Abstract
The tumor suppressor p53, a master regulator of the cellular response to stress, is tightly regulated by the E3 ubiquitin ligase MDM2 via an autoregulatory feedback loop. In addition to its well-established role in tumorigenesis, p53 has also been associated with aging in mice. Several mouse models with aberrantly increased p53 activity display signs of premature aging. However, the relationship between dysfunction of the MDM2/p53 axis and human aging remains elusive. Here, we have identified an antiterminating homozygous germline mutation in MDM2 in a patient affected by a segmental progeroid syndrome. We show that this mutation abrogates MDM2 activity, thereby resulting in enhanced levels and stability of p53. Analysis of the patient’s primary cells, genome-edited cells, and in vitro and in vivo analyses confirmed the MDM2 mutation’s aberrant regulation of p53 activity. Functional data from a zebrafish model further demonstrated that mutant Mdm2 was unable to rescue a p53-induced apoptotic phenotype. Altogether, our findings indicate that mutant MDM2 is a likely driver of the observed segmental form of progeria.
Keywords: Aging, Genetics
Keywords: DNA repair, P53, Ubiquitin-proteosome system
Introduction
Two compelling lines of evidence establish p53 as a major tumor suppressor in humans. First, heterozygous germline mutations in the p53-encoding gene TP53 cause Li-Fraumeni syndrome, an autosomal-dominant and highly penetrant early-onset cancer predisposition syndrome (1). Second, inactivating somatic TP53 mutations have been identified in the majority of human tumors (2). The p53 protein is a DNA sequence–dependent transcriptional regulator of myriad genes whose products mediate diverse physiological pathways involved in cell-cycle progression, apoptosis, senescence, autophagy, genomic stability, fertility, stem cell differentiation, and cellular metabolism, many of which can be linked to tumor suppression (3, 4). Under basal conditions, p53 is maintained at very low cellular levels. However, in response to exogenous and endogenous stressors, p53 is rapidly stabilized and activated, leading to one or more of the above-mentioned cellular outcomes. Maintenance of low p53 levels or deactivation of p53 during recovery from stress is mostly attributable to the function of the E3 ubiquitin ligase MDM2 that binds primarily to a region within the p53 N-terminal transactivation domain. MDM2 controls cellular p53 levels and activity through at least 3 mechanisms: (a) direct binding and blockage of p53 transactivation ability; (b) monoubiquitylation leading to export of p53 out of the nucleus; and (c) polyubiquitylation leading to proteasomal degradation of p53 (5, 6). In fact, MDM2 is itself a direct transcriptional target of p53 and thereby forms an autoregulatory feedback loop, such that p53 and MDM2 tightly regulate each other’s cellular levels and activities (7, 8).
While it is well established that p53 loss of function increases the risk for cancer, the relationship between the MDM2/p53 axis and human aging remains a matter of debate (9). This is an interesting question, especially in light of the envisaged therapeutic use of MDM2 inhibitors in oncology (10). For example, p53 protein levels accumulate in aged, “near-senescent” human fibroblasts (11), but decline in aged mouse populations (12). The p53 SNP p.Arg72Pro has been associated with increased longevity and survival (13), but paradoxically also confers reduced apoptotic potential (14, 15) and modest increases in cancer susceptibility (16). In addition, some human premature aging disorders (segmental forms of progeria) have been linked to p53 signaling and may yield insights into the broader aging phenomenon (17, 18).
The putative relevance of p53 action in aging is also reflected in mouse models that modulate the MDM2/p53 axis directly. Mice with N-terminal truncations of p53 in the presence of functional WT p53 (19, 20) and mice with N-terminal p53 mutations that mimic phosphorylation and are unable to be regulated by MDM2 have constitutively active p53 activity and display accelerated aging phenotypes (21). On the contrary, some mouse models with altered p53 or MDM2 expression, i.e., “super-p53” or hypomorphic MDM2 mice, display no signs of premature aging (22, 23), suggesting that simple elevation of p53 levels is insufficient to cause premature aging, as p53 is still tightly regulated in these cases. These seemingly conflicting results may be reconciled by positing that deregulating the MDM2-p53 feedback loop might be critical for abnormal aging processes.
Here, we present a patient affected by a segmental progeroid syndrome in whom we identified a homozygous germline MDM2 mutation. This antiterminating MDM2 mutation results in constitutive stabilization of p53 and MDM2, leading to altered p53 activity and dysfunction of the MDM2/p53 axis.
Results
Characteristics of the studied patient.
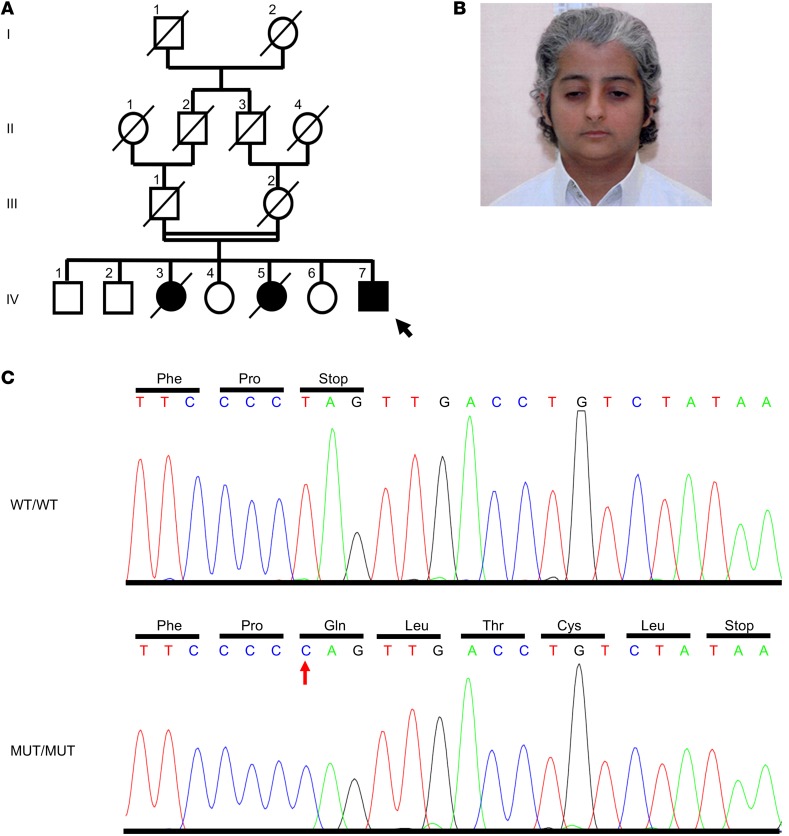
We studied a patient who was referred to the International Registry of Werner Syndrome (www.wernersyndrome.org) with the putative diagnosis of a Werner syndrome–like segmental progeroid disorder. The index patient (IV:7, Figure 1, A and B) is the seventh child of unaffected, consanguineous parents of Saudi Arabian origin. At the age of 19 years, he presented with short stature (151 cm), pinched facial features, prematurely gray hair, scleroderma-like skin changes, high-pitched voice, hypogonadism, sparse pubic hair, small kidneys and consecutive kidney failure, followed by severe arterial hypertension (Figure 1B and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI92171DS1). Two of his sisters, IV:3 and IV:5, were reported to have had similar clinical features. In more detail, individual IV:3 had short stature, prematurely gray hair, hypertension, and kidney failure and died at the age of 31 years from myocardial infarction. Individual IV:5 also had short stature and prematurely gray hair and died suddenly at the age of 23 years, possibly from an epileptic attack. Their parents, III:1 and III:2, died at an unknown age from myocardial infarction and respiratory infection, respectively. No DNA from the deceased individuals was available for genetic testing. Conventional Sanger sequencing revealed no mutation in the known disease genes for progeroid syndromes (24).
Figure 1. Identification of a causative mutation in MDM2.
(A) Pedigree of a consanguineous family (black and white circles and squares denote affected and healthy individuals, respectively; arrow indicates the index patient; diagonal lines indicate deceased status; double line shows parental consanguinity). (B) Facial image of the index patient IV:7 at the age of 19 years. Note the patient’s short stature (151 cm on the scale), prematurely gray hair, pinched facial features with a narrow nasal bridge, and small mouth. (C) Sequence chromatogram shows the c.1492T>C (p.*498Qext5) mutation (red arrow).
Identification of the causative mutation.
To unravel the cause of this disorder with suspected autosomal-recessive inheritance, we first performed SNP array analysis of the index patient. In line with parental consanguinity, we identified 11 extended genomic regions of homozygosity with a maximum reachable LOD score (Supplemental Figure 1). We then performed whole-exome sequencing in patient IV:7. Bioinformatic filtering identified only a single homozygous variant not present in public databases that has a severe impact on protein structure in the above-mentioned putatively linked genomic regions (refer to the Supplemental text and Supplemental Table 2), namely, a homozygous antiterminating mutation, c.1492T>C, in MDM2. This mutation removes the physiological MDM2 stop codon and is predicted to extend the protein for 5 further erroneous amino acids, p.*498Qext5 (amino acids: QLTCL, Figure 1C). Previous cell-based transfection studies have shown that the highly conserved extreme C-terminus of MDM2 is essential for its oligomerization and E3 ubiquitin ligase activity (25, 26). In addition, a more recent study showed that the same MDM2 variant, when ectopically expressed, is unable to degrade coexpressed p53 in vitro (27).
Regulation of cellular p53 levels.
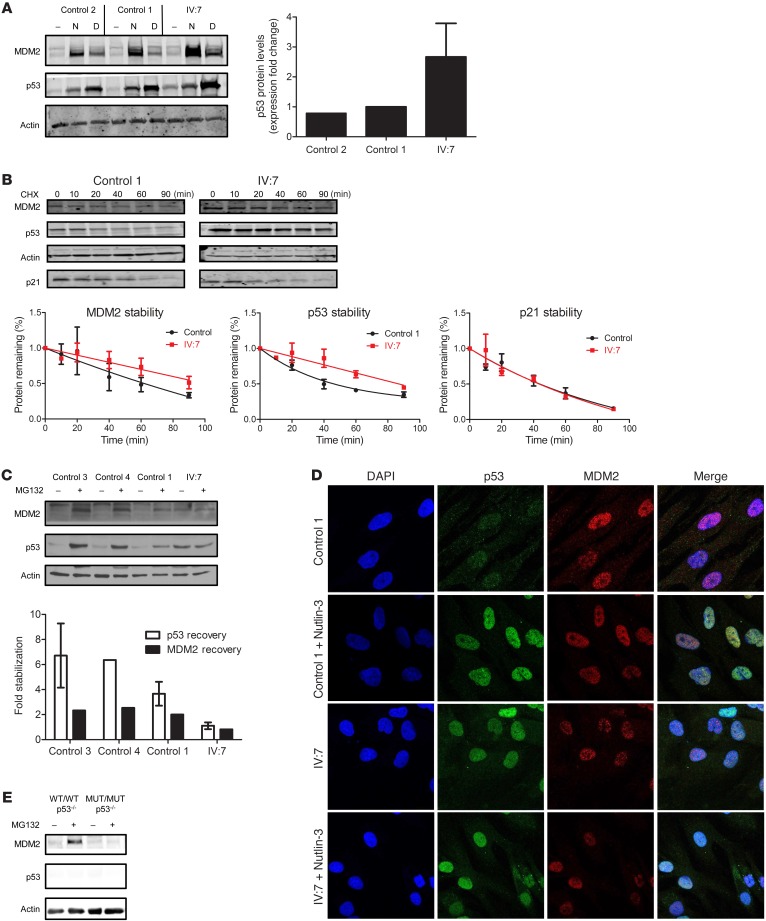
To corroborate our genetic findings, we analyzed p53 levels in primary dermal fibroblasts from patient IV:7. When compared with fibroblasts from healthy control individuals, we observed both elevated basal p53 protein levels and pronounced induction of p53 protein in the patient’s fibroblasts after treatment with 2 well-studied p53 inducers, i.e., Nutlin-3, a small-molecule inhibitor of MDM2-p53 interaction, and the anthracycline daunorubicin, a topoisomerase II inhibitor (Figure 2A). Markedly elevated p53 and MDM2 protein levels in lymphoblastoid cell lines (LCLs) from patient IV:7 as compared with LCLs from an unaffected individual confirmed these findings (Supplemental Figure 2A). Notably, p53 activation by Nutlin-3 treatment suggests that mutant MDM2 has residual p53 degradation activity. Taken together, these data indicate a compromised MDM2-p53 negative feedback loop as the likely pathogenic mechanism.
Figure 2. The antiterminating MDM2 mutation is defective in its regulation of p53.
(A) Protein levels of MDM2, p53, and actin in control and IV:7 fibroblasts treated with vehicle DMSO (–), Nutlin-3 (N, 10 μM), or daunorubicin (D, 0.1 mg/ml) for 24 hours. Immunoblot with the indicated antibodies. Graph shows the fold change of basal p53 levels over actin levels from 5 independent experiments. (B) Protein degradation rates of MDM2 and p53 in fibroblasts via CHX chase. Immunoblots show results of control 1 and IV:7 fibroblasts harvested at the indicated times (minutes) after CHX (100 μg/ml) treatment. Graphs show protein quantification normalized to actin (using ImageJ and GraphPad Prism software) and the half-lives of MDM2 (control 1: 62 min; IV:7: 92 min) and p53 (control 1: 45 min; IV:7: 82 min). (C) Effects of proteasome inhibition on MDM2 and p53 levels. Immunoblot of control fibroblasts (controls 3, 4, and 1) and IV:7 fibroblasts treated with vehicle (DMSO) or MG132 (25 μM) for 6 hours. Graph shows protein quantification (using ImageJ) and normalization to actin. Fold recovery was calculated by comparing p53 or MDM2 values from treated cells with those of untreated cells. (D) Subcellular localization of MDM2 and p53 in control 1 and IV:7 fibroblasts treated with vehicle (DMSO) or Nutlin-3 (10 μM) for 24 hours. Original magnification, ×63. (E) U2OS genome-edited cells expressing WT/WT and MUT/MUT MDM2 with no p53 were treated with vehicle (DMSO) or the proteasome inhibitor MG132 (25 μM) for 6 hours before harvesting and immunoblotting. In A and B, error bars represent mean ± standard deviation. In C, error bars represent mean ± SEM.
Consistent with the study mentioned above (27), we showed that ectopically expressed MDM2 with the antiterminating mutation is defective in its ability to degrade both ectopic and endogenous p53 in U2OS cells (Supplemental Figure 2, B and C). In fact, expression of mutant MDM2 led to increased levels of p53 protein in either case. Additionally, ectopic mutant MDM2 accumulated to markedly higher levels than did WT MDM2 when introduced into U2OS cells or H1299 cells (a p53-null cell line), indicating that its increased stability is an intrinsic property of the protein itself and not a result of p53 transcriptional activity (Supplemental Figure 2D). When conditions were calibrated such that mutant MDM2 and WT MDM2 were expressed at equivalent levels, mutant MDM2 still dramatically stabilized coexpressed p53 levels (Supplemental Figure 2E). These findings prompted us to further investigate MDM2-p53 homeostasis in fibroblasts from patient IV:7. Inhibition of protein synthesis by cycloheximide (CHX) revealed that both MDM2 and p53 were significantly stabilized compared with fibroblasts from an unaffected individual (Figure 2B), and the relative stabilities of transiently coexpressed mutant MDM2 and p53 were also markedly pronounced compared with WT MDM2 and p53 (Supplemental Figure 2F). Further, inhibition of the proteasome by MG132 confirmed the increased stability of p53 and MDM2 in the patient’s fibroblasts, since their levels were just minimally increased after treatment as compared with levels in fibroblasts from unaffected individuals (Figure 2C). Together, these data indicate that the identified mutation leads to impaired MDM2 E3 ubiquitin ligase activity. This was confirmed by transient transfection experiments showing that p53 coexpressed with mutant MDM2 was underubiquitinated when compared with WT MDM2 (Supplemental Figure 2G) and, relatedly, that MG132 treatment only very modestly increased ectopic p53 levels in the presence of mutant MDM2 (Supplemental Figure 2H). Finally, immunocytochemistry revealed that the patient’s untreated fibroblasts expressed relatively higher levels of nuclear p53, as was seen with fibroblasts from an unaffected individual treated with Nutlin-3 (Figure 2D). Transient transfection experiments again confirmed these observations, showing that p53 was found in both the nucleus and cytoplasm when coexpressed with WT MDM2 but was exclusively nuclear when coexpressed with mutant MDM2 (Supplemental Figure 2I).
In order to further analyze this mutation in a controlled background, we generated U2OS cell lines bearing 1 or 2 copies of mutant MDM2, heterozygous and homozygous, respectively, at its endogenous locus using TALENs. It is noteworthy that, despite the generation of multiple clones of other endogenous MDM2 mutations (data not shown), we were only able to obtain a single homozygous clone expressing the antiterminating MDM2 mutation, suggesting that cells expressing high levels of p53 could not be expanded during the selection process. In fact, basal and Nutlin-3–induced levels of p53 protein were similar among cells expressing WT/WT, WT/mutant (WT/MUT), and MUT/MUT MDM2 (Supplemental Figure 3), indicating that the genome-edited MUT/MUT MDM2 cells may have acquired a compensating mechanism to control basal p53 levels. To further determine the stability of mutant MDM2 in the absence of complications due to the presence of WT p53, we used CRISPR technology to generate p53-null versions in both parental and homozygous TALEN-generated cells (Figure 2E). Proteasome inhibition demonstrated that the MUT/MUT MDM2 could not be further stabilized when p53 was knocked out, suggesting that it is inherently more stable than its WT counterpart.
Cellular consequences of the MDM2 mutation.
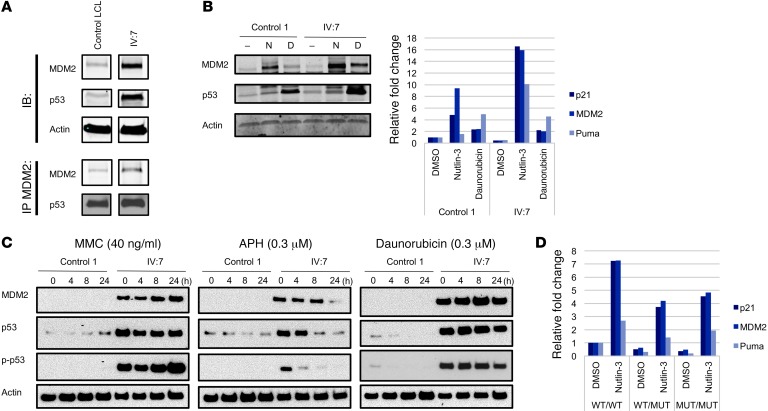
Since the primary sites of interaction between MDM2 and p53 are within their respective N-termini, we expected the mutant MDM2 to retain binding to p53. This was confirmed in the patient’s LCLs (Figure 3A) and in cells ectopically expressing mutant MDM2 and p53 (Supplemental Figure 4). Therefore, it is likely that the mutant MDM2 still retains its ability to repress the transcriptional transactivation ability of p53 through direct binding (28), potentially explaining why this mutation does not lead to embryonic lethality in humans. However, we repeatedly observed that p53 in the patient’s cells could still be highly activated by treatments that released p53 from MDM2 (Figure 2A), leading to a pronounced induction of the RNA expression levels of p53 target genes (p21, MDM2, NOXA, and PUMA) (Figure 3B). Notably, the mRNA induction of p53 target genes after Nutlin-3 treatment was markedly higher as compared with cells treated with daunorubicin. Although control and patient IV:7 fibroblasts expressed comparative total relative RNA levels upon daunorubicin treatment, the fold change upon daunorubicin induction varied between cell lines because of differing basal target gene expression. It is possible that the aging phenotype is mediated through pathways distinct from those activated by daunorubicin. Additionally, it is well known that the dynamic behavior of p53 varies on the basis of numerous factors such as the duration of the drug treatment and recovery period and the concentration of the drug used (29). In this experiment, we used a specific time point (24 hours) to investigate whether p53 could be activated in the patient’s fibroblasts. To investigate this further, we used different stressors, namely adriamycin, daunorubicin, mitomycin C (MMC), and aphidicolin (APH), and analyzed protein expression after different recovery periods (0, 4, 8, and 24 hours). Indeed, each treatment resulted in enhanced and prolonged p53 protein levels with concomitant p53 serine 15 phosphorylation in the patient’s fibroblasts (Figure 3C and Supplemental Figure 5). Additionally, with the caveat that a single homozygous clone was used, our data with genome-edited cells also showed that p53 was hyperactivatable after Nutlin-3 treatment, as evidenced by a much greater fold increase over basal RNA levels expressed from a set of p53 target genes (Figure 3D). Also important, we found that basal expression of these p53 targets was markedly lower in homozygous MUT/MUT cells than in the parental cells, with mRNA expression in the heterozygous cells at intermediate levels (Figure 3D). Altogether, these data indicate that mutant MDM2 is able to bind and repress the transcriptional activity of basal p53, but upon stress leads to p53 hyperactivation.
Figure 3. The antiterminating mutation results in abnormal p53 functional responses.
(A) MDM2 and p53 could be coimmunoprecipitated. Proteins were harvested from control and IV:7 LCLs and detected after coimmunoprecipitation with anti-MDM2 antibody (H-221) and subsequent immunoblotting. All samples were run on the same gel but were noncontiguous. (B) Control 1 and IV:7 fibroblasts were treated for 24 hours with DMSO, Nutlin-3 and daunorubicin. Cells were harvested and used for protein or mRNA analysis. Cells were lysed and used for immunoblotting with the indicated antibodies. Quantitative reverse transcription PCR (qRT-PCR) analysis revealed relative mRNA levels of p53 target genes after treatment. Bar graph is representative of 3 independent experiments and shows no statistical significance. (C) Time course of p-p53, p53, and MDM2 protein levels after treatment with MMC, APH, and daunorubicin. Protein levels of p-p53 (Ser15), p53, and MDM2 in fibroblasts treated with MMC (40 ng/ml), APH (0.3 μM), or daunorubicin (0.1 μM). Fibroblasts from control 1 (lanes 1–4) and IV:7 (lanes 5–8) were treated for 24 hours with the indicated stressors. Cells were harvested either right after treatment (lanes 1 and 5), or were further cultured for 4 (lanes 2 and 6), 8 (lanes 3 and 7), or 24 hours (lanes 4 and 8) in stress-free medium. Cell lysates were used for immunoblotting with p-p53 (Ser15); p53 (AF1355); MDM2 (N20); and anti-actin. (D) qRT-PCR analysis showing relative mRNA levels of p53 target genes after treatment with DMSO or Nutlin-3 for 24 hours in WT/WT, WT/MUT, and MUT/MUT U2OS genome-edited cells. Bar graph is representative of 3 independent experiments and shows no statistical significance for Nutlin-3–treated p21 WT/MUT and MUT/MUT cell lines as compared with WT/WT cell lines.
These findings prompted us to perform chromosome breakage analysis and cytokinesis-block micronucleus (CBMN) assays, the latter of which enables simultaneous monitoring of genomic stability and cellular proliferation in LCLs under basal and stressed conditions. In contrast to many other segmental progeroid syndromes (30), LCLs from the index patient had no genomic instability but rather showed a certain level of protection against ionizing radiation and MMC, in accordance with the pronounced p53 stabilization after MMC treatment (Figure 4, A and B). This protection, however, seemed to occur at the cost of cell viability, as calculated by the nuclear division index (NDI) (Figure 4A), a finding compatible with a decreased regenerative capacity and the development of signs of accelerated aging on an organismal level. Additionally, the patient’s fibroblasts had reduced replicative capacity and entered replicative senescence already at passage 28, whereas the control fibroblasts stopped replicating at passage 43, comparable to the findings in other progeroid syndromes (31). This was further confirmed by senescence-associated β-gal staining (Figure 4C) and loss of lamin B1 (LMNB1) (Figure 4D).
Figure 4. Functional consequences of the identified mutation in the patient’s cells.
(A) CBMN assays in LCLs. Effect of γ-irradiation (1 and 2 Gy) on the induction of micronuclei (MN) in control and IV:7 (left y axis) LCLs and on cell viability in the presence of CytB, indicated by the NDI (right y axis). Data represent 3 independent experiments. P = 0.2; 1 Gy: P = 0.0006; 2 Gy: P = 0.0001 (2-tailed Student’s t test). NT, nontreated cells. (B) Chromosomal stability in LCLs. Graph shows the mean number of aberrations per cell observed in 100 metaphases of an unaffected individual (Control LCLs) and the index patient (IV:7 LCLs). LCLs were left untreated or treated with 80 ng/ml MMC for 48 hours. Data represent 2 independent experiments. P values are relative to control LCLs for each treatment. #P = 0.0002 (2-sample Poisson tests). (C) Senescence phenotype of IV:7 fibroblasts. Control 1 and IV:7 fibroblasts at passage 28 (P28) were stained for β-gal. Original magnification ×20. (D) Protein levels of LMNB1 and actin in primary fibroblasts from an unaffected individual (Control 1) and the index patient IV:7 at passages 18 (both), 31 (control 1), and 27 (index patient).
Impaired Mdm2 function in zebrafish.
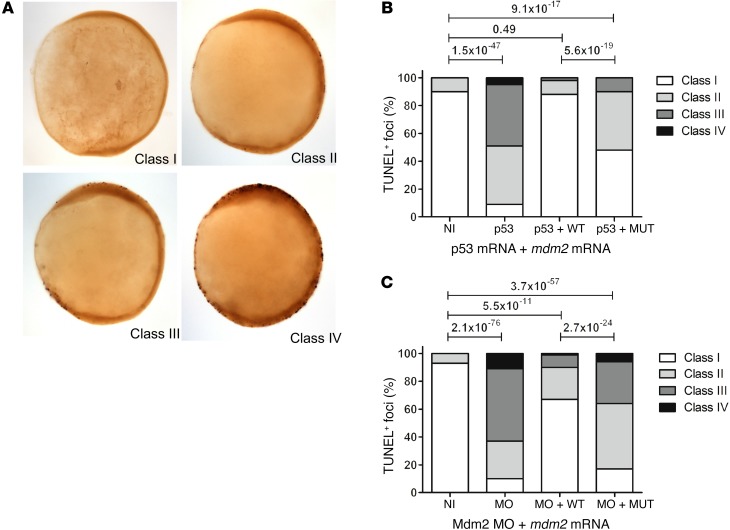
In order to confirm the impact of the identified MDM2 mutation on cellular outcomes at a physiological level in vivo, we used an established zebrafish model. As expected from previous work (32), embryos either deficient in Mdm2 or overexpressing p53 displayed a severe apoptotic phenotype. This phenotype could be rescued upon coinjection of WT mdm2 mRNA, but not by an equimolar amount of mutant mdm2 mRNA bearing an extension of the 5 amino acids identified in the index patient (Figure 5). These findings confirmed the evolutionarily conserved role of the Mdm2 C-terminal tail and the pathogenic nature of the identified mutation.
Figure 5. Impaired function of the antiterminating Mdm2 mutation in a zebrafish model.
Complementation assay in 10 hpf zebrafish embryos (tailbud stage). (A) Representative images of the lateral view of 10 hpf zebrafish embryos stained with TUNEL. Embryos were classified by the number of TUNEL-positive foci as follows: class I (0–10); class II (10–20); class III (20–40); and class IV (>40). (B and C) Percentage of TUNEL-positive foci of various apoptosis classes of embryos injected with 3 ng Mdm2 morpholino and coinjections of 40 pg mdm2 mRNAs and of embryos injected with p53 RNA and coinjections of 40 pg mdm2 mRNAs (right panel). NI, noninjected embryos; MO, embryos injected with Mdm2 morpholino; p53, embryos injected with p53 mRNA; + WT, coinjection of mdm2-WT; + MUT, coinjection of mdm2 bearing the antiterminating mutation. Data represent 3 independent experiments (2-sample Poisson tests).
Discussion
The human organism is under constant exposure to both endogenous and exogenous stressors, such as oxidative stress, environmental toxic substances, and UV light. If not dealt with, these threats can lead to DNA damage and cellular lesions that have the potential to harm the organism and limit survival. In order to cope with these pervasive threats, numerous cellular systems have evolved that can sensitively detect stressors and initiate adequate cellular responses to limit potential damage. In this respect, p53 plays an integrating and pivotal role in coordinating cellular reactions to stress signals, mainly by activating cell-cycle arrest, apoptosis, or senescence (2, 33). The beneficial role of p53 as a tumor suppressor and the consequences of loss of p53 activity in human cancers have been intensively studied. It therefore seems logical that upregulation of WT p53 could, in principle, represent an attractive therapeutic approach (34). Indeed, numerous studies report the identification of drugs that specifically target the MDM2/p53 axis for cancer therapy, with clinical trials already underway (35–37). Since the potential effects of increased p53 activation on an organismal level in humans are still unknown, the detailed elucidation of the possible effects of p53 activation on MDM2 inhibition is of growing biomedical importance. In this study, we have described the identification and characterization of a germline antiterminating MDM2 mutation in a patient with a segmental progeroid syndrome. Although we describe a single patient and his family, the combination of (a) disease gene locus refinement by homozygosity mapping, (b) the systematic analysis of all homozygous variants by exome sequencing and, especially, (c) the demonstration of altered regulation of and by p53 in cells from this specific patient fulfill previously proposed criteria for identification of novel disease genes in single patients (38). Our studies indicate that dysfunctional regulation of p53 by MDM2 can lead to aberrant p53 activity that poses deleterious effects on human aging processes.
Until now, the strongest evidence linking aging and dysregulated p53 has come from engineered mouse models. Nonetheless, it is not a given that any genetic variant in mice will be phenocopied in humans. Two studies involving mice with N-terminal truncated p53 in the presence of functional WT p53 revealed accelerated aging phenotypes (19, 20). These mice appear to have aberrant MDM2 regulation of p53, and mouse embryonic fibroblasts from these mouse models express normal p53 protein levels but have constitutively active p53 (39). Further, mice with mutations in key N-terminal phosphorylation sites that can inhibit interaction with MDM2 also display an aging phenotype (40). Yet other studies have argued against a role for p53 in aging. In 1 case, super-p53–transgenic mice harboring an extra copy of DNA spanning the p53 genomic locus appear to age normally (23). Similarly, mice that are hypomorphic for MDM2 have no signs of early aging (22), while other mice bearing extra copies of both p53 (super-p53) and ARF actually experience delayed onset of aging (41). Notably, the above-mentioned mice without signs of premature aging have intact p53/MDM2 circuitry, indicating that p53 in these mice is regulated by MDM2 normally. In contrast, the mutant p53 mice showing premature signs of aging collectively feature abnormal MDM2/p53 regulation due to mutant p53 alleles that cannot interact with MDM2. Relatedly, conditional mice with targeted deletion of Mdm2 in the epidermis show characteristics of aging skin and have stable p53 (42).
Engineered mice with altered MDM2 have also greatly contributed to our understanding of the MDM2/p53 axis. Mice null for MDM2 expression or expressing a key MDM2 RING domain mutation that abrogates E3 ligase activity (p.Cys462Ala) display early embryonic lethal phenotypes unless the mice are generated in a p53-null background (43–45). These data indicate that MDM2 requires functional E3 ubiquitin ligase activity to restrain p53 for normal development. The extreme C-terminus of MDM2 plays a critical role in RING-RING interactions and is required for MDM2 oligomerization and E3 ligase activity in human cells (25, 26). Yet mice engineered with a mutation in the extreme C-terminus of MDM2 (p.Tyr487Ala) that also affects RING-RING interactions develop and age normally (46). Reports that defective E3 ligase activity due to mutations or extensions in the extreme C terminus of MDM2 can be partially rescued by coexpression of the MDM2 homolog MDMX could explain the viability of the Tyr487Ala mouse in comparison with the Cys462Ala mouse (25–27). However, since MDM2 variants with mutations in the last 5 amino acids of MDM2 share characteristic properties with those of antiterminating MDM2 mutations, it is surprising that the Tyr487Ala mouse does not display some segmental progeroid characteristics (25-27). We speculate that key differences between our index patient and Tyr487Ala mice could account for the different phenotypes. Specifically, the enhanced MDM2 stability, levels of basal p53 target genes, and hyperactivation of the p53 transcriptional program in the patient’s cells differ from the reported characteristics of the Tyr487Ala mouse. Additionally, laboratory mice could be spared exposure to many naturally occurring stresses or environmental stimuli, which could be important cofactor(s) in the normal or pathological aging process.
We postulate that either the ability of p53 to be hyperactivated or its prolonged activation due to the impaired ability of the antiterminating mutant MDM2 in downregulating p53 after stress might be critical factors contributing to the observed clinical phenotype in the index patient. Thus, we hypothesize that in the index patient, certain everyday stresses generate a chronic condition, whereby pervasive and pronounced deleterious activation of p53 downstream targets induces a program of accelerated aging. Indeed, elevated levels of p53 with concomitant activation of the p53 pathway have been previously identified in several other segmental progeroid syndromes, such as ataxia telangiectasia, Hutchinson-Gilford progeria, and Cockayne syndrome (18, 21, 47). Further, we should consider the possibility that basally underexpressed p53 target genes in the patient-derived and engineered cells are also contributing to one or more of the characteristics of the patient, as basal p53 has been implicated in survival functions and regulation of longevity-associated processes (3). In addition, p53 crosstalk with other pathways adds another layer of complexity to delineating the underlying mechanism(s) linking p53 and aging (48–50). Nor can we exclude the possibility that p53-independent activities of MDM2 might also contribute to the observed phenotype (6, 51).
Indeed, p53 activation is stressor dependent and may lead to abnormally enhanced levels of autophagy, cell-cycle arrest, apoptosis, or senescence that can ultimately cause tissue and organ dysfunction resulting in accelerated aging symptoms (52). Two common hallmarks in both segmental forms of progeria and physiological aging are genomic instability and cellular senescence (49, 53). However, unlike the situation in other progeroid syndromes that derive from defects in genome maintenance proteins and thus exhibit diminished genomic integrity, we observed increased chromosomal stability in the patient’s cells. The latter is largely in accordance with prolonged p53 hyperactivation, which could potentially promote DNA repair mechanisms and enhance genome stability (3). On the other hand, we observed premature senescence in the patient’s fibroblasts. Although we acknowledge that it is almost impossible to exactly define which of the numerous cellular outcomes contribute to the progeroid phenotype, as the p53 response is cell-, tissue-, and stress-type dependent, our data implicate senescence as playing a role in facilitating the aging symptoms in the patient.
In the family studied here, idiopathic, early-onset kidney failure followed by arterial hypertension and the family’s predisposition to myocardial infarction seem to be the most malicious outcomes at the organismal level. Intriguingly, in addition to being essential for maintenance of the nephron progenitor niche (53), studies in mice have suggested that the MDM2/p53 axis specifically regulates kidney-specific cell healing, development, and the response to inflammation (54, 55). Our data, even though based on a single patient, may indicate a potentially increased risk of side effects for systemic, long-term therapies with MDM2 inhibitors. Special emphasis on the parameters of kidney function may therefore be advisable for the clinical trials already underway involving synthetic small molecules that interfere with MDM2-p53 interaction.
In summary, we report on a patient who developed — at a rather early age — several symptoms that are typically observed in the elderly, compatible with the diagnosis of a segmental progeroid syndrome (56). By combining up-to-date genetic approaches including genome-wide linkage mapping and massively parallel sequencing with in-depth functional characterization of patient-derived primary cell lines with complementary in vitro and in vivo analyses, we identified a homozygous germline antiterminating MDM2 mutation as the genetic cause of his condition. Taken together, our evidence provides strong support for a role for functional disturbance of the MDM2/p53 axis in promoting a premature aging phenotype in humans.
Methods
Cell lines.
EBV-transformed LCLs and primary dermal fibroblasts were established as previously described (57). Previously used LCLs from an unaffected individual, AG1010 (24), termed control LCLs, were used in all experiments. In experiments with primary dermal fibroblasts, a previously used cell line from a healthy individual, 82-6 (30), termed control fibroblasts 1, was used for all experiments. To corroborate the findings in some experiments, additional cell lines from unaffected individuals (IMR-90, NHDF, and 1101-SK) were used and are termed control fibroblasts 2, 3, and 4, respectively. 88-1 normal fibroblasts were also used. Primary dermal fibroblasts were maintained in DMEM supplemented with 10% FBS. Human osteosarcoma (U2OS) and non–small-cell lung cancer (H1299) cell lines were also maintained in DMEM supplemented with 10% FBS. LCLs were maintained in RPMI medium supplemented with 10% FBS. Cell lines were obtained from Junko Oshima, Lyubomir Vassilev (Serono Research and Development Institute, Billerica, MA, USA), and ATCC. For replicative capacity studies, primary dermal fibroblasts from the index patient and control fibroblasts 1 were thawed at passage 6 and passaged at 80% confluency. Fibroblasts were not allowed to become fully confluent at any time in order to maintain maximal growth rates.
Homozygosity mapping and candidate gene sequencing.
Genome-Wide Human SNP Array 6.0 (Affymetrix) was used to exclude the possibility of genomic rearrangements and map putative homozygous regions. Genome-wide homozygosity scores were produced by HomozygosityMapper, a Web-based approach to homozygosity mapping (58). Data handling, evaluation, and statistical analysis were previously described in detail (30). To confirm the homozygous antiterminating mutation, c.1492T>C, in MDM2, we designed primers, 5´- gagggctttgatgttcctga-´3 and 5´-ggagttggtgtaaaggatgagc-´3, and PCR amplified it by using genomic DNA from the affected individual. PCR products were sequenced on an ABI 3730 DNA Analyzer with BigDye Chemistry, version 3.1 (Applied Biosystems). Sequence traces were assembled, aligned, and analyzed with Seqman software (DNASTAR Lasergene).
Exome-sequencing analysis.
Exome sequencing of the proband was performed on 2 lanes of an Illumina GAIIx Sequencer using a single-read 150-bp protocol after enrichment of exonic and splice-site sequences with the Agilent SureSelect Human All Exon 50 Mb Kit. More than 194 million reads were mapped to the hg19 human reference genome. Approximately 93% of target sequences had at least 10-fold coverage and 75% had 30-fold coverage, with a mean coverage of approximately 91. Data analysis of filter-passed reads was performed with the in-house pipeline V1.3 using BWA-short in combination with SAMTOOLS pileup 0.1.7 for the detection of SNPs and short insertions and deletions (indels). In-house–developed scripts were applied to detect protein changes, affected splice sites, and overlaps to known variations, with filtering against dbSNP build 138, the 1000 Genomes Project data build November 2014, ExAC Browser (status from April 2015), and the in-house database of exome variants (with data from >200 exomes of individuals affected by different disorders). The criteria for a variation to be taken into account were: more than 6 reads; a phred scaled quality score of greater than 15; a population allele frequency of less than 1% seen fewer than 10 times in our in-house database; and more than 15% of the reads supporting the allele.
Plasmids.
Full-length zebrafish mdm2 and p53 and human MDM2 cDNAs were cloned into pCS2+ and pcDNA3.1 (Invitrogen, Thermo Fisher Scientific), respectively. Mutants were cloned using PCR amplification and restriction enzyme digestion and recombination. Site-directed mutagenesis was performed by PCR to introduce the desired mutations. The correctness of the DNA sequence was verified by sequencing.
Genome-edited cell lines.
Two TALEN plasmids were designed to target nucleotides within the intron between exons 10 and 11 (taccttagacatagcaaa[G]ttgctagcattcctgtgactgagcagtta, where [G] indicates a TALEN cutting site, and the 2 TALEN binding sites are underlined bold), as recommended by ZiFiT Targeter software (http://zifit.partners.org/ZiFiT/TALZiFiTNuclease.aspx). A donor vector with two 1-kb homologous arms plus an FNF selection cassette (Frt-EM7- NeoR-Frt) was also constructed. The FNF selection cassette was inserted into the intron between exons 10 and 11 of the mdm2 gene to facilitate clone selection as well as a unique HindIII site in exon 11. A T to C mutation was introduced into the donor vector at the stop codon TGA by site-directed mutagenesis. This resulted in an MDM2 variant with a 5–amino acid extension. The donor and TALEN plasmids were transfected into U2OS cells, followed by selection with G418 (800 μg/ml) for 2 to 3 weeks. The resistant clones were expanded and examined for FNF insertion on the mdm2 locus by PCR. The corrected clones were confirmed by sequencing to ensure that the allele carrying the mutation was corrected. Both heterozygous and homozygous clones were obtained. To serve as controls, isogenic WT clones were also obtained through the same selection process.
Mutant MDM2-p53–KO cells were generated using CRISPR/Cas9 genome editing technology. Cells (7 × 105) were transfected with 2 μg p53 CRISPR/Cas9–KO plasmid (Santa Cruz Biotechnology Inc.). Two days later, cells were treated with Nutlin-3 (10 μM) for six days to inhibit the proliferation of cells with WT p53, thereby enriching for p53-KO cells. Single-cell clones were selected via limiting dilution, and p53-KO clones were confirmed by Western blotting using FL-393 (Santa Cruz Biotechnology Inc.) rabbit polyclonal antibody.
Immunoblotting.
Whole-cell lysates were analyzed by standard immunoblotting procedures. Protein concentration was measured using the Bio-Rad protein assay (Life Science Research). Equal amounts of total protein were resolved on 8% or 12% polyacrylamide gels (for ectopic and fibroblast/LCL work, respectively), transferred to a nitrocellulose membrane, and then blocked for 30 minutes in PBS containing 0.1% Tween-20 (PBST) (Sigma-Aldrich) and 5% nonfat dry milk. The membrane was incubated overnight at 4°C with primary antibodies in 1% milk in PBST. The commercial primary antibodies used include: anti-FLAG (M2; Sigma-Aldrich); anti-HA (6B12; Covance); anti-p53 (AF1355; R&D Systems); anti-MDM2 (N20 or H-221; Santa Cruz Biotechnology Inc.); anti–phosphorylated p53 (anti–p-p53) (Ser15) (Cell Signaling Technology); anti–lamin B1 (S20; Santa Cruz Biotechnology Inc.); anti-p21 (C-19; Santa Cruz Biotechnology Inc.); anti-GFP (B2; Santa Cruz Biotechnology Inc.); anti–γ-tubulin (T5192; Sigma-Aldrich); and anti-actin (A2066; Sigma-Aldrich). The monoclonal antibodies used as in-house–produced hybridoma supernatants include: anti-p53 (1801/DO-1) and anti-MDM2 (2A9, 3G5, 4B11, and 5B10). After 3 washes with PBST, membranes were incubated with secondary antibodies in 1% milk in PBST for 30 minutes at room temperature (RT). The secondary antibodies used include goat anti-mouse or goat anti-rabbit conjugated to HRP (Sigma-Aldrich) or fluorescent green goat anti-mouse and fluorescent red donkey anti-rabbit (IRDye 800CW and IRDye 680LT; LI-COR Biosciences). Membranes were visualized using either ECL (GE Healthcare or Thermo Fisher Scientific) or the Odyssey Imaging System (LI-COR Biosciences).
Drugs.
The following drugs were used: Nutlin-3 (a racemic mixture of active Nutlin-3a and inactive Nutlin-3b; Sigma-Aldrich); CHX (Calbiochem); daunorubicin (Sigma-Aldrich); MG132 (Calbiochem); MMC (Sigma-Aldrich); cytochalasin B (CtyB) (Sigma-Aldrich); APH (Sigma-Aldrich); and adriamycin (Sigma-Aldrich).
Immunofluorescence analyses.
U2OS, control 1, and IV:7 primary fibroblasts were grown on coverslips in 35-mm Petri dishes. Cells were washed twice with PBS and fixed with 4% paraformaldehyde solution in PBS for 20 minutes at RT. Following 2 additional washes, fixed cells were permeabilized with 0.5% Triton X-100 in PBS at RT for 1.5 minutes, washed twice, blocked with 0.5% BSA in PBS for 30 minutes at RT, incubated with primary antibodies at RT for 1 hour in blocking solution, washed thrice, incubated with a mixture of secondary antibodies conjugated with fluorescent dye (Alexa Fluor 488 and Alexa Fluor 594; Life Technologies, Thermo Fisher Scientific) at RT for 1 hour in the dark, washed thrice, and then mounted with VECTASHIELD mounting medium for fluorescence with DAPI (Vector Laboratories). The following primary antibodies were used: anti–MDM2 N20 (Santa Cruz Biotechnology Inc.); anti–p53 1801/DO-1 (produced in-house); anti-HA (16B12; Covance); and anti-FLAG (M2; Sigma-Aldrich). Images were taken on a Zeiss LSM 700 confocal microscope.
Quantitative reverse transcription PCR.
RNA was isolated from cultured cells using the RNeasy Mini Kit (QIAGEN) and then converted into cDNA using the QuantiTect Reverse Transcription Kit (QIAGEN). PCR was performed using the StepOne Real-time PCR system with Power SYBR Green PCR Master Mix (Applied Biosystems). Relative mRNA levels were calculated by the ΔΔCt method and normalized against control L32 mRNA levels.
Cytokinesis-block micronucleus assay.
The CBMN assay was performed with cells from patient IV:7 and AG 1011 cells. Cells (2.5 million) were cultivated in 10 ml RPMI medium irradiated with 1 Gy or 2 Gy γ rays using a Cs-137 source (Gammacell 2000; Nuclear Data). After irradiation, CytB was added to the cultures at a final concentration of 2 μg/ml. Cultures were harvested 41 hours later by centrifugation, treated with a hypotonic solution (0.56% KCl), and fixed once with methanol/glacial acetic acid (5 + 1) mixed with an equal amount of 0.9% NaCl. Cells were then fixed 3 times with methanol/glacial acetic acid (5 + 1). Air-dried slides were stained with acridine orange (60 μg/ml in phosphate buffer). The frequency of micronucleated cells was determined by analyzing 1,000 binuclear cells from coded slides. Toxicity was measured using the nuclear division index (NDI), which was calculated from 500 cells according to the formula: NDI = (M1 + 2M2 + 3M3 + 4M4 + 5M5)/N, where M1 – M5 indicates the number of cells with 1 to5 nuclei and N the total number of cells scored.
Chromosome analysis.
Exponentially growing LCLs were reseeded into culture flasks at a density of 2 × 105/ml, left untreated for an additional 24 hours, or incubated with 80 ng/ml MMC for 48 hours. Cells were exposed to 100 ng/ml colcemid for 2 hours, treated with hypotonic solution (NaCl/sodium-citrate, 0.4 %) for 15 minutes, and fixed with 3:1 methanol/acetic acid. Slides were stained with Giemsa, and 100 metaphase spreads were scored for chromosomal aberrations in 2 independent experiments. Metaphase spreads were observed using a Leitz Laborlux S microscope and captured using CytoVision (Applied Imaging). Data for chromosomal breaks in LCLs were compared (untreated and MMC-treated) using Poisson regressions with log link functions, with the number of observed metaphases as an offset variable. AG1010 LCLs served as the reference. P values were not adjusted for multiple testing.
Detection of senescence-associated β-gal activity.
Endogenous mammalian senescence-associated β-gal activity (SAβ-gal) was evaluated using the Senescence β-Galactosidase Staining Kit (Cell Signaling Technology) according to the manufacturer’s guidelines.
Zebrafish maintenance and manipulation.
WT Tupfel long fin (TL) zebrafish were maintained following standard protocols. mdm2 RNA (3 ng) antisense morpholino oligonucleotides (MO) or 40 pg capped RNA, transcribed with the mMessage mMachine Kit (Ambion) from linearized plasmids, were injected into the blastodisc prior to the first cleavage. Embryos were kept at 28.5°C up to the tailbud stage (10 high-power fields [hpf]) and analyzed and processed. Concentrations of injected MO and RNA were calculated, and volumes were adjusted in mineral oil for reproducible injection volumes prior to microinjections. RNA quality was analyzed prior to injections by using 1.5% agarose gel electrophoresis. Phenol red (0.5%) and rhodamine-dextrane (0.5%) were coinjected in order to monitor the equal distribution of the solution during injection. Six hours after injection, embryos were sorted for homogeneity of the injected solution using a fluorescence stereoscope. A previously established splice-site mdm2 MO (5′-TGTTAAGAGATTCAGTACGCACCGC-3′) (59) was used to generate loss-of-function zebrafish. Detection of apoptotic cells by TUNEL staining at 10 hpf was performed using the In Situ Cell Death Detection Kit (Roche) as described previously (60).
Statistics.
Statistical evaluation was performed depending on the experiment by using a 2-tailed Student′s t test or 2-sample Poisson tests, as specifically indicated for each experiment in the figure legends. A P value of less than 0.05 was considered significant.
Study approval.
All biological samples and photographs were obtained following written informed consent from the patient. The International Registry of Werner Syndrome has been recruiting patients suspected of Werner syndrome since 1988. The study has ongoing approval from the IRB of the University of Washington. This study was performed in accordance with Declaration of Helsinki protocols.
Author contributions
DL, DW, C. Kubisch, CP, YZ, DT, IL, TR, MH, C. Katz, KR, IG, PS, and GS designed and performed the experiments. JH performed statistical analyses. HT, GN, and PN performed homozygosity mapping and exome sequencing. CT, MKA, BS, FMH, GMM, and JO provided the patient’s samples and information. DL, DW, CP, and C. Kubisch wrote the manuscript.
Supplementary Material
Acknowledgments
We are thankful to the family members for their participation. This work was supported by grants from the NIH (CA58316, to CP, and R24AG042328, to GMM and JO) and the German Research Foundation (DFG) in the framework of the Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (to C. Kubisch). DW was supported by a National Science Foundation Graduate Research Fellowship (DGE-11-44155). Any opinion, findings, and conclusions or recommendations expressed here are those of the authors(s) and do not necessarily reflect the views of the National Science Foundation.
Version 1. 08/24/2017
Electronic publication
Version 2. 09/20/2017
Print issue publication
Footnotes
TR’s present address is: MRC Centre for Inflammation Research, Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, United Kingdom.
YZ’s present address is: Department of Biological Sciences, St. John’s University, Queens, New York, USA.
Conflict of interest: P. Nürnberg is a founder, CEO, and shareholder of ATLAS Biolabs GmbH.
Reference information: J Clin Invest. 2017;127(10):3598–3608.https://doi.org/10.1172/JCI92171.
Contributor Information
Davor Lessel, Email: d.lessel@uke.de.
Danyi Wu, Email: dw2506@columbia.edu.
Carlos Trujillo, Email: cjtrujillo@gmail.com.
Thomas Ramezani, Email: thomas.ramezani@gmail.com.
Ivana Lessel, Email: ivana.lessel@gmail.com.
Bidisha Saha, Email: bidishasaha@gmail.com.
Katrin Rading, Email: k.rading@uke.de.
Ingrid Goebel, Email: ingridgoebel@gmx.de.
Petra Schütz, Email: petra.schuetz@uni-ulm.de.
Günter Speit, Email: g.speit@mailbox.org.
Josef Högel, Email: josef.hoegel@uni-ulm.de.
Holger Thiele, Email: holger.thiele@uni-koeln.de.
Matthias Hammerschmidt, Email: mhammers@uni-koeln.de.
Yan Zhu, Email: yz2134@gmail.com.
David R. Tong, Email: drt2118@columbia.edu.
Chen Katz, Email: ck2563@columbia.edu.
George M. Martin, Email: gmmartin@u.washington.edu.
Junko Oshima, Email: picard@u.washington.edu.
Carol Prives, Email: clp3@columbia.edu.
Christian Kubisch, Email: c.kubisch@uke.de.
References
- 1.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460(7259):1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17(1):86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20(5):299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 8.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12(2):461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B, Golding BT, Hardcastle IR. Small-molecule MDM2-p53 inhibitors: recent advances. Future Med Chem. 2015;7(5):631–645. doi: 10.4155/fmc.15.13. [DOI] [PubMed] [Google Scholar]
- 11.Kulju KS, Lehman JM. Increased p53 protein associated with aging in human diploid fibroblasts. Exp Cell Res. 1995;217(2):336–345. doi: 10.1006/excr.1995.1095. [DOI] [PubMed] [Google Scholar]
- 12.Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci U S A. 2007;104(42):16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ørsted DD, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J Exp Med. 2007;204(6):1295–1301. doi: 10.1084/jem.20062476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergamaschi D, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet. 2006;38(10):1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- 15.Dumont P, Leu JI, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 16.van Heemst D, et al. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol. 2005;40(1-2):11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312(5776):1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varela I, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437(7058):564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 19.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 20.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18(3):306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, et al. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol. 2010;12(10):993–998. doi: 10.1038/ncb2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendrysa SM, et al. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20(1):16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Cao I, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21(22):6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lessel D, et al. POLD1 Germline Mutations in Patients Initially Diagnosed with Werner Syndrome. Hum Mutat. 2015;36(11):1070–1079. doi: 10.1002/humu.22833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poyurovsky MV, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26(1):90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26(1):102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolezelova P, Cetkovska K, Vousden KH, Uldrijan S. Mutational analysis of Mdm2 C-terminal tail suggests an evolutionarily conserved role of its length in Mdm2 activity toward p53 and indicates structural differences between Mdm2 homodimers and Mdm2/MdmX heterodimers. Cell Cycle. 2012;11(5):953–962. doi: 10.4161/cc.11.5.19445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-R. [DOI] [PubMed] [Google Scholar]
- 29.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336(6087):1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessel D, et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat Genet. 2014;46(11):1239–1244. doi: 10.1038/ng.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tivey HS, Brook AJ, Rokicki MJ, Kipling D, Davis T. p38 (MAPK) stress signalling in replicative senescence in fibroblasts from progeroid and genomic instability syndromes. Biogerontology. 2013;14(1):47–62. doi: 10.1007/s10522-012-9407-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langheinrich U, Hennen E, Stott G, Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol. 2002;12(23):2023–2028. doi: 10.1016/S0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- 33.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoo KH, Hoe KK, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13(3):217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q, Zeng SX, Lu H. Targeting p53-MDM2-MDMX loop for cancer therapy. Subcell Biochem. 2014;85:281–319. doi: 10.1007/978-94-017-9211-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreeff M, et al. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin Cancer Res. 2016;22(4):868–876. doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner AJ, et al. Phase I trial of the human double minute 2 inhibitor MK-8242 in patients with advanced solid tumors. J Clin Oncol. 2017;35(12):1304–1311. doi: 10.1200/JCO.2016.70.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med. 2014;211(11):2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore L, Lu X, Ghebranious N, Tyner S, Donehower LA. Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity. Mech Ageing Dev. 2007;128(11-12):717–730. doi: 10.1016/j.mad.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donehower LA. Using mice to examine p53 functions in cancer, aging, and longevity. Cold Spring Harb Perspect Biol. 2009;1(6):a001081. doi: 10.1101/cshperspect.a001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448(7151):375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 42.Gannon HS, Donehower LA, Lyle S, Jones SN. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Dev Biol. 2011;353(1):1–9. doi: 10.1016/j.ydbio.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12(4):355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378(6553):206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 45.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 46.Tollini LA, Jin A, Park J, Zhang Y. Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell. 2014;26(2):235–247. doi: 10.1016/j.ccr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latini P, et al. CSA and CSB proteins interact with p53 and regulate its Mdm2-dependent ubiquitination. Cell Cycle. 2011;10(21):3719–3730. doi: 10.4161/cc.10.21.17905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poyurovsky MV, Prives C. P53 and aging: A fresh look at an old paradigm. Aging (Albany NY) 2010;2(7):380–382. doi: 10.18632/aging.100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campisi J. Fragile fugue: p53 in aging, cancer and IGF signaling. Nat Med. 2004;10(3):231–232. doi: 10.1038/nm0304-231. [DOI] [PubMed] [Google Scholar]
- 51.Li Q, Lozano G. Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clin Cancer Res. 2013;19(1):34–41. doi: 10.1158/1078-0432.CCR-12-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26(9):1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 53.Hilliard SA, Yao X, El-Dahr SS. Mdm2 is required for maintenance of the nephrogenic niche. Dev Biol. 2014;387(1):1–14. doi: 10.1016/j.ydbio.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulay SR, Thomasova D, Ryu M, Anders HJ. MDM2 (murine double minute-2) links inflammation and tubular cell healing during acute kidney injury in mice. Kidney Int. 2012;81(12):1199–1211. doi: 10.1038/ki.2011.482. [DOI] [PubMed] [Google Scholar]
- 55.El-Dahr S, Hilliard S, Aboudehen K, Saifudeen Z. The MDM2-p53 pathway: multiple roles in kidney development. Pediatr Nephrol. 2014;29(4):621–627. doi: 10.1007/s00467-013-2629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin GM. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig Artic Ser. 1978;14(1):5–39. [PubMed] [Google Scholar]
- 57.Huang S, et al. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27(6):558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seelow D, et al. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37(Suppl 2):W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parant JM, George SA, Holden JA, Yost HJ. Genetic modeling of Li-Fraumeni syndrome in zebrafish. Dis Model Mech. 2010;3(1-2):45–56. doi: 10.1242/dmm.003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cole LK, Ross LS. Apoptosis in the developing zebrafish embryo. Dev Biol. 2001;240(1):123–142. doi: 10.1006/dbio.2001.0432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.