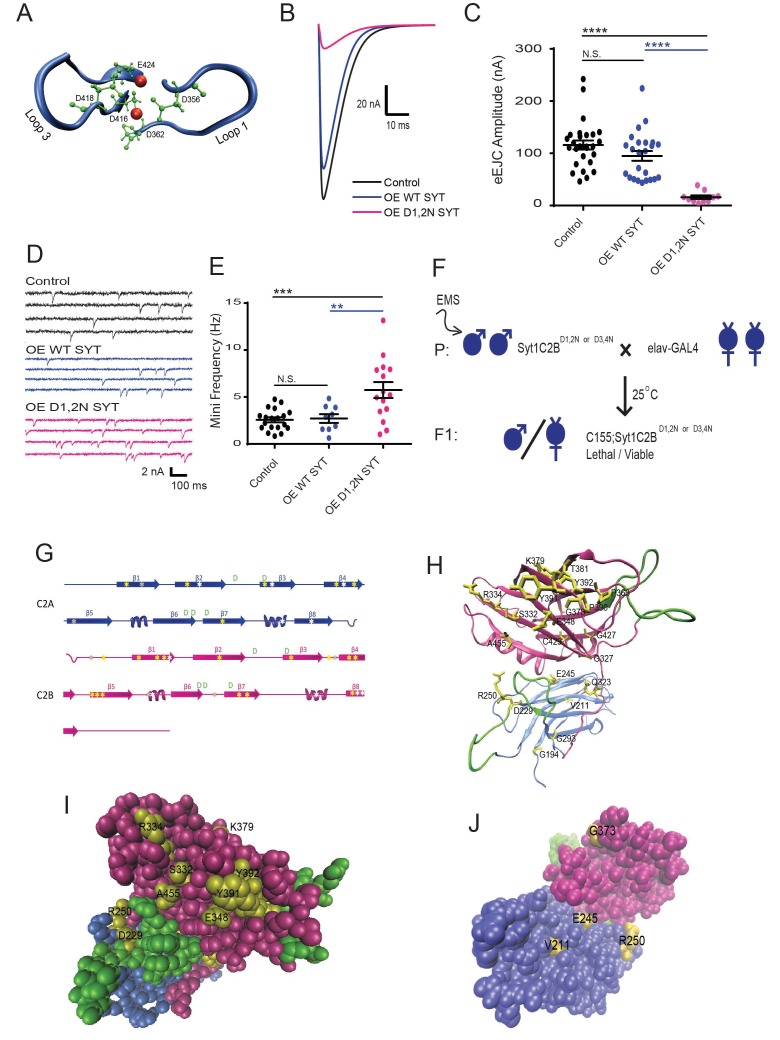
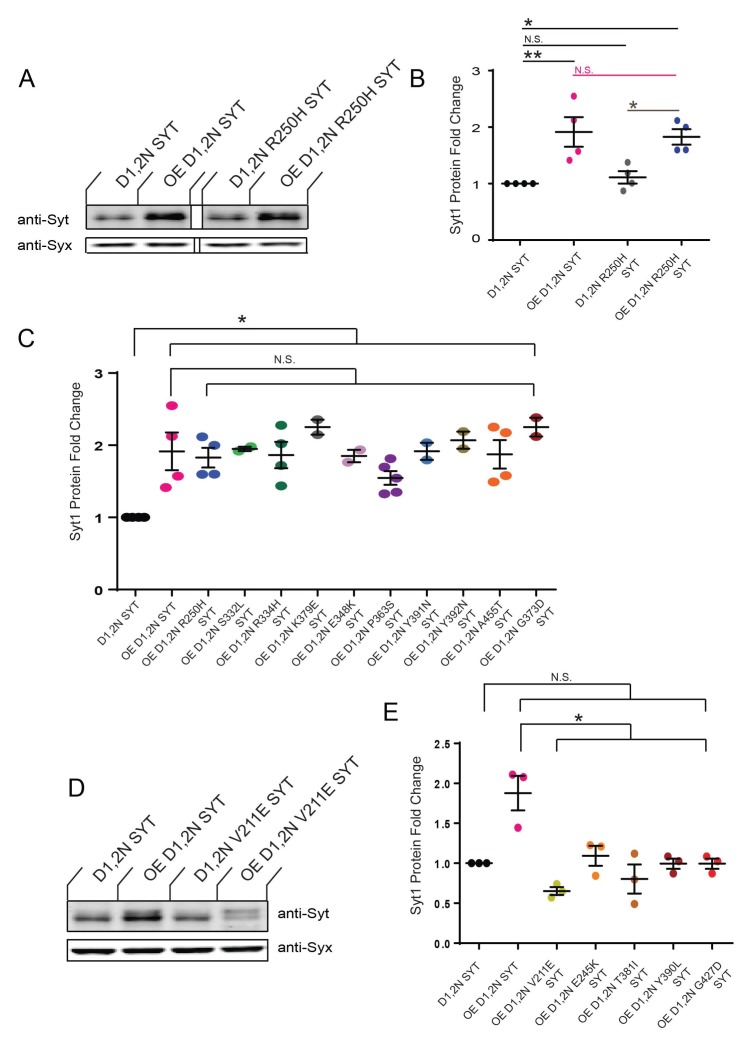
Figure 1. An intragenic suppressor screen for mutations that disrupt the lethality induced by Syt1C2BD1,2N or Syt1C2BD3,4N expression.
(A) View of key residues in the Syt1 C2B Ca2+ binding pocket. The Ca2+ binding loops 1 and 3 are highlighted in blue, the negatively charged Ca2+-binding residues in green, and Ca2+ ions in red. Neuronal overexpression of Syt1C2BD416,D418N (D1,2N) or Syt1C2BD356,D362N (D3,4N) results in pharate adult lethality. (B) Representative evoked excitatory junctional currents (eEJCs) recorded in 0.2 mM extracellular Ca2+ in control white larvae (black trace), elavC155-GAL4; UAS-Syt1 wildtype (OE WT SYT, blue trace) and elavC155-GAL4; UAS-Syt1C2BD1,2N (OE D1,2N SYT, magenta trace). (C) Quantification of mean eEJC amplitude in the indicated genotypes: control, 116.0 ± 8.7 nA, n = 27; OE WT Syt1, 95.0 ± 9.4 nA, n = 24; OE D1,2N Syt1, 15.9 ± 3.5 nA, n = 10. (D) Representative postsynaptic current recordings of spontaneous release in the indicated genotypes. (E) Quantification of average mini frequency for the indicated genotypes: control, 2.6 ± 0.3 Hz, n = 19; OE WT Syt1, 2.7 ± 0.5 Hz, n = 9; OE D1, 2N Syt1, 5.7 ± 0.8 Hz, n = 15. (F) Crossing scheme used for EMS screening of suppressors of Syt1C2BD1,2N or D3,4N-induced lethality. (G) Location of identified suppressors (point mutant - yellow asterisks, stop codon - white asterisks, deletion - grey asterisks) on the Syt1 secondary structure. C2A is indicated blue and C2B in magenta. (H) View of identified point mutant alleles (yellow) in the DSyt1 homology model. For all remaining structural images, the C2A domain is colored blue and the C2B domain is colored magenta, while the Ca2+ binding loops are highlighted in green. (I) Suppressor point mutants (yellow) located in a space-filling model of DSyt1. (J) Suppressor mutations (yellow) located on the opposite face of the DSyt1 structure compared to panel I. Statistical significance was determined using one-way ANOVA (nonparametric) with post hoc Sidak’s multiple comparisons test. N.S. = no significant change, *p<0.05, **p<0.01, ***p≤0.0005, ****p<0.0001. All error bars are standard error of the mean (SEM).