Abstract
The family Aristolochiaceae, comprising about 600 species of eight genera, is a unique plant family containing aristolochic acids (AAs). The complete chloroplast genome sequences of Aristolochia debilis and Aristolochia contorta are reported here. The results show that the complete chloroplast genomes of A. debilis and A. contorta comprise circular 159,793 and 160,576 bp-long molecules, respectively and have typical quadripartite structures. The GC contents of both species were 38.3% each. A total of 131 genes were identified in each genome including 85 protein-coding genes, 37 tRNA genes, eight rRNA genes and one pseudogene (ycf1). The simple-sequence repeat sequences mainly comprise A/T mononucletide repeats. Phylogenetic analyses using maximum parsimony (MP) revealed that A. debilis and A. contorta had a close phylogenetic relationship with species of the family Piperaceae, as well as Laurales and Magnoliales. The data obtained in this study will be beneficial for further investigations on A. debilis and A. contorta from the aspect of evolution, and chloroplast genetic engineering.
Keywords: Aristolochia debilis, Aristolochia contorta, chloroplast genome, molecular structure, phylogenetic analyses
1. Introduction
The traditional Chinese medicine plants, Aristolochia debilis and Aristolochia contorta, are herbaceous climbers in the family Aristolochiaceae. Aristolochiae fructus originates from the mellow fruit of the two species, while Aristolochiae herba originates from their dried aerial parts. Aristolochiae fructus and Aristolochiae herba have been recorded as traditional herbal medicines which can clear lung-heat to stop coughing and activate meridians to stop pain, respectively [1]. Modern pharmacology studies have shown that the primary chemical constituents of the two species are aristolochic acid analogues including aristolochic acids (AAs) and aristolactams (ALs) [2,3]. AAs and ALs have been found among species from the family Aristolochiaceae [4]. Previous researches have revealed that AAs are able to react with DNA to form covalent dA-aristolactam (dA-AL) and dG-aristolactam (dG-AL) adducts [5,6]. With further research, current evidence from studies of AAs has demonstrated that AAs can cause nephrotoxicity, carcinogenicity, and mutagenicity [7,8,9,10], especially after prolonged low-dose or shortdated high-dose intake [11,12]. Some nephropathy and malignant tumours including renal interstitial fibrosis, Balkan endemic nephropathy, and upper tract urothelial carcinomas are caused by AAs [13,14,15]. Currently, there are different degrees of restrictions on the sale and use of AAs-containing herbal preparations in many countries.
Chloroplasts are key and semi-autonomous organelles for photosynthesis and biosynthesis in plant cells [16,17,18]. The chloroplast genome, one of three major genetic systems (the other two are nuclear and mitochondrial genomes), is a circular molecule with a typical quadripartite structure of 115 to 165 kb in length [19,20]. All chloroplast genomes of land plants, apart from several rare exceptions, are highly conserved in terms of size, structure, gene content, and gene [21,22,23]. Due to its self-replication mechanism and relatively independent evolution, the genetic information from the chloroplast genome has been used in studies of molecular markers, barcoding identification, plant evolution and phylogenetic [24,25,26]. In 1976, Bedbrook and Bogorad produced the first chloroplast physical mapping of Zea mays by digestion with multiple restriction enzymes [27]. Subsequently, the first complete chloroplast genome sequence of Nicotiana tabacum was determined [28]. With the development of sequencing technology and bioinformatics, research into the chloroplast genome has increased rapidly. By now, the number of chloroplast genome sequence recorded in the National Center for Biotechnology Information (NCBI) has reached more than 1,500 plant species [29].
About eight genera and 600 species are classified within Aristolochiaceae, and are primarily distributed in tropical and subtropical regions. Of these plants, there are four genera (one endemic) and 86 species (69 endemic) distributed widely in China. The genus Aristolochia L., comprising about 400 species (45 species in China), is the largest and most representative genus of Aristolochiaceae [30]. However, there are no reports on the chloroplast genomes of the family Aristolochiaceae at present, and this has hindered our understanding and progress in the research of the evolution, phylogeny, species identification, and genetic engineering of Aristolochiaceae.
In this study, we determined the complete chloroplast genome sequences of A. debilis and A. contorta, which are the first two sequenced members of the family Aristolochiaceae. Furthermore, to reveal the phylogenetic positions of the two species, we conducted a phylogenetic tree using the maximum parsimony (MP) method based on common protein-coding genes from 37 species. Overall, the results provide basic genetic information on the chloroplast of A. debilis and A. contorta, and the role of the two species in plant systematics.
2. Results and Discussion
2.1. The Chloroplast Genome Structures of A. debilis and A. contorta
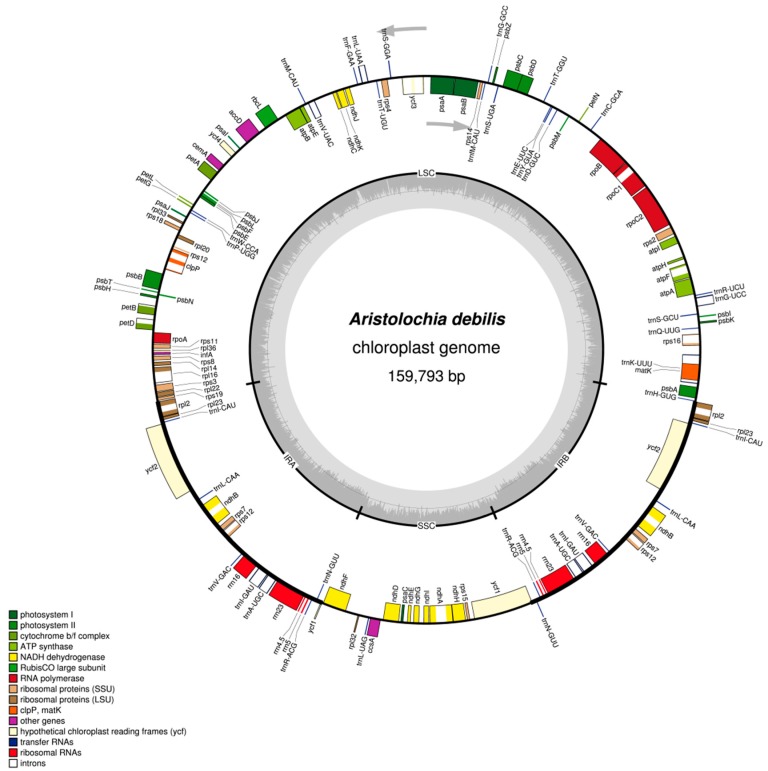
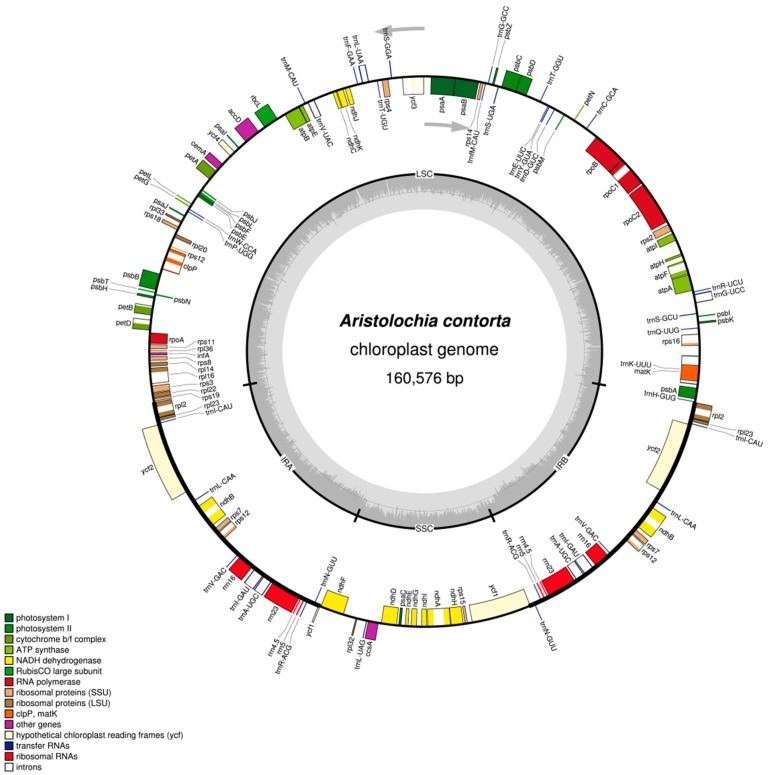
Both species displayed a typical quadripartite structure, and the corresponding regions were of similar lengths. The complete chloroplast genome of A. debilis is a circular molecule of 159,793 bp in length comprising a large single-copy (LSC) region of 89,609 bp and a small single-copy (SSC) region of 19,834 bp separated by a pair of inverted repeats (IRs), each 25,175 bp in length (Figure 1, Table 1). The complete chloroplast genome of A. contorta is 160,576 bp in length, which is divided into one LSC (89,781 bp), one SSC (19,877 bp) and two IRs, each 25,459 bp in length (Figure 2, Table 1).
Figure 1.
Gene map of the complete chloroplast genome of A. debilis. Genes on the inside of the circle are transcribed clockwise, while those outside are transcribed counter clockwise. The darker gray in the inner circle corresponds to GC content, whereas the lighter gray corresponds to AT content.
Table 1.
Base composition in the chloroplast genomes of A. debilis and A. contorta.
| Species | Regions | Positions | T(U) (%) | C (%) | A (%) | G (%) | Length (bp) |
|---|---|---|---|---|---|---|---|
| A. debilis | LSC | - | 32.2 | 18.7 | 31.2 | 17.9 | 89,609 |
| SSC | - | 34.0 | 17.4 | 33.2 | 15.5 | 19,834 | |
| IRa | - | 28.4 | 22.4 | 28.3 | 21.0 | 25,175 | |
| IRb | - | 28.3 | 21.0 | 28.4 | 22.4 | 25,175 | |
| Total | - | 31.2 | 19.5 | 30.5 | 18.8 | 159,793 | |
| CDS | - | 30.9 | 18.1 | 30.2 | 20.8 | 78,717 | |
| - | 1st position | 23.5 | 18.8 | 30.5 | 27.2 | 26,239 | |
| - | 2nd position | 32.2 | 20.5 | 29.2 | 18.1 | 26,239 | |
| - | 3rd position | 36.9 | 15.1 | 31.1 | 17.0 | 26,239 | |
| A. contorta | LSC | - | 32.2 | 18.7 | 31.2 | 17.8 | 89,781 |
| SSC | - | 33.9 | 17.4 | 33.3 | 15.4 | 19,877 | |
| IRa | - | 28.4 | 22.4 | 28.2 | 21.0 | 25,459 | |
| IRb | - | 28.2 | 21.0 | 28.4 | 22.4 | 25,459 | |
| Total | - | 31.2 | 19.5 | 30.6 | 18.8 | 160,576 | |
| CDS | - | 30.9 | 18.1 | 30.3 | 20.7 | 78,765 | |
| - | 1st position | 23.5 | 18.8 | 30.5 | 27.2 | 26,255 | |
| - | 2nd position | 32.2 | 20.6 | 29.2 | 18.1 | 26,255 | |
| - | 3rd position | 37.0 | 15.0 | 31.1 | 16.9 | 26,255 |
* CDS: protein-coding regions.
Figure 2.
Gene map of the complete chloroplast genome of A. contorta. Genes on the inside of the circle are transcribed clockwise, while those outside are transcribed counter clockwise. The darker gray in the inner circle corresponds to GC content, whereas the lighter gray corresponds to AT content.
The analysis results revealed that both species had a GC content of 38.3%. However, this was unevenly distributed across the whole chloroplast genome. In both species, the GC content exhibited the highest values of the IR regions across the complete chloroplast genome, 43.4% both in A. debilis and A. contorta. The high GC content in the IR regions was the result of four rRNA genes (rrn16, rrn23, rrn4.5 and rrn5) that occur in this region [31]. In addition, the LSC regions have GC contents of 36.6% and 35.5%, as well as the lowest values of 32.9% and 32.8% are seen in SSC regions in A. debilis and A. contorta, respectively. Within the protein-coding regions (CDS) of chloroplast genome of A. debilis, the percentage of AT content for the first, second and third codon positions were 54%, 61.4% and 68%, respectively (Table 1). A bias towards a higher AT representation at the third codon position has also been observed in other land plant chloroplast genomes [32,33,34].
A total of 131 genes were identified from each genome including 85 protein-coding genes, 37 tRNAs, eight rRNAs, and one pseudogene (ycf1) (Table 2). The functional ycf1 copy existed encompassing IR-SSC boundary and the other pseudogene ycf1 copy was on the other IR region. Six protein-coding genes, seven tRNA genes, and all rRNA genes were duplicated in the IR regions. Coding regions including protein-coding genes (CDS), tRNAs, and rRNAs constituted 56.7% and 56.4% in the chloroplast genomes of A. debilis and A. contorta, respectively; while the non-coding regions including introns, pseudogenes, and intergenic spacers constituted 43.3% and 43.6% of the genome, respectively.
Table 2.
Gene contents in the chloroplast genomes of A. debilis and A. contorta.
| No. | Group of Genes | Gene names | Amount |
|---|---|---|---|
| 1 | Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| 2 | Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 |
| 3 | Cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | 6 |
| 4 | ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | 6 |
| 5 | NADH dehydrogenase | ndhA *, ndhB *(×2)1, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12(1) |
| 6 | RubisCO large subunit | rbcL | 1 |
| 7 | RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | 4 |
| 8 | Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12 **(×2), rps14, rps15, rps16 *, rps18, rps19 | 14(2) |
| 9 | Ribosomal proteins (LSU) | rpl2 *(×2), rpl14, rpl16 *, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 | 11(2) |
| 10 | Proteins of unknown function | ycf1, ycf2(×2), ycf3 **, ycf4 | 5(1) |
| 11 | Transfer RNAs | 37 tRNAs (6 contain an intron, 7 in the IRs) | 37(7) |
| 12 | Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn23(×2) | 8(4) |
| 13 | Other genes | accD, clpP **, matK, ccsA, cemA, infA | 6 |
* Gene contains one intron; ** gene contains two introns; (×2) indicates the number of the repeat unit is 2.
Introns play an important role in the regulation of gene expression and can enhance the expression of exogenous genes at specific sites and specific times of the plant [35]. The intron content of genes reserved in the chloroplast genomes of A. debilis and A. contorta are maintained in other angiosperms [31,36]. Data revealed the presence of 18 genes containing introns in each chloroplast genome, including atpF, rpoC1, ycf3, rps12, rpl2, rpl16, clpP, petB, petD, rps16, ndhA, ndhB, and six tRNA genes (Table 3). In addition, the ycf3 gene and rps12 gene each contain two introns and three exons. The ycf3 gene is located in LSC region and the rps12 gene is a special trans-splicing gene, the 5′ exon is located in LSC, while the 3′ exon is located in IR, which is similar to that in Aquilaria sinensis [25], Panax ginseng [36] and Cistanche deserticola [37].
Table 3.
Genes with introns in the chloroplast genomes of A. debilis and A. contorta as well as the lengths of the exons and introns.
| Species | Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|---|
| A. debilis | atpF | LSC | 145 | 805 | 410 | - | - |
| clpP | LSC | 71 | 781 | 292 | 678 | 255 | |
| ndhA | SSC | 552 | 1090 | 540 | - | - | |
| ndhB | IR | 777 | 705 | 756 | - | - | |
| petB | LSC | 6 | 214 | 642 | - | - | |
| petD | LSC | 6 | 485 | 476 | - | - | |
| rpl16 | LSC | 8 | 1065 | 403 | - | - | |
| rpl2 | IR | 391 | 657 | 431 | - | - | |
| rpoC1 | LSC | 430 | 776 | 1622 | - | - | |
| rps12 | LSC | 114 | - | 232 | 536 | 26 | |
| rps16 | LSC | 46 | 853 | 191 | - | - | |
| trnA-UGC | IR | 38 | 809 | 35 | - | - | |
| trnG-UCC | LSC | 24 | 761 | 48 | - | - | |
| trnI-GAU | IR | 37 | 937 | 35 | - | - | |
| trnK-UUU | LSC | 37 | 2658 | 35 | - | - | |
| trnL-UAA | LSC | 35 | 521 | 50 | - | - | |
| trnV-UAC | LSC | 39 | 597 | 37 | - | - | |
| ycf3 | LSC | 126 | 777 | 228 | 753 | 147 | |
| A. contorta | atpF | LSC | 145 | 771 | 410 | - | - |
| clpP | LSC | 71 | 821 | 292 | 664 | 255 | |
| ndhA | SSC | 552 | 1091 | 540 | - | - | |
| ndhB | IR | 777 | 716 | 756 | - | - | |
| petB | LSC | 6 | 214 | 642 | - | - | |
| petD | LSC | 7 | 485 | 476 | - | - | |
| rpl16 | LSC | 8 | 1088 | 403 | - | - | |
| rpl2 | IR | 391 | 657 | 431 | - | - | |
| rpoC1 | LSC | 430 | 776 | 1619 | - | - | |
| rps12 | LSC | 114 | - | 232 | 536 | 26 | |
| rps16 | LSC | 46 | 832 | 221 | - | - | |
| trnA-UGC | IR | 38 | 809 | 35 | - | - | |
| trnG-UCC | LSC | 24 | 751 | 48 | - | - | |
| trnI-GAU | IR | 37 | 938 | 35 | - | - | |
| trnK-UUU | LSC | 37 | 2648 | 35 | - | - | |
| trnL-UAA | LSC | 35 | 552 | 50 | - | - | |
| trnV-UAC | LSC | 39 | 605 | 37 | - | - | |
| ycf3 | LSC | 126 | 764 | 228 | 760 | 147 |
2.2. IR Contraction and Expansion
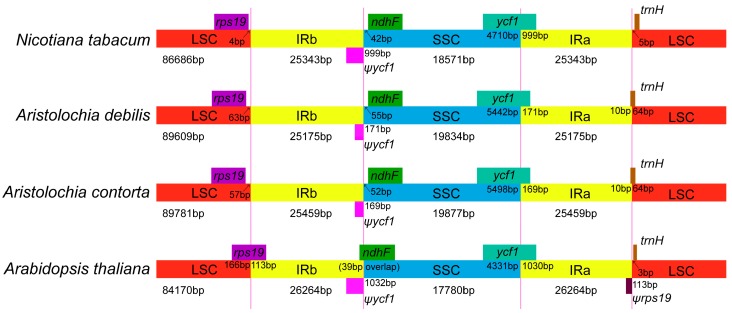
Although genomic structure and size were highly conserved in Angiosperms chloroplast genomes, the IR/SC boundary regions still varied slightly (Figure 3). The contraction and expansion at the borders of the IR regions are common evolutionary events and represent the main reasons for size variation of the chloroplast genomes [33,38]. From Figure 3, the junctions of the IR and LSC regions of four species including Arabidopsis thaliana (accession number: NC_000932), Nicotiana tabacum (NC_001879), as well as two Aristolochia species were compared. The IRb/SSC border extended into the ycf1 genes to cause long ycf1 pseudogenes in all species; however, compared with A. thaliana and N. tabacum, the length of ycf1 pseudogene of two Aristolochia species were only 171 and 169 bp, respectively. The IRa/SSC border was located in the CDS of the ycf1 gene and expanded the same length into the 5′ portion of ycf1 gene as IRb expanded in the four chloroplast genomes. The trnH genes were located in the LSC regions in Nicotiana tabacum, Sesamum indicum, Arabidopsis thaliana, and Salvia miltiorrhiza [31], while this gene was usually located in the IR region in the monocot chloroplast genomes [39]. Interestingly, the IRa/LSC borders were located in the coding region of trnH genes in the two Aristolochia species.
Figure 3.
Comparison of the borders of LSC, SSC and IR regions among four chloroplast genomes. Number above the gene features means the distance between the ends of genes and the borders sites. The IRb/SSC border extended intothe ycf1 genes to create various lengths of ycf1 pseudogenes among four chloroplast genomes. These features are not to scale.
2.3. Codon Usage and RNA Editing Sites
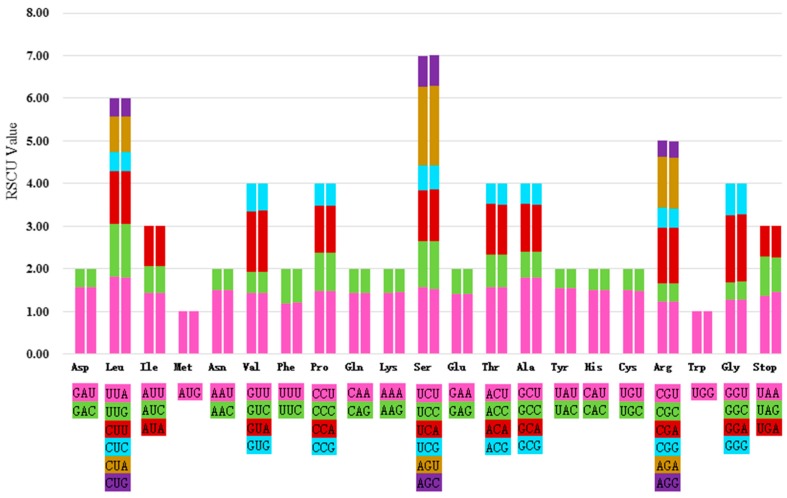
All the protein-coding genes were composed of 26,239 and 26,255 codons in the chloroplast genomes of A. debilis and A. contorta, respectively. Among these codons, 2737 encode leucine and 315 encode cysteine, respectively, the most and least universal amino acids in the A. debilis chloroplast genome. The codon usages of protein-coding genes in the A. debilis and A. contorta chloroplast genomes are deduced and summarized in Figure 4 and Table S1. Figure 4 shows that the relative synonymous codon usage (RSCU) value increased with the quantity of codons that code for a specific amino acid. Most of the amino acid codons have preferences except for methionine and tryptophan. The results presented here are similar in codon usage with the chloroplast genomes of species within the genus Ulmus [40] and Aq. sinensis [25]. In addition, potential RNA editing sites were predicted for 35 genes of the chloroplast genomes of two species. A total of 92 RNA editing sites were identified (Table S2). The amino acid conversion S to L occurred most frequently, while P to S and R to W occurred least. Seventy-six common RNA editing sites were shared in genes of the two species.
Figure 4.
Codon content of 20 amino acid and stop codons in all protein-coding genes of the chloroplast genomes of two Aristolochia species. The histogram on the left-hand side of each amino acid shows codon usage within the A. debilis chloroplast genome, while the right-hand side illustrates the genome of A. contorta.
2.4. Repeat Structure and Simple Sequence Repeats Analyses
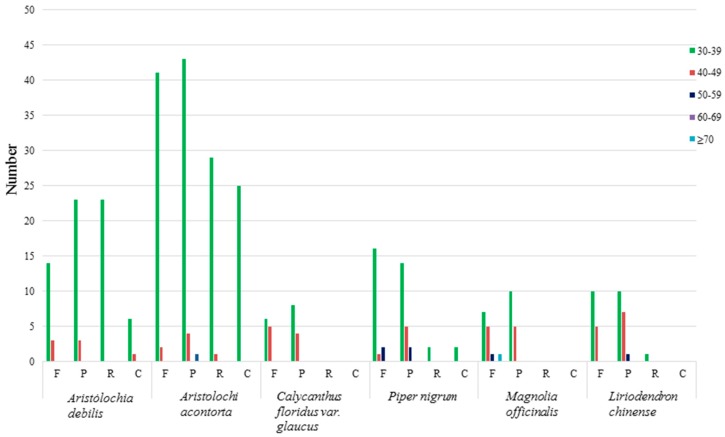
The repeats were mostly distributed in the intergenic spacer (IGS) and intron sequences. Figure 5 shows the repeat structure analyses of six species. The results revealed that the repeats of chloroplast genome of A. contorta had the greatest number, comprising of 41 forward, 43 palindromic, 29 reverse, and 25 complement repeats. Followed by A. debilis, contained 14 forward, 23 palindromic, 23 reverse, and six complement repeats. Simple sequence repeats (SSRs), which are ubiquitous throughout the genomes and are also known as microsatellites, are tandemly repeated DNA sequences that consist of 1–6 nucleotide repeat units [41]. SSRs are widely used for molecular markers in species identification, population genetics, and phylogenetic investigations based on their high level of polymorphism [42,43,44]. A total of 129 and 156 SSRs were identified using the microsatellite identification tool (MISA) in the chloroplast genomes of A. debilis and A. contorta, respectively (Table 4; Tables S3 and S4). In these SSRs, mononucletide repeats were largest in number, which were found 81 and 96 times in A. debilis and A. contorta, respectively. A/T mononucleotide repeats (96.3% and 94.8%, respectively) were the most common, while the majority of dinucleotide repeat sequences comprised of AT/TA repeats (100% and 92.8%, respectively). This result agreed with the previous studies where proportions of polyadenine (polyA) and polythymine (polyT) were higher than polycytosine (polyC) and polyguanine (polyG) within chloroplast SSRs in many plants [24].
Figure 5.
Repeat sequences in six chloroplast genomes. REPuter was used to identify repeat sequences with length ≥30 bp and sequence identified ≥90% in the chloroplast genomes. F, P, R, and C indicate the repeat types F (forward), P (palindrome), R (reverse), and C (complement), respectively. Repeats with different lengths are indicated in different colours.
Table 4.
Types and amounts of SSRs in the A. debilis and A. contorta chloroplast genomes.
| SSR Type | Repeat Unit | Amount | Ratio (%) | ||
|---|---|---|---|---|---|
| A. debilis | A. contorta | A. debilis | A. contorta | ||
| Mono | A/T | 78 | 91 | 96.3 | 94.8 |
| C/G | 3 | 5 | 3.7 | 5.2 | |
| Di | AC/GT | 0 | 1 | 0 | 3.6 |
| AG/CT | 0 | 1 | 0 | 3.6 | |
| AT/TA | 19 | 26 | 100 | 92.8 | |
| Tri | AAC/GTT | 1 | 1 | 10 | 8.3 |
| AAG/CTT | 1 | 1 | 10 | 8.3 | |
| ATC/ATG | 1 | 0 | 10 | 0 | |
| AAT/ATT | 7 | 10 | 70 | 83.4 | |
| Tetra | AAAC/GTTT | 2 | 2 | 16.7 | 14.3 |
| AAAT/ATTT | 4 | 5 | 33.3 | 35.7 | |
| AATC/ATTG | 1 | 1 | 8.3 | 7.1 | |
| AGAT/ATCT | 2 | 1 | 16.7 | 7.1 | |
| AATT/AATT | 0 | 1 | 0 | 7.1 | |
| ACAT/ATGT | 0 | 1 | 0 | 7.1 | |
| AACT/AGTT | 1 | 1 | 8.3 | 7.1 | |
| AATG/ATTC | 2 | 2 | 16.7 | 14.3 | |
| Penta | AATAT/ATATT | 2 | 2 | 33.3 | 50 |
| AAATT/AATTT | 1 | 0 | 16.7 | 0 | |
| AAATC/ATTTG | 1 | 0 | 16.7 | 0 | |
| AACAT/ATGTT | 0 | 1 | 0 | 25 | |
| AAAAT/ATTTT | 2 | 1 | 33.3 | 25 | |
| Hexa | AAATAG/ATTTCT | 0 | 1 | 0 | 50 |
| ACATAT/ATATGT | 0 | 1 | 0 | 50 | |
| ACTGAT/AGTATC | 1 | 0 | 100 | 0 | |
2.5. Comparative Genomic Analysis
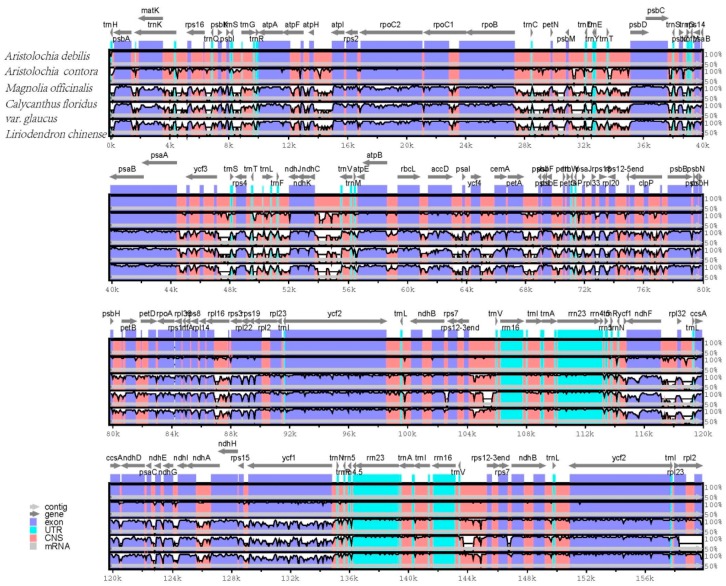
The whole chloroplast genome sequences of A. debilis and A. contorta were compared to those of Calycanthus floridus var. glaucus (accession number: NC_004993), Magnolia officinalis (NC_020316), and Liriodendron chinense (NC_030504) using the mVISTA program (Figure 6). The comparison showed that the two IR regions were less divergent than the LSC and SSC regions. The four rRNA genes were the most conserved, while the most divergent coding regions were ndhF, rpl22, ycf1, rpoC2 and ccsA. Additionally, the results revealed that non-coding regions exhibited a higher divergence than coding regions, and the most divergent regions localized in the intergenic spacers among the five chloroplast genomes.
Figure 6.
Sequence identity plot comparing the five chloroplast genomes with A. debilis as a reference by using mVISTA. Grey arrows and thick black lines above the alignment indicate genes with their orientation and the position of the IRs, respectively. A cut-off of 70% identity was used for the plots, and the Y-scale represents the percent identity ranging from 50% to 100%.
2.6. Phylogenetic Analyses
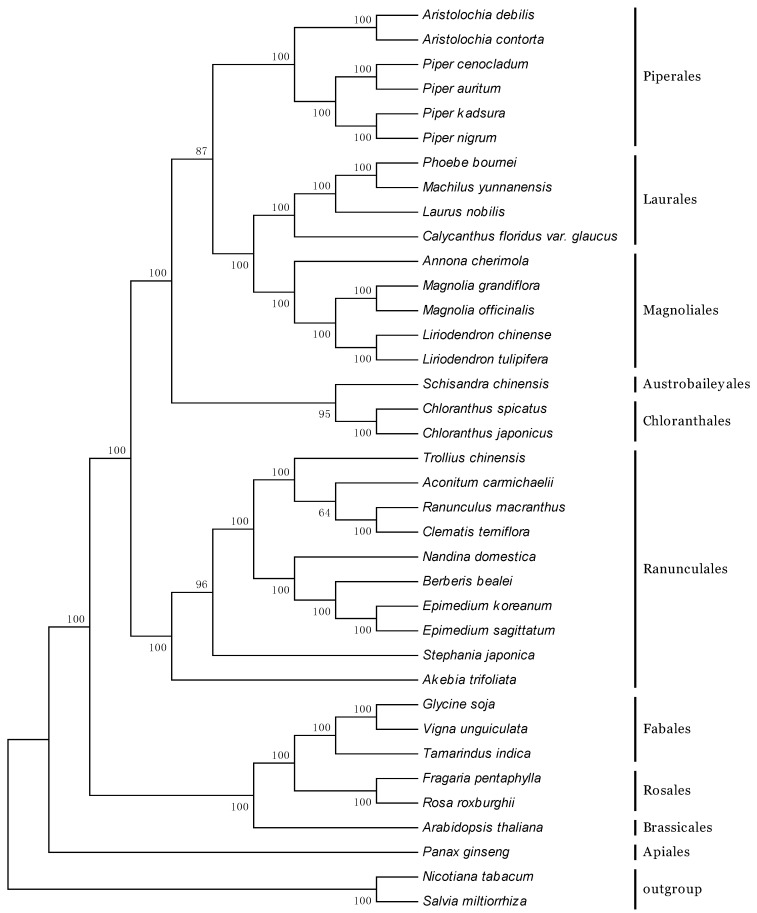
Chloroplast genomes provide abundant resources, which are significant for evolutionary, taxonomic, and phylogenetic studies [31,45,46]. The whole chloroplast genomes and protein-coding genes have been successfully used to resolve phylogenetic relationships at almost any taxonomic level during the past decade [31,37]. Aristolochia, consisting of nearly 400 species, is the largest genus in the family Aristolochiaceae [30]. Phylogenetic analyses employing one or several genes have been performed in previous studies [47,48,49]; however, these analyses were restricted to the species of Aristolochiaceae and included few species from other families. In this study, to identify the phylogenetic positions of A. debilis and A. contorta within Angiosperms, 60 protein-coding genes commonly present in 37 species from Piperales, Laurales, Magnoliales, Ranunculales, Fabales, Rosales, Chloranthales, as well as two Aristolochia species were used to construct the phylogenetic tree using the Maximum parsimony (MP) method (Figure 7). All the nodes in the MP trees have high bootstrap support values, and 30 out of 34 nodes with 100% bootstrap values were found. The result illustrated that two Aristolochia species were sister taxa with respect to four Piper species (Piperaceae), and these species were grouped with four species from Laurales and five species from Magnoliales. Additionally, all species are clustered within a lineage distinct from the outgroup. This result (inferred from the chloroplast genome data) obtained high support values, which suggested that the chloroplast genome could effectively resolve the phylogenetic positions and relationships of this family. Nevertheless, to accurately illustrate the evolution of the family Aristolochiaceae, it is necessary to use more species to analyze the phylogeny. This study will also provide a reference for species identification among Aristolochia and other genus using the chloroplast genome.
Figure 7.
Phylogenetic tree constructed using Maximum parsimony (MP) method based on 60 protein-coding genes from different species. Numbers at nodes are values for bootstrap support.
3. Materials and Methods
3.1. Plant Material, DNA Extraction, and Sequencing
Fresh plants of A. debilis and A. contorta were collected from Lichuan City in Hubei Province and Tonghua City in Jilin Province, respectively. All samples were identified by Professor Yulin Lin, who is based at the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences (CAMS) and Peking Union Medical College (PUMC). The voucher specimens were deposited in the herbarium of the IMPLAD. The leaves were cleansed and preserved in a −80 °C refrigerator. Total genomic DNA was extracted from approximately 100 mg of samples using DNeasy Plant Mini Kit with a standard protocol (Qiagen Co., Hilden, Germany). Final DNA quality was assessed based on spectrophotometry and their integrity was examined by electrophoresis in 1% (w/v) agarose gel. The DNA was used to construct shotgun libraries with insert sizes of 500 bp and sequenced according to the manufacturer’s manual for the Illumina Hiseq X. Approximately 6.3 Gb of raw data from A. debilis and 5.8 Gb from A. contorta were produced with 150 bp pair-end read lengths.
3.2. Chloroplast Genome Assembly and Annotation
First, we used the software Trimmomatic (v0.36, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany) [50] to trim the low-quality reads. After quality control, the clean reads were used to assemble the chloroplast genome. All chloroplast genomes of plants recorded in the National Center for Biotechnology Information (NCBI) were used to construct a reference database. Next, the clean reads were mapped to the database on the basis of their coverage and similarity, and the mapped reads extracted. Extracted reads were assembled to contigs using SOAPdenovo (v2, BGI HK Research Institute, Hong Kong, China) [51], and the resulting contigs were combined and extended to obtain a complete chloroplast genome sequence. To verify the accuracy of assembly, four boundaries of single copy (SC) and inverted repeat (IR) regions of the assembled sequences were confirmed by PCR amplification and Sanger sequencing using the primers listed in Table S5.
We used the online program Dual Organellar GenoMe Annotator (DOGMA), (University of Texas at Austin, Austin, TX, USA) [52] and the software Chloroplast Genome Annotation, Visualization, Analysis, and GenBank Submission (CPGAVAS), (Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) [53] coupled with manual corrections to perform the preliminarily gene annotation of chloroplast genomes of two species. The tRNA genes were identified using the software tRNAscan-SE (v2.0, University of California, Santa Cruz, CA, USA) [54] and DOGMA [52]. The gene map was drawn using the Organellar Genome DRAW (OGDRAW) (v1.2, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany) [55] with default settings and checked manually. The complete and correct chloroplast genome sequences of the two species were deposited in GenBank, accession numbers of A. debilis and A. contorta are MF539928 and MF539927, respectively.
3.3. Genome Structure Analyses and Genome Comparison
The distribution of codon usage was investigated using the software CodonW ( University of Texas, Houston, TX, USA) with the RSCU ratio [56]. Thirty-five protein-coding genes of the chloroplast genomes of two species were used to predict potential RNA editing sites using the online program Predictive RNA Editor for Plants (PREP) suite [57] with a cutoff value of 0.8. GC content was analyzed using Molecular Evolutionary Genetics Analysis (MEGA v6.0, Tokyo Metropolitan University, Tokyo, Japan) [58]. REPuter ( University of Bielefeld, Bielefeld, Germany) [59] to identify the size and location of repeat sequences, including forward, palindromic, reverse, and complement repeats in the chloroplast genomes of six species C. floridus var. glaucus, M. officinalis, L. chinense and Piper nigrum (NC_034692). For all repeat types, the minimal size was 30 bp and the two repeat copies had at least 90% similarity. Simple sequence repeats (SSRs) were detected using MISA software [60] with parameters set the same as Li et al. [61]. The whole-genome alignment for the chloroplast genomes of the five species including A. debilis, A. contorta, C. floridus var. glaucus, M. officinalis, and L. chinense were performed and plotted using the mVISTA program [62].
3.4. Phylogenetic Analyses
A total of 35 complete chloroplast genomes were downloaded from the NCBI Organelle Genome Resources database (Table S6). The 60 protein-coding gene sequences commonly present in 37 species, including the two species in this study, were aligned using the Clustal algorithm [63]. To determine the phylogenetic positions of A. debilis, and A. contorta, we analyzed the chloroplast genomes of these 60 protein-coding genes. Maximum parsimony (MP) analysis was performed with PAUP*4.0b10 [64], using a heuristic search performed with the MULPARS option, the random stepwise addition with 1000 replications and tree bisection-reconnection (TBR) branch swapping. Bootstrap analysis was also performed with 1,000 replicates with TBR branch swapping.
4. Conclusions
The complete chloroplast genomes of A. debilis and A. contorta, the first two sequenced members of the family Aristolochiaceae, were determined in this study. The genome structure and gene content were relatively conserved. The phylogenetic analyses illustrated that these two Aristolochia species were positioned close to four species from the family Piperaceae and had a close phylogenetic relationship with Laurales and Magnoliales. The results provided the basis for the study of the evolutionary history of A. debilis and A. contorta. All the data presented in this paper will facilitate the further investigation of these two medicinal plants.
Acknowledgments
This work was supported by Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (No. 2016-I2M-3-016), Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2014ZX09304307001) and The Key Projects in the National Science and Technology Pillar Program (No. 2011BAI07B08).
Abbreviations
| LSC | Large single copy |
| SSC | Small single copy |
| IR | Inverted repeat |
| MP | Maximum parsimony |
| SSR | Simple sequence repeats |
| ATP | Adenosine triphosphate |
| NADH | Nicotinamide adenine dinucleotide |
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/9/1839/s1.
Author Contributions
Jianguo Zhou, Xinlian Chen, and Yingxian Cui, performed the experiments; Jianguo Zhou, Wei Sun, and Jingyuan Song, assembled sequences and analyzed the data; Jianguo Zhou wrote the manuscript; Yonghua Li, and Yu Wang, collected plant material; Hui Yao conceived the research and revised the manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chinese Pharmacopoeia Commission . The Chinese Pharmacopoeia. Chemical Industry Press; Beijing, China: 2015. pp. 51–52. [Google Scholar]
- 2.Chen C.X. Studies on the chemical constituents from the fruit of Aristolochia debilis. J. Chin. Med. Mater. 2010;33:1260–1261. [PubMed] [Google Scholar]
- 3.Xu Y., Shang M., Ge Y., Wang X., Cai S. Chemical constituent from fruit of Aristolochia contorta. Chin. J. Chin. Mater. Medica. 2010;35:2862. [PubMed] [Google Scholar]
- 4.Mix D.B., Guinaudeau H., Shamma M. The aristolochic acids and aristolactams. J. Nat. Prod. 1982;45:657–666. doi: 10.1021/np50024a001. [DOI] [Google Scholar]
- 5.Arlt V.M., Stiborova M., Schmeiser H.H. Aristolochic acid as a probable human cancer hazard in herbal remedies: A review. Mutagenesis. 2002;17:265. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- 6.Schmeiser H.H., Janssen J.W., Lyons J., Scherf H.R., Pfau W., Buchmann A., Bartram C.R., Wiessler M. Aristolochic acid activates RAS genes in rat tumors at deoxyadenosine residues. Cancer Res. 1990;50:5464–5469. [PubMed] [Google Scholar]
- 7.Chen L., Mei N., Yao L., Chen T. Mutations induced by carcinogenic doses of aristolochic acid in kidney of big blue transgenic rats. Toxicol. Lett. 2006;165:250–256. doi: 10.1016/j.toxlet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Cheng C.L., Chen K.J., Shih P.H., Lu L.Y., Hung C.F., Lin W.C., Yesong G.J. Chronic renal failure rats are highly sensitive to aristolochic acids, which are nephrotoxic and carcinogenic agents. Cancer Lett. 2006;232:236–242. doi: 10.1016/j.canlet.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Cosyns J.P., Goebbels R.M., Liberton V., Schmeiser H.H., Bieler C.A., Bernard A.M. Chinese herbs nephropathy-associated slimming regimen induces tumours in the forestomach but no interstitial nephropathy in rats. Arch. Toxicol. 1998;72:738–743. doi: 10.1007/s002040050568. [DOI] [PubMed] [Google Scholar]
- 10.Hoang M.L., Chen C.H., Sidorenko V.S., He J., Dickman K.G., Yun B.H., Moriya M., Niknafs N., Douville C., Karchin R. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balachandran P., Wei F., Lin R.C., Khan I.A., Pasco D.S. Structure activity relationships of aristolochic acid analogues: Toxicity in cultured renal epithelial cells. Kidney Int. 2005;67:1797. doi: 10.1111/j.1523-1755.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsai D.M., Kang J.J., Lee S.S., Wang S.Y., Tsai I., Chen G.Y., Liao H.W., Li W.C., Kuo C.H., Tseng Y.J. Metabolomic analysis of complex Chinese remedies: Examples of induced nephrotoxicity in the mouse from a series of remedies containing aristolochic acid. Evid. Based Compl. Alt. 2013;2013:263757. doi: 10.1155/2013/263757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grollman A.P., Shibutani S., Moriya M., Miller F., Wu L., Moll U., Suzuki N., Fernandes A., Rosenquist T., Medverec Z., et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl. Acad. Sci. USA. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lord G.M., Tagore R., Cook T., Gower P., Pusey C.D. Nephropathy caused by Chinese herbs in the UK. Lancet. 1999;354:481–482. doi: 10.1016/S0140-6736(99)03380-2. [DOI] [PubMed] [Google Scholar]
- 15.Vanherweghem J.L., Tielemans C., Abramowicz D., Depierreux M., Vanhaelen-Fastre R., Vanhaelen M., Dratwa M., Richard C., Vandervelde D., Verbeelen D., et al. Rapidly progressive interstitial renal fibrosis in young women: Association with slimming regimen including Chinese herbs. Lancet. 1993;341:387–391. doi: 10.1016/0140-6736(93)92984-2. [DOI] [PubMed] [Google Scholar]
- 16.Dong W., Xu C., Cheng T., Lin K., Zhou S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013;5:989–997. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leliaert F., Smith D.R., Moreau H., Herron M.D., Verbruggen H., Delwiche C.F., Clerck O.D. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012;31:1–46. doi: 10.1080/07352689.2011.615705. [DOI] [Google Scholar]
- 18.Raman G., Park S. Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. longicalyncinus, from a comparative genomics perspective. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0141329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen R.K., Raubeson L.A., Boore J.L., Depamphilis C.W., Chumley T.W., Haberle R.C., Wyman S.K., Alverson A.J., Peery R., Herman S.J. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- 20.Wolfe K.H., Li W.H., Sharp P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNA. Proc. Natl. Acad. Sci. USA. 1988;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith D.R., Keeling P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA. 2015;112:10177–10184. doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tonti-Filippini J., Nevill P.G., Dixon K., Small I. What can we do with 1,000 plastid genomes? Plant J. 2017;90:808–818. doi: 10.1111/tpj.13491. [DOI] [PubMed] [Google Scholar]
- 23.Wicke S., Schneeweiss G.M., Müller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant. Mol. Boil. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuang D.Y., Wu H., Wang Y.L., Gao L.M., Zhang S.Z., Lu L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome. 2011;54:663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Zhan D.F., Jia X., Mei W.L., Dai H.F., Chen X.T., Peng S.Q. Complete chloroplast genome sequence of Aquilaria sinensis (lour.) gilg and evolution analysis within the Malvales order. Front. Plant Sci. 2016;7:280. doi: 10.3389/fpls.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu F.H., Chan M.T., Liao D.C., Chentran H., Yiwei L., Daniell H., Duvall M.R., Lin C.S. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedbrook J.R., Bogorad L. Endonuclease recognition sites mapped on Zea mays chloroplast DNA. Proc. Natl. Acad. Sci. USA. 1976;73:4309–4313. doi: 10.1073/pnas.73.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayshida N., Matsubayasha T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome. EMBO J. 1986;4:111–148. doi: 10.1007/BF02669253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.NCBI, Genome. [(accessed on 30 June 2017)]; Available online: https://www.ncbi.nlm.nih.gov/genome/browse/?report=5.
- 30.The Editorial Committee of Flora of China . Flora of China. Volume 5. Science Press; Beijing, China: Missouri Botanical Garden Press; St. Louis, MO, USA: 2003. pp. 246–269. [Google Scholar]
- 31.Qian J., Song J., Gao H., Zhu Y., Xu J., Pang X., Yao H., Sun C., Li X., Li C. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clegg M.T., Gaut B.S., Learn G.H., Jr., Morton B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA. 1994;91:6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M., Zhang X., Liu G., Yin Y., Chen K., Yun Q., Zhao D., Al-Mssallem I.S., Yu J. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.) PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi D.K., Kim K.J. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0035872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J., Feng D., Song G., Wei X., Chen L., Wu X., Li X., Zhu Z. The first intron of rice EPSP synthase enhances expression of foreign gene. Sci. China Life Sci. 2003;46:561–569. doi: 10.1360/02yc0120. [DOI] [PubMed] [Google Scholar]
- 36.Kim K.J., Lee H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 37.Li X., Zhang T.C., Qiao Q., Ren Z., Zhao J., Yonezawa T., Hasegawa M., Crabbe M.J., Li J., Zhong Y. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae) PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0058747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raubeson L.A., Peery R., Chumley T.W., Dziubek C., Fourcade H.M., Boore J.L., Jansen R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huotari T., Korpelainen H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 2012;508:96–105. doi: 10.1016/j.gene.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 40.Zuo L.H., Shang A.Q., Zhang S., Yu X.Y., Ren Y.C., Yang M.S., Wang J.M. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0171264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell W., Morgante M., McDevitt R., Vendramin G.G., Rafalski J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao Y., Jia H., Li X., Chai M., Jia H., Chen Z., Wang G., Chai C., Weg E.V.D., Gao Z. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra) BMC Genomics. 2012;13:201. doi: 10.1186/1471-2164-13-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue J., Wang S., Zhou S.L. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae) Am. J. Bot. 2012;99:240–244. doi: 10.3732/ajb.1100547. [DOI] [PubMed] [Google Scholar]
- 44.Yang A.H., Zhang J.J., Yao X.H., Huang H.W. Chloroplast microsatellite markers in Liriodendron tulipifera (Magnoliaceae) and cross-species amplification in L. chinense. Am. J. Bot. 2011;98:123–126. doi: 10.3732/ajb.1000532. [DOI] [PubMed] [Google Scholar]
- 45.Jansen R.K., Cai Z., Raubeson L.A., Daniell H., Depamphilis C.W., Leebens-Mack J., Müller K.F., Guisinger-Bellian M., Haberle R.C., Hansen A.K. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore M.J., Bell C.D., Soltis P.S., Soltis D.E. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murata J., Ohi T., Wu S., Darnaedi D., Sugawara T., Nakanishi T., Murata H. Molecular phylogeny of Aristolochia (Aristolochiaceae) inferred from matK sequences. Apg Acta Phyto. Geo. 2001;52:75–83. [Google Scholar]
- 48.Ohi-Toma T., Sugawara T., Murata H., Wanke S., Neinhuis C., Jin M. Molecular phylogeny of Aristolochia sensu lato (Aristolochiaceae) based on sequences of rbcL, matK, and phyA genes, with special reference to differentiation of chromosome numbers. Syst. Bot. 2006;31:481–492. doi: 10.1600/036364406778388656. [DOI] [Google Scholar]
- 49.Silva-Brandão K.L., Solferini V.N., Trigo J.R. Chemical and phylogenetic relationships among Aristolochia L. (Aristolochiaceae) from southeastern Brazil. Biochem. Syst. Ecol. 2006;34:291–302. doi: 10.1016/j.bse.2005.10.011. [DOI] [Google Scholar]
- 50.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 53.Liu C., Shi L., Zhu Y., Chen H., Zhang J., Lin X., Guan X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012;13:715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohse M., Drechsel O., Bock R. Organellargenomedraw (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 56.Sharp P.M., Li W.H. The codon Adaptation Index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mower J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Comput. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. Reputer: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Misa-Microsatellite Identification Tool. [(accessed on 2 June 2017)]; Available online: http://pgrc.ipk-gatersleben.de/misa/
- 61.Li X.W., Gao H.H., Wang Y.T., Song J.Y., Henry R., Wu H.Z., Hu Z.G., Hui Y., Luo H.M., Luo K. Complete chloroplast genome sequence of Magnolia grandiflora and comparative analysis with related species. Sci. China Life Sci. 2013;56:189–198. doi: 10.1007/s11427-012-4430-8. [DOI] [PubMed] [Google Scholar]
- 62.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swofford D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates; Sunderland, MA, USA: 2002. Version 4.0b10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.