Abstract
Herpes simplex virus 2 (HSV2) is a human herpesvirus found worldwide that causes genital lesions and more rarely causes encephalitis. This pathogen is most common in Africa, and particularly in central and east Africa, an area of particular significance for the evolution of modern humans. Unlike HSV1, HSV2 has not simply co-speciated with humans from their last common ancestor with primates. HSV2 jumped the species barrier between 1.4 and 3 MYA, most likely through intermediate but unknown hominin species. In this article, we use probability-based network analysis to determine the most probable transmission path between intermediate hosts of HSV2, from the ancestors of chimpanzees to the ancestors of modern humans, using paleo-environmental data on the distribution of African tropical rainforest over the last 3 million years and data on the age and distribution of fossil species of hominin present in Africa between 1.4 and 3 MYA. Our model identifies Paranthropus boisei as the most likely intermediate host of HSV2, while Homo habilis may also have played a role in the initial transmission of HSV2 from the ancestors of chimpanzees to P.boisei.
Keywords: network analysis, human evolution, infectious disease, virology, archaeology
1. Introduction
Herpes simplex virus 2 (HSV2) is a sexually transmitted human pathogen that causes genital lesions and, rarely, encephalitis (Tang et al. 2003), and is associated with increased risk of HIV acquisition (Freeman et al. 2006). After primary infection, the virus adopts a life cycle of latency punctuated by periods of lytic replication when new hosts can be infected through genital contact. The virus is related to the human oral pathogen herpes simplex virus 1 (HSV1). Both HSV1 and 2 are alphaherpesviruses, which are found in many primates (Wertheim et al. 2014). HSV1 primarily causes infection, and sporadic lesions, within the oral cavity and establishes latency in the trigeminal ganglia, while HSV2 is associated with infection of the genitalia and surrounding skin, and establishes latency in the sacral ganglia (Whitley et al. 2007). Both simplex viruses can infect either body cavity, although there is viral shedding data from co-infected individuals to suggest that HSV1 is a more successful oral and HSV2 a more successful genital pathogen (Kim et al. 2006). The genetic basis of this difference in tropism isn’t fully understood, some studies of recombinant HSV1 × HSV2 strains have highlighted the importance of the latency-associated transcript in how successfully the two simplex viruses reactivate from latency within different nerve types (Bertke et al. 2007). HSV1 and 2 also differ in the length of the glycoprotein G (US4) open reading frame, which may play a role in tropism (Baines and Pellett 2007).
HSV2 was originally thought to have co-speciated with humans when our lineage diverged from that of the ancestors of chimpanzees and bonobos (anc-chimps). Comparisons of the HSV1, HSV2 and chimpanzee herpes virus 1 (ChHV1) genomes (Tang et al. 2003) suggest that HSV2 is more closely related to ChHV1 than HSV1 (Wertheim et al. 2014). This analysis also found that ChHV1 and HSV2 diverged from one another between 1.4 and 3 MYA, and the authors inferred that an unknown hominin (or hominins) was infected with HSV2 before it switched host to the ancestors of modern humans.
We hypothesise that by combining fossil data on when and where different hominin species were likely to be present in Africa, the geographical range of modern chimpanzees and bonobos, and the reconstructed distribution of tropical rainforest habitat as a proxy for the past range of the chimpanzee/bonobo ancestor (anc-chimps) it will be possible to develop a model to statistically infer the species that facilitated the host-switch of HSV2 in the modern human lineage defined here as beginning with Homo erectus (Anton et al. 2016). Before H.erectus it is unclear which hominins are the direct ancestors of anatomically modern humans (AMH). However, once HSV2 has reached H.erectus, no further host-switches are required for HSV2 to be considered present in the ancestors of modern humans.
HSV2 is found in all living populations (Looker et al. 2015), is accepted as having an African origin (Burrel et al. 2017; Koelle et al. 2017), and has patterns of genetic divergence consistent with having diverged with human populations as they spread out of Africa (Koelle et al. 2017). Current genetic, archaeological and fossil evidence suggests 100 KYA as a plausible (although not universally accepted) earliest date for AMH to have left Africa (Mirazón Lahr et al. 2016). We therefore infer that HSV2 must have been in the population of AMH before they left Africa, to take it with them when migrating to the rest of the world.
2. Methods
The process of identifying the most probable intermediary species that transmitted proto-HSV2 (henceforth HSV2) from anc-chimps to modern humans involves four steps: (1) data acquisition, which involves collecting information on the prevalence of HSV2 in Africa, fossil records of potential intermediary hominins, their geographical range and time-period, and the extent of tropical rainforest data, (2) network generation, where a network of possible transmission paths of HSV2 from anc-chimps to humans through different hominins is created, (3) probabilistic network analysis, which involves assigning weights to different transmission paths on the graph network based on a certain probability distribution, and (4) optimal path traversal, which evaluates the shortest path from anc-chimps to modern humans based on the probabilistic weights assigned to different paths.
2.1 HSV2 prevalence data, hominin fossil data and chimpanzee and tropical rainforest geographic range data
HSV2 is most closely related to ChHV1 which infected the ancestors of modern chimpanzees. However, only one ancestral-chimpanzee fossil, dated to c. 500 KYA, is currently known (McBrearty and Jablonski 2005). This means that the habitat range of anc-chimps is not directly measurable from the fossil record but is inferred to be a larger geographical range than that of modern Pan. We can use the Geographic Information System (GIS) data on the range of modern Pan troglodytes and Pan paniscus, provided by the International Union for Conservation of Nature (IUCN) Red List (Oates et al. 2008), shown in Fig. 1, to make some inferences about the range of anc-chimps but a measure of paleo-tropical rainforest range is likely to be a better proxy for the range of anc-chimps (Elton 2008). Therefore, we have used the paleo-tropical rainforest range during the period of 1.4–3 MYA from the Koppen–Geiger climate classification dataset (Peel et al. 2007) as a proxy for the ancient range of chimpanzees and combined this with data based on modern great ape distribution patterns and range size estimates (Myers Thompson 2003). This is shown in Fig. 1.
Figure 1.
Map showing the distribution of extant chimpanzee (P.troglodytes) and bonobo (P.paniscus) populations [IUCN redlist http://maps.iucnredlist.org/map.html?id=15933, http://maps.iucnredlist.org/map.html?id=15932]; the locations of hominin fossils [Supplementary Table S1] are shown with markers. The colour of the marker indicates the hominin genus; the symbol represents the species. The map also shows the location of hominin fossils relative to ancient minimum and maximum rainforest distributions [Peel et al. 2007]. This figure is available interactively: https://wadhamite.github.io/hsv2-map/.
To identify which species could have been involved in the host-switch of HSV2 from the ancestors of chimpanzees to the ancestors of modern humans, we collated spatio-temporal data on African hominin fossil species extant between 100 KYA and 3 MYA. Latitude and longitude of site location was used to provide a spatial data point for each species. Temporally, published fossil dating evidence was used to provide a first appearance datum and last appearance datum (LAD) for each species (Supplementary Table S1).
Data on the prevalence of HSV2 between 2000 and 2015 CE was taken from the supplementary materials of Looker et al. (2015) and plotted to demonstrate the distribution of HSV2 across Africa (Supplementary Fig. S1).
2.2 Network analysis
To establish the most probable transmission path of HSV2 from anc-chimps to modern humans, it is important to identify potential species that may have been intermediary hosts. Initially, all hominins in Africa with temporal ranges within the 1.4–3 MYA confidence window (Wertheim et al. 2014) of the chimp-hominin transmission were identified (Supplementary Table S1). Their distance to ancient tropical rainforest was calculated, and only those species whose fossil remains were found within 400 km of tropical rainforest were considered as putative species for initial ancestral–chimp-hominin HSV2 transmission. This initial threshold on the distance reflects the distance that would have been covered by hominins employing three possible strategies: (1) a broadly omnivorous subsistence strategy based on scavenging, (2) hunting using carnivore and herbivore movement patterns, and (3) modern hunter-gatherer range sizes (Foley 1978; Grant et al. 1992). A matrix of spatio-temporal distances was then calculated to map the distances between the nearest neighbours of each species and also to calculate the temporal overlap between species using the fossil record (Supplementary Tables S2 and S3). Fossils from four genera (Ardipithecus, Kenyanthropus, Orrorin, and Sahelanthropus) were excluded from the analysis on the basis that there is no fossil evidence that they persisted after 3 MYA.
A network of possible transmission paths of HSV2 from anc-chimps to humans through different hominins was developed as a directed acyclic graph (DAG) G = (V, E) comprising of a set of nodes (V), representing potential intermediary hosts, and edges (E) connecting the nodes, which represents the direction of transmission between species (Figs 1 and 2). The DAG comprised of the anc-chimps as the start node, H.erectus as the target node, and other potential species forming secondary nodes in the graph. The edges are typically weighted; in this analysis, the weights are based on the inverse probability of transmission (1 − P, where P is the probability of transmission).
Figure 2.
The A* shortest path path(s) for the Infection Prevalence model. The lines with arrows are the possible transmission paths. The values on the lines are the edge costs (inverse probability of transmission). This model predicts that the host-switch of HSV2 occurred through the path anc-chimps -> P.boisei -> H.erectus (highlighted in bold). The path remained unchanged in the sensitivity analysis.
If HSV2 was transmitted to H.erectus, no further cross-species transmission event is needed to explain the infection of modern humans. Simple vertical mother-to-child or horizontal (sexual) transmission of the virus through the genus Homo from this point would be sufficient as the ancestor-descendent path from H.erectus to Homo sapiens is relatively secure (Maslin et al. 2015).
2.2.1 Bayesian inference
A Bayesian network or a belief network is a graphical structure which allows us to represent and reason about an uncertain domain. In a Bayesian network, the species (nodes) are variables, and the transmission paths (edges) represent direct links between species. The process of conditioning (also called probability propagation or inference) is performed via a ‘flow of decisions’ through the network, which involves computing the posterior probability distribution for a set of query nodes (hominins which could have transmitted HSV2), given values for some evidence (or observation) nodes (anc-chimps and H.erectus). The presence of HSV2 in the common ancestors of modern humans, and anc-chimps being the source species of the virus, provides the evidence of the presence of HSV2 in anc-chimps and H.erectus, and these species form the observation nodes in the belief network. The process of conditioning involves computing the probability of different nodes (hominins) for the acquisition and transmission of HSV2 through all possible transmission paths in the network, such that the observations of HSV2 in anc-chimps (origin node) and H.erectus (target node) are true. An important consideration in all Bayesian-based methods is the choice of a prior. An empirical Bayesian method that estimates the likelihood of HSV2 infection using a prior beta distribution is adopted, which is an approximation to a hierarchical Bayesian approach (Murphy 2012; Farine and Strandburg-Peshkin 2015).
Bayesian networks provide full representations of probability distributions over their variables, which allows us to infer upon any subset of variables. A Bayesian network is created using the DAG described earlier. Each node on the graph represents a potential intermediary host that has a probability of transmitting HSV2. The probability of infection transmission is represented as a beta distribution, with shape parameters alpha representing the time-period of the species in 100,000 years and beta representing the distance to the neighbour in kilometres. A conditional probability table (CPT) is generated for all possible combinations of a dichotomous outcome for each variable (thus HSV2 infection is true or false).
For example, to evaluate the CPT of node Paranthropus boisei, we identified its parent nodes on the network graph as the following species: anc-chimps, Australopithecus africanus, Homohabilis, and H.rudolfensis. To calculate the probability of transmission from anc-chimps to P.boisei, we assume a beta distribution. The parameter α represents the time-period overlap between the two species, in this case 700 Kyr and β represents the distance between the two species (35 kilometers between the fossils of P.boisei and the extent of anc-chimps range). Based on this beta distribution, Monte Carlo simulations are run to sample from the distribution. This yields a probability of P.boisei acquiring HSV2 from anc-chimps of 0.954. Similarly, the probability of other hominins infecting P.boisei are evaluated for A.africanus, H.habilis, and H.rudolfensis as 0.096, 0.996, and 0.34, respectively. A CPT is constructed for the node P.boisei (Supplementary Table S4).
CPTs are evaluated for each node on the graph network. The Bayesian inference analysis was performed on the network using the Artificial Intelligence space decision network tool (Poole and Mackworth 2010) (http://aispace.org/bayes/). The ‘Combined Inference’ approach is adopted to evaluate the probability of intermediary hosts transmitting HSV2, by conditioning the anc-chimp and H.erectus nodes for the presence of HSV2 (Korb and Nicholson 2003), i.e. they are treated as being infected by HSV2.
2.2.2 Optimal path traversal
A* is an informed search algorithm that searches through all possible paths to the target that yields the smallest cost (Hart et al. 1968), which allows us to identify the most probable HSV2 transmission path. This is done by combining information by favouring vertices that are close to the starting point and to the target. At each time-step, the A* algorithm selects the path at a given vertex ‘n’ that has the lowest cost f(n) = g(n) + h(n), where ‘g(n)’ represents the exact cost of the path from the starting point to any vertex ‘n’, and ‘h(n)’ represents the heuristic estimated cost from vertex ‘n’ to the goal. A* balances the two as it moves from the starting node to the target node. The most probable transmission path is evaluated by minimising the traversal costs based on the edge cost and nodal heuristics of the transmission network graph. A Python script with the NetworkX package was used to implement the A* pathfinding algorithm on the DAGs to identify the most probable (optimal) HSV2 transmission path (see https://gist.github.com/wadhamite/e8991aff7d8d3a48d5bc846c747de6d8).
Two species, anc-chimps and H.erectus, formed the start node and the target node of the DAG. The edges represent the direction of HSV2 transmission and are weighted based on the inverse probability of transmission between the species (nodes). The weights (edge costs) are determined based on two probability models: Infection Prevalence (HSV2-IP) and Infection Transmission (HSV2-IT) based on the temporal and geographic range of the species. More details about the probability models are discussed in the next section. A CPT is used to determine the nodal heuristics.
2.2.2.1 HSV2-IP model
The Infection Prevalence model is a local model of probability, which assumes each transmission path and species to be independent: the probability of a species being infected by HSV2 is affected by how long the species persisted and where the species was located, not the probability that other species were infected.
The model uses a beta distribution to determine the probability of a species transmitting/being infected by HSV2 based on the proximity of the species to the rainforest habitat and the duration (difference between the first and the LAD) of the species. Both the probability of transmission and infection are described using beta distributions. The probability density function (PDF) for a beta distribution is defined as:
Where, Γ defines a gamma function,is the time-period of existence of the species in 100,000 years, and is the distance of the fossil from the rainforest in kilometers. The probability of transmission through a particular path is determined as the combined probability of the species (nodes) forming the edge. For example, the probability of transmission path from anc-chimps (P(A)) to P.boisei (P(B)) is the combined probability of both these species (P(A*B)). The probability of transmission from anc-chimps is evaluated using a beta distribution with as 6,000 (corresponding to 6 million years of existence) and as 100 (distance to the rainforest in km). The beta density functions of HSV2 transmission/infection for anc-chimps and P.boisei are shown in Supplementary Fig. S2. The probability of P.boisei being infected also follows the same beta distribution with the parameters as 700 (corresponding to 700,000 years of existence) and as 35 (distance to the rainforest in km). The edge costs were estimated by Monte Carlo simulations of the combined probabilities of two species (nodes), forming the edge, by sampling from their respective beta distributions. A typical run of a Monte Carlo simulation is shown as a blue line in Supplementary Fig. S2, where the probability of transmission by anc-chimps (P(A) = 0.97) and the probability of P.boisei being infected (P(B) = 0.9375), evaluates the combined probability of the transmission path, for that run, as 0.909. Monte Carlo simulation results are averaged over 1,000 simulation runs. Monte Carlo simulations evaluating combined probabilities P(A*B) give an edge cost of 0.937 for the edge (path) connecting anc-chimps to P.boisei, i.e. the probability of HSV2 being transmitted from anc-chimps to P.boisei is 0.937.
2.2.2.2 Stochastic modelling of infection transmission
Infection transmission in epidemiology can be predicted using mathematical models: a popular approach is the SIR model (Chen et al. 2008). In the stochastic version of the SIR (Susceptible-Infected-Recovered) model, the continuous variables are replaced by discrete numbers, and the process rates are replaced by process probabilities. At time ‘t’ the probability that a new susceptible host is infected is modelled as an exponential distribution, which is epidemiologically incorrect for most diseases (Sartwell et al. 1950; Bailey 1975; Wearing et al. 2005), i.e. the rate of leaving the exposed is independent of the time spent on the host. Wearing et al. (2005) suggested a more realistic distribution of latent and infectious periods, with a stronger central tendency:
More realistic distributions can be obtained by choosing a PDF of the infectious period, p(t) to be a gamma PDF (Blythe and Anderson 1988).
2.2.2.3 HSV2-IT model
The proposed Infection Transmission model considers the history of transmission, and the probability of paths (edges) and species (nodes) are dependent on the parent nodes and paths. The probability of a parent node being infected with HSV2 affects the probability of a child node being infected with HSV2 in addition to the proximity and time-period overlap between the parent and child nodes.
The model utilises the temporal overlap between hominin species and their geographic proximity to one another to determine the probability of a transmission path, using a gamma distribution. The PDF of a gamma distribution is given as:
in terms of shape α and rate β. The shape parameter α is defined as the ratio of the time-period in 1,000 years/distance in kilometres and the rate parameter β is defined as the normalised time-period ‘Y/x’, where ‘x’ is the time-period of the species in 1,000 years and ‘Y’ is the time-period of anc-chimps. The probability density distribution of transmission of HSV2 from anc-chimps is shown in Supplementary Fig. S3. Monte Carlo simulations were performed to evaluate the conditional probability of transmission between species, as mutually exclusive events, considering the parent nodes by sampling from the respective probability distributions.
For example, the probability of P.boisei transmitting HSV2 to H.erectus depends on the probabilities of P.boisei being infected by the anc-chimps and/or H.habilis and/or H.rudolfensis. The probability of transmission follows the gamma distribution. The gamma distribution for anc-chimp is defined with a shape parameter α of 6, and a rate parameter β of 10. Similarly, the gamma probability distribution is evaluated for other parent nodes of P.boisei (i.e. H.habilis and/or H.rudolfensis) based on their time-period overlap and distance. The path from A.afarensis is ignored in this analysis due to its low probability of transmitting HSV2. A conditional probability of transmitting HSV2 from P.boisei to H.erectus, given the probability distributions of all possible paths capable of infecting P.boisei that is P(C | A U B U C), is evaluated as 0.152.
2.3 Sensitivity analysis
Sensitivity analysis is performed to identify the impact of the probability distribution models in estimating the edge costs and in turn the optimal path of transmission. A variance-based global sensitivity analysis was performed using the Saltelli method, a variation of the Sobol technique, to evaluate the sensitivity indices of the transmission model (Sobol′ 2001; Saltelli et al. 2010). The sensitivity of each input is represented by a numeric value called the sensitivity index. These indices are used to estimate the influence of individual variables or groups of variables on the model output, by decomposing the variance of the model output into fractions. Although spatial and temporal range is often presented as a hard number in biology the complexities found on the ground are much more nuanced both in terms of daily interactions and evolutionary trajectories (Benton and Hitchin 1996). These complexities are, naturally, further compounded when dealing with a fragmentary fossil record. To allow for natural uncertainty to be introduced into the models the level of variation for sensitivity analysis was set at 10 per cent to mirror diffusion between temporal and spatial borders. A parametric space of edge costs was generated for the Sobol analysis by sampling 50,000 times from the probabilistic distribution for each edge. This generated a total of 2.3 million analyses for the sensitivity analysis for each transmission model. The first-, second-, and the total-order indices were measured using a Python script with the SALib package.
3. Results
The results from the Bayesian inference and the optimal path traversal using the Infection Prevalence and Infection Transmission models are presented later, beginning with the Bayesian inference analysis, the optimal path traversal results and finally the sensitivity analysis of the different models.
3.1 Bayesian inference
By conditioning anc-chimps and H.erectus for the presence of HSV2, the combined Bayesian inference of the DAG reveals A.afarensis (90%), H.habilis (67.8%), P.boisei (50%), and H.rudolfensis (55.8%) to be the likely intermediary hosts for the transmission of HSV2. These are the most probable hosts to have acquired HSV2, and a transmission path through these hosts is more likely.
3.2 Optimal path traversal
The optimal traversal path of the DAG for HSV2 transmission was modelled using the Infection Prevalence and the Infection Transmission models. The A* algorithm using the weighted DAGs from both models identified P.boisei as the most probable intermediary hosts that transmitted HSV2 from anc-chimps to the ancestors of modern humans (Fig. 2). In addition to the direct transmission of HSV2 from anc-chimps to P.boisei, the Infection Transmission model also identified H.habilis as an intermediary host that transmitted HSV2 to P.boisei, which subsequently transmitted to the ancestors of modern humans (Fig. 3). Table 1 shows the probable HSV2 intermediary paths and their rankings based on the edge weights from the probabilistic models.
Figure 3.
The A* shortest path for the Infection Transmission model. The lines with arrows are the possible transmission paths. The values on the lines are the edge costs (inverse probability of transmission). This model predicts that the host-switch of HSV2 occurred through the primary path of anc-chimps -> H.habilis -> P.boisei -> H.erectus (shown in bold). Sensitivity analysis revealed the primary path occurred in 60 per cent of cases, as opposed to the secondary path of anc-chimps -> P.boisei -> H.erectus (which occurred the remaining 40% of cases, shown as dotted lines) when time-period overlaps and distance were varied by ± 10 per cent.
Table 1.
Probable HSV2 transmission paths.
| Disease transmission path | HSV2-IP path ranking (normalised path cost)a | HSV2-IT path ranking (normalised path cost)a |
|---|---|---|
| anc-chimp → P.boisei → H.erectus | 1 (1.0) | 3 (1.14) |
| anc-chimp → H.habilis → P.boisei → H.erectus | 4 (1.43) | 1 (1.00) |
| anc-chimp → H.habilis → H.erectus | 2 (1.20) | 2 (1.05) |
| anc-chimp → H.rudolfensis → H.erectus | 3 (1.24) | 4 (1.54) |
aThe most probable path has a cost of 1. The path costs are normalised to the shortest path. The optimal path is shown in bold.
3.3 Sensitivity analysis
We performed sensitivity analysis allowing the input parameters of the probabilistic distributions for both the models to vary by 10 per cent. A total of 2.3 million analyses were performed for each model by sampling 50,000 times from the respective probability distributions for each edge. The cost of transmission of HSV2 along all possible paths (path costs) from anc-chimps to H.erectus were calculated. The path cost represents the ease of transmitting HSV2 through the path. A path cost is calculated as an arithmetic sum of the inverse probability of transmission through each edge in a given transmission path. The most probable transmission path is always the one that has the lowest cost (i.e. the sum of the probabilities of all of the edges in the path is the highest).
3.3.1 HSV2-IP model
The input parameters to this model were distance to the rainforest and the overlap in the time-period, allowed to vary by 10 per cent. Sensitivity analysis of HSV2-IP model always predicted the transmission path as ‘anc-chimps -> P.boisei -> H.erectus’. The path costs show a uniform distribution between 0.526 and 0.649, with an average path cost of 0.587 (Supplementary Fig. S5). The total- and first-order Sobol indices of the ‘anc-chimp -> P.boisei’ path is 4.53E-2 and 4.53E-2, respectively. The ‘P.boisei -> H.erectus’ path has a total index of 9.55E-1 and a first-order index of 9.55E-1. All other paths had negligible or zero index values. The frequency of occurrence of different transmission paths from the sensitivity analyses are summarised in Supplementary Table S5. When the critical path, defined as the path that costs the least to traverse, through P.boisei is not available, the transmission path from anc-chimps to H.erectus was through H.habilis (normalised path costs, defined as the ratio of the path cost to the critical path cost in the graph, 1.08–1.32 of the critical path cost) for 67 per cent of the cases and through H.rudolfensis (normalised path costs of 1.11–1.32 of the critical path cost) for the remaining 33 per cent.
3.3.2 HSV2-IT model
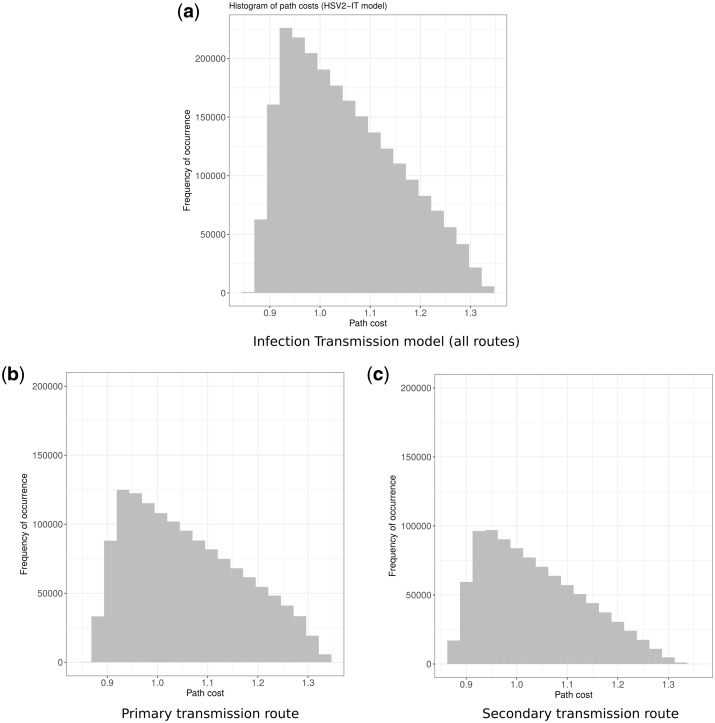
In this analysis, the input parameters of the gamma distribution were the proximity of species and the time-period overlap between species, varied by 10 per cent. The distribution of individual path costs used to populate the edge costs of the DAG used in the sensitivity analysis are shown in Fig. 4. Sensitivity analysis predicted a primary transmission path (60% of the cases) of anc-chimps -> H.habilis -> P.boisei -> H.erectus and a secondary transmission path (remaining 40%) through anc-chimps -> P.boisei -> H.erectus. The distribution of the shortest path costs for both the primary and secondary transmission paths are shown in Fig. 5. The Sobol indices for the HSV2-IT model are presented in Table 2.
Figure 4.
Distribution of selected edge costs for Infection Transmission model. Edge costs represent the inverse probability of transmission along that edge (path). The frequency of occurrence is shown on a log-scale. Each histogram shows the distribution of edge costs for the IT model of HSV2 transmission. The lowest edge costs are seen for anc-chimp -> H.habilis, anc-chimp -> P.boisei, and P.boisei-> H.erectus.
Figure 5.
Path costs distribution for Infection Transmission model. The path cost is the arithmetic sum of the inverse probability of transmission through all the edges connecting a given path. Each histogram shows the distribution of path costs for the IT model of HSV2 transmission, and the frequency of occurrence of all path costs observed in the sensitivity analysis. The optimal transmission path is anc-chimp -> H.habilis -> P.boisei -> H.erectus. (a) Total distribution of path costs for HSV2-IT model, (b) distribution path costs for the primary transmission path (anc-chimp -> H.habilis -> P.boisei -> H.erectus), and (c) distribution path costs for the secondary transmission path (anc-chimp -> P.boisei -> H.erectus).
Table 2.
Sobol indices of transmission paths for HSV2-IT model using a gamma distribution.
| Disease transmission path | Sobol total-order indices | Sobol first-order indices |
|---|---|---|
| anc-chimp → P.boisei | 0.640 | 0.454 |
| anc-chimp → H.habilis | 0.519 | 0.334 |
| P.boisei → H.erectus | 0.0262 | 0.0262 |
4. Discussion
4.1 Transmission of HSV2
The combined inference analysis of a Bayesian Network, considering the presence of HSV2 in both anc-chimps and H.erectus, revealed A.afarensis (90%), H.habilis (67.8%), P.boisei (50%), and H.rudolfensis (55.8%) to be the likely intermediary hosts. Although A.afarensis had a close proximity to the geographical range of anc-chimps, the only hominin it could transmit HSV2 to was A.africanus, and A.africanus has a mere 3 per cent chance of being infected by HSV2 due its geographic location. This limits the possibility of HSV2 transmission to H.habilis, P.boisei, and H.rudolfensis either directly or as intermediary hosts.
The HSV2-IP is a local model that describes the probability of HSV2 infection based on the proximity to the rainforest habitat and the duration a species persisted in the fossil record using a beta distribution. In a beta distribution, the parameters α (time-period) and β (proximity to the rainforest) are weighted equally, i.e. an increase in the distance to the rainforest by 1 km has the same effect on probability as a decrease of 1,000 years in the time-period. Thus the beta distribution assumes a constant rate of infection transmission. The HSV2-IP model considers each transmission as an independent event, i.e. the probability of a species transmitting HSV2 is independent of previous events leading to the HSV2 infection of that species.
The A* algorithm using the weighted DAG with the HSV2-IP model predicted the transmission of HSV2 from anc-chimps to H.erectus through P.boisei with an average path cost of 0.587. Sensitivity analysis did not show any variation in the path (i.e. varying the location and the time-period of the probable species by 10 per cent did not affect the optimal path). Non-zero values of sensitivity indices were observed only along the critical transmission path of ‘anc-chimp -> P.boisei -> H.erectus’. Although, the ‘P.boisei -> H.erectus’ path accounted for 95.5 per cent variation in the path costs, the path cost of ‘anc-chimp -> P.boisei’ was the critical value that controlled the transmission path. Hence, the optimal path through P.boisei remained unaffected. Further analysis of the path revealed a significant influence (88–90% of the variation in the results) of P.boisei (its proximity and duration) on the path cost in contrast to the effect of proximity and duration of anc-chimps and H.erectus on the path costs. The infection prevalence model uses a beta distribution, which assumes equal weights for the shape parameters: alpha and beta. As expected, Sobol analysis showed equal influence for the distance and time-period parameters (a first-order Sobol indices of 0.493 and 0.492, respectively) on the path costs. Due to the nature of beta distribution, care should be taken in defining the rate of transmission (ratio of the shape parameters, alpha and beta), as the probability value estimated from the distribution is dependent on the ratio of the shape parameters. The intermediary host identified as P.boisei is only 35 kilometers from the rainforest, which results in high probability of transmission, and hence results in the lowest cost of transmission through this species. HSV2-IP is a local model, which is independent of other events and does not consider the history of transmission and the associated probabilities.
The HSV2-IT model assigns probability values (edge costs) to the DAG utilising the temporal overlap between hominin species and their geographic proximity to one another. The HSV2-IT model is history dependent, i.e. the probability of a child node transmitting a disease depends on the probability of the parent nodes infecting the child node in a DAG. The Infection Transmission model predicts an initial transmission of HSV2 from anc-chimps to H.habilis. Further, H.habilis transmitted the virus to P.boisei, which then infected H.erectus. The average shortest path cost was estimated as 1.05. Sensitivity analysis of the HSV2-IT model revealed two transmission paths: (1) primary transmission path (60% of the cases) of ‘anc-chimps -> H.habilis -> P.boisei -> H.erectus’ and (2) a secondary transmission path (remaining 40% of the cases) through ‘anc-chimps -> P.boisei -> H.erectus’. However, the distribution of path costs through both the paths were similar, showing that both transmission paths could have been possible. Sensitivity analysis shows that the ‘anc-chimp -> H.habilis’ and ‘anc-chimp -> P.boisei’ have quite a significant effect on the transmission path (total path cost). About 70 per cent the variation in the path of ‘anc-chimp -> H.habilis’ is due to the direct effect of edge weight determined by the gamma distribution in comparison to 64 per cent of variation for ‘anc-chimp -> P.boisei’ path. Secondary interactions (interactions between different paths) accounts for 30–35 per cent of the overall routing. The probability of a species transmitting, and another being infected had similar effects (similar Sobol indices), which explains that the model did not have a bias either towards the proximity or the temporal overlap between species. Because the HSV2-IT model considers the history of transmission, it is a better predictor than the HSV2-IP model.
Our analysis suggests that P.boisei was the most critical intermediate host for transmitting HSV2 between anc-chimps and the ancestors of H.sapiens. The transmission of HSV2 from anc-chimps to P.boisei could have happened directly or through H.habilis. However, our analysis did not predict H.habilis transmitting HSV2 directly to H.erectus. Our results suggest the initial transmission to P.boisei could have happened from H.habilis 60 per cent of the time (most likely through direct sexual contact rather than consumption of infected tissue) and the remaining 40 per cent was directly transmitted from anc-chimps via hunting or scavenging.
P aranthropus boisei would have been well placed to act as an intermediate host for HSV2. It most likely contracted the infection through hunting or more likely scavenging infected ancestral-chimpanzee meat. Processing (with or without tools) and consumption of raw meat would act as a simple path for ChHV1 to have crossed into P.boisei via open cuts or sores. Tropical refugia during hot dry periods may have driven chimpanzees into higher concentrations in certain areas, driving them into contact and competition with P.boisei and H.habilis as the margins of tropical forest blended into more open savannah-like habitats (Julier et al. 2017). Violent confrontation or hunting/scavenging and butchery practices would have provided a viable path of transmission for HSV2. Homo habilis remains have been recovered from the same layers as stone tools and bones carrying evidence of butchery, supporting a possible transmission–through-hunting/scavenging hypothesis for the initial anc-chimp to H.habilis transmission (Clarke 2012). Paranthropus aethiopicus, P.boisei, and P.robustus are associated with the Oldowan stone tool complex (De Heinzelin et al. 1999), and P.boisei explicitly with butchery (Domínguez-Rodrigo et al. 2013) lending support to the hypothesis that bushmeat hunting/scavenging and butchery may have led to the initial transmission of HSV2 to the hominins.
Both H.erectus and P.boisei are known from sites around Lake Turkana in Kenya that are contiguous in age (Wood and Constantino 2007; Anton et al. 2016) and it is likely that close contact between the species would have been relatively common especially around water sources. The appearance of H.erectus 2.0 MYA is accompanied by evidence of active hunting and butchery, and from 1.76 MYA increasingly sophisticated stone tools (Cachel and Harris 1998). This behavioural shift towards active hunting displayed by H.erectus, combined with direct archaeological evidence of hunting, provides a credible path for direct transmission of HSV2 from P.boisei to H.erectus through contact and/or consumption of infected material processed from P.boisei carcasses.
In this study, H.erectus is chosen as the target node for the transmission of HSV2 for a number of reasons. Morphological adaptation for bipedal locomotion is used as the primary trait for assigning fossils to the hominin sub-family, especially between 7.0 and 4.5 MYA, but is not an effective tool for determining patterns of ancestor-descendant relationships in the fossil record. The hominin sub-family currently contains seven genera but the exact taxonomic relationship between each genus is not clearly definable because of the fragmentary nature of the fossil record. Similarly, patterns of intra- and inter-species variation are difficult to define (Foley 2016). The adaptive radiation of the genus Australopithecus between 4.5 and 2.0 MYA represents the first morphologically coherent group of fossil hominin species but its relationship to the genus Homo is not yet clear. The appearance of H.erectus circa 2.0 MYA in East Africa marks the first appearance of recognisably ‘human’ morphology, life history and brain development and represents a secure most recent common ancestor for all subsequent Homo species (with the possible exception of Homo floresiensis (Anton et al. 2016; Argue et al. 2017)). Therefore, the ancestor-descendant relationship between H.erectus at c. 2.0 MYA and H.sapiens c. 200 KYA is an evolutionarily secure path of transmission for HSV2 to leave Africa as a modern human-borne virus.
Although ChHV1 causes outbreaks of oral and pharyngeal lesions in chimpanzees in a manner similar to HSV1, in hominins contracting HSV2, the oral niche was already occupied by HSV1. This may have protected the hominin first infected with HSV2: pre-existing infection with HSV1 reduces the likelihood that subsequent infection with HSV2 will be symptomatic (Langenberg et al. 1999), and also reduces the risk of HSV2 meningitis (Aurelius et al. 2012). HSV2 may have been forced to adapt to a different mucosal niche to reduce competition from the co-evolved, native HSV1. However, both simplexviruses remain capable of infecting both the oral and genital niche in modern humans (Kim et al. 2006; Whitley et al. 2007), and of causing co-infection in both niches (e.g. 10–15% of herpes labialis (oral lesions) is caused by HSV2 (Glick and Siegel 1999)).
We suggest that the mode of transmission of HSV2 to hominins was most likely through hunting injuries (e.g. chimpanzee bites or cuts sustained during meat processing), although onwards transmission into the ancestors of H.sapiens could have been sexual (horizontal) or a result of hunting injuries (vertical). There are reports of transmission of B virus (Cercopithecine herpesvirus 1), the cercopith homolog of HSV1 and ChHV1, to humans, where disease ranges from mild to fatal. Transmission has occurred from bites and scratches, needle sticks and even scratches from cage bars that are contaminated with B virus-positive bodily fluids (Huff and Barry 2003). Onwards transmission between humans has been reported to occur (Centers for Disease Control (CDC) 1987). HSV1 can infect other primates, from gorillas (Gilardi et al. 2014) to owl monkeys (Melendez et al. 1969), typically causing fatal disease in species more distantly related to H.sapiens, while causing oral lesions and milder disease in great apes such as Gorilla beringei graueri (Gilardi et al. 2014). We therefore infer that herpes simplex-like viruses spread relatively easily between individuals even across species barriers, increasing the chances of transmission between hominins and other primates from close contact such as hunting, butchery, inter-personal violence, or sexual contact. Evidence for close hominin–hominid contact is also found in other ‘heirloom’ human pathogens (Houldcroft and Underdown 2016; Houldcroft et al. 2017).
The high prevalence of HSV2 in central and eastern Africa (Looker et al. 2015) (Supplementary Fig. S1 and interactive maps at https://wadhamite.github.io/hsv2-map) is consistent with the limited genetic data available from African HSV2 isolates. A study from (Burrel et al. 2015) showed that HSV2 can be divided into African and worldwide lineages on the basis of diversity in gene UL30. Furthermore, two groups found evidence of gene flow from HSV1 into HSV2, and speculated that the flow of HSV1 loci into the worldwide HSV2 lineage may have helped this lineage of HSV2 to further adapt to human hosts, and so spread more successfully around the world from around 41 KYA (Burrel et al. 2017; Koelle et al. 2017). Recent studies have significantly increased the number of whole HSV2 genomes available for analysis, contributing to our knowledge of HSV2 diversity (Szpara et al. 2014; Kolb et al. 2015); but there is conflicting evidence on whether the most basal HSV2 genotypes are from west and central (Burrel et al. 2017) or east (Koelle et al. 2017) Africa. Our analyses predict that individuals from east Africa are likely to carry the most ancient HSV2 lineages.
The time-depth of ancient DNA analysis is dually limited by technology and preservation of DNA. Similarly, the archaeological and fossil records suffer from differential rates of preservation (Allentoft et al. 2012; Kistler et al. 2017) and gaps that can never be filled because the material has simply not survived. Our analysis has allowed the reconstruction of hominin/human-disease interaction well beyond the horizon of ancient DNA and at a level that is invisible to the fossil and archaeological records. There are other ancient human pathogens that have switched between different primate and hominin hosts over the last 6 million years and these transmission paths could be further explored with this methodology. For example, human pubic lice (Pthirus pubis) were introduced by an unknown hominin through contact with the ancestor of gorillas around 3.3 MYA (Reed et al. 2007). There is also evidence of Neanderthal to human transmission of human papillomavirus genotypes (Pimenoff et al. 2017), and of hominin to human transmission of body louse genotypes (Reed et al. 2004). The studies and the findings presented here demonstrate the potential for using modern disease genetics to understand better the evolutionary interaction between humans and disease in deep time.
Data availability
The distribution maps were created with custom JavaScript codes using Leaflet.js library [http://leafletjs.com/] and interactive maps can be accessed at https://github.com/wadhamite/hsv2-map.
Author contributions
S.J.U. and C.H. conceived the study and contributed data. S.J.U., K.K., and C.H. performed the analyses. S.J.U., K.K., and C.H. wrote the article. All authors approved the publication of the article.
Supplementary Material
Acknowledgements
The authors thank C. Ruis (University College London), and J.B. Ramond and R.F. Rifkin (University of Pretoria) for helpful discussion. S.J.U. was funded by Oxford Brookes University. K.K. and C.H. were funded by the University of Cambridge. K.K. is a college research associate at King’s College, Cambridge. C.H. is a post-doctoral affiliate at Christ’s College, Cambridge.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
References
- Allentoft M. E. et al. (2012) ‘The Half-Life of DNA in Bone: Measuring Decay Kinetics in 158 Dated Fossils’, Proceedings of the Royal Society. Series B, Biological sciences, 279: 4724–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton S. C. et al. (2016) ‘Morphological Variation in Homo erectus and the Origins of Developmental Plasticity’, Philosophical Transactions of the Royal Society. Series B, 371: 20150236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argue D. et al. (2017) ‘The Affinities of Homo floresiensis Based on Phylogenetic Analyses of Cranial, Dental, and Postcranial Characters’, Journal of Human Evolution, 107: 107–33. [DOI] [PubMed] [Google Scholar]
- Aurelius E. et al. (2012) ‘Long-Term Valacyclovir Suppressive Treatment After Herpes Simplex Virus Type 2 Meningitis: A Double-Blind, Randomized Controlled Trial’, Clinical Infectious Diseases, 54: 1304–13. [DOI] [PubMed] [Google Scholar]
- Bailey N. T. J. (1975) The Mathematical Theory of Infectious Diseases and Its Applications, 2nd edn. Bucks, UK: Charles Griffin & Company Ltd.
- Baines J. D., Pellett P. E. (2007) Genetic Comparison of Human Alphaherpesvirus Genomes. Cambridge, UK: Cambridge University Press. [PubMed] [Google Scholar]
- Benton M. J., Hitchin R. (1996) ‘Testing the Quality of the Fossil Record by Groups and by Major Habitats’, Historical Biology: An International Journal of Paleobiology, 12: 111–57. [Google Scholar]
- Bertke A. S., Patel A., Krause P. R. (2007) ‘Herpes Simplex Virus Latency-Associated Transcript Sequence Downstream of the Promoter Influences Type-Specific Reactivation and Viral Neurotropism’, Journal of Virology, 81: 6605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe S. P., Anderson R. M. (1988) ‘Distributed Incubation and Infectious Periods in Models of the Transmission Dynamics of the Human Immunodeficiency Virus (HIV)’, IMA Journal of Mathematics Applied in Medicine and Biology, 5: 1–19. [DOI] [PubMed] [Google Scholar]
- Burrel S. et al. (2017) ‘Ancient Recombination Events Between Human Herpes Simplex Viruses’, Molecular Biology and Evolution, 25: 1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrel S. et al. (2015) ‘Genetic Diversity Within Alphaherpesviruses: Characterization of a Novel Variant of Herpes Simplex Virus 2’, Journal of Virology, 89: 12273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachel S., Harris J. (1998) ‘The Lifeways of Homo erectus Inferred from Archaeology and Evolutionary Ecology: A Perspective from East Africa’, Early Human Behaviour in Global Context, pp. 108–32. Oxford, UK: Routledge. [Google Scholar]
- Centers for Disease Control (CDC) (1987) ‘B-Virus Infection in Humans–Pensacola, Florida’, Morbidity and Mortality Weekly Report, 36: 289–90, 295–6. [PubMed] [Google Scholar]
- Chen Y. et al. (2008) ‘Finding a Better Immunization Strategy’, Physical Review Letters, 101: 58701. [DOI] [PubMed] [Google Scholar]
- Clarke R. J. (2012) ‘A Homo habilis Maxilla and Other Newly-Discovered Hominid Fossils from Olduvai Gorge, Tanzania’, Journal of Human Evolution, 63: 418–28. [DOI] [PubMed] [Google Scholar]
- De Heinzelin J. et al. (1999) ‘Environment and Behavior of 2.5-Million-Year-Old Bouri Hominids’, Science (80-), 284111185: 625–9. [DOI] [PubMed] [Google Scholar]
- Domínguez-Rodrigo M. et al. (2013) ‘First Partial Skeleton of a 1.34-Million-Year-Old Paranthropus boisei from Bed II, Olduvai Gorge, Tanzania. Curnoe D (ed.)’, PLoS One, 8: e80347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton S. (2008) ‘The Environmental Context of Human Evolutionary History in Eurasia and Africa’, Journal of Anatomy, 212: 377–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farine D. R., Strandburg-Peshkin A. (2015) ‘Estimating Uncertainty and Reliability of Social Network Data Using Bayesian Inference’, Royal Society Open Science, 2: 150367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley R. A. (1978) ‘Incorporating Sampling into Initial Research Design: Some Aspects of Spatial Archaeology’ in: Cherry J.F., Gamble C., Shennan S. (eds.) Sampling in Contemporary British Archaeology, pp. 49–66. Oxford, UK: BAR British Series. [Google Scholar]
- Foley R. A. (2016) ‘Mosaic Evolution and the Pattern of Transitions in the Hominin Lineage’, Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 371: 20150244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman E. E. et al. (2006) ‘Herpes Simplex Virus 2 Infection Increases HIV Acquisition in Men and Women: Systematic Review and Meta-Analysis of Longitudinal Studies’, AIDS, 20: 73–83. [DOI] [PubMed] [Google Scholar]
- Gilardi K. et al. (2014) ‘Human Herpes Simplex Virus Type 1 in Confiscated Gorilla’, Emerging Infectious Diseases, 20: 1883–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick M., Siegel M. A. (1999) ‘Viral and Fungal Infections of the Oral Cavity in Immunocompetent Patients’, Infectious Disease Clinics of North America, 13: 817–31. [DOI] [PubMed] [Google Scholar]
- Grant J. W. A., Chapman C. A., Richardson K. S. (1992) ‘Defended Versus Undefended Home Range Size of Carnivores, Ungulates and Primates’, Behavioral Ecology and Sociobiology, 31: 149–61. [Google Scholar]
- Hart P., Nilsson N., Raphael B. (1968) ‘A Formal Basis for the Heuristic Determination of Minimum Cost Paths’, IEEE Transactions on Systems Science and Cybernetics, 4: 100–7. [Google Scholar]
- Houldcroft C. J. et al. (2017) ‘Migrating Microbes: What Pathogens Can Tell Us About Population Movements and Human Evolution’, Annals of Human Biology, 44: 397–407. [DOI] [PubMed] [Google Scholar]
- Houldcroft C. J., Underdown S. J. (2016) ‘Neanderthal Genomics Suggests a Pleistocene Time Frame for the First Epidemiologic Transition’, American Journal of Physical Anthropology, 160: 379–88. [DOI] [PubMed] [Google Scholar]
- Huff J. L., Barry P. A. B. (2003) ‘Virus (Cercopithecine herpesvirus 1) Infection in Humans and Macaques: Potential for Zoonotic Disease’, Emerging Infectious Diseases, 9: 246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier A. et al. (2017) ‘Characterisation and Differentiation of the Modern Pollen-Vegetation Relationships of Sites Within a Forest-Savannah Mosaic Landscape in Tropical West Africa (Ghana)’, Palynology, pp. 1–15. Doi: 10.1080/01916122.2017.1356392 (in press). [Google Scholar]
- Kim H. N. et al. (2006) ‘Oral Herpes Simplex Virus Type 2 Reactivation in HIV‐Positive and ‐Negative Men’, Journal of Infectious Diseases, 194: 420–7. [DOI] [PubMed] [Google Scholar]
- Kistler L. et al. (2017) ‘A New Model for Ancient DNA Decay Based on Paleogenomic Meta-Analysis’, Nucleic Acids Research, 5: e17092.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle D. M. et al. (2017) ‘Worldwide Circulation of HSV-2 × HSV-1 Recombinant Strains’, Scientific Reports, 7: 44084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A. W. et al. (2015) ‘Genomic, Phylogenetic, and Recombinational Characterization of Herpes Simplex Virus 2 Strains’, Journal of Virology, 89: 6427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb K. B., Nicholson A. E. (2003) Bayesian Artificial Intelligence. Florida, USA: CRC Press, Inc.
- Langenberg A. G. M. et al. (1999) ‘A Prospective Study of New Infections with Herpes Simplex Virus Type 1 and Type 2’, New England Journal of Medicine, 341: 1432–8. [DOI] [PubMed] [Google Scholar]
- Looker K. J. et al. (2015) ‘Global Estimates of Prevalent and Incident Herpes Simplex Virus Type 2 Infections in 2012’, PLoS One, 10. doi:10.1371/journal.pone.0114989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- doi: 10.1098/rstb.2014.0064. Maslin, M. et al. (2015) ‘A synthesis of the theories and concepts of early human evolution’, Philos Trans R Soc B Biol Sci; 370: 20140064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrearty S., Jablonski N. G. (2005) ‘First Fossil Chimpanzee’, Nature, 437: 105–8. [DOI] [PubMed] [Google Scholar]
- Melendez L. V. et al. (1969) ‘Natural Herpes Simplex Infection in the Owl Monkey (Aotus trivirgatus)’, Laboratory Animal Science, 19: 38–45. [PubMed] [Google Scholar]
- Mirazón Lahr M. et al. (2016) ‘The Shaping of Human Diversity: Filters, Boundaries and Transitions’, Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 371: 62–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. P. (2012) Machine Learning: A Probabilistic Perspective, 25 Cambridge, MA, USA: MIT Press. [Google Scholar]
- Myers Thompson J. A. (2003) ‘A Model of the Biogeographical Journey from Proto-Pan to Pan paniscus’, Primates, 44: 191–7. [DOI] [PubMed] [Google Scholar]
- Oates J. F. et al. (2008) Pan troglodytes (Chimpanzee, Common Chimpanzee, Robust Chimpanzee). International Union for Conservation of Nature Red List Threat Species. <http://www.iucnredlist.org/details/15933/0> accessed 02 Feb 2017.
- Peel B. L., Finlayson B. L., McMahon T. A. (2007) ‘Updated World Map of the Köppen-Geiger Climate Classification’, Hydrology and Earth System Sciences, 11: 1633–44. [Google Scholar]
- Pimenoff V. N., de Oliveira C. M., Bravo I. G. (2017) ‘Transmission Between Archaic and Modern Human Ancestors During the Evolution of the Oncogenic Human Papillomavirus 16’, Molecular Biology and Evolution, 34: 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole D. L., Mackworth A. K. (2010) Artificial Intelligence: Foundations of Computational Agents. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Reed D. L. et al. (2007) ‘Pair of Lice Lost or Parasites Regained: The Evolutionary History of Anthropoid Primate Lice’, BMC Biology, 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D. L. et al. (2004) ‘Genetic Analysis of Lice Supports Direct Contact between Modern and Archaic Humans’. Nick Barton (ed.), PLoS Biology, 2: e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltelli A. et al. (2010) ‘Variance Based Sensitivity Analysis of Model Output. Design and Estimator for the Total Sensitivity Index’, Computer Physics Communications, 181: 259–70. [Google Scholar]
- Sartwell P. E., and other (1950) ‘The Distribution of Incubation Periods of Infectious Disease’, American Journal of Hygiene, 51: 310–8. [DOI] [PubMed] [Google Scholar]
- Sobol′ I. (2001) ‘Global Sensitivity Indices for Nonlinear Mathematical Models and Their Monte Carlo Estimates’, Mathematics and Computers in Simulation, 55: 271–80. [Google Scholar]
- Szpara M. L. et al. (2014) ‘Evolution and Diversity in Human Herpes Simplex Virus Genomes’, Journal of Virology, 88: 1209–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. W. et al. (2003) ‘Brain Stem Encephalitis Caused by Primary Herpes Simplex 2 Infection in a Young Woman’, Journal of Neurology, Neurosurgery, and Psychiatry, 74: 1323–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearing H. J., Rohani P., Keeling M. J. (2005) ‘Appropriate Models for the Management of Infectious Diseases’, PLoS Medicine, 2: e174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J. O. et al. (2014) ‘Evolutionary Origins of Human Herpes Simplex Viruses 1 and 2’, Molecular Biology and Evolution, 31: 2356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R., Kimberlin D. W., Prober C. G. (2007) Pathogenesis and Disease. Cambridge, UK: Cambridge University Press. [PubMed] [Google Scholar]
- Wood B., Constantino P. (2007) ‘Paranthropus boisei: Fifty Years of Evidence and Analysis’, American Journal of Physical Anthropology, 134: 106–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The distribution maps were created with custom JavaScript codes using Leaflet.js library [http://leafletjs.com/] and interactive maps can be accessed at https://github.com/wadhamite/hsv2-map.