Abstract
Juvenile myelomonocytic leukemia (JMML) is a pediatric myeloproliferative neoplasm that bears distinct characteristics associated with abnormal fetal development. JMML has been extensively modeled in mice expressing the oncogenic KrasG12D mutation. However, these models have struggled to recapitulate the defining features of JMML due to in utero lethality, nonhematopoietic expression, and the pervasive emergence of T cell acute lymphoblastic leukemia. Here, we have developed a model of JMML using mice that express KrasG12D in multipotent progenitor cells (Flt3Cre+ KrasG12D mice). These mice express KrasG12D in utero, are born at normal Mendelian ratios, develop hepatosplenomegaly, anemia, and thrombocytopenia, and succumb to a rapidly progressing and fully penetrant neonatal myeloid disease. Mutant mice have altered hematopoietic stem and progenitor cell populations in the BM and spleen that are hypersensitive to granulocyte macrophage–CSF due to hyperactive RAS/ERK signaling. Biased differentiation in these progenitors results in an expansion of neutrophils and DCs and a concomitant decrease in T lymphocytes. Flt3Cre+ KrasG12D fetal liver hematopoietic progenitors give rise to a myeloid disease upon transplantation. In summary, we describe a KrasG12D mouse model that reproducibly develops JMML-like disease. This model will prove useful for preclinical drug studies and for elucidating the developmental origins of pediatric neoplasms.
Keywords: Hematology
Keywords: Genetic diseases, Hematopoietic stem cells, Leukemias
Introduction
Juvenile myelomonocytic leukemia (JMML) is a pediatric myeloproliferative neoplasm (MPN) caused by somatic mutations in the RAS/MEK/ERK pathway signaling genes, including KRAS, NRAS, PTPN11, NF1, and c-CBL (1). These mutations result in a hypersensitivity of hematopoietic progenitors to granulocyte macrophage–CSF (GM-CSF) and lead to monocytosis, anemia, thrombocytopenia, hepatosplenomegaly, and infiltration of peripheral tissues with histiocytes (2–4). Compared with other pediatric hematologic malignancies, the prognosis of patients with JMML is very poor. Allogeneic hematopoietic stem cell (HSC) transplantation is the only curative therapy, which nonetheless has a 5-year overall survival rate of only 52% (5).
The majority of JMML cases result from a mutation in a single gene (6–8). As such, disease models using the most common JMML-initiating mutations have been readily generated (9–11). The Mx1Cre KrasG12D mouse was the first conditional animal model of JMML and continues to be studied extensively (12, 13). However, these mice succumb with MPN that can be exacerbated by T cell leukemia/lymphoma (T-ALL) and that is confounded by nonhematopoietic KrasG12D expression (14–18). While the use of inducible Mx1Cre serves to limit oncogene expression until after birth, in utero KrasG12D expression owing to spontaneous Mx1Cre activity was not assessed in this model. Moreover, in utero KrasG12D expression induced by LysMCre or VavCre led to lung adenocarcinoma or prenatal lethality, respectively (19, 20). Thus, existing KrasG12D models do not directly address the fetal origin of JMML and do not reliably recapitulate the myeloid-restricted nature of the disease.
Converging clinical evidence suggests that the origin of JMML is closely associated with fetal development. Patients present very young with a median age of less than 2 years, and retrospective analyses indicate that the somatic disease–initiating mutation is frequently present at birth (6, 21, 22). Furthermore, BM progenitors of most patients exhibit a fetal-like gene expression signature, which correlates with an inferior prognosis (23). These findings strongly implicate a developmental origin of JMML and imply that disease-initiating mutations occur within a specific spatial and temporal context.
Fetal hematopoietic progenitors are functionally distinct from adult progenitors. Murine fetal progenitors have greater engraftment efficiency, biased lineage differentiation, and altered susceptibility to transformation compared with adult counterparts (24–27). Analogous studies in humans showed that fetal and cord blood CD34+ cells are more proliferative and have a greater propensity to form myeloid colonies in methylcellulose culture than do adult cells (28, 29). These characteristics of fetal progenitors suggest a mechanism through which they may evoke clinical features of JMML when challenged with a somatic oncogenic mutation.
We hypothesized that temporal expression of KrasG12D during fetal hematopoiesis that was limited functionally to the hematopoietic progenitor population would produce a JMML-like disease. Recently, the expression pattern of Flt3Cre has been extensively studied using lineage-tracing methods (30–32). Robust Flt3Cre activity begins in multipotent progenitors (MPPs) at E10.5 and is subsequently observed in more than 90% of mature leukocytes. We now demonstrate that KrasG12D is sufficient to produce a perinatal MPN when expressed in fetal hematopoietic progenitors using Flt3Cre and have thereby defined the crucial cellular and developmental environment needed to produce authentic features of KrasG12D-induced JMML.
Results and Discussion
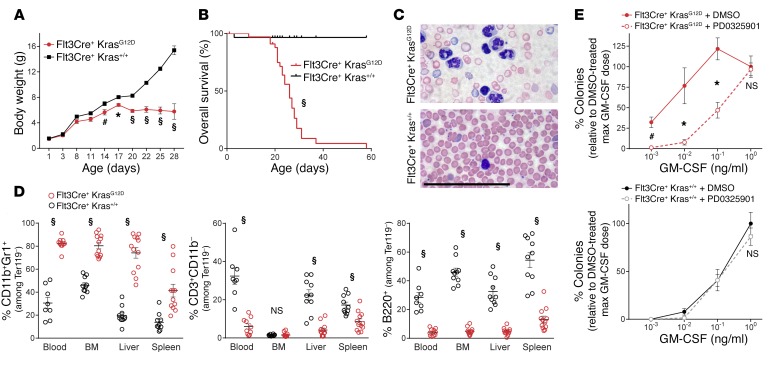
We mated Flt3Cre+ ROSAmTmG/mTmG studs with dams bearing a conditional Lox-STOP-Lox KrasG12D/+ allele (LSL-KrasG12D/+)to generate Flt3Cre+ ROSAmTmG/+ LSL-KrasG12D/+ mice (hereafter referred to as Flt3Cre+ KrasG12D mice), in which oncogene expression could be monitored by a switch from Tomato to GFP expression. Flt3Cre+ KrasG12D mutants were born at expected Mendelian ratios and had weight gain comparable to that of their littermates until 2 weeks of age (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI94031DS1). Mutants and littermates had equivalent activity of Flt3Cre, as measured by the percentage of GFP+ cells, and LSL-KrasG12D recombination in mutant mice was confirmed by PCR (Supplemental Figure 1, B and C). After 2 weeks, Flt3Cre+ KrasG12D mice showed progressive weight loss, leukocytosis, anemia, thrombocytopenia, and hepatosplenomegaly and died at a median age of 26 days (Figure 1, A–C, Supplemental Figure 2, and Supplemental Figure 3). Histological organ examination revealed a histiocytic infiltrate in the spleen, liver, lung, and intestines (Supplemental Figure 4), and a markedly increased frequency of CD11b+Gr1+ cells in the blood, BM, liver, and spleen was confirmed by flow cytometry (Figure 1D and Supplemental Figure 5). Notably, the frequency of CD3+ T lymphocytes and B220+ B lymphocytes was decreased, and Flt3Cre+ KrasG12D mice had an atrophied thymus compared with that seen in the littermates (Figure 1D, Supplemental Figure 3, and Supplemental Figure 4C). Consistent with a faithful model of hyperactive RAS-induced JMML, BM progenitors from Flt3Cre+ KrasG12D animals demonstrated hypersensitivity to GM-CSF in colony-forming assays, which was corrected by MEK inhibition (Figure 1E and Supplemental Figure 6).
Figure 1. Flt3Cre+ KrasG12D mice develop a JMML-like disease.
(A) Weight gain from birth (n = 12 mutants and 19 controls). (B) Overall survival (statistical analysis by Mantel-Cox test). (C) Peripheral blood smear (n = 5). Scale bar: 100 μm. (D) Flow cytometric quantification of tissue leukocytes. (E) Seven-day BM colony formation with 100 nM PD0325901 or 0.1% DMSO (n = 3 biological replicates/group). All analyses were performed on 3- to 4-week-old moribund Flt3Cre+ KrasG12D mice and age-matched littermates. *P < 0.05, #P < 0.01, and §P < 0.001, by unpaired, 2-tailed Student’s t test (A, D, and E).
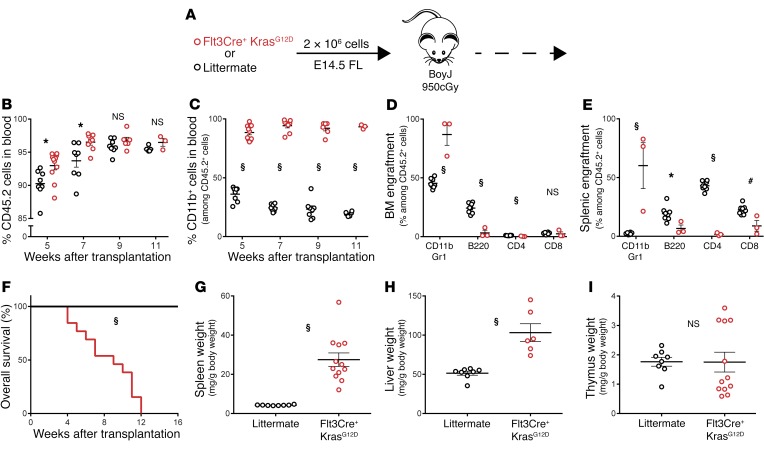
To confirm that the disease in Flt3Cre+ KrasG12D animals was initiated in utero and could be propagated autonomously in vivo, we transplanted E14.5 fetal liver (FL) cells into adult BoyJ animals (Figure 2A). Progenitors from mutant donors showed robust engraftment and rapidly contributed to monocytosis, anemia, and thrombocytopenia (Figure 2, B and C, and Supplemental Figure 7). Mutant progenitors gave rise to expanded myeloid cell populations in the BM and spleen, leading to hepatosplenomegaly and a median survival of 9 weeks (Figure 2, D–H). In stark contrast to other KrasG12D models, primary recipients of Flt3Cre+ KrasG12D progenitors showed no signs of T-ALL (Figure 2I). Upon secondary transplantation with 4 × 106 primary BM cells, Flt3Cre+ KrasG12D mutant cells engrafted, and 5 of 6 recipients rapidly succumbed with monocytosis, splenomegaly, and thymic atrophy (Supplemental Figure 8). These findings indicate that temporal expression of KrasG12D in utero transforms fetal hematopoietic progenitors into transplantable JMML-initiating cells.
Figure 2. Fetal Flt3Cre+ KrasG12D progenitors initiate a JMML-like disease upon transplantation.
(A) Schematic of FL transplants (n = 12 mutant and 8 control recipients). Analysis of donor cell (B) engraftment and (C) myeloid contribution in peripheral blood. (D and E) Flow cytometric quantification of donor leukocytes in BM and spleens of moribund mutant recipients and control recipients 16 weeks after transplantation. max, maximum. (F) Overall survival following transplantation (statistical analysis by Mantel-Cox test). (G–I) Normalized tissue weights of analyzed animals. *P < 0.05, #P < 0.01, and §P < 0.001, by unpaired, 2-tailed Student’s t test (B–E and G–I).
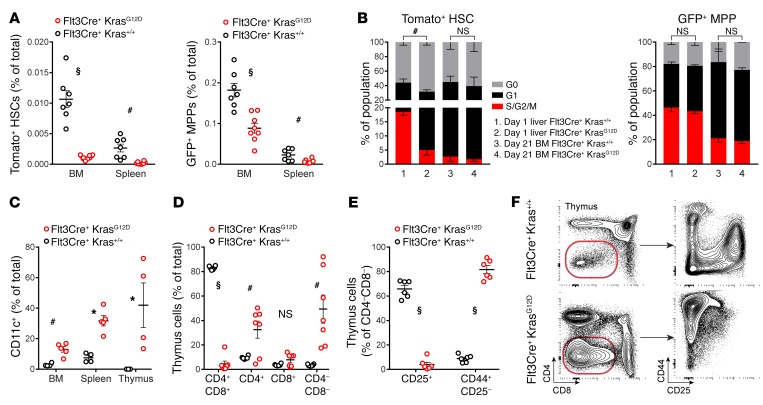
We proceeded to analyze the effect of fetal KrasG12D expression on the frequency and distribution of HSCs and progenitors. In contrast to Mx1Cre+ KrasG12D animals (16), we observed a reduction of HSCs (Tomato+ LSK CD150+CD48–) and MPPs (GFP+ LSK CD150–CD48+) in both the BM and spleen of moribund Flt3Cre+ KrasG12D mice (Figure 3A). This reduction corresponded with increased quiescence among HSCs in Flt3Cre+ KrasG12D mice (Figure 3B and Supplemental Figure 9). These effects were mediated non–cell autonomously, since LSL-KrasG12D was not recombined in HSCs (Supplemental Figure 10). We proceeded to analyze the progeny of Flt3Cre+ KrasG12D progenitors and found that BM cells cultured in cytokine-free medium gave rise to histiocytes that expressed CD11c and CD135 (Supplemental Figure 11). This finding is reminiscent of reports describing DC-like tumor cells in patients with JMML (2, 33, 34) and prompted us to analyze DC populations in Flt3Cre+ KrasG12D animals. We observed a marked increase in the frequency of CD11c+ cells in the BM and spleen (Figure 3C). Strikingly, the CD11c+ cell expansion was particularly prominent in the atrophied thymus, where we saw a concomitant deficit of CD4–CD8–CD25+ committed T cell progenitors and CD4+CD8+ double-positive cells (Figure 3, C–F, and Supplemental Figure 12). We also observed this propensity for preferential DC differentiation in recipients of Flt3Cre+ KrasG12D FL progenitors (Supplemental Figure 13).
Figure 3. Analysis of Flt3Cre+ KrasG12D progenitor frequency and differentiation.
(A) Frequency of HSCs and MPPs in the BM and spleens of mutants and littermates. (B) Cell-cycle analysis of HSCs and MPPs from 1-day-old liver (n = 3 WT and 4 mutants) and 21-day-old BM (n = 2/group); representative gating is shown in Supplemental Figure 9. (C) Flow cytometric quantification of tissue DCs. (D–F) Flow cytometric quantification and representative gating of thymic cells. Error bars represent the SEM. Cell-cycle statistical analyses were performed using a χ2 test, and other analyses were performed using an unpaired, 2-tailed Student’s t test. *P < 0.05, #P < 0.01, and §P < 0.001.
We present the first KrasG12D model to our knowledge to unify JMML disease–defining features: an in utero origin; viability at birth followed by a failure to thrive; anemia; thrombocytopenia; monocytosis; hepatosplenomegaly; and infiltration of tissues with histiocytes. All Flt3Cre+ KrasG12D mice succumbed to a myeloid disease that could be recapitulated following FL transplantation. This is in contrast to existing KrasG12D models, whose MPN is exacerbated by nonhematopoietic oncogene expression and by the unpredictable coemergence of T-ALL (15–20).
Flt3Cre has an expression pattern markedly different from that of Mx1Cre, which may explain the observed respective myeloid versus lymphoid disease outcomes. Mx1Cre is used to target adult progenitors, whereas Flt3Cre becomes active in fetal MPPs. As such, Flt3Cre initiates KrasG12D expression within an in utero progenitor that is more proliferative, has greater repopulating ability, and has enhanced myeloid cell production compared with adult progenitors targeted by Mx1Cre (24, 27). This context emulates studies of JMML patients that highlighted the fetal origins of this disease: the causative somatic mutation commonly occurs before birth, and BM cells have a gene expression signature that is characteristic of fetal progenitors (22, 23). Therefore, in contrast to Mx1Cre, Flt3Cre targets KrasG12D expression to hematopoietic progenitors at the appropriate developmental stage to recapitulate the origin of JMML.
The identity of the JMML-initiating cell has been controversial. On the one hand, case reports have shown that CD34+CD38– phenotypic HSCs express disease-initiating mutations (6, 35) and that xenotransplantation of patients’ progenitors gives rise to mutated myeloid, B, and T cells with a common clonal origin (36). On the other hand, circulating T lymphocytes from most patients do not express the disease-initiating mutation (7, 37), suggesting that JMML is initiated within a MPP that undergoes a differentiation block during T lymphocyte commitment. Consistent with this hypothesis, case reports suggest that patients with JMML have decreased T cell frequencies in the BM and spleen (38, 39). These findings parallel our Flt3Cre+ KrasG12D model, which has a paucity of T cells, an atrophied thymus, and abnormal T cell differentiation.
An earlier study found similarly skewed T lymphocyte development when KrasG12D expression was restricted to DCs in p53–/– mice (40). Our results advance these findings to show that KrasG12D expression in multipotent progenitors results in widespread tissue infiltration with DCs that are distinct from the concomitantly expanded neutrophils (Supplemental Figure 13B). Importantly, case reports have equally noted that JMML patients’ tissues are infiltrated by atypical histiocytes (2, 41) and that children with the DC disorder juvenile xanthogranuloma are at increased risk of JMML (42). Additionally, Langerhans cell histocytosis and Erdheim-Chester disease, two aggressive DC disorders, are also characterized by hyperactive RAS signaling (43). These reports, along with our findings from Flt3Cre+ KrasG12D mice, suggest that a more formal investigation of DC involvement in JMML is warranted.
A unique feature of Flt3Cre+ KrasG12D mice is that their HSCs do not express the oncogene (Supplemental Figure 10). The full penetrance of a MPN in our model is therefore consistent with the hypothesis that the HSC is the cell of origin for KrasG12D-evoked T-ALL (16, 17). Notably, the quiescence of nononcogene-expressing HSCs in Flt3Cre KrasG12D mice demonstrates a profound non–cell-autonomous effect of this mutation. Our finding supports the conclusions of Sabnis et al., who noted that residual nonrecombined LSK Flt3– cells in Mx1Cre KrasG12D mice did not expand to compensate for diminishing oncogene-expressing HSCs (16). Our results suggest that KrasG12D-expressing hematopoietic cells induce an aberrant BM microenvironment that stifles the expansion of normal neighboring HSCs. This yields the provocative hypothesis that patients with JMML relapse following allogenic transplantation as a result of an adverse niche that impedes the proliferation of donor HSCs.
In summary, we describe what to our knowledge is the first KrasG12D mouse model that recapitulates defining features of JMML. Flt3Cre+ KrasG12D mice are viable, develop monocytosis, anemia, thrombocytopenia, and hepatosplenomegaly and die from a fully penetrant myeloid disease. This model further emulates underappreciated features of JMML such as a paucity of mature T lymphocytes and an expansion of DCs and thereby hints at potential new therapeutic strategies. Flt3Cre+ KrasG12D mice will prove useful for preclinical drug studies targeting the RAS/MEK/ERK signaling pathway and will help elucidate the developmental origins of JMML and pediatric leukemias.
Methods
Detailed methods, including all flow cytometry antibodies (Supplemental Table 1), are described in the Supplemental Methods.
Study approval.
Animal studies were approved by the IACUC of the Indiana University School of Medicine. Animals were genotyped using primers outlined in Supplemental Table 2 of Supplemental Methods.
Statistics.
P values comparing mutant and littermate groups were calculated using 2-tailed Student’s t tests, Mantel-Cox log-rank tests, or χ2 tests, as indicated in the figure legends. P values of less than 0.05 were considered significant. All error bars represent the SEM.
Author contributions
SPT conceived the study, designed, performed, and analyzed experiments, and wrote the manuscript. MK designed and performed experiments. RJC and MCY conceived the study, designed and analyzed experiments, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank Momoko Yoshimoto (University of Texas Health Science Center, Houston, Texas, USA) for their helpful discussions. The authors are grateful to Slava Epelman (University of Toronto, Toronto, Canada) for the gift of Flt3Cre+ ROSA26mTmG/mTmG mice. This work was supported by the Riley Children’s Foundation and the NIH (F30 HL128011, to SPT, and R21 CA202296, to RJC and MCY). We appreciate the technical assistance provided by Karen Pollok and Tony Sinn of the Indiana University In Vivo Therapeutics Core and Susan Rice of the Indiana University Flow Cytometry Resource Facility (supported by NIH grant P30 CA082709). The authors gratefully acknowledge the administrative assistance of Tracy Winkle and Tiffany Lewallen (Indiana University School of Medicine, Indianapolis, Indiana, USA).
Version 1. 08/21/2017
Electronic publication
Version 2. 09/20/2017
Print issue publication
Version 3. 11/14/2017
Michihiro Kobayashi was added to the author list.
Footnotes
MK’s present address is: Center for Stem Cell and Regenerative Medicine, the Brown Institute of Molecular Medicine, Houston, Texas, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(10):3652–3656.https://doi.org/10.1172/JCI94031.
References
- 1.Chan RJ, Cooper T, Kratz CP, Weiss B, Loh ML. Juvenile myelomonocytic leukemia: a report from the 2nd International JMML Symposium. Leuk Res. 2009;33(3):355–362. doi: 10.1016/j.leukres.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng CS, et al. Juvenile chronic myeloid leukemia. A malignancy of S-100 protein-positive histiocytes. Am J Clin Pathol. 1988;90(5):575–582. doi: 10.1093/ajcp/90.5.575. [DOI] [PubMed] [Google Scholar]
- 3.Locatelli F, Niemeyer CM. How I treat juvenile myelomonocytic leukemia. Blood. 2015;125(7):1083–1090. doi: 10.1182/blood-2014-08-550483. [DOI] [PubMed] [Google Scholar]
- 4.Loh ML. Recent advances in the pathogenesis and treatment of juvenile myelomonocytic leukaemia. Br J Haematol. 2011;152(6):677–687. doi: 10.1111/j.1365-2141.2010.08525.x. [DOI] [PubMed] [Google Scholar]
- 5.Locatelli F, et al. Analysis of risk factors influencing outcomes after cord blood transplantation in children with juvenile myelomonocytic leukemia: a EUROCORD, EBMT, EWOG-MDS, CIBMTR study. Blood. 2013;122(12):2135–2141. doi: 10.1182/blood-2013-03-491589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stieglitz E, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet. 2015;47(11):1326–1333. doi: 10.1038/ng.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakaguchi H, et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet. 2013;45(8):937–941. doi: 10.1038/ng.2698. [DOI] [PubMed] [Google Scholar]
- 8.Caye A, et al. Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet. 2015;47(11):1334–1340. doi: 10.1038/ng.3420. [DOI] [PubMed] [Google Scholar]
- 9.Xu D, et al. Non-lineage/stage-restricted effects of a gain-of-function mutation in tyrosine phosphatase Ptpn11 (Shp2) on malignant transformation of hematopoietic cells. J Exp Med. 2011;208(10):1977–1988. doi: 10.1084/jem.20110450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan G, et al. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 2009;113(18):4414–4424. doi: 10.1182/blood-2008-10-182626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le DT, et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood. 2004;103(11):4243–4250. doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- 12.Braun BS, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101(2):597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan IT, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113(4):528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kindler T, et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood. 2008;112(8):3373–3382. doi: 10.1182/blood-2008-03-147587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staffas A, Karlsson C, Persson M, Palmqvist L, Bergo MO. Wild-type KRAS inhibits oncogenic KRAS-induced T-ALL in mice. Leukemia. 2015;29(5):1032–1040. doi: 10.1038/leu.2014.315. [DOI] [PubMed] [Google Scholar]
- 16.Sabnis AJ, et al. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol. 2009;7(3):e59. doi: 10.1371/journal.pbio.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009;113(6):1304–1314. doi: 10.1182/blood-2008-01-134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong G, et al. Notch1 gene mutations target KRAS G12D-expressing CD8+ cells and contribute to their leukemogenic transformation. J Biol Chem. 2013;288(25):18219–18227. doi: 10.1074/jbc.M113.475376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507(7491):190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang P, Gao C, Li A, Aster J, Sun L, Chai L. Differential roles of Kras and Pten in murine leukemogenesis. Leukemia. 2013;27(5):1210–1214. doi: 10.1038/leu.2012.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kratz CP, et al. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood. 2005;106(6):2183–2185. doi: 10.1182/blood-2005-02-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuda K, et al. Quantitative assessment of PTPN11 or RAS mutations at the neonatal period and during the clinical course in patients with juvenile myelomonocytic leukaemia. Br J Haematol. 2010;148(4):593–599. doi: 10.1111/j.1365-2141.2009.07968.x. [DOI] [PubMed] [Google Scholar]
- 23.Helsmoortel HH, et al. LIN28B overexpression defines a novel fetal-like subgroup of juvenile myelomonocytic leukemia. Blood. 2016;127(9):1163–1172. doi: 10.1182/blood-2015-09-667808. [DOI] [PubMed] [Google Scholar]
- 24.Copley MR, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15(8):916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335(6073):1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter SN, et al. Fetal and neonatal hematopoietic progenitors are functionally and transcriptionally resistant to Flt3-ITD mutations. Elife. 2016;5:e18882. doi: 10.7554/eLife.18882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest. 2006;116(10):2808–2816. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lansdorp PM, Dragowska W, Mayani H. Ontogeny-related changes in proliferative potential of human hematopoietic cells. J Exp Med. 1993;178(3):787–791. doi: 10.1084/jem.178.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broxmeyer HE, et al. Growth characteristics and expansion of human umbilical cord blood and estimation of its potential for transplantation in adults. Proc Natl Acad Sci U S A. 1992;89(9):4109–4113. doi: 10.1073/pnas.89.9.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer SW, Schroeder AV, Smith-Berdan S, Forsberg EC. All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell. 2011;9(1):64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epelman S, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40(1):91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaudin AE, et al. A Transient developmental hematopoietic stem cell gives rise to innate-like B and T cells. Cell Stem Cell. 2016;19(6):768–783. doi: 10.1016/j.stem.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longoni D, et al. Commitment of juvenile myelo-monocytic (JMML) leukemic cells to spontaneously differentiate into dendritic cells. Hematol J. 2002;3(6):302–310. doi: 10.1038/sj.thj.6200192. [DOI] [PubMed] [Google Scholar]
- 34.Estrov Z, et al. Characterization of malignant peripheral blood cells of juvenile chronic myelogenous leukemia. Cancer Res. 1986;46(12 Pt 1):6456–6461. [PubMed] [Google Scholar]
- 35.Busque L, et al. Clonality in juvenile chronic myelogenous leukemia. Blood. 1995;85(1):21–30. [PubMed] [Google Scholar]
- 36.Nakamura Y, et al. Engraftment of NOD/SCID/gammac(null) mice with multilineage neoplastic cells from patients with juvenile myelomonocytic leukaemia. Br J Haematol. 2005;130(1):51–57. doi: 10.1111/j.1365-2141.2005.05578.x. [DOI] [PubMed] [Google Scholar]
- 37.Flotho C, et al. RAS mutations and clonality analysis in children with juvenile myelomonocytic leukemia (JMML) Leukemia. 1999;13(1):32–37. doi: 10.1038/sj.leu.2401240. [DOI] [PubMed] [Google Scholar]
- 38.Krombholz CF, et al. Long-term serial xenotransplantation of juvenile myelomonocytic leukemia recapitulates human disease in Rag2-/-γc-/- mice. Haematologica. 2016;101(5):597–606. doi: 10.3324/haematol.2015.138545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira AF, Tansini A, Vidal DO, Lopes LF, Metze K, Lorand-Metze I. Characteristics of the phenotypic abnormalities of bone marrow cells in childhood myelodysplastic syndromes and juvenile myelomonocytic leukemia. Pediatr Blood Cancer. 2017;64(4):e26285. doi: 10.1002/pbc.26285. [DOI] [PubMed] [Google Scholar]
- 40.Böttcher JP, Zelenay S, Rogers NC, Helft J, Schraml BU, Reis e Sousa C. Oncogenic transformation of dendritic cells and their precursors leads to rapid cancer development in mice. J Immunol. 2015;195(10):5066–5076. doi: 10.4049/jimmunol.1500889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozono S, et al. Juvenile myelomonocytic leukemia characterized by cutaneous lesion containing Langerhans cell histiocytosis-like cells. Int J Hematol. 2011;93(3):389–393. doi: 10.1007/s12185-011-0787-x. [DOI] [PubMed] [Google Scholar]
- 42.Paulus S, Koronowska S, Fölster-Holst R. Association between juvenile myelomonocytic leukemia, juvenile xanthogranulomas and neurofibromatosis type 1: Case report and review of the literature. Pediatr Dermatol. 2017;34(2):114–118. doi: 10.1111/pde.13064. [DOI] [PubMed] [Google Scholar]
- 43.Emile JF, et al. Recurrent RAS and PIK3CA mutations in Erdheim-Chester disease. Blood. 2014;124(19):3016–3019. doi: 10.1182/blood-2014-04-570937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.