Mattick J. S., Mesirov J. P., Cech T. R., Chang H. Y., RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zappulla D. C., Cech T. R., RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 71, 217–224 (2006). [DOI] [PubMed] [Google Scholar]
- 158.Bassett A. R., Akhtar A., Barlow D. P., Bird A. P., Brockdorff N., Duboule D., Ephrussi A., Ferguson-Smith A. C., Gingeras T. R., Haerty W., Higgs D. R., Miska E. A., Ponting C. P., Considerations when investigating lncRNA function in vivo. eLife 3, e03058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., Lim W. A., Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tseng Y.-Y., Moriarity B. S., Gong W., Akiyama R., Tiwari A., Kawakami H., Ronning P., Reuland B., Guenther K., Beadnell T. C., Essig J., Otto G. M., O’Sullivan M. G., Largaespada D. A., Schwertfeger K. L., Marahrens Y., Kawakami Y., Bagchi A., PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A., Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666–670 (2007). [DOI] [PubMed] [Google Scholar]
- 162.Shamovsky I., Ivannikov M., Kandel E. S., Gershon D., Nudler E., RNA-mediated response to heat shock in mammalian cells. Nature 440, 556–560 (2006). [DOI] [PubMed] [Google Scholar]
- 163.Bumgarner S. L., Neuert G., Voight B. F., Symbor-Nagrabska A., Grisafi P., van Oudenaarden A., Fink G. R., Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell 45, 470–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maamar H., Cabili M. N., Rinn J., Raj A., linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 27, 1260–1271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]


