Abstract
Recent advances allow access to human cell-based intestinal organoids that recreate human physiology to levels not possible with conventional 2D cell cultures. Despite their huge potential, there are many challenges that remain. This review will cover recent bioengineering approaches to improve organoid maturation, scale up, reproducibility and analysis. The first section covers the advances in engineering the culture environment, followed by the section on tools for micro-manipulation and analysis of organoids. The last section reviews the computational models developed to guide the use of engineered materials and tools, and to interpret observed results as well. The ability to use organoids for discovery research, and the need to both exert exquisite experimental control and obtain quantitative measurements from organoid models means that the field is ripe for collaborative efforts between biologists, engineers, clinicians and industry.
Graphical Abstract
Introduction
Because of the importance of the gastrointestinal (GI) tract to health and disease, many in vitro and in vivo animal models and human tissue models of the GI tract have been developed.[1–3] Conventional in vitro models that rely heavily on 2D cell cultures, however, do not recapitulate the complex in vivo cell and tissue organization and are often representative of cancerous tissue. Animal models also fail to recapitulate much of human physiology and disease.[1] A breakthrough emerged in 2009 when Sato et al.[4] successfully cultured murine epithelial organoid from single Lgr5+ intestinal stem cell (ISC) in vitro. Subsequently, human pluripotent stem cell(hPSC)-derived intestinal organoid (HIO)[5] and organoids derived from human normal and diseased biopsy samples [6,7] were developed. These 3-D, self-organized, physiologically-relevant cellular structures can be grown long-term and remain genomically stable. These systems have already proven useful in studies of GI tract development, homeostasis, human-microbiome symbiosis and pathogenesis.[1,3,8]
The speed at which organoid technologies have been developed and implemented in biomedical research is also accompanied by technical challenges and new opportunities. This short article highlights bioengineering solutions to solve unmet needs in intestinal organoid research, such as improving scalability and reproducibility, producing organoids with more mature or adult phenotypes, developing versatile functional readouts, and providing mathematical frameworks for analyses.
Engineering the culture environment of intestinal organoids
The Extracellular matrix (ECM)
The extracellular matrix (ECM) interacts with cells and tissue, providing both mechanical support and biochemical cues. Naturally-derived ECMs are readily available and widely used for intestinal organoid production and growth, with Matrigel (or similar products) being the most common commercially available products used.[4,5,9] However, there are several limitation with these ECM products; they are often derived from tumor cell lines and have ill-defined properties, batch-to-batch variability, and non-intestine specific origin, better-defined Matrigel-alternative materials are actively sought after.[10,11]
Two additional examples of naturally-derived ECMs include Collagen I and decellularized intestinal ECM, which is the compositionally complex but intestine-specific. Collagen I has been used to culture both human and murine reconstituted intestinal organoids.[12,13] In a comparative analysis, gene expression in human epithelial organoids after 1 week of co-culture with intestinal subepithelial myofibroblasts embedded in Collagen I was relatively similar to Matrigel control[13] but cell monolayers were also formed in Collagen I, suggesting that the growth properties in different matrices are altered. Other differences in collagen I cultures include formation of a smoother epithelium and decreased budding.[14] In this context, it is interesting to note that matrix stiffness has recently been linked with maintenance of the stem cell identity, with softer matrices being required for cellular differentiation.[10] Recent work placed both murine colonic crypt and human rectal crypts on top of neutralized Collagen I rather than embedding in the gel, which formed 2D monolayers containing both proliferative and differentiated cells. These monolayers were capable of converting to 3D organoids when embedded in Matrigel and the opposite was also possible, up to five passages in murine colonic monolayers in particular.[15]
One perceived advantage of using decellularized ECM is that the 3D microstructure of the intestine and intestine-specific ECM molecules are preserved. [16,17] Interestingly, reseeding the decellularized human or porcine ECM with HIOs was shown to be possible; however, reseeded matrices did not assume the normal crypt-villus structure of the intestine, and, reseeded scaffolds did not persist when transplanted into immunocompromised mice.[16] In contrast, simply transplanting hPSC-derived intestinal organoids into mice led to remarkable engraftment and growth into crypt-villus structures.[18]
Synthetic ECMs have been designed to customize mechanical and biochemical properties, and create controlled environments. DiMarco et al.[19] developed an elastin-based engineered-ECM (eECM), where stiffness and adhesion site density could be independently controlled. Mouse organoid formation efficiency was similar to collagen I with the best efficiency observed with an eECM of 180 Pa mechanical stiffness and 3.2 mM of Arg-Gly-Asp (RGD) cell-binding sites. (Figure 1A) A minimal yet cell-supportive, defined ECM was engineered by the Lutolf lab using a chemically inert polyethylene glycol (PEG) polymer with adjustable stiffness as the base material and varying densities of key cell-adhesive components including RGD peptide and laminin-111.[10] This study showed that different matrix parameters are preferred at different stages of organoid formation; stiffer matrix was required during initial stage of ISC expansion then subsequent softening of the matrix was essential for continued expansion and organoid formation, and only laminin-111 supported budding of organoids whereas laminin-111-derived peptides did not.(Figure 1B) Interestingly, the reported optimal ECM mechanical stiffness for organoid formation is similar for both the elastin-based eECM[19] and the PEG-based synthetic ECM[10]. The use of artificial matrices with minimal components allows the deconvolution of relevant environmental cues, otherwise unachievable using naturally-derived matrices. While tubular scaffold of polyglycolic/poly-L-lactic acid (PGA/PLLA) demonstrated the potential to guide expansion of HIOs mimicking human adult small intestinal architecture when implanted to mice[16], synthetic ECMs when used in vitro have yet to match Matrigel for their ability to support long-term growth and differentiation.
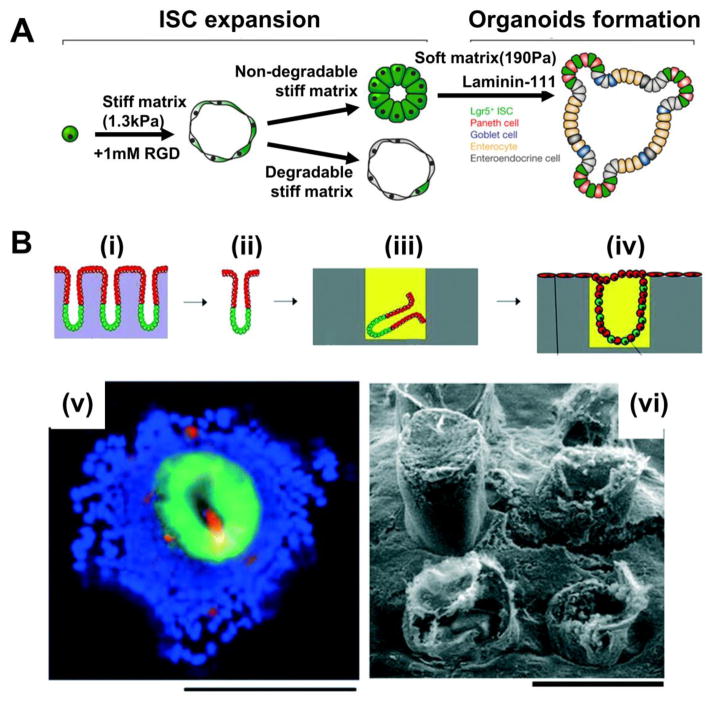
Figure 1. Engineering the culture environment of intestinal organoids.
(A) Recombinantly engineered ECM with lower stiffness and high concentration of cell-binding sites increased organoid formation efficiency to levels comparable to collagen I cultures. Adapted from [19] with permission. Scale bar=50 μm.
(B) Optimal mechanical and biochemical properties of synthetic ECM should be modulated depending on the organoid formation stage (i) and confocal images of differentiated cells in the enteroid, grown in synthetic ECM (ii). Adapted from [10] with permission
(C) Schematics illustrating the experimental procedure for generating colonic epithelium using microstructured scaffolds: (i,ii) isolation of colonic crypt, (iii) embedding the crypt in Matrigel-filled microwell, and (iv) in vitro 2D/3D colonic epithelium with polarized proliferative and non-proliferative architecture. Immunofluorescence image (green = GFP, red = MUC2, blue = nuclei stain)(v) and scanning electron microscopy(iv) of the epithelium. Adapted from [24] with permission. Scale bars=200 μm.
Microfabrication-assisted 3D culture
The intestine has villi and crypts that not only increase surface area but also form niches for stem cell self-renewal and differentiation. This has inspired man-made, intestinal villus and crypt-like, 3D microstructured culture substrates, which have also been used, either alone or in combination with ECM material to culture intestine cells. Initial demonstrations used immortalized intestinal cell lines[20–22] but the strategy also translates to culture of primary cells. These crypt-like domains in colonic organoids derived from both mouse and human; however, do not have discrete proliferative and non-proliferative regions, as would be the case in vivo.[7,23] Wang et al.[24] attributed this lack of spatial organization partially to the current colonic organoid expansion protocol not precisely recapitulating in vivo microenvironment, particularly the microstructure. An effort to combine ECM guidance cues together with topography led to the development of polydimethylsiloxane (PDMS) microwell arrays filled with Matrigel. Isolated individual crypts cultured in these hybrid microstructures expanded while maintaining a stem cell population within the Matrigel-filled microwells, while the exposed top surface between adjacent crypts became confluent with a differentiated epithelium (Figure 1C).
Micropatterned culture platforms have also been adopted to mimic the early development of embryonic stem cells [25,26]. For example, spatially confining stem cells cultured on a 2D surface into small islands leads to more homogenous self-organized germ layer differentiation.[25] Using a similar micropatterning approach, a xenogeneic-free protocol for generating HIOs was recently reported.[27] In contrast to conventional methods used to generate HIOs, which relies on chemical cues used in a stepwise manner to mimic different stages of embryonic development in vitro [5], the xenogeneic-free protocol utilized stochastic differentiation, which led to all of the germ layers to being incorporated into the organoids. Unique features of this method include spatially-segregated formation of more uniformly-sized HIOs and presence of mesoderm-derived smooth muscle cells and ectoderm-derived enteric neurons that collectively create peristalsis-like motility. Drawbacks to this method include the relatively inefficient generation of intestinal organoids, since many non-intestinal 3D structures are generated due the stochastic differentiation procedure used; gene expression profiles were highly varied within organoids owing partly due to a more complex cellular composition.
Instrumenting intestinal organoids
Modulating and measuring the organoid environment
Intestinal motility and luminal flow are key intestinal functions required for efficient nutrient absorption, waste secretion and sustaining stable host-microbe interactions.[28] While flow and some motility is relatively straightforward to incorporate in epithelial cell-lined microfluidic devices[29–32], it is much more difficult to perform long-term luminal perfusion of intestinal organoids while preserving organoid structure and function. Since postnatal intestinal development and pathophysiology require host-microbiota-virus interaction, inoculating the intestinal organoids with bacteria and viruses is crucial part of many studies.[3]
Epithelium-only human organoids can be readily transferred from a 3D organoid culture onto 2D Transwell cultures where cells form a polarized monolayer.[33] With both apical and basal side of the epithelium exposed to fluid that is readily accessed by simple pipetting, co-culture with bacteria and measurement of transepithelial electrical resistance (TEER) is straight forward. (Figure 2A) [17,33,34] TEER measurements of primary intestinal cells are reported to exhibit physiological values whereas traditional cell culture lines, such as polarized Caco-2 cell monolayers typically have TEER values exceeding physiological values.[17]
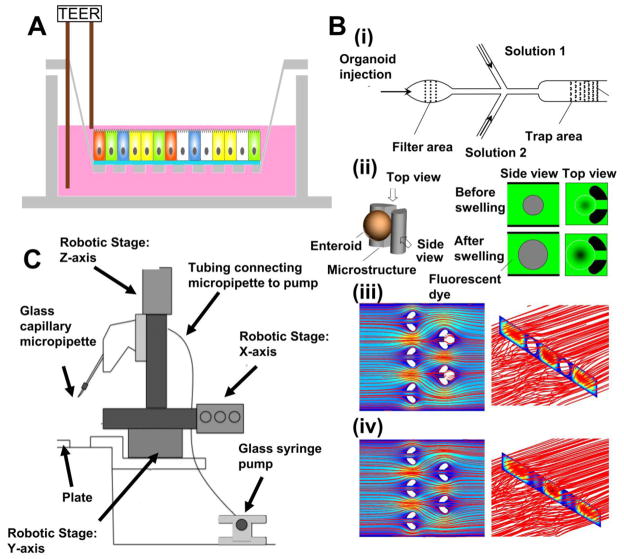
Figure 2. Instrumenting intestinal organoids.
(A)TEER measurement across a monolayer created from enteroids.
(B) (i) Microfluidic device for trapping enteroids filtered based on size. (ii) Enteroid swelling indirectly measured by imaging the external fluid volume change in a fixed field-of-view. Numerical simulations of fluid flow patterns in the device with microstructure-trapped enteroids (iii) and without them (iv). Adapted from [37] with permission.
(C) Instrumentation for automated high-throughput sorting of intestinal organoids. Adapted from [39] with permission.
Unlike epithelium-only patient-derived organoids, HIOs possess a mesenchymal layer and do not readily spread into clean epithelial monolayers on Transwells (our unpublished data). Thus, luminal side bacteria interaction requires microinjection of bacteria into the organoid.[35,36] As long as microinjection procedures are performed carefully, bacteria leakage is minimized.[35] The process is, however, labor intensive and inherently inefficient when a large number of organoids are involved.
High-throughput batch processing
Conventional methods of passaging, sorting, and analyzing intestinal organoids take time and require practiced skills. A number of recent efforts have aimed to overcome this limitation to enable higher-throughput handling. An example is the microfluidic filtering and trapping device designed for epithelium-only organoids.(Figure 2Bi)[37] In this work, Jin et al. performed multiplex measurements of epithelial swelling in response to external osmolality changes or secretory diarrhea-inducing viral toxins. Here, the change in external fluorescent fluid volume within a fixed field-of-view was monitored to indirectly measure the epithelial volume change held in place by PDMS pillars. This approach circumvents the potential error arising from a platform dependent on microinjection into organoids embedded within ECM. Such error can arise from optical path perturbations by the ECM thereby creating imaging errors, and from pressure build-up due to the surrounding ECM limiting expansion due to stiffness mismatches at the ECM and tissue interface. (Figure 2Bii)[37]
Another organoid arraying device was recently described to facilitate single cell-level spatio-temporal tracking of ISCs and Paneth cells (PCs).[38] In a polystyrene-coated PDMS microwells, cells in suspension are centrifuged to have a small population of cells per microwell (<7cells/well). Wells were subsequently filled with Matrigel and culture media. Cell-containing wells are tile-scan imaged over time. With this approach, a large number of replicated data were collected in a single data acquisition cycle and analyzed based on initial cellular composition, intercellular distance over time, and morphology evolution as the cells start to form organoids. For gene expression analysis, each cell/organid-containing magnetic polystyrene raft was released from the PDMS wells. These types of regularly patterned microdevices enable multiple data points from every well of the array with the assistance of computer interface. These throughput capabilities enhance the likelihood that intestinal organoid systems can be scaled appropriately for industry applications.
Process engineering tools were also developed to sort hindgut spheroids based on morphological features, such as size and inner mass of the spheroids. The relationship between these features and maturation of the spheroids to intestinal organoids was used to optimize the paramaters for selecting spheroids.[39] The microscope used for the automated capillary-based sorting was instrumented with an image processing unit, a 3D stage control, and a harvesting unit. (Figure 2C) After the image processing unit determined which spheroid to harvest based on morphological features, the 3D stage control positions the tip of a glass capillary close to the spheroid of interest and pulls it up. The speed of the whole procedure is reported to be one spheroid per minute. Using this sorting method, the portion of spheroids that matured to be intestinal organoids were enriched from 13% to 20%, by size exclusion of spheroid diameter smaller than 75 μm, between day 5 and day 10. This approach may be adopted to culture more consistent intestinal organoid using various morphological features.
Computational modeling of intestinal organoid systems
Modeling morphogenesis
Several computational models of the morphological development of the gut have been developed. Buske et al.[40] modeled how proliferation-induced local shape and mechanical property fluctuation in epithelial intestinal organoids induce crypt-like domains where stem cells are localized. The model provides an explanation for how geometric and mechanical features of the culture environment is critical and why flat hard plastic dish culture may be unable to support organoid formation. Shyer et al.[41] simulated how villus morphogenesis is mechanically-coupled with muscle layer development during embryonic chick gut development. Circumferential compression and successive longitudinal compression applied to the epithelium by surrounding muscle layers result in the formation of longitudinal ridges and the formation of zigzag patterns and villi, respectively, within 2 weeks of fertilization. (Figure 3A) On the other hand, due to a thicker epithelium, it has been put forward that pattern formation in the mouse gut requires localized biochemical signals in addition to mechanical forces.[42–44]
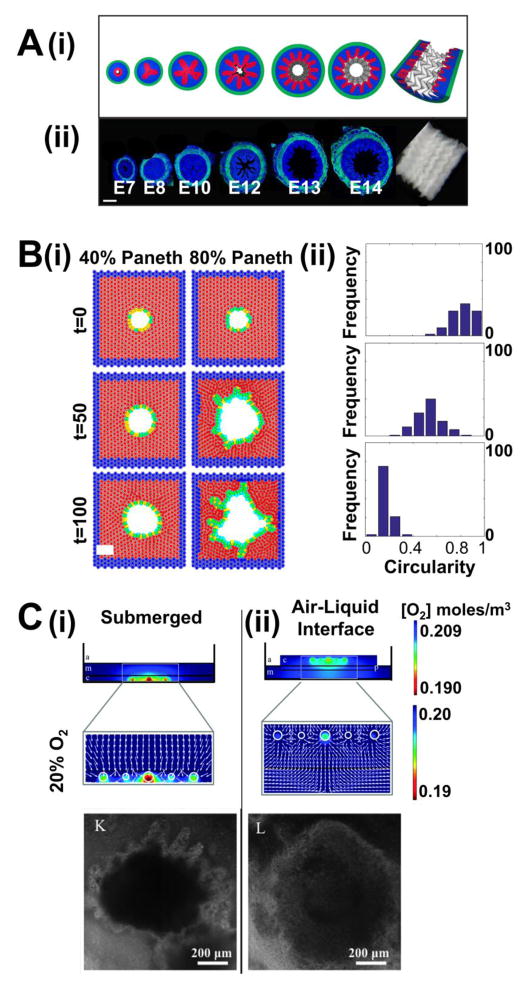
Figure 3. Computation modeling of the intestinal organoid systems.
(A) Computational simulation of villification assisted by circumferential and longitudinal compression (green=muscle layer; blue=mesenchyme; red=endoderm) (i) and corresponding chick intestine dissections (blue = DAPI nuclei stain; green = αSMA smooth muscle actin stain) (ii). Adapted from [41] with permission. Scale bar=100 μm.
(B) (i) Mathematical modeling showing increased proportion of stiff Paneth cells in epithelium lead to buckling of the epithelium, hence crypt fission. (Blue=non-epithelial cells; red=Matrigel or stromal cells; yellow=Lgr5+ cells; green=Paneth cells) (ii) Histograms of final circularity index distribution with respect to Paneth cell composition. Adapted from [49] with permission.
(C) Numerical simulations for the oxygenation of tissue-derived intestinal organoids embedded in collagen (i, top) and cultured at an air-liquid interface (ALI) (ii, top) under ambient air. Brightfield images of organoids grown in submerged culture (i, bottom) and ALI culture (ii, bottom) for 2 weeks. Adapted from [51] with permission.
After crypts have formed, crypt fission becomes a critical process for tissue growth and maintenance.[45,46] Computer-aided simulations of these processes have been reported.[47–49] In a recent study, Langlands et al.[49] studied the mechanisms of crypt fission from a cell population and biomechanical perspective using mouse the small intestine and organoids as the experimental test-bed. In situ imaging of mouse small intestinal crypt fission show Lgr5+ stem cells at the bifurcation sites with Paneth cells just adjacent to them. Weaker attachment to the basement membrane of Lgr5+ cells compared to that of Paneth cells, led to preferential buckling at stem cell clusters. Mouse epithelium-only organoid cultures allowed precise modulation of the number of Paneth cells relative to the number of stem cells. Experimentally, the authors found that having more Paneth cells created organoids with fewer crypt-like domains and changed the crypt shapes to be more spherical. In their computational model, however, where only the mechanical stiffness difference between the two cell types were assumed, increased relative population of Paneth cells led to increased budding of crypt-like domains. (Figure 3B) This disagreement between experiment and model may be explained by formation of more Lgr5+ cell clusters via mitosis in the experiments; something not included in the model.[49] Temporal patterns of crypt budding has also been analyzed and modeled using epithelium-only organoids. Crypt budding in organoids were observed to occur every 12 hours. Although the physiological relevance of this finding is still controversial, computational models suggest that the circadian rhythm synchronizes this rhythmic cell cycle entry.[50]
Currently, modeling efforts related to organoid morphogenesis have focused more on mechanical mechanisms. As biomechanical interactions and biochemical signals are intimately coupled, morphogenesis models that integrate both aspects may provide further insights.
Analysis of the microenvironment
Microscale culture platforms limit the use of conventional macroscopic physical sensors for static and dynamic environment measures near tissue constructs. The in vivo intestinal oxygen (O2) environment is very characteristic; however, little work has been done to understand the effect of O2 microenvironments on in vitro intestinal organoid cultures. A recent study compared the O2 gradient surrounding tissue-derived organoids in submerged or air-liquid interface (ALI) using a numerical simulation tool.[51] Combining measurement of O2 concentrations right beneath the organoids, and Michaelis-Menten kinetics, oxygenation was visualized in different culture platforms. Organoids in ALI platform generally experience higher O2 tension and smaller O2 gradient than those in submerged cultures. Within 2 weeks of culture the organoids grew up to 1 mm and exhibit more circular morphology in ALI culture. (Figure 3C) It is still unclear whether such environment creates more physiologically relevant organoids. For organoid cultures with a fluidic component, flow patterns surrounding the tissue construct can be simulated.[17,37] Such an approach allowed the visualization of flow pattern change in response to tissue entrapment (Figure 2Biii,iv)[37], and shear stress calculations made to match experiments to the in vivo fluid flow in the luminal environment[17]. While these advances are encouraging, much work remains for microenvironment simulation technologies to be fully exploited for in-depth physiologic interpretations.
Conclusion
This short review highlights recent advances in bioengineering tools to create well-defined intestinal organoid cultures. Compared to the tremendous pace of scientific advances in the field of intestinal organoid biology, advances in bioengineering tools and methods for organoid manufacture, maturation, quality control, and analysis are less developed. Further integration of experiments with computational models should synergistically enhance intestinal organoid cultures to provide more biological insights as well as significantly reducing cost and time. Many opportunities exist for bioengineers to contribute, and the key will be the close communication and collaboration between engineers, biologists, and end-users of the intestinal organoids such as pharmaceutical companies.
Highlights.
Synthetic extracellular matrix with appropriate cues can form intestinal organoids
Microtechnology allows growing intestinal organoids in physiologic microenvironment
High-throughput batch processing tools enable efficient organoid handling
Simulation of morphogenesis and culture environment can complement experimental data
Acknowledgments
This work is supported by the National Institutes of Health (U19AI116482).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- •1.Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid models of human gastrointestinal development and disease. Gastroenterology. 2016;150:1098–1112. doi: 10.1053/j.gastro.2015.12.042. This paper comprehensively reviews the types of organoid systems modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aurora M, Spence JR. hPSC-derived lung and intestinal organoids as models of human fetal tissue. Dev Biol. 2016;420:230–238. doi: 10.1016/j.ydbio.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill DR, Spence JR. Gastrointestinal organoids. understanding the molecular basis of the host–microbe interface. Cell Mol Gastroenterol Hepatol. 2017;3:138–149. doi: 10.1016/j.jcmgh.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 5.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van Den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 8.Zachos NC, Kovbasnjuk O, Foulke-abel J, In J, Blutt SE, De Jonge HR, Estes MK. Human enteroids/colonoids and intestinal organoids functionally recapitulate and pathophysiology. J Biol Chem. 2016;291:3759–3766. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••10.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. This paper highlighted the design principles of synthetic extracellular matrix to grow intestinal organoids from intestinal stem cells; dynamic and controllable modulation of mechanical and biochemical properties is critical. [DOI] [PubMed] [Google Scholar]
- 11.Czerwinski M, Spence JR. Hacking the matrix. Cell Stem Cell. 2017;20:9–10. doi: 10.1016/j.stem.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jabaji Z, Sears CM, Brinkley GJ, Lei NY, Joshi VS, Wang J, Lewis M, Stelzner M, Martín MG, Dunn JCY. Use of collagen gel as an alternative extracellular matrix for the in vitro and in vivo growth of murine small intestinal epithelium. Tissue Eng Part C Methods. 2013;19:961–9. doi: 10.1089/ten.tec.2012.0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jabaji Z, Brinkley GJ, Khalil HA, Sears CM, Lei NY, Lewis M, Stelzner M, Martín MG, Dunn JCY. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS One. 2014;9:e107814. doi: 10.1371/journal.pone.0107814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastuła A, Middelhoff M, Brandtner A, Tobiasch M, Hohl B, Nuber AH, Demir IE, Neupert S, Kollmann P, Mazzuoli-Weber G, et al. Three-dimensional gastrointestinal organoid culture in combination with nerves or fibroblasts: a method to characterize the gastrointestinal stem cell niche. Stem Cells Int. 2016;2016:3710836. doi: 10.1155/2016/3710836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, DiSalvo M, Gunasekara DB, Dutton J, Proctor A, Lebhar MS, Williamson IA, Speer J, Howard RL, Smiddy NM, et al. A self-renewing monolayer of primary colonic or rectal epithelial cells. Cell Mol Gastroenterol Hepatol. 2017 doi: 10.1016/j.jcmgh.2017.02.011. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finkbeiner SR, Freeman JJ, Wieck MM, El-Nachef W, Altheim CH, Tsai Y-H, Huang S, Dyal R, White ES, Grikscheit TC, et al. Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids. Biol Open. 2015;4:1462–1472. doi: 10.1242/bio.013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweinlin M, Wilhelm S, Schwedhelm I, Hansmann J, Rietscher R, Jurowich C, Walles H, Metzger M. Development of an advanced primary human in vitro model of the small intestine. Tissue Eng Part C Methods. 2016;22:873–883. doi: 10.1089/ten.TEC.2016.0101. [DOI] [PubMed] [Google Scholar]
- 18.Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai Y-H, Chin AM, Mahe MM, Watson CL, Freeman JJ, et al. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Reports. 2015;4:1140–1155. doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •19.DiMarco RL, Dewi RE, Bernal G, Kuo C, Heilshorn SC. Protein-engineered scaffolds for in vitro 3D culture of primary adult intestinal organoids. Biomater Sci. 2015;3:1376–1385. doi: 10.1039/c5bm00108k. This work used elasin-like polypeptide hydrogels as extracelular matrix to culture intestinal organoid, and showed that simultaneous modulation of mechanical and biochemical properties results in different organoid formation efficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee M, Dunn JCY, Wu BM. Scaffold fabrication by indirect three-dimensional printing. Biomaterials. 2005;26:4281–4289. doi: 10.1016/j.biomaterials.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Murthy SK, Fowle WH, Barabino GA, Carrier RL. Influence of micro-well biomimetic topography on intestinal epithelial Caco-2 cell phenotype. Biomaterials. 2009;30:6825–6834. doi: 10.1016/j.biomaterials.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 22.Sung JH, Yu J, Luo D, Shuler ML, March JC. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip. 2011;11:389–392. doi: 10.1039/c0lc00273a. [DOI] [PubMed] [Google Scholar]
- 23.Ramalingam S, Daughtridge GW, Johnston MJ, Gracz AD, Magness ST. Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. AJP Gastrointest Liver Physiol. 2012;302:G10–G20. doi: 10.1152/ajpgi.00277.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Wang Y, Ahmad AA, Sims CE, Magness ST, Allbritton NL. In vitro generation of colonic epithelium from primary cells guided by microstructures. Lab Chip. 2014;14:1622–1631. doi: 10.1039/c3lc51353j. This work seeded intestinal crypts into microwell scaffold to create in vitro colonic epithelium with proliferative domain inside the well and 2D non-proliferative monolayer at the top surface connecting the microwells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etoc F, Metzger J, Ruzo A, Kirst C, Yoney A, Ozair MZ, Brivanlou AH, Siggia ED. A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev Cell. 2016;39:302–315. doi: 10.1016/j.devcel.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchida H, Machida M, Miura T, Kawasaki T, Okazaki T, Sasaki K, Sakamoto S, Ohuchi N, Kasahara M, Umezawa A, et al. A xenogeneic-free system generating functional human gut organoids from pluripotent stem cells. JCI Insight. 2017;2:G260–G273. doi: 10.1172/jci.insight.86492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin AM, Hill DR, Aurora M, Spence JR. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jäger C, Seguin-Devaux C, et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cremer J, Segota I, Yang C-Y, Arnoldini M, Sauls JT, Zhang Z, Gutierrez E, Groisman A, Hwa T. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proc Natl Acad Sci. 2016;113:11414–11419. doi: 10.1073/pnas.1601306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon C, VanDussen KL, Miyoshi H, Stappenbeck TS. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 2013;7:818–828. doi: 10.1038/mi.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, Spence JR. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forbester JL, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, Mukhopadhyay S, Dougan G. Interaction of Salmonella enterica Serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun. 2015;83:2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •37.Jin B-J, Battula S, Zachos N, Kovbasnjuk O, Fawlke-Abel J, In J, Donowitz M, Verkman AS. Microfluidics platform for measurement of volume changes in immobilized intestinal enteroids. Biomicrofluidics. 2014;8:24106. doi: 10.1063/1.4870400. This work incorporated microfluidic technology to simultaneously measure morphological change of multiple epithelium-only intestinal organoids in response to external fluid flow. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gracz AD, Williamson IA, Roche KC, Johnston MJ, Wang F, Wang Y, Attayek PJ, Balowski J, Liu XF, Laurenza RJ, et al. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arora N, Alsous JI, Guggenheim JW, Mak M, Munera J, Wells JM, Kamm RD, Asada HH, Shvartsman SY, Griffith LG. A process engineering approach to increase organoid yield. Development. 2017 doi: 10.1242/dev.142919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buske P, Przybilla J, Loeffler M, Sachs N, Sato T, Clevers H, Galle J. On the biomechanics of stem cell niche formation in the gut - modelling growing organoids. FEBS J. 2012;279:3475–3487. doi: 10.1111/j.1742-4658.2012.08646.x. [DOI] [PubMed] [Google Scholar]
- 41.Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ, Mahadevan L. Villification: how the gut gets its villi. Science. 2013;342:212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walton KD, Freddo AM, Wang S, Gumucio DL. Generation of intestinal surface: an absorbing tale. Development. 2016;143:2261–2272. doi: 10.1242/dev.135400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walton KD, Whidden M, Kolterud AK, Shoffner S, Czerwinski MJ, Kushwaha J, Parmar N, Chandhrasekhar D, Freddo AM, Schnell S, et al. Villification in the mouse: Bmp signals control intestinal villus patterning. Development. 2016;143:427–436. doi: 10.1242/dev.130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freddo AM, Shoffner SK, Shao Y, Taniguchi K, Grosse AS, Guysinger MN, Wang S, Rudraraju S, Margolis B, Garikipati K, et al. Coordination of signaling and tissue mechanics during morphogenesis of murine intestinal villi: a role for mitotic cell rounding. Integr Biol. 2016;8:918–928. doi: 10.1039/c6ib00046k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Booth C, Potten CS. Gut instincts: thoughts on intestinal epithelial stem cells. J Clin Invest. 2000;105:1493–1499. doi: 10.1172/JCI10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cummins AG, Catto-Smith AG, Cameron DJ, Couper RT, Davidson GP, Day AS, Hammond PD, Moore DJ, Thompson FM. Crypt fission peaks early during infancy and crypt hyperplasia broadly peaks during infancy and childhood in the small intestine of humans. J Pediatr Gastroenterol Nutr. 2008;47:153–157. doi: 10.1097/MPG.0b013e3181604d27. [DOI] [PubMed] [Google Scholar]
- 47.Pin C, Parker A, Gunning AP, Ohta Y, Johnson IT, Carding SR, Sato T. An individual based computational model of intestinal crypt fission and its application to predicting unrestrictive growth of the intestinal epithelium. Integr Biol. 2015;7:213–228. doi: 10.1039/c4ib00236a. [DOI] [PubMed] [Google Scholar]
- 48.Dunn S-J, Osborne JM, Appleton PL, Näthke I. Combined changes in Wnt signaling response and contact inhibition induce altered proliferation in radiation-treated intestinal crypts. Mol Biol Cell. 2016;27:1863–1874. doi: 10.1091/mbc.E15-12-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •49.Langlands AJ, Almet AA, Appleton PL, Newton IP, Osborne JM, Näthke IS. Paneth cell-rich regions separated by a cluster of Lgr5+ cells initiate crypt fission in the intestinal stem cell niche. PLOS Biol. 2016;14:e1002491. doi: 10.1371/journal.pbio.1002491. This work combined experimental observations of crypt fission in murine epithelium-only intestinal organoids and the simulation model to explain the phenomena based on mechanical microenvironment, then explained potential cause for the discrepancies in final crypt morphologies between in vitro and computational models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsu-ura T, Dovzhenok A, Aihara E, Rood J, Le H, Ren Y, Rosselot AE, Zhang T, Lee C, Obrietan K, et al. Intercellular coupling of the cell cycle and circadian clock in adult stem cell culture. Mol Cell. 2016;64:900–912. doi: 10.1016/j.molcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiMarco RL, Su J, Yan KS, Dewi R, Kuo CJ, Heilshorn SC. Engineering of three-dimensional microenvironments to promote contractile behavior in primary intestinal organoids. Integr Biol. 2014;6:127–142. doi: 10.1039/c3ib40188j. [DOI] [PMC free article] [PubMed] [Google Scholar]