Abstract
Many studies manifested miRNA-100 was deregulated in various cancers, which indicated that miRNA-100 might be a potential biomarker of cancer diagnosis and prognosis. However, the role of miRNA-100 was still uncertain. We searched for qualified studies using PubMed, EMBASE, Web of Science, Cochrane library and CNKI databases. The diagnostic effect was evaluated by the pooled sensitivity, specificity, and other indexes. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) for overall survival (OS) were calculated to assess the prognostic value. This meta-analysis included 7 and 19 studies about diagnosis and prognosis, respectively. The results of pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) were 0.75 (95%CI: 0.71-0.78), 0.74 (95%CI: 0.69-0.78), 2.61 (95%CI: 1.81-3.76), 0.33 (95%CI: 0.24-0.45), 8.46 (95%CI: 4.85-14.77), respectively. And, the area under SROC curve (AUC) was 0.8141. We also found that lower expression of miRNA-100 in cancer tissues could significantly predict poorer prognosis in overall cancer (HR = 0.59, 95%CI: 0.39-0.90), especially in genital system tumors (HR = 0.42, 95%CI: 0.27-0.66, P = 0.431), bladder cancer (HR = 0.21, 95%CI: 0.06-0.73, P = 0.143) and esophageal squamous cell carcinoma (HR = 0.26, 95%CI: 0.13-0.52, P = 0.164). Our studies concluded that miRNA-100 has a certain value in diagnosis and it may indicate a poor prognosis of cancers.
Keywords: miRNA-100, diagnosis, prognosis, meta-analysis
INTRODUCTION
Cancer is always a fearsome disease because of its high mortality. It was estimated by GLOBOCAN that there were about 14.1 million new cancer cases and 8.2 million deaths occurring in 2012 and about 57% of cases and 65% of cancer deaths in developed countries worldwide [1]. Therefore, cancer has become a compelling health problem, and early diagnosis is particularly important in the treatment of cancer, but it is difficult because of the limitations in present diagnostic methods. Imaging examination and biopsy have the disadvantages of their invasive and harmful procedure, and many current biomarkers lack high accuracy in clinical diagnosis. In addition, it’s difficult to predict the clinical outcomes of cancer, which significantly varied in different people. At present, the research of biomarkers has made rapid development [2]. It’s highly needed to seek for new biomarkers that can exert on detection or diagnosis in early-age or estimate the prognosis of patients.
With a length of 19-25 nucleotides, microRNAs (miRNAs) are small non-coding RNAs which could regulate gene expression by blinding 3’ untranslated region (3’UTR) of their target mRNA and inhibiting gene translation. These miRNAs are considered as gene regulators at post transcriptional gene level [3]. As a member of miRNA-99a family, miRNA-100 is located on chromosome11 at 11q24.1 (Gene ID: 406892) and has been demonstrated to play a potential role in cell proliferation, tumorigenesis, angiogenesis and differentiation [4, 5]. Dysregulated expression of miRNA-100 is correlated with cancer diagnosis and prognosis [6]. Many studies suggested miRNA-100 as an oncogene or a tumor suppress gene. However, their conclusions remain controversial. Recent studies demonstrated obviously down-regulated expression of miRNA-100 in many tumor tissues, such as bladder cancer [7, 8], lung cancer [9, 10], esophageal squamous cell carcinoma [11, 12], epithelial ovarian cancer [13, 14], and other cancers [15-19, 20], indicating that it may have a relationship with poorer prognosis in cancer patients. But, evidence from some other studies showed opposite results in several types of cancer [21-25]. In addition, the diagnostic accuracy and the prognostic significance of miRNA-100 remain unclear. With due consideration of the limitations of a single study, we performed this systematic review and meta-analysis to evaluate the diagnostic and prognostic value of miRNA-100 in various cancers.
RESULTS
Literature search
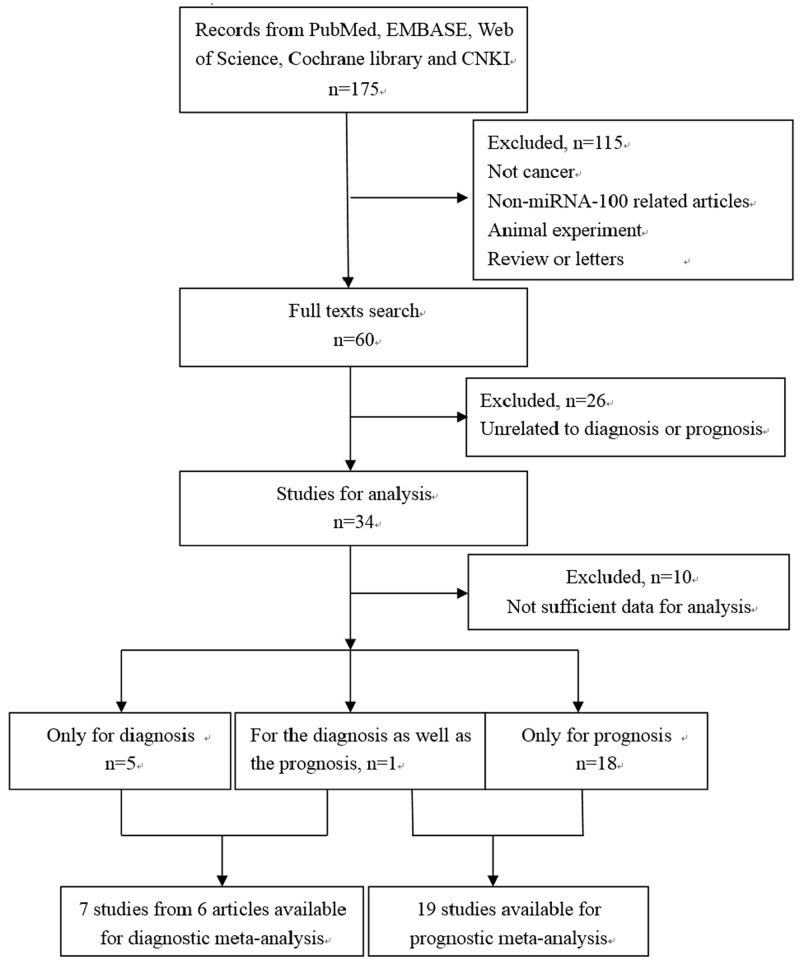
A total of 175 studies from a primary literature were searched in PubMed, EMBASE, Web of Science, Cochrane library and CNKI. After reviewing titles and abstracts manually, some studies were excluded due to their irrelevance to the analysis, or because they were review articles, duplicate studies, letters, animal experiments or laboratory studies. Then, we reviewed full texts and omitted 26 studies that were unrelated to diagnosis or prognosis and 10 studies without sufficient data to obtain the crucial data for analysis. Finally, 24 available articles were included. Among those articles, one article researched the diagnostic as well as the prognostic value of miRNA-100, meanwhile, it was divided into two studies because of its different investigations in plasma and tissue for diagnosis [16]. One study for prognosis was omitted because of its investigation in serum alone [26]. Finally, we enrolled 19 eligible prognostic studies and 7 eligible diagnostic studies from 6 articles in this meta-analysis (Figure 1).
Figure 1. The flow diagram of the study selection process.
Diagnostic meta-analysis
Study characteristics
7 eligible articles of cancer diagnosis were published from 2010 to 2016, involving a total of 883 participants. These participants were from China, Egypt, Poland, and Mexico. Various types of tumors contain bladder cancer, gastric cancer, endometrioid endometrial carcinoma, esophageal squamous cell carcinoma, acute lymphoblastic leukemia and prostate cancer. Specimens contain serum/plasma, tissue, and urine. And, all studies adopted the approach of quantitative reverse transcription polymerase chain reaction (qRT-PCR) to measure the expression of miRNA-100. The main characteristics of these eligible studies were listed in Table 1 [16, 27-31]. The quality of the studies according to QUADAS-2 tool was good, which was summarized in Figure 2.
Table 1. Main characteristics of eligible studies in diagnostic systematic review.
| Author | Year | Country | Tumor type | Patients | Controls | Specimen | Method | AUC | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tarek et al | 2016 | Egypt | BC | 70 | 62 | serum | qRT-PCR | 0.823(0.728-0.917) | 63 | 21 | 7 | 41 |
| Wang et al | 2014 | China | GC | 50 | 47 | serum | qRT-PCR | 0.71(0.61-0.82) | 36 | 20 | 15 | 27 |
| Anna et al | 2012 | Poland | EEC | 34 | 14 | plasma | qRT-PCR | 0.740(0.592-0.857) | 22 | 3 | 12 | 11 |
| 73 | 31 | tissue | qRT-PCR | 0.652(0.548-0.746) | 63 | 16 | 10 | 16 | ||||
| Zhang et al | 2010 | China | ESCC | 149 | 100 | serum | qRT-PCR | 0.817(0.763-0.870) | 95 | 19 | 54 | 81 |
| Menha et al | 2016 | Egypt | ALL | 85 | 25 | serum/plasma | qRT-PCR | 0.87(0.779–0.934) | 70 | 0 | 15 | 25 |
| Alberto et al | 2016 | Mexico | prostate cancer | 73 | 70 | urine | qRT-PCR | 0.738(0.652-0.823) | 51 | 13 | 22 | 57 |
BC = bladder cancer, GC = gastric cancer, EEC = endometrioid endometrial carcinoma, ESCC = esophageal squamous cell carcinoma, ALL = acute lymphoblastic leukemia, qRT-PCR = quantitative reverse transcription polymerase chain reaction, AUC = the area under the SROC curve, TP = true-positive, FP = false-positive, FN = false-negative, TN = true negative.
Figure 2. Details of quality assessment by the QUADAS-2 tool.
“-” in red and “+” in green mean high risk and low risk respectively. “?” in yellow means unclear risk.
Diagnostic accuracy and threshold analysis
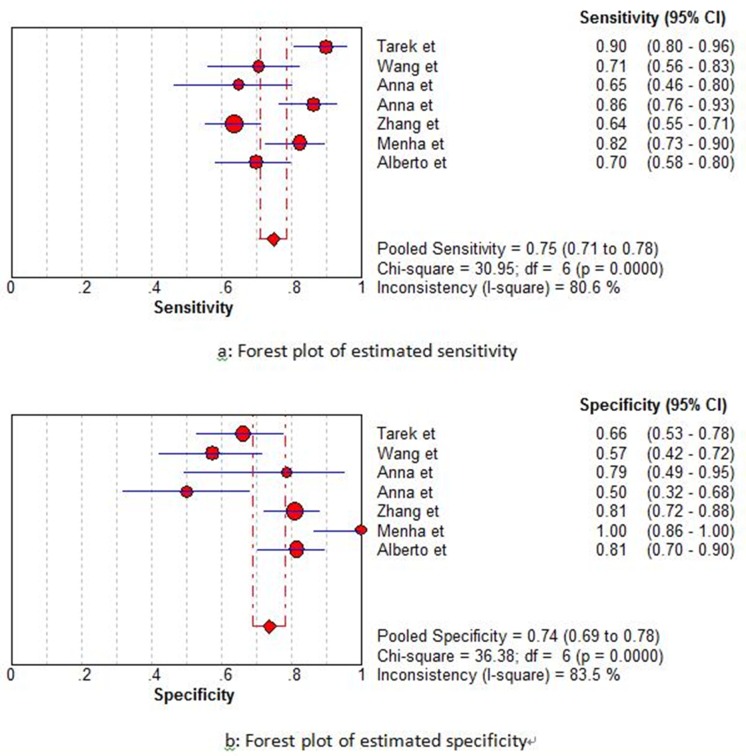
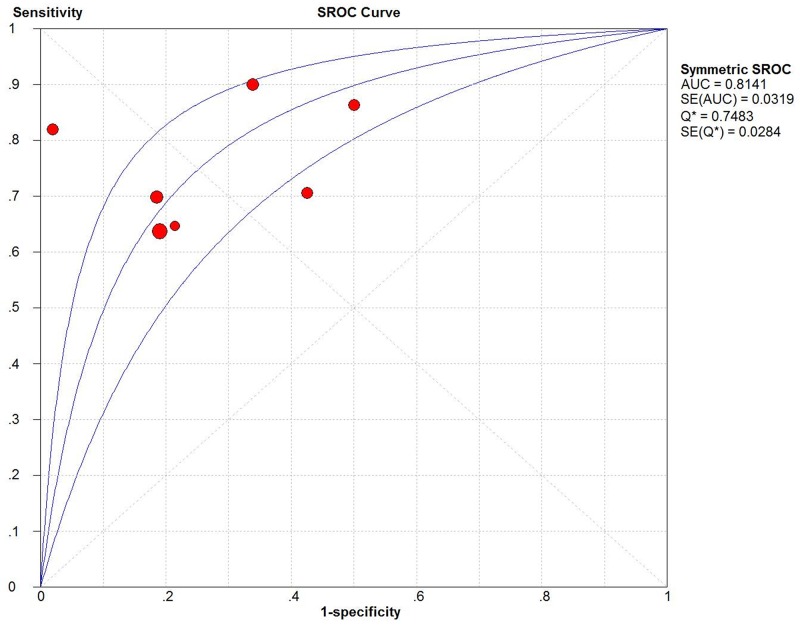
Firstly, we used the receiver operating characteristic curve (ROC) to identify whether it exist threshold effect. The result showed that there was no heterogeneity from threshold effect. What’s more, the Spearman’s correlation coefficient in this meta-analysis was 0.393 (P= 0.383), which confirmed the result was objective. According to results of the inconsistency index (I2), we chose the random-effect model to calculate all indexes. The results of the pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) were 0.75 (95%CI: 0.71-0.78), 0.74 (95%CI: 0.69-0.78), 2.61 (95%CI: 1.81-3.76), 0.33 (95%CI: 0.24-0.45), 8.46 (95%CI: 4.85-14.77), respectively (Figure 3). Moreover, as shown in Figure 4, the area under curve (AUC) was 0.8141, suggesting that miRNA-100 had a certain value in diagnosis.
Figure 3. Forest plots of estimated sensitivity (a) and specificity (b) for miRNA-100 in the diagnostic analysis.
Figure 4. Summary receiver operating characteristic (SROC) Curves of miRNA-100.
Prognostic meta-analysis
Study characteristics
A total of 19 studies with 2009 patients were included in this prognostic meta-analysis. Among those, patients in 16 studies were from China [7-12, 14, 15, 17-21, 23-25] and the other 3 studies were from Germany [22], Poland [16], and Iran [13]. All the studies were published from 2012 to 2016. The tumors types involved to colorectal cancer (n=2), lung cancer (n=3), bladder cancer (n=2), esophageal squamous cell carcinoma (n=2), acute leukemia (n=2), breast cancer (n=1), pancreatic ductaladeno carcinoma (n=1), hepatocellular carcinoma (n=1), renal cell carcinoma (n=1), endometrioid endometrial carcinoma (n=1), small cell carcinoma of the cervix (n=1), and epithelial ovarian cancer (n=2). The numbers of patients ranged from 44 to 204. The expression level of miRNA-100 was measured by qRT-PCR, and HRs and 95%CIs for OS was extracted from each studies. There were 10 studies based on univariate analysis and 9 studies with multivariate analysis. The main characteristics of the eligible studies were listed in Table 2, which also included the scores according to the Newcastle-Ottawa scale (NOS).
Table 2. Main characteristics of eligible studies in prognostic systematic review.
| Author | Year | Country | Tumor type |
Sample size | Specimen | Method | Cutoff | Outcomes | Follow-up (months) | Survival analysis | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Susan et al | 2016 | Iran | EOC | 55 | tissue | qRT-PCR | - | OS | 40(7-90) | U | 8 |
| Zhang et al | 2015 | China | breast cancer | 204 | tissue | TCGA database | - | OS | 10-170 | U | 8 |
| Zhang et al | 2015 | China | CRC | 172 | tissue | qRT-PCR | median | OS | 41 | U | 7 |
| Sameer et al | 2015 | Germany | PDAC | 98 | tissue | qRT-PCR | 5 | OS | 0-120 | U,M | 8 |
| Luo et al | 2015 | China | NSCLC | 48 | tissue | qRT-PCR | median | OS | 18 | U | 7 |
| Cao et al | 2015 | China | BC | 92 | tissue | qRT-PCR | - | OS | 0-50 | U,M | 7 |
| Zhou et al | 2014 | China | ESCC | 120 | tissue | qRT-PCR | median (1.77) | OS | 22.62(2.63-76.87) | U,M | 8 |
| Chen et al | 2014 | China | CRC | 138 | tissue | qRT-PCR | median (1.26) | OS | 5-60 | U.M | 8 |
| Li et al | 2013 | China | ALL | 111 | bone marrow | qRT-PCR | - | OS | 0-60 | U | 8 |
| Chen et al | 2013 | China | HCC | 134 | tissue | qRT-PCR | - | OS | 0-60 | U,M | 7 |
| Wang et al | 2013 | China | RCC | 96 | tissue | qRT-PCR | median (5.5) | OS | 81.8(25.2–133.6) | U,M | 8 |
| Sun et al | 2013 | China | ESCC | 61 | tissue | qRT-PCR | - | OS | 0-100 | U | 7 |
| Wang et al | 2012 | China | NSCLC | 92 | tissue | qRT-PCR | median (0.02) | OS | 6 (1-33) | U,M | 7 |
| Anna et al | 2012 | Poland | EEC | 104 | tissue | qRT-PCR | - | OS | 10-150 | U | 7 |
| Wang et al | 2012 | China | BC | 126 | tissue | qRT-PCR | - | OS | 36 | U,M | 7 |
| Huang et al | 2012 | China | SCCC | 44 | tissue | qRT-PCR | 6.515 | OS | 23.6(2-70) | U,M | 7 |
| Peng et al | 2012 | China | EOC | 98 | tissue | qRT-PCR | median (0.14) | OS | 0-60 | U | 8 |
| Liu et al | 2012 | China | NSCLC | 110 | tissue | qRT-PCR | - | OS | 0-65 | U | 7 |
| Bai et al | 2012 | China | AML | 106 | bone marrow | qRT-PCR | median (10.8) | OS | 35(10-86) | U | 8 |
M = multivariate, U = univariate, qRT-PCR = quantitative reverse transcription polymerase chain reaction, CRC = colorectal cancer, PDAC = pancreatic ductal adenocarcinoma, NSCLC = non small cell lung cancer, BC = bladder cancer, EEC = endometrioid endometrial carcinoma, ESCC = esophageal squamous cell carcinoma, AML = acute myelocytic leukemia, ALL = acute lymphoblastic leukemia, HCC = hepatocellular carcinoma, RCC = renal cell carcinoma, GC = gastric cancer, SCCC = small cell carcinoma of the cervix, EOC = epithelial ovarian cancer, OS = overall survival, NOS = Newcastle-Ottawa scale.
Meta-analysis and subgroup analysis
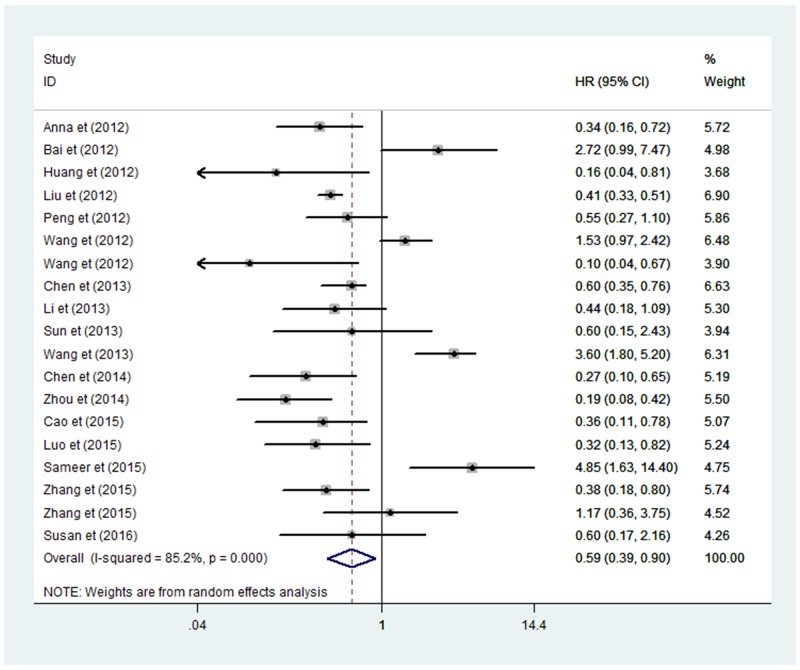
Obvious heterogeneity was found among these 19 studies for the correlation between the expression of miRNA-100 and overall survival (OS) (I2 = 85.2%), so we used the random-effect model to combine hazard ratio (HR) value and 95%CI. With a pooled HR for OS of 0.59 (95%CI: 0.39-0.90), our findings demonstrated that decreased expression of miRNA-100 in tissue predicted a poor clinical outcome (Figure 5). Likewise, the subgroup analysis was integrated into the investigation of heterogeneous sources and the relationship between HRs value and other variables, including sample size, types of cancers, methods, and countries (Table 3). Apparently, we found a significant relationship with lower HR in genital system tumors (HR=0.42, 95%CI: 0.27-0.66 P=0.431), bladder cancer (HR = 0.21, 95%CI: 0.06-0.73, P = 0.143) and esophageal squamous cell carcinoma (HR = 0.26, 95%CI: 0.13-0.52, P = 0.164). Moreover, the results showed there was obvious statistical significance among Chinese subjects (HR = 0.55, 95%CI: 0.35-0.86) and for studies with larger sample sizes (>100 subjects) (HR = 0.44, 95%CI: 0.30-0.64). Considering the difference between analysis methods, we conducted subgroup analysis by analysis methods, showing that the results were meaningful among the studies used univariate analyses (HR = 0.56, 95%CI: 0.38-0.82, shown in Table 3).
Figure 5. Forrest plots of studies evaluating HRs of high miRNA-100 expression as compared to low expression for cancer.
CI = confidence interval, HR = hazard ratio.
Table 3. Main results of the pooled analysis.
| Survival | Variables | No. of studies | Rondom-effects model or fixed-effects model | Heterogeneity | |||
|---|---|---|---|---|---|---|---|
| No. of patients | Pooled HR | 95%CI | I2 | P | |||
| OS | All | 19 | 2009 | 0.59 | 0.39-0.90 | 85.20% | 0.000 |
| Type | |||||||
| genital system tumors | 4 | 301 | 0.42 | 0.27-0.66 | 0.00% | 0.431 | |
| digestive system | 6 | 723 | 0.65 | 0.29-1.47 | 80.40% | 0.000 | |
| respiratory system | 3 | 250 | 0.60 | 0.23-1.62 | 92.60% | 0.000 | |
| urinary system | 3 | 314 | 0.54 | 0.04-4.74 | 93.90% | 0.000 | |
| others | 3 | 421 | 0.74 | 0.23-2.37 | 80.9% | 0.005 | |
| Sample | |||||||
| >100 | 10 | 1325 | 0.44 | 0.30-0.64 | 67.70% | 0.001 | |
| <100 | 9 | 684 | 0.83 | 0.41-1.70 | 84.40% | 0.000 | |
| Country | |||||||
| China | 16 | 1752 | 0.55 | 0.35-0.86 | 85.80% | 0.000 | |
| Other countries | 3 | 257 | 0.98 | 0.19-5.10 | 87.20% | 0.000 | |
| Method | |||||||
| Univariate | 10 | 1069 | 0.56 | 0.38-0.82 | 52.40% | 0.032 | |
| Multivariate | 9 | 940 | 0.57 | 0.28-1.17 | 89.80% | 0.000 | |
Meta aggression and sensitivity analysis
We conducted the meta-regression based on publication year, country, sample size, analysis method, tumor type and follow-up period, with an intention of exploring the potential source of heterogeneity in our analysis. However, there was no obvious evident revealed from the results that either of the above covariates in this meta-regression contributed to heterogeneity (shown in Table 4).
Table 4. Meta-regression analyses of potential source of heterogeneity.
| Heterogeneity factors | Coefficient | SE | Z | p | 95% CI | |
|---|---|---|---|---|---|---|
| LL | UL | |||||
| Publication year | 0.029 | 0.181 | 0.16 | 0.875 | -0.354 | 0.412 |
| Country | 0.565 | 0.661 | 0.85 | 0.404 | -0.829 | 1.960 |
| Number of patients | -0.001 | 0.006 | -0.16 | 0.876 | -0.144 | 0.012 |
| Analysis method | 0.400 | 0.472 | 0.85 | 0.408 | -0.595 | 1.396 |
| Tumor types | -0.106 | 0.140 | -0.76 | 0.459 | -0.402 | 0.189 |
| Follow-up | 0.006 | 0.006 | 1.07 | 0.299 | -0,006 | 0.018 |
SE = standard error, CI = confidence interval, LL = lower limit, UL = upper limit.
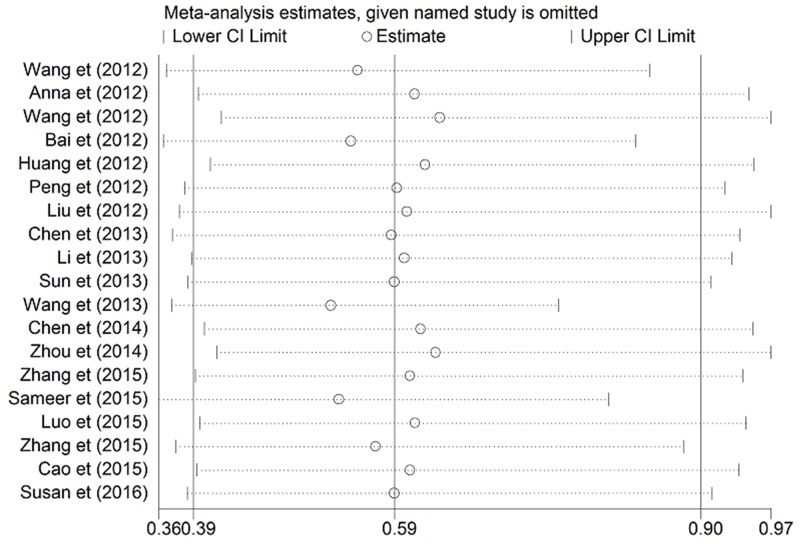
Meanwhile, we performed sensitivity analysis on the pooled HR for OS about the expression of miRNA-100 in patients. The selected studies were sequentially removed to investigate whether any single study could have an influence on the pooled HRs. As displayed in Figure 6, the results were stable and not significantly affected by each individual study.
Figure 6. Sensitivity analysis on the pooled hazard ratio for miRNA-100 and overall survival of patients.
Publication bias
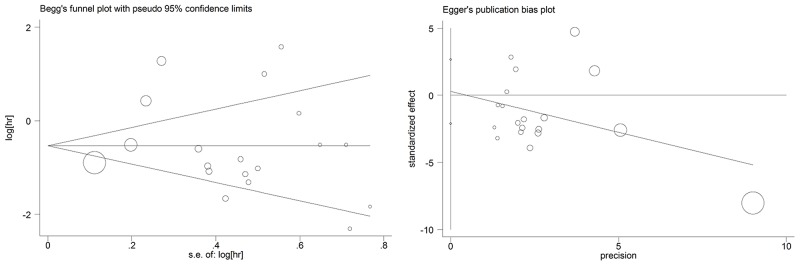
Begg’s and Egger’s tests were used to evaluate the publication bias of the included studies (Figure 7). Begg’s funnel plot did not reveal any evidence of significant asymmetry. With the P value of Egger’s test of being 0.800, it indicated no significant existence of publication bias.
Figure 7. Begg’s and egger’s funnel plots for all of the included studies reported with overall survival.
DISCUSSION
Many investigators reported miRNA-100 in various cancers as a novel molecular target. And, mammalian target of rapamycin (mTOR) gene and insulin-like growth factor 1 receptor (IGF1R) are direct target of miRNA-100 in bladder cancer, acute myelocytic leukemia, endometrioid endometrial carcinoma and so on [12, 16, 20, 32]. MiRNA-100 could suppress the related proteins of the IGF/mTOR signaling cascade in different cancers. Overexpression of miR-100 inhibited the expressions of IGF1R and mTOR by targeting its 3′-UTR at posttranscriptional gene level, so that interfer cell proliferation and survival signaling in some types of tumors. And, miRNA-100 can also exert as a tumor suppressor in many cancers by targeting polo-like kinase 1 (PLK1) [9, 14, 19]. MiRNA-100 was found to significantly inhibit the expression of PLK1 and other proteins, which has a vital effect on cell growth, apoptosis, development and drug resistance. In addition, several studies reported that miR-100 regulated apoptosis in gastric tumor cells and breast cancer cells [33, 34], and they declared miR-100 antagonism triggers apoptosis by inhibiting ubiquitination-mediated p53 degradation [35]. Besides, Cyr61 and RBSP3 was discovered a potential target of miRNA-100 for regulation. Hence, these miRNA-100 related cellular and molecular pathways may provide some ideas for new therapeutic targets in many types of cancers.
The present meta-analysis for diagnosis showed us a pooled sensitivity of 0.75, a pooled specificity of 0.74, DOR of 8.46, AUC of 0.1841, and a PLR of 2.61, which illustrated that there was an approximately 3-fold higher possibility of being miRNA-100 positive for patients with cancer in comparison to those without. And, a NLR of 0.33 mean the probability of miRNA-100 negative patients having cancers was 33%, which suggested that the diagnosis of miRNA-100 existed a certain degree of accuracy but not high enough. But it still had a great advantage compared to other traditional serum-based biomarkers, such as the sensitivities for lung cancer of CEA, Cyfra21-1, SCC and NSE were 46.2%, 40.0%, 43.1%, and 46.2%, respectively. The clinical significance of single biomarker was not ideal. So it may achieve a better diagnostic accuracy through uniting other biomarkers or clinical examinations.
For prognostic value, some studies indicated the low miR-100 expression in bladder cancer predicts unfavorable prognosis and it might regulate tumor metastasis or other related processes about tumorigenesis by inhabiting mTOR [36-41]. A study in EEC observed that decreased miRNA-100 in EEC tissues and up-regulated miRNA-100 in plasma by targeting mTOR making it as a promising biomarker for diagnosis and prognosis [16]. Some studies on EOC, SCCC and NSCLC found miR-100 was significantly decreased in cancer tissues in comparison to healthy people, which appeared that low miR-100 was a poor prognostic biomarker by targeting PLK1 in patients [9, 11, 13, 14, 19, 42] and some researches in HCC, CRC and ESCC hold the similar view [11, 12, 17, 43-45]. But, there are also some studies had different opinions in some types of cancer. Some studies showed miRNA-100 was up-regulated in cancer tissues of AML, RCC, PDAC and NSCLC, causing the result was quite opposite [21-23, 46]. Then, we conducted this meta-analysis for prognosis, and we found lower miRNA-100 expression may predict a poorer outcome in various cancers, and the predictive efficacy was more significant in genital system tumors, BC and ESCC. HRs were significant for studies in Chinese subjects, larger sample sizes (>100 subjects) and by univariate analyses. So we think miRNA-100 may be a potential biomarker for prognostic. However, as the results of sensitivity analysis and meta-regression, we couldn’t find the resource of heterogeneity, so we summarized the data using the random-effect model. Considering the source of heterogeneity, we speculated that the heterogeneity may be caused by the cut-off value of miRNA-100 expression, which was not been reported explicit values in many articles, and multiple factors may influence the heterogeneity together, such as the difference of selection criteria for patients in various tumor types and studies, the diversified clinicopathological characteristics of patients in different studies, the specific method of randomization and blind, the random errors in studies and so on.
Recently, a meta-analysis was reported about prognostic value of miRNA-100 in cancers. They suggested that patients with lower expression of miRNA-100 in cancer tissue had poorer survival in a variety of carcinomas, which was similar to our result in prognostic meta-analysis [47]. There have many differences between us, such as published time, sample size, statistical software, data processing and so on. Firstly, six studies published before October 2013 were included in that study, while we extracted data from 19 available studies published before October 2016 for prognostic and 7 studies for diagnostic meta-analysis. Secondly, in the measures of data processing, they only calculated the pooled HR for OS and investigated publication bias in their analysis, without exploring the source of heterogeneity in that study. However, we carried out the subgroup analysis by some variables and discovered that there were significant results especially in genital system tumors, bladder cancer and esophageal squamous cell carcinoma. Meanwhile, with an aim of probing deeply into the source of heterogeneity, we executed the meta-regression, subgroup analysis and sensitivity analysis. What’s more, the diagnostic meta-analysis was also referred to the investigation of the diagnostic and prognostic significance ofmiRNA-100 in various cancers. So, we have more advantages in comparison with previous study. Our study is the first meta-analysis to research the diagnostic and prognostic value of miRNA-100 in various cancers. In addition, our study may be more comprehensive and abundant due to our efforts in subgroup analysis, meta-regression and so on. Besides, many new studies have been incorporated into our article, which contributed to a more reliable conclusion.
Nevertheless, this study still exists some limitations. Firstly, research and sample size in single tumor type was relatively small, which probably influenced the research in single cancer. Secondly, our studies have a very high ratio of data in Chinese patients, which may limits its application to global range. Thirdly, some HRs could not be extracted from primary studies and needed to calculate from the Kaplan-Meier survival curves using indirectly method, which may cause a certain calculation error. Finally, it exists an obvious heterogeneity in our meta-analysis, according to the sensitivity analysis and meta-regression, and we could just supposed the resource of heterogeneity. Therefore, further studies in the future are expected to draw a more definitive conclusion.
In summary, we concluded miRNA-100 had a certain value in diagnosis despites its diagnostic accuracy was not high enough, and it had a significant value as a prognostic biomarker. Therefore, investigating the expression of miRNA-100 in various cancers may provide a new thinking into cancer prevention and therapeutic strategy, and the different expression level of miRNA-100 in cancers may indicate different endings of patients.
METHODS
Search strategy and selection criteria
This meta-analysis was conducted following the guidelines of the Meta-analysis of Observational Studies in Epidemiology and Preferred Reporting Items for Systematic Reviews and Meta-Analyses groups [48]. We carefully searched for the relevant articles in PubMed, Web of Science, EMBASE and CNKI (up to October 31, 2016) assessing the diagnostic accuracy and the prognostic significance of miRNA-100 in various types of cancers. The keywords such as microRNA-100/miRNA-100/miR-100, cancer/carcinomas, prognosis and diagnosis were used. Moreover, the reference articles from all associated articles were also found and scanned manually to retrieve any additional eligible studies.
The eligible studies must fit the following inclusion criteria: (i) the study investigates the diagnostic or prognostic value of miRNA-100 in patients with various carcinomas; (ii) for diagnosis, they must provide enough information that we could obtain the crucial data directly or through calculation, such as true-positive (TP), false-positive (FP), false-negative (FN), and true negative (TN); (iii) for prognosis, they must provide enough information so that we could extract directly or indirectly HRs with 95% CIs for OS; (iv) for prognosis, they measured the expression of miRNA-100 in tumor tissues.
Articles will be excluded by following criteria: (i) duplicate studies; (ii) review articles or letters; (iii) non-original articles; (iv) animal experiments and laboratory studies. Two reviewers independently searched and identified all articles, resolving disagreements by consensus in research group.
Data extraction
Two investigators independently made judgments and extraction of the relevant data, settling disagreements through consensus adjudication by research group. The extracted data included name of the first author, publication year, number of patients, cancer types, specimen, test method, diagnostic results (AUC, TP, FP, TN, FN) and related data for prognostic (cut-off, follow-up, HR, 95%CI). If not obtaining diagnostic results directly, we calculate the data using their sensitivity and specificity. We collect HRs and their 95% CIs preferentially from multivariate or univariate analyses in the original article, and HR>1 means higher expression of miRNA-100 in tumor tissues that may have a poorer prognosis in cancer patients. If not available, we calculate HRs with corresponding 95% CIs from Kaplan–Meier curves through Engauge 4.0 software. Meanwhile, study quality of studies in diagnostic meta-analysis was rated by Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) assessment tool. We also use the Newcastle-Ottawa (NOS) scale to evaluate the quality of each included study, which score ranges from “0” to “9” and a score ≥6 indicates high quality.
Statistical analysis
As for diagnostic meta-analysis, we calculated and combined sensitivity, specificity, PLR, NLR, DOR, and corresponding 95% 95% CIs based on the key data (TP, FP, FN, TN), ROC and Spearman correlation coefficient were applied to verify a threshold effect. We measured the heterogeneity by the I2. I2> 50% indicated that significant heterogeneity exists in studies, then we implemented the random-effect model to calculate the related indexes (DerSimonian-Laird method), otherwise, the fixed-effect model was selected (Mantel-Haenszel method). Simultaneously, the diagnostic accuracy was assessed by the area under the SROC curve (AUC) from summary receiver operative curve (SROC). In addition, in the case of two-sided p values across the board, P < 0.05 was considered statistical significant. The diagnostic meta-analysis were performed with Meta- Disc software, version 1.4 (Unit of Clinical Biostatistics, Ramony 94 Cajal Hospital, Madrid, Spain) [49].
For prognostic meta-analysis, HRs and 95%CIs of OS were weighted and pooled to estimate the contact between expression of miRNA-100 and prognostic significance in various cancer patients. Higgins I-squared statistic was used to measure statistical heterogeneity, we adopted random-effect model for severe heterogeneity with I2>50%, while fixed-effect model for the absence of heterogeneity with I2<50% [50]. The confounder contribution to heterogeneity was explored through the approach of subgroup analysis, meta-regression and sensitivity analysis. Besides, we adopted Begg’s and Egger’s test to study whether it exists publication bias. All the results were considered statistical significant at two-sided P-value of 0.05. The prognostic meta-analysis was conducted with STATA statistical software, version 14.0 (Stata Corporation, College Station, TX, USA).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 3.Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol. 2011;3:140–55. [PMC free article] [PubMed] [Google Scholar]
- 4.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berardi E, Pues M, Thorrez L, Sampaolesi M. miRNAs in ESC differentiation. Am J Physiol Heart Circ Physiol. 2012;303:H931–9. doi: 10.1152/ajpheart.00338.2012. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao YH, Zhang HH, Xu HF, Duan YJ, Li Q, Huang B. Prognostic role of microRNA-100 in patients with bladder cancer. Genet Mol Res. 2015;14:15948–54. doi: 10.4238/2015.December.7.6. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Xue S, Dai YQ, Yang JF, Chen ZJ, Fang XW, Zhou WS, Wu W, Li QW. Reduced expression of microRNA-100 confers unfavorable prognosis in patients with bladder cancer. Diagn Pathology. 2012;7:159. doi: 10.1186/1746-1596-7-159. Artn 15910.1186/1746-1596-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Lu KH, Liu ZL, Sun M, De W, Wang ZX. MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1. BMC Cancer. 2012;12:519. doi: 10.1186/1471-2407-12-519. doi: Artn 51910.1186/1471-2407-12-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo J, Chen B, Ji XX, Zhou SW, Zheng D. Overexpression of miR-100 inhibits cancer growth, migration, and chemosensitivity in human NSCLC cells through fibroblast growth factor receptor 3. Tumour Biol. 2015 Aug 28; doi: 10.1007/s13277-015-3850-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao Y, Sun N, Xu X, Shao K, He J. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med Oncol. 2013;30:411. doi: 10.1007/s12032-012-0411-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S, Yang B, Zhao Y, Xu S, Zhang H, Li Z. Prognostic value of microRNA-100 in esophageal squamous cell carcinoma. J Surg Res. 2014;192:515–20. doi: 10.1016/j.jss.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Azizmohammadi S, Azizmohammadi S, Safari A, Kosari N, Kaghazian M, Yahaghi E, Seifoleslami M. The role and expression of miR-100 and miR-203 profile as prognostic markers in epithelial ovarian cancer. Am J Transl Res. 2016;8:2403–10. [PMC free article] [PubMed] [Google Scholar]
- 14.Peng DX, Luo M, Qiu LW, He YL, Wang XF. Prognostic implications of microRNA-100 and its functional roles in human epithelial ovarian cancer. Oncol Rep. 2012;27:1238–44. doi: 10.3892/or.2012.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, Zhao R, He Y, Fu X, Fu L, Zhu Z, Fu L, Dong JT. MicroRNA 100 sensitizes luminal A breast cancer cells to paclitaxel treatment in part by targeting mTOR. Oncotarget. 2015;7:5702–14. doi: 10.18632/oncotarget.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, Zamboni G, Maciejewski R. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer. 2012;12:369. doi: 10.1186/1471-2407-12-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P, Xi Q, Wang Q, Wei P. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in colorectal cancer. Med Oncol. 2014;31:235. doi: 10.1007/s12032-014-0235-x. [DOI] [PubMed] [Google Scholar]
- 18.Chen P, Zhao X, Ma L. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in hepatocellular carcinoma. Mol Cell Biochem. 2013;383:49–58. doi: 10.1007/s11010-013-1753-0. [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Lin JX, Yu YH, Zhang MY, Wang HY, Zheng M. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS One. 2012;7:e33762. doi: 10.1371/journal.pone.0033762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XJ, Luo XQ, Han BW, Duan FT, Wei PP, Chen YQ. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J Cancer. 2013;109:2189–98. doi: 10.1038/bjc.2013.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Chen L, Meng J, Chen M, Zhuang L, Zhang L. Overexpression of microRNA-100 predicts an unfavorable prognosis in renal cell carcinoma. Int Urol Nephrol. 2013;45:373–9. doi: 10.1007/s11255-012-0374-y. [DOI] [PubMed] [Google Scholar]
- 22.Dhayat SA, Abdeen B, Kohler G, Senninger N, Haier J, Mardin WA. MicroRNA-100 and microRNA-21 as markers of survival and chemotherapy response in pancreatic ductal adenocarcinoma UICC stage II. Clin Epigenetics. 2015;7:132. doi: 10.1186/s13148-015-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai J, Guo A, Hong Z, Kuai W. Upregulation of microRNA-100 predicts poor prognosis in patients with pediatric acute myeloid leukemia. Onco Targets Ther. 2012;5:213–9. doi: 10.2147/OTT.S36017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Lv M, Shen S, Zhou S, Wang P, Chen Y, Liu B, Yu L, Hou Y. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS One. 2012;7:e43268. doi: 10.1371/journal.pone.0043268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Yuan W, Tang W, Xu C, Ma J. Expression of microRNA-100 and its relation with prognosis of colorectal cancer. [Article in Chinese] Chin J Oncol. 2015;37:603–8. [PubMed] [Google Scholar]
- 26.Adusumilli PS, Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, Zhang Y, Chen X, Wang J, Zen K, Zhang CY, Zhang C. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9:e92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motawi TK, Rizk SM, Ibrahim TM, Ibrahim IA. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem Funct. 2016;34:142–8. doi: 10.1002/cbf.3171. [DOI] [PubMed] [Google Scholar]
- 28.Salido-Guadarrama AI, Morales-Montor JG, Rangel-Escareno C, Langley E, Peralta-Zaragoza O. Cruz Colin JL, Rodriguez-Dorantes M. Urinary microRNA-based signature improves accuracy of detection of clinically relevant prostate cancer within the prostate-specific antigen grey zone. Mol Med Rep. 2016;13:4549–60. doi: 10.3892/mmr.2016.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swellam M, El-Khazragy N. Clinical impact of circulating microRNAs as blood-based marker in childhood acute lymphoblastic leukemia. Tumour Biol. 2016;37:10571–6. doi: 10.1007/s13277-016-4948-7. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, Liu L, Ling R, Yi J, Wang L, Bian J, Chen J, Li N, et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31:298. doi: 10.1007/s12032-014-0298-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 32.Liu MX, Li J, Geng YL, Wang YC, Li J, Chen YJ, Ali G, Tarver SL, Wen YF, Sun WJ. Correlation study of knowledge and behavior regarding breast care among female undergraduate students in China. Asian Pac J Cancer Prev. 2014;15:10943–7. doi: 10.7314/apjcp.2014.15.24.10943. [DOI] [PubMed] [Google Scholar]
- 33.Yang G, Gong Y, Wang Q, Wang Y, Zhang X. The role of miR-100-mediated Notch pathway in apoptosis of gastric tumor cells. Cell Signal. 2015;27:1087–101. doi: 10.1016/j.cellsig.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Gong Y, He T, Yang L, Yang G, Chen Y, Zhang X. The role of miR-100 in regulating apoptosis of breast cancer cells. Sci Rep. 2015;5:11650. doi: 10.1038/srep11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang G, Gong Y, Wang Q, Wang L, Zhang X. miR-100 antagonism triggers apoptosis by inhibiting ubiquitination-mediated p53 degradation. Oncogene. 2017;36:1023–37. doi: 10.1038/onc.2016.270. [DOI] [PubMed] [Google Scholar]
- 36.Xu C, Zeng Q, Xu W, Jiao L, Chen Y, Zhang Z, Wu C, Jin T, Pan A, Wei R, Yang B, Sun Y. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol Cancer Ther. 2013;12:207–19. doi: 10.1158/1535-7163.MCT-12-0273. [DOI] [PubMed] [Google Scholar]
- 37.Ratert NM, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I. MiRNA profiling identifies candidate miRNAs for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013;15:695–705. doi: 10.1016/j.jmoldx.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira JC, Brassesco MS, Morales AG, Pezuk JA, Fedatto PF, da Silva GN, Scrideli CA, Tone LG. MicroRNA-100 acts as a tumor suppressor in human bladder carcinoma 5637 cells. Asian Pac J Cancer Prev. 2011;12:3001–4. [PubMed] [Google Scholar]
- 39.Dip N, Reis ST, Timoszczuk LS, Viana NI, Piantino CB, Morais DR, Moura CM, Abe DK, Silva IA, Srougi M, Dall’Oglio MF, Leite KR. Stage, grade and behavior of bladder urothelial carcinoma defined by the microRNA expression profile. J Urol. 2012;188:1951–6. doi: 10.1016/j.juro.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Xue S, Dai Y, Yang J, Chen Z, Fang X, Zhou W, Wu W, Li Q. Reduced expression of microRNA-100 confers unfavorable prognosis in patients with bladder cancer. Diagn Pathol. 2012;7:159. doi: 10.1186/1746-1596-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng B, Wang R, Chen LB. MiR-100 resensitizes docetaxel-resistant human lung adenocarcinoma cells (SPC-A1) to docetaxel by targeting Plk1. Cancer Lett. 2012;317:184–91. doi: 10.1016/j.canlet.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 42.Xiao F, Bai Y, Chen Z, Li Y, Luo L, Huang J, Yang J, Liao H, Guo L. Downregulation of HOXA1 gene affects small cell lung cancer cell survival and chemoresistance under the regulation of miR-100. Eur J Cancer. 2014;50:1541–54. doi: 10.1016/j.ejca.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Shi ZL, Yang X, Yin ZF. Targeting of circulating hepatocellular carcinoma cells to prevent postoperative recurrence and metastasis. World J Gastroenterol. 2014;20:142–7. doi: 10.3748/wjg.v20.i1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen P, Zhao X, Ma L. Downregulation ofmicroRNA-100 correlates with tumor progression and poor prognosis in hepatocellular carcinoma. Mol Cell Biochem. 2013;383:49–58. doi: 10.1007/s11010-013-1753-0. [DOI] [PubMed] [Google Scholar]
- 45.Tovar V, Alsinet C, Villanueva A, Hoshida Y, Chiang DY, Sole M, Thung S, Moyano S, Toffanin S, Minguez B, Cabellos L, Peix J, Schwartz M, et al. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. J Hepatol. 2010;52:550–9. doi: 10.1016/j.jhep.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Zheng B, Wang C, Chen Y, Du C, Zhao G, Zhou Y, Shi Y. Prognostic role of microRNA-100 in various carcinomas: evidence from six studies. Tumour Biol. 2014;35:3067–71. doi: 10.1007/s13277-013-1398-3. [DOI] [PubMed] [Google Scholar]
- 48.Zeng X, Zhang Y, Kwong JS, Zhang C, Li S, Sun F, Niu Y, Du L. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evid Based Med. 2015;8:2–10. doi: 10.1111/jebm.12141. [DOI] [PubMed] [Google Scholar]
- 49.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]