Abstract
Tumors adapt to an unfavorable microenvironment by controlling the balance between cell proliferation and cell motility, but the regulators of this process are largely unknown. Here, we show that an alternatively spliced isoform of syntaphilin (SNPH), a cytoskeletal regulator of mitochondrial movements in neurons, is directed to mitochondria of tumor cells. Mitochondrial SNPH buffers oxidative stress and maintains complex II–dependent bioenergetics, sustaining local tumor growth while restricting mitochondrial redistribution to the cortical cytoskeleton and tumor cell motility. Conversely, introduction of stress stimuli to the microenvironment, including hypoxia, acutely lowered SNPH levels, resulting in bioenergetics defects and increased superoxide production. In turn, this suppressed tumor cell proliferation but increased tumor cell invasion via greater mitochondrial trafficking to the cortical cytoskeleton. Loss of SNPH or expression of an SNPH mutant lacking the mitochondrial localization sequence resulted in increased metastatic dissemination in xenograft or syngeneic tumor models in vivo. Accordingly, tumor cells that acquired the ability to metastasize in vivo constitutively downregulated SNPH and exhibited higher oxidative stress, reduced cell proliferation, and increased cell motility. Therefore, SNPH is a stress-regulated mitochondrial switch of the cell proliferation-motility balance in cancer, and its pathway may represent a therapeutic target.
Keywords: Cell Biology, Oncology
Keywords: Cancer, Cell migration/adhesion, Mitochondria
Introduction
Tumors must cope with an unpredictable microenvironment depleted of oxygen and nutrients but saturated in oxidative radicals, toxins, and immune-inflammatory mediators (1). This requires a process of adaptation, or “plasticity” (2), that improves tumor fitness via genetic and nongenetic changes (3, 4), buffers stress signals (5), and reprograms metabolism for tailored bioenergetics needs (6).
A key decision in tumor adaptation is how best to allocate limited resources in the microenvironment (1) to either support cell proliferation, and therefore local tumor growth; or, conversely, stimulate cell motility (7), thus enabling the dissemination of transformed cells to distant organs, or metastasis (8). Both responses are intensely energy-intensive, and it has been speculated that tumors can undergo dynamic cycles of each process, but not both at the same time in a mechanism called cell proliferation-motility dichotomy (9), or phenotype-switching (10). Although this process may dictate metastatic competence and ultimately influence disease outcome, its mechanistic underpinnings (9) have remained elusive (11), with only a handful of regulators of cell cycle transitions (12), transcriptional (13) and translational (14) programs, and membrane dynamics of cell motility (15) potentially implicated in phenotype switching (16).
In this context, a role for metabolic reprogramming in phenotype switching has not been widely investigated. We know that most tumors adopt a mostly glycolytic metabolism, the so-called Warburg effect (6), but recent evidence has reinforced a central role of mitochondrial biology as a driver of cancer traits (17, 18), in particular metastatic competence (19), even under microenvironment stress (20). Several mechanisms have been implicated in this process, including modulation of oxidative stress (21); cycles of organelle fusion and fission, i.e. dynamics (22); and horizontal transfer of respiration-competent mitochondria from stromal cells (23, 24). Recently, an additional mechanism of active mitochondrial trafficking to the cortical cytoskeleton has been linked to greater tumor cell invasion and metastasis (25, 26), potentially by providing a concentrated, “regional” energy source to fuel membrane dynamics of chemotaxis and tumor cell movements. Unexpectedly, a key regulator of this pathway was identified as syntaphilin (SNPH), a cytoskeletal protein known in neurons for arresting mitochondrial movement at sites of high energy demands (27, 28). Instead, SNPH was found to be expressed in cancer, but downregulated in advanced disease settings, blocking mitochondrial trafficking to the cortical cytoskeleton and suppressing cell motility and invasion (29).
In this study, we explored the complexity of the SNPH pathway (29) as a potential regulator of proliferation-motility decisions in tumors (7). We identified an alternatively spliced isoform of SNPH that localizes to mitochondria of tumor cells and orchestrates bioenergetics, buffering of ROS, and differential modulation of tumor cell proliferation versus cell invasion and dissemination, in vivo.
Results
Identification of alternatively spliced SNPH isoforms.
A genome-wide shRNA screen recently identified SNPH as a regulator of mitochondrial trafficking in tumors and potential metastasis suppressor (29). Here, inspection of the human SNPH locus indicated the presence of at least two SNPH transcripts, potentially originated by alternative splicing of the 5′ end of the SNPH gene (Figure 1A). This process generated a previously unrecognized short SNPH isoform (S-SNPH) of 494 amino acids containing a mitochondrial localization sequence (MLS) embedded in a new amino-terminal region (Figure 1, A and B). In contrast, a long SNPH isoform (L-SNPH) of 538 amino acids corresponded to previously described “neuronal” SNPH (27, 28), containing an extra 44-amino-acid NH2-proline–rich region and no MLS (Figure 1, A and B). Downstream of the first 44 residues, the two SNPH proteins were identical (Figure 1, A and B).
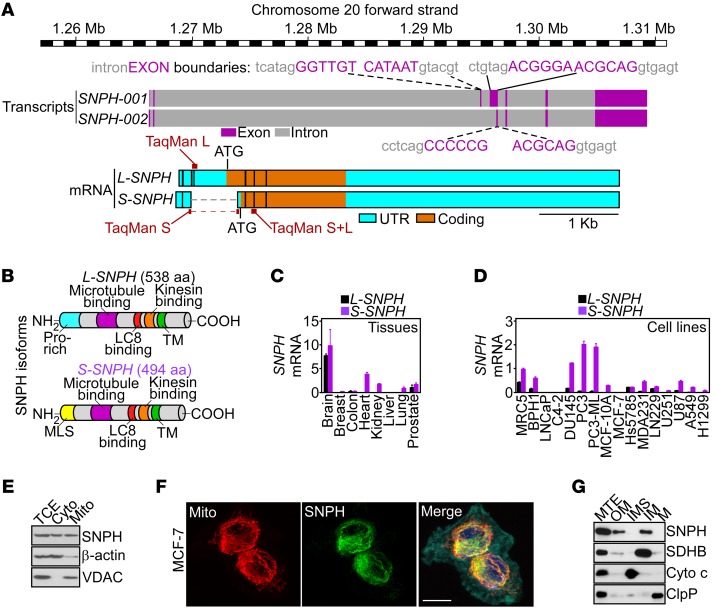
Figure 1. SNPH isoforms.
(A) Schematic diagram of the human SNPH locus (based on the Vertebrate Genome Annotation [Vega] repository; http://vega.archive.ensembl.org/index.html). The position and sequences of intron-exon boundaries, long (L) or short (S) SNPH transcripts, and TaqMan gene expression assays utilized for mRNA amplification of the two SNPH isoforms are indicated. (B) Schematic diagram of L-SNPH or S-SNPH protein isoforms. Pro, proline. (C and D) The indicated normal human tissues (C), normal diploid (MRC5) cells, or tumor cell types (D) were analyzed for L-SNPH or S-SNPH mRNA copy number, and normalized to 1,000 molecules of β-actin. Mean ± SEM (n = 3 per tissue or cell line examined). (E) PC3 cells were fractionated in cytosol (Cyto) or mitochondrial (Mito) extracts and analyzed by Western blotting. TCE, total cell extracts. (F) MCF-7 cells devoid of endogenous SNPH as in D were transfected with SNPH cDNA and analyzed by fluorescence microscopy. Merge image includes the F-actin channel (cyan). Scale bar: 5 μm. (G) PC3 cells were fractionated in sub-mitochondrial extracts containing outer membrane (OM), inter-membrane space (IMS), inner membrane (IM), or matrix (M) and analyzed by Western blotting. The expression of SDHB, cytochrome c (Cyto c), or ClpP was used as a markers for each fraction. MTE, unfractionated mitochondrial extracts.
We next used gene expression assays that individually detect L-SNPH or S-SNPH (Figure 1A) to map the distribution and absolute abundance of the two isoforms in human tissues and cell lines. L-SNPH was expressed in normal brain but mostly undetectable in all other tissues examined, including breast, colon, heart, kidney, liver, and lung, and present at a low level in the prostate (Figure 1C). Unexpectedly, S-SNPH was expressed at levels comparable to or higher than those of L-SNPH in the brain, and was present in other tissues, including heart, kidney, lung, and prostate (Figure 1C). S-SNPH was also the dominant isoform in normal and tumor cell lines, whereas L-SNPH was present at a low level or undetectable (Figure 1D). Similar results were obtained with analysis of public databases, with broad expression of SNPH in all human tumors examined, albeit at different levels (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI93172DS1), as well as human cell lines representative of disparate tumor types (Supplemental Figure 1B).
Consistent with a predicted MLS (Figure 1B), S-SNPH (hereafter referred to as SNPH) was detected by Western blotting in both cytosol and mitochondria of prostate adenocarcinoma PC3 cells (Figure 1E). Similarly, SNPH transfected in breast adenocarcinoma MCF-7 cells, which are devoid of endogenous SNPH (Figure 1D; see complete unedited blots in the supplemental material.), localized to mitochondria, by fluorescence microscopy (Figure 1F). In terms of submitochondrial distribution, endogenous SNPH localized to both the inner and outer mitochondrial membranes of tumor cells, whereas matrix and inter-membrane space were unreactive (Figure 1G).
Mitochondrial SNPH regulates bioenergetics.
To probe the function of SNPH in mitochondria, we next used multiple independent siRNA sequences that silence the expression of SNPH mRNA (Supplemental Figure 1C) and protein (Supplemental Figure 1D) in tumor and normal cell types. As an additional, alternative approach, we generated two independent clones of PC3 cells with stable knockdown of SNPH by short hairpin RNA (shRNA) (Supplemental Figure 1E). As cellular models for the targeting experiments, we focused on PC3 and glioblastoma LN229 cells, representative of neuroendocrine and CNS origin, respectively, with high endogenous SNPH expression (Figure 1D). SNPH siRNA silencing did not affect total mitochondrial mass, compared with control transfectants (Supplemental Figure 2A). Conversely, shRNA-mediated loss of SNPH decreased oxygen consumption rates (OCR), a marker of oxidative metabolism (Figure 2A), and reduced overall adenosine triphosphate (ATP) production in PC3 cells (Figure 2B). Glycolytic metabolism was also affected, with a modest, but significant, decrease in glucose consumption (Supplemental Figure 2B) and lactate production (Supplemental Figure 2C) after shRNA-SNPH knockdown. To validate these results, we next carried out reconstitution experiments in which SNPH-depleted PC3 cells were transduced with adenovirus expressing SNPH (Ad-SNPH) (Supplemental Figure 2, D and E). Under these conditions, reexpression of SNPH restored OCR (Figure 2C) and ATP production (Figure 2D), whereas Ad-LacZ (Supplemental Figure 2, D and E) had no effect (Figure 2, C and D).
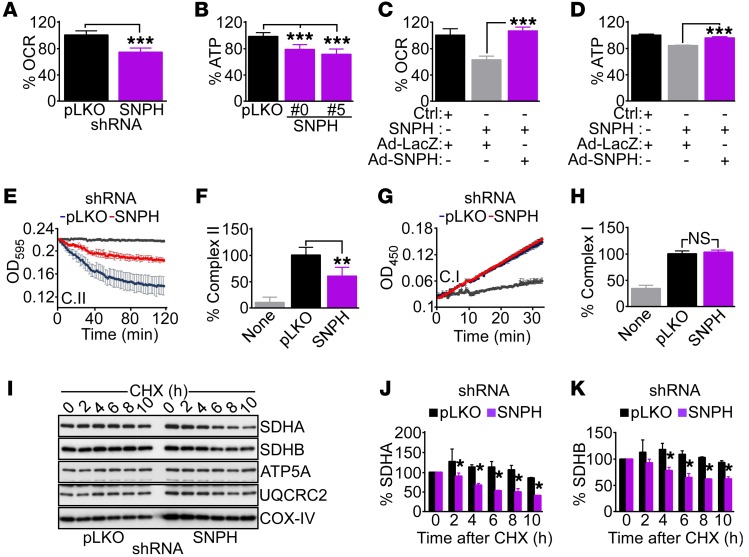
Figure 2. SNPH regulation of mitochondrial bioenergetics.
(A and B) PC3 cells stably transduced with control pLKO or shRNA-SNPH (clone 0 or 5) were analyzed for OCR (A) or ATP production (B). Data are expressed as mean ± SD of replicates of a representative experiment (n = 3). ***P < 0.0001, by 2-tailed Student’s t test (A). ***P < 0.001 by ANOVA and Bonferroni’s post-test (B). (C and D) PC3 cells transfected with control siRNA (Ctrl) or siRNA-SNPH were transduced with control adenovirus (Ad-LacZ) or SNPH-directed adenovirus (Ad-SNPH), and analyzed for OCR (C) or ATP production (D). Data are expressed as mean ± SD of replicates of a representative experiment (n = 3). ***P < 0.001 by ANOVA and Bonferroni’s post-test. (E and F) PC3 cells transduced with pLKO or shRNA-SNPH were analyzed for oxidative phosphorylation complex II (C.II) activity (E) and normalized to citrate synthase activity (F). Gray tracing, blank reaction. Data are expressed as mean ± SD of replicates of a representative experiment (n = 3). **P < 0.01 by ANOVA and Bonferroni’s post-test. (G and H) The experimental conditions were as in E and F, except that transduced PC3 cells were analyzed for oxidative phosphorylation complex I (C.I) activity (G) and normalized to citrate synthase activity (H). Gray tracing, blank reaction. Data are expressed as mean ± SD of replicates of a representative experiment (n = 3). NS, not significant (P > 0.05) by ANOVA and Bonferroni’s post-test. (I) PC3 cells transduced with pLKO or shRNA-SNPH were treated with CHX, and aliquots of cell extracts harvested at the indicated time intervals after release (h) were analyzed by Western blotting. (J and K) Protein bands from the experiment in I were quantified by densitometric scanning after CHX release. Changes in SDHA (J) or SDHB (K) protein bands in pLKO or shRNA-SNPH are shown. Data are expressed as mean ± SD (n = 4). The statistical analyses are as follows: SDHA (J), 2 hours, P = 0.05; 4 hours, P < 0.0001; 6 hours, P = 0.0001; 8 hours, P = 0.0001; 10 hours, P < 0.0001; SDHB (K), 2 hours, P = 0.14; 4 hours, P = 0.0007; 6 hours, P < 0.0001; 8 hours, P < 0.0001; 10 hours, P < 0.0001, by 2-tailed Student’s t test.
To further characterize the role of SNPH in mitochondrial metabolism, we next looked at the activity of individual oxidative phosphorylation complexes. Stable depletion of SNPH in PC3 cells reduced the activity of mitochondrial complex II (Figure 2, E and F), but not complex I (Figure 2, G and H), compared with control transfectants. This was associated with decreased expression of the complex II subunits succinate dehydrogenase A (SDHA) and B (SDHB) (Supplemental Figure 2F), whereas SDHC or complex III or complex V subunits were not affected (Supplemental Figure 2F and Supplemental Figure 1E). Consistent with these findings, shRNA-SNPH knockdown resulted in accelerated degradation of SDHA and SDHB (Figure 2I), and significantly shortened the half-life of both proteins, as quantified in cycloheximide (CHX) block experiments (Figure 2, J and K). Other subunits of oxidative phosphorylation complexes, including ATP5A (complex V) and UQCRC2 (complex III), were not affected (Figure 2I), and the half-life of COX-IV (complex IV) was unchanged with or without SNPH knockdown (Supplemental Figure 2G).
Mitochondrial SNPH regulation of oxidative stress.
Consistent with defective mitochondrial bioenergetics, shRNA-SNPH knockdown in PC3 cells resulted in increased production of total ROS (Figure 3A). Specifically, loss of SNPH was associated with heightened generation of mitochondria-derived superoxide, compared with control transfectants (Figure 3B). Known cellular markers of oxidative stress, including hyperoxidation of peroxiredoxin 3 (Prx3) (Supplemental Figure 3B), and increased NAD+/NADH ratio (Figure 3C) were also elevated in these settings. In rescue experiments, transfection of shRNA-SNPH–depleted cells with cDNA encoding the antioxidant SOD2 or Prx3 (Supplemental Figure 3B) reversed the increase in total cellular ROS (Figure 3A), as well as mitochondrial superoxide production (Supplemental Figure 3C).
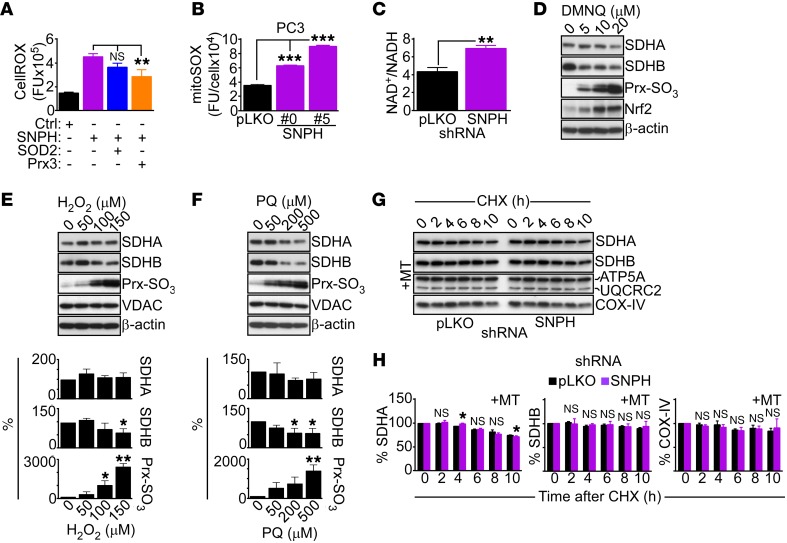
Figure 3. Effect of SNPH on mitochondrial oxidative stress.
(A) LN229 cells expressing control siRNA (Ctrl) or siRNA-SNPH were transfected with cDNA encoding antioxidant SOD2 or Prx3 and analyzed for total ROS production by fluorescence microscopy. Data are expressed as mean ± SD of single-cell determinations (Ctrl, n = 109; SNPH, n = 75; SOD2, n = 74; and Prx3, n = 55). **P < 0.01 by ANOVA and Bonferroni’s post-test. CellROX, ROS sensor. (B) PC3 cells transduced with pLKO or shRNA-SNPH (clones 0 and 5) were analyzed for mitochondrial superoxide production (mitoSOX) by fluorescence microscopy. Data are expressed as mean ± SD of single cell determinations (pLKO, n = 202; SNPH #0, n = 296; SNPH #5, n = 297). ***P < 0.001 by ANOVA and Bonferroni’s post-test. FU, fluorescence units. (C) PC3 shRNA-SNPH #0 cells were analyzed for NAD+/NADH ratio. Data are expressed as mean ± SD of replicates of a representative experiment (n = 3). **P = 0.001, by 2-tailed Student’s t test. (D–F) PC3 cells were treated with the indicated increasing concentrations of the oxidative stimuli DMNQ (D), H2O2 (E), and paraquat (PQ, F) and analyzed by Western blotting. Bottom panels (E and F): Densitometric quantification of SDHA, SDHB, or Prx-SO3 protein bands. Data are expressed as mean ± SD (n = 3). *P < 0.05; **P < 0.01; by ANOVA and Bonferroni’s post-test. (G) PC3 shRNA-SNPH #0 cells were treated with the mitochondrial superoxide scavenger MT and incubated with CHX, and cell extracts harvested at the indicated time intervals after release (h) were analyzed by Western blotting. (H) Densitometric quantification of SDHA, SDHB, or COX-IV proteins bands from the experiment in G. Data are expressed as mean ± SD (n = 4). The statistical analyses are as follows: SDHA, 2 hours, NS; 4 hours, P = 0.001; 6 hours, NS; 8 hours, NS; 10 hours, P = 0.033; SDHB, NS for all time points; COX-IV, NS for all time points by 2-tailed Student’s t test.
Based on these observations, we next asked whether oxidative damage affected the stability and/or function of mitochondrial oxidative phosphorylation complex subunits. Consistent with this possibility, treatment of PC3 cells with the oxidative stress stimulus 2,3-dimethoxy-1,4-naphthoquinone (DMNQ) resulted in a concentration-dependent decrease in SDHA and SDHB expression (Figure 3D and Supplemental Figure 3D). Consistent with oxidative damage, this was associated with Prx3 hyperoxidation and potent induction of the oxidative stress–regulated, cytoprotective transcription factor Nrf2 (30) in treated cells (Figure 3D and Supplemental Figure 3D). Similar results were obtained after exposure of PC3 cells to hydrogen peroxide (Figure 3E) or paraquat (Figure 3F), a redox cycler that stimulates superoxide production, resulting in concentration-dependent loss of SDHB and, to a lesser extent, SDHA expression (Figure 3, E and F). Conversely, selective scavenging of mitochondrial superoxide with MitoTempo (MT) inhibited the increase in mitochondrial ROS induced by SNPH knockdown (Supplemental Figure 3F), and restored SDHA and SDHB levels in these settings (Figure 3G). Accordingly, MT treatment normalized the half-life of both complex II subunits, SDHA and SDHB, in CHX block experiments (Figure 3, G and H), whereas the half-life of COX-IV was unchanged with or without MT (Figure 3H).
SNPH regulation of oxidative stress controls mitochondrial trafficking and tumor cell motility.
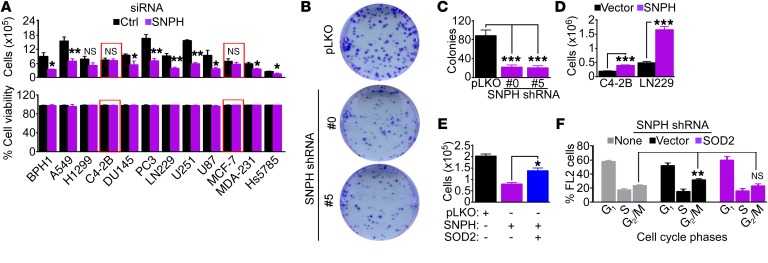
Next, we asked whether SNPH regulation of mitochondrial oxidative stress was important for tumor behavior. For these experiments, we used the model of LN229 glioblastoma cells, recently utilized to characterize the SNPH pathway in tumors (29), as well as prostate adenocarcinoma C4-2B cells, which have undetectable levels of endogenous SNPH (Figure 1D). Using these cellular models, forced expression of SNPH was sufficient to suppress mitochondrial trafficking to the cortical cytoskeleton (Figure 4A and Supplemental Figure 4A), which fuels membrane dynamics of chemotaxis and cell invasion (29).
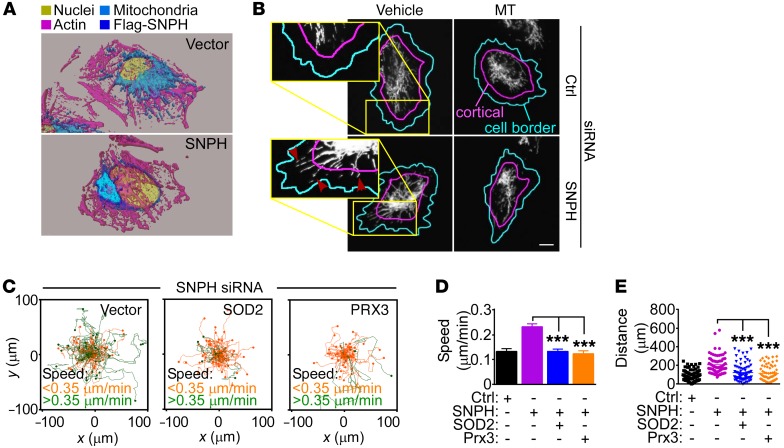
Figure 4. SNPH regulation of mitochondrial oxidative stress controls tumor cell motility.
(A) LN229 cells were transfected with vector or Flag-SNPH cDNA and analyzed for subcellular mitochondrial localization by confocal fluorescence microscopy. Three-dimensional isosurface renderings of representative cells are shown. (B) LN229 cells transfected with control siRNA or siRNA-SNPH were treated with vehicle or MT (200 μM) and analyzed by fluorescence microscopy. Masks are superimposed to the mitochondrial fluorescence channel (MTC02) to indicate the cell border (cyan lines, based on the actin channel) and the cortical region (area between the cyan and magenta lines). Arrowheads, cortical mitochondria. Scale bar: 10 μm. (C–E) LN229 cells transfected with siRNA-SNPH were transfected with vector, SOD2, or Prx3 cDNA and analyzed for cell motility in a 2D chemotaxis chamber (C), with quantification of speed of cell migration (D) and distance traveled by individual cells (E). Each tracing in C and symbol in E corresponds to an individual cell. Data are expressed as mean ± SEM (n = 91–112). ***P < 0.0001, by ANOVA and Bonferroni’s post-test. The cutoff velocities for each condition in 2D chemotaxis experiments (C) are indicated.
Mechanistically, treatment of SNPH-depleted LN229 cells with MT, alone or in combination with the ROS scavenger N-acetyl-cysteine (NAC) suppressed mitochondrial trafficking to the cortical cytoskeleton (Figure 4B and Supplemental Figure 4, B and C). In turn, MT treatment, with or without NAC, was sufficient to suppress the increased chemotaxis associated with SNPH depletion (Supplemental Figure 4C), reducing the speed of cell movements (Supplemental Figure 4D) and the distance traveled by individual cells (Supplemental Figure 4E). Similar results were obtained with an independent experimental approach, where transfection of SNPH-depleted LN229 cells with antioxidant SOD2 or Prx3 also reversed the increased chemotaxis induced by SNPH loss (Figure 4C), with comparable inhibition of the speed of cell migration (Figure 4D) and distance traveled per cell (Figure 4E) to the levels of control cultures.
Mitochondrial SNPH differentially regulates tumor cell proliferation.
In addition to increased cell motility (Figure 4C and Supplemental Figure 4D), SNPH depletion blocked tumor cell proliferation (Figure 5A, top) without loss of cell viability (Figure 5A, bottom). Importantly, tumor cells lacking endogenous SNPH, including C4-2B or breast adenocarcinoma MCF-7 cells (Supplemental Figure 5A), were not affected (red boxes in Figure 5A, top). Consistent with these data, loss of SNPH also suppressed colony formation in semisolid medium (Figure 5, B and C). In reciprocal experiments, forced expression of SNPH was sufficient to increase tumor cell proliferation, compared with control transfection (Figure 5D). The requirement(s) of SNPH regulation of tumor cell proliferation was next investigated. SNPH depletion did not appreciably affect BrdU incorporation (Supplemental Figure 5B), a measure of S-phase, but significantly increased the percentage of cells with G2/M DNA content (Supplemental Figure 5C), suggestive of mitotic arrest. Consistent with this possibility, SNPH depletion was associated with increased levels of mitotic cyclin B1, whereas the expression of cyclin A or cyclin D1 was unchanged (Supplemental Figure 5D). Mechanistically, transfection of SNPH-depleted cells with antioxidant SOD2 cDNA restored tumor cell proliferation (Figure 5E) and reversed the cell cycle arrest at G2/M (Figure 5F), compared with control transfection.
Figure 5. Mitochondrial SNPH supports tumor cell proliferation.
(A) The indicated tumor cells transfected with control siRNA (Ctrl) or siRNA-SNPH were analyzed by direct cell counting (top) or cell viability by trypan blue exclusion (bottom) after 72 hours. The same number of cells were seeded at time 0. Data are expressed as mean ± SEM (n = 4). Red boxes indicate two cell types (C4-2B and MCF-7) with low to undetectable levels of endogenous SNPH. The statistical analyses per each cell type are as follows: BPH1, P = 0.03; A549, P = 0.004; H1299, NS; C4-2B, NS; DU145, P = 0.03; PC3, P = 0.002; LN229, P = 0.002; U251, P < 0.0001; U87, P = 0.03; MCF-7, NS; MDA-231, P = 0.01; Hs578T, P = 0.02 by 2-tailed Student’s t test. (B and C) PC3 cells transduced with pLKO or shRNA-SNPH (clones 0 and 5) were analyzed in a colony formation assay, and crystal violet–stained colonies (B) were counted after 10 days (C). Data are expressed as mean ± SEM (n = 3). ***P < 0.001 by ANOVA and Bonferroni’s post-test. (D) The indicated tumor cell types were transfected with vector or SNPH cDNA and analyzed by direct cell counting after 72 hours. Data are expressed as mean ± SD (n = 3). ***P < 0.0001, by 2-tailed Student’s t test. (E) PC3 cells transduced with pLKO or shRNA-SNPH were transfected with SOD2 cDNA and analyzed by direct cell counting. Data are expressed as mean ± SD (n = 3). *P < 0.01, by ANOVA and Bonferroni’s post test. (F) PC3 cells transduced with shRNA-SNPH were reconstituted with vector or SOD2 cDNA and analyzed by propidium iodide staining and flow cytometry. The cellular fractions in the indicated cell cycle phases are indicated. Data are expressed as mean ± SD (n = 3). **P = 0.01 by 2-tailed Student’s t test.
Mitochondrial SNPH modulates cell proliferation–cell motility decisions in cancer.
Next, we asked whether the mitochondrial localization of this SNPH isoform was required for tumor functions. Reconstitution of shRNA-SNPH–silenced PC3 cells with full-length (FL) SNPH (Supplemental Figure 6A) reversed the bioenergetics defects in oxygen consumption (Figure 6A) and ATP production (Figure 6B) associated with SNPH depletion. Conversely, reconstitution of these cells with a SNPH mutant lacking the MLS (Δ-MLS) was ineffective (Figure 6, A and B, and Supplemental Figure 6A). In these studies, FL SNPH and Δ-MLS SNPH were expressed at comparable levels in reconstituted PC3 cells (Supplemental Figure 6A). Next, we examined the requirement of mitochondrial localization in SNPH regulation of mitochondrial movement and cell motility-proliferation. Consistent with the data above, forced expression of FL SNPH in C4-2B cells (Supplemental Figure 6B) inhibited mitochondrial trafficking to the cortical cytoskeleton (Figure 6C) and promoted increased cell proliferation (Figure 6D), compared with control transfection. Conversely, expression of Δ-MLS SNPH in C4-2B cells (Supplemental Figure 6B) was considerably less effective, partially reducing mitochondrial trafficking (Figure 6C) and modestly increasing cell proliferation (Figure 6D). Similar results were obtained in LN229 cells, where FL SNPH inhibited mitochondrial trafficking (Supplemental Figure 6C) and promoted cell proliferation (Supplemental Figure 6D), whereas Δ-MLS SNPH had a limited effect (Supplemental Figure 6, C and D). Finally, reconstitution of shRNA-SNPH–depleted PC3 cells with FL SNPH abolished the increase in tumor cell invasion induced by SNPH loss, whereas Δ-MLS SNPH had no effect (Figure 6E and Supplemental Figure 6E).
Figure 6. Mitochondrial SNPH regulation of metastasis.
(A and B) siRNA-SNPH PC3 cells were reconstituted with vector (pCMV), FL SNPH, or an SNPH mutant deleted in the MLS (Δ-MLS), and analyzed for OCR (A) or ATP production (B). Data are mean ± SD (n = 3). **P < 0.01; ***P < 0.001 by ANOVA and Bonferroni’s post-test. (C and D) C4-2B cells were transfected with pCMV6, FL SNPH, or Δ-MLS SNPH and analyzed for mitochondrial trafficking to the cortical cytoskeleton (C) or direct cell counting (D). For C, data are mean ± SEM (vector, n = 89; FL SNPH, n = 96; Δ-MLS SNPH, n = 87). **P < 0.01; ***P < 0.001 by ANOVA and Bonferroni’s post-test. For D, data are mean ± SD (n = 6). (E) shRNA-SNPH PC3 cells were reconstituted with pCMV6, FL SNPH, or Δ-MLS SNPH and analyzed for Matrigel invasion. Data are mean ± SEM (n = 3). ***P < 0.001, by ANOVA and Bonferroni’s post-test. (F) Yumm1.7 cells expressing mCherry and stably transfected with pCMV6, FL SNPH, or Δ-MLS SNPH were injected subcutaneously into syngeneic C57BL/6 mice, and mCherry-positive cells (insets) disseminated to the lungs were detected by immunocytochemistry. Scale bar: 50 μm. (G) Quantification of mCherry-positive Yumm1.7 cells transfected as in F in lungs of reconstituted animals. Each symbol corresponds to the mean number of disseminated cells per lung of an individual animal. DTC, disseminated tumor cells. *P < 0.05; ***P < 0.001, by ANOVA and Bonferroni’s post-test.
Based on these observations, we next asked whether the localization of SNPH to mitochondria was required to modulate tumor cell dissemination in vivo. For these experiments, we utilized the mCherry-labeled Yumm1.7 cell line, derived from a genetically engineered mouse model of invasive melanoma carrying the genotype BrafV600E; Cdkn2a–/–;Pten–/– (31) and utilized in recent studies (32). For these experiments, Yumm1.7 cells expressing negligible levels of endogenous SNPH (Supplemental Figure 6F) were stably transfected with vector or SNPH variants and first analyzed for cell motility/proliferation responses in culture. Transfection of FL SNPH suppressed Yumm1.7 cell invasion across Matrigel, compared with vector control transfection (Supplemental Figure 6G) and in agreement with the data above. In contrast, expression of Δ-MLS SNPH had no effect in Yumm1.7 cells (Supplemental Figure 6G). In these experiments, expression of FL SNPH or Δ-MLS SNPH did not significantly modulate proliferation of Yumm1.7 cells (Supplemental Figure 6H). When reconstituted subcutaneously in syngeneic C57BL/6 mice, Yumm1.7 cells expressing vector disseminated to the lung, as quantified by immunocytochemistry of mCherry reactivity (Figure 6, F and G), and in agreement with recent observations (32). Under these conditions, expression of FL SNPH abolished metastatic seeding of Yumm1.7 cells to the lungs, whereas transfection of Δ-MLS SNPH was associated with increased tumor cell dissemination in vivo (Figure 6, F and G).
Reciprocal regulation of tumor growth and metastasis by SNPH.
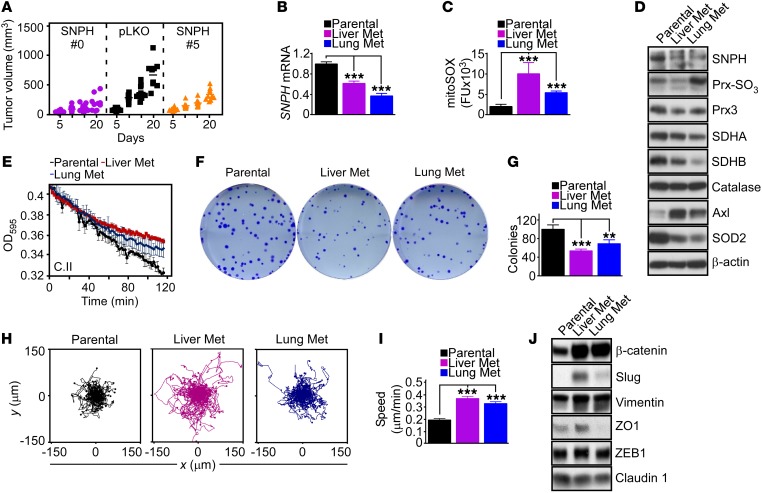
We next carried out a reciprocal experiment in vivo and analyzed the xenograft growth of PC3 cells stably transduced with two independent shRNA-SNPHs in immunocompromised mice. In these experiments, stable depletion of SNPH suppressed the growth of superficial PC3 xenograft tumors, compared with pLKO transfection (Figure 7A). To further investigate the role of this pathway in metastasis, we isolated pLKO-transfected cells that had spontaneously metastasized to the lung or liver of engrafted animals. These lung or liver metastatic cell lines showed constitutive downregulation of SNPH (Figure 7B) and markers of oxidative stress, including greater mitochondrial superoxide production (Figure 7C) and Prx hyperoxidation and reduced SOD2 levels (Figure 7D). In line with these findings, analysis of public databases revealed that metastatic breast cancer (Supplemental Figure 7A) or metastatic prostate cancer (Supplemental Figure 7B) had reduced levels of SNPH compared with the corresponding primary tumor. Phenocopying the effect of SNPH silencing, loss of endogenous SNPH in metastatic tumor cells was associated with decreased expression of the complex II subunits SDHA and SDHB (Figure 7D and Supplemental Figure 7C), diminished complex II activity (Figure 7E and Supplemental Figure 7D), and lower OCR (Supplemental Figure 7E), compared with parental, pLKO transfection.
Figure 7. SNPH regulation of cell proliferation-motility in vivo.
(A) PC3 cells transduced with pLKO or shRNA-SNPH (clones 0 and 5) were injected subcutaneously in immunocompromised mice (5 mice per group; 2 tumors/mouse), and tumor volume was quantified with a caliper at the indicated time intervals. Each symbol corresponds to an individual tumor. On day 20, pLKO vs. SNPH #0, P < 0.001; pLKO vs. SNPH #5, P < 0.05, by ANOVA and Bonferroni’s post-test. (B) pLKO-transduced PC3 cells or pLKO-transduced PC3 cells isolated from a liver or lung metastatic site (Met) from the experiment in A were analyzed for SNPH mRNA levels by qPCR. Data are expressed as mean ± SD (n = 3). ***P < 0.001 by ANOVA and Bonferroni’s post-test. (C and D) The metastatic cell lines in B were analyzed for mitochondrial superoxide production by mitoSOX reactivity and fluorescence microscopy (C) or Western blotting (D). Data in C are expressed as mean ± SEM of single-cell determinations (n = 76–200). ***P < 0.001 by ANOVA and Bonferroni’s post-test. (E) The indicated metastatic cell lines were analyzed for mitochondrial oxidative phosphorylation complex II (C.II) activity at the indicated time intervals. (F and G) The indicated metastatic cell lines were analyzed in a colony formation assay (F), and crystal violet–stained colonies were quantified after 10 days (G). Data are expressed as mean ± SD (n = 3). **P < 0.01; ***P < 0.001 by ANOVA and Bonferroni’s post-test. (H and I) The indicated metastatic cell lines were analyzed for cell motility in a 2D chemotaxis chamber (H) with quantification of speed of cell migration (I). Data are expressed as mean ± SEM (n = 104–106). ***P < 0.0001, by ANOVA and Bonferroni’s post-test. (J) The indicated metastatic cell lines were analyzed by Western blotting.
When analyzed for changes in the cell proliferation-motility balance, lung or liver metastatic cells showed reduced colony formation (Figure 7, F and G) but increased 2D chemotaxis (Figure 7H), characterized by greater speed of cell migration (Figure 7I) and longer distance traveled by individual cells (Supplemental Figure 7F). Finally, these metastatic cells expressed markers of epithelial-mesenchymal transition (EMT), including β-catenin, Slug, and vimentin (Figure 7J), whereas ZO1 modulation was observed in lung but not liver metastatic cells, and claudin 1 expression was unchanged (Figure 7J).
Stress-regulated exploitation of the SNPH pathway in cancer.
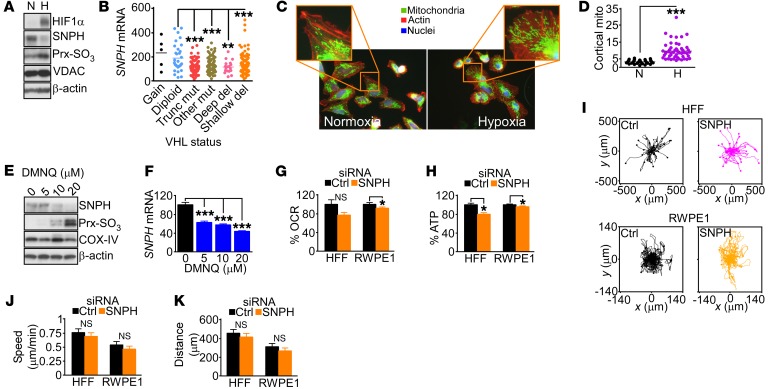
Next, we asked whether stress conditions of the tumor microenvironment affected SNPH levels in tumors. Exposure of PC3 cells to hypoxia (1% O2 for 24 hours) acutely downregulated SNPH levels (Figure 8A) as early as 3 hours after treatment (Supplemental Figure 8A), and was associated with Prx hyperoxidation and HIF1α stabilization (Figure 8A). Similar findings were observed in cases of clear cell renal cell carcinoma in vivo, where HIF1α stabilization due to mutations or deletions in its negative regulator von Hippel–Lindau (VHL) (33) (Figure 8B) or decreased VHL copy number (Supplemental Figure 8B) correlated with reduced levels of SNPH. Functionally, loss of SNPH in hypoxic cells was associated with increased mitochondrial trafficking to the cortical cytoskeleton (Figure 8, C and D) and greater tumor cell motility, as determined in a wound closure assay (Supplemental Figure 8, C and D). Oxidative stress generated by DMNQ treatment also lowered SNPH protein (Figure 8E) and mRNA (Figure 8F) expression in PC3 cells.
Figure 8. Hypoxic and oxidative stress regulation of SNPH.
(A) PC3 cells were exposed to normoxia (N) or hypoxia (H; 1% O2 for 24 hours) and analyzed by Western blotting. (B) Cases of clear cell renal cell carcinoma in the TCGA database were stratified for SNPH mRNA expression and VHL mutational status. Mut, mutations; Trunc, truncated; del, deletions. Each symbol corresponds to an individual tumor. **P < 0.01; ***P < 0.001 by ANOVA and Bonferroni’s post-test. (C and D) LN229 cells exposed to normoxia or hypoxia as in A were analyzed for mitochondrial trafficking to the cortical cytoskeleton by fluorescence microscopy (C), and cortical mitochondria (mito) were quantified (D). Each symbol corresponds to an individual cell. ***P < 0.0001, by 2-tailed Student’s t test. The representative images displayed in C are brightness- and contrast-enhanced to highlight cortical mitochondria. Quantification was done in unsaturated images. (E and F) PC3 cells were treated with the indicated concentrations of DMNQ and analyzed by Western blotting (E) or qPCR amplification of SNPH mRNA (F). Data are expressed as mean ± SD (n = 3). ***P < 0.001, by ANOVA and Bonferroni’s post-test. (G and H) Normal diploid HFFs or normal human prostate epithelial cells (RWPE1) were transfected with control siRNA or siRNA-SNPH and analyzed for OCR (G) or ATP production (H). Data are mean ± SD (HFFs, n = 3; RWPE1, n = 12). *P = 0.001 to P < 0.0001; NS, not significant, by 2-tailed Student’s t test. (I) HFFs (top) or RWPE1 cells (bottom) transfected as in G were analyzed for cell motility in a 2D chemotaxis chamber. Each trace corresponds to the movements of an individual cell. (J and K) HFFs or RWPE1 cells transfected as in G were analyzed by 2D chemotaxis, and speed of cell migration (J) and distance traveled by individual cells (G) were quantified. Data are expressed as mean ± SD (HFFs, n = 35–42; RWPE1 cells, n = 50–52). NS, 2-tailed Student’s t test.
Finally, we asked whether the SNPH pathway was selectively exploited in cancer, compared with normal cells. Silencing of SNPH in primary human HFFs or normal prostate epithelial RWPE1 cells had marginal effects on cellular bioenergetics, with a modest decrease in OCR (Figure 8G) and ATP production (Figure 8H). Similarly, SNPH silencing in HFF cultures did not generate oxidative stress, with no Prx3 hyperoxidation observed in these settings (Supplemental Figure 8E). Finally, depletion of SNPH in either HFFs or RWPE1 cells did not affect 2D chemotaxis (Figure 8I), with no changes observed in the speed of cell migration (Figure 8J) or distance traveled per cell (Figure 8K), compared with control transfectants.
Discussion
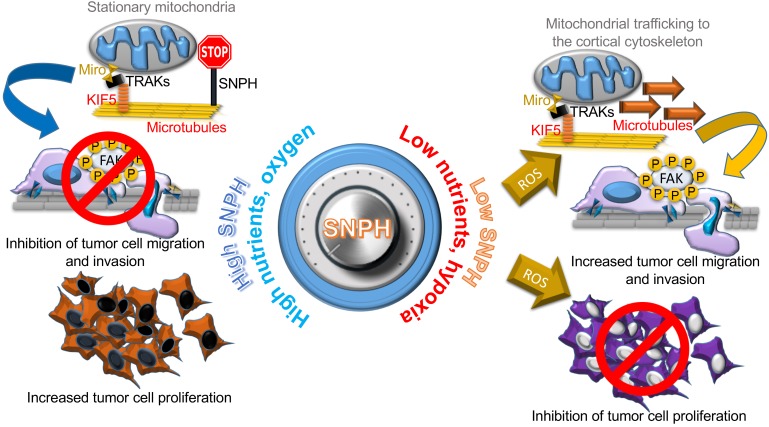
In this study, we have identified an alternatively spliced isoform of SNPH (27, 28) that localizes to mitochondria and functions as a stress-regulated switch for proliferation-motility decisions in cancer. High levels of SNPH maintain efficient, “non-leaky” mitochondrial bioenergetics, which sustains tumor cell proliferation while suppressing cell motility (Figure 9). Reciprocally, downregulation of SNPH due to oxidative or hypoxic stress impairs mitochondrial metabolism, shuts off cell proliferation and stimulates greater tumor cell motility and invasion (Figure 9). A critical signaling regulator of this response was mitochondria-produced superoxide, which promoted oxidative degradation of the complex II subunits SDHA and SDHB, cell cycle arrest at G2/M, and heightened mitochondrial trafficking to the cortical cytoskeleton to fuel membrane dynamics of cell motility (Figure 9). Altogether, this pathway functioned as a major switch to regulate the metastatic propensity in vivo, as depletion of SNPH or expression of a SNPH mutant lacking the MLS dramatically increased metastatic dissemination in syngeneic as well as xenograft tumor models in vivo. Accordingly, tumor cell lines established from lung or liver metastatic sites in vivo exhibited lower levels of SNPH compared with their primary sites, higher oxidative stress, reduced cell proliferation, and heightened cell motility.
Figure 9. A mitochondrial SNPH “rheostat” for cell proliferation-motility decisions in cancer.
An adequate supply of oxygen and nutrients in the tumor microenvironment maintains high levels of SNPH in mitochondria for efficient, “non-leaky” oxidative bioenergetics and low ROS generation. These conditions support continued tumor cell proliferation while halting mitochondrial trafficking to the cortical cytoskeleton as a “regional” energy source to fuel membrane dynamics of cell motility and invasion. Conversely, the emergence of an oxidative and hypoxic microenvironment, typical of advanced tumors, acutely lowers SNPH levels, compromising oxidative phosphorylation complex II integrity and mitochondrial bioenergetics. The resulting increase in ROS inhibits tumor cell proliferation, while promoting increased mitochondrial trafficking to the cortical cytoskeleton and focal adhesion kinase–dependent (FAK-dependent) tumor cell migration and invasion. KIF5, kinesin family member 5; TRAK, trafficking kinesin-binding protein 1.
Previously considered “neuron-specific” (27), SNPH has been known as a negative regulator of mitochondrial trafficking, tethering mitochondria to cytoskeletal microtubules and halting their movements at axonal regions with high energy demands (28). Unexpectedly, a similar pathway was recently uncovered in cancer (29), where SNPH prevented the repositioning of mitochondria to the cortical cytoskeleton, a process that fuels chemotaxis and cell invasion (25, 26), thus suppressing tumor cell movements (29). Here, we uncovered a mechanistic underpinning of the SNPH pathway in cancer, centered on the expression of a previously unrecognized, alternatively spliced SNPH isoform, characterized by a unique mitochondrial localization. At variance with a canonical, long SNPH isoform, which corresponds to previously described “neuronal” SNPH (27), the new, short SNPH isoform was broadly expressed, albeit at wide-ranging levels, in both neuronal and non-neuronal tissues, including primary human tissues and tumor cell lines. A detailed molecular understanding of how the SNPH gene is differentially processed, including the potential generation of additional isoform(s) from this locus, remains to be elucidated. However, it is intriguing that analysis of public databases points to a potential role of stress-regulated transcriptional mechanisms in SNPH gene expression, in keeping with the stress-regulated modulation of SNPH levels observed here in tumor cells.
Consistent with its predicted mitochondrial localization, the new, short SNPH isoform accumulated in mitochondria of tumor cells, sorting to both the inner and outer organelle membranes. It seems plausible that the putative transmembrane (TM) domain embedded in the COOH-terminus of SNPH (27) provides membrane insertion at both mitochondrial locations, and, consistent with this possibility, deletion of the TM domain disrupted mitochondrial tethering to cytoskeletal microtubules (27) and impaired mitochondrial support of tumor cell proliferation (our unpublished observations). Together, these data suggest the possibility of a differential spatial organization for the diverse SNPH functions in mitochondria, where outer membrane–associated SNPH provides organelle attachment to the cytoskeleton (27, 29), whereas inner membrane SNPH sustains efficient, non-leaky oxidative metabolism. Mechanistic data presented reinforce the absolute requirement of mitochondrial localization for SNPH function in tumors, as an SNPH Δ-MLS mutant lacked the ability to affect oxidative bioenergetics, mitochondrial trafficking to the cortical cytoskeleton, and cell proliferation.
Consistent with this view, mitochondrial SNPH emerged here as a regulator of organelle bioenergetics, preserving the stability and function of the complex II subunits SDHA and SDHB against oxidative damage. Although the structural arrangement of complex II is known (34), how subunit integrity is preserved, especially during oxidative (35) or proteotoxic (36) stress, has not been clearly delineated. We know that pharmacologic or genetic targeting of complex II causes electron leakage (37) and increased superoxide production (38). Here, we identified mitochondrial superoxide as a potent stimulus for SDHA and SDHB degradation, potentially via protein oxidation, further impairing energy production under stress. How mitochondrial SNPH prevents superoxide-induced SDHA or SDHB oxidation and degradation remains to be elucidated. One possibility is that SNPH operates in concert with other regulators of complex II (39), including survivin (26), to preserve subunit assembly and optimal bioenergetics, especially under stress conditions. In fact, it is intriguing that compared with other mitochondrial respiration complexes, complex II appears especially vulnerable to stress stimuli, including defective mitochondrial protein folding (40–42) and oxidative damage (the present study).
Although it has long been known that tumors produce more mitochondrial superoxide than normal tissues, a role of this pathway in disease progression is controversial, and likely context dependent (43). Low levels of ROS may contribute to tumorigenesis via increased DNA damage and activation of oncogene signaling (44), whereas higher generation of ROS may trigger cytotoxicity and suppress metastasis (45). Here, the heightened production of mitochondrial superoxide in response to SNPH depletion triggered a dual phenotype of inhibition of tumor cell proliferation via G2/M arrest and stimulation of cell motility by increasing mitochondrial trafficking to the cortical cytoskeleton (25). This is consistent with a long-held tenet that ROS mediate cell motility (46), contributing to the acquisition of an EMT phenotype (47) and metastatic competence (21). Conversely, a role for ROS signaling in subcellular mitochondrial trafficking and a pathway of “regional” bioenergetics of cell motility (25) have not been previously described.
In terms of disease relevance, we have shown here that expression of mitochondria-localized SNPH is sufficient to suppress tumor cell dissemination from xenograft tumors as well as in syngeneic models of early metastatic seeding in immunocompetent animals, defining this pathway as a conceptually novel metastasis suppressor. Conversely, we found that SNPH becomes transcriptionally downregulated in cells that have acquired the ability to metastasize in vivo, correlating with a cellular phenotype of heightened oxidative stress, reduced cell proliferation, and increased cell motility. The acquisition of metastatic competence reflects a complex and multifaceted transcriptional and posttranscriptional program (48), including modulation of intrinsic antioxidant mechanisms (45). How SNPH participates in this process remains to be fully elucidated, but data collected from analysis of public databases and primary patient cohorts revealed that downregulation or loss of SNPH is a common feature of tumor progression, correlating with metastatic disease and worse outcome (29). Our data indicating that stress conditions of the tumor microenvironment, such as hypoxia and oxidative damage (1), acutely lower SNPH levels may explain the silencing of this pathway in advanced disease, coinciding with the emergence of a restrictive and unfavorable microenvironment, typically depleted of oxygen and nutrient (1).
Based on these findings, we propose a model in which SNPH functions as a stress-regulated mitochondrial “rheostat” for allocation of often limited resources of the microenvironment in proliferation-motility decisions (9), or phenotype-switching (10). According to this model, downregulation of SNPH due to hypoxic or oxidative stress (1) turns the rheostat toward greater cell motility at the expense of cell proliferation (Figure 9). Fueled by increased mitochondrial accumulation at the cortical cytoskeleton (25), the resulting heightened cell motility may provide an “escape” mechanism for tumor cells to evade an unfavorable ecosystem and colonize alternative, distant tissue sites (49). Conversely, sustained levels of oxygen and nutrients maintain high SNPH expression in mitochondria. This turns the rheostat toward continued tumor cell proliferation enabled by low ROS production and efficient oxidative metabolism, while restricting mitochondrial trafficking and cell movements (Figure 9).
Although metastatic disease is the primary cause of death for cancer patients, there is a paucity of therapeutic targets to interfere with the process(es) of tumor cell dissemination to distant organs (50). In line with the renewed emphasis of mitochondria in metastasis (19, 21, 29), molecules in the SNPH pathway may provide fresh therapeutic opportunities to target adaptive mechanisms of cell proliferation-motility in progressive disease. As this pathway is selectively exploited in cancer, as opposed to normal tissues, modulators of mitochondrial trafficking may constitute a favorable strategy and be uniquely suited to disrupt the metabolic requirements of metastatic cells (29).
Methods
Antibodies and reagents.
A custom rabbit polyclonal antibody against human SNPH (aa 207–221) was produced by NEO Group Inc. and purified by antigen affinity. The anti-SNPH custom antibody was validated in knockdown cells (Supplemental Figure 1, D and E) and in cells overexpressing SNPH cDNA (Supplemental Figure 2D) at a 1:1,000 dilution. Antibodies to Prx3 (Santa Cruz Biotechnology Inc., clone 4G10, catalog sc-59663, diluted 1:3,000), 2-Cys Prx-SO3 (Thermo Fisher Scientific Inc., clone 10A1, catalog LF-MA0088, diluted 1:1,000), FLAG (Sigma-Aldrich, clone M-2, catalog F1804, diluted 1:5,000), SDHA (Abcam, clone 2E3GC12FB2AE2, catalog 14715, diluted 1:5,000), SDHB (Abcam, clone 21A11AE7, catalog 14714, diluted 1:2,000), oxidative phosphorylation antibody cocktail (MitoSciences, Abcam catalog s110411, diluted 1:2,000), citrate synthase (CS, clone D7V8B, catalog 14309), SOD2 (clone D3X8F, catalog 13141), catalase (clone D4P7B, catalog 12980), Axl (clone C89E7, catalog 8661), COX-IV (clone 4D11-B3-E8, catalog 11967), Nrf2 (clone D1Z9C, catalog 12721), caveolin 1 (clone D46G3, catalog 3267), VDAC (catalog 4866), HIF1α (catalog 3716), β-catenin (clone D10A8, catalog 8480), Slug (clone C19G7, catalog 9585), ZO1 (clone D7D12, catalog 8193), claudin 1 (clone D5H1D, catalog 13255), ZEB1 (clone D80D3, catalog 3396), vimentin (clone D21H3, catalog 5741) (Cell Signaling Technology, all diluted 1:1,000), and β-actin (Sigma-Aldrich, clone AC-15, cat#A5441, diluted 1:100,000) were used for Western blotting. Antibodies to paxillin (Upstate Biotechnologies, clone 5H11, Millipore catalog 05-417), β-tubulin (Sigma-Aldrich, clone AA2, catalog T8328), Tom20 (Santa Cruz Biotechnology Inc., clone FL-145, catalog sc11415), and MTC02 (Abcam, clone MTC02, catalog 79479) were used for immunofluorescence. DMNQ, FCCP, CHX, MT (>98% TLC), and N-acetyl-cysteine (NAC, >99% HPLC) were from Sigma-Aldrich. MitoTracker Green, phalloidin Alexa Fluor 488, CellLight Mito-RFP BacMam 2.0, and secondary antibodies for immunofluorescence were from Molecular Probes.
Cell culture.
Human glioblastoma (LN229, U251, and U87), prostate adenocarcinoma (LNCaP, C4-2B, DU145, and PC3), breast epithelial (MCF10A), breast adenocarcinoma (MCF7, Hs578T, and MDA-MB-231), lung adenocarcinoma (A549 and H1299), human diploid fibroblasts (MRC5), prostate epithelial cells (RWPE-1), and mouse fibroblasts (NIH 3T3) were obtained from ATCC, and maintained in culture according to the supplier’s specifications. Benign prostate hyperplasia (BPH1) cells were a gift from Simon Hayward (Vanderbilt University, Nashville, Tennessee, USA). A highly metastatic clone of PC3 cells colonizing the bone (PC3-ML) was a gift from Alessandro Fatatis (Drexel University College of Medicine, Philadelphia, Pennsylvania, USA). Primary HFFs were a gift from Meenhard Herlyn (Wistar Institute). Yumm1.7 cells were a gift from Marcus Bosenberg (Yale University, New Haven, Connecticut). Hypoxic treatment was carried out using an enclosed chamber (BioSpherix) flushed with a nitrogen and CO2 gas. The O2 and CO2 concentrations in the chamber were maintained at 1% and 5%, respectively, using an oxygen and carbon dioxide controller (BioSpherix). These conditions were maintained constant throughout the course of the experiments.
Plasmids, mutagenesis, and transfections.
TrueORF pCMV6-Entry-myc-Flag plasmids encoding the short isoform of SNPH (NM_014723, catalog RC207749), SOD2 (catalog RC202330), and Prx3 (catalog RC205080) were from Origene. The human ORF for the long isoform of SNPH from the Kazusa DNA Research Institute was obtained as HaloTag(R) in pFN21A (NM_001318234.1, Promega). Short SNPH mutants lacking the microtubule-binding domain (Δ-MTB, Δ86–159 aa), TM domain (Δ-TM, Δ425–444 aa), or MLS (Δ-MLS, Δ1–20 aa) were generated using a Stratagene QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) and confirmed by DNA sequencing. Cells were transfected with 2 μg pcDNA plus 4 μl X-treme gene HP (Roche) for 24 hours in complete medium, washed, and subjected to the indicated treatments.
Adenoviral vectors expressing SNPH were produced using Gateway technology (Thermo Fisher Scientific). Briefly, SNPH or LacZ cDNA was inserted into pDONR221 vector and recombined into the adenovirus expression vector pAd/CMV/V5-DEST. The plasmids were digested with PacI restriction enzyme and transfected in 293A cells for production of adenoviruses. The cells containing adenoviruses were collected at 7 days after transfection, according to the manufacturer’s instructions.
Gene silencing.
Gene knockdown experiments by siRNA were carried out as described previously (20). The following sequences were used: control, ON-TARGETplus Non-targeting siRNA pool (Dharmacon, D-001810), or human SNPH siRNA (Dharmacon, L-020417, or Santa Cruz Biotechnology Inc., sc-41369). Tumor cells were transfected with the individual siRNA pools at 30 nM in Lipofectamine RNAiMAX (Invitrogen) at a 1:1 ratio (vol siRNA 20 μM/vol Lipofectamine RNAiMAX). After 48 hours, the various transfected cells were validated for target protein knockdown by Western blotting and processed for subsequent experiments. Alternatively, two independent shRNA sequences were used for targeting the 3′ UTR of human SNPH: TRCN 0000147900 and TRCN 0000128545 (Wistar Molecular Screening Shared Resource). An empty pLKO-based lentivirus was used as control. PC3 cells stably expressing shRNA targeting SNPH were generated by infection with lentiviral particles, followed by a 2-week selection in the presence of puromycin at 2 μg/ml.
Protein analysis.
For Western blotting, protein lysates were prepared in Triton X-100 lysis buffer (20 mM Tris HCl, pH 7.5, 137 mM NaCl, 1% Triton X-100, 10% glycerol) containing EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich) and Phosphatase Inhibitor Cocktail PhosSTOP (Roche), sonicated, and precleared by centrifugation at 14,000 g for 10 minutes at 4°C. Equal amounts of protein lysates were separated by SDS gel electrophoresis, transferred to PVDF membranes, blocked in 5% low-fat milk diluted in TBST buffer (20 mM Tris HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20), and further incubated with primary antibodies of various specificities diluted 1:1,000 in 5% BSA/TBST for 18 hours at 4°C. After washing in TBST, membranes were incubated with HRP-conjugated secondary antibodies (1:1,000–1:5,000 dilution in 5% BSA/TBST) for 1 hour at 22°C and washed with TBST, and protein bands were visualized by enhanced chemiluminescence. For analysis of protein stability and determination of protein half-life, PC3 cells were incubated in the presence of 100 μg/ml of the protein synthesis inhibitor CHX, with or without the mitochondrial superoxide scavenger MT, and released in complete medium, and aliquots of cell lysates collected at increasing time intervals after release (2–10 hours) were analyzed by Western blotting.
Mitochondrial isolation.
Mitochondrial fractions were prepared from PC3 cells using a mitochondrial isolation kit for cultured cells (Fisher Scientific). Briefly, PC3 cells were homogenized by 70 strokes using a Dounce grinder in isolation buffer A plus protease inhibitor cocktail. Cell extracts were collected into equal volumes of isolation buffer C with buffer A. Cell debris and nuclei were removed by centrifugation at 700 g for 10 minutes, and mitochondrial fractions were collected by centrifugation at 3,000 g for 25 minutes. For submitochondrial fractionation experiments, initial mitochondrial extracts were further centrifuged at 12,000 g for 10 minutes in an equal volume of isolation buffer C. The resulting samples were further processed by sequential centrifugation in fractions containing outer membrane (OM), inner membrane (IM), inter-membrane space (IMS), and matrix, as described previously (42).
mRNA quantification.
Absolute mRNA levels for human SNPH were determined by qPCR. Briefly, RNA was extracted with a PureLink RNA Mini Kit (Life Technologies) following the in-column DNA digestion protocol. For mouse tissues, RNA was extracted from 25 mg of tissue by homogeneization in TRI Reagent, followed by purification with a Direct-zol RNA Mini Kit (Zymo Research). RNA from 8 normal human tissues was obtained from BioChain and digested with RNAse-free DNAse I (Thermo Scientific). Five micrograms of RNA was reversed transcribed using a combination of oligo(dT) and an SNPH-specific reverse primer (RNAse-free HPLC purified, CTGGCGGTCACCACAGAC) for 1 hour at 53°C using the ThermoScript RT-PCR system (Life Technologies). One microliter of cDNA diluted 1:5 was used as template for qPCR reactions with TaqMan Gene Expression assays. Pre-designed Taqman assays were: mouse Snph (Mm01243855_m1), human long+short (L+S) SNPH transcripts (Hs00920132_m1), ACTB (Hs99999903_m1), GAPDH (Hs99999905_m1), and eukaryotic 18S rRNA (4352930E).
Custom isoform-specific TaqMan gene expression assays to detect Long human SNPH transcript (NM_001318234.1, L-SNPH), or short human SNPH transcript (NM_014723, S-SNPH) were used (Figure 1A). Custom L-SNPH assay primers and probe were: forward, TCAGGGTTGTTGAGAGGAGTCA; reverse, CCAGTTGGCCCGTGGTT; probe, ATAATACGGGAAGCCCC. Custom S-SNPH assay primers and probe were: forward, AGTGGTGCGAGCCG; reverse, GGTGGGATGGGCGGTATC; probe, CAGTGGACTCAGCCCCC. A standard synthetic gBlock containing the target amplicons for common SNPH (S+L), short SNPH, long SNPH, actin, and GAPDH in tandem (CSLAG) was purchased from IDT. The efficiency of amplification (Ex) of the 3 assays (long, short, and common SNPH) was determined using the Ct slope method with 6 concentrations of CSLAG standard covering a 5-log range, and found to be identical (P > 0.05 for all comparisons). The mean and SD were: Ex long = 82.33 ± 8.069; Ex short = 80.90 ± 2.700; Ex common = 84.82 ± 7.179. The mean and SD for the correlation coefficients were: r2 long = 0.9975 ± 0.002500; r2 short = 0.9875 ± 0.01250; r2 common = 0.9970 ± 0.001000. All r2 values were identical (P > 0.05 for all comparisons). Absolute copy number for each transcript was determined against a standard curve of CSLAG that was run in parallel with the cDNA samples. For relative quantitation, the ΔΔCt method was used.
Analysis of SNPH mRNA expression in public databases.
The NCBI’s Gene Expression Omnibus (GEO) genomics data repository was interrogated for SNPH mRNA expression in primary versus metastatic cancer using the GEO Profiles Database (https://www.ncbi.nlm.nih.gov/geoprofiles/). The “Metastatic prostate cancer” dataset GDS2545 (51, 52) containing 65 primary prostate and 15 metastases to regional and distal lymph nodes was downloaded. The study contained one probeset for SNPH (4117_at, GPL8300: [HG_U95Av2] Affymetrix Human Genome U95 Version 2 Array). The “Human epidermal growth factor receptor 2–positive breast cancer brain metastases” dataset GDS5306 (53) containing 19 HER+ breast primary non-metastatic and 19 brain metastases was downloaded. The study contained 5 probesets for SNPH: each probe was searched in the GPL1352: [U133_X3P] Affymetrix Human X3P Array and matched to gene bank ID, and the sequence was downloaded from the NCBI nucleotide collection and blasted against the Human Genomic Plus Transcript (Human G+T). Four probes (Hs.323833.1.S1_3p_a_at, Hs.323833.1.S1_3p_at, 215917_3p_at, and Hs.323833.1.S1_3p_x_at) were primarily covering a neighbor gene, RAD21L1 (NM_001136566.2 at 61% coverage), with a lower coverage for SNPH isoforms (38% for either isoform). These 4 probes were discarded from the analyses. Only one probe (ID: g7662081_3p_at) was a bona fide probe for SNPH (both isoforms were detected; 100% coverage for NM_001318234.1 and 89% coverage for NM_014723.3). Based on this probe, primary versus metastatic tumors were compared with a 2-tailed unpaired Student’s t test using GraphPad Prism 6.0 software.
For the Cancer Cell Line Encyclopedia (CCLE) (54), mRNA expression for SNPH was accessed through the cBioPortal for Cancer Genomics (http://www.cbioportal.org/) (55, 56) and downloaded. Individual cancer cell lines were grouped by primary tissue of origin according to the CCLE classification and plotted with GraphPad Prism 6.0 software. The TCGA tumor expression data for SNPH mRNA (RNA-seq values) were downloaded from the cBioPortal and plotted with GraphPad Prism 6.0 software.
For the SNPH versus VHL status analysis, the Kidney Renal Clear Cell Carcinoma study (The Cancer Genome Atlas [TCGA; https://cancergenome.nih.gov/], Provisional) was downloaded through the cBioPortal (55, 56), and the levels of SNPH mRNA were plotted against the copy number variation/mutation status of VHL. Outliers were tested with Grubbs test and removed from the dataset, and the multiple groups were tested with 1-way ANOVA and Bonferroni’s post-test for pairwise comparisons. A correlation between SNPH mRNA levels and linear copy number alteration (CNA) for VHL were was examined using Spearman’s test using GraphPad Prism 6.0 software.
Immunofluorescence.
Tumor cells were fixed in formalin/PBS (4% final concentration) for 15 minutes at 22°C, permeabilized in 0.1% Triton X-100/PBS for 5 minutes, washed, and incubated in 5% normal goat serum (NGS, Vector Laboratories) diluted in 0.3 M glycine/PBS for 60 minutes. Primary antibodies against Tom20 (diluted 1:300), β-tubulin (diluted 1:200), SNPH (diluted 1:500), and MTC02 (diluted 1:500) were added in 5% NGS/0.3 M glycine/PBS and incubated for 18 hours at 4°C. After 3 washes in PBS, secondary antibodies conjugated to Alexa Fluor 488, TRITC, or Alexa Fluor 633 were diluted 1:500 in 5% NGS/0.3 M glycine/PBS and added to cells for 1 hour at 22°C. Where indicated, F-actin was stained with phalloidin Alexa Fluor 488 (1:200 dilution) for 30 minutes at 22°C. Slides were washed and mounted in DAPI-containing ProLong Gold mounting medium (Invitrogen). At least 7 random fields were analyzed by fluorescence microscopy in a Nikon i80 microscope.
Cortical mitochondria and total mitochondrial mass quantification.
Mitochondria/F-actin composite images were analyzed in ImageJ (NIH), as described previously (25). The F-actin channel was used to manually label the cell boundary, and a belt extending from the boundary toward the inside of the cell was marked as “cortical mask” (see Figure 4B). This cortical mask was subsequently applied to the mitochondrial channel to measure intensity at the cortical region, which was normalized to total mitochondrial intensity per cell and cell area. For quantification of total mitochondrial mass, composite images were analyzed in ImageJ. The cell border was manually traced on the F-actin channel, and this “cell mask” was subsequently applied to the mitochondria channel to measure the total mitochondria signal per cell. Maximum intensity was monitored to ensure no pixel saturation (e.g., maximum intensity <256 for 8-bit images). Mitochondrial mass was normalized to total cell area. A minimum of 30 cells was analyzed in each independent experiment to obtain mean values.
Mitochondrial ROS quantification in live cells.
Mitochondrial superoxide production was analyzed as described previously (25). Briefly, 1.5 × 104 cells were grown on high-optical-quality 8-well μ-slides (Ibidi) and stained with MitoSOX Red mitochondrial superoxide indicator (Life Technologies, 5 μM, 10 minutes) in complete medium, followed by washes in warm medium. Stained cells were imaged with a 40× objective on a Nikon TE300 inverted time-lapse microscope equipped with a video system containing an Evolution QEi camera and a time-lapse video cassette recorder. The atmosphere was equilibrated to 37°C and 5% CO2 in an incubation chamber. Phase and red fluorescence (TRITC filter cube, excitation wavelength 532–554 nm, and emission wavelength 570–613 nm) images were captured. For quantification, files were imported into ImageJ, and masks were manually created around the periphery of the cell based on the phase image and subsequently applied to the TRITC channel to measure intensity. A minimum of 100 cells was analyzed in each independent experiment to obtain mean values.
Analysis of bioenergetics.
Cells were analyzed for ATP generation (BioChain catalog Z5030041) or OCR (Enzo Life Sciences catalog ENZ-51045-1), according to the manufacturer’s specifications. In some experiments, the culture medium was exchanged with dialyzed FBS containing growth medium and incubated for 2 hours, followed by analysis of lactate production (Abcam, catalog ab65331). For glucose consumption, cells were grown for 24 hours at 37°C, and aliquots of the culture supernatant were collected and processed using a glucose assay kit (eEnzyme).
Mitochondrial respiration complex activity.
Extracts from PC3 cells stably transduced with pLKO or SNPH-directed shRNA were analyzed for changes in oxidative phosphorylation complex activity using Abcam reagents (for complex I, ab109721; for complex II, ab109908). Twenty micrograms of cell lysates was assayed in parallel for citrate synthase (CS) activity (ScienCell Research Laboratories). Aliquots of lysates with comparable CS activity were applied for quantification of mitochondrial oxidative phosphorylation complex activity. Relative complex activities were calculated by determining the change in absorbance over time in the linear range of the measurements.
Cell motility analysis.
2D tumor cell motility experiments were carried out as described previously (25). Briefly, 1 × 104 cells under the various conditions tested were seeded in 4-well Ph+ chambers (Ibidi) in complete medium and allowed to attach overnight. Videomicroscopy was performed over 10 hours, with a time-lapse interval of 10 minutes. Stacks were imported into ImageJ for analysis. Images were aligned according to subpixel intensity registration with the StackReg plugin for ImageJ. At least 30 cells were tracked using the Manual Tracking plugin for ImageJ, and the tracking data from 4 independent time-lapse experiments were pooled and exported into Chemotaxis and Migration Tool v2.0 (Ibidi) for graphing and calculation of mean and SD of speed, accumulated distance, and Euclidean distance of movement. For cell migration using a wound closure assay, a monolayer of PC3 cells was incubated at 1% O2 for 24 hours, wounded using a 10-μl pipette tip, immediately returned to the hypoxia chamber, and incubated for an additional 20 hours in 1% O2. Cells were imaged by phase-contrast microscopy,and images were imported into FIJI software (http://fiji.sc/) and processed to measure the area within the wound. The percentage of wound closure was calculated based on the maximum initial area for each well.
Cell proliferation.
For direct cell counting experiments, tumor cells were plated in triplicate on 6-MW plates (4 × 104 cells/well) and counted at increasing time intervals at 37°C. In parallel, cell viability was measured by Trypan blue exclusion. Where indicated, ROS scavengers, MT (50 μM), or NAC (10 μM), alone or in combination, were added to the medium, and fresh medium containing drugs was added every 2 days. For colony formation, 200 cells were plated in triplicate onto 6-MW plates and allowed to grow for 10–14 days, with fresh medium added every 2–3 days. Colonies were stained with 0.5% w/v crystal violet/methanol for 30 minutes at 22°C and quantified by ImageJ. Cell proliferation experiments were done by labeling cells in 1:1,000 dilution BrdU (Amersham Pharmacia Biotech) in culture medium for 1 hour and analysis by multiparametric flow cytometry with quantitation of BrdU+ cells. Cell cycle analysis was carried out in ethanol-fixed cells, stained for 10 minutes with 2.5 μl/ml propidium iodide in the presence of ribonuclease A. Twenty thousand events were acquired on a FACSCalibur flow cytometer and quantified using CellQuest Pro software (Becton Dickinson).
Animal studies.
Groups of 6- to 8-week-old male NOD SCID γ (NSG, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) immunocompromised mice (Jackson Laboratory) (5 mice per group) were injected s.c. with PC3 cells stably transfected with pLKO or two independent SNPH-directed shRNA sequences (clones 0 and 5), and superficial tumor growth (2 tumors/mouse) was quantified with a caliper over a 2-week interval. At the end of the experiment, animals bearing PC3-pLKO (puromycin-resistant) tumors were euthanized, and the liver and lungs were dissected and washed in PBS. Metastatic nodules were excised and cut into 1-mm sections, washed in PBS, and plated in RPMI 1640 medium containing puromycin. Tumor cells were allowed to attach to the plate overnight, and the next day any residual tissue fragments were removed. Four animals were used to generate 4 independent cell lines from liver and lung metastatic sites. Parental PC3-pLKO cells were used as control for these experiments.
For a syngeneic model of metastasis in vivo, Yale University Mouse Melanoma 1.7 (Yumm1.7) 1.7 cells derived from a genetically engineered mouse model of invasive melanoma with the genotype BrafV600E; Cdkn2a–/–;Pten–/– were used (31). Yumm1.7 cells stably expressing mCherry were described previously (32). Cells were transiently transfected with empty pCMV6 vector, or cDNAs encoding FL short SNPH or ΔMLS SNPH mutant, and selected with G418 at 400 μg/ml for 15 days. Stably transfected cells (2.5 × 105) were injected into the flanks of syngeneic 8-week-old male C57BL/6NCr (NCI Inbred mice, Charles River strain code 556). One to 3 weeks later, tumor cells disseminated to lungs were identified and quantitated based on expression of the mCherry transgene by IHC (see below).
IHC.
Lungs were fixed in neutral formalin (Fisher Scientific, SF93-4) for 36 hours, transferred to 70% ethanol for 3 days, and then paraffin embedded. Five-micrometer sections were stained with a rabbit anti-mCherry polyclonal antibody (Novus, NBP2-25157) as follows. Slides were warmed at 50°C for 30 minutes; deparaffinized in xylene for 20 minutes, then xylene/ethanol 1:1 for 5 minutes; and rehydrated in alcohol series (100%, 95%, 90%, 70%, 50%, 30% ethanol and dH2O, 5 minutes each). Antigen retrieval was done in citrate-based solution (Vector Laboratories, H-3300) at pH 6.0 in a pressure cooker for 5 minutes, followed by cooling to room temperature. Next, slides were washed once in PBS for 5 minutes and incubated in 3% hydrogen peroxide for 20 minutes. Slides were then washed 3 times with PBS (5 minutes each) and blocked in 10% normal goat serum/PBS for 1 hour at room temperature. Primary antibody was diluted 1:500 in 10% normal goat serum/PBS and incubated in a humidified chamber overnight. The next day, slides were washed 3 times with PBS for 5 minutes each, incubated with anti-rabbit HRP-labeled polymer (Dako, K4002) at room temperature for 30 minutes, and washed 3 times with PBS for 5 minutes each. Slides were developed with a DAB+ substrate chromogen system (Dako, K3467) for 30 minutes, rinsed in dH2O, and stained with Mayer’s hematoxylin solution (Sigma-Aldrich, MHS16) for 10 seconds. Slides were dehydrated in dH2O, 30%, 50%, 70%, 90%, 95%, and 100% ethanol (5 minutes each); immersed in xylene for 15 minutes; and mounted with Permount mounting medium (Fisher Scientific, SP15-100).
Quantification of disseminated tumor cells to the lungs.
Five lungs per group were stained for mCherry as described above and scanned for the presence of mCherry+ cells in a Nikon i80 upright microscope. Each mCherry+ cell was photographed at 40× magnification and manually counted using ImageJ software. For each animal, the average number of mCherry+ cells per lung was calculated and presented. ANOVA with Bonferroni’s post-test was used to compare the means between the groups and derived pairwise comparison P values.
Statistics.
Dare expressed as mean ± SEM or mean ± SD of multiple independent experiments or replicates of representative experiments out of a minimum of 2 or 3 independent determinations. Two-tailed Student’s t test or Wilcoxon rank-sum test was used for 2-group comparative analyses. For multiple-group comparisons, ANOVA or Kruskal-Wallis test with Bonferroni’s post-hoc procedure was applied. All statistical analyses were performed using GraphPad software package (Prism 6.0) for Windows. A P value less than 0.05 was considered as statistically significant.
Study approval.
Studies involving vertebrate animals (rodents) were carried out in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). Protocols were approved by the IACUC of the Wistar Institute (protocol 112625).
Author contributions
MCC, JHS, and DCA conceived the project; MCC performed experiments on mitochondrial trafficking, chemotaxis, tumor cell invasion, the xenograft mouse model of localized and metastatic tumor growth, the syngeneic model of metastasis, and analysis of SNPH expression in public databases; JHS performed experiments on tumor cell proliferation, mitochondrial bioenergetics, and ROS production; YW performed experiments on cortical mitochondria; YW and DBR performed experiments on cell cycle progression; ETK prepared and characterized Ad-LacZ and Ad-SNPH; ATW provided the Yumm1.7 cells stably expressing the mCherry transgene; MCC, JHS, YW, DBR, DIG, LRL, and DCA analyzed data; and MCC, JHS, and DCA wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank Frederick Keeney (Wistar Institute) for assistance with time-lapse microscopy. This work was supported by NIH grants P01 CA140043 (DCA, DIG, and LRL), R01 CA78810 and CA190027 (DCA), R01 CA089720 (LRL), and F32 CA177018 (MCC); the Office of the Assistant Secretary of Defense for Health Affairs through the Prostate Cancer Research Program under award W81XWH-13-1-0193 (DCA); and a Challenge Award from the Prostate Cancer Foundation (PCF) to MCC, DIG, LRL, and DCA. Support for Core Facilities utilized in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to the Wistar Institute.
Version 1. 09/08/2017
Electronic publication
Version 2. 09/20/2017
Print issue publication
Funding Statement
Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute
Footnotes
MCC’s present address is: Pharmacology Department, University of Colorado Denver AMC, Denver, Colorado, USA.
DBR’s present address is: University of Pittsburgh Medical Center, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(10):3755–3769.https://doi.org/10.1172/JCI93172.
Contributor Information
M. Cecilia Caino, Email: ccaino@wistar.org.
Jae Ho Seo, Email: jseo@wistar.org.
Yuan Wang, Email: yuwang@wistar.org.
Dmitry I. Gabrilovich, Email: dgabrilovich@wistar.org.
Eui Tae Kim, Email: kime2@chop.edu.
Lucia R. Languino, Email: lucia.languino@jefferson.edu.
Dario C. Altieri, Email: daltieri@wistar.org.
References
- 1.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 2.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brock A, Chang H, Huang S. Non-genetic heterogeneity — a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10(5):336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 4.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14(9):581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 6.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao CF, et al. Proliferation and invasion: plasticity in tumor cells. Proc Natl Acad Sci U S A. 2005;102(30):10528–10533. doi: 10.1073/pnas.0504367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Hatzikirou H, Basanta D, Simon M, Schaller K, Deutsch A. ‘Go or grow’: the key to the emergence of invasion in tumour progression? Math Med Biol. 2012;29(1):49–65. doi: 10.1093/imammb/dqq011. [DOI] [PubMed] [Google Scholar]
- 10.Kemper K, de Goeje PL, Peeper DS, van Amerongen R. Phenotype switching: tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014;74(21):5937–5941. doi: 10.1158/0008-5472.CAN-14-1174. [DOI] [PubMed] [Google Scholar]
- 11.Hecht I, Natan S, Zaritsky A, Levine H, Tsarfaty I, Ben-Jacob E. The motility-proliferation-metabolism interplay during metastatic invasion. Sci Rep. 2015;5:13538. doi: 10.1038/srep13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venere M, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med. 2015;7(304):304ra143. doi: 10.1126/scitranslmed.aac6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsdale R, et al. The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci Signal. 2015;8(390):ra82. doi: 10.1126/scisignal.aab1111. [DOI] [PubMed] [Google Scholar]
- 14.Falletta P, et al. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 2017;31(1):18–33. doi: 10.1101/gad.290940.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang SD, et al. EphB2 receptor controls proliferation/migration dichotomy of glioblastoma by interacting with focal adhesion kinase. Oncogene. 2012;31(50):5132–5143. doi: 10.1038/onc.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim IS, et al. Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat Commun. 2017;8:14343. doi: 10.1038/ncomms14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell. 2016;61(5):667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vyas S, Zaganjor E, Haigis MC. Mitochondria and cancer. Cell. 2016;166(3):555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeBleu VS, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caino MC, et al. Metabolic stress regulates cytoskeletal dynamics and metastasis of cancer cells. J Clin Invest. 2013;123(7):2907–2920. doi: 10.1172/JCI67841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porporato PE, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8(3):754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32(40):4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong LF, et al. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. Elife. 2017;6:e22187. doi: 10.7554/eLife.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan AS, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21(1):81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Caino MC, et al. PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion. Proc Natl Acad Sci U S A. 2015;112(28):8638–8643. doi: 10.1073/pnas.1500722112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivadeneira DB, et al. Survivin promotes oxidative phosphorylation, subcellular mitochondrial repositioning, and tumor cell invasion. Sci Signal. 2015;8(389):ra80. doi: 10.1126/scisignal.aab1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang JS, et al. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132(1):137–148. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng ZH. Mitochondrial trafficking and anchoring in neurons: new insight and implications. J Cell Biol. 2014;204(7):1087–1098. doi: 10.1083/jcb.201312123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caino MC, et al. A neuronal network of mitochondrial dynamics regulates metastasis. Nat Commun. 2016;7:13730. doi: 10.1038/ncomms13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshmukh P, Unni S, Krishnappa G, Padmanabhan B. The Keap1-Nrf2 pathway: promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys Rev. 2017;9(1):41–56. doi: 10.1007/s12551-016-0244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 2016;29(5):590–597. doi: 10.1111/pcmr.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur A, et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532(7598):250–254. doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123(9):3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun F, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121(7):1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Quinlan CL, Orr AL, Perevoshchikova, Treberg JR, Ackrell BA, Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem. 2012;287(32):27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin YF, Haynes CM. Metabolism and the UPR(mt) Mol Cell. 2016;61(5):677–682. doi: 10.1016/j.molcel.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemarie A, Huc L, Pazarentzos E, Mahul-Mellier AL, Grimm S. Specific disintegration of complex II succinate:ubiquinone oxidoreductase links pH changes to oxidative stress for apoptosis induction. Cell Death Differ. 2011;18(2):338–349. doi: 10.1038/cdd.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong LF, et al. Alpha-tocopheryl succinate induces apoptosis by targeting ubiquinone-binding sites in mitochondrial respiratory complex II. Oncogene. 2008;27(31):4324–4335. doi: 10.1038/onc.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang MS, Rohlena J, Dong LF, Neuzil J, Grimm S. Powerhouse down: Complex II dissociation in the respiratory chain. Mitochondrion. 2014;19 Pt A:20–28. doi: 10.1016/j.mito.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Chae YC, et al. Landscape of the mitochondrial Hsp90 metabolome in tumours. Nat Commun. 2013;4:2139. doi: 10.1038/ncomms3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cole A, et al. Inhibition of the mitochondrial protease clpp as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2015;27(6):864–876. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seo JH, et al. The mitochondrial unfoldase-peptidase complex clpxp controls bioenergetics stress and metastasis. PLoS Biol. 2016;14(7):e1002507. doi: 10.1371/journal.pbio.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14(11):709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 45.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hurd TR, DeGennaro M, Lehmann R. Redox regulation of cell migration and adhesion. Trends Cell Biol. 2012;22(2):107–115. doi: 10.1016/j.tcb.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radisky DC, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436(7047):123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanharanta S, Massagué J. Origins of metastatic traits. Cancer Cell. 2013;24(4):410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pienta KJ, Robertson BA, Coffey DS, Taichman RS. The cancer diaspora: metastasis beyond the seed and soil hypothesis. Clin Cancer Res. 2013;19(21):5849–5855. doi: 10.1158/1078-0432.CCR-13-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandran UR, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 53.McMullin RP, et al. A BRCA1 deficient-like signature is enriched in breast cancer brain metastases and predicts DNA damage-induced poly (ADP-ribose) polymerase inhibitor sensitivity. Breast Cancer Res. 2014;16(2):R25. doi: 10.1186/bcr3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrentina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.