Abstract
Low molecular mass protein (LMP) gene performs a critical role in the foreign antigen processing machine via the major histocompatibility complex-I (MHC-I) complex CD8+ cytotoxic T lymphocytes (CTL) pathway. Recent studies have reported the association of LMP2-60 G>A (rs17587) and LMP7-145 C>A (rs2071543) polymorphisms with various types of cancers, but the outcomes remained inconsistent. To obtain a reliable conclusion, we summarized available data and conducted a meta-analysis involving a total of 19 published studies. Evidences were obtained from the PubMed, Google Scholar, Web of Science and Chinese National Knowledge Infrastructure (CNKI) databases. The results demonstrated that the rs17587 and rs2071543 polymorphisms were associated with an increased cancer risk in the recessive and homozygote models. Stratified analyses by ethnicity indicated a significant association only in Asian population. Furthermore, rs17587 showed a greater susceptibility to gynecological cancers, while rs2071543 increased the risk of gastrointestinal and gynecological cancers. Our results indicate that the LMP2 rs17587 and LMP7 rs2071543 polymorphisms may act as risk factors for cancer, especially for Asian populations. Additional larger-scale multicenter studies should be performed to validate our results.
Keywords: LMP2, LMP7, polymorphism, meta-analysis, cancer
INTRODUCTION
Currently, cancer is a major cause of human death and a public health problem that seriously threatens human health worldwide [1]. Large epidemiological and biological investigations have indicated that genetic and environmental factors contribute to tumourigenesis. However, the exact mechanisms of carcinogenesis have not been fully illuminated.
In recent years, many researches have confirmed that the elimination of cancer cell is promoted by the classical MHC-I-restricted T lymphocytes pathway [2, 3]. In this pathway, the CD8+ CTL could recognize the peptide antigen of cancer cell which is processed by LMP2/LMP7 molecules (GenBank Accession: X66401.1 GI: 34634) and presented by TAP1/TAP2 molecules (TAP, transporter associated with antigen presentation) [4]. The LMP/TAP system could recognize cancer antigen and act a pivotal role in immune surveillance via MHC-I molecule and CTL in the human host protective immunity [5, 6]. Therefore, the integrality of LMP2/LMP7 function played a restrictive effect on the processing of cancer cell antigen.
Previous researches indicated that the polymorphisms of LMP2- 60(G>A) and LMP7-145(C>A) would give rise to the functional alteration, and then impaired the capacity of antigen processing machine. As a consequence, LMP2/LMP7 gene variants were associated with development, occurrence and prognosis of many types of cancers, such as colorectal cancer, cervical cancer, gastric carcinoma and so on [7-16]. However, no consensus has yet been achieved, which was partially due to heterogeneity between cancer types, different ethnicities of patient cohorts, diverse genotyping methods and relatively small sample sizes. Accordingly, we conducted this meta-analysis to derive a more precise and up-to-date estimation of associations of LMP gene polymorphisms with cancer risk.
RESULTS
Literature search and study characteristics
The flow chart of study selection process is shown in Figure 1. We identified 163 records, among which 10 publications appeared to be eligible and were retrieved in full texts [7-16]. Eventually, a total of 19 case-control studies from 10 publications (4360 cases and 4987 controls) were included in this study, and details of each study were recorded in Table 1. The eligible studies presented data for several different cancer types, including gastric cancer, cervical cancer, ovarian cancer, hematological malignancy, colorectal cancer and esophageal squamous cell carcinoma. Among these 19 studies, 11 were based on Asian populations [7-9, 14-16], 8 on Caucasian populations [10-13]. Furthermore, the genotyping methods utilized in the studies included PCR-RFLP (RFLP, restriction fragment length polymorphism), Taqman, Sequencing and ARMS-PCR (ARMS, amplification refractory mutation system). Quality assessment with Newcastle-Ottawa Scale (NOS) showed that 17 studies had a quality score higher than 7 points, and 2 studies had a quality score equal to 7 points (Table 1).
Figure 1. The flow diagram of retrieval for this study.
CNKI: China National Knowledge Infrastructure.
Table 1. Characteristics of studies included in the meta-analysis.
| SNP | Study | Ethnicity | Genotyping method | Source of control | Cancer type | Case | Control | P value of HWE | NOS points | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | ||||||||
| LMP2 rs17587 | Ma(2015) | Asian | PCR-RFLP | HB | GC | 338 | 151 | 13 | 335 | 155 | 12 | 0.228 | 8 |
| Mehta(2015) | Asian | Taqman | PB | CC | 82 | 90 | 21 | 79 | 77 | 10 | 0.117 | 9 | |
| Song(2014) | Asian | PCR-RFLP | HB | OC | 129 | 72 | 34 | 208 | 86 | 44 | 0.000 | 8 | |
| Gerceker(2013) | Caucasian | PCR-RFLP | PB | HM | 46 | 52 | 34 | 49 | 46 | 35 | 0.001 | 9 | |
| Fellerhoff(2011) | Caucasian | ARMS-PCR | HB | CRC | 87 | 71 | 16 | 86 | 68 | 11 | 0.617 | 8 | |
| Deshpande(2008) | Caucasian | Sequencing | PB | CC | 206 | 90 | 21 | 246 | 155 | 16 | 0.162 | 9 | |
| Mehta(2007) | Caucasian | Taqman | HB | CC | 66 | 52 | 9 | 65 | 54 | 5 | 0.126 | 8 | |
| Cao(2006) | Asian | PCR-RFLP | PB | GC | 93 | 47 | 5 | 95 | 55 | 2 | 0.053 | 7 | |
| Cao(2005) | Asian | PCR-RFLP | PB | ESCC | 167 | 83 | 15 | 239 | 102 | 16 | 0.234 | 8 | |
| LMP7 rs2071543 | Ma(2015) | Asian | PCR-RFLP | HB | GC | 310 | 169 | 23 | 349 | 141 | 12 | 0.612 | 8 |
| Mehta(2015) | Asian | Taqman | PB | CC | 173 | 18 | 1 | 141 | 22 | 1 | 0.888 | 9 | |
| Song(2014) | Asian | PCR-RFLP | HB | OC | 120 | 86 | 29 | 249 | 76 | 13 | 0.243 | 8 | |
| Gerceker(2013) | Caucasian | PCR-RFLP | PB | HM | 111 | 18 | 3 | 112 | 17 | 1 | 0.692 | 9 | |
| Fellerhoff(2011) | Caucasian | ARMS-PCR | HB | CRC | 97 | 70 | 7 | 145 | 20 | 0 | 0.407 | 8 | |
| Deshpande(2008) | Caucasian | Sequencing | PB | CC | 268 | 60 | 2 | 351 | 69 | 1 | 0.208 | 9 | |
| Mehta(2007) | Caucasian | Taqman | HB | CC | 96 | 31 | 0 | 78 | 43 | 3 | 0.297 | 8 | |
| Yang(2007) | Asian | PCR-RFLP | PB | ESCC | 137 | 25 | 6 | 194 | 84 | 5 | 0.228 | 8 | |
| Cao(2006) | Asian | PCR-RFLP | PB | GC | 63 | 69 | 13 | 59 | 85 | 8 | 0.001 | 7 | |
| Cao(2005) | Asian | Sequencing | PB | ESCC | 130 | 114 | 21 | 210 | 130 | 17 | 0.583 | 8 | |
PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; ARMS: amplification refractory mutation system; HB: hospital-based studies; PB: population-based studies. GC: gastric cancer; CC: cervical cancer; OC: ovarian cancer; HM: hematological malignancy; CRC: colorectal cancer; ESCC: esophageal squamous cell carcinoma; HWE: Hardy–Weinberg equilibrium, P > 0.05 indicates that the participants in the control group met the HWE. A: the major allele; B: the minor allele; NOS: Newcastle-Ottawa Scale.
Meta-analysis of the LMP polymorphisms and cancer risk
The main results of this meta-analysis are listed in Table 2. Nine studies involving 2,090 cases and 2,351 controls were included for rs17587. As indicated in Figure 2 and Table 2, we observed an increased cancer susceptibility associated with the rs17587 polymorphism under homozygote and recessive models [AA vs. GG: OR = 1.36, (95% CI: 1.07-1.74), P = 0.931, I2 = 0.0%; AA vs. GA/GG: OR = 1.31; (95% CI: 1.03-1.65), P = 0.787, I2 = 0.0%]. Subgroup analysis by ethnicity showed a significant association only in the Asian populations [AA vs. GG: OR = 1.38, (95% CI: 1.00-1.91), P = 0.750, I2 = 0.0%] (Figure 2B, Table 2). In addition, stratified analysis by design of study showed a significant relationship in population-based studies [AA vs. GG: OR = 1.43, (95% CI: 1.02-2.01), P = 0.679, I2 = 0.0%; AA vs. GA/GG: OR = 1.38; (95% CI: 1.00-1.90), P = 0.446, I2 = 0.0%] (Figure 2C, Table 2). In the cancer-specific analysis, the results showed significant correlations between rs17587 and risk of gynecological cancers in different comparison models. [AA vs. GG: OR = 1.49, (95% CI: 1.06-2.10), P = 0.766, I2 = 0.0%; AA vs. GA/GG: OR = 1.46; (95% CI: 1.05-2.03), P = 0.572, I2 = 0.0%] (Figure 2D, Table 2).
Table 2. Meta-analysis results of association between LMP2/LMP7 polymorphisms and cancer risk.
| LMP2-60 G>A (rs17587) | LMP7-145 C>A (rs2071543) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA vs GG | AA vs GG/GA | AA vs CC | AA vs CC/CA | |||||||
| Variables | N | OR(95%CI) | P/I2(%) | OR(95%CI) | P/I2(%) | N | OR(95%CI) | P/I2(%) | OR(95%CI) | P/I2(%) |
| Total | 9 | 1.36(1.07-1.74) | 0.931/0.0 | 1.31(1.03-1.65) | 0.787/0.00 | 10 | 2.38(1.72-3.29) | 0.215/24.8 | 2.17(1.57-2.99) | 0.506/0.00 |
| Ethnicity | ||||||||||
| Asian | 5 | 1.38(1.00-1.91) | 0.750/0.00 | 1.31(0.95-1.79) | 0.705/0.00 | 6 | 2.37(1.68-3.36) | 0.344/11.2 | 2.16(1.54-3.04) | 0.679/0.00 |
| Caucasian | 4 | 1.34(0.92-1.95) | 0.770/0.00 | 1.31(0.92-1.86) | 0.466/0.00 | 4 | 2.39(0.93-6.13) | 0.096/52.6 | 2.20(0.83-5.83) | 0.161/41.7 |
| Source of control | ||||||||||
| PB | 5 | 1.43(1.02-2.01) | 0.679/0.00 | 1.38(1.00-1.90) | 0.446/0.00 | 6 | 1.83(1.14-2.93) | 0.976/0.00 | 1.82(1.15-2.88) | 0.987/0.00 |
| HB | 4 | 1.29(0.90,1.84) | 0.900/0.00 | 1.23(0.87-1.74) | 0.850/0.00 | 4 | 3.00(1.90-4.73) | 0.032/65.9 | 2.54(1.62-4.00) | 0.087/54.4 |
| Cancer type | ||||||||||
| Gastrointestinal | 4 | 1.35(0.87-2.07) | 0.825/0.00 | 1.33(0.87-2.03) | 0.808/0.00 | 5 | 2.15(1.44-3.20) | 0.517/0.00 | 2.02(1.36-2.99) | 0.706/0.00 |
| Gynecological | 4 | 1.49(1.06-2.10) | 0.766/0.00 | 1.46(1.05-2.03) | 0.572/0.00 | 4 | 2.91(1.62-5.21) | 0.070/57.4 | 2.46(1.38-4.40) | 0.153/43.0 |
| Hematological | 1 | - | - | - | - | 1 | - | - | - | - |
N: number of studies; P: P value of Q test for heterogeneity; PB: population-based; HB: hospital-based.
Figure 2. Forest plots of cancer risk associated with LMP2-60 G>A (rs17587) polymorphism under homozygote model.
(A) Overall result; (B) subgroup analysis by ethnicity; (C) stratified analysis by design of study; (D) subgroup analysis by cancer types.
The association of LMP7 rs2071543 polymorphism with cancer risk was investigated in 10 studies involving 2,270 cases and 2,636 controls. This polymorphism was associated with an increased cancer susceptibility in the overall population under the two models [AA vs. CC: OR = 2.38, (95% CI: 1.72-3.29), P = 0.215, I2 = 24.8%; AA vs. CC/CA: OR = 2.17, (95% CI: 1.57-2.99), P = 0.506, I2 = 0.0%] (Figure 3A, Table 2). In the ethnicity-specific analysis, a significant association was observed in the Asian populations [AA vs. CC: OR = 2.37, (95% CI: 1.68-3.36), P = 0.344, I2 = 11.2%; AA vs. CC/CA: OR = 2.16, (95% CI: 1.54-3.04), P = 0.679, I2 = 0.0%], but not in the Caucasian populations (Figure 3B, Table 2). Additionally, subgroup analysis by the source of control showed a significant correlation in population-based and hospital-based studies (Figure 3C, Table 2). Furthermore, in analysis stratified by cancer types, the results showed significant correlations between rs2071543 and the risk of gastrointestinal cancers [AA vs. CC: OR = 2.15, (95% CI: 1.44-3.20), P = 0.517, I2 = 0.0%; AA vs. CC/CA: OR = 2.02, (95% CI: 1.36-3.04), P = 0.679, I2 = 0.0%] and gynecological cancers [AA vs. CC: OR = 2.37, (95% CI: 1.68-3.36), P = 0.344, I2 = 11.2%; AA vs. CC/CA: OR = 2.16, (95% CI: 1.54-3.04), P = 0.679, I2 = 0.0%] (Figure 3D, Table 2).
Figure 3. Forest plots of cancer risk associated with LMP7-145 C>A (rs2071543) polymorphism under homozygote model.
(A) Overall result; (B) subgroup analysis by ethnicity; (C) stratified analysis by design of study; (D) subgroup analysis by cancer types.
Test of heterogeneity
In this study, Chi-squared-based Q-statistic test was used to evaluate between-study heterogeneity that resulted from methodological or clinical dissimilarity across studies. As a result, heterogeneity between studies was not identified for rs17587 and rs2071543 under the homozygote and recessive models (P > 0.1) (Table 2).
Sensitivity and publication bias analysis
To confirm the reliability of results, we further conducted sensitivity analyses by repeating the meta-analysis while sequentially omitting the studies included (one deleted at a time). The analysis outcomes demonstrated that no single study greatly influenced the overall cancer risk estimations with respect to the LMP polymorphisms. (Figure 4A–4B), suggesting that our results were robust.
Figure 4.
Sensitivity analysis of cancer risk associated with rs17587 (A) and rs2071543 (B) polymorphisms under homozygous model.
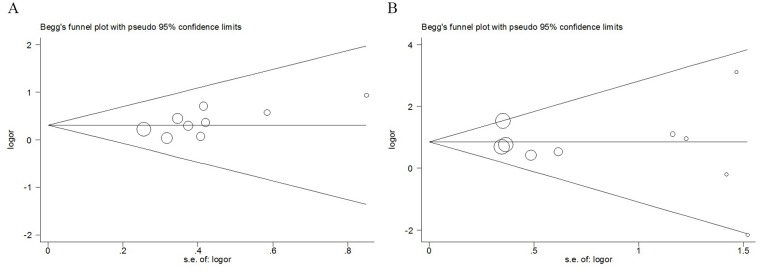
We also performed Begg’s and Egger’s test to assess the possible publication bias. The funnel plots of Begg’s test were symmetrical inverted (Figure 5A–5B), which suggested no significant publication bias. In addition, the results Egger’s test also showed no evidence of publication bias for LMP2/LMP7 polymorphisms. Therefore, the outcomes revealed that publication bias was not significant in this meta-analysis.
Figure 5.
Begg’s funnel plot for publication bias test of LMP polymorphisms: rs17587 (A), rs2071543 (B), under homozygous model.
DISCUSSION
Cancer cells can escape immune recognition via insufficient expression of peptides presented by the MHC, because HLA-mediated presentation of immunogenic cancer peptides class I is a prerequisite of a successful antitumor immune response [17]. Recent studies indicated a participation of the antigen processing machinery in carcinogenesis, such as in esophagus cancer [18], malignant melanoma [19], and hepatocellular carcinoma [20]. Two groups of proteins that participate in the antigen processing are LMPs and TAPs. Genetic polymorphisms of immunoproteasome subunits (LMP2, LMP7) and of transporter subunits (TAP1 and TAP2) are documented [21-23].
As a significant cause of population diversity, SNP is the most common type of human genetic variation correlated with cancer susceptibility [24]. Recently, much effort has been directed toward illuminating the role of LMP gene polymorphisms and their impacts on susceptibility to and progression of various diseases [7-16, 25]. Clarifying the association between LMP gene SNPs and cancer risk will help further illuminate the mechanisms underlying the carcinogenesis, which will in turn provide novel biomarkers for screening high-risk populations for cancer and improve the development of molecular-targeted therapy.
The current study is the very first meta-analysis of the association between LMP2/LMP7 polymorphisms and cancer risk. Analysis among all subjects suggested a significant increase in the risk of cancer associated with LMP2/LMP7. In our meta-analysis, the genetic heterogeneity between the selected studies was evaluated, and no significant heterogeneity was observed. All studies included were found homogeneous without any study disproportionately driving the combined estimates. In subgroup analysis of ethnicity, our results showed that both rs17587 and rs2071543 were correlated with cancer risk in Asian populations, but not in Caucasian populations. The differences between Asians and other races may be partly attribute to different genetic backgrounds, lifestyles or environments. Besides, when the stratification analyses were conducted by cancer types, we identified a significantly increased susceptibility to gynecological cancer for rs17587 polymorphism, to gastrointestinal and gynecological cancer for rs2071543.
Meta-analysis is a powerful method for analyzing cumulative data of studies where the single sample size is small and the statistical power is low [26]. However, several limitations of this meta-analysis should be acknowledged. To begin with, this analysis was based on unadjusted ORs because of a lack of information for several potential confounding variables. Secondly, due to the limited scope of databases, we cannot exclude the possibility of missing data. Additionally, more studies from all over the world should be performed to make our conclusions more persuasive.
Generally speaking, our meta-analysis manifests that LMP2/LMP7 polymorphisms (rs17587, rs2071543) are risk factors for cancer, and presence of the two polymorphisms in Asian population will increase their susceptibility to cancer. However, larger-scale multicenter studies are needed to further clarify the possible roles of the polymorphisms in the development of cancers.
MATERIALS AND METHODS
Primary search strategy
The PubMed, Web of Science, Google Scholar and the China National Knowledge Infrastructure (CNKI) databases were searched for relevant studies up to April 14, 2017 in both English and Chinese through with the following terms and their combinations: “LMP2”, “LMP7”, “low molecular mass protein”, “polymorphism” or “variant”, “rs17587”, “rs2071543” and “cancer” or “tumor”. The reference lists of the retrieved studies were also screened to prevent the loss of any important data. All studies involved had to satisfy the following criteria: (a) case-control design was utilized; (b) studies focused on the association of LMP2/LMP7 polymorphisms with the risk of cancer; (c) published data must have been sufficient to allow OR estimation an odds ratio (OR) with a 95% CI. (d) for publications reporting the same data or overlapping data, only the largest or latest one was selected.
Data extraction
Initially, all the following information was extracted independently by three investigators (Y Wu, D Liu and J Zhang) and recorded in a standardized form. We extracted the following items from each article: including: first author’s name, year of publication, ethnicity of each study population, genotyping method, source of controls, cancer types, number of cancer cases and controls, allele frequencies, genotype distributions of LMP2/LMP7 polymorphisms, and Hardy-Weinberg equilibrium (HWE) results (Table 1). Disagreements were resolved through discussions involving a senior investigator (K Jiang).
Quality assessment
The studies quality was evaluated utilizing Newcastle-Ottawa Scale (NOS) for nonrandomized studies, including case-control and cohort studies [27]. NOS awards eight points to each case-control study (four for quality of selection, one for comparability, and three for quality of exposure). A study can be awarded a maximum of one star for each point within the selection and exposure categories, and a maximum of two stars for comparability. We considered studies with scores of more than 7 as high-quality studies, and those with scores of 7 or less as low-quality studies.
Statistical analysis
All analyses were performed using the Stata software, version 12.0 (Stata Corp., College Station, TX, USA). All P values were 2-sided and a P value <0.5 was regarded as statistically significant.
To evaluate the cancer risk associated with LMP2/LMP7 polymorphisms, the pooled odds ratios (ORs) and 95% CIs were calculated. Heterogeneity between studies was evaluated using the I2 test, with a higher I2 value indicating a higher level of heterogeneity (I2 = 0-25%: no heterogeneity; I2 = 25-50%: moderate heterogeneity; I2 = 50-75%: great heterogeneity; I2 = 75-100%: extreme heterogeneity;). Meanwhile, if heterogeneity P value was higher than 0.10, the fixed effects model (the Mantel-Haenszel method) was used [28]. Otherwise, the random-effects model (The DerSimonian-Laird method) was used [29]. Then, we performed the stratified analyses according to ethnicity, source of controls and cancer types.
A sensitivity analysis was performed to assess the stability of final results by calculating the outcomes again by omitting one single study at a time. Begg’s funnel plots and Egger’s linear regression were adopted to evaluate the publication bias [30]. HWE was checked by the goodness-of-fit chi-square test and a P < 0.05 was regarded as statistically significant [31].
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81572337, 81672449 and 81402489), the Innovation Capability Development Project of Jiangsu Province (BM2015004), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801).
Footnotes
CONFLICTS OF INTEREST
The authors have declared that no competing interests exist.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Morse MA, Secord AA, Blackwell K, Hobeika AC, Sinnathamby G, Osada T, Hafner J, Philip M, Clay TM, Lyerly HK, Philip R. MHC class I-presented tumor antigens identified in ovarian cancer by immunoproteomic analysis are targets for T-cell responses against breast and ovarian cancer. Clin Cancer Res. 2011;17:3408–3419. doi: 10.1158/1078-0432.CCR-10-2614. [DOI] [PubMed] [Google Scholar]
- 3.Nesbeth YC, Martinez DG, Toraya S, Scarlett UK, Cubillos-Ruiz JR, Rutkowski MR, Conejo-Garcia JR. CD4+ T cells elicit host immune responses to MHC class II-negative ovarian cancer through CCL5 secretion and CD40-mediated licensing of dendritic cells. J Immunol. 2010;184:5654–5662. doi: 10.4049/jimmunol.0903247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driscoll J, Brown MG, Finley D, Monaco JJ. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 5.Xu C, Qi S, Gao L, Cui H, Liu M, Yang H, Li K, Cao B. Genetic polymorphisms of LMP/TAP gene and hepatitis B virus infection risk in the Chinese population. J Clin Immunol. 2007;27:534–541. doi: 10.1007/s10875-007-9095-x. [DOI] [PubMed] [Google Scholar]
- 6.Maeurer MJ, Martin DM, Storkus WJ, Lotze MT. TCR usage in CTLs recognizing melanoma/melanocyte antigens. Immunol Today. 1995;16:603–604. doi: 10.1016/0167-5699(95)80084-0. [DOI] [PubMed] [Google Scholar]
- 7.Ma X, Yang C, Tang R, Xu Z, Zhang Z, Wang Y, Zhang J, Yang LI. Association between LMP2 and LMP7 gene polymorphisms and the risk of gastric cancer: A case-control study. Oncol Lett. 2015;10:509–517. doi: 10.3892/ol.2015.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta AM, Spaans VM, Mahendra NB, Osse EM, Vet JN, Purwoto G, Surya IG, Cornian S, Peters AA, Fleuren GJ, Jordanova ES. Differences in genetic variation in antigen-processing machinery components and association with cervical carcinoma risk in two Indonesian populations. Immunogenetics. 2015;67:267–275. doi: 10.1007/s00251-015-0834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song L, Ma N, Han L, Yan H, Yan B, Yuan Z, Cao B. Association between LMP2/LMP7 genetic variability and the metastasis risk of ovarian cancer in Chinese women in Beijing. Hum Immunol. 2014;75:239–244. doi: 10.1016/j.humimm.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Ozbas-Gerceker F, Bozman N, Kok S, Pehlivan M, Yilmaz M, Pehlivan S, Oguzkan-Balci S. Association of an LMP2 polymorphism with acute myeloid leukemia and multiple myeloma. Asian Pac J Cancer Prev. 2013;14:6399–6402. doi: 10.7314/apjcp.2013.14.11.6399. [DOI] [PubMed] [Google Scholar]
- 11.Fellerhoff B, Gu S, Laumbacher B, Nerlich AG, Weiss EH, Glas J, Kopp R, Johnson JP, Wank R. The LMP7-K allele of the immunoproteasome exhibits reduced transcript stability and predicts high risk of colon cancer. Cancer Res. 2011;71:7145–7154. doi: 10.1158/0008-5472.CAN-10-1883. [DOI] [PubMed] [Google Scholar]
- 12.Deshpande A, Wheeler CM, Hunt WC, Peyton CL, White PS, Valdez YE, Nolan JP. Variation in HLA class I antigen-processing genes and susceptibility to human papillomavirus type 16-associated cervical cancer. J Infect Dis. 2008;197:371–381. doi: 10.1086/524300. [DOI] [PubMed] [Google Scholar]
- 13.Mehta AM, Jordanova ES, van Wezel T, Uh HW, Corver WE, Kwappenberg KM, Verduijn W, Kenter GG, van der Burg SH, Fleuren GJ. Genetic variation of antigen processing machinery components and association with cervical carcinoma. Genes Chromosomes Cancer. 2007;46:577–586. doi: 10.1002/gcc.20441. [DOI] [PubMed] [Google Scholar]
- 14.Cao BW, Dai Y, Gai YH, Chai QB, Wang X. No association between LMP2/LMP7 gene polymorphisms or haplotypes and gastric cancer. World Chin J Digestol. 2006;14:3471–3476. [Google Scholar]
- 15.Cao B, Tian X, Li Y, Jiang P, Ning T, Xing H, Zhao Y, Zhang C, Shi X, Chen D, Shen Y, Ke Y. LMP7/TAP2 gene polymorphisms and HPV infection in esophageal carcinoma patients from a high incidence area in China. Carcinogenesis. 2005;26:1280–1284. doi: 10.1093/carcin/bgi071. [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Chen L, Sun ZZ, Zhang HY, Ren T, Qi Y, Li HA, Jiang JF, Liang WH, Qin JM, Li F. Study on LMP7 gene polymorphism in kazakh esophageal squamous cell carcinomas in Xinjiang. J Nongken Med. 2007;29:249–253. [Google Scholar]
- 17.Lee YM, Leu SY, Chiang H, Fung CP, Liu WT. Human papillomavirus type 18 in colorectal cancer. J Microbiol Immunol Infect. 2001;34:87–91. [PubMed] [Google Scholar]
- 18.Cabrera CM, Jimenez P, Concha A, Garrido F, Ruiz-Cabello F. Promyelocytic leukemia (PML) nuclear bodies are disorganized in colorectal tumors with total loss of major histocompatibility complex class I expression and LMP7 downregulation. Tissue Antigens. 2004;63:446–452. doi: 10.1111/j.0001-2815.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 19.Dissemond J, Goette P, Moers J, Lindeke A, Goos M, Ferrone S, Wagner SN. Immunoproteasome subunits LMP2 and LMP7 downregulation in primary malignant melanoma lesions: association with lack of spontaneous regression. Melanoma Res. 2003;13:371–377. doi: 10.1097/00008390-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Shen YQ, Zhang JQ, Xia M, Miao FQ, Shan XN, Xie W. Low-molecular-weight protein (LMP)2/LMP7 abnormality underlies the downregulation of human leukocyte antigen class I antigen in a hepatocellular carcinoma cell line. J Gastroenterol Hepatol. 2007;22:1155–1161. doi: 10.1111/j.1440-1746.2006.04421.x. [DOI] [PubMed] [Google Scholar]
- 21.Deng GY, Muir A, Maclaren NK, She JX. Association of LMP2 and LMP7 genes within the major histocompatibility complex with insulin-dependent diabetes mellitus: population and family studies. AmJ Hum Genet. 1995;56:528–534. [PMC free article] [PubMed] [Google Scholar]
- 22.Faucz FR, Probst CM, Petzl-Erler ML. Polymorphism of LMP2, TAP1, LMP7 and TAP2 in Brazilian Amerindians and Caucasoids: implications for the evolution of allelic and haplotypic diversity. Eur J Immunogenet. 2000;27:5–16. doi: 10.1046/j.1365-2370.2000.00186.x. [DOI] [PubMed] [Google Scholar]
- 23.Lim JK, Hunter J, Fernandez-Vina M, Mann DL. Characterization of LMP polymorphism in homozygous typing cells and a random population. Hum Immunol. 1999;60:145–151. doi: 10.1016/s0198-8859(98)00106-2. [DOI] [PubMed] [Google Scholar]
- 24.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–51. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 25.El Awady MK, Omran MH, Ibrahim MK, Moustafa AM, Dawood RM, Bader El Din NG, Elsharkawy A, Abdel Aziz MS, El Shenawy R, El Abd YS, Abdel Aziz AO. Low molecular mass polypeptide 7 single nucleotide polymorphism is associated with the progression of liver fibrosis in patients infected with hepatitis C virus genotype 4. Clin Lab. 2016;62:381–387. doi: 10.7754/clin.lab.2015.150710. [DOI] [PubMed] [Google Scholar]
- 26.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Li X, You R, Wang X, Liu C, Xu Z, Zhou J, Yu B, Xu T, Cai H, Zou Q. Effectiveness of prophylactic surgeries in BRCA1 or BRCA2 mutation carriers: a meta-analysis and systematic review. Clin Cancer Res. 2016;22:3971–3981. doi: 10.1158/1078-0432.CCR-15-1465. [DOI] [PubMed] [Google Scholar]
- 28.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 29.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 30.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 31.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]