Abstract
Macrophage-lineage cells are indispensable to vertebrate homeostasis and immunity. In turn, macrophage development is largely regulated through colony-stimulating factor-1 (CSF1) binding to its cognate receptor (CSF1R). To study amphibian monopoiesis, we identified and characterized the X. laevis CSF1R cDNA transcript. Quantitative analysis revealed that CSF1R tissue gene expression increased with X. laevis development, with greatest transcript levels detected in the adult lung, spleen and liver tissues. Notably, considerable levels of CSF1R mRNA were also detected in the regressing tails of metamorphosing animals, suggesting macrophage involvement in this process, and in the adult bone marrow; corroborating the roles for this organ in Xenopus monopoiesis. Following animal infections with the ranavirus Frog Virus 3 (FV3), both tadpole and adult X. laevis exhibited increased kidney CSF1R gene expression. Conversely, while FV3-infected tadpoles increased their spleen and liver CSF1R mRNA levels, the FV3-challenged adults did not. Notably, FV3 induced elevated bone marrow CSF1R expression, and while stimulation of tadpoles with heat-killed E. coli had no transcriptional effects, bacterial stimulation of adult frogs resulted in significantly increased spleen, liver and bone marrow CSF1R expression. We produced the X. laevis CSF1R in recombinant form (rXlCSF1R) and determined, via in vitro cross-linking studies, that two molecules of rXlCSF1R bound the dimeric rXlCSF1. Finally, administration of rXlCSF1R abrogated the rXlCSF1-induced tadpole macrophage recruitment and differentiation as well as bacterial and FV3-elicited peritoneal leukocyte accumulation. This work marks a step towards garnering greater understanding of the unique mechanisms governing amphibian macrophage biology.
Keywords: Xenopus, monopoiesis, CSF1, macrophage development, ranavirus
Introduction
Macrophage-lineage cells are indispensable to homeostasis and host defenses of all vertebrate species. Monopoiesis, or the development and differentiation of macrophage lineage cells is chiefly mediated by the colony stimulating factor-1 (CSF1; macrophage colony-stimulating factor, MCSF) cytokine and growth factor (Garceau et al., 2010; Hanington et al., 2007; Pixley and Stanley, 2004; Wang et al., 2008). CSF1 functions as a homo-dimer, ligating the high-affinity tyrosine kinase CSF1 receptor (CSF1R) (Dai et al., 2002), the cell surface expression of which is restricted predominantly to mononuclear phagocytes and their derivative cell populations (Guilbert and Stanley, 1980; Lichanska et al., 1999).
In mammals, the expression levels of the CSF1R increase progressively with macrophage development, from lower levels by myeloid precursors to greater levels by monocytes and further elevated levels by terminally differentiated macrophages (Stanley et al., 1997). Furthermore, the ligation of CSF1R by CSF1 is critical not only to macrophage proliferation, differentiation and survival, but also for effective antimicrobial and anti-tumor host responses orchestrated by these cells (Bober et al., 1995; Karbassi et al., 1987; Munn and Cheung, 1995; Sweet and Hume, 2003). Thus, CSF1R gene expression serves as a reliable marker in the study of monopoiesis during animal development and host immune responses.
Despite the fact that more ancestral vertebrate species possess the CSF1/CSF1R monopoietic systems, the evolutionary origins of these pathways remain poorly understood. CSF1 is present in birds (Garceau et al., 2010), amphibians (Grayfer and Robert, 2013), and bony fish (Hanington et al., 2007); and CSF1Rs have been identified, in birds (Garceau et al., 2010) and teleosts (Barreda et al., 2005; Honda et al., 2005; How et al., 1996; Parichy et al., 2000; Pettersen et al., 2008). However, it appears that the bony fish CSF1 ligand-receptor axis is distinct to that of mammals (Hanington et al., 2007; Wang et al., 2008; Wang et al., 2014). Indeed, fish species possess multiple distinct gene copies of both the CSF1 ligand and receptor, which contrasts to single copies seen across higher vertebrates. Owing to the key position occupied by amphibians in vertebrate evolution, it would be interesting to determine whether the Xenopus CSF1 ligand-receptor system is more reminiscent of those seen in fish, mammals or alternatively a hybrid intermediate. Moreover, amphibian macrophage development remains to be fully characterized. Notably, monopoiesis of most vertebrate species occurs within designated hematopoietic tissues/organs such as the bone marrow of birds and mammals (Garceau et al., 2010; Tushinski et al., 1982) and the teleost head kidney (Belosevic et al., 2006; Neumann et al., 2000). By contrast, although the Xenopus liver periphery clearly functions as primary site of hematopoiesis (Hadji-Azimi et al., 1987; Hadji-Azimi et al., 1990; Lane and Sheets, 2002), our recent findings indicate that rather than the subcapsular liver, the bone marrow functions as the primary site of X. laevis macrophage development and contains CSF1 responsive macrophage precursors (Grayfer and Robert, 2013).
Infections of amphibians by ranaviruses (family Iridoviridae) and the resulting population die-offs are now believed to be a significant contributing factor to amphibian declines (Chinchar, 2002; Chinchar et al., 2009; Williams et al., 2005). A growing body of literature indicates that amphibian macrophage-lineage cells are critical players in both anti-ranaviral immunity and the ranavirus infection strategies (reviewed in Grayfer et al., 2012). Thus, greater insight into amphibian macrophage development may contribute to bridging the gap in our understanding of the evolution of vertebrate monopoiesis and to studying ranaviruses and developing preventative measures against these emerging pathogens.
Accordingly, to begin to delineate the roles of CSF1R in amphibian macrophage development and anti-RV immunity, we identified the X. laevis CSF1R, characterized the expression of this gene in developing, bacterially and virally challenged X. laevis and assessed the biological efficacies of a recombinant form of the X. laevis CSF1R protein. This manuscript marks the first characterization of an amphibian colony-stimulating factor-1 receptor.
Results
Identification and amino acid alignment analysis of the X. laevis CSF1R
To investigate amphibian macrophage development, we identified the full cDNA transcript encoding the X. laevis principal macrophage growth factor receptor, the colony-stimulating factor-1 receptor (CSF1R) by means of conventional and RACE PCR. The extracellular portion of the putative X. laevis CSF1R exhibits low amino acid sequence identity with the CSF1R molecules of other vertebrate species. However, all of these molecules have in common conserved and structurally important cysteine residues and the presence of five putative immunoglobulin-like domains (D1–D5; Fig. 1). Akin to the other vertebrate CSF1Rs, the putative X. laevis receptor possesses a disrupted intracellular domain composed of an ATP binding motif, a kinase insert domain as well as a major tyrosine kinase catalytic domain (Fig. 1). Notably, in contrast to the poor amino acid sequence conservation across the extracellular portions of the vertebrate CSF1Rs (including that of X. laevis), the intracellular components of these respective proteins exhibit many stretches of evolutionarily conserved residues, particularly throughout the tyrosine kinase catalytic domains (Fig. 1).
Fig. 1. Amino acid alignment of X. laevis CSF1R with other vertebrate CSF1R protein sequences.
Protein alignments were performed using ClustalW2 server. Fully conserved residues are indicated by an asterisk (*), partially conserved and semi-conserved substitutions are represented by “:” and “.”, respectively. The X. laevis CSF1R signal peptide is in bold, the immunoglobulin-like domains are indicated as D1–D5, the evolutionarily conserved structural cysteines are in white text within black boxes and the transmembrane domain is also in bold face. As indicated overhead; the X. laevis CSF1R ATP binding and tryrosine kinase major catalystic domains are in white text within black boxes while the kinase insert domain is indicated by an overhead line.
Phylogenetic analysis of vertebrate CSF1R proteins
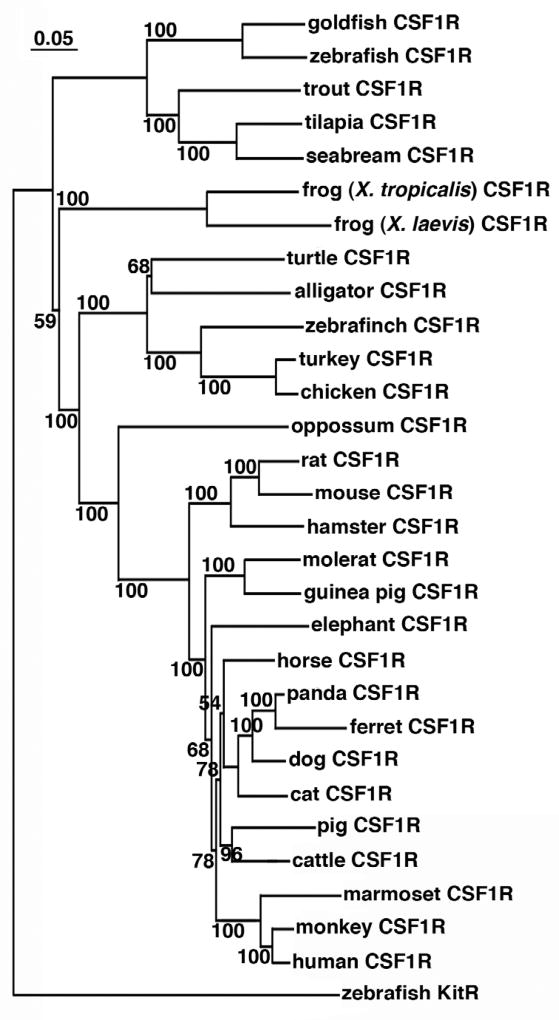
Phylogenetic analysis of the putative vertebrate CSF1R protein sequences indicated a clear evolutionary relationship, wherein the fish CSF1Rs formed a separate clade, ancestral to the amphibian CSF1Rs, which in turn branched independently and ancestral to other vertebrate CSF1Rs (Fig. 2). The orthology of the Xenopus sequences is supported by high boostrap values (Fig. 2). Notably, the avian and reptilian CSF1R molecules branched closer together, in a separate clade from the marsupial and mammalian CSF1R proteins (Fig. 2). The zebrafish KitR, which is structurally related to CSF1R, was used as an out-group to root the tree (Fig. 2).
Fig. 2. Phylogenetic analysis of teleost, amphibian, reptile, avian and mammalian colony-stimulating factor-1 receptor (CSF1R) molecules.
The phylogenetic tree was constructed from multiple deduced amino acid sequence alignments using the neighbor joining method and bootstrapped 10,000 times (denoted as %). The zebrafish KitR was used as outgroup to root the tree.
Quantitative analysis of CSF1R gene expression in tissues of tadpoles, metamorphic and adult X. laevis
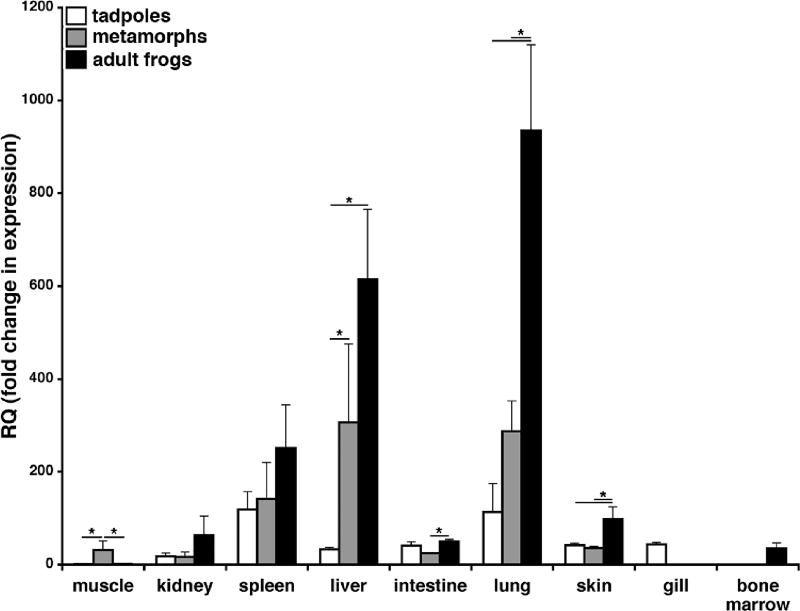
Since CSF1R is primarily expressed by macrophage-lineage cells, we next wanted to examine the quantitative gene expression of the X. laevis CSF1R in tadpoles (Stg. 54; (Thors et al., 1982a; Thors et al., 1982b)), metamorphs (Stg. 64) and adults (2 years old) and hence delineate the distribution of macrophage populations during development (Fig. 3). CSF1R gene expression in tadpole and adult muscle was low, whereas CSF1R mRNA levels were significantly greater in regressing tail-muscle tissues of metamorphosing animals (Fig. 3), consistent with the accumulation of active macrophages involved in tail reabsorption (Nishikawa et al., 1998). The spleen, liver and lung CSF1R gene expression increased with development, where the metamorph and adult liver and lung CSF1R transcript levels were significantly greater than those seen in the respective tadpole tissues (Fig. 3). Interestingly, the lung CSF1R mRNA levels were also significantly greater than those see in tadpoles and metamorphs (Fig. 3). Finally, marginal but detectable levels of CSF1R transcripts were detected in the adult bone marrow (Fig. 3).
Fig. 3. Quantitative colony-stimulating factor-1 receptor (CSF1R) tissue gene expression analysis of tadpoles (Stg. 54), metamorphic (Stg. 64) and adult (2 years-old) frogs.
Tissues from three individuals (N=3) were used for all expression studies, the expression was performed via the delta^delta CT method using X. laevis CSF1R-specific primers. The expression examined relative to the GAPDH endogenous control and normalized against the tadpole muscle tissue expression. Results are means ± SEM, N=4 and the (*) above lines denotes significant difference between tissues indicated by the lines, P<0.05.
Quantitative analysis of CSF1R gene expression in tissue of X. laevis tadpoles and adult frogs immunologically challenged with FV3 and heat-killed E. coli
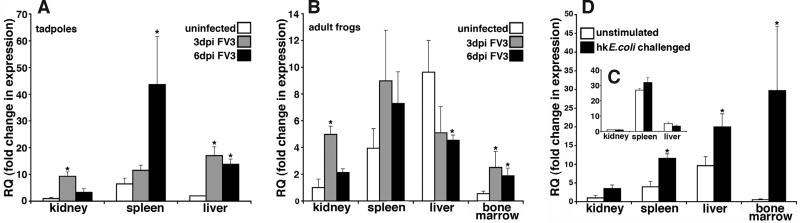
To infer on macrophage distribution upon immunological challenge, we examined the expression of CSF1R following infection with the ranavirus Frog Virus 3 (FV3; Iridoviridae) or after injections with heat-killed E. coli (Fig. 4). In tadpoles, the kidney, which is the central site of FV3 replication, exhibited a modest but significant increase in the CSF1R gene expression at 3 days post FV3 infection (dpi), followed by a decrease at 6 dpi (Fig. 4A). In contrast, a substantial increase of CSFR1 gene expression was observed in tadpole spleens (the primary amphibian immune tissue) at 6 (but not 3) dpi (Fig. 4A). The hematopoietic tadpole liver exhibited significant transcriptional increases in CSF1R at both 3 and 6 dpi (Fig. 4A). Interestingly, challenge of tadpoles with heat-killed E. coli had no bearing on CSF1R gene expression (Fig. 4C).
Fig. 4. Quantitative analysis of colony-stimulating factor-1 receptor (CSF1R) tissue gene expression in Frog Virus 3 (FV3) and heat-killed E. coli challenged tadpoles and adult X. laevis.
Tadpoles (Stg. 54) and adult frogs (2 years-old) were infected by ip injections with 1×104 and 5×106 PFU of FV3, respectively. Alternatively, tadpoles and adult frogs were injected respectively with 10µl and 100µl of heat-killed (hk) E. coli. (A) FV3-infected tadpole CSF1R gene expression. (B) FV3-infected tadult frog CSF1R gene expression. (C) Heat-killed E. coli-stimulated tadpole CSF1R gene expression. (D) Heat-killed E. coli-stimulated adult frog CSF1R gene expression. All gene expression analysis was performed via the delta^delta CT method using X. laevis CSF1R-specific primers, the expression examined relative to the GAPDH endogenous control and normalized against respective uninfected kidney expression. Results are means ± SEM, N=4 and the (*) above lines denotes significant difference between tissues indicated by the lines, P<0.05.
As in tadpoles, FV3 infection of adult frogs resulted in increased kidney CSF1R gene expression at 3 dpi and a subsequent decrease at 6 dpi (Fig. 4B). In contrast to the increased CSF1R gene expression in tadpole spleens following FV3 infection, no significant increase of CSF1R gene expression was observed in adult spleens following ranaviral challenge (Fig. 4B). Furthermore, the adult liver CSF1R gene expression decreased following FV3 infection, significantly so at 6 dpi (Fig. 4B). The bone marrow CSF1R gene expression was significantly increased at both 3 and 6 dpi (Fig. 4B), supporting our previous findings that the amphibian bone marrow may serve as a site of monopoiesis (Grayfer and Robert, 2013),
Intriguingly and again in contrast to the tadpole expression patterns; six days following heat-killed E. coli stimulation of adult frogs, these animals possessed significantly elevated spleen, liver and bone marrow CSF1R gene expression, where the magnitude of the bone marrow upregulation of this gene far exceeded any other CSF1R transcriptional changes reported here (Fig. 4C).
In vitro rXlCSF1 and rXlCSF1R binding studies
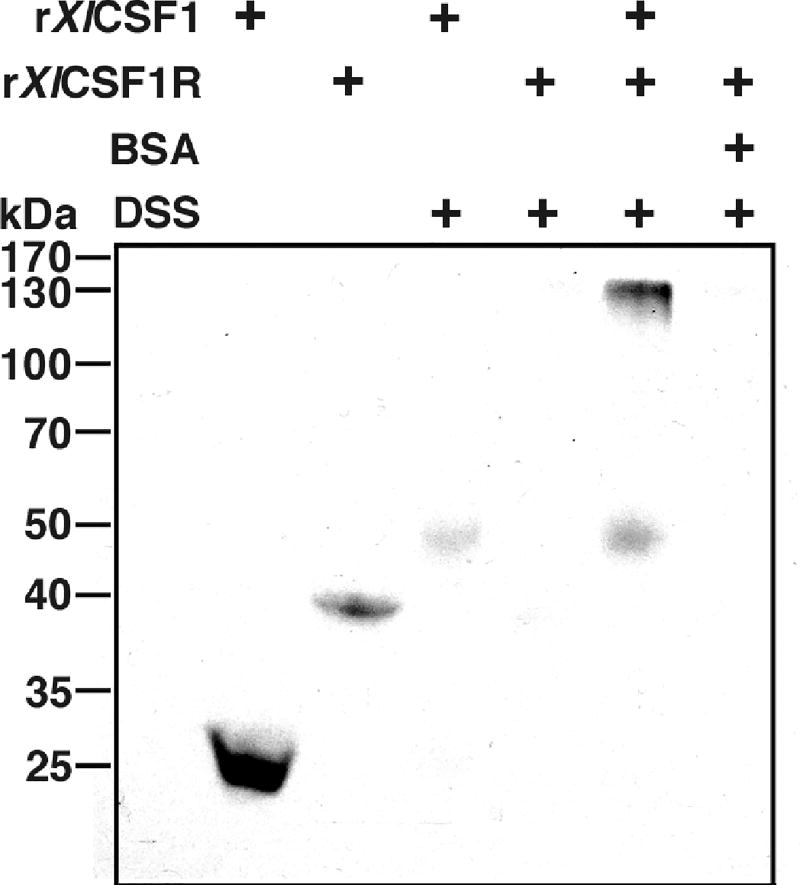
To confirm that the Xenopus CSF1R is the cognate receptor for the monopoietic CSF1 ligand, we produced recombinant rXlCSF1R and rXlCSF1 molecules using an insect expression system, and assessed ligand-receptor binding in vitro using the disuccinimidyl suberate (DSS, 2.5 mM final concentration) crosslinker to stabilize protein interactions and western blot analysis against the V5 tags on these recombinant proteins (Fig. 5). Since the mammalian CSF1 interacts with the D2 and D3 domains of its cognate CSF1R (Chen et al., 2008), we engineered the (rXl)CSF1R to comprise of the D2 and D3 extracellular domains of the X. laevis receptor. Western blot analysis of the crosslinked 25 kDa rXlCSF1 ligand revealed a band shift to 50 kDa; indicative of rXlCSF1 dimerization (Fig. 5). Following DSS crosslinking, rXlCSF1R could no longer be detected by western blot, which is typical of non-interacting proteins (Fig. 5). Notably, when the rXlCSF1 and rXlCSF1R were coincubated and crosslinked with DSS, a band shift to 130 kDa was observed, indicating that the dimerized rXlCSF1 interacted with two 40 kDa molecules of rXlCSF1R in solution (Fig. 5). The crosslinking of rXlCSF1R in the presence of bovine serum albumin (BSA) did not yield a banding pattern, indicating lack of non-specific interactions (Fig. 5).
Fig. 5. In vitro rXlCSF1 and rXlCSF1R cross-linking studies.
One microgram each of rXlCSF1, rXlCSF1R, rXlCSF1 + rXlCSF1R and BSA + rXlCSF1R were incubated in APBS (100µl final volume) for 30 min and cross-linked for 30 min using 2.5 mM DSS, final concentration. Cross-linking reactions were terminated by addition of 50 mM Tris (final concentration). Reactions were resolved and visualized using SDS-PAGE and western blot against the V5 epitopes on the recombinant proteins.
rXlCSF1R abrogates the rXlCSF1-mediated recruitment and differentiation of tadpole peritoneal macrophages
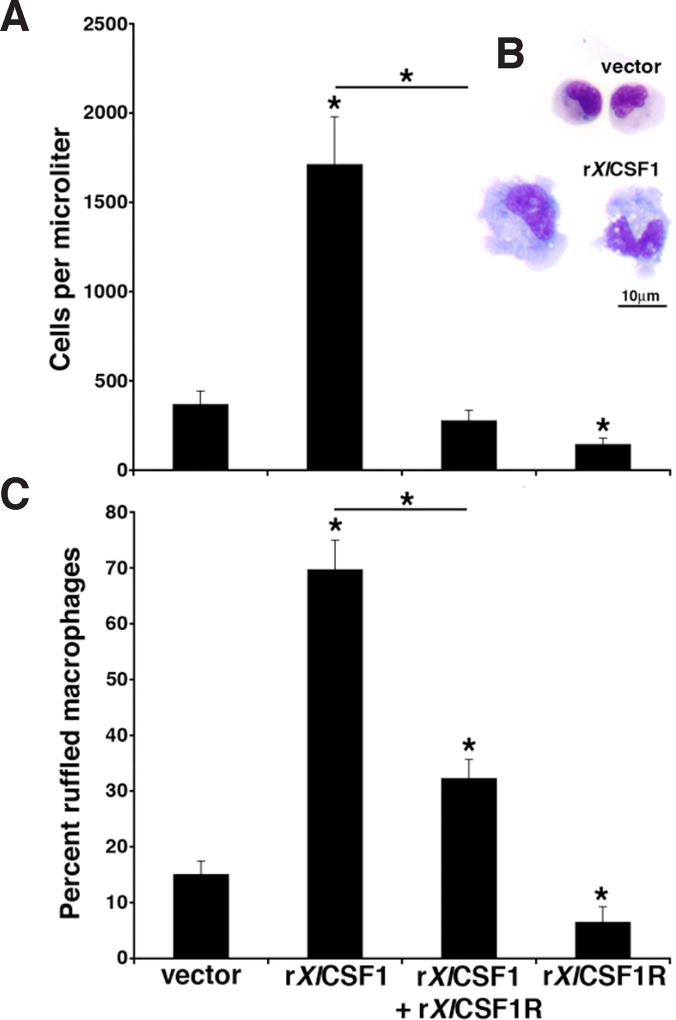
To confirm the biological roles of rXlCSF1 and rXlCSF1R, we examined the ability of the recombinant ligand to elicit and differentiate tadpole peritoneal macrophages, and the capacity of the CSF1R D2-D3 extracellular domain-containing rXlCSF1R to abrogate this process by antagonizing the effects induced by rXlCSF1 (Fig. 6). Twenty-four hours following intraperitoneal injection of tadpoles with the rXlCSF1 resulted in significant accumulation of peritoneal macrophages (Fig. 6A). In contrast, administration of rXlCSF1R induced a reduction in the numbers of tadpole resident peritoneal phagocyte populations (Fig. 6A). Furthermore, a four-fold excess of rXlCSF1R co-injected with rXlCSF1 abrogated the rXlCSF1-mediated macrophage elicitation (Fig. 6A).
Fig. 6. The rXlCSF1R abrogates the rXlCSF1-mediated tadpole macrophage recruitment and differentiation.
(A) Tadpoles were injected with vector control, 250ng of rXlCSF1, 1000ng of rXlCSF1R or a combination of rXlCSF1 (250ng) and rXlCSF1R (10000ng). After 24hrs peritoneal phagocytes were lavaged and enumerated. Results are means ± SEM, N=6 and the (*) above lines denotes significant difference between treatment groups indicated by the lines, P<0.05. (B) Morphological analysis of Giemsa-stained vector-control and rXlCSF1R derived cultures. Scale bar = 10µm. (C) Cultures from (A) were Giemsa-stained, enumerated for the presence of morphologically differentiated macrophages as exemplified in (B) by the rXlCSF1R-derived cells. Results are expressed as percent means ± SEM from ten random fields of view. The (*) above lines denotes significant difference between treatment groups indicated by the lines, P<0.05.
Microscopy analysis of cytospin preparations stained by Giemsa revealed that peritoneal leukocytes obtained from tadpoles injected with vector control were comprised primarily of smaller mononuclear phagocytes, whereas rXlCSF1 treatment resulted in the accumulation/differentiation of considerably larger, ruffled and highly vacuolated macrophages (Fig. 6B). On average, cells from vector control administered animals consisted of only 14.9% large, ruffled and vacuolated macrophages, whereas these larger cells comprised 69.6% of the rXlCSF1-elicited tadpole peritoneal phagocytes (Fig. 6C). Notably, peritoneal phagocytes derived from animals that had been co-injected with rXlCSF1 and rXlCSF1R had significantly lower proportions (32.1%) of these morphologically distinct macrophage populations as compared to the rXlCSF1-cultures (Fig. 6C). Interestingly, peritoneal phagocytes isolated from tadpoles administered rXlCSF1R alone had significantly lower numbers of large, ruffled macrophages (6.4%) than even those seen in the vector control cultures (Fig. 6C), suggesting that resident peritoneal macrophages also rely on native CSF1 for survival.
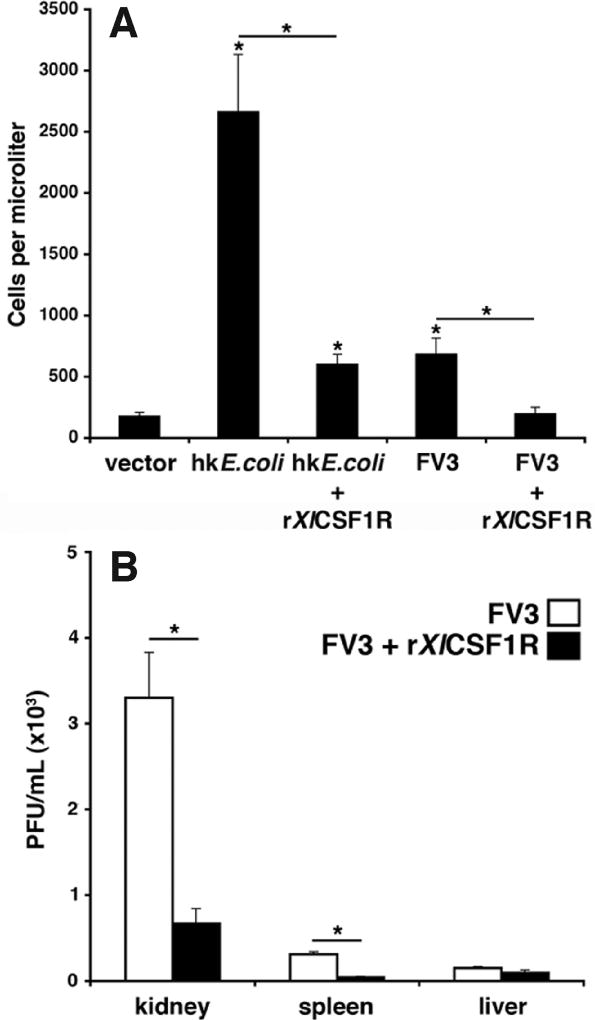
Given that the administration of rXlCSF1R significantly affected the differentiation and abundance of resident tadpole peritoneal macrophages (see Fig. 6), we next assessed how rXlCSF1R treatment would impact the tadpoles’ capacity to recruit leukocytes into the peritoneum following heat-killed E. coli or FV3 challenge (Fig. 7A). Notably, the numbers of peritoneal leukocytes elicited one day after stimulation with heat-killed E. coli were dramatically decreased by co-injection with rXlCSF1R (Fig. 7A). Similarly, macrophage infiltration induced one day after FV3 infection was significantly decreased by co-injecting rXlCSF1R with the FV3 (Fig. 7A). These findings suggest that the macrophage-suppressive effects conferred by rXlCSF1R administration (Fig. 6) culminate in decreased capacities of resident mononuclear phagocytes to recruit additional leukocyte populations in response to immune stimuli.
Fig. 7. The rXlCSF1R abrogates heat-killed E. coli and Frog Virus 3 elicited recruitment of tadpole peritoneal leukocytes and reduces FV3 dissemination.
(A) Tadpoles were injected with vector control, heat-killed (hk) E. coli, FV3 (104 PFU), a combination of hk E. coli and rXlCSF1 (1µg) or a combination of FV3 (104 PFU) and rXlCSF1R (1µg). After 24hrs peritoneal phagocytes were lavaged and enumerated. Results are means ± SEM, N=4. (*) denotes statistical difference from the vector control and the (*) above lines denotes significant difference between treatment groups indicated by the lines, P<0.05. (B) Tadpoles were injected with 1×104 PFU of FV3 alone, or in combination with 1µg of rXlCSF1R. Following 24 hrs, tadpoles were sacrificed; their tissues were isolated and examined for infectious FV3 burdens by plaque assays. Results are means ± SEM, N=3. (*) above lines denotes significant difference between treatment groups indicated by the lines, P<0.05.
Since we have shown that X. laevis macrophages derived by the recombinant ligand rXlCSF1 increases tadpoles susceptibility to FV3 (Grayfer and Robert, 2014) and that converging evidence indicates that ranaviruses rely on macrophages to disseminate inside their hosts (reviewed in Grayfer et al., 2012), we postulated that ablation of tadpole resident peritoneal macrophages by rXlCSF1R should also reduce FV3 dissemination into tadpole organs. Accordingly, tadpoles were injected with FV3 alone, or in combination with rXlCSF1R and virus loads in various tissues were assessed the following day (Fig. 7B). In support of our previous findings, the rXlCSF1R administration significantly reduced FV3 loads in kidney and spleen, but not liver (Fig. 7B). This suggests that peritoneal CSF1-dependent phagocytes are critical for the dissemination of this virus.
Discussion
This manuscript represents the first characterization of an amphibian CSF1R. Despite the poor conservation of the overall amino acid sequence among vertebrate CSF1R molecules, all CSFR1 gene products, including the X. laevis, share hallmark features including 5 putative immunoglobulin domains, structurally conserved cysteine residues as well as a disrupted tyrosine kinase domain. The extracellular portions of CSF1R molecules exhibit more divergence, possibly reflecting evolutionary drift to facilitate the binding of respective cognate CSF1 ligands, which also display low amino acid sequence conservation (Grayfer and Robert, 2013). By contrast, the intracellular catalytic tyrosine kinase domains of these respective CSF1R proteins are remarkably well conserved, perhaps marking the evolutionary functional necessity for retaining these protein sequences. The evolutionarily diverged CSF1R amino acid sequences of distinct vertebrate classes are also reflected in their phylogenetic relationships, wherein the fish, amphibian, avian, reptilian and mammalian receptors all branched in respective separate clades. Indeed the CSF1R catalytic domains have been evolutionarily conserved across vertebrates, suggesting conservation in downstream CSF1R cell signaling and presumably the resulting biological outcomes. However, many other aspects of lower vertebrate macrophage biology appear to be distinct from what has been documented in mammals. This includes varying teleost CSF1 ligand and receptor gene copy numbers (Hanington et al., 2007; Wang et al., 2008; Wang et al., 2014; Williams et al., 2002) and unique physiological localization of amphibian monopoiesis (Grayfer and Robert, 2013). It remains to be determined whether these differences arise from and/or are dependent on CSF1-CSF1R functions which are different to those of mammals. Thus, it is possible that the monopoietic roles conferred by CSF1-CSF1R of distinct lower vertebrate species are at least partially unique to those described in mammals.
Whereas birds and mammals possess a single CSF1 gene expressing alternatively spliced transcripts, fish possess at least two distinct CSF1 genes that do not appear to produce alternatively spliced products (Hanington et al., 2007; Wang et al., 2008; Wang et al., 2014). Presently, it is unknown whether the functions of the different fish CSF1 genes correspond to those mediated by the respective alternatively spliced mammalian transcript products. Furthermore, at least some fish species, including Fugu, possess 2 distinct CSF1R genes (Williams et al., 2002). Therefore, it is quite possible that similar to the fish type II IFN system (Aggad et al., 2012; Grayfer and Belosevic, 2009; Grayfer et al., 2010; Shibasaki et al., 2013; Yabu et al., 2011), the multiple teleost CSF1 receptor and ligand gene products may exhibit complex interactions, distinct from the single ligand, single receptor strategy of higher vertebrates. In our recent study of the X. tropicalis and X. laevis CSF1 genes (Grayfer and Robert, 2013), we did not identify additional CSF1 gene copies in X. laevis and X. tropicalis genomes, nor did we detect additional CSF1R genes during the studies described here. It is noteworthy that despite our best efforts, using both conventional and RACE PCR approaches, we were unable to identify alternatively spliced X. laevis CSF1 transcripts (data not shown). Although more investigation are needed, it appears that alternatively spliced CSF1 and/or CSF1R genes are absent or at least of minor importance in Xenopus. It is possibly that the unique Xenopus monopoietic strategy requires a single non-alternatively spliced Xenopus CSF1 and a single CSF1R.
The highest level of CSF1R gene expression was observed in the lung and liver of X. laevis adults. Presumably these expression patterns reflect the presence of alveolar macrophages (Lin et al., 1989) and Kupffer cells (Amemiya et al., 2011), wherein the mammalian counterparts of both of these macrophage-lineage populations express high CSF1R levels. Notably, CSF1R gene expression increased with X. laevis development in the majority of tissues examined, including kidney, spleen, liver, lung and skin. This may represent the accumulation of resident macrophages, reflecting complex growing biological necessities for these cells with frog development and maturation. Alternatively, it is possible that the level of CSF-1R gene expression per cell increases during development. In addition, the CSF1R gene expression increased in the muscle tissue of regressing tails from metamorphosing animal. Indeed, macrophages have been previously demonstrated to play crucial roles in tail and body muscle remodeling during metamorphosis (Nishikawa et al., 1998), where our present observations corroborate with these earlier findings. It will be interesting to examine the differences in CSF1 responsiveness of macrophage precursors in tadpoles and adult X. laevis, since they are both CSF1-responsive (Grayfer and Robert, 2013; Grayfer and Robert, 2014), whereas macrophage development in adult involves the bone marrow that is absent in tadpoles (Grayfer and Robert, 2013).
Following FV3 infection, both tadpoles and adults exhibited increased kidney CSF1R gene expression, which is interesting considering that the kidney is the primary site of FV3 replication (Gantress et al., 2003; Robert et al., 2007). In addition, macrophages are intimately involved in both immunity and the infection strategy of FV3 (Morales et al., 2010; Grayfer et al., 2012). Therefore, it seems reasonable to speculate that CSFR gene expression increases due to the infiltration of kidney tissues by macrophage-lineage cells. Intriguingly, the spleen and liver CSF1R gene expression patterns induced by FV3 infection were markedly different between tadpoles and adults, possibly underlining their distinct monopoietic strategies. In light of our recent findings that the X. laevis bone marrow serves as the prime source of macrophage precursors (Grayfer and Robert, 2013), together with our present observation that FV3 and heat-killed E. coli both elicit increased bone marrow CSF1R expression may indicate that in response to immunological challenges, adult Xenopus increase monopoietic activity at the level of the bone marrow. By contrast, tadpoles do not possess bone marrow and thus, presumably orchestrate monopoiesis in the hematopoietic liver. This would explain the increased CSF1R gene expression in FV3-infected tadpole, but not adult liver tissues. In comparison to adults, the tadpole spleen may likewise be more prominently involved in macrophage development and immunity as reflected in substantially more upregulated CSF1R gene expression within this tadpole tissue following FV3 challenge. It is interesting that heat-killed E. coli did not elicit significant CSF1R expression changes in any of the tadpole tissues examined, whereas following this inflammatory stimulus adults exhibited substantially more robust CSF1R gene expression increases in the spleen, liver and bone marrow than observed following FV3 infection. These disparities may reflect differences in pathogen pattern recognition receptor expression between tadpoles and adults, and a physiological necessity for more prominent adult monopoietic responses to inflammatory, rather than viral challenges. A greater understanding of Xenopus macrophage development and immune responses will no doubt shed light on this present enigma. We emphasize that in our present work E. coli was used as an inflammatory stimulus rather than a direct point of comparison with the more relevant FV3 viral challenge. Future X. laevis infection studies using relevant bacterial pathogens will lend to our understanding of the differences in macrophage involvement during different host responses.
The mammalian dimeric CSF1 binds exclusively to the D2 and D3 domains of the CSF1R, dimerizing the receptor (Chen et al., 2008). Our in vitro binding studies indicate that the D2 and D3 domains of a recombinant amphibian CSF1R are also sufficient to bind the recombinant homodimerized CSF1 ligand. More detailed studies will be needed to elucidate the modalities and stoichiometry of these interactions. Notably, the rXlCSF1R very effectively inhibited rXlCSF1-mediated tadpole elicitation and development of macrophages into cells morphologically resembling mature, differentiated populations. Furthermore, rXlCSF1R administration also ablated the recruitment of peritoneal leukocytes elicited by heat-killed E. coli and the FV3. Since tadpoles administered with rXlCSF1R exhibited diminishing numbers of mature resident peritoneal macrophages, the compromised capacities of rXlCSF1R-treated tadpoles to recruit leukocytes into the peritoneum upon ip immune challenges likely reflects a functional impairment of peritoneal macrophages, which would normally chemo-attract additional immune populations in response to immune stimuli. Notably, teleost fish appear to have evolved an additional regulatory mechanism of monopoiesis and inflammation whereby they produce an alternatively spliced, soluble CSF1R (Barreda et al., 2005; Rieger et al., 2014a; Rieger et al., 2014b). Intriguingly, this moiety only possesses the D1 and D2 domains of the membrane-bound CSF1R and yet this molecule is highly biologically active and inhibits fish macrophage proliferation and inflammatory responses (Barreda et al., 2005; Rieger et al., 2014a; Rieger et al., 2014b). It will be interesting to learn which specific domains are involved in the interactions of the respective teleost CSF1 ligand(s), soluble and membrane bound CSF1Rs and whether a similar system is present in amphibian species.
Converging evidence indicate that macrophage-lineage cells are utilized by ranaviruses as a means of dissemination within their hosts (Grayfer et al., 2012). In addition, CSF1-derived macrophages render tadpoles more susceptible to FV3 (Grayfer and Robert, 2014). Here, we show that intraperitoneal administration of rXlCSF1R reduces resident phagocyte numbers and impairs some of their functions such as leukocyte recruitment. The treatment with rXlCSF1R also results in decreased dissemination of FV3 from the inoculation site of the peritoneum to the kidney (central site of ranavirus replication) and the spleen (central immune organ), whereas it does not affect low viral dissemination into the liver. These observations underline the complexity of the roles of mononuclear phagocytes as both mediators of anti-RV immune responses and culprits in the progression of these infections.
The CSF1-CSF1R axis represents the focal point of vertebrate monopoiesis and it has become apparent that evolutionarily more ancestral species such as teleosts and amphibians may possess varying strategies for CSF1-mediated macrophages development, distinct from those described in mammalian species. Further research into CSF1 and CSF1R macrophage biology of lower vertebrates will yield new insights into the evolutionary basis of monopoiesis and permit the development of more effective preventative measures against pathogens that infiltrate macrophage-lineage cells of more ancient species.
Materials and Methods
Animals
Outbred pre-metamorphic (stage 54, (Thors et al., 1982a; Thors et al., 1982b)) tadpoles, metamorphic (stage 64) and adult (2 years old) frogs were obtained from our X. laevis research resource for immunology at the University of Rochester (http://www.urmc.rochester.edu/mbi/resources/Xenopus-laevis/). All animals were handled under strict laboratory and University Committee on Animal Research regulations (Approval number 100577/2003-151).
Identification and analysis of X. laevis colony-stimulating factor-1 receptor
The identification of the X. laevis CSF1 was described previously (Grayfer and Robert, 2013). A fragment of the X. laevis CSF1R cDNA transcript was identified using primers against the predicted X. tropicalis CSF1R sequence (Acc. no.: BC082504). Subsequently, RACE PCR was performed in accordance with manufacturers’ directions (Clonetech) to identify the complete X. laevis CSF1R cDNA transcript (Acc. No.: KM400585). All primer sequences are available upon request.
Protein sequence alignments were performed using the Clustal W software (http://www.ebi.ac.uk/clustalw/). Signal peptide regions were identified using the SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/). Protein motif and domain predictions were achieved using the ELM (http://elm.eu.org/) and SMART (http://smart.embl-heidelberg.de/) servers. Phylogenetic analysis was performed by Clustal X software using the neighbor joining method and bootstrapped 10,000 times, with values expressed as percentages.
Production of rXlCSF1 and rXlCSF1R
The production of the rXlCSF1 has been described previously (Grayfer and Robert, 2013). The rXlCSF1R was produced by transfecting Sf9 insect cells (cellfectin II, Invitrogen) with the pMIB/V5 HisA insect expression vector (Invitrogen) containing the X. laevis CSF1R sequence corresponding to the IG2 and IG3 domains of the extracellular portion of the protein. Transfected Sf9 supernatants were confirmed for rXlCSF1R expression, positive transfectants were scaled up to 500 mL in liquid cultures and grown for 6 days in blasticidin (10 µg/mL)-containing medium. Resulting supernatants were dialyzed overnight at 4/C against 150 mM sodium phosphate, concentrated against polyethylene glycol flakes (8 kDa), dialyzed overnight at 4CC against 150 mM sodium phosphate and passed through Ni-NTA agarose columns (Qiagen). Columns were washed with 2×10 volumes of high stringency wash buffer (0.5% Tween 20; 50 mM Sodium Phosphate; 500 mM Sodium Chloride; 100 mM Imidazole) and 5× with low stringency wash buffer (as above, but with 40 mM Imidazole). The rXlCSF1R was eluted in fractions using 400 mM Imidazole. The purity of the eluted fractions was confirmed by silver-stain and the presence of rXlCSF1R was assessed by western blot against the V5 epitope on the recombinant protein. Fractions containing the rXlCSF1R were pooled and the protein concentration determined by the Bradford Protein Assay (BioRad). A protease inhibitor cocktail (Roche) was added to the purified protein, which was then aliquoted and stored at −20°C until use.
The vector control was produced by transfecting Sf9 cells with an empty expression vector and treating the resulting supernatants akin to and in parallel to the rXlCSF1 production.
Cell culture media
The amphibian (ASF) culture medium used in these studies has been previously described (Robert et al., 2007). All cell cultures were established using ASF supplemented with 10% fetal bovine serum, 2.5% heat-inactivated X. laevis serum, 20 µg/mL kanamycin and 100 U/mL penicillin / 100 µg/mL streptomycin (Gibco). Amphibian PBS (APBS) has been previously described (Robert et al., 2007).
Isolation of rXlCSF1-elicited tadpole macrophages
Tadpoles at developmental stages 54 (Stg. 54; (Thors et al., 1982a; Thors et al., 1982b)) were intraperitoneally (ip) injected with 10 µL of the vector control, rXlCSF1 (250 ng), rXlCSF1R (1000 ng) or a combination of rXlCSF1 (250 ng) and rXlCSF1R (1000 ng) in 10 µL volumes. After 24 hrs, peritoneal macrophages were collected by lavage with 50µL volumes of APBS. Cells were enumerated using trypan blue exclusion, collected on glass slides by cytospin and stained with Giemsa (Fluka).
Frog Virus 3 stocks and animal infections
Fathead minnow cells (FHM; American Type Culture Collection, ATCC No.: CCL-42) were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), penicillin (100 U/mL) and streptomycin (100 µg/mL) with 5% CO2 at 30°C. FV3 was grown by a single passage on FMH cells, purified by ultracentrifugation on a 30% sucrose cushion and quantified by plaque assay on BHK monolayer under an overlay of 1% methylcellulose (Morales et al., 2010).
Tadpoles and adult frogs were infected by ip injections with previously-established (Gantress et al., 2003; Hanington et al., 2007; Robert et al., 2005) infection doses of 1×104 and 5×106 plaque forming units (PFU) of FV3, respectively. Three and six days post infection (dpi) animals were euthanized by immersion in 0.5% tricaine methane sulfonate (MS-222), tissues removed and processed for RNA isolation.
For determination of FV3 loads, tadpoles were ip injected with 1×104 PFU of FV3 alone, or in combination with 1µg of rXlCSF1R. After 24 hrs, tadpoles were sacrificed, their tissues excised and processed for plaque forming assay analysis to determine respective infectious FV3 burdens.
Heat-killed Escherichia coli stimulation
E. coli (XL1-blue, Strategene, La Jolla, Ca.) cultured overnight at 37°C, were boiled for 1 hour, pelleted by centrifugation and resuspended in one-tenth of the initial volume (approximately 1 × 108 bacteria/mL) of APBS. Tadpoles and adult frogs were injected ip with 10 and 100 µl of this heat-killed (hk) bacterial preparation.
Quantitative gene expression analysis
Total RNA was extracted from frog tissues using the Trizol reagent following the manufacturer’s directions (Invitrogen). All cDNA synthesis was performed using the iScript cDNA synthesis kit according to manufacturers’ directions (Bio-Rad, Hercules, CA) using 500 ng of total DNAse treated (Ambion) RNA.
Relative qRT-PCR gene expression analyses was performed via the delta^delta CT method using validated X. laevis CSF1R-specific primers, with expression examined relative to the GAPDH endogenous control and normalized against the lowest observed expression. All experiments were performed using the ABI 7300 real-time PCR system and PerfeCTa® SYBR Green FastMix, ROX (Quanta). Expression analysis was performed using ABI sequence detection system software (SDS). All primers were validated prior to use. All primer sequences are available upon request.
In vitro rXlCSF1 and rXlCSF1R cross-linking studies
One microgram each of rXlCSF1, rXlCSF1R, rXlCSF1 + rXlCSF1R and BSA + rXlCSF1R were incubated in APBS (100µl final volume) for 30 min, cross-linked for 30 min using 2.5 mM disuccinimidyl suberate (DSS, final concentration, Thermo Scientific). Cross-linking was terminated for 15 min by the addition of 50 mM Tris (final concentration). The reactions were then resolved and visualized using SDS-PAGE and western blot against the V5 epitopes on the recombinant proteins and developed using ECL (Pierce) on X-ray film (Eastman Kodak Co.)
Statistical analysis
Statistical analysis was performed using a one-way analysis of variance (ANOVA) and Tukey’s post hoc test, using Vassar Stat (http://faculty.vassar.edu/lowry//anova1u.html) statistical program. Probability level of P<0.05 was considered significant.
Acknowledgments
This work was supported by National Institute of Health (R24-AI-059830) and National Science Foundation (IOB-074271) grants to J.R. L.G. was supported by a National Sciences and Engineering Research Council of Canada Postdoctoral Fellowship and a Life Sciences Research Fellowship from the Howard Hughes Medical Institute. We thank Tina Martin for animal husbandry. This manuscript was improved by the insightful suggestions of one anonymous reviewer.
Abbreviations used in this paper
- CSF-1
colony-stimulating factor-1
- FV3
frog virus 3
References
- Aggad D, Stein C, Sieger D, Mazel M, Boudinot P, Herbomel P, Levraud JP, Lutfalla G, Leptin M. In vivo analysis of Ifn-gamma1 and Ifn-gamma2 signaling in zebrafish. J Immunol. 2012;185:6774–6782. doi: 10.4049/jimmunol.1000549. [DOI] [PubMed] [Google Scholar]
- Amemiya H, Kono H, Fujii H. Liver regeneration is impaired in macrophage colony stimulating factor deficient mice after partial hepatectomy: the role of M-CSF-induced macrophages. J Surg Res. 2011;165:59–67. doi: 10.1016/j.jss.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Barreda DR, Hanington PC, Stafford JL, Belosevic M. A novel soluble form of the CSF-1 receptor inhibits proliferation of self-renewing macrophages of goldfish (Carassius auratus L.) Dev Comp Immunol. 2005;29:879–894. doi: 10.1016/j.dci.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Belosevic M, Hanington PC, Barreda DR. Development of goldfish macrophages in vitro. Fish Shellfish Immunol. 2006;20:152–171. doi: 10.1016/j.fsi.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Bober LA, Grace MJ, Pugliese-Sivo C, Rojas-Triana A, Sullivan LM, Narula SK. The effects of colony stimulating factors on human monocyte cell function. Int J Immunopharmacol. 1995;17:385–392. doi: 10.1016/0192-0561(95)00025-w. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci USA. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchar VG. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch Virol. 2002;147:447–470. doi: 10.1007/s007050200000. [DOI] [PubMed] [Google Scholar]
- Chinchar VG, Hyatt A, Miyazaki T, Williams T. Family Iridoviridae: poor viral relations no longer. Curr Top Microbiol Immunol. 2009;328:123–170. doi: 10.1007/978-3-540-68618-7_4. [DOI] [PubMed] [Google Scholar]
- Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- Gantress J, Maniero GD, Cohen N, Robert J. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology. 2003;311:254–262. doi: 10.1016/s0042-6822(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Garceau V, Smith J, Paton IR, Davey M, Fares MA, Sester DP, Burt DW, Hume DA. Pivotal Advance: Avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J Leukoc Biol. 2010;87:753–764. doi: 10.1189/jlb.0909624. [DOI] [PubMed] [Google Scholar]
- Grayfer L, Andino Fde J, Chen G, Chinchar GV, Robert J. Immune evasion strategies of ranaviruses and innate immune responses to these emerging pathogens. Viruses. 2012;4:1075–1092. doi: 10.3390/v4071075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Belosevic M. Molecular characterization of novel interferon gamma receptor 1 isoforms in zebrafish (Danio rerio) and goldfish (Carassius auratus L.) Mol Immunol. 2009;46:3050–3059. doi: 10.1016/j.molimm.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Grayfer L, Garcia EG, Belosevic M. Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.) J Biol Chem. 2010;285:23537–23547. doi: 10.1074/jbc.M109.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Colony-stimulating factor-1-responsive macrophage precursors reside in the amphibian (Xenopus laevis) bone marrow rather than the hematopoietic subcapsular liver. J Innate Immun. 2013;5:531–542. doi: 10.1159/000346928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Divergent antiviral roles of amphibian (Xenopus laevis) macrophages elicited by colony-stimulating factor-1 and interleukin-34. J Leukoc Biol. 2014 doi: 10.1189/jlb.4A0614-295R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbert LJ, Stanley ER. Specific interaction of murine colony-stimulating factor with mononuclear phagocytic cells. J Cell Biol. 1980;85:153–159. doi: 10.1083/jcb.85.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadji-Azimi I, Coosemans V, Canicatti C. Atlas of adult Xenopus laevis laevis hematology. Dev Comp Immunol. 1987;11:807–874. doi: 10.1016/0145-305x(87)90068-1. [DOI] [PubMed] [Google Scholar]
- Hadji-Azimi I, Coosemans V, Canicatti C. B-lymphocyte populations in Xenopus laevis. Dev Comp Immunol. 1990;14:69–84. doi: 10.1016/0145-305x(90)90009-4. [DOI] [PubMed] [Google Scholar]
- Hanington PC, Wang T, Secombes CJ, Belosevic M. Growth factors of lower vertebrates: characterization of goldfish (Carassius auratus L.) macrophage colony-stimulating factor-1. J Biol Chem. 2007;282:31865–31872. doi: 10.1074/jbc.M706278200. [DOI] [PubMed] [Google Scholar]
- Honda T, Nishizawa T, Uenobe M, Kohchi C, Kuroda A, Ototake M, Nakanishi T, Yokomizo Y, Takahashi Y, Inagawa H, Soma G. Molecular cloning and expression analysis of a macrophage-colony stimulating factor receptor-like gene from rainbow trout, Oncorhynchus mykiss. Mol Immunol. 2005;42:1–8. doi: 10.1016/j.molimm.2004.07.002. [DOI] [PubMed] [Google Scholar]
- How GF, Venkatesh B, Brenner S. Conserved linkage between the puffer fish (Fugu rubripes) and human genes for platelet-derived growth factor receptor and macrophage colony-stimulating factor receptor. Genome Res. 1996;6:1185–1191. doi: 10.1101/gr.6.12.1185. [DOI] [PubMed] [Google Scholar]
- Karbassi A, Becker JM, Foster JS, Moore RN. Enhanced killing of Candida albicans by murine macrophages treated with macrophage colony-stimulating factor: evidence for augmented expression of mannose receptors. J Immunol. 1987;139:417–421. [PubMed] [Google Scholar]
- Lane MC, Sheets MD. Primitive and definitive blood share a common origin in Xenopus: a comparison of lineage techniques used to construct fate maps. Dev Biol. 2002;248:52–67. doi: 10.1006/dbio.2002.0717. [DOI] [PubMed] [Google Scholar]
- Lichanska AM, Browne CM, Henkel GW, Murphy KM, Ostrowski MC, Mckercher SR, Maki RA, Hume DA. Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU.1. Blood. 1999;94:127–138. [PubMed] [Google Scholar]
- Lin HS, Lokeshwar BL, Hsu S. Both granulocyte-macrophage CSF and macrophage CSF control the proliferation and survival of the same subset of alveolar macrophages. J Immunol. 1989;142:515–519. [PubMed] [Google Scholar]
- Morales HD, Abramowitz L, Gertz J, Sowa J, Vogel A, Robert J. Innate immune responses and permissiveness to ranavirus infection of peritoneal leukocytes in the frog Xenopus laevis. J Virol. 2010;84:4912–4922. doi: 10.1128/JVI.02486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Cheung NK. Antibody-independent phagocytosis of tumor cells by human monocyte-derived macrophages cultured in recombinant macrophage colony-stimulating factor. Cancer Immunol Immunother. 1995;41:46–52. doi: 10.1007/BF01788959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann NF, Barreda DR, Belosevic M. Generation and functional analysis of distinct macrophage sub-populations from goldfish (Carassius auratus L.) kidney leukocyte cultures. Fish Shellfish Immunol. 2000;10:1–20. doi: 10.1006/fsim.1999.0221. [DOI] [PubMed] [Google Scholar]
- Nishikawa A, Murata E, Akita M, Kaneko K, Moriya O, Tomita M, Hayashi H. Roles of macrophages in programmed cell death and remodeling of tail and body muscle of Xenopus laevis during metamorphosis. Histochem Cell Biol. 1998;109:11–17. doi: 10.1007/s004180050197. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Ingerslev HC, Stavang V, Egenberg M, Wergeland HI. A highly phagocytic cell line TO from Atlantic salmon is CD83 positive and M-CSFR negative, indicating a dendritic-like cell type. Fish Shellfish Immunol. 2008;25:809–819. doi: 10.1016/j.fsi.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Rieger AM, Hanington PC, Belosevic M, Barreda DR. Control of CSF-1 induced inflammation in teleost fish by a soluble form of the CSF-1 receptor. Fish Shellfish Immunol. 2014a;41:45–51. doi: 10.1016/j.fsi.2014.03.035. [DOI] [PubMed] [Google Scholar]
- Rieger AM, Konowalchuk JD, Havixbeck JJ, Robbins JS, Smith MK, Lund JM, Barreda DR. A soluble form of the CSF-1 receptor contributes to the inhibition of inflammation in a teleost fish. Dev Comp Immunol. 2014b;39:438–446. doi: 10.1016/j.dci.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Robert J, Abramowitz L, Gantress J, Morales HD. Xenopus laevis: a possible vector of Ranavirus infection? J Wildl Dis. 2007;43:645–652. doi: 10.7589/0090-3558-43.4.645. [DOI] [PubMed] [Google Scholar]
- Robert J, Morales H, Buck W, Cohen N, Marr S, Gantress J. Adaptive immunity and histopathology in frog virus 3-infected Xenopus. Virology. 2005;332:667–675. doi: 10.1016/j.virol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shibasaki Y, Yabu T, Araki K, Mano N, Shiba H, Moritomo T, Nakanishi T. Peculiar monomeric interferon gammas, IFNgammarel 1 and IFNgammarel 2, in ginbuna crucian carp. FEBS J. 2013;281:1046–56. doi: 10.1111/febs.12666. [DOI] [PubMed] [Google Scholar]
- Stanley ER, Berg KL, Einstein DB, Lee PS, Pixley FJ, Wang Y, Yeung YG. Biology and action of colony--stimulating factor-1. Mol Reprod Dev. 1997;46:4–10. doi: 10.1002/(SICI)1098-2795(199701)46:1<4::AID-MRD2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Sweet MJ, Hume DA. CSF-1 as a regulator of macrophage activation and immune responses. Arch Immunol Ther Exp (Warsz) 2003;51:169–177. [PubMed] [Google Scholar]
- Thors F, De Kort EJ, Nieuwenhuys R. On the development of the spinal cord of the clawed frog, Xenopus laevis. I. Morphogenesis and histogenesis. Anat Embryol (Berl) 1982a;164:427–441. doi: 10.1007/BF00315763. [DOI] [PubMed] [Google Scholar]
- Thors F, De Kort EJ, Nieuwenhuys R. On the development of the spinal cord of the clawed frog, Xenopus laevis. II. Experimental analysis of differentiation and migration. Anat Embryol (Berl) 1982b;164:443–454. doi: 10.1007/BF00315764. [DOI] [PubMed] [Google Scholar]
- Tushinski RJ, Oliver IT, Guilbert LJ, Tynan PW, Warner JR, Stanley ER. Survival of mononuclear phagocytes depends on a lineage-specific growth factor that the differentiated cells selectively destroy. Cell. 1982;28:71–81. doi: 10.1016/0092-8674(82)90376-2. [DOI] [PubMed] [Google Scholar]
- Wang T, Hanington PC, Belosevic M, Secombes CJ. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J Immunol. 2008;181:3310–3322. doi: 10.4049/jimmunol.181.5.3310. [DOI] [PubMed] [Google Scholar]
- Wang T, Kono T, Monte MM, Kuse H, Costa MM, Korenaga H, Maehr T, Husain M, Sakai M, Secombes CJ. Identification of IL-34 in teleost fish: differential expression of rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor. Mol Immunol. 2014;53:398–409. doi: 10.1016/j.molimm.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Williams H, Brenner S, Venkatesh B. Identification and analysis of additional copies of the platelet-derived growth factor receptor and colony stimulating factor 1 receptor genes in fugu. Gene. 2002;295:255–264. doi: 10.1016/s0378-1119(02)00736-9. [DOI] [PubMed] [Google Scholar]
- Williams T, Barbosa-Solomieu V, Chinchar VG. A decade of advances in iridovirus research. Adv Virus Res. 2005;65:173–248. doi: 10.1016/S0065-3527(05)65006-3. [DOI] [PubMed] [Google Scholar]
- Yabu T, Toda H, Shibasaki Y, Araki K, Yamashita M, Anzai H, Mano N, Masuhiro Y, Hanazawa S, Shiba H, Moritomo T, Nakanishi T. Antiviral protection mechanisms mediated by ginbuna crucian carp interferon gamma isoforms 1 and 2 through two distinct interferon gamma-receptors. J Biochem. 2011;150:635–648. doi: 10.1093/jb/mvr108. [DOI] [PubMed] [Google Scholar]