Abstract
The mammalian target of rapamycin complex 1 (mTORC1) kinase promotes cell growth by activating biosynthetic pathways and suppressing catabolic pathways, particularly that of macroautophagy. A prerequisite for mTORC1 activation is its translocation to the lysosomal surface. Deregulation of mTORC1 has been associated with the pathogenesis of several diseases, but its role in skeletal disorders is largely unknown. Here, we show that enhanced mTORC1 signaling arrests bone growth in lysosomal storage disorders (LSDs). We found that lysosomal dysfunction induces a constitutive lysosomal association and consequent activation of mTORC1 in chondrocytes, the cells devoted to bone elongation. mTORC1 hyperphosphorylates the protein UV radiation resistance–associated gene (UVRAG), reducing the activity of the associated Beclin 1–Vps34 complex and thereby inhibiting phosphoinositide production. Limiting phosphoinositide production leads to a blockage of the autophagy flux in LSD chondrocytes. As a consequence, LSD chondrocytes fail to properly secrete collagens, the main components of the cartilage extracellular matrix. In mouse models of LSD, normalization of mTORC1 signaling or stimulation of the Beclin 1–Vps34–UVRAG complex rescued the autophagy flux, restored collagen levels in cartilage, and ameliorated the bone phenotype. Taken together, these data unveil a role for mTORC1 and autophagy in the pathogenesis of skeletal disorders and suggest potential therapeutic approaches for the treatment of LSDs.
Keywords: Bone Biology, Therapeutics
Keywords: Autophagy, Collagens, Lysosomes
Introduction
Endochondral ossification is the coordinated process by which long bones develop (1). In humans, it begins during embryogenesis and is completed by puberty. Skeletal elements are composed of 2 distinct tissue types, cartilage and bone, and 3 distinct cell types: chondrocytes (in cartilage) and osteoblasts and osteoclasts (in bone) (1). Chondrocytes reside within articular cartilages and growth plates. Articular cartilage is found on the articular surfaces of bones and provides a lubricated surface for articulation. The growth plate, through the sequence of chondrocyte proliferation, hypertrophic differentiation, and replacement of the cartilaginous extracellular matrix (ECM) by an osseous one, is responsible for the longitudinal growth of the skeleton. In bone, osteoblasts produce and mineralize the matrix that forms the osseous tissue. Osteoclasts are multinucleated cells of myeloid origin deputed to the resorption of the bone matrix produced by osteoblasts. The balance between bone formation and bone resorption determines bone mass. The ECM, produced by chondrocytes and osteoblasts, is particularly enriched in collagens, type II (COL II) in cartilage and type I (COL I) in bones. Collagens are fundamental components of the ECM and confer biomechanical proprieties to tissues and contribute to bone elongation and maintenance. Consistently, defects in collagen synthesis and secretion cause conditions such as osteogenesis imperfecta, chondrodysplasias, and Ehlers-Danlos syndrome (2).
The lysosome is a catabolic organelle devoted to the degradation of biological substrates through the coordinated activity of hydrolases that reside within its lumen (3). Defects in lysosomal function cause lysosomal storage diseases (LSDs), a group of approximately 50 rare inherited metabolic disorders (4). Although each disorder results from mutations in different genes, they all share a common biochemical characteristic: abnormal accumulation of substances in the lysosome resulting in lysosomal dysfunction. Bone growth retardation and skeletal dysostosis are present in approximately 50% of the patients affected by LSDs, suggesting a critical role for the lysosome during bone development and maintenance (5). To date, the biological mechanisms at the basis of the skeletal phenotype observed in LSDs have not been elucidated, nor has an effective therapy been developed.
Recently, the lysosome has emerged as a key signaling hub that regulates, and is in turn regulated by, the activity of signaling molecules (6). The crosstalk between the lysosome and the serine/threonine protein kinase mTORC1, a master regulator of energy metabolism and cell growth, represents a notable example. Amino acids, the most potent activators of mTORC1, mediate the translocation of mTORC1 from the cytoplasm to the lysosomal surface, where the lysosome-anchored GTPase Rheb promotes the kinase activity of mTORC1 and enables substrate phosphorylation (7). Activated mTORC1 suppresses lysosome catabolism by inhibiting the nuclear translocation of TFEB and TFE3, the master transcriptional regulators of lysosome biogenesis and function (8–10). In addition, mTORC1 limits the delivery of substrates to the lysosomes by suppressing (macro)autophagy, an evolutionarily conserved trafficking pathway that relies on the biogenesis of autophagic vacuoles (AVs) that sequester intracellular cargoes and deliver them to the lysosome upon AV-lysosome fusion (11). Briefly, mTORC1 inhibits AV biogenesis by phosphorylating ULK1 and ATG13, members of the ULK1 autophagy initiation kinase complex (12). In addition, mTORC1 suppresses AV-lysosome fusion via the phosphorylation of UV radiation resistance–associated gene (UVRAG) protein (13). Together with Beclin 1, Vps34, and Vps15, UVRAG forms the endosomal class III–Vps34 complex that catalyzes the production of phosphatidylinositol 3-phosphate (PI3P) required for the fusion of vesicles and (auto)phagosomes with lysosomes (14–17). UVRAG also regulates AV-lysosome fusion by interacting with members of the homotypic fusion and vacuole protein–sorting (HOPS) complex that tethers membranes to promote fusion events (18). mTORC1 phosphorylates UVRAG at serine 497 and enhances its affinity for RUN domain Beclin 1–interacting and cysteine-rich–containing protein (RUBICON), which antagonizes UVRAG activity (13).
In recent years, the lysosomal autophagy pathway has emerged as a key pathway through which misfolded and aggregated proteins from the ER are disposed of as an alternative to ER-associated degradation (ERAD) (19). We recently demonstrated that autophagy in chondrocytes, the main cells deputed to bone growth, removes defective and excessive collagen molecules from the ER, thereby clearing folding and assembly machineries in the secretory pathway (20). Mice that lack an active autophagy pathway in chondrocytes (in which the Atg7 gene has been deleted) display defective collagen secretion, characterized by intracellular accumulation of type II procollagen (PC2) and lower COL II levels in the ECM and resulting in impaired postnatal bone growth (20).
To date, however, whether an impairment in the autophagy pathway may play a role in the pathogenesis of skeletal disorders is still unknown. Therefore, we decided to investigate the role of mTORC1 and autophagy in the pathogenesis of the skeletal features observed in LSDs.
Results
Enhanced mTORC1 signaling in LSD chondrocytes.
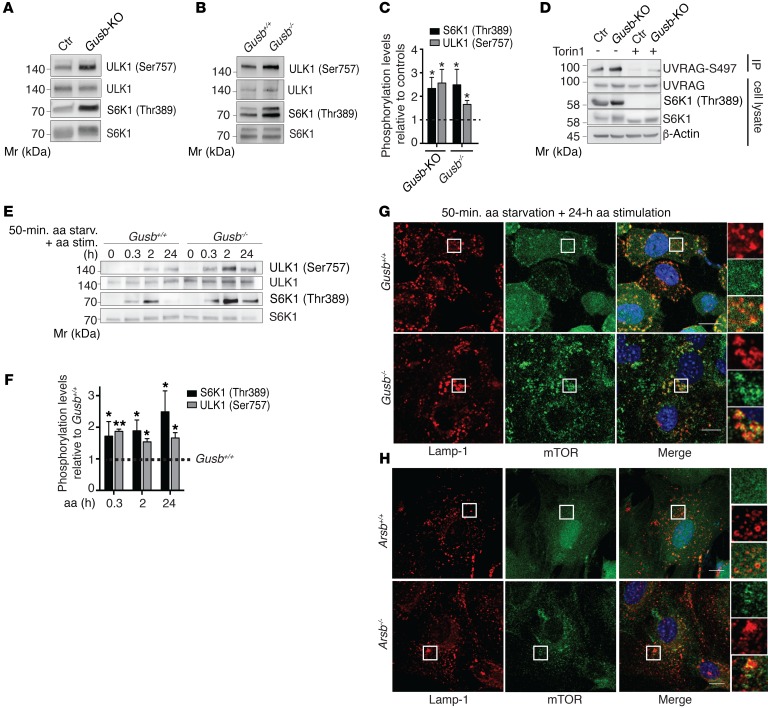
Mucopolysaccharidoses (MPSs) are the most prevalent subfamily of LSDs with skeletal abnormalities (21). They are caused by deficiency of a specific lysosomal enzyme responsible for the breakdown of glycosaminoglycans. We generated a chondrocyte cell line model of mucopolysaccharidosis type VII (MPSVII), in which the lysosomal β-glucuronidase gene (Gusb) was deleted through CRISPR/Cas9-mediated genome editing (Gusb-KO) (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI94130DS1). We observed lysosomal vacuolization in Gusb-KO chondrocytes (Supplemental Figure 1C), suggesting that they recapitulate the characteristic feature found in primary cells isolated from LSD models (Supplemental Figure 1D) (22). Interestingly, we observed increased mTORC1 activity in Gusb-KO cells compared with that seen in control parental cells, as measured by the phosphorylation levels of P70S6K and ULK1 (Figure 1A and quantification in Figure 1C). Furthermore, the phosphorylation level of the UVRAG protein at serine 497 was increased in an mTORC1-dependent manner in Gusb-KO chondrocytes compared with control chondrocytes (Figure 1D). Similar findings were observed in primary chondrocytes isolated from the Gusb-KO mouse (Gusb–/–) (Figure 1, B, C, E, and F), from the mouse model of mucopolysaccharidosis type VI (MPSVI) (23) due to mutations in the lysosomal sulfatase arylsulfatase B (Arsb–/–) (Supplemental Figure 2A), and in chondrocyte precursor cells isolated from mucopolysaccharidosis type I (MPSI) human patients (IDUA–/–) (Supplemental Figure 2B). Notably, despite increased mTORC1 activity, the nuclear localization of TFEB and TFE3 transcription factors was increased in Gusb–/– chondrocytes compared with control chondrocytes (Supplemental Figure 2, C and D), suggesting that in this cellular system TFEB and TFE3 nuclear accumulation is mTORC1 independent. However, we did not observe increased mTORC1 activity in fibroblasts isolated from MPSVII mice or in a HeLa cell line in which the ARSB gene was deleted through CRISPR/Cas9-mediated genome editing (Arsb-KO) (Supplemental Figure 2, E and F).
Figure 1. Enhanced mTORC1 signaling in LSD chondrocytes.
(A) Western blot analysis of mTORC1 substrates in control (Ctr) and Gusb-KO RCS cells (A) and in primary chondrocytes from Gusb+/+ and Gusb–/– mice (B). (C) Quantification of phosphorylation levels relative to control levels (dashed line). n = 3. (D) Representative blot from a UVRAG IP assay in control and Gusb-KO RCS chondrocytes with or without Torin1 treatment (1 μM; 6 h). n = 3. (E) Western blot analysis of mTORC1 signaling on a time course of amino acid stimulation in Gusb+/+ and Gusb–/– primary chondrocytes. Cells were starved (starv.) of amino acids for 50 minutes and then stimulated (stim.) with amino acids for the indicated durations. (F) Bar graph displays phosphorylation levels of the indicated mTORC1 substrates (normalized to their total levels). Values are expressed as the fold increase compared with Gusb+/+ (represented by a dashed line). n = 3. (G and H) Gusb–/– (G) and Arsb–/– (H) primary chondrocytes were starved of amino acids for 50 minutes and then stimulated with amino acids for the indicated durations. Cells were then processed in an immunofluorescence assay to detect mTOR and Lamp-1, costained with DAPI for DNA content, and imaged. Insets show zoom ×3 and single-color channels of the boxed areas. Scale bars: 10 μm. Quantification of Lamp-1–mTOR colocalization is shown in Supplemental Figure 3. Data represent the mean values derived from the indicated number of independent experiments. Error bars indicate the SEM. *P ≤ 0.05 and **P ≤ 0.005, by unpaired Student’s t test. Mr, marker.
Because the lysosome is the site of mTORC1 activation (24), we tested whether the increase in activity correlates with altered mTOR localization on lysosomes. We found that mTOR was persistently associated with lysosomes, even in the absence of nutrients, in both Gusb–/– and Arsb–/– chondrocytes compared with their respective controls (Figure 1, G and H and Supplemental Figure 3).
Enhanced mTORC1 signaling inhibits AV-lysosome fusion in MPS chondrocytes.
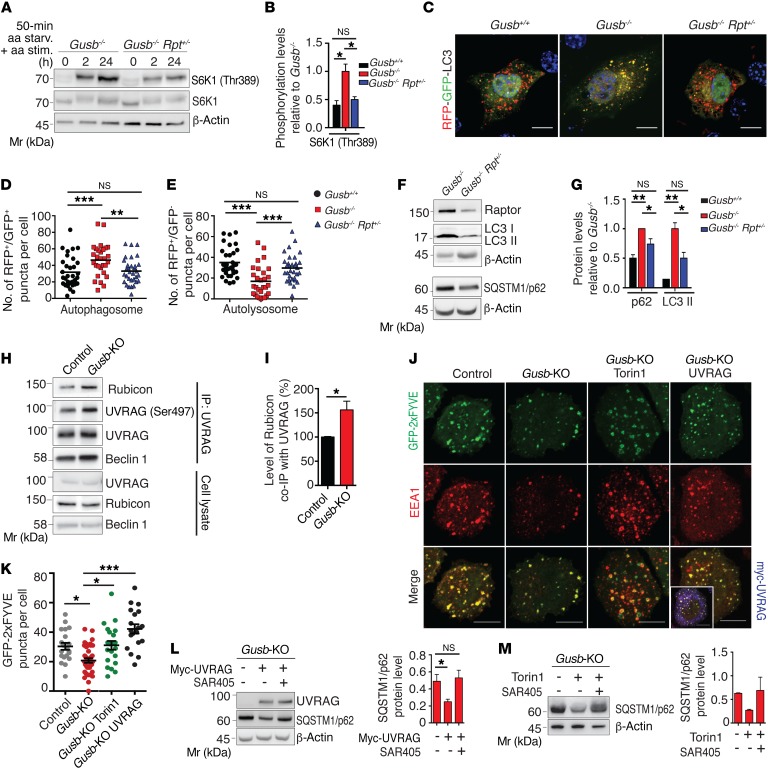
Next, we asked whether altered mTORC1 signaling plays a role in the pathogenesis of LSD. Despite the increase in ULK1 phosphorylation, AV biogenesis was largely unaffected in Gusb-KO and Gusb–/– chondrocytes, as demonstrated by our analysis of WIPI-2 puncta formation (Supplemental Figure 4, A and B) (25) and time-course assessment of LC3-II lipidation in the presence of vinblastine, which inhibits AV-lysosome fusion by preventing microtubule polymerization, or in the presence of the lysosomal inhibitor bafilomycin A1 (Baf.A1) (Supplemental Figure 4, C–F) (26). Conversely, autophagosome maturation (AV-lysosome fusion) was severely impaired in Gusb-KO and Gusb–/– chondrocytes compared with that observed in control cells, as demonstrated by reduced AV-lysosome colocalization and accumulation of AVs and the SQSTM1/p62 autophagy substrate (Supplemental Figure 4, G–M). We also confirmed the decreased rate of AV-lysosome fusion in Gusb-KO compared with control cells by expressing LC3 protein tandem tagged with red fluorescent protein–green fluorescent protein (RFP-GFP) (27). Autophagosomes (GFP- and RFP-positive) are discriminated from autophagolysosomes (GFP-negative and RFP-positive) because of quenching of the GFP signal (but not of RFP) inside the acidic lysosomal compartment (Supplemental Figure 4, N–O). To test whether the block in autophagy flux in LSD chondrocytes was caused by mTORC1, we isolated primary chondrocytes from Gusb–/– mice lacking 1 allele of the mTORC1 subunit Raptor (Gusb–/– Rpt+/– mice) (28). mTORC1 activity in primary chondrocytes isolated from Gusb–/– Rpt+/– mice was reduced compared with chondrocytes from Gusb–/– mice and comparable to those from WT mice (Gusb+/+), demonstrating that Raptor haploinsufficiency in Gusb–/– mice normalized mTORC1 signaling in chondrocytes (Figure 2, A and B). We observed a complete rescue of the autophagic flux in Gusb–/– Rpt+/– compared with Gusb–/– primary chondrocytes, as demonstrated by enhanced delivery of AVs to lysosomes (Figure 2, C–E) and reduced levels of LC3-II and SQSTM1/p62 (Figure 2, F and G). Notably, Gusb–/– Rpt+/– chondrocytes showed a rate of AV biogenesis similar to that of Gusb–/– chondrocytes (Supplemental Figure 4, P and Q), suggesting that reducing mTORC1 activity rescued autophagy by promoting AV-lysosome fusion.
Figure 2. Hyperactivation of mTORC1 in MPS chondrocytes suppresses autophagy via a Beclin 1–Vps34–UVRAG kinase complex.
(A) Western blot analysis of mTORC1 signaling upon amino acid stimulation in Gusb–/– and Gusb–/– Rpt+/– primary chondrocytes. (B) Quantification of S6K1 phosphorylation in primary chondrocytes isolated from mice of the indicated genotypes. n = 3. (C) Representative images of an RFP-GFP-LC3 assay in primary chondrocytes from mice of the indicated genotypes. Scale bars: 10 μm. (D and E) Quantitative analysis of RFP+GFP+ puncta (D) and RFP+GFP– puncta (E). Horizontal bars indicates the mean value. n = 3 chondrocytes donors/phenotype; n = 30 cells analyzed. (F) Western blot analysis of the indicated proteins in Gusb–/– and Gusb–/– Rpt+/– primary chondrocytes. β-Actin was used as a loading control. (G) Quantification of protein levels in cells from mice of the indicated genotypes. n = 3. (H) Representative blot of a UVRAG IP assay testing UVRAG interaction with Rubicon in control and Gusb-KO RCS chondrocytes. (I) Quantification of Rubicon co-IP with UVRAG. n = 3. (J) GFP-2xFYVE transfection in control and Gusb-KO RCS chondrocytes. Gusb-KO cells were treated with Torin1 (1 μM; 6 h) or cotransfected with Myc-UVRAG. Cells were costained for endosomes (EEA1). Boxed area in the bottom right-hand panel shows positive myc staining. Scale bars: 10 μm. (K) Quantitative analysis of PI3P puncta. n = 3; n = 25 cells analyzed. (L and M) Western blot analysis of SQSTM1/p62 in Gusb-KO cells transfected with Myc-UVRAG (L) or treated with Torin1 (1 μM, 6 h) and SAR405 (10 μM, 6 h) (M). β-Actin was used as a loading control. Bar graphs show quantification of protein levels. n = 3 (L) and n = 2 (M). Data represent the mean values derived from the indicated number of independent experiments. Error bars indicate the SEM. *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005, by ANOVA followed by Tukey’s post-hoc test (B, D, E, G, and K) and unpaired Student’s t test (I and L).
mTORC1 phosphorylates UVRAG and suppresses Beclin 1–Vps34 complex activity in MPS chondrocytes.
We next tested whether the enhanced phosphorylation of UVRAG by mTORC1 contributes to the defective AV maturation observed in LSD chondrocytes. The phosphorylation of mTORC1 on serine 497 enhances the affinity of UVRAG for its inhibitor Rubicon (13). Consistent with this model, the interaction between UVRAG and Rubicon was increased in Gusb-KO cells compared with that detected in control cells (Figure 2, H and I). As part of the Beclin 1–Vps34 complex, UVRAG promotes PI3P production on endolysosomal membranes (29). We observed that PI3P levels, as indicated by 2xFYVE-GFP–labeled puncta (17), were significantly reduced on endosomes of Gusb-KO chondrocytes compared with levels in control chondrocytes and that this phenotype was rescued by the overexpression of UVRAG (Figure 2, J and K). More important, UVRAG overexpression enhanced autophagic flux in Gusb–/– chondrocytes, as demonstrated by decreased levels of SQSTM1/p62 protein (Figure 2L and Supplemental Figure 5, A and B). This rescue was prevented by treatment with the Vps34 inhibitor SAR405 (Figure 2L) (30). PI3P levels were also increased in Gusb–/– Rpt+/– chondrocytes compared with Gusb–/– primary chondrocytes (Supplemental Figure 5, C and D). Similarly, mTORC1 inhibition with Torin1 rescued endosomal PI3P levels (Figure 2, J and K) and autophagic flux in Gusb-KO and Gusb–/– chondrocytes (Figure 2M and Supplemental Figure 5, E and F). Also in this case, Torin1-mediated rescue of the autophagic flux in Gusb-KO chondrocytes required the activity of Vps34, since this improvement of autophagic flux was prevented by treatment with SAR405 (Figure 2M and Supplemental Figure 5G). Notably, SAR405 treatment in WT chondrocytes induced an LSD-like phenotype (Supplemental Figure 5, H and I). Altogether, these data suggest that mTORC1 inhibits AV-lysosome fusion in MPS chondrocytes by inhibiting Beclin 1–Vps34–UVRAG complex activity and PI3P production.
Decrease in mTORC1 signaling improves collagen levels and bone phenotype in MPSVII mice.
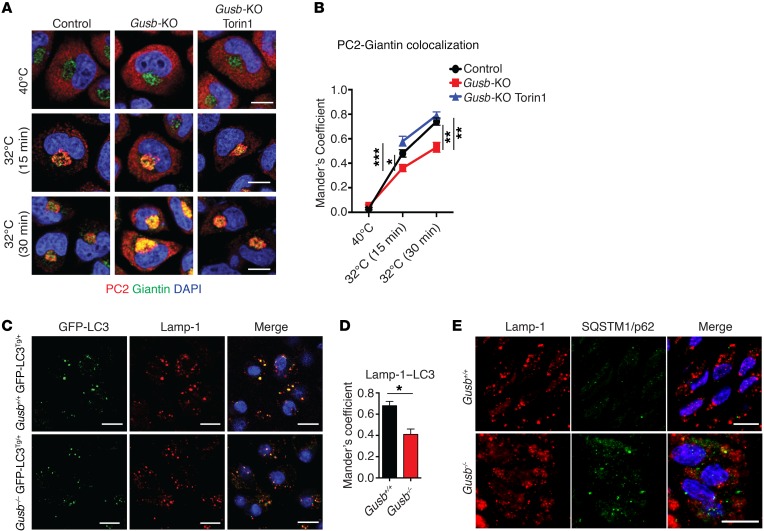
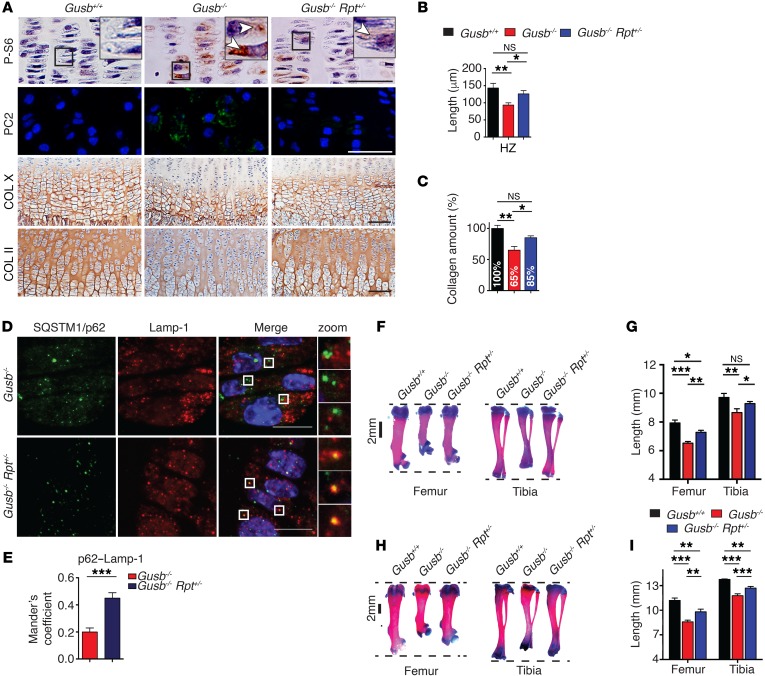
Consistent with a defective autophagic pathway, Gusb-KO chondrocytes showed PC2 trafficking defects characterized by a prolonged retention of PC2 in the ER compared with control cells (Figure 3, A and B). Importantly, this phenotype was rescued by Torin1 treatment (Figure 3, A and B). In vivo, Gusb–/– mice expressing the fluorescent autophagy reporter GFP-LC3 protein (Gusb–/– GFP-LC3Tg/+) (31) showed defective AV-lysosome fusion (Figure 3, C and D) and accumulation of SQSTM1/p62 (Figure 3E) compared with controls (Gusb+/+ GFP-LC3Tg/+). Consistent with the in vitro observations, femoral growth plate sections isolated from Gusb–/– mice showed increased mTORC1 signaling compared with Gusb+/+ littermate mice, as measured by phosphorylation levels of ribosomal protein S6 (P-S6) (Figure 4A). In addition, Gusb–/– mice showed intracellular accumulation of PC2 molecules in chondrocytes and significantly lower levels of collagen in femoral cartilages compared with levels in control mice (Figure 4, A–C). Conversely, femoral growth plate sections isolated from Gusb–/– Rpt+/– mice showed reduced mTORC1 signaling (Figure 4A) and increased delivery of SQSTM1/p62 to lysosomes (Figure 4, D and E), indicating enhanced AV-lysosome fusion compared with that observed in Gusb–/– mice. Furthermore, Gusb–/– Rpt+/– mice showed an almost normal level of collagen in the femoral growth plates (Figure 4, A–C). Remarkably, Gusb–/– Rpt+/– mice had a significant improvement in femur and tibia size at both P15 (Figure 4, F and G) and P30 (Figure 4, H and I). These data demonstrate that mTORC1 and autophagy play an important role in the pathogenesis of the bone phenotype in MPSVII mice and suggest that pharmacological modulation of this pathway may represent a new approach to treating LSD bone features. To test this hypothesis, we administered the mTORC1 inhibitor rapamycin to control and Gusb–/– mice from P0 for 15 days and measured femur and tibia lengths. Unlike what we observed in Rpt+/– mice, we found that rapamycin administration inhibited bone growth in control mice, suggesting that the systemic administration of rapamycin may have altered additional pathways that are converging on bone growth, consistent with previously published results (Supplemental Figure 6, A and B and ref. 32).
Figure 3. Altered collagen trafficking and autophagy dysfunction in growth plates of MPSVII mice.
(A) PC2 secretion was synchronized for 3 hours at 40°C and then shifted to 32°C (ER block release) for the indicated durations (min). Representative images of PC2 localization (red) in ER and Golgi area (Giantin, green) in control chondrocytes and vehicle- or Torin1-treated (6 h, 1 μM) Gusb-KO chondrocytes, 15 and 30 minutes after the ER block release of PC2. Scale bars: 10 μm. (B) Quantification of PC2-Giantin colocalization over time. n = 3; n = 50 cells for each time point. (C) Representative images of GFP-LC3 puncta (autophagosomes) and Lamp-1 (lysosomes) immunostaining in femoral growth plates from Gusb+/+ GFP-LC3Tg/+ and Gusb–/– GFP-LC3Tg/+ mice at P6. Scale bars: 10 μm. (D) Quantification of Lamp-1–LC3 colocalization. n = 3 mice per group. (E) Representative images of SQSTM1/p62 and Lamp-1 immunofluorescence in femoral growth plates from mice of the indicated genotypes. Scale bars: 10 μm. n = 3 mice per group. Data represent the mean values derived from the indicated number of independent experiments. Error bars indicate the SEM. *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005, by ANOVA followed by Tukey’s post-hoc test (B) and unpaired Student’s t test (D).
Figure 4. Normalization of mTORC1 signaling restores collagen trafficking and rescues bone phenotype in MPS VII mice.
(A) Representative images of femoral growth plate sections isolated from Gusb+/+, Gusb–/–, and Gusb–/– Rpt+/– mice at P15. P-S6 immunostaining (arrows, brown, top row), intracellular PC2 (green, second row), COL X (third row), and COL II (brown, bottom row). Nuclei were counterstained with hematoxylin (pink, top row and bottom row) or DAPI (blue, second row). Scale bars: 100 μm. Insets show magnification ×3. (B) Bar graphs show the length of the HZ (COL X+ area). n = 5 mice per genotype. (C) Quantification of collagen isolated from femoral and tibia cartilages from mice of the indicated genotypes and expressed as the percentage of Gusb+/+ mice. n ≥ 5 mice per genotype. (D) Representative images of SQSTM1/p62 and Lamp-1 immunofluorescence in femoral growth plates isolated from Gusb–/– and Gusb–/– Rpt+/– mice at P15. n = 3 mice per group. Scale bars: 10 μm; zoom, ×5. (E) Quantification of P62–Lamp-1 colocalization. (F) Representative images of Alcian blue and alizarin red staining of femurs and tibiae from P15 Gusb+/+, Gusb–/–, and Gusb–/– Rpt+/– mice. (G) Femur and tibia lengths for mice of the indicated genotypes. n = 6 mice per genotype. (H) Representative images of Alcian blue and alizarin red staining of femurs and tibiae from P30 Gusb+/+, Gusb–/–, and Gusb–/– Rpt+/– mice. (I) Femur and tibia lengths for mice of the indicated genotypes. n = 6 mice per genotype. Data represent the mean values derived from the indicated number of mice. Error bars indicate the SEM. *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005, by ANOVA followed by Tukey’s post-hoc test (B, C, G, and I) and unpaired Student’s t test (E).
Targeting Beclin 1 activity rescues AV-lysosome fusion and ameliorates bone growth retardation in MPSVII and MPSVI mice.
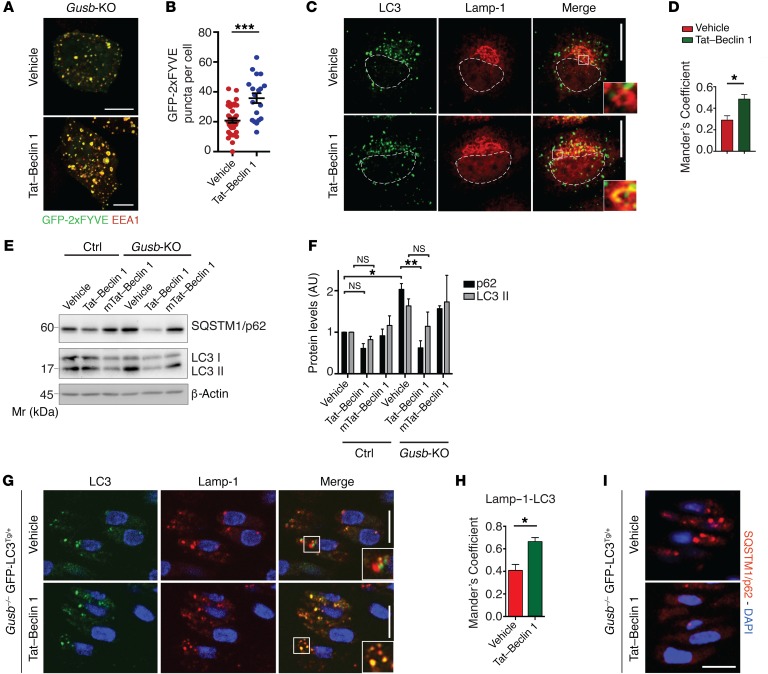
Beclin 1 is a member of the Vps34-UVRAG complex (16), and its activity can be pharmacologically enhanced both in vitro and in vivo using the Tat–Beclin 1 peptide (33). We tested whether enhancing Beclin 1–Vps34–UVRAG had an effect similar to that of Raptor haploinsufficiency on MPS chondrocytes both in vitro and in vivo. Tat–Beclin 1, but not a mutated version, rescued endosomal PI3P levels (Figure 5, A and B) and in turn normalized the autophagic flux in Gusb-KO chondrocytes, as demonstrated by increased AV-lysosome fusion and decreased levels of LC3II and SQSTM1/p62 (Figure 5, C–F). When administered to mice, Tat–Beclin 1 is able to activate autophagy in growth plate chondrocytes (20). We observed that Tat–Beclin 1 administration to WT mice did not affect bone length, growth plate morphology, or chondrocyte function (Supplemental Figure 6, C–E), consistent with previous results (20). More important, daily injection of Tat–Beclin 1 peptide into both control Gusb+/+ GFP-LC3Tg/+ and Gusb–/– GFP-LC3Tg/+ mice enhanced AV-lysosome fusion and SQSTM1/p62 degradation in the femoral growth plate (Figure 5, G–I, and Supplemental Figure 6, F–I). In addition, Gusb–/– mice treated daily with Tat–Beclin 1 showed rescued levels of collagen secretion (Figure 6, A–C). As a consequence, we measured a significant recovery of femur and tibia growth retardation in Tat–Beclin 1–treated Gusb–/– mice at P15 (Figure 6, D and E). We obtained similar results with the MPSVI mouse model (Figure 6, F and G), indicating that our findings may also be extended to other LSDs. These data suggest that the pharmacological modulation of autophagy may represent a future therapy for the treatment of bone phenotype in LSDs.
Figure 5. Autophagy induction with Tat–Beclin 1 rescues the AV-lysosome fusion defect in MPS chondrocytes.
(A) Gusb-KO cells were transfected with GFP-2xFYVE and treated or not with Tat–Beclin 1 (10 μM, 2 h). Cells were costained for endosomes (EEA1). Scale bars: 10 μm. (B) Quantitative analysis of PI3P puncta. n = 3 independent transfections; n = 20 cells analyzed. (C) Immunofluorescence of Lamp-1 and LC3 in Gusb-KO cells treated with Tat–Beclin 1 peptide (10 μM; 2 h). Insets show colocalization in selected areas at higher magnification (zoom ×3.5). Scale bars: 10 μm. (D) Quantification of Lamp-1–LC3 colocalization. n = 3 independent treatments; n = 30 cells analyzed. (E) Western blot analysis of the indicated proteins in control and Gusb-KO cells treated with vehicle, Tat–Beclin 1 (20 μM; 2 h), or with an inactive form of Tat–Beclin 1 (mTat-Beclin 1) (20 μM; 2 h). β-Actin was used as a loading control. (F) Quantification of protein levels. n = 3. (G and I) Representative images of GFP–LC3 puncta (AVs), Lamp-1 (G), and SQSTM1/p62 (I) immunostaining in femoral growth plates from Gusb–/– GFP-LC3Tg/+ mice at P6. Tat–Beclin 1 peptide was administered as indicated (2 mg/kg, daily for 6 d). Scale bar: 10 μm (zoom ×2). (H) Quantification of Lamp-1–LC3 colocalization. n = 3 mice per group. Data represent the mean values derived from the indicated number of mice per independent experiment. Error bars indicate the SEM. *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005, by paired Student’s t test (B, D, and H) and ANOVA followed by Tukey’s post-hoc test (F).
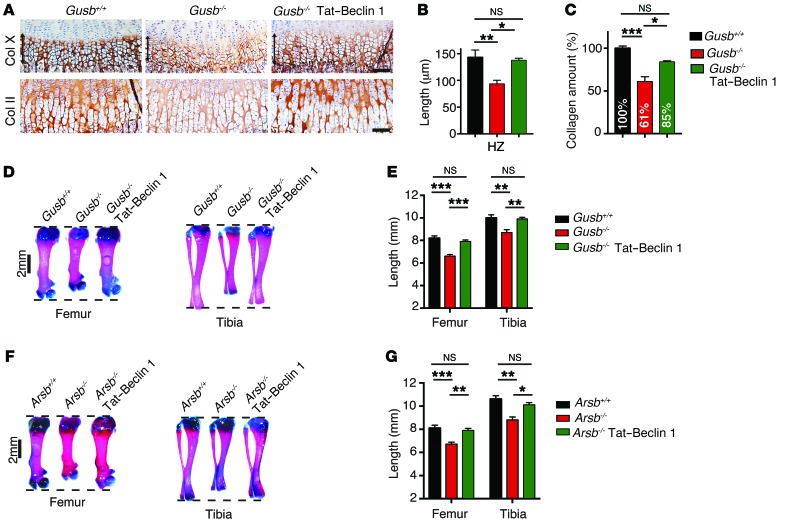
Figure 6. Autophagy induction with Tat–Beclin 1 improves bone phenotype in MPS mice.
(A) Representative images of femoral growth plate sections from Gusb+/+, Gusb–/–, and Gusb–/– Tat–Beclin 1–treated mice (2 mg/kg daily for 15 d) immunostained with COL X and COL II. n = 3 mice per group. Scale bars: 100 μm. (B) Length of HZ. n = 3 mice per group. (C) Quantitative measurement of collagen isolated from the growth plates of mice treated as in A. n = 8. (D) Representative images of femurs and tibiae from P15 mice treated as in A. (E) Femur and tibia lengths. n = 8 mice per group. (F) Representative images of femurs and tibiae from Arsb+/+, Arsb–/–, and Arsb–/– Tat–Beclin 1–treated mice (2 mg/kg daily for 15 days). (G) Femur and tibia lengths. n = 6 mice per group. Data represent the mean values derived from the indicated number of mice. Error bars indicate the SEM. *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005, by ANOVA followed by Tukey’s post-hoc test.
Discussion
The lysosome is emerging as a critical regulator of mTORC1 signaling (34). However, whether this regulation is physiologically relevant has not yet been determined. In this study, we identified mTORC1 hyperactivation as a main pathological mechanism through which lysosomal storage impairs chondrocyte function and bone growth (Figure 7). Our data demonstrate a critical role of the lysosome as a physiological regulator of mTORC1.
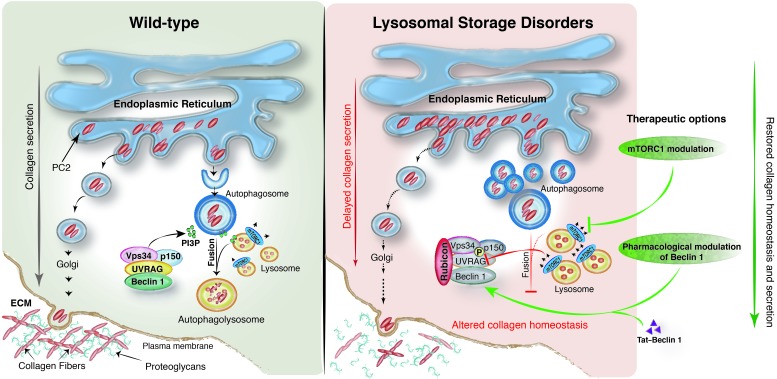
Figure 7. Proposed pathogenetic mechanism in LSD chondrocytes.
Left: In WT chondrocytes, collagen homeostasis is reached through equilibrium between the pool that needs to be secreted and the pool that needs to be degraded. Within this context, fine-tuned mTORC1 activity is particularly important for autophagy regulation, autophagosome biogenesis, AV-lysosome fusion, and, in turn, PC2 homeostasis. A key protein complex required for AV-lysosome fusion is represented by Beclin 1–Vps34–UVRAG, a class III–PI3K complex that produces a pool of PI3P required for vesicle fusion. Right: Lysosomal impairment in LSD is responsible for abnormal mTORC1 signaling. Among the autophagy targets of mTORC1, UVRAG activity is particular sensitive to mTORC1 phosphorylation. Thus, in LSD, a stable complex between UVRAG and Rubicon is seen, and insufficient PI3P is produced. This primarily affects AV-lysosome fusion, leading to AV accumulation and a subsequent delay in procollagen trafficking. To overcome the blockage of AV-lysosome fusion and restore proper collagen homeostasis in LSD, therapeutic options can be directed toward either mTORC1 modulation (repression) or pharmacological modulation of the Beclin 1–Vps34 complex (induction).
Notably, it has been demonstrated that an acute (pharmacological) inhibition of lysosome function impairs mTORC1 activation by nutrients in different experimental conditions (35). Unexpectedly, in this work we observed that lysosomal storage enhances, rather than inhibits, mTORC1 signaling in LSD chondrocytes. Similar observations were recapitulated pharmacologically by others (36), suggesting a chondrocyte-specific response of mTORC1 to lysosomal dysfunction.
The role of mTORC1 during longitudinal bone growth has recently begun to emerge. Disruption of mTORC1 signaling through the biallelic deletion of Raptor in chondrocytes inhibits chondrocyte functions, leading to impaired skeletal growth (37). In addition, genetic inactivation of the mTORC1 inhibitor TSC1 gene in chondrocytes causes chondrodysplasia in mice (38), suggesting that the fine-tuning of mTORC1 signaling is essential for longitudinal bone growth.
We demonstrate that decreasing levels of mTORC1 signaling or inducing autophagy by Tat–Beclin 1 improves collagen levels in cartilages and rescues bone growth retardation in MPS mice. Notably, recent findings showed that feeding mice a low-protein diet that induces autophagy reduced the intracellular accumulation of misfolded collagen in osteoblasts and improved bone matrix mineralization in a mouse model of osteogenesis imperfecta (39). Taken together, these observations suggest that the modulation of autophagy may represent a possible therapeutic approach for the treatment of skeletal disorders that are associated with defective collagen trafficking and secretion.
Consistent with this hypothesis, our data show that amelioration of the bone features in MPS mice upon autophagy activation occurs without reverting the lysosomal phenotype in chondrocytes (Supplemental Figure 6J), suggesting that targeting secondary pathways that are altered as a consequence of lysosomal storage may represent an appealing therapeutic approach for the treatment of LSD features that are currently unresponsive to therapy.
Notably, autophagy dysfunction has been reported in other LSDs (40), however, the role mTORC1 signaling plays in these cases remains to be determined. Interestingly, the analysis of craniofacial and dental abnormalities in a LSD mouse model, namely mucolipidosis II (MLII), showed a loss of collagen fibril organization (41), which strikingly resembles the one observed in the cartilage of mice lacking ATG7 in chondrocytes (20).
In conclusion, our data suggest a role for mTORC1 and autophagy in the pathogenesis of genetic disorders affecting the skeleton and hence extend the list of diseases that may benefit from the development of therapeutic agents targeting these 2 pathways.
Methods
Animals.
The Gusb–/– mouse strain carrying the missense E536Q mutation was shared by W. Sly (St. Louis University, St. Louis, Missouri, USA) (42). The Raptorfl/fl mouse strain was purchased from The Jackson Laboratory (stock no. 013188). The CMV-Cre mouse line was provided by A. Ballabio. Raptorfl/fl mice were mated with CMV-Cre mice to obtain heterozygous mice that were subsequently mated with Gusb+/– mice. Double-heterozygous Gusb+/– Raptor+/– mice were then mated with Gusb+/– mice in order to obtain Gusb–/– Raptor+/– mice. The Arsb–/– mouse line was from A. Auricchio (TIGEM). The GFP-LC3 mouse line was obtained from N. Mizushima (University of Tokyo, Tokyo, Japan). All mice were maintained on a C57BL/6 background. The number of mice used in each experiment is specified in the figure legends. The sex of the mice was not taken into account until P15. Mice were randomly assigned to the treatment groups. The investigators were not blinded to treatment allocation during the experiments or outcome assessment.
Skeletal staining.
Skeletons were fixed in 95% ethanol overnight and stained with Alcian blue and alizarin red (Sigma-Aldrich) according to standard European Mouse Phenotyping Resource of Standardised Screens (EMPReSS) protocols (http://empress har.mrc.ac.uk/browser/). Measurement of bone length was performed using a Leica M205 A stereo microscope equipped with LAS X Software.
Tissue histology, immunohistochemistry, and immunofluorescence.
Histology was performed according to standard EMPReSS procedures (http://empress.har.mrc.ac.uk/browser/). Briefly, femurs were fixed overnight in 4% (wt/vol) paraformaldehyde (PFA) and then demineralized in 10% EDTA (pH 7.4) for 48 hours. Specimens were then dehydrated, embedded in paraffin, sectioned at 7-μm thickness, and stained with H&E.
COL II, COL X, and phosphorylated P-S6.
Proteolytic-induced epitope retrieval (PIER) was performed as an antigen retrieval method. Deparaffinized sections were treated with 1 mg/ml pepsin at 37°C for 20 minutes, followed by treatment with 2 mg/ml testicular hyaluronidase (Sigma-Aldrich) at 37°C for 60 minutes for COL X, or with chondroitinase ABC (Sigma-Aldrich) for 2 hours at 37°C for COL II, and then washed in PBS and incubated in 3% H2O2 in methanol. Sections were incubated with diluted normal blocking serum for 20 minutes and overnight at 4°C with a monoclonal antibody against COL X (clone X53; Quartett); COL II (II-II6B3; Hybridoma Bank); or phosphorylated P-S6 (p–P-S6) (Ser240/Ser244) (5364; Cell Signaling Technology).
SQSTM1/p62 and intracellular COLII immunofluorescence.
For immunofluorescence, femurs were dissected from euthanized mice and fixed with buffered 4% PFA overnight at 4°C, washed with PBS and cryoprotected in successive sucrose solutions diluted with PBS (10% for 2 hours, 20% for several hours, and 30% overnight at 4°C; all wt/vol), and finally embedded in OCT (Sakura). Cryostat sections were cut at 10-μm thickness. Sections were blocked and permeabilized in 3% (wt/vol) BSA and 5% FBS in PBS plus 0.3% Triton X-100 for 3 hours and then incubated overnight with the primary antibody SQSTM1/p62 (GP62-C; Progen) or with the primary antibody COLII (II-II6B3;Hybridoma Bank). Sections were washed 3 times with 3% BSA in PBS plus 0.3% Triton X-100 and then incubated for 3 hours with secondary antibodies conjugated with Alexa Fluor (goat anti-rabbit A11011/A11008 or goat anti-mouse A11001/A11004; Life Technologies, Thermo Fisher Scientific).
Growth plate measurement.
Individual zonal heights were measured for the morphologically defined proliferative and hypertrophic zones of the growth plate. The proliferative zone (PZ) is defined as the region of a chondrocyte showing a flattened-disc morphology. The hypertrophic zone (HZ) is defined as the region beginning with the consistent cylindrical cellular profile and extending to the metaphyseal chondro-osseous junction. We used CellSens software (Olympus Life Science; manual measurement) to estimate PZ and HZ heights by measuring the length between the initial and final proliferating and hypertrophic chondrocytes, respectively.
Collagen quantification and analysis.
Colorimetric assays were performed using the Soluble Collagen Assay Sircol (Biocolor) following the manufacturer’s protocol. Briefly, femoral cartilages were isolated from 2 to 3 mice with the same genotype, pooled and homogenized in 0.5 ml of 1 mg/ml cold (4°C) pepsin in 0.2 M NaCl, 0.5 M acetic acid to pH 2.1 with HCl, and then digested at 4°C for 24 hours, twice. The pellet was discarded, and an equal volume (1 ml) of 4 M NaCl in 1 M acetic acid was added to precipitate collagen. The pellet was then resuspended in 0.8 ml of 0.2 M NaCl in 0.5 M acetic acid and was precipitated again 2 times. After the last precipitation, the pellet was washed twice with 70% EtOH to remove residual NaCl and then complexed with Sircol dye. Absorbance was measured at 555 nm, and the concentration was calculated using a standard curve. Values were normalized to DNA levels calculated measuring the absorbance at 260 nm.
Transmission electron microscopy.
For electron microscopy (EM) analysis, growth plates were fixed in 1% glutaraldehyde in 0.2 M HEPES buffer. Small blocks of growth plates were then post-fixed in uranyl acetate and OsO4. After dehydration through a graded series of ethanol, tissue samples were cleared in propylene oxide, embedded in epoxy resin (Epon 812), and polymerized at 60°C for 72 hours. For pre-embedding, immuno-EM rat chondrosarcoma (RCS) cells were fixed, permeabilized, and labeled as described previously (43). Anti-human Lamp-1 antibody was purchased from Sigma-Aldrich (catalog N-L1418). From each sample, thin sections were cut with a Leica EM UC6 ultramicrotome, and images were acquired using a FEI Tecnai-12 electron microscope (FEI; Thermo Fisher Scientific) equipped with a VELETTA CCD digital camera (Soft Imaging Systems GmbH).
Cell culture, transfections, and plasmids.
Primary cultured chondrocytes were prepared from rib cartilage of P5 mice. Rib cages were first incubated in DMEM (Euroclone) using 0.2% collagenase D (Roche), and after adherent connective tissue was removed (1.5 h), the specimens were washed and incubated in fresh collagenase D solution (3 mg/ml) for a further 4.5 hours. Isolated chondrocytes were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen, Thermo Fisher Scientific). Adult fibroblasts were prepared from the skin of P5 mice. Samples, cut into small pieces using a razor blade, were first incubated with collagenase (30 min) and then washed and incubated in trypsin for 20 minutes. After centrifugation, the cell pellet was resuspended in complete medium. MPSI undifferentiated mesenchymal stromal cells were provided by M. Serafini (University of Milano-Bicocca). The Arsb-KO HeLa cell line was provided by J. Monfregola (TIGEM). RCS cells were a Swarm rat chondrosarcoma chondrocyte line (44). For amino acid starvation and stimulation experiments, the cell culture medium was amino acid–free RPMI 1640 (US Biological) supplemented with 10% dialyzed FBS (Invitrogen, Thermo Fisher Scientific). Cells were rinsed twice with starvation medium and then kept in a full volume of starvation medium for 50 minutes. At the 50-minute time point, a standard amino acid mixture composed of MEM nonessential amino acid solution, MEM essential amino acid solution, and L-glutamine (Invitrogen, Thermo Fisher Scientific), was added back to a final concentration of 3×. Unless otherwise specified, to synchronize mTORC1 signaling before performing the experiments, primary chondrocytes and RCS cells were stimulated with amino acids for 24 hours and 6 hours, respectively. For Tat–Beclin 1 peptide treatment, cells were washed with PBS and treated with peptides (20 μM, 2 h) or vehicle dissolved in Opti-MEM (Gibco, Thermo Fisher Scientific) acidified with 0.15% (v/v) 6N HCl. Cells were transfected with Lipofectamine LTX and Plus Reagent (Invitrogen, Thermo Fisher Scientific) following a reverse transfection protocol. The tandem mRFP–GFP–LC3 plasmid was provided by T. Yoshimori (Osaka University, Osaka, Japan). The Myc-UVRAG plasmid was provided by D.H. Kim (University of Minnesota, Minneapolis, USA). The GFP-2xFYVE plasmid was provided by S. Tooze (The Francis Crick Institute, London, United Kingdom).
Cell immunofluorescence.
Chondrocytes were fixed for 10 minutes in 4% PFA in PBS and permeabilized for 30 minutes in 0.05% (w/v) saponin, 0.5% (w/v) BSA, 50 mM NH4Cl, and 0.02% NaN3 in PBS (blocking buffer). For the detection of endogenous LC3, cells were methanol fixed. The cells were incubated for 1 hour with primary antibodies against Lamp-1 (sc-19992; Santa Cruz Biotechnology Inc.); SQSTM1/p62 (GP62-C; Progen); mTOR (2983; Cell Signaling Technology); LC3 (NB100-2220; Novus Biologicals); WIPI-2 (ab105459; Abcam); c-Myc (sc-40; Santa Cruz Biotechnology Inc.); and EEA1 (3288; Cell Signaling Technology); washed 3 times in PBS; incubated for 1 hour with the secondary antibody (Alexa Fluor–labeled goat anti-rat A11077, goat anti-guinea pig A11073, goat anti-rabbit A11011/A11008, and goat anti-mouse A11001, A11004; Life Technologies, Thermo Fisher Scientific); washed 3 times in PBS; incubated for 20 minutes with 1 μg/ml Hoechst 33342; and finally mounted in Mowiol (Sigma-Aldrich). For the detection of TFEB and TFE3, cells were permeabilized for 30 minutes in 0.2% Triton X-100 in PBS and blocked for 1 hour with 0.1% Triton X-100 and 10% goat serum in PBS. The cells were incubated overnight with primary antibodies against TFEB (MBS120432; MyBioSource) and TFE3 (HPA023881; Sigma-Aldrich). All confocal experiments showing colocalization were acquired with a slice thickness of 0.5 μm using an LSM 710 confocal microscope equipped with a 63 × 1.4 numerical aperture oil objective. Colocalization analysis was performed using ImageJ software (JaCoP plug-in; NIH).
Western blot analysis.
Cells were washed twice with PBS and then scraped in RIPA lysis buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate) in the presence of PhosSTOP and EDTA-free protease inhibitor tablets (Roche). Cell lysates were incubated on ice for 30 minutes, and then the soluble fraction was isolated by centrifugation at 16,000 g for 20 minutes at 4°C. The total protein concentration in cellular extracts was measured using a colorimetric BCA protein assay kit (Pierce Chemical). Protein extracts, separated by SDS-PAGE and transferred onto membranes, were probed with antibodies against phosphorylated p70S6K (p-p70S6K) (Thr389) (9234; Cell Signaling Technology); p70S6K (9292; Cell Signaling Technology); p-ULK1 (Ser757; Ser555; Ser317) (6888; 5869; 12753; Cell Signaling Technology); ULK1 (BK8054; Cell Signaling Technology); Raptor (2280; Cell Signaling Technology); Lamp-1 (ab24170; Abcam); LC3B (NB100-2220; Novus Biologicals); β-actin (NB600-501; Novus Biologicals); and SQSTM1/p62 (H00008878-M01; Abnova). Proteins of interest were detected with HRP-conjugated goat anti-mouse or anti-rabbit IgG antibody (8125, 8114; Cell Signaling Technology) and visualized with the Super Signal West Dura substrate (Thermo Fisher Scientific), according to the manufacturer’s protocol. Western blot images were acquired using the ChemiDoc-lt imaging system (UVP), and band intensity was calculated with ImageJ software using the Gels and Plot Lanes plug-in. See complete unedited blots in the supplemental material.
Generation of Gusb-KO clone.
To create Gusb gene disruption through CRISPR/Cas9 technology, an RCS chondrocyte cell line was used. Cells (1 × 106) were transfected with an all-in-one vector containing the sgRNA of interest (target site sequence: CACCTTGAGTTCCCGCGAAG, with predicted no off-target sites), whose expression was driven by the U6 promoter, a recombinant form of Cas9 protein under the control of the CMV promoter and an mCherry reporter gene under the control of the SV40 promoter (Genecopoeia). Cells were transfected with Lipofectamine LTX and Plus Reagent (Invitrogen, Thermo Fisher Scientific) following a reverse transfection protocol. Forty-eight hours after transfection, putative positive clones were FACS sorted for mCherry expression using the BD FACSAria flow cytometer. Sorted cells were kept in culture until confluence and then subjected to PCR analysis, followed by Sanger sequencing to identify mutations. Positive clones were screened for β-glucuronidase activity. Briefly, cell pellets were lysed in extraction buffer (50 mM NaHPO4, pH 7.0, 10 mM 2-mercaptoethanol, 10 mM Na2 EDTA, 0.1% sodium lauryl sarcosine,0.1% Triton X-100 [100 ml]), and protein concentration was measured using a colorimetric BCA Protein Assay Kit (Pierce Chemical). Proteins (200 and 400 μg) were incubated with 200 μl of the fluorogenic substrate, 4-methylumbelliferyl-β-D-glucuronide (2 mM) (Sigma-Aldrich), for 0.5 and 1 hours at 37°C. The reaction was stopped by adding 200 μl of the carbonate stop buffer (0.5 M NaHCO3/0.5 M Na2CO3, pH 10.7), and the fluorescence of the 4-methylumbelliferone liberated was measured in a fluorimeter (GloMax-Multi Detection System; Promega) using 365 nm excitation and 460 nm emission. Absolute fluorescence of WT RCS was used as a control. The Gusb-KO clone as well as WT RCS were kept in medium containing 2 mg/ml chondroitin sulphate (Sigma-Aldrich) for 48 hours before any experiment was performed.
Co-IP experiments.
For co-IP experiments, whole-cell extracts were prepared in 0.3% CHAPS-containing lysis buffer (40 mM HEPES, pH 7.4, 120 mM NaCl, 1 mM EDTA, 50 mM NaF, 1.5 mM Na3VO4, 10 mM β-glycerophosphate, 0.3% CHAPS, and EDTA-free protease inhibitors) as described previously (28), and immunoprecipitated with anti-UVRAG antibody (M160-3; MBL). The precipitated proteins were washed 5 times with 0.3% CHAPS lysis buffer, separated by SDS–PAGE, transferred onto membranes, and probed with antibodies against p-UVRAG (Ser497) (gift of D.H. Kim) (13), Beclin 1 (3495; Cell Signaling Technology), UVRAG (M160-3; MBL), and Rubicon (8465; Cell Signaling Technology).
Procollagen secretion assay.
Synchronized PC2 secretion was obtained after incubating chondrocytes for 3 hours at 40°C to block PC2 in the ER and then shifting the temperature to 32°C (ER block release) for 15 minutes. Cells were then fixed in 4% PFA for 10 minutes at room temperature and processed for immunofluorescence. Cells were incubated for 1 hour with primary antibodies against PC2 (CII-C1; Hybridoma Bank) and Giantin (ab37266; Abcam).
Treatments in vivo.
Newborn mice were intraperitoneally injected daily with Tat–Beclin 1 peptide at 2 mg/kg resuspended in PBS. Control mice were injected with vehicle only. Mice were sacrificed after 6 or 15 days, as specified in the figure legends. Rapamycin was dissolved in pure ethanol as a 20-mg/ml stock solution, diluted in injection buffer (4% ethanol, 5% Tween-80, 5% PEG400 in dH2O) at 1 mg/ml, aliquoted, and stored at –20°C. Mice were weighed daily throughout the experiment, and the dosage (0.375 mg/kg) was adjusted accordingly. Control mice received a similar volume of injection buffer without drug.
Chemicals.
Tat–Beclin 1 peptide (D–amino acid peptide sequence: RRRQRRKKRGYGGDHWIHFTANWV) and its mutated version mTat-Beclin 1 (L–amino acid peptide sequence: YGRKKRRQRRRGGVGNDFFINHETTGFATEW) were synthesized at the Protein Chemistry Technology Center (UT Southwestern Medical Center). Bafilomycin A1 (200 nM), vinblastine (200 nM) were from Sigma-Aldrich. Torin1 (1 μM) was from Cell Signaling Technology. Protein G sepharose was from GE Healthcare. SAR405 (10 μM) was from ApexBio Technology. Rapamycin (sirolimus) was from LC Laboratories.
Statistics.
A 2-tailed, paired Student’s t test was performed when comparing the same cell population with 2 different treatments. A 2-tailed, unpaired Student’s t test was performed when comparing 2 groups of mice or different primary chondrocyte preparations. A 1-way ANOVA and Tukey’s post-hoc test were performed when comparing more than 2 groups relative to a single factor (treatment or genotype). A P value of 0.05 or less was considered statistically significant.
Study approval.
Experiments were conducted in accordance with the guidelines of the Animal Care and Use Committee of Cardarelli Hospital in Naples, Italy, and approved by the Italian Ministry of Health.
Author contributions
RB and CS designed the study, analyzed the data, and wrote the manuscript. RB, LC, and CDL performed most of the experiments. AF performed Lamp-1–mTOR colocalization analysis. ACS, EN, and CL provided technical support for the in vivo studies. JM and EDG generated and analyzed Arsb-KO cells. MS and IA generated human mesenchymal cells. BL and AB provided reagents and participated in the preparation of the manuscript.
Supplementary Material
Acknowledgments
We thank A. Auricchio, G. Napolitano, and G. Diez Roux (from TIGEM) for critical reading of the manuscript; S. Strollo for immunohistochemistry; L. Polishchuk for EM analysis; A. Carissimo for statistical analysis; N. Mizushima, W. Sly (from St. Louis University School of Medicine, St. Louis, USA), and A. Auricchio for sharing the mouse lines used in this work; and D.H. Kim for sharing materials used in this work. This work was supported by grants to CS: Italian Telethon Foundation (TCP12008) and TIGEM institutional Grant, Marie Curie (PCIG13-GA-2013-618805), STAR/Banco di Napoli, Italian Ministry of Research (FIRB RBFR13LH4X), European Research Council starting grant (714551), Penn Orphan Disease Center (MPSI-16-004-01) and National MPS Society; by the NIH U19AI199725 to BL; by the European Research Council Advanced Investigator grant no. 694282 and U.S. National Institutes of Health (R01-NS078072) to AB.
Version 1. 09/01/2017
Electronic publication
Version 2. 09/15/2017
Text color corrected in PDF
Version 3. 09/20/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(10):3717–3729.https://doi.org/10.1172/JCI94130.
Contributor Information
Rosa Bartolomeo, Email: bartolomeo@tigem.it.
Laura Cinque, Email: l.cinque@tigem.it.
Chiara De Leonibus, Email: c.deleonibus@tigem.it.
Alison Forrester, Email: a.forrester@tigem.it.
Jlenia Monfregola, Email: j.monfregola@tigem.it.
Emanuela De Gennaro, Email: e.degennaro@tigem.it.
Edoardo Nusco, Email: nusco@tigem.it.
Isabella Azario, Email: isabella.azario@live.it.
Carmela Lanzara, Email: c.lanzara@tigem.it.
Marta Serafini, Email: serafinim72@gmail.com.
Beth Levine, Email: beth.levine@utsouthwestern.edu.
Andrea Ballabio, Email: ballabio@tigem.it.
Carmine Settembre, Email: settembr@tigem.it.
References
- 1.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 2.Bateman JF, Boot-Handford RP, Lamandé SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat Rev Genet. 2009;10(3):173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 3.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8(8):622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 4.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5(7):554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 5.Clarke LA, Hollak CE. The clinical spectrum and pathophysiology of skeletal complications in lysosomal storage disorders. Best Pract Res Clin Endocrinol Metab. 2015;29(2):219–235. doi: 10.1016/j.beem.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14(5):283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vega-Rubin-de-Celis S, Peña-Llopis S, Konda M, Brugarolas J. Multistep regulation of TFEB by MTORC1. Autophagy. 2017;13(3):464–472. doi: 10.1080/15548627.2016.1271514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 13.Kim YM, et al. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell. 2015;57(2):207–218. doi: 10.1016/j.molcel.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 15.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X, et al. Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17. Mol Cell. 2017;65(6):1029–1043.e5. doi: 10.1016/j.molcel.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10(7):776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchberger A. ERQC autophagy: Yet another way to die. Mol Cell. 2014;54(1):3–4. doi: 10.1016/j.molcel.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 20.Cinque L, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528(7581):272–275. doi: 10.1038/nature16063. [DOI] [PubMed] [Google Scholar]
- 21. Neufeld EF and Muenzer J. The Mucopolysaccharidoses. The Online Metabolic and Molecular Bases of Inherited Disease. https://doi.org/10.1036/ommbid.165 Accessed August 1, 2017.
- 22.Settembre C, et al. Proteoglycan desulfation determines the efficiency of chondrocyte autophagy and the extent of FGF signaling during endochondral ossification. Genes Dev. 2008;22(19):2645–2650. doi: 10.1101/gad.1711308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evers M, et al. Targeted disruption of the arylsulfatase B gene results in mice resembling the phenotype of mucopolysaccharidosis VI. Proc Natl Acad Sci U S A. 1996;93(16):8214–8219. doi: 10.1073/pnas.93.16.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polson HE, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6(4):506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 26.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 28.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 29.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19(17):4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronan B, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10(12):1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Álvarez-García Ó, et al. Growth hormone improves growth retardation induced by rapamycin without blocking its antiproliferative and antiangiogenic effects on rat growth plate. PLoS ONE. 2012;7(4):e34788. doi: 10.1371/journal.pone.0034788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietrocola F, et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell. 2016;30(1):147–160. doi: 10.1016/j.ccell.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim CY, Zoncu R. The lysosome as a command-and-control center for cellular metabolism. J Cell Biol. 2016;214(6):653–664. doi: 10.1083/jcb.201607005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newton PT, Vuppalapati KK, Bouderlique T, Chagin AS. Pharmacological inhibition of lysosomes activates the MTORC1 signaling pathway in chondrocytes in an autophagy-independent manner. Autophagy. 2015;11(9):1594–1607. doi: 10.1080/15548627.2015.1068489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Long F. mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development. 2014;141(14):2848–2854. doi: 10.1242/dev.108811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan B, et al. mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat Commun. 2016;7:11151. doi: 10.1038/ncomms11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mertz EL, et al. Makings of a brittle bone: Unexpected lessons from a low protein diet study of a mouse OI model. Matrix Biol. 2016;52-54:29–42. doi: 10.1016/j.matbio.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy. 2012;8(5):719–730. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koehne T, et al. Mannose 6-phosphate-dependent targeting of lysosomal enzymes is required for normal craniofacial and dental development. Biochim Biophys Acta. 2016;1862(9):1570–1580. doi: 10.1016/j.bbadis.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 42.Tomatsu S, et al. Missense models [Gustm(E536A)Sly, Gustm(E536Q)Sly, and Gustm(L175F)Sly] of murine mucoTopolysaccharidosis type VII produced by targeted mutagenesis. Proc Natl Acad Sci U S A. 2002;99(23):14982–14987. doi: 10.1073/pnas.232570999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polishchuk EV, et al. Wilson disease protein ATP7B utilizes lysosomal exocytosis to maintain copper homeostasis. Dev Cell. 2014;29(6):686–700. doi: 10.1016/j.devcel.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King KB, Kimura JH. The establishment and characterization of an immortal cell line with a stable chondrocytic phenotype. J Cell Biochem. 2003;89(5):992–1004. doi: 10.1002/jcb.10571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.