Abstract
Developmental genetic studies of Antirrhinum majus demonstrated that two transcription factors from the MYB gene family, RADIALIS (RAD) and DIVIRICATA (DIV), interact through antagonism to regulate floral dorsoventral asymmetry. Interestingly, similar antagonistic interaction found among proteins of FSM1 (RAD-like) and MYBI (DIV-like) in Solanum lycopersicum is involved in fruit development. Here, we report the reconstruction of the phylogeny of I-box-like and R-R-type clades, where RAD- and DIV-like genes belong, respectively. We also examined the homology of these antagonistic MYB proteins using these phylogenies. The results show that there are likely three paralogs of RAD-/I-box-like genes, RAD1, RAD2, and RAD3, which originated in the common ancestor of the core eudicots. In contrast, R-R-type sequences fall into two major clades, RR1 and RR2, the result of gene duplication in the common ancestor of both monocots and dicots. RR1 was divided into clades RR1A, RR1B, and RR1C, while RR2 was divided into clades RR2A/DIV1, RR2B/DIV2, and RR2C/DIV3. We demonstrate that among similar antagonistic interactions in An. Majus and So. lycopersicum, RAD-like genes originate from the RAD2 clade, while DIV-like genes originate from distantly related paralogs of the R-R-type lineage. The phylogenetic analyses of these two MYB clades lay the foundation for future comparative studies including testing the evolution of the antagonistic relationship of proteins.
Keywords: RADIALIS-like genes, DIVIRICATA-like genes, gene duplication, angiosperms, phylogeny, antagonism of proteins, MYB gene family
1. Introduction
The MYB gene family comprises three members, A-, B- and c-MYB [1,2], found in many vertebrates, that are involved in the regulation of cell proliferation, differentiation, and apoptosis [3]. Homologs of MYB genes have also been identified in insects, fungi, and slime molds [4]. The first plant MYB gene, C1, was isolated from Zea mays, and it encodes a c-MYB-like transcription factor involved in anthocyanin biosynthesis [5]. Plant MYB proteins were found to be involved in the regulation of many developmental processes including the biosynthesis of anthocyanin and flavonoids, trichome differentiation, the determination of cell shapes, and the regulation of cell proliferation and cell cycles [5,6,7,8,9].
In plants, the MYB genes have also been found in the regulation of the development of floral symmetry in the Lamiales [10]. In the zygomorphic flowers of Antirrhinum majus L., the two dorsal petals are significantly enlarged compared to the lateral and ventral petals, and the single dorsal stamen is aborted [11]. Two genes, CYCLOIDEA (CYC) and DICHOTOMA (DICH), belonging to the CYC/TB1 clade of the TCP transcription factor family, were found to promote the dorsal identity of zygomorphic flowers [11,12,13,14]. RADIALIS (RAD), a member of the MYB gene family, was found to be the downstream target of CYC and DICH [15,16,17]. Plants of the double cyc/dich or the single rad mutants produce flowers that have entirely or partially lost their dorsal identity [11,16]. The dorsal petals assume the ventral petal identity and the aborted dorsal stamen becomes functional [11]. DIVARICATA (DIV), a member of a different MYB lineage, promotes ventral floral identity [18]. A single div mutant causes the loss of the ventral petal identity [18,19]. In the cyc/dich/div triple mutant, where the function of both the dorsal and ventral identity genes was lost, all petals resume the lateral petal identity [18,19].
Recently, antagonism involving three MYB-like proteins was found to be a mechanism regulating floral symmetry in the flowers of Antirrhinum [10]. Despite the role of DIV in controlling ventral petal identity, its mRNA is transcribed across the floral meristem [18,19]. RAD was found to be the dorsal factor inactivating DIV, but not at the transcriptional level [10,19]. Interestingly, it was found that RAD and DIV do not directly interact with each other, but compete for their protein target, DIV-and-RAD-interacting-factors (DRIFs), also members of the MYB family [10]. In particular, DIV and DRIFs show overlapping expression patterns and can form heterodimer complexes that bind to the DNA of DIV, suggesting the regulation of its transcription. RAD inhibits the interaction between DIV and DRIFs in the dorsal regions of the flowers of Antirrhinum by either binding directly to a DRIF protein in the nucleus or/and by sequestering the DRIF proteins in the cytoplasm [10]. Therefore, RAD acts as the antagonist that blocks the binding of DIV, the agonist, with the DRIFs, which is required for regulating ventral symmetry in the flowers of Antirrhinum.
Similar antagonistic relationships involving three MYB homologs were reported in the fruit development of Solanum lycopersicum L. [20]. The fruit SANT/MYB binding protein1 (FSB1), a DRIF homolog, was found to form a protein complex with the transcription factor MYBI, a DIV homolog. The fruit SANT/MYB-like (FSM1) protein, a RAD homolog, competes for FSB1 with MYBI. The function of FSM1 is to reduce fruit size and preferentially restrict differential cell expansion [20]. Ectopic expression of FSM1 results in a reduction in organ size by negatively affecting cell expansion. In contrast, FSB1 positively regulates differential cell expansion through a physical interaction with MYBI [20]. This is analogous with the competition between RAD and the DIV-DRIF complex in the dorsal regions of the flowers of Antirrhinum. The function for the FSM1–FSB1–MYBI complex in tomato controls cell expansion, while RAD–DRIF–DIV similarly also controls cell expansion by regulating dorsoventral flower asymmetry in snapdragon [10,20].
Previous works indicated frequent gene duplications during the evolution of RAD- and DIV-like genes [21,22]. Three paralogs of the RAD lineage, RAD1, RAD2, and RAD3, as well as three paralogs of the DIV lineage, DIV1, DIV2, and DIV3, are recognized [21,22]. The gene duplications that gave rise to these paralogs were predicted to have occurred around the diversification of the Pentapetalae. Therefore, there may exist antagonistic relationships among the homologs of RAD–DRIF–DIV in diverse lineages of the core eudicots. DRIFs, one of the three factors involved in this antagonistic interaction, belong to an ancient MYB-like gene family with several homologs also found in the moss Physcomitrella patens [10]. Two paralogs of DRIFs resulting from gene duplication at least in the common ancestor of monocots and dicots are named Group 1 and 2 [10]. The DRIF1 and DRIF2 of An. majus belong to Group 1, while the only DRIF-like protein (SlFSB1) found in So. lycopersicum belongs to Group 2. Therefore, in the antagonized systems in An. majus and So. lycopersicum, the DRIF homologs involved belong to two paralogous clades.
Here, we report on the evolution of the I-box-like and R-R-type lineages where RAD- and DIV-like genes belong, respectively, and aim to (1) reconstruct the phylogeny of the two MYB lineages using the maximum likelihood (ML) and Bayesian algorithms, (2) clarify the phylogenetic relationships of the paralogs, and (3) identify the homology of RAD- and DIV-like genes that form the antagonistic relationships in An. majus and So. lycopersicum. We also focus on RAD-like gene evolution in Solanaceae, where lineage-specific gene duplications were identified. We demonstrate that, among similar antagonistic interactions in An. Majus and So. lycopersicum, RAD-like genes originate from the closely related ortholog, while DIV-like genes originate from distantly related paralogs. Furthermore, the phylogenies of the I-box-like and R-R-type lineages generated in this study will guide future works in understanding the functional divergence of these MYB lineages.
2. Results
2.1. RAD-Like Genes from Solanaceae
Sixteen sequences of RAD-like genes were discovered in this study (GenBank numbers MF398572-MF398587) (Table 1). We show that our cloning method can recover all of the RAD2 paralogs identified from the genome data of P. hybrida and So. lycopersicum (Table 1).
Table 1.
Species sampled for the RAD2 clade with collection locations, voucher information, sequence name, phylogenetic placement, and number of clones sequenced.
| Species | Family | Location | Voucher | Sequence Names | Clades | # of Clones Sequenced |
|---|---|---|---|---|---|---|
| Petunia sp. | Solanaceae | VCU Greenhouse | Zhang_Lab_23 (VCU) | Petunia sp RAD1 | RAD2A | 12 |
| Petunia sp RAD2 | RAD2A | 20 | ||||
| Petunia sp RAD3 | RAD2B | 8 | ||||
| Lycium ruthenicum Murray. | Solanaceae | Taxkorgan Tajik Autonomous County, Xinjiang, China | CPG13183 (PE) | Lycium ruthenicum Murr RAD | RAD2A | 20 |
| Atropa belladonna L. | Solanaceae | Hotel Elites, Nathia Gali, Northwest Frontier Province, Pakistan | CPG13594 (PE) | Atropa belladonna Linn RAD | RAD2B | 20 |
| Schizanthus pinnatus Ruiz & Pav. | Solanaceae | VCU Greenhouse | Zhang_Lab_20 (VCU) | Schizanthus pinnatus RAD1 | RAD2B | 21 |
| Schizanthus pinnatus RAD2 | RAD2A | 22 | ||||
| Schizanthus grahamii Gillies | Solanaceae | VCU Greenhouse | Zhang_Lab_19 (VCU) | Schizanthus grahamii RAD1 | RAD2A | 21 |
| Schizanthus grahamii RAD2 | RAD2B | 19 | ||||
| Nicotiana obtusifolia M.Martens & Galeotti. | Solanaceae | VCU Greenhouse | Zhang_Lab_11 (VCU) | Nicotiana obtusifolia RAD1 | RAD2A | 14 |
| Nicotiana obtusifolia RAD2 | RAD2A | 16 | ||||
| Solanum lycopersicum L. | Solanaceae | VCU Greenhouse | Zhang_Lab_21 (VCU) | Solanum lycopersicum microtom RAD1 | RAD2A | 17 |
| Solanum lycopersicum microtom RAD2 | RAD2B | 13 | ||||
| Evolvulus sp. | Convolvulaceae | VCU Greenhouse | Zhang_Lab_18 (VCU) | Evulupus sp RAD1 | RAD2B | 20 |
| Evulupus sp RAD2 | RAD2A | 17 | ||||
| Ipomoea tricolor Cav. | Convolvulaceae | VCU Greenhouse | Zhang_Lab_22 (VCU) | Ipomoea tricolor RAD1 | RAD2A | 20 |
Virginia Commonwealth University (VCU) is in Richmond, VA, USA. VCU, Virginia Commonwealth University Herbaria; PE, Institute of Botany, Chinese Academy of Sciences Herbarium, Beijing, China.
2.2. Diversity and Phylogeny of I-Box-Like MYB Genes
A total of 274 RAD-like coding DNA sequences (CDSs) were found in 101 species representing 28 families and 15 orders of dicots (Solanales, Vitales, Brassicales, Malvales, Malpighiales, Ranunculales, Lamiales, Saxifragales, Rosales, Fabales, Proteales, Cucurbitales, Myrtales, Dipsacales, and Sapindales) and monocots (Table S1). Among these sequences, 79 CDSs belong to 17 species of seven genera of Solanaceae, which includes the FSM1 from So. lycopersicum [20]. For Arabidopsis, six RAD homologs, At4g39250 (Arabidopsis_thaliana_RL1, NM_120086.2), At2g21650 (Arabidopsis_thaliana_RL2, NM_127736.3), At4g36570 (Arabidopsis_thaliana_RL3, BT011255.1), DQ395345 (Arabidopsis_thaliana_RL4, NM_001084443.1), At1g19510 (Arabidopsis_thaliana_RL5, NM_101808.4), and At1g75250 (Arabidopsis_thaliana_RL6, NM_001084356.2), were included.
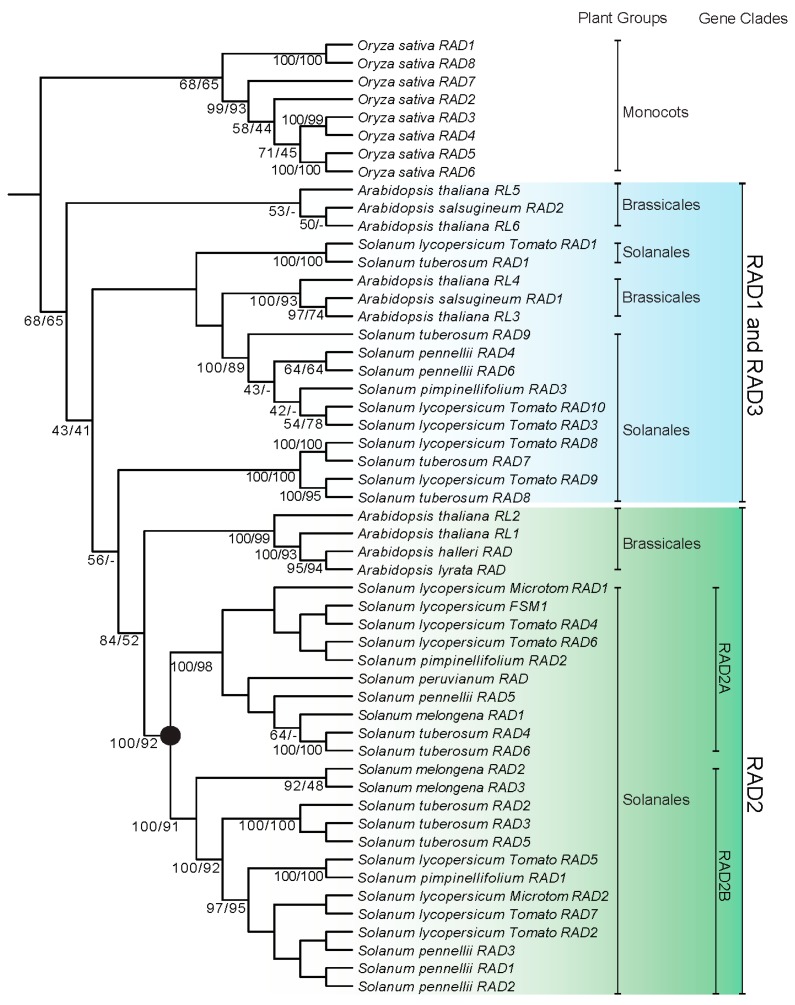
A phylogeny of RAD-like genes was constructed based on 53 sequences from four species of Arabidopsis (A. thaliana, A. halleri, A. lyrata, and A. salsuginea), six species of Solanum (So. melongena So. pennellii, So. lycopersicum, So. pimpinellifolium, So. peruvianum, and So. tuberosum), and Oryza sativa (Figure 1). The phylogeny indicated that sequences from O. sativa form a monophyletic clade. However, the phylogenetic relationships among the three previously identified RAD1, RAD2, and RAD3 clades [21] were not fully resolved. The RAD2 clade is likely monophyletic while the RAD1 and RAD3 clades are not (Figure 1, also see below). The RAD2 clade consists of Arabidopsis thaliana RL1 and Arabidopsis thaliana RL2 and species of Solanum, which were further divided into two Solanum-specific clades, RAD2A and RAD2B. The FSM1 of So. lycopersicum was placed in the RAD2A clade. It is unclear, however, how the other sequences of Solanum should be placed within the RAD1 clade represented by Arabidopsis thaliana RL3 and Arabidopsis thaliana RL4 and with the RAD3 clade represented by Arabidopsis thaliana RL5 and Arabidopsis thaliana RL6 (Figure 1) [21].
Figure 1.
Phylogeny of I-box-binding/RADIALIS-like genes of four species of Arabidopsis, six species of Solanum, and Oryza sativa based on Bayesian and maximum likelihood (ML) inferences. All sequences from O. sativa formed a monophyletic clade was used to root the phylogeny. Based on the clade defined by Boyden et al. (2012), [21], only the RAD2 clade was monophyletic and contained sequences from Arabidopsis and Solanum. There are two paralogs in the RAD2 clade, i.e., RAD2A and RAD2B, which resulted from a gene duplication that Arabidopsis was not involved. RAD1 and RAD3 are paraphyletic. Bayesian posterior probabilities and bootstrap frequencies ≥40% depicted close to the branches, respectively.
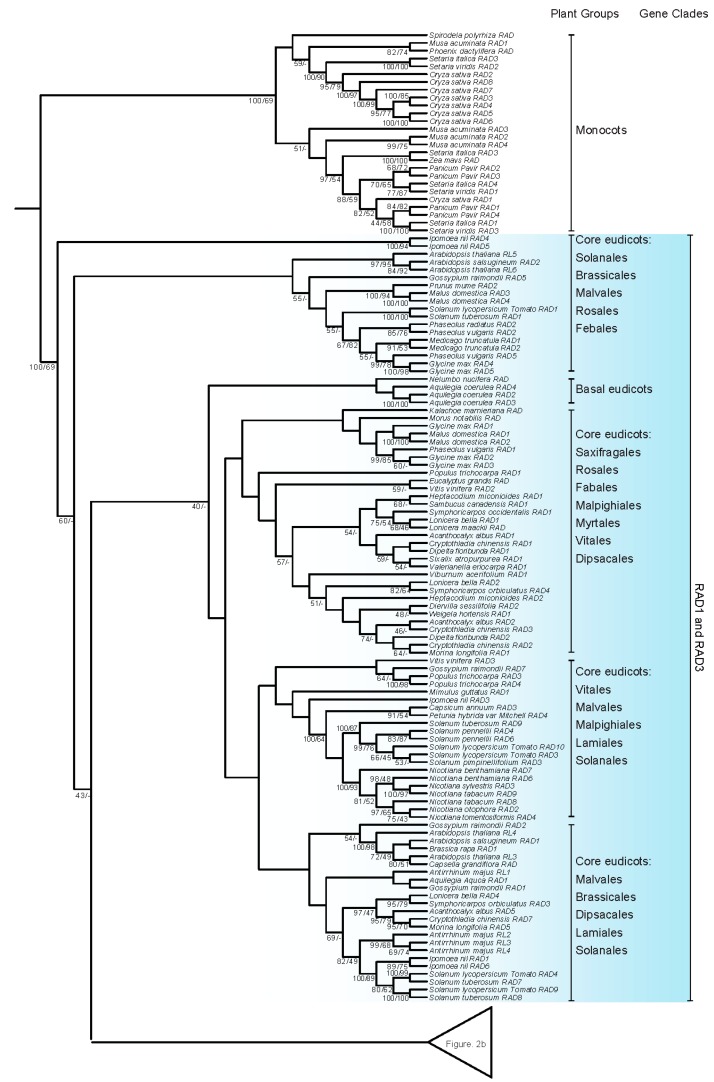
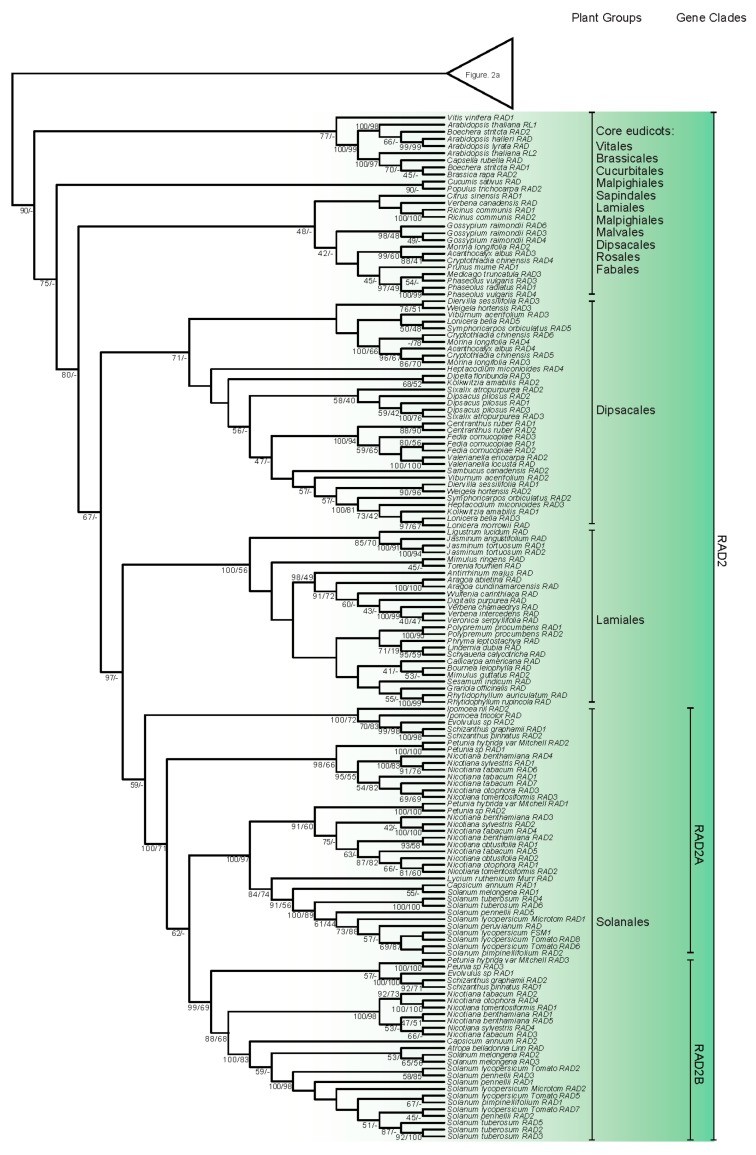
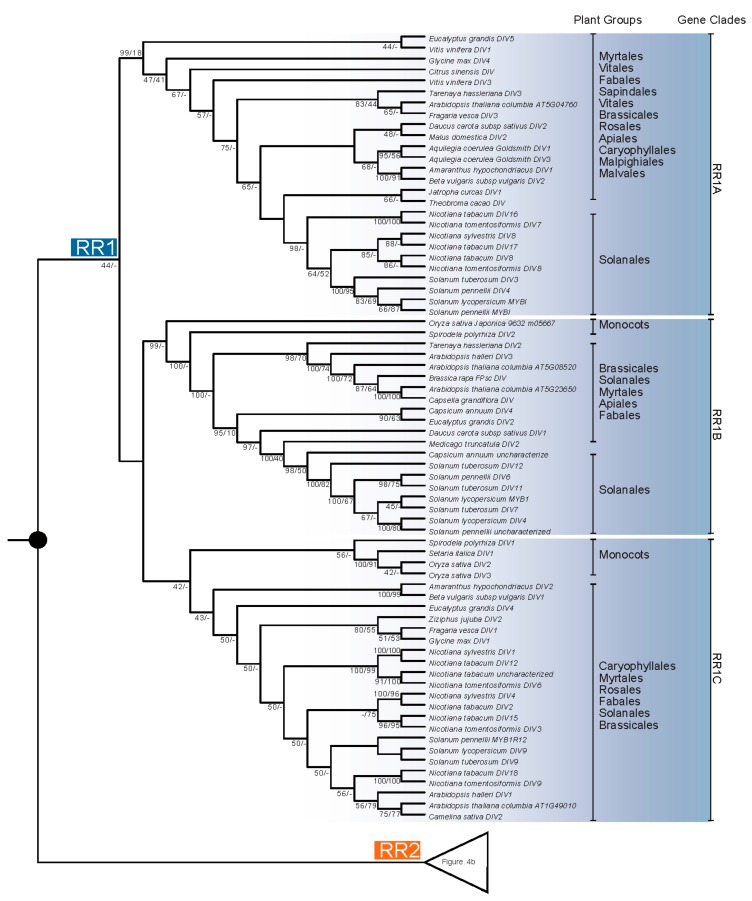
Another phylogeny of RAD-like genes was reconstructed based on 274 CDSs, including 258 from blast results and 16 in this study (Figure 2, Table S1). All eight species from seven families of monocots form a monophyletic clade and were used to root the phylogeny. RAD2 forms a monophyletic clade, while both RAD1 and RAD3 were not fully resolved (Figure 1 and Figure 2). RAD2 comprises representatives from eleven orders: Vitales, Rosales, Malvales, Fabales, Cucurbitales, Sapindales, Malpighiales, Brassicales, Solanales, Lamiales, and Dipsacales (Figure 2 and Table S1). Most of the solanaceous and convolvulaceous RAD-like sequences fell into the RAD2 clade, which is further divided into two clades, RAD2A and RAD2B (Figure 1 and Figure 2; Figure S1). The unrooted phylogeny including only RAD2 of Solanaceae and Convolvulaceae further indicates that two paralogs have been likely formed at least in the common ancestor of the two families (Figure S1). Further gene duplication and gene losses likely also occurred, which led to Nicotiana and Petunia having additional paralogs in RAD2A (Figure 2, Figure S1). RAD2 sequences from the two species of Schizanthus, the first branching clade of Solanaceae [23], are more closely related to the sequences from Convolvulaceae, which might be due to the limited sampling. The FSM1 of So. lycopersicum expressed in fruit is grouped in the RAD2A clade, while the RAD of A. majus is also in the RAD2 clade.
Figure 2.
Phylogeny of I-box-binding/RADIALIS-like genes based on Bayesian and ML inferences. 274 CDSs of I-box-binding/RADIALIS-like genes from both monocots and dicots were analyzed. All sequences from monocots formed a monophyletic group was used to root the phylogeny. RAD2 formed a monophyletic clade. At least one gene duplication was identified in the common ancestor of Solanaceae and Convolvulaceae. RAD1 and RAD3 clades are paraphyletic. Bayesian posterior probabilities and bootstrap frequencies ≥40% depicted close to the branches, respectively.
2.3. Diversity and Phylogeny of R-R-Type MYB Genes
One thousand and seventy-five CDSs that represent both R-R-type and CCA1-like genes from 109 species representing 34 different families from 22 orders of plants (16 of dicots, four of monocots, and two of mosses) were recovered (Table S2). For A. thaliana, the BLASTn results nine R-R-type, i.e., At1g49010 (AY519528.1), At2g38090 (AY519529.1), At3g11280 (AY550308.1), At5g01200 (AY519530.1), At5g05790 (AY519531.1), At5g08520 (AY519532.1), At5g58900 (AY519533.1), At5g23650 (DQ056685.1), and At5g04760 (AB493736.1) and one CCA1-like gene, i.e., At3g16350 (AY519512.1) [24]. For Solanaceae, we recovered 124 CDSs, namely DIV-, MYB- or MYB1R1-like genes, from 12 species in four genera, including the MYBI of So. lycopersicum.
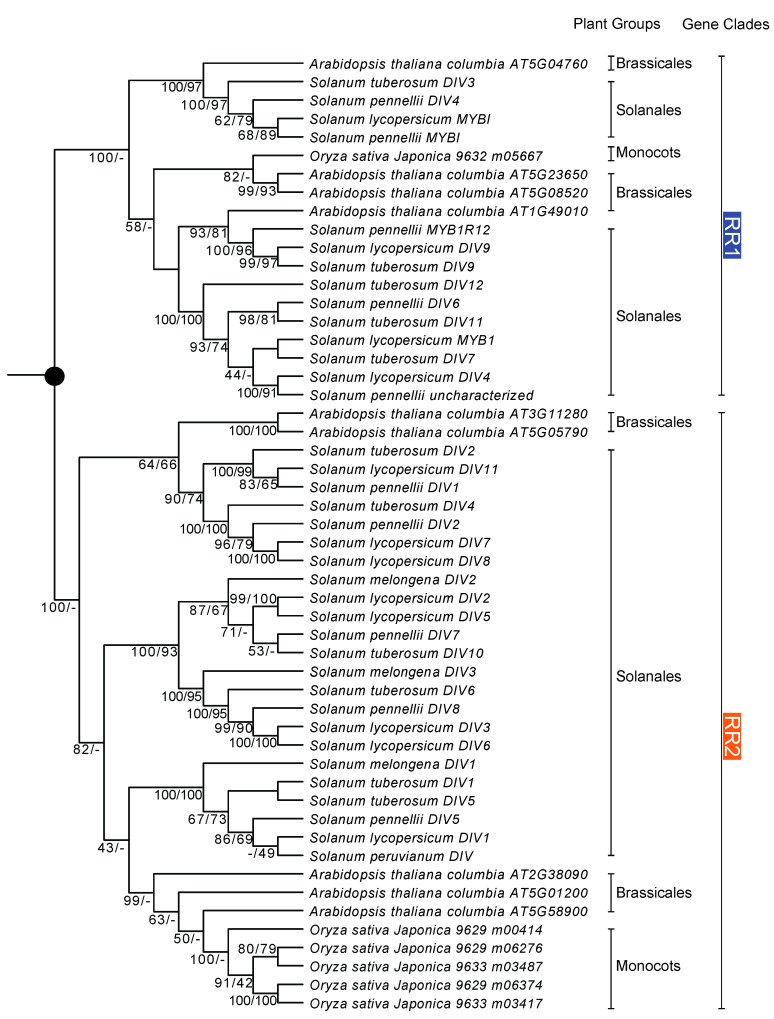
An R-R-type gene phylogeny was first reconstructed based on 52 CDSs from O. sativa japonica, A. thaliana, and five species of Solanum (So. melongena, So. lycopersicum, So. pennellii, So. peruvianum, and So. tuberosum) (Figure 3). All sequences fell into two clades, RR1 and RR2/DIV. The RR2/DIV clade represented the DIV clade identified by Howarth and Donoghue [22]. Each of these two clades contained sequences from O. sativa, A. thaliana, and Solanum.
Figure 3.
Phylogeny of R-R-type genes of five species of Solanum, Arabidopsis thaliana, and Oryza sativa based on Bayesian and ML inferences. Two major clades, RR1 and RR2, were identified, each of which includes sequences from Arabidopsis, Oryza, and Solanum. Bayesian posterior probabilities and bootstrap frequencies ≥40% depicted close to the branches, respectively.
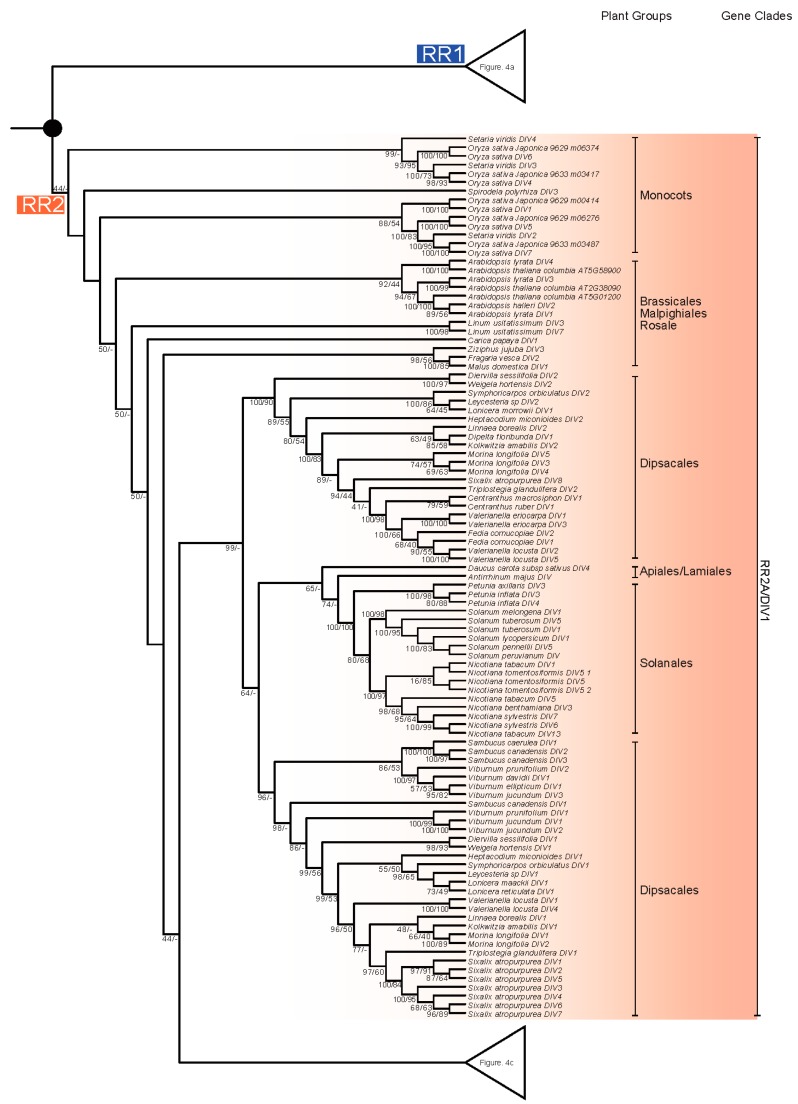
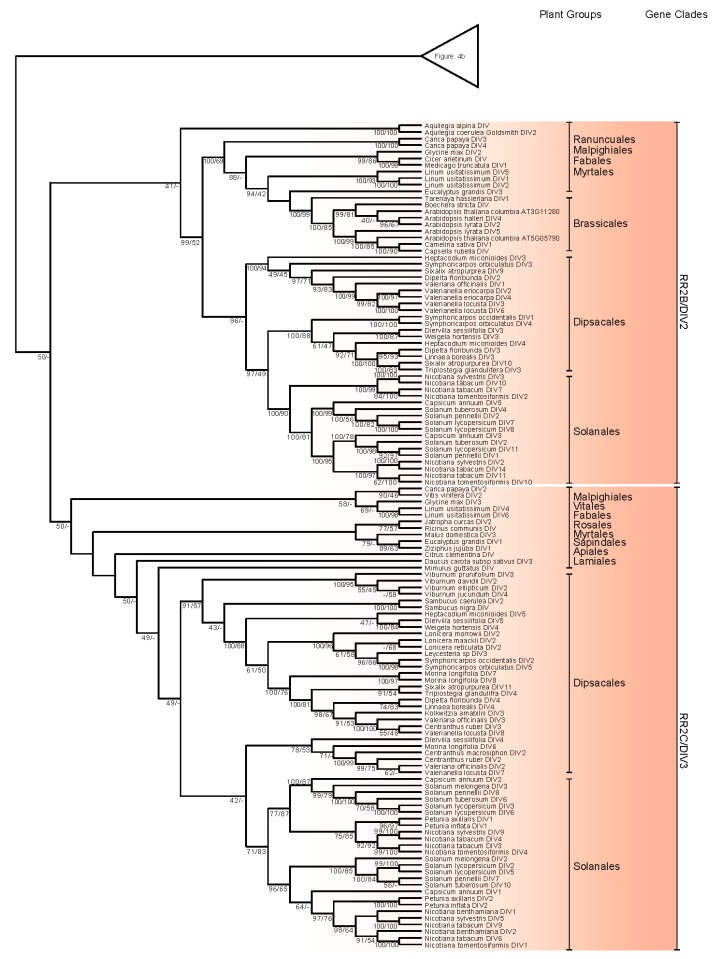
The R-R-type gene phylogeny was also reconstructed based on 298 CDSs from 75 species of 23 families (Figure 4, Table S2). The unrooted tree indicated that the RR1 and RR2/DIV clades were monophyletic (Figure 4). RR1 was further divided into three clades i.e., RR1A, RR1B, and RR1C. The RR1A clade included sequences from 12 orders of dicots (Myrtales, Fabales, Sapindales, Vitales, Brassicales, Rosales, Malvales, Malpighiales, Ranunculales, Caryophyllales, Apiales, and Solanales). The RR1B clade had representatives from monocots and five orders of dicots (Myrtales, Brassicales, Fabales, Apiales, and Solanales). RR1C clade had representatives from monocots and six orders of dicots (Caryophyllales, Myrtales, Brassicales, Rosales, Fabales, and Solanales). For Arabidopsis, AT5g04760 was placed in the RR1A clade, AT5G08520 and At5g23650 in the RR1B clade, and AT1G49010 in the RR1C clade. For the RR2/DIV clade, previously identified DIV2 and DIV3 clades formed monophyletic clades [22]. The sequences of A. thaliana, At2g38090, At5g01200, At5g58900, belonged to DIV1, while At3g11280 and At5g05790 belonged to DIV2. Arabidopsis lacked the DIV3 copy based on previous work [22]. The MYBI of So. lycopersicum expressed in fruit was grouped in the RR1A of RR1 clade, while the DIV of A. majus was likely in RR2A/DIV1 of the RR2 clade.
Figure 4.
Phylogeny of R-R-type genes based on Bayesian and ML inferences. Two hundred and ninety-eight CDSs of R-R-type genes from both monocots and dicots were analyzed. They formed two major clades, RR1 and RR2/DIV, each of which contained sequences from monocots and dicots. The RR1 clade was further divided into three groups, RR1A, RR1B, and RR1C. For the three RR2/DIV clades identified by Howarth and Donoghue [22] only the DIV2 and DIV3 are monophyletic. The Arabidopsis sequences include AT2G38090, AT5G01200, and AT5G58900 identified as DIV1, which is not a clade in this phylogeny. Bayesian posterior probabilities and bootstrap frequencies ≥40% depicted close to the branches, respectively.
2.4. Testing the Tree Topology for R-R-Type Genes
We further examined whether either of the two clades of R-R-type genes, RR1 including RR1A, RR1B, and RR1C; and RR2 including RR2A, RR2B, and RR2C, are monophyletic. Our results indicate that the tree topology number one, of which the subclades RR1A, RR1B, and RR1C formed a monophyletic RR1 clade, and the subclades RR2A, RR2B, and RR2C formed a monophyletic RR2 clade, is the most likely phylogeny [p > 0.5; Kishino-Hasegawa test (KH) = 0.794, Shimodaira-Hasegawa test (SH) = 0.997, and Approximately Unbiased test (AU) = 0.872] (Figure S2, Table 2). All the other tree topologies, except for the tree topology number nine, which have the RR2A subclade grouped within the RR1 clade, were rejected. However, the tree topology number nine is not strongly supported (0.2 < p < 0.5; KH = 0.206, SH = 0.343, and AU = 0.213) compared to the tree topology number one (Figure S2, Table 2).
Table 2.
Comparison of the statistics of the phylogenetic hypotheses.
| Tree Topology | l | δ | p Values | ||
|---|---|---|---|---|---|
| KH | SH | AU | |||
| 1: [A-B-C] [D-E-F] | −34754.03 | 0.00 | 0.794 | 0.997 | 0.872 |
| 2: [B-C] [A-D-E-F] | −34775.47 | 21.45 | 0.037 * | 0.284 | 0.048 * |
| 3: [A-C] [B-D-E-F] | −34801.25 | 47.22 | <0.001 * | 0.029 | <0.001 * |
| 4: [A-B] [C-D-E-F] | −34808.97 | 54.95 | <0.001 * | 0.014 | <0.001 * |
| 5: [A] [B-C-D-E-F] | −34820.49 | 66.46 | <0.001 * | 0.003 | <0.001 * |
| 6: [B] [A-C-D-E-F] | −34820.49 | 66.46 | <0.001 * | 0.003 | <0.001 * |
| 7: [C] [A-B-D-E-F] | −34820.49 | 66.46 | <0.001 * | 0.003 | <0.001 * |
| 8: [A-B-C-D-E-F] | −34820.49 | 66.46 | <0.001 * | 0.003 | <0.001 * |
| 9: [A-B-C-D] [E-F] | −34774.05 | 20.02 | 0.206 | 0.343 | 0.213 |
| 10: [A-B-C-E] [D-F] | −34818.22 | 64.19 | <0.001 * | 0.004 | <0.001 * |
| 11: [A-B-C-F] [D-E] | −34818.03 | 64.00 | <0.001 * | 0.007 | <0.001 * |
| 12: [A-B-C-D-E] [F] | −34820.49 | 66.46 | <0.001 * | 0.003 | <0.001 * |
| 13: [A-B-C-D-F] [E] | −34820.49 | 66.46 | <0.001 * | 0.003 | <0.001 * |
| 14: [A-B-C-E-F] [D] | −34820.49 | 66.46 | <0.001 * | 0.003 | <0.001 * |
A = RR1A, B = RR1B, C = RR1C, D = RR2A, E = RR2B, and F = RR2C. l = Log-likelihood scores. δ = the log-likelihood differences to the best tree. * denotes statistical significance at the 0.05 level.
2.5. Motif Analyses
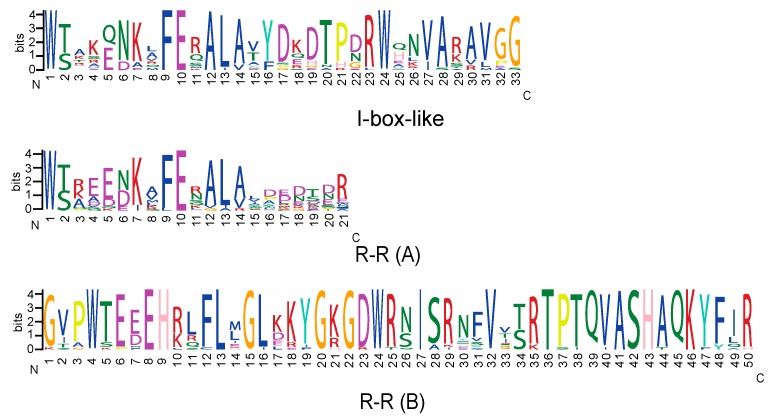
We also analyzed the nucleotide sequences, which cover the diverse lineages of the I-box-like and R-R-type from A. thaliana and O. sativa, and the representatives from An. majus and So. lycopersicum, to identify protein motifs. Our results largely agree with the study of Chen et al. [24], which analyzed the motifs for the I-box-like, R-R-type, and CCA1-like MYB genes. We found that the I-box-like genes have only one motif, while the R-R-type genes have two motifs, i.e., R-R (A), and R-R (B). R-R (A) locates at the N-terminal of the R-R-type genes, and shows high similarity to the I-box-like genes (Figure 5). R-R (B) locates at the C-terminal of the R-R-type genes, and is distinct in amino acid sequences compared to the R-R (A) and the only motif of I-box-like genes.
Figure 5.
Motif analyses of I-box-like and R-R-type genes of the MYB family. One motif was identified for the I-box-like genes and two motifs, i.e., R-R (A) and R-R (B), were found for the R-R-type genes. The x-axis indicates the sequence of the amino acids from the N- to C- terminal for each motif; the y-axis provides the information at each position in the motif; and the highly conserved positions in the motif show a large proportion of the bits for a particular amino acid.
On the other hand, the results of our motif analyses also show differences compared to the results of Chen et al. [24]. For I-box-like genes, our analyses identified a single motif that is 33 amino acids in length. In contrast, the same motif based on Chen et al. [24] contains 56 amino acids, which has eight and 15 extra amino acids at the N- and C-terminal, respectively. For R-R-type genes, our results indicate that R-R (A) is 21 amino acids in length, while Chen′s results [24] include 59 amino acids for the same motif, which has eight and 30 extra amino acids at the N- and C-terminal, respectively. Our results suggest that R-R (B) is 50 amino acids in length. However, the same motif based on the work of Chen et al. [24] has 53 amino acids, which has five extra amino acids at the N-terminal but lacks two amino acids at the C-terminal. One possible reason for the discrepancy between the two studies is that the Multiple Expectation maximizations for the Motif Elicitation (MEME) methods applied in our study only consider the continued amino acid sequences for a motif and no gap in the sequences is allowed.
3. Discussion
3.1. Phylogenetic Positions of RAD- and DIV-Like Genes in the Plant MYB Lineage.
MYB proteins contain a conserved MYB domain, which usually comprises one to three imperfect repeats, namely R1, R2, R3 [4,24]. Each of these repeats comprises about 52 amino acid residues that encode a helix-loop-helix structure involved in DNA binding [4,25]. MYB genes have been found in all eukaryotes [4,26].
Phylogenetic analysis indicates that the MYB genes of plants, which is sister to all animal MYB genes, form a monophyletic clade [25]. MYB genes in plants are structurally and functionally more variable compared to MYB genes in vertebrates [25,27]. Based on the MYB domain structures, the MYB proteins of plants can be classified into three major groups: R1R2R3-MYB with three adjacent repeats, R2R3-MYB with two adjacent repeats, and MYB-related proteins, a heterogeneous group, often containing a single MYB repeat [7,24,25,28,29,30]. The R2R3-MYB group is thought to be derived from the R1R2R3-MYB group, which occurs in all major lineages of land plants [27]. Based on the phylogenetic analysis and the protein domain structure, MYB-related proteins were further divided into five subfamilies: CCA1-like, CPC-like, TBP-like, I-box-binding-like (abbreviated I-box-like), and R-R-type [24,30]. Based on Chen et al. [24], A. thaliana has five I-box-like genes, i.e., At1g75250, At1g19510, At2g21650, At4g39250, and At4g36570, and nine R-R-type genes, i.e., At1g49010, At2g38090, At3g11280, At5g01200, At5g05790, At5g08520, At5g58900, At5g23650, and At5g04760. Boyden, Donoghue, and Howarth [21] indicated that RAD-like genes belong to the I-box-like clade. Our analyses further indicate that the I-box-like lineage is synonymous with RAD-like genes (Figure 1 and Figure 2). Furthermore, Howarth and Donoghue [22] focused on the evolution of DIV-like genes in core eudicots especially in Dipsacales, and indicated that the DIV-like genes belong to an R-R-type lineage. Our analysis of R-R-type genes showed that the gene duplication occurred at least in the common ancestor of dicots and monocots, giving rise to two paralogs, the RR1 and RR2 clades (Figure 3 and Figure 4), of which the RR2 clade is synonymous with the DIV-like lineage [22].
3.2. Evolution of the I-Box-Like Subfamily
Boyden, Donoghue, and Howarth [21] indicated that RAD-like genes consist of three major clades: RAD1, RAD2, and RAD3, which were speculated to result from genome duplications associated with the origin of core eudicots. The RAD1 clade has Arabidopsis AT4G36570 and DQ395345 of Clade I, defined in Reference [24], and RAD2 and RAD3 have the Arabidopsis sequences from Clade III (AT2G21650 and AT4G39250 belong to the RAD2, and AT1G19510 and AT1G75250 belong to RAD3). Our analysis recognized RAD2 as a monophyletic clade (Figure 1 and Figure 2). Furthermore, there are two RAD2 paralogs involving Solanaceae and Convolvulaceae, RAD2A and RAD2B, which likely resulted from a gene duplication at least in the common ancestor of these two plant families. On the other hand, the RAD1 and RAD3 clades were not fully resolved based on our analyses. Our phylogenetic analyses indicated that the RAD of A. majus belongs to the RAD2 clade, while FSM1 is placed in the RAD2A clade, suggesting that RAD and FSM1 belong to the same orthologous lineage.
3.3. Evolution of the R-R-Type Subfamily
The R-R-type genes have two imperfect repeats of the MYB domain, namely R-R (A) and R-R (B) [24]. The N-terminal MYB repeat R-R (A) was found to be closely related to the MYB repeats of the I-box-like genes, and the C-terminal MYB repeat R-R (B) was closely related to those of certain CCA1-like genes based on the positions of the introns and shared motifs [24]. The phylogeny of R-R-type genes based on nine sequences of A. thaliana and seven of O. sativa japonica suggests several gene duplications in the common ancestor of the monocots and dicots, but the phylogenetic relationships of the predicted paralogs were unresolved in that study [24]. The work by Howarth and Donoghue [22] focused on the evolution of DIV-like genes in core eudicots, especially in Dipsacales, which showed duplications giving rise to three DIV-like clades in the core eudicots, DIV1, DIV2, and DIV3. Our blast and phylogenetic analyses indicated that most of the sequences named DIV-like genes belong to the R-R-type subfamily, while most of the sequences named as MYB1R1-like genes belong to the CCA1-like gene family (Table S2). Each of the two R-R-type subclades, RR1 and RR2, was further divided into three paralogs, which likely resulted from genome duplication in the common ancestor of core eudicots [22]. RR1 consists of RR1A, RR1B, and RR1C, while RR2/DIV is composed of RR2A/DIV1, RR2B/DIV2, and RR2C/DIV3 (Figure 3 and Figure 4) [22]. We found that the DIV of An. majus belongs to the DIV1 of the RR2/DIV clade [22], while the MYBI of tomato belongs to the RR1A of the RR1 clade.
3.4. Evolution of the Antagonism among RAD-DRIF-DIV and FSM1-FSB1-MYBI in An. majus and So. lycopersicum, Respectively
Based on an analysis of amino acid sequences, the two MYB domains of DIV had different functions with the C-terminal domain similar to known DNA binding MYB proteins, while the N-terminal domain was associated with protein-protein interactions (Figure 5) [19,31]. In contrast, RAD has a single MYB domain that is predicted to act through a mechanism involving protein–protein interactions (Figure 5) [16]. As the members of MYB-related subfamilies, I-box-like and R-R-type genes were previously placed in the same clade by Riechmann and Ratcliffe [30], which suggested that they might be closely related paralogs. One possible hypothesis proposed for the evolution of these two MYB-related subfamilies is that I-box-like genes evolved through the loss of the MYB domain at the C-terminal end [24,32]. RAD-DRIF-DIV and FSM1-FSB1-MYBI therefore represent the recruitment of homologous genes from similar MYB lineages in the development of floral zygomorphy in An. majus, and the development of fruit in So. lycopersicum [10].
In summary, I-box-like and R-R-type lineages have experienced extensive gene duplication that predated the diversification of the core eudicots. Our work further clarified the evolution of these two MYB subfamilies, which will help the future inquiry into the functional studies of the paralogs of the I-box-like and R-R-type genes that may have been involved in the evolution of molecular antagonism.
4. Materials and Methods
4.1. Cloning RAD-Like Genes from Species of Solanaceae and Convolvulaceae
Primers incorporated with degenerate polymorphic sites based on the alignment of RAD-like sequences, especially the RAD2 clade from Solanaceae and Lamiales, were used for amplifying the genes from species of Solanaceae and representatives of Convolvulaceae. The locations of our primers referred to the study by Boyden, Donoghue, and Howarth [21]. These primers, i.e., forward primer 5′-AACAAGGCITTTGARARGGCWTYRGC-3′, and reverse primer 5′-GGRAARGGBAYIMYACCAIDITCAAT-3′, successfully amplified RAD-like genes from both the basal and derived clades of Solanaceae (Schizanthus pinnatus Ruiz & Pav, Schizanthus grahamii Gillies, Petunia sp., Nicotiana obtusifolia M. Martens & Galeotti, Solanum lycopersicum L., Lycium ruthenicum Murray, and Atropa belladonna L.) and species of Convolvulaceae (Evolvulus sp. and Ipomoea tricolor Cav.) (Table 1). PCR reactions were performed using GoTaq® G2 Hot Start Polymerase (Promega, Madison, WI, USA), as follows: 95 °C for 5 min, 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 1 min and 30 s, repeated for 39 cycles, with a final step at 72 °C for 10 mins. PCR products were then purified through gel extraction using Wizard SV Gel and PCR Clean-Up System from Promega. The purified PCR products were used as a template for the second round of PCR following the same PCR program described above. The purified second round PCR products were used in ligation and transformation with pGEM-T Easy Vector System I from Promega. At least 50 clones were screened for each species. The sequences of the clones were determined using Sanger sequencing by GENEWIZ (115 Corporate Boulevard, South Plainfield, NJ, USA).
4.2. Gene Mining
The RAD- and DIV-like genes were obtained through blasting RAD and DIV CDSs of A. majus (GenBank accession numbers: AY954971.1 and AY077453.1, respectively) against the databases, including NCBI BLASTn (available online: http://www.ncbi.nlm.nih.gov/ BLAST/), Phytozome 11 (available online: https://phytozome.jgi.doe.gov), Sol Genomics Network (available online: https://solgenomics.net), and Rice Genome Annotation Project (available online: http://rice.plantbiology.msu.edu).
4.3. Alignment and Phylogenetic Analyses
The DNA matrices of the coding sequences were aligned using Geneious version 7.1.9 (PO Box 5677, Wellesley St, Auckland 1010, New Zealand). The MUSCLE algorism that refers to the protein sequence alignment for building nucleotide sequence alignment was applied. Each DNA matrix was analyzed by using the Bayesian and ML inferences, which were implemented in RAxML_HPC2, and MrBayes version 3.2.6 on XSEDE, respectively, at the CIPRES Science Gateway V. 3.3. [33,34,35,36]. For ML analyses, a random seed value for rapid ML bootstrapping was estimated on each dataset. The GTRCAT model was chosen for the bootstrapping analysis based on the program recommendation because GTRCAT shows lower computational costs and memory consumption for the ML method [34]. The models used for the Bayesian analyses were estimated using jmodeltest 2.1.10 [37,38]. The Akaike Information Criterion (AIC) [39] was used to determine the best-fit model for each DNA sequence matrix, i.e., K80 (K2P) + g model for the I-box-like/RAD gene phylogeny including Arabidopsis, Solanum, and Oryza alone, JC + g model for the large RAD phylogeny, GTR + i + g model for the R-R-type gene phylogeny including Arabidopsis, Solanum, and Oryza alone, and GTR + i + g model for the large R-R-type gene phylogeny. We used the Metropolis-coupled Markov chain Monte Carlo method as implemented in MrBayes to run four chains. We ran five million generations for each chain, and sampled every 1000 generations with a burn-in of the first 2000 trees.
4.4. Phylogeny Assessment for R-R-Type Genes
We generated 14 tree topologies manually based on our Bayesian tree to test the alternate hypotheses. To produce these topologies, first the RR2A clade was set as monophyletic. Second, we collapsed the relationships among the subclades in RR1 and RR2 clades. Finally, the subclades were subsequently moved around as indicated in Figure S2. The log-likelihoods for each tree topology were calculated using TREE-PUZZLE (ver. 5.3.rc16) [40] with the HKY model of evolution [41] and four rate categories for the discrete Gamma distribution. The log-likelihoods information estimated for the 14 topologies from TREE-PUZZLE was then entered into CONSEL (ver. 0.20) [42] to generate the bootstrap replicates for each tested tree. The p-values of KH [43], SH [44], and AU tests [45] were subsequently calculated based on the bootstrap samples [42]. The confidence of the 14 trees was then assessed by the p-values [42]. If the p-value estimated for a tree was < 0.05, the topology was rejected; if the p value >0.5, the topology was preferred [45,46].
4.5. Motif Analyses
Motif analyses based on nucleotide sequences were carried out for I-box-like and R-R-type genes. For I-box-like genes, we included six sequences of A. thaliana, i.e., At4g39250 (Arabidopsis_thaliana_RL1, NM_120086.2), At2g21650 (Arabidopsis_thaliana_RL2, NM_127736.3), At4g36570 (Arabidopsis_thaliana_RL3, BT011255.1), DQ395345 (Arabidopsis_thaliana_RL4, NM_001084443.1), At1g19510 (Arabidopsis_thaliana_RL5, NM_101808.4), and At1g75250 (Arabidopsis_thaliana_RL6, NM_001084356.2); eight sequences of O. sativa, i.e., (Oryza_sativa_RAD1, LOC_Os01g44390.2), 9640.m03280 (Oryza_sativa_RAD2, LOC_Os12g33950), 9631.m01422 (Oryza_sativa_RAD3, LOC_Os03g14810), 9631.m06332 (Oryza_sativa_RAD4, LOC_Os03g63890), 9633.m03415 (Oryza_sativa_RAD5, LOC_Os05g37040), 9633.m03416 (Oryza_sativa_RAD6, LOC_Os05g37050), 9635.m02514 (Oryza_sativa_RAD7, LOC_Os07g26150.1), and 9640.m03280 (Oryza_sativa_RAD8, LOC_Os12g33950); one sequence of An. majus, i.e., RAD; and one sequence of So. Lycopersicum, i.e., FSM1.
For R-R-type genes, we included nine sequences of A. thaliana, i.e., At1g49010 (AY519528.1), At2g38090 (AY519529.1), At3g11280 (AY550308.1), At5g01200 (AY519530.1), At5g05790 (AY519531.1), At5g08520 (AY519532.1), At5g58900 (AY519533.1), At5g23650 (DQ056685.1), and At5g04760 (AB493736.1); seven sequences of O. sativa, i.e., 9632.m05667 (LOC_Os04g58020), 9629.m00414 (LOC_Os01g04930), 9629.m06276 (LOC_Os01g63460), 9629.m06374 (LOC_Os01g64360), 9633.m03417 LOC_Os05g37060), 9633.m03487 (LOC_Os05g37730), and 9631.m06132 (LOC_Os03g62100); one sequence of An. majus, i.e., DIV; and one sequence of So. lycopersicum, i.e., MYBI.
The nucleotide sequences of these CDSs were translated into amino acid sequences by Mesquite version 3.2 [47]. We used the MEME algorithm, which extends the Expectation Maximization (EM) algorithm for identifying motifs in unaligned amino acid sequences [48]. The MEME algorithm is designed to discover novel and ungapped motifs in a set of homologous sequences [48]. To use this function, we uploaded and analyzed the I-box-like and R-R-type amino acid sequences at http://meme-suite.org/index.html [49]. For the MEME options, we set the numbers of motifs to be found as three, and each motif occurred only one time in each testing sequence.
Acknowledgments
We thank David E. Boufford and the three anonymous reviewers. This work was funded by Virginia Commonwealth University and National Science Foundation Grant DEB-1355109.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/9/1961/s1.
Author Contributions
Ao Gao, Jingbo Zhang, and Wenheng Zhang conceived and designed the experiments; Ao Gao performed the experiments; Ao Gao, Jingbo Zhang, and Wenheng Zhang analyzed the data; Ao Gao, Jingbo Zhang, and Wenheng Zhang wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Klempnauer K.H., Gonda T.J., Bishop J.M. Nucleotide-sequence of the retroviral leukemia gene v-MYB and its cellular progenitor c-MYB-the architecture of a transduced oncogene. Cell. 1982;31:453–463. doi: 10.1016/0092-8674(82)90138-6. [DOI] [PubMed] [Google Scholar]
- 2.Nomura N., Takahashi M., Matsui M., Ishii S., Date T., Sasamoto S., Ishizaki R. Isolation of human cDNA clones of MYB-related genes, a-MYB and b-MYB. Nucleic Acids Res. 1988;16:11075–11089. doi: 10.1093/nar/16.23.11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weston K. MYB Proteins in life, death and differentiation. Curr. Opin. Genet. Dev. 1998;8:76–81. doi: 10.1016/S0959-437X(98)80065-8. [DOI] [PubMed] [Google Scholar]
- 4.Lipsick J.S. One billion years of MYB. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 5.Paz-Ares J., Ghosal D., Wienand U., Peterson P.A., Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to MYB proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cominelli E., Tonelli C. A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Res. 2009;19:1231–1232. doi: 10.1038/cr.2009.123. [DOI] [PubMed] [Google Scholar]
- 7.Martin C., PazAres J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/S0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 8.Oppenheimer D.G., Herman P.L., Sivakumaran S., Esch J., Marks M.D. A MYB gene required for leaf trichome differentiation in arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhai R., Wang Z.M., Zhang S.W., Meng G., Song L.Y., Wang Z.G., Li P.M., Ma F.W., Xu L.F. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (pyrus bretschneideri REHD.) J. Exp. Bot. 2016;67:1275–1284. doi: 10.1093/jxb/erv524. [DOI] [PubMed] [Google Scholar]
- 10.Raimundo J., Sobral R., Bailey P., Azevedo H., Galego L., Almeida J., Coen E., Costa M.M.R. A subcellular tug of war involving three MYB-like proteins underlies a molecular antagonism in Antirrhinum flower asymmetry. Plant J. 2013;75:527–538. doi: 10.1111/tpj.12225. [DOI] [PubMed] [Google Scholar]
- 11.Luo D., Carpenter R., Vincent C., Copsey L., Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 12.Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/S0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- 13.Howarth D.G., Donoghue M.J. Phylogenetic analysis of the “ECE” (cyc/tb1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA. 2006;103:9101–9106. doi: 10.1073/pnas.0602827103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cubas P., Lauter N., Doebley J., Coen E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999;18:215–222. doi: 10.1046/j.1365-313X.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 15.Baxter C.E.L., Costa M.M.R., Coen E.S. Diversification and co-option of rad-like genes in the evolution of floral asymmetry. Plant J. 2007;52:105–113. doi: 10.1111/j.1365-313X.2007.03222.x. [DOI] [PubMed] [Google Scholar]
- 16.Corley S.B., Carpenter R., Copsey L., Coen E. Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc. Natl. Acad. Sci. USA. 2005;102:5068–5073. doi: 10.1073/pnas.0501340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa M.M.R., Fox S., Hanna A.I., Baxter C., Coen E. Evolution of regulatory interactions controlling floral asymmetry. Development. 2005;132:5093–5101. doi: 10.1242/dev.02085. [DOI] [PubMed] [Google Scholar]
- 18.Almeida J., Rocheta M., Galego L. Genetic control of flower shape in Antirrhinum majus. Development. 1997;124:1387–1392. doi: 10.1242/dev.124.7.1387. [DOI] [PubMed] [Google Scholar]
- 19.Galego L., Almeida J. Role of divaricata in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev. 2002;16:880–891. doi: 10.1101/gad.221002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machemer K., Shaiman O., Salts Y., Shabtai S., Sobolev I., Belausov E., Grotewold E., Barg R. Interplay of MYB factors in differential cell expansion, and consequences for tomato fruit development. Plant J. 2011;68:337–350. doi: 10.1111/j.1365-313X.2011.04690.x. [DOI] [PubMed] [Google Scholar]
- 21.Boyden G.S., Donoghue M.J., Howarth D.G. Duplications and expression of radialis-like genes in dipsacales. Int. J. Plant Sci. 2012;173:971–983. doi: 10.1086/667626. [DOI] [Google Scholar]
- 22.Howarth D.G., Donoghue M.J. Duplications and expression of divaricata-like genes in dipsacales. Mol. Biol. Evol. 2009;26:1245–1258. doi: 10.1093/molbev/msp051. [DOI] [PubMed] [Google Scholar]
- 23.Särkinen T., Bohs L., Olmstead R.G., Knapp S. A phylogenetic framework for evolutionary study of the nightshades (solanaceae): A dated 1000-tip tree. BMC Evol. Biol. 2013;13:214. doi: 10.1186/1471-2148-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y.H., Yang X.Y., He K., Liu M.H., Li J.G., Gao Z.F., Lin Z.Q., Zhang Y.F., Wang X.X., Qiu X.M., et al. The MYB transcription factor superfamily of arabidopsis: Expression analysis and phylogenetic comparison with the rice myb family. Plant Mol. Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 25.Rosinski J.A., Atchley W.R. Molecular evolution of the MYB family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998;46:74–83. doi: 10.1007/PL00006285. [DOI] [PubMed] [Google Scholar]
- 26.Kranz H.D., Denekamp M., Greco R., Jin H., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., et al. Towards functional characterisation of the members of the R2R3-MYB gene family from arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- 27.Kranz H., Scholz K., Weisshaar B. C-MYB Oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 2000;21:231–235. doi: 10.1046/j.1365-313x.2000.00666.x. [DOI] [PubMed] [Google Scholar]
- 28.Jin H.L., Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999;41:577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- 29.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 30.Riechmann J.L., Ratcliffe O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000;3:423–434. doi: 10.1016/S1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- 31.Rose A., Meier I., Wienand U. The tomato i-box binding factor lemybi is a member of a novel class of MYB-like proteins. Plant J. 1999;20:641–652. doi: 10.1046/j.1365-313X.1999.00638.x. [DOI] [PubMed] [Google Scholar]
- 32.Stevenson C.E.M., Burton N., Costa M.M.R., Nath U., Dixon R.A., Coen E.S., Lawson D.M. Crystal structure of the MYB domain of the rad transcription factor from Antirrhinum majus. Protein Struct. Funct. Biol. 2006;65:1041–1045. doi: 10.1002/prot.21136. [DOI] [PubMed] [Google Scholar]
- 33.Huelsenbeck J.P., Ronquist F. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 34.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 35.Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 36.Miller M.A., Pfeiffer W., Schwartz T. Creating the cipres science gateway for inference of large phylogenetic trees; Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 37.Darriba D., Taboada G.L., Doallo R., Posada D. Jmodeltest 2: More models, new heuristics and parallel computing. Nat. Meth. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov B.N., Csaki F., editors. Proceedings of the Second International Symposium in Information Theory; Tsahkadsor, Armenia. 2–8 September 1971; Budapest, Hungary: Akademiai Kiado; 1973. pp. 267–281. [Google Scholar]
- 40.Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. Tree-puzzle: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 41.Hasegawa M., Kishino H., Yano T.A. Dating of the human ape splitting by a molecular clock of mitochondrial-DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 42.Shimodaira H., Hasegawa M. Consel: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 43.Kishino H., Hasegawa M. Evaluation of the maximum-likelihood estimate of the evolutionary tree topologies from DNA-sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 44.Shimodaira H., Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 45.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 46.Susko E., Leigh J., Doolittle W.F., Bapteste E. Visualizing and assessing phylogenetic congruence of core gene sets: A case study of the gamma-proteobacteria. Mol. Biol. Evol. 2006;23:1019–1030. doi: 10.1093/molbev/msj113. [DOI] [PubMed] [Google Scholar]
- 47.Maddison W., Maddison D. Mesquite: A Modular System for Evolutionary Analysis. [(accessed on 11 January 2017)]; Version 3.2. Available online: http://mesquiteproject.org.
- 48.Bailey T.L., Elkan C. Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Mach. Learn. 1995;21:51–80. doi: 10.1007/BF00993379. [DOI] [Google Scholar]
- 49.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.