Abstract
White spot disease caused by white spot syndrome virus (WSSV) is a lethal disease for shrimp. Envelope structural proteins play a major role in viral attachment and are believed to be the initial molecules to interact with the host cell. Thus, the envelope proteins have been preferred as a potential molecular target for drug discovery. In the present investigation, molecular docking and simulation analysis were performed to predict the binding efficiency of phytocompounds identified from Phyllanthus amarus with major envelope proteins, VP26, VP28, and VP110, and a nucleocapsid protein VP664 of WSSV. The docking result reveals that the compounds 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate and 1,4-benzenediamine, N,N′-diphenyl exhibited highest binding energy with the envelope proteins. The mobility of protein–ligand binding complex at various time intervals was validated by molecular dynamics and simulation study. Therefore, P. amarus phytocompounds were found to be most suitable inhibitors for the antiviral treatment for WSSV infection.
Keywords: WSSV, Envelope proteins, Phytocompounds, Docking, Molecular dynamics
Introduction
Shrimp culture covers a major portion of the aquaculture industry and its export forms the main source of revenue of many countries including India. The white spot syndrome virus (WSSV) has emerged globally as one of the most prevalent, widespread, and lethal disease for shrimp populations that leads to huge economic loss to shrimp industries. It is an enveloped rod-shaped double-stranded DNA virus of approximately 300 kbp in size, which belongs to the family Nimaviridae, genus Whispovirus (Sundaram et al. 2016). WSSV can cause huge mortalities up to 100% within 3–10 days in farmed shrimp (Leu et al. 2009). The signs and symptoms of WSSV-infected shrimp are white spots appearing on exoskeleton, loosening of cuticle, reduced food consumption, lethargy, and red discoloration throughout the body (Arturo Sanchez-Paz 2010). Many species of arthropods have been reported as carriers or hosts for WSSV, which includes shrimp, crayfish, crabs, and lobsters (Sundaram et al. 2016). The virus entry into the host cells occurs through the interactions of viral envelope proteins with the host cellular receptors. These interactions are so specific that inhibitors that could prevent these interactions show promising antiviral activity (Joseph et al. 2017). Virus envelope structural proteins play a major role in viral attachment and are believed to be the initial molecules to interact with the host cell (Arturo Sanchez-Paz 2010). VP26 and VP28 are the major envelope proteins present approximately 60% in the envelope of WSSV. Recently, both VP26 and VP28 were found to naturally form projected trimers in the viral envelope and may have an important role on the infective mode of interaction between the viral envelope membrane and the host cell receptors (Tang et al. 2007). VP28 played a major role in the interaction of WSSV attachment by interacting with Penaeus monodon Ras-related nuclear protein Rab7 (PmRab7). Rab7 is a shrimp membrane protein, which facilitates the entry of virus into host cells (Sivakumar et al. 2016). Similarly the envelope protein VP110 has been reported to contain a cell attachment signature, termed as RGD motif, characterized by an Arg-Gly-Asp sequence, which seems to be involved in virus binding-to-host cell surface integrins (Tang et al. 2007). The nucleocapsid protein VP664 of WSSV was believed to be involved in the process of assembly and morphogenesis of the virion. VP664 is the largest protein identified in viruses; it had a sequence of 18,234 nucleotides, which encodes 6077 amino acids (Leu et al. 2005). These structural envelope proteins of WSSV have been preferred as a potential molecular target for drug discovery. However, at present there is no effective drug or treatment strategy available to control the spread of WSSV in farmed shrimp. Shrimp lack adaptive immune system and they rely on innate immune system to fight against invading microbes; due to this, developing a vaccine against pathogens was not possible to prevent the disease in crustaceans. But in laboratory scale few of them were observed that the mortality caused due to WSSV was delayed by administrating plants extracts, probiotics, DNA vaccines or antibodies raised against WSSV envelope proteins (Musthaq and Kwang 2015).
Plant-derived natural products play a significant role by being a lead molecule in the development of novel drug candidates for many diseases. Herbal extracts represent the primary form of health care for a major proportion of the world population and are the important sources of single-molecule drug leads (Chakraborty et al. 2014). Nowadays, there is a growing interest in natural products used to cure or prevent life-threatening diseases by improving the host immune system. For example, many traditional medicinal plants (Cynodon dactylon, Aegle marmelos, Tinospora cordifolia, Picrorhiza kurroa, Eclipta alba, and Pongamia pinnata) have been reported for antiviral activity against WSSV in shrimp P. monodon ( Sanchez-Paz 2010). Drugs derived from terrestrial and marine plants have been reported for anti-WSSV activity (Chakraborty et al. 2014; Ghosh et al. 2014). Our previous study has identified that the acetone extract of Phyllanthus amarus had potent antiviral effect against WSSV in freshwater crabs Paratelphusa hydrodromous (Sundaram et al. 2016). In the present investigation, attempts have been made to explore the inhibition mode of phytocompounds identified from acetone extract of P. amarus against major envelope proteins, VP26, VP28, and VP110, and a nucleocapsid protein VP664 of WSSV using molecular docking and simulation approaches for the development of new drugs to prevent or treat shrimp from WSSV infection.
Materials and methods
Gas chromatography–mass spectrometry (GC–MS)
GC–MS analysis of the P. amarus acetone extract was performed using a Perkin Elmer GC Clarus 680 system and gas chromatograph interfaced to a mass spectrometer Clarus 600 system equipped with Elite-5MS column (30.0 m, 0.25 mm ID, 250 µm df). For GC–MS detection, an electron ionization energy system with ionization energy of 70 eV was used. Helium gas (99.999%) was used as the carrier gas at a constant flow rate of 1 mL/min and an injection volume of 1 μL was employed (split ratio of 10:1). Injector temperature was 250 °C. The initial oven temperature was programmed from 60 °C for 2 min, with an increase of 10 °C/min to 300 °C, holding for 6 min. Mass spectra were taken at 70 eV; a scan interval of 0.5 s and fragments from 50 to 600 Da. Total GC running time was 32 min. The relative percentage amount of each component was calculated by comparing its average peak area to the total area. Software adopted to handle mass spectra and chromatograms was a TurboMass Version 5.4.2. Compound identification was obtained by comparing the retention times with those of authentic compounds and the spectral data obtained from library database, National Institute of Standards and Technology (NIST) of the corresponding compounds.
Toxicity prediction of the phytocompounds
The toxicity analysis for the phytocompounds was verified using ECOSAR (ecological structure activity relationships) toxicity testing software tool (Cash 1998). The tool predicts the toxicity of neutral organics, organic chemicals, and surfactants to the aquatic organisms. The median lethal concentration LC50 for the aquatic organisms, such as fish, daphnid, mysid, and other organisms, was predicted. The SMILES notations of the phytocompounds were submitted to the tool and the toxicity for the fishes and daphnid was analyzed.
Evaluation of in vitro toxicity using brine shrimp (Artemia salina)
Brine shrimp lethality bioassay was carried out to determine the general toxic property of the P. amarus acetone extract (Meyer et al. 1982; Islam et al. 2003; Rahmatullah et al. 2010; Al-Hazmi 2010). For bioassay, Artemia salina eggs were hatched in natural seawater obtained from local shrimp hatchery, Chennai, India. The brine shrimp egg was added to seawater in a glass-hatching chamber and kept under constant aeration. The chamber was kept illuminated. After 24 h incubation at room temperature (26–30 °C), the larvae (nauplii) were collected using plastic Pasteur pipette by attracting them to one side of the vessel using a light source.
The P. amarus acetone extract was prepared at ten different concentrations ranging from 100 to 1000 µg mL−1 using seawater and was poured in six-well plates (tissue culture plate, TARSONS Pvt. Ltd, India). Ten newly hatched nauplii were added into each well having 5 mL of seawater with plant extract at different concentrations and were incubated for 24 h at room temperature. After 24 h, total number of live and dead nauplii was counted using magnifying lens. Control group was included without the addition of plant extract and the test was carried out in triplicates to determine the toxicity.
Preparation of small molecules
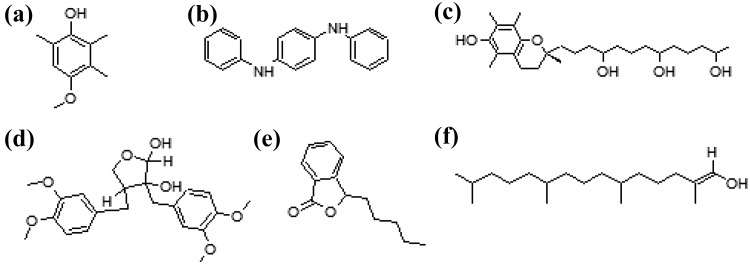
The SMILES (simplified molecular input line entry system) notations of the bioactive compounds identified from the leaves of P. amarus were converted to PDB coordinate using CORINA server. The structures were optimized and energy minimized using AM1 forcefield in Arguslab 4.0.1 (Thompson 2004) (Fig. 1).
Fig. 1.
Structures of phytocompounds isolated from P. amarus. a phenol, 4-methoxy-2,3,6-trimethyl, b 4-benzenediamine, N,N′-diphenyl, c 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate, d carissanol dimethyl ether, e N,N-butylpthalimide and f phytol
Preparation of protein target structure
The initial coordinates of the envelope proteins VP26 (Acc.no. 2EDM) and VP28 (Acc.no. 2ED6) were retrieved from Protein Data Bank (Berman et al. 2000). The VP26 protein was assembled into a trimer using PDBePISA server (Berman et al. 2000). The sequences of VP110 (Acc. no: A0A0K2JN67) and VP664 (Acc. no: A0A0K2JP33) were retrieved from UniProtKB/Swiss-Prot (The Uniprot Consortium 2015). The structures for the other two proteins VP110 and VP664 were modeled using I-TASSER server (Yang et al. 2015). The stereo-chemical analysis for the best model was predicted using PROCHECK program (Laskowski et al. 1993). The 1D–3D profile of the best model was assessed using Verify3D program (Eisenberg et al. 1997). The proteins were subjected to energy minimization using Gromacs 4.3 (Berendsen et al. 1995). The parameters defined were as follows: emtol was set to less than 1000 kJ/mol/nm, maximum number of iterations was set to 50,000, the long-range electrostatic interactions were treated with particle mesh Ewald (PME) algorithm, and the minimization was carried out using steepest descendant algorithm.
Molecular docking analysis
Molecular docking studies were carried out to investigate the binding efficacy of the phytocompounds with the envelope proteins using Autodock 4.2 (Morris et al. 2009). Blind docking was carried out to enable the ligand to choose their preferential binding sites. The partial charges for the protein and ligands were computed using Gasteiger–Marsili and Kollman charges, respectively. Since blind docking was carried out, the grid box was set over the entire protein and the grid space was set to 0.375 nm. Lamarckian genetic algorithm was used to generate the conformers of the ligands bound to the protein. The number of runs was set to 10 leaving other settings to default values. The protein–ligand conformation with lowest binding energy was selected as stable conformation. The best protein–ligand complex was further subjected to molecular dynamics and simulation studies.
Molecular dynamics simulation
Eight independent molecular dynamics (MD) simulations (4—only protein and 4—protein and ligand complex) were performed in Gromacs 4.3 using grooms53a6 force-field. The ligand coordinates were generated using PRODRG server (SchuÈttelkopf and Van Aalten 2004). The protein and the protein–ligand complex systems were solvated using explicit SPC water molecules extended about 10 nm from the protein and the protein–ligand complex in a cubic periodic box. Sodium and chloride ions were added to neutralize the systems. The systems were energetically minimized with 50,000 steps and heated to 310 K over a period of 1 ns. A non-constrained MD simulation was performed at constant pressure (1 atm) and constant temperature (310 K) for 5 ns. The long-range electrostatic interactions were monitored using PME algorithm. For each system, the trajectory snap shots were saved every 2 ps for further analysis.
Results
GC–MS analysis
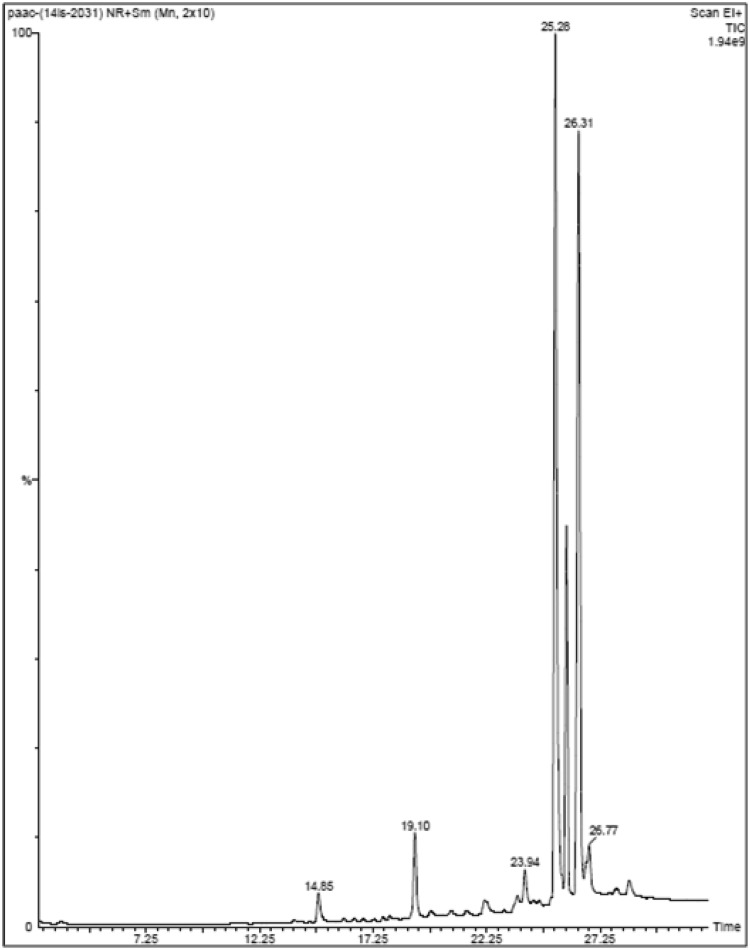
GC–MS analysis was performed for P. amarus acetone extract; the chromatogram is shown in Fig. 2. Based on the NIST library search, the molecular weight, molecular formula, and retention time for the phytocompounds present in the extract are given in Table 1. There were more than six compounds identified in the extract. The prevailing compounds were carissanol dimethyl ether, phenol, 4-methoxy-2,3,6-trimethyl, phytol, 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate and 1,4-benzenediamine, N,N′-diphenyl, N,N-butylpthalimide.
Fig. 2.
Chromatogram obtained from GC–MS analysis of P. amarus acetone extract
Table 1.
Phytocompounds identified in acetone extract of P. amarus
| S. no | Retention time | Name of the compound | Molecular formula | Molecular weight |
|---|---|---|---|---|
| 1 | 14.84 | N,N-Butylpthalimide | C12H13NO2 | 203 |
| 2 | 19.40 | Phytol | C20H40O | 296 |
| 3 | 23.61 | Phenol, 4-methoxy-2,3,6-trimethyl | C10H14O2 | 166 |
| 4 | 23.94 | 1,4-Benzenediamine, N,N′-diphenyl | C18H16N2 | 260 |
| 5 | 25.28 | Carissanol dimethyl ether | C22H28O7 | 404 |
| 6 | 26.77 | 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate | C31H52O3 | 472 |
Toxicity analysis of the phytocompounds against shrimps
The toxicity analysis of the phytocompounds identified from P. amarus was verified using the ECOSAR tool. The results revealed that the phytocompounds did not exhibit any toxicity against any aquatic organisms, as the LC50 values of the compounds were found to be very minimal and below the threshold criteria for causing any lethality to the aquatic organisms. However, the toxicity analysis against the aquatic organisms for the three compounds, such as phenol, 4-methoxy-2,3,6-trimethyl, carissanol dimethyl ether, and phytol, were not predicted by the ECOSAR, as the compounds are yet to be tested for the toxicity. Table 2 summarizes the LC50 values of the phytocompounds against the aquatic organisms.
Table 2.
Toxicity prediction based on ECOSAR class
| Compounds | ECOSAR class | Fish LC50 96 ha | Daphnid LC50 96 hb |
|---|---|---|---|
| Phenol, 4-methoxy-2,3,6-trimethyl | Phenol | – | – |
| 1,4-Benzenediamine, N,N′-diphenyl | Neutral organics | 3.150 | 2.160 |
| 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate | Phenol | 1.37e-006 | 3.84e-005 |
| Carissanol dimethyl ether | Neutral organics | – | – |
| N,N-Butylpthalimide | Imides | 3.494 | 3.479 |
| Phytol | Allyl alcohol | – | – |
aFish LC50 96 h is the dose (mg L−1) required to kill half the members of fish after 96 h
bDaphnid LC50 96 h is the dose (mg L−1) required to kill half the members of fish after 96 h
In vitro toxicity analysis in brine shrimp
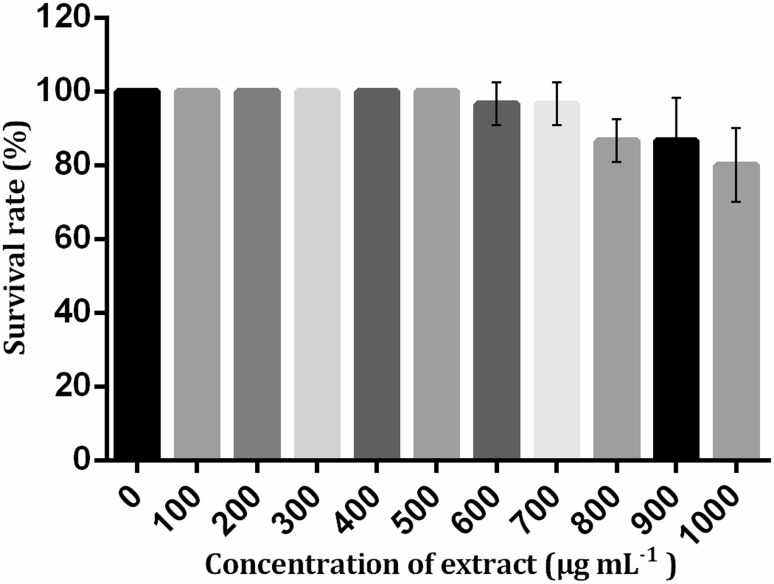
It was observed that the extract did not exhibit any toxicity against Artemia nauplii, as the LC50 value of the extract was considered as >1000 µg mL−1. 80% of the nauplii had survived after 24 h in the highest concentration of the extract (1000 µg mL−1) and revealed that the toxicity of the extract was very minimal. There was no death of a single nauplius after 24 h in the concentrations up to 500 µg mL−1 (Fig. 3). Therefore, the extract was non-toxic as well as safe for shrimp.
Fig. 3.
Survival graph of brine shrimp toxicity assay
Structural validation of VP110 and VP664 proteins
The best I-TASSER model for the VP110 and VP664 proteins was assessed using confidence score (C score). C score denotes the significance of the ab initio method of protein modeling. The high C score indicates the structure with high confidence. The best model had a score of −1.80 and −3.00 for VP110 and VP664, respectively. The stereo-chemical properties of the predicted models were validated using PROCHECK and verify 3D programs. The Ramachandran plot for the VP110 protein predicted by PROCHECK program revealed 90.8% of residues falling in allowed region and 9.2% of residues falling in outlier zone. Similarly, the Ramachandran plot of VP664 revealed overall 97.5% residues falling within allowed region and 5.5% of residues lying in the outlier region. The Verify 3D plot predicted around 57.00 and 60.22% of residues to have an average score of <0.2 for VP110 and VP664, respectively, revealing that majority of the residues were satisfied in its environment. The above results further validate the quality of the predicted models of VP110 and VP664.
Binding mode analysis of phytocompounds using molecular docking
The six phytocompounds identified from the P. amarus were docked with the envelope proteins, VP26, VP28, and VP110, and VP664 to monitor the binding efficiency of phytocompounds with the proteins. Table 3 summarizes the binding energy of the compounds with the envelope proteins.
Table 3.
Binding energy of compounds with the envelope proteins
| Compounds | VP26 (kcal mol−1) | VP28 (kcal mol−1) | VP110 (kcal mol−1) | VP664 (kcal mol−1) |
|---|---|---|---|---|
| Phenol, 4-methoxy-2,3,6-trimethyl | −5.25 | −5.57 | −5.37 | −5.87 |
| 1,4-Benzenediamine, N,N′-diphenyl | −6.63 | −8.39 | −7.24 | −7.55 |
| 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate | −6.71 | −8.94 | −6.86 | −5.83 |
| Carissanol dimethyl ether | −6.57 | −8.39 | −5.49 | −5.36 |
| N,N-Butylpthalimide | −6.56 | −6.79 | −6.01 | −7.04 |
| Phytol | −5.05 | −6.49 | −5.12 | −5.74 |
Compounds having high binding capacity are given in bold
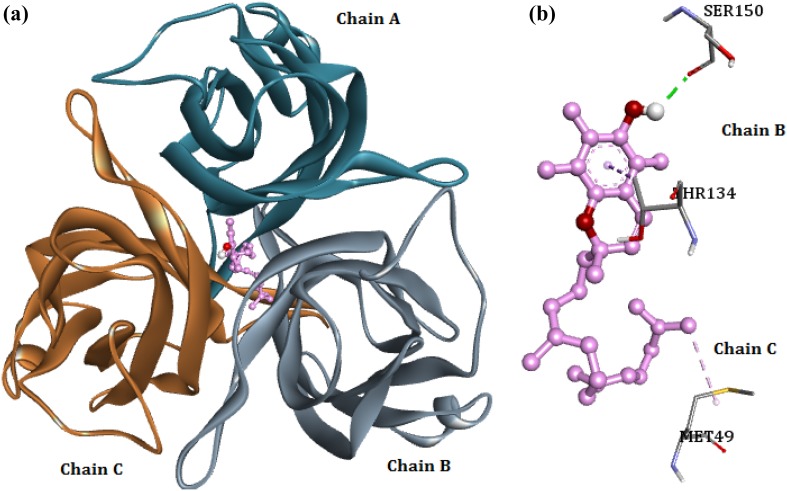
The molecular docking analysis of envelope protein VP26 with the phytocompounds showed that majority of the compounds bind to VP26 with favorable binding energy ranged between −6.71 and −5.05 kcal/mol. When the binding energy of phytocompounds was investigated, the compound 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate exhibited highest binding energy of −6.71 kcal/mol (Fig. 4). Binding pattern of the compound showed the formation of a hydrogen bond with Ser150 of chain B. The ligand forms two hydrophobic interactions with Met49 (alkyl bond) of chain C and Thr134 (pi–sigma bond) of chain B with VP26.
Fig. 4.
a Binding of 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate with VP26. b Interaction between VP26 residues and the ligand; green dashed lines—hydrogen bonds, pink dashed lines—hydrophobic interactions
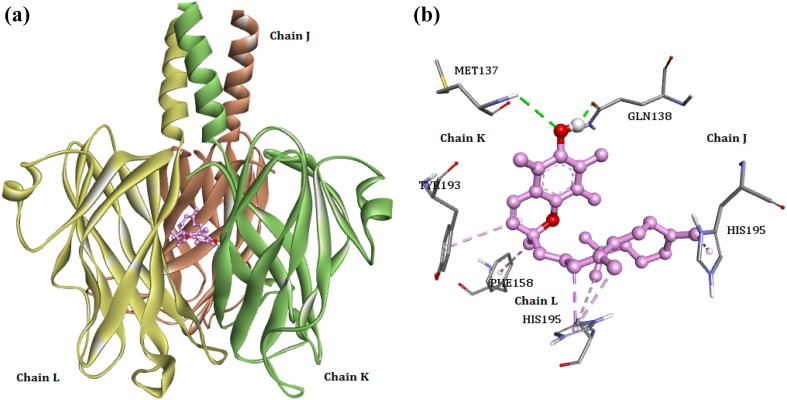
Binding energy of the envelope protein VP28 with the phytocompounds ranged between −8.94 and −5.57 kcal/mol. Similar to VP26, the VP28 showed preference to the compound 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12 trimethyltridecyl)-acetate with highest binding energy of −8.94 kcal/mol. When the interaction pattern of the compound with VP28 was investigated, we noted that the ligand forms two hydrogen bonds with Met137 of K chain and GLN138 of J chain (Fig. 5). The ligand also forms hydrophobic interactions with Phe158 (pi–alkyl bond) of L chain, Tyr193 (pi–alkyl bond) of K chain, and His195 of J (pi–alkyl) and L chains (pi–sigma bond).
Fig. 5.
a Binding of 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate with VP28. b The interaction patterns of the ligand with VP28 residues; green dashed lines—hydrogen bonds, pink dashed lines—hydrophobic interactions
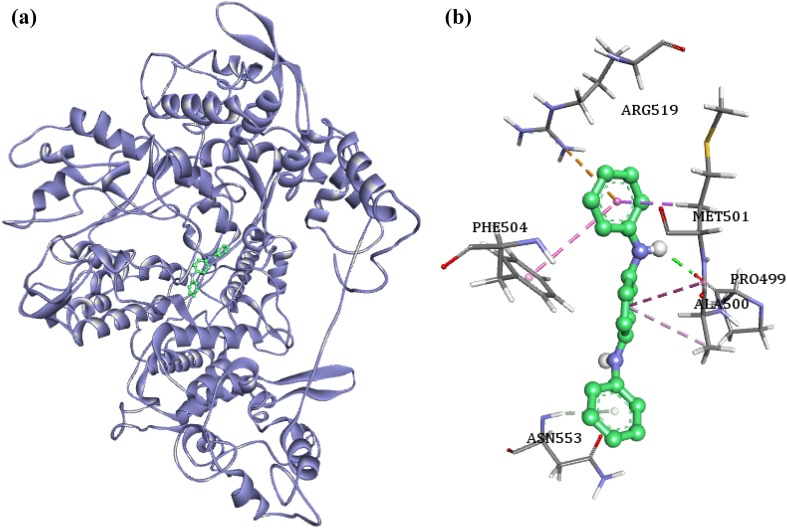
The binding energy of the envelope protein VP110 with the phytocompounds ranged between −7.24 and –5.12 kcal/mol. The highest binding energy of −7.24 kcal/mol was reported by the compound 1,4-benzenediamine, N,N′-diphenyl, when docked with VP110. The binding pattern analysis showed that the compound formed hydrogen bonds with Pro499 and Asn553 (Fig. 6). The compound also formed hydrophobic interactions with Met501 (pi–sigma), Thr550 (pi–sigma), Phe504 (pi–pi), and Ala500 (pi–alkyl). An amide bond is observed between the compound and Ala500, and Met501 (amide–pi). The ligand is also involved in the formation of electrostatic interaction with Arg519 (pi–cation).
Fig. 6.
a Binding mode of 4-benzenediamine, N,N′-diphenyl with VP110 protein. b The binding pattern of the ligand with the VP110 residues; green dashed lines—hydrogen bonds, pink, orange and indigo dashed lines—hydrophobic interactions
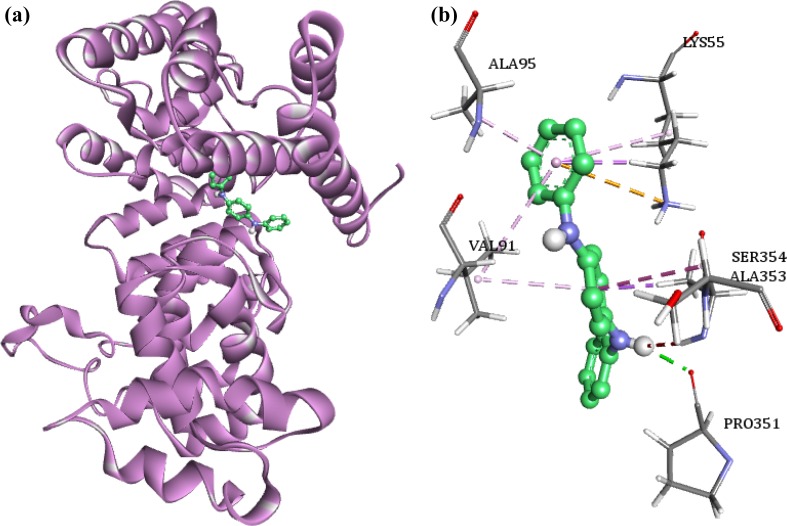
The binding energy of the nucleocapsid protein VP664 with the phytocompounds ranged between −7.55 and −5.36 kcal/mol. Similar to VP110, the compound 1,4-benzenediamine N,N′diphenyl, N,N-butylpthalimide is found to bind with high binding energy of −7.55 kcal/mol with VP664. The interaction pattern analysis of the compound with VP664 revealed the presence of a hydrogen bond between Pro351 and the ligand (Fig. 7). Hydrophobic interactions were reported between the ligand and Lys55 (pi–sigma), Ala95 (pi–sigma), Ala353 (pi–sigma), Val191 (pi–alkyl), Lys55 (pi–alkyl), and Val 91 (pi–alkyl) residues of the protein. An electrostatic interaction was observed between the Lys55 of VP664 and the compound. We also observed an amide bond (amide–pi) between Ala353 and Ser354 with the ligand.
Fig. 7.
a Binding pattern of 4-benzenediamine, N,N′-diphenyl with VP664 protein. b The interaction of VP664 residues with the ligand; green dashed lines—hydrogen bonds, pink, indigo and orange dashed lines—hydrophobic interactions
Comparative analysis of the MD trajectories
To understand the impact of best binding ligands on the protein-free protein and protein–ligand complex simulations were carried out. The influence of the ligands over the protein structure was assessed by comparing the 5 ns MD trajectories of the protein and the ligand bound complex. The parameters, such as RMSD (root mean square deviation), RMSF (root mean square fluctuations), and number of hydrogen bonds, were used to assess the effects exhibited by the inhibitor on the protein.
Root mean square deviation (RMSD)
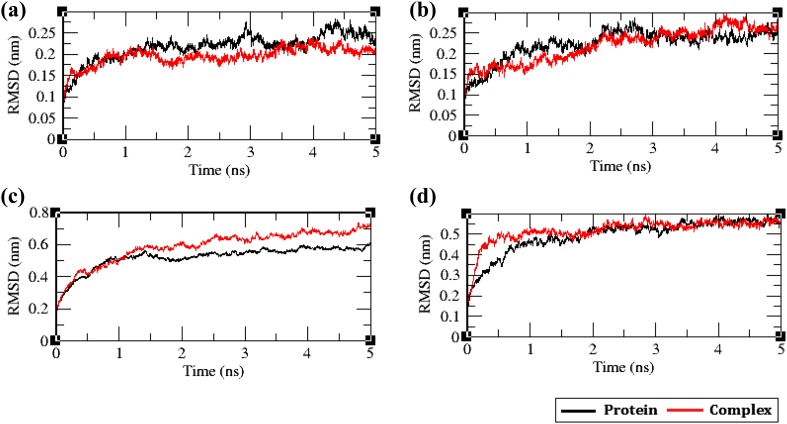
The protein stability and the protein–ligand complex stability during the 5 ns simulation time were monitored using the time evolution of the RMSD. Eight 5-ns MD simulations were carried out for the envelope proteins and their respective protein–ligand complexes.
The RMSDs of the envelope proteins with and without their respective ligands were calculated against the simulation time and the results are shown in Fig. 8. In VP26 protein and the complex, the RMSD of both the systems remained similar until 1.5 ns, which later diverged into two different paths before the end of simulation time period. The RMSD of the VP26–ligand complex was lower (0.2 nm), when compared to the free protein VP26 (0.225 nm). The decreased RMSD of the complex reveals increased rigidity of the protein upon ligand binding. The RMSD of the VP28–ligand complex was lower, when compared to the VP28 protein before 3 ns; however, the VP28–ligand complex showed more deviation before 4.5 ns. The VP28 protein and the VP28–ligand complex converged together to 0.25 nm at 5 ns revealing that the ligand has induced fluctuations during the simulation time imparting flexibility in the VP28 protein by the ligand. When the RMSD of the VP110 protein and VP110–protein ligand complex was investigated, we observed the RMSD of VP110–protein ligand complex was higher, when compared to VP110 protein system. The RMSD of both the systems differed by 0.1 nm. The binding of the ligand led to the loss of structural stability of VP110 protein, which is evident from the RMSD plot. The investigation of RMSD of the VP664 systems showed that both the systems became stable after 2 ns revealing that the ligand binding to VP664 protein did not induce any structural alterations in the VP664 protein making the protein rigid (Fig. 8).
Fig. 8.
The root mean square deviation (RMSD) plot of the free envelope proteins along with protein bound with their respective ligands; a VP26, b VP28, c VP110, and d VP664
Root mean square fluctuation (RMSF)
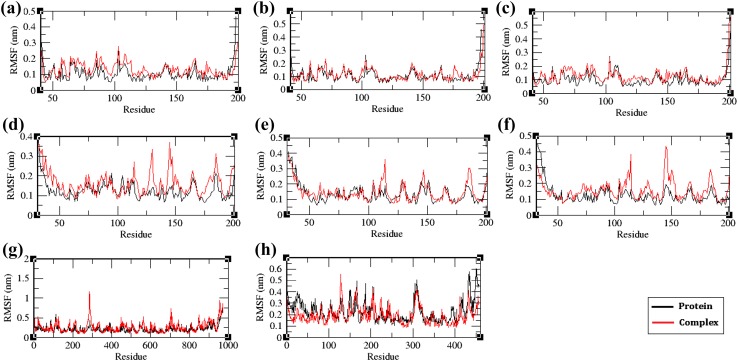
To monitor the mobility of residues in the protein contributed by ligand binding, the root mean square fluctuations (RMSF) of the residues were compared between the protein and protein–ligand complex systems (Fig. 9). In VP26, VP28, and VP110 proteins, and protein–ligand complexes, the residual motilities in the complex system were higher when compared to the free protein system, indicating that the ligand binding has disrupted the protein structure. However, in VP664 protein and protein–ligand complex, the protein showed higher fluctuations in majority of the residues when compared to the complex system, indicating that the binding of the ligand has restricted the motions of the residues involved in the bonding of the ligand.
Fig. 9.
The root mean square fluctuation (RMSF) plot of the unbound envelope proteins and ligand bound protein complex; a VP26—chain A, b VP26—chain B, c VP26—chain C, d VP28—chain J, e VP28—chain K, f VP28—chain L, g VP110, and h VP664
H-bond analysis
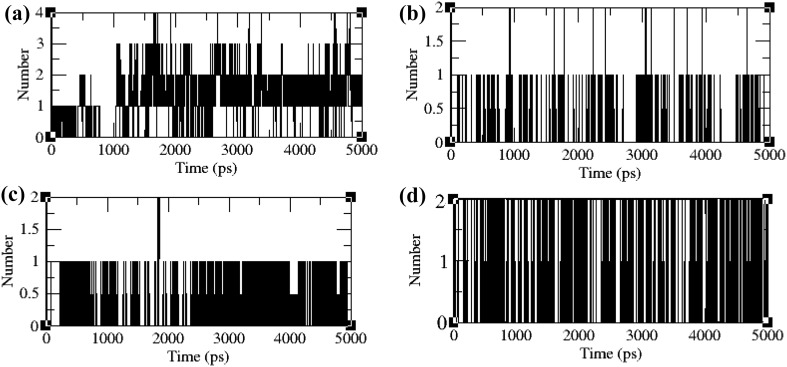
The H-bond network between the envelope proteins and their respective ligands was computed to assess the stability of the protein–ligand complex (Fig. 10). In VP26 protein–ligand complex system, a maximum of one hydrogen bond and a minimum of four hydrogen bonds were formed during the simulation time period. In VP28 protein–ligand complex system, a minimum of two hydrogen bonds and maximum of one hydrogen bond were observed. Similarly in VP110 protein–ligand complex system, a minimum of two hydrogen bonds and maximum of one hydrogen bond were maintained. In VP664 protein–ligand complex, maximum of two hydrogen bonds and a minimum of one hydrogen bond were maintained. In all the four systems, a minimum of one hydrogen bond was maintained throughout the entire simulation time validating the stability of the ligand complex.
Fig. 10.
Hydrogen bonding patterns in the ligand bound protein complex; a VP26, b VP28, c VP110, and d VP664
Discussion
Worldwide, the WSSV still continues to be the major viral pathogen affecting the farmed shrimp and creates huge economic losses to the shrimp industries. Due to several known and unknown vectors, there is no effective treatment to control the spread of WSSV infection in shrimp farms (Sivakumar et al. 2016). Therefore, development of an effective drug against WSSV infection is urgently needed to overcome this problem. WSSV envelope structural proteins (VP26, VP28, and VP110) play a major role in viral attachment and are believed to be the initial molecules to interact with the host cell for replication (Tang et al. 2007; Sanchez-Paz 2010). VP664 is an important nucleocapsid protein of WSSV, which is believed to be involved in the process of virus assembly and morphogenesis. Recently, both VP26 and VP28 were found to naturally form projected trimers in the viral envelope and may have an important role on the infective mode of interaction between the viral envelope membrane and the host cell receptors (Tang et al. 2007). These WSSV viral proteins have been preferred as a potential molecular target for the development of new drugs.
Medicinal plants are considered as one of the most significant resources for the isolation of bioactive phytocompounds. The growing interest in these medicinal plants has increased worldwide, because they are less toxic and relatively low cost compared to other synthetic drugs. P. amarus is an important and easily available well-known ayurvedic medicinal plant, which has wide range of pharmacological activities including antiviral, antibacterial, anticancer, anti-inflammatory, etc. (Patel et al. 2011). P. amarus had potent antiviral activity against WSSV, in vivo antiviral bioassay of the extract which was determined in the freshwater crab showed 100% survival; it has been reported in our previous report (Sundaram et al. 2016). Sivakumar et al. (2016) have made an attempt to identify the potential drug candidate from marine origin against WSSV using VP28 as a target by employing in silico docking and molecular dynamic simulations; they extracted 388 marine bioactive compounds from the reports published in Marine Drugs for the study, and they identified the drug (30797199) preferred to be a potential compound for WSSV VP28 inhibition and can act as a drug candidate for WSSV infection in shrimp. In this direction, we made an attempt to develop a non-toxic drug from a known anti-WSSV plant P. amarus. Since, the crude extract of P. amarus showed antiviral effect against WSSV, further we want to know the active phytocompounds responsible for antiviral property that is more important. Therefore, the current research is aimed to identify the various phytocompounds from P. amarus using GC–MS for understanding the binding mode of various phytocompounds against major envelope proteins, VP26, VP28, and VP110, and a nucleocapsid protein VP664 of WSSV. The toxicity analysis of the phytocompounds was evaluated using ECOSAR tool, which exhibits no significant toxicity against aquatic organisms. For in vitro toxicity analysis, we followed brine shrimp lethality bioassay. Brine shrimp is widely used in the evaluation of toxicity of heavy metals, pesticides, medicines especially natural plant extracts, etc. (Price et al. 1974; Sorgeloos et al. 1978). Our in vitro brine shrimp lethality assay also exhibits that the toxicity of the plant extract was very minimal and safe for treating shrimp against WSSV. The docking analysis of phytocompound 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-acetate exhibited highest binding energy to the envelope proteins VP26 and VP28. Sudharsana et al. (2016) also reported a similar kind of molecular docking and simulation studies using an antiviral compound 3-(1-chloropiperidin-4-yl)-6-fluoro benzisoxazole 2 docked with WSSV envelope proteins VP26 and VP28. Similarly, the other compound 1,4-benzenediamine, N,N′-diphenyl from our study exhibited highest binding energy to VP110 and VP664 proteins of WSSV.
Conclusion
Phytocompounds from plant source is an alternative approach to the modern medication against several pathogenic diseases. According to the recent research, usage of plant phytocompounds is the most reliable and efficient method with therapeutic significance. Our in vitro primary studies are looking for the phytocompounds from P. amarus which have antiviral properties against WSSV in shrimp as well as are less toxic to the aquatic animals. Hence, phytocompounds from the crude extract were identified using GC–MS and these identified compounds were further subjected to in silico analysis to understand the molecular mechanism of antiviral activity.
Computational studies provide an insight into developing novel inhibitor against WSSV infection in shrimp. The computational analysis reveals that the phytocompounds 2H-1-benzopyran-6-ol, 3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8, 12-trimethyltridecyl)-acetate and 1, 4-benzenediamine, N,N′-diphenyl identified from P. amarus could be the potential molecules for the treatment of white spot disease in shrimp culture. These findings may be helpful to develop a safe antiviral drug against WSSV to prevent rapid mortality as well as huge economic losses to the aquaculture industry. Further wet lab experiment of identified phytocompounds is necessary to validate these results and is more necessary for the development of promising antiviral drug for WSSV infection.
Acknowledgements
The authors are thankful to Prof. A.S. Sahul Hameed of C. Abdul Hakeem College, Tamil Nadu, India, for providing the virus stock to perform the studies. The authors are grateful to the management of VIT University for providing necessary facilities to carry out this study.
References
- Al-Hazmi NA. Fungal isolates and their toxicity from different ecosystems in Jeddah, Saudi Arabia. Afr J Biotechnol. 2010;9(34):5590–5598. [Google Scholar]
- Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash GG. Prediction of chemical toxicity to aquatic organisms: ECOSAR vs. Microtox® assay. Environ Toxicol. 1998;3(3):211–216. [Google Scholar]
- Chakraborty S, Ghosh U, Balasubramanian T, Das P. Screening, isolation and optimization of anti-white spot syndrome virus drug derived from marine plants. Asian Pac J Trop Biomed. 2014;4:S107–S117. doi: 10.12980/APJTB.4.2014C1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Ghosh U, Chakraborty S, Balasubramanian T, Das P. Screening, isolation and optimization of anti–white spot syndrome virus drug derived from terrestrial plants. Asian Pac J Trop Biomed. 2014;4:S118–S128. doi: 10.12980/APJTB.4.2014C1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam AKMN, Ali MA, Sayeed A, Salam SMA, Islam A, Rahman M, Khan GRMA, Khatun S. An antimicrobial terpenoid from Caesalpinia pulcherrima Swartz: its characterisation, antimicrobial and cytotoxic activities. Asian J Plant Sci. 2003;2(17–24):1162–1165. [Google Scholar]
- Joseph J, Bhaskaran R, Kaliraj M, Muthuswamy M, Suresh A. Molecular Docking of Phytoligands to the viral protein receptor P. monodon Rab7. Bioinformation. 2017;13(4):116–121. doi: 10.6026/97320630013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26(2):283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Leu JH, Tsai JM, Wang HC, Wang AHJ, Wang CH, Kou GH, Lo CF. The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found. J Virol. 2005;79(1):140–149. doi: 10.1128/JVI.79.1.140-149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JH, Yang F, Zhang X, Xu X, Kou GH, Lo CF. Whispovirus. Curr Top Microbiol Immunol. 2009;328:197–227. doi: 10.1007/978-3-540-68618-7_6. [DOI] [PubMed] [Google Scholar]
- Meyer BN, Ferrign RN, Putnam JE, Jacobson LB, Nicholas DE, McLaughlin JL. Brine shrimp: a convenient general bioassay for active plant constituents. Planta Med. 1982;45:31–34. doi: 10.1055/s-2007-971236. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musthaq SKS, Kwang J. Reprint of “Evolution of specific immunity in shrimp—a vaccination perspective against white spot syndrome virus”. Dev Comput Immunol. 2015;48(2):342–353. doi: 10.1016/j.dci.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Patel JR, Tripathi P, Sharma V, Chauhan NS, Dixit VK. Phyllanthus amarus: ethnomedicinal uses, phytochemistry and pharmacology: a review. J Ethnopharmacol. 2011;138(2):286–313. doi: 10.1016/j.jep.2011.09.040. [DOI] [PubMed] [Google Scholar]
- Price KS, Waggy GT, Conway RA. Brine shrimp bioassay and seawater BOD of petrochemicals. J Water Pollut Control Fed. 1974;46:63–77. [PubMed] [Google Scholar]
- Rahmatullah M, Sadeak SMI, Bachar SC, Hossain MT, Abdullah-al-Mamun, Montaha Jahan N, Chowdhury MH, Jahan R, Nasrin D, Rahman M, Rahman S. Brine shrimp toxicity study of different Bangladeshi medicinal plants. Adv Nat Appl Sci. 2010;4(2):163–173. [Google Scholar]
- Sanchez-Paz A. White spot syndrome virus: an overview on an emergent concern. Vet Res. 2010;41(6):43. doi: 10.1051/vetres/2010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SchuÈttelkopf AW, Van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Cryst. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Sivakumar KC, Sajeevan TP, Singh IB. Marine derived compounds as binders of the White spot syndrome virus VP28 envelope protein: in silico insights from molecular dynamics and binding free energy calculations. Comput Biol Chem. 2016;64:359–367. doi: 10.1016/j.compbiolchem.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Sorgeloos P, Remiche-Van Der Wielen C, Persoone G. The use of Artemia nauplii for toxicity tests—a critical analysis. Ecotoxicol Environ Saf. 1978;2:249–255. doi: 10.1016/S0147-6513(78)80003-7. [DOI] [PubMed] [Google Scholar]
- Sudharsana S, Rajashekar Reddy CB, Dinesh S, Rajasekhara Reddy S, Mohanapriya A, Itami T, Sudhakaran R. Molecular docking and simulation studies of 3-(1-chloropiperidin-4-yl)-6-fluoro benzisoxazole 2 against VP26 and VP28 proteins of white spot syndrome virus. J Fish Dis. 2016;39:1231–1238. doi: 10.1111/jfd.12454. [DOI] [PubMed] [Google Scholar]
- Sundaram D, Kesavan K, Kumaravel H, Mohammed RF, Tohru M, Toshiaki I, Raja S. Protective efficacy of active compounds from Phyllanthus amarus against white spot syndrome virus in freshwater crab (Paratelphusa hydrodomous) Aquac Res. 2016;47:2061–2067. doi: 10.1111/are.12660. [DOI] [Google Scholar]
- Tang X, Wu J, Sivaraman J, Hew CL. Crystal structures of major envelope proteins VP26 and VP28 from white spot syndrome virus shed light on their evolutionary relationship. J Virol. 2007;81(12):6709–6717. doi: 10.1128/JVI.02505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M (2004) Molecular docking using ArgusLab, an efficient shape-based search algorithm and the AScore scoring function. In: ACS meeting. Philadelphia. 172, CINF 42: PA
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]