The powerful tools of genome editing are rapidly making their way toward the clinic. Zinc-finger nucleases, TALENs, and CRISPR-Cas have all been used in conjunction with somatic cell therapies, and in vivo approaches are being tested. Both excitement and concern have been elicited by the prospects for gene editing in human embryos, a procedure that could eliminate the causes of particular genetic diseases in the treated individual and all of his/her descendants. A recent Nature article by Ma et al. provides a proof-of-principle demonstration of genome editing technologies to correct germline mutations, reporting targeted correction of the heterozygous MYBPC3 gene mutation that is responsible for hypertrophic cardiomyopathy in human preimplantation embryos (1).
All of the genome editing platforms rely on the ability to design molecules that will make breaks in chromosomal DNA efficiently and specifically at a chosen target (2–5). In the case of CRISPR, the Cas9 protein acts as a nuclease when it is guided to its target by a specific single guide RNA (sgRNA). Breaks made by any of the platforms are recognized by cells as potentially lethal damage, and they are repaired by two alternative pathways: nonhomologous end joining (NHEJ), which often introduces new mutations at the break, and homology-dependent repair (HDR), which can use endogenous sequences or experimenter-provided DNA as a template.
What was done?
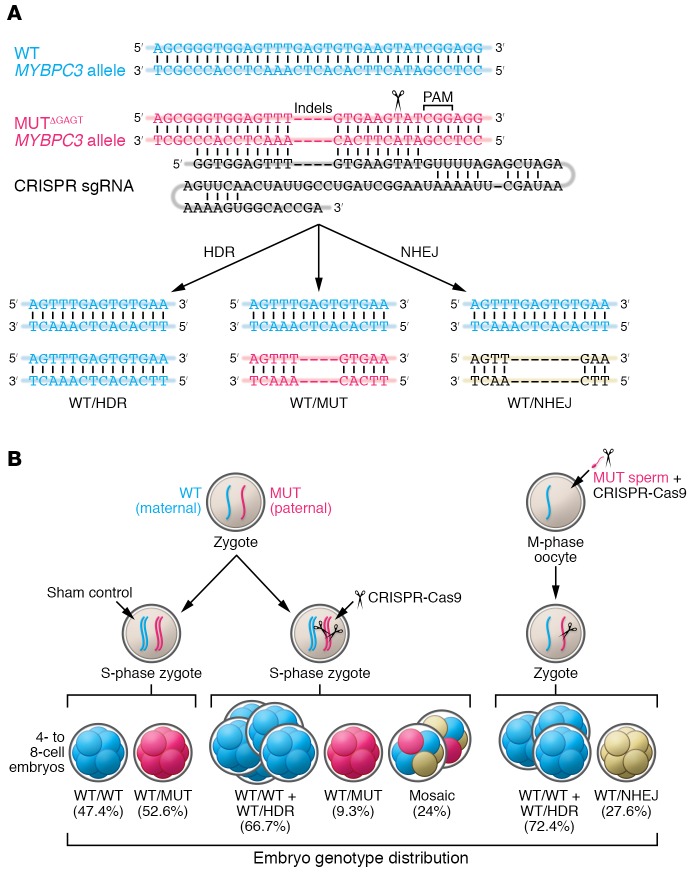
Ma et al. — including scientists at the Oregon Health Sciences University; the Center for Genome Engineering in Seoul, Korea; the Salk Institute; and several sites in China — focused their efforts on a mutation in the MYBPC3 gene that is implicated in inherited hypertrophic cardiac myopathy (HCM) (1). They fertilized oocytes from normal healthy donors with sperm from a single male patient, who carried a heterozygous-dominant 4-bp deletion in the MYBPC3 gene (Figure 1A).
Figure 1. Genome editing of the MYBPC3ΔGAGT mutation by CRISPR-Cas9 and sgRNA injection into human embryos.
(A) (Top) Diagram of the WT and mutant (MUT) alleles and the sgRNA that specifically targets the mutant sequence. (Bottom) Possible genotypes from the editing of the paternal MYBPC3ΔGAGT mutant allele using CRISPR-Cas9 with and without ssODNs. (B) Blastomere genotype distribution in 4- to 8-cell embryos, arising after injection of CRISPR-Cas9 either into human S-phase zygotes or into M-phase MII oocytes. Because the sperm donor was heterozygous and the egg donors WT, about half the cells in control embryos were fully WT and half heterozygous. The paternal mutant allele is preferentially corrected using the homologous WT maternal chromosome in human embryos rather than the ssODNs provided by the investigator. Figure modified with permission from Nature (1). HDR, repaired allele resulting from homologous recombination that is indistinguishable from WT; NHEJ, insertions and deletions resulting from nonhomologous end joining; PAM, protospacer adjacent motif; sgRNA, single guide RNA.
In the initial experiments, Cas9 protein, sgRNA and a single-stranded oligodeoxyribonucleotide (ssODN) template were injected 18 hours after fertilization. Three days after fertilization, individual cells from the 4- to 8-cell embryos were isolated and evaluated. The expectation was that some mutant alleles would be corrected using the ssODN, and some would carry a new mutation due to inaccurate repair of the Cas9-induced DNA break. The results showed clear evidence that the mutant allele had been corrected in a number of the treated embryos; however, this was done using the maternal WT allele as the template, not the injected template DNA (Figure 1). In addition, there were cells that were unaffected by Cas9 and some that had novel NHEJ mutations.
About a quarter of the embryos in this experiment were mosaic — i.e., they contained cells with more than one genotype, indicating that Cas9 activity continued beyond the one-cell stage. This outcome is not desirable because correction could be incomplete, both in the affected tissue and in the germline. To address this issue in a second experiment, Ma et al. injected the CRISPR reagents into metaphase II–stage (MII-stage) oocytes along with the sperm, so that Cas9 was present from the moment of fertilization (Figure 1B) (1). Analysis of individual cells 3 days later revealed no mosaic embryos and a higher proportion of fully WT embryos. Again, all correction appeared to occur from the maternal allele and none from the ssODN donor.
The treated embryos developed normally during the brief incubation period, but a key concern in genome editing is whether changes to the genome are limited to the intended target. Ma et al. performed an extensive search for off-target mutations in a small number of embryos and identified none that could be attributed to CRISPR treatment (1).
What was learned?
This study represents the first report of CRISPR editing in human embryos in work done largely in the US, and it produced several important conclusions. First, coinjecting the CRISPR reagents at the time of fertilization eliminates unwanted mosaicism without impairing editing activity. This approach was only possible with embryos that were created specifically for use in this research, which is not allowed in the US with federal funding. Surplus embryos from in vitro fertilization (IVF) treatments will have advanced well beyond the one-cell stage. Second, at the one-cell stage, the homologous chromosome is used effectively as a repair template by HDR. It is somewhat difficult to see how this occurs, since the parental genomes normally do not see each other until after DNA replication and pronuclear fusion. Nonetheless, repair from the homologue can be a good thing when a WT allele is present, as in the case of the heterozygous embryos in this study. Third, the injected ssODN donor was apparently not used for HDR. The failure to incorporate sequences from the synthetic donor means, unfortunately, that investigator-designed changes may be more difficult to achieve than previously thought. Fourth, the editing can be very specific; in this case, no induced off-target mutations were detected. However, caution is still warranted as prior studies, using different embryos and experimental protocols, reported high levels of off-target effects (6). Fifth, as the authors emphasize, the efficiency of editing they achieved is not adequate for current use. Alternative strategies, such as preimplantation genetic diagnosis (PGD), are currently more reliable. We do not find compelling the authors’ suggestion that the modest increase they see in homozygous WT embryos could be used to reduce the number of embryos screened by PGD.
It is clear that these conclusions need to be verified by others and additional work needs to be done to resolve remaining issues. Because the offending mutation in the MYBPC3 gene was a 4-bp deletion, it was easy to produce an sgRNA that targeted the mutant, but not the WT allele (Figure 1A). This will often not be the case, for example, when the disease mutation is a single bp substitution. Although HDR from the homologue was rather efficient, NHEJ mutations were still produced at the cleavage site; these need to be suppressed. Other forms of the exogenous donor DNA — perhaps double-stranded linear or circular molecules — should be tested to see if they are more effective in competing with the homologue as a template for repair. Experiments are also warranted to examine what happens when the maternal allele is mutant and the paternal allele WT to see if HDR goes both directions between homologues. The fundamental issue of off-target mutations will need to be addressed with each new sgRNA and examination of larger numbers of embryos.
What is next?
Our view is that reproductive human genome editing will eventually happen. It is possible that the first such pregnancies have already been initiated somewhere in the world. The responsibility of research scientists is to work toward making the editing process in embryos sufficiently safe and effective for medical uses and to engage in the broad discussion of what specific uses the technology should be directed toward. It would be tragic if the first attempts had disastrous consequences. We fully subscribe to the admonition put forward by the National Academies of Sciences and Medicine committee report that germline editing should be used only in cases of serious diseases and when a sensible alternative is not available (7). We acknowledge that there will be considerable pressure from patients, families, and other advocates for the editing option to be available, even when safe alternatives like PGD are available. In addition, there will certainly be people who want to attempt cosmetic, performance, and other enhancements. In a wealth-driven society like ours, it will be difficult to prevent this. As yet, we need not fear “designer” babies, since we are very far from living in a world of Gattaca.
Version 1. 08/24/2017
Electronic publication
Version 2. 09/20/2017
Print issue publication
Footnotes
Conflict of interest: S. Chandrasegaran owns stock in Sangamo Therapeutics. DC receives license royalties from Sangamo Therapeutics.
Reference information: J Clin Invest. 2017;127(10):3588–3590. https://doi.org/10.1172/JCI96962.
Contributor Information
Srinivasan Chandrasegaran, Email: chandra@jhmi.edu.
C. Korin Bullen, Email: korin23@gmail.com.
Dana Carroll, Email: dana@biochem.utah.edu.
References
- 1.Ma H, et al. Correction of a pathogenic gene mutation in human embryos [published online ahead of print August 2, 2017] Nature. doi: 10.1038/nature23305. https://doi.org/10.1038/nature23305 [DOI] [PubMed] [Google Scholar]
- 2.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21(1):289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang P, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. National Academies of Sciences, Engineering, and Medicine. Human Genome Editing: Science, Ethics, and Governance. Washington, DC, USA: The National Academies Press; 2017. https://doi.org/10.17226/24623. [PubMed]