Abstract
CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) is caused by mutations in the NOTCH3 gene, affecting the number of cysteines in the extracellular domain of the receptor, causing protein misfolding and receptor aggregation. The pathogenic role of cysteine-sparing NOTCH3 missense mutations in patients with typical clinical CADASIL syndrome is unknown. The aim of this article is to describe these mutations to clarify if any could be potentially pathogenic. Articles on cysteine-sparing NOTCH3 missense mutations in patients with clinical suspicion of CADASIL were reviewed. Mutations were considered potentially pathogenic if patients had: (a) typical clinical CADASIL syndrome; (b) diffuse white matter hyperintensities; (c) the 33 NOTCH3 exons analyzed; (d) mutations that were not polymorphisms; and (e) Granular osmiophilic material (GOM) deposits in the skin biopsy. Twenty-five different mutations were listed. Four fulfill the above criteria: p.R61W; p.R75P; p.D80G; and p.R213K. Patients carrying these mutations had typical clinical CADASIL syndrome and diffuse white matter hyperintensities, mostly without anterior temporal pole involvement. Cysteine-sparing NOTCH3 missense mutations are associated with typical clinical CADASIL syndrome and typical magnetic resonance imaging (MRI) findings, although with less involvement of the anterior temporal lobe. Hence, these mutations should be further studied to confirm their pathological role in CADASIL.
Keywords: CADASIL, cysteine, NOTCH3, mutation, temporal pole
1. Introduction
CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) is an inherited systemic arterial vessel disease caused by mutations in the NOTCH3 gene, located on the 19p13 chromosome [1,2], which encodes a transmembrane receptor that is mainly expressed in the smooth muscle cells of blood vessels and pericytes [3].
This receptor has three domains: (1) a large extracellular domain (ECD) with 34 epidermal growth factor (EGF)-like repeats encoded by exons 2–24, where NOTCH3 mutations are typically located; (2) a transmembrane domain; and (3) an intracellular domain (ICD). Each EGF-like repeat contains six cysteines that form three disulfide bonds, which contribute significantly to the protein’s tertiary structure.
Pathogenic mutations in CADASIL are dominant; the presence of only one mutation in one of both alleles is the cause of the pathology. The pathogenic mutations are associated with changes in the number of cysteines, these changes cause an odd number of cysteines, leading to a misfolding of the receptor, enhanced formation of oligomers and ECD aggregation, facilitated by crosslinking of sulfhydryl groups [4,5,6]. This ECD aggregation is considered to be the main pathogenic mechanism of the disease. In terms of NOTCH3 activity, classic mutations have been described with normal or decreased NOTCH3 activity. Nevertheless, its role in the pathogenic mechanism is controversial, and several authors have shown that altered NOTCH3 function is not the primary determinant of the disease [7,8,9].
The main symptoms of CADASIL are migraine, psychiatric disorders, recurrent small subcortical infarctions, and dementia [10]. The average age at stroke onset is approximately 46 years, although recent studies have described an older onset, at 52 years for males and 57 for females [11]; for dementia, the average age at onset is 55 [12].
Magnetic resonance imaging (MRI) and histopathology show small subcortical infarctions and confluent white matter hyperintensities (WMH), mostly in periventricular locations and with additional involvement of the anterior temporal pole and the external capsule [13].
It has been described elsewhere that WMH in the temporal pole have 89–95% sensitivity and 80–86% specificity for CADASIL diagnosis in Caucasian patients [14], and up to 95% of these patients have this alteration [15]. Nevertheless, in Asian populations the prevalence of temporal pole involvement is only between 44.8–71% [16].
Additionally, it is typical to find granular osmiophilic material (GOM), consisting of parts of the extracellular domain of NOTCH3 [17], in the extracellular space and located close to the cell surface in smooth-muscle cells [18]. The presence of GOM has 45–96% sensitivity and 100% specificity for CADASIL diagnosis [15,19], but the pathological role of GOM is still debated.
Recent studies have found that mutations that do not affect the number of cysteines (unlike the typical mutations) seem to be associated with clinical CADASIL syndrome. However, the pathogenic role of these mutations is uncertain.
Therefore, this is a systematic review of those studies in which patients with a clinical suspicion of CADASIL and cysteine-sparing NOTCH3 missense mutations have been described. The aims of this article are to: (1) find data in the literature and in databases relating to cysteine-sparing NOTCH3 missense mutations observed in patients with typical clinical CADASIL syndrome; (2) describe the epidemiological characteristics of these patients; (3) determine whether these mutations could be considered potentially pathogenic.
There is not a consensus to determine whether a mutation can be considered pathogenic. Depending on the disease the presence of a missense mutation that has not been described previously in healthy subjects and with bioinformatic data supporting the pathogenicity is enough to consider a mutation the cause of the disease [20]. However this is not the case for other diseases and more analysis should be performed. For instance in neurodegenerative disorders there are studies that have analyzed the conservation of the residue during the evolution or have analyzed the presence of described mutations involving the same Wild-type amino acid [21,22].
In the case of CADASIL, the presence of GOM is very important because this finding indicates that the patient has CADASIL. The other critical point is to analyze all the exons of the NOTCH3 gene to find the mutation that causes the pathology. In our systematic review we have considered these two analyzes among others to identify a pathogenic mutation.
2. Results
Of the 244 studies reviewed, 16 fulfilled the inclusion/exclusion criteria and 34 index cases were described, with 25 different mutations. Twelve out of the 18 patients were female (67%) and six were male (33%); 17 of the total number of patients were European (50%), 14 were Asian (41%), two patients were from Oceania and one was North-American (Table 1).
Table 1.
Mutations found that do not affect cysteine amino acids.
| Number | Mutation | Replaced | Substitution | Exon | Sequencing | MAF (ExAC) | MAF (1000 Genomes) | Author |
|---|---|---|---|---|---|---|---|---|
| 1 | p.R61W | Arg | Trp | 2 | 33 exons | 0.00007471 | - | Brass [23] |
| 2 | p.R75P | Arg | Pro | 3 | Exon: 3, 4, 11, 18; intron | 0 | 0 | Kim [24] |
| 3 | p.R75P | Arg | Pro | 3 | Exon: 3, 4, 11, 18; intron | 0 | 0 | Kim [24] |
| 4 | p.R75P | Arg | Pro | 3 | Exon: 3, 4, 11, 18; intron | 0 | 0 | Kim [24] |
| 5 | p.R75P | Arg | Pro | 3 | 33 exons, promotor | 0 | 0 | Mizuno [25] |
| 6 | p.R75P | Arg | Pro | 3 | 33 exons, promotor | 0 | 0 | Mizuno [25] |
| 7 | p.R75P | Arg | Pro | 3 | Exon: 2–24 | 0 | 0 | Wang [26] |
| 8 | p.D80G | Asp | Gly | 3 | 33 exons | 0 | 0 | Wollenweber [27] |
| 9 | p.R107W | Arg | Trp | 3 | Exon: 2–23 | 0.00001750 | - | Ungaro [28] |
| 10 | p.G149V | Gly | Val | 4 | 33 exons, intron | 0 | 0 | Ge [29] |
| 11 | p.Q151E | Gln | Glu | 4 | Exon: 2–23 | 0.00005703 | - | Ungaro [28] |
| 12 | p.Q151E | Gln | Glu | 4 | Exon: 2–6, 8, 11, 14, 18, 19, 22, 23; intron | 0.00005703 | - | Ampuero [30] |
| 13 | p.H170R | His | Arg | 4 | Exon: 2–6, 8, 11, 14, 18, 19, 22, 23; intron | 0.001917 | 0.0014 | Ampuero [30] |
| 14 | p.H170R | His | Arg | 4 | Exon: 2, 3, 4, 11, 18, 19 | 0.001917 | 0.0014 | Roy [31] |
| 15 | p.A198T | Ala | Thr | 4 | Exon: 2–23 | 0.00002513 | - | Ungaro [28] |
| 16 | p.A202V | Ala | Val | 4 | Exon: 2, 3, 4, 11, 18, 19 | 0.00001672 | - | Roy [31] |
| 17 | p.R207H | Arg | His | 4 | Exon: 2–23 | 0.00001664 | 0.0004 | Ungaro [28] |
| 18 | p.R213K | Arg | Lys | 4 | - | 0 | 0 | Uchino [32] |
| 19 | p.R213K | Arg | Lys | 4 | 33 exons | 0 | 0 | Santa [33] |
| 20 | p.V237M | Val | Met | 5 | - | 0.0002239 | 0.0008 | Uchino [32] |
| 21 | p.V252M | Val | Met | 5 | Exon: 2–23; intron | 0.00002493 | - | Abramycheva [34] |
| 22 | p.E309K | Glu | Lys | 6 | Exon: 2–23 | 0 | 0 | Ungaro [28] |
| 23 | p.S497L | Ser | Leu | 9 | Exon: 2–23; intron | 0.01234 | 0.0074 | Abramycheva [34] |
| 24 | p.T577A | Thr | Ala | 11 | - | 0.00001669 | 0 | Ferreira [35] |
| 25 | p.R592S | Arg | Ser | 11 | Exon: 2–23 | 0.00006714 | - | Ungaro [28] |
| 26 | p.V644D | Val | Asp | 12 | Exon: 2–23 | 0.0007013 | 0.0002 | Ungaro [28] |
| 27 | p.S978R | Ser | Arg | 18 | - | 0.0004606 | 0.0004 | Ferreira [35] |
| 28 | p.A1020P | Ala | Pro | 19 | - | 0.07318 | 0.110 | Scheid [36] |
| 29 | p.A1020P | Ala | Pro | 19 | - | 0.07318 | 0.110 | Scheid [36] |
| 30 | p.T1098S | Thr | Ser | 20 | Exon: 2–24 | 0 | 0 | Wang [26] |
| 31 | p.H1133Q | His | Gln | 21 | Exon: 2–23, intron | 0.01022 | 0.0030 | Abramycheva [34] |
| 32 | p.H1235L | His | Leu | 22 | Exon: 2–23 | 0.003990 | 0.0012 | Ungaro [28] |
| 33 | p.L1515P | Leu | Pro | 25 | 33 exons, intron | 0 | 0 | Fouillade [37] |
| 34 | p.V1762M | Val | Met | 29 | 33 exons | 0.0002146 | - | Bersano [38] |
This table shows the main genetic features of the 34 index patients: mutation, amino acid replaced, amino acid substituent, exon where mutation is located, exons sequenced, minor allele frequency (MAF) according to Exome Aggregation Consortium (ExAC), MAF according to 1000 Genomes Project, author and reference.
2.1. Mutation Features
Twenty-five different cysteine-sparing NOTCH3 missense mutations were identified from the 34 patients described in the 18 papers analyzed (Table 1). A complete evaluation of the NOTCH3 exons was carried out in 29% (8 out of 28) of the patients; therefore, in the other cases it was not possible to rule out mutations involving cysteine residues. The mutations with the 33 exons analyzed were: p.R61W [23]; p.R75P [25]; p.D80G [27]; p.G149V [29]; p.R213K [33]; p.L1515P [37]; and p.V1762M [38] (Table 1).
Amino acid changes involving arginine were present in 44% of the patients, while those involving proline were present in 26%. Only two mutations were found in exons that do not encode for the ECD: p.L1515P [37] and p.V1762M [38], which encoded for the ICD.
The search for all mutations in Exome Aggregation Consortium (ExAC) and the 1000 Genomes Project showed that only p.H1235L [28], p.H170R [30,31], p.S497L [34], p.A1020P [36], and p.H1133Q [34] were polymorphisms or subpolymorphic variants, as their MAF was >0.1%.
2.2. Clinical Features
Of the 34 probands described in the 18 articles, 33% presented cardiovascular risk factors such as smoking, hypertension, and/or dyslipidemia. Clinical symptoms included migraine (93%), stroke (100%), seizures (60%), psychiatric disturbances (100%), pseudobulbar palsy (80%), and dementia (94%), and gait disturbance was specified in four cases. A family history of stroke and/or dementia was observed in 96% of the probands (Table 2).
Table 2.
Clinical and demographic data of the patients with cysteine-sparing NOTCH3 mutations.
| Numb. | Mutation | Origin | Sex | Smoker (n=7) | HT (n = 15) | DM (n = 13) | Dyslip (n = 11) | Clin. Onset (Years) | Migraine (n = 15) | Stroke (n =17) | Seizure (n = 5) | Psych. Disturb (n = 8) | Pseudb. Palsy (n = 5) | Demen (n = 18) | Gait Distur (n = 4) | FH a (n = 25) | LETP b (n = 11) | GOM (n = 10) | Author |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | p.R61W | USA | - | Yes | No | No | Yes | 20 | Yes | Yes | - | - | - | - | - | Yes | No | * | Brass |
| 2 | p.R75P | Korea | M | - | Yes | No | No | 53 | - | Yes | - | - | - | Yes | - | Yes | - | Yes | Kim |
| 3 | p.R75P | Korea | F | No | No | No | No | 47 | - | Yes | - | - | - | - | - | Yes | - | - | Kim |
| 4 | p.R75P | Korea | M | No | Yes | No | No | 65 | - | Yes | - | - | - | Yes | - | Yes | - | - | Kim |
| 5 | p.R75P | Japan | F | - | - | - | - | - | - | Yes | Yes | Yes | Yes | - | - | Yes | No | - | Mizuno |
| 6 | p.R75P | Japan | F | - | - | - | - | - | - | Yes | Yes | - | Yes | - | - | Yes | No | Yes | Mizuno |
| 7 | p.R75P | China | M | - | - | - | - | 34 | - | Yes | - | Yes | - | - | - | No | No | Yes | Wang |
| 8 | p.D80G | Germany | F | No | No | No | No | - | No | Yes | - | Yes | - | Yes | Yes | Yes | Yes | * | Wollenweber |
| 9 | p.R107W | Germany | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 10 | p.G149V | China | F | No | No | No | No | 39 | - | Yes | - | - | - | - | - | Yes | No | - | Ge |
| 11 | p.Q151E | Italy | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 12 | p.Q151E | Spain | - | - | - | - | - | - | - | Yes | - | - | - | - | - | - | - | - | Ampuero |
| 13 | p.H170R | Spain | - | - | - | - | - | - | - | Yes | - | - | - | - | - | - | - | - | Ampuero |
| 14 | p.H170R | Oceania | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Roy |
| 15 | p.A198T | Italy | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 16 | p.A202V | Oceania | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Roy |
| 17 | p.R207H | Italy | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 18 | p.R213K | Japan | M | - | No | No | No | 63 | Yes | Yes | No | - | Yes | Yes | Yes | Yes | - | - | Uchino |
| 19 | p.R213K | Japan | M | - | No | No | No | 10 | Yes | Yes | - | Yes | Yes | Yes | Yes | Yes | - | Yes | Santa |
| 20 | p.V237M | Japan | F | - | No | No | No | 71 | - | Yes | No | - | No | Yes | Yes | Yes | - | - | Uchino |
| 21 | p.V252M | Russia | - | - | No | No | - | - | - | - | - | - | - | - | - | - | - | - | Abramycheva |
| 22 | p.E309K | Italy | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 23 | p.S497L | Russia | - | - | No | No | - | - | - | - | - | - | - | - | - | - | - | - | Abramycheva |
| 24 | p.T577A | Portugal | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Ferreira |
| 25 | p.R592S | Italy | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 26 | p.V644D | Italy | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 27 | p.S978R | Portugal | F | - | - | - | - | - | - | Yes | Yes | Yes | - | Yes | - | - | - | - | Ferreira |
| 28 | p.A1020P | Germany | F | - | Yes | - | - | Adolesc | Yes | - | - | - | - | Yes | - | Yes | No | Yes | Scheid |
| 29 | p.A1020P | Germany | F | - | Yes | - | - | - | - | - | - | Yes | - | - | - | Yes | No | No | Scheid |
| 30 | p.T1098S | China | M | - | - | - | - | 39 | - | Yes | - | Yes | - | Yes | - | Yes | No | Yes | Wang |
| 31 | p.H1133Q | Russia | - | - | No | No | - | - | - | - | - | - | - | - | - | - | - | - | Abramycheva |
| 32 | p.H1235L | Italy | - | - | - | - | - | - | Yes | - | - | - | - | Yes | - | Yes | - | - | Ungaro |
| 33 | p.L1515P | France | F | No | No | No | No | 35 | Yes | Yes | - | - | - | - | - | Yes | No | No | Fouillade |
| 34 | p.V1762M | Italy | F | Yes | - | - | Yes | Childhd | Yes | - | - | Yes | - | No | - | Yes | No | No | Bersano |
| Perc | - | - | - | 29% | 27% | 0% | 18% | - | 93% | 100% | 60% | 100% | 80% | 94% | 100% | 96% | 9% | 40% | - |
Numb: Number; Smoker: current/past smoker; HT: hypertension; DM: diabetes mellitus; Dyslip: dyslipidaemia; Clin. Onset: Clinical Onset; Psych.distur: psychiatric disturbance; Pseudb.palsy: pseudobulbar palsy; Dem.: dementia; Gait distur: gait disturbance; GOM: Granular osmiophilic material; M: male; F: female; Adolesc: adolescence; Childhd: childhood; Perc: percentage; a FH: family history; b LE TP: leukoencephalopathy with anterior temporal pole involvement; * GOM found in proband’s siblings, who were symptomatic.
The neuroimaging data revealed that all probands had severe WMH, although 91% did not have anterior temporal pole involvement.
GOM deposits were found in six out of nine probands (67%). In two of these cases, they were also found in the proband’s siblings. Specifically, mutations with GOM were: p.R61W [23] in the symptomatic proband’s siblings, who had the same mutation; p.R75P [24,25,26]; p.D80G [30] in the symptomatic proband’s siblings, who had the same mutation; p.R213K [33], p.A1020P [36] (in one of the two probands); and p.T1098S [26]. However, three out of nine probands had cysteine-sparing NOTCH3 missense mutations not associated with GOM: p.A1020P [33] (in one of two probands), p.L1515P [34] and p.V1762M [35]; the latter two were mutations located in the ICD.
A summary of the main characteristics of cysteine-sparing NOTCH3 missense mutations is given in Table 3.
Table 3.
Summary of the main characteristics of cysteine-sparing NOTCH3 missense mutations.
| Number | Mutation | Typical Clinical CADASIL Syndrome | WMH | Whole Exon Analysis | Mutation | GOM | Author |
|---|---|---|---|---|---|---|---|
| 1 | p.R61W | Yes | Yes | Yes | Yes | * | Brass |
| 2 | p.R75P | Yes | Yes | No | Yes | Yes | Kim |
| 3 | p.R75P | Yes | Yes | No | Yes | - | Kim |
| 4 | p.R75P | Yes | Yes | No | Yes | - | Kim |
| 5 | p.R75P | Yes | Yes | Yes | Yes | - | Mizuno |
| 6 | p.R75P | Yes | Yes | Yes | Yes | Yes | Mizuno |
| 7 | p.R75P | Yes | Yes | No | Yes | Yes | Wang |
| 8 | p.D80G | Yes | Yes | Yes | Yes | * | Wollenweber |
| 9 | p.R107W | Yes | Yes | No | Yes | - | Ungaro |
| 10 | p.G149V | Yes | Yes | Yes | Yes | - | Ge |
| 11 | p.Q151E | Yes | Yes | No | Yes | - | Ungaro |
| 12 | p.Q151E | Yes | Yes | No | Yes | - | Ampuero |
| 13 | p.H170R | Yes | Yes | No | No | - | Ampuero |
| 14 | p.H170R | Yes | NS | No | No | - | Roy |
| 15 | p.A198T | Yes | Yes | No | Yes | - | Ungaro |
| 16 | p.A202V | Yes | NS | No | Yes | - | Roy |
| 17 | p.R207H | Yes | Yes | No | Yes | - | Ungaro |
| 18 | p.R213K | Yes | Yes | NS | Yes | - | Uchino |
| 19 | p.R213K | Yes | Yes | Yes | Yes | Yes | Santa |
| 20 | p.V237M | Yes | Yes | NS | Yes | - | Uchino |
| 21 | p.V252M | Yes | Yes | No | Yes | - | Abramycheva |
| 22 | p.E309K | Yes | Yes | No | Yes | - | Ungaro |
| 23 | p.S497L | Yes | NS | No | No | - | Abramycheva |
| 24 | p.T577A | NS | NS | NS | Yes | - | Ferreira |
| 25 | p.R592S | Yes | Yes | No | Yes | - | Ungaro |
| 26 | p.V644D | Yes | Yes | No | Yes | - | Ungaro |
| 27 | p.S978R | Yes | Yes | NS | Yes | - | Ferreira |
| 28 | p.A1020P | Yes | Yes | NS | No | Yes | Scheid |
| 29 | p.A1020P | Yes | Yes | NS | No | No | Scheid |
| 30 | p.T1098S | Yes | Yes | No | Yes | Yes | Wang |
| 31 | p.H1133Q | Yes | NS | No | No | - | Abramycheva |
| 32 | p.H1235L | Yes | Yes | No | No | - | Ungaro |
| 33 | p.L1515P | Yes | Yes | Yes | Yes | No | Fouillade |
| 34 | p.V1762M | Yes | Yes | Yes | Yes | No | Bersano |
WMH: white matter hyperintensities; GOM: Granular osmiophilic material; NS: not specified; * GOM were found in proband’s sibling, who was symptomatic.
The table not only summarizes the main characteristics of cysteine-sparing NOTCH3 missense mutations, but also shows the five criteria required for mutations to be considered potentially pathogenic: type of mutation, typical clinical CADASIL syndrome, diffuse WMH, whole exon analysis, mutations that were not polymorphisms and GOM deposition, and the author.
Therefore, we considered that p.R61W [23], p.R75P [25], p.D80G [27], andp.R213K [33] could be potentially pathogenic mutations, because they were associated with typical clinical CADASIL syndrome and extensive WMH in MRI, the 33 NOTCH3 exons were analyzed and no other potential pathogenic mutations were found; they had an MAF < 0.1% (ExAC, 1000 Genomes Project) (confirming that they are not common polymorphisms) and GOM deposits were observed in the skin biopsy. (The criteria for considering mutations as potentially pathogenic are explained in the Material and Methods section).
In relation to co-segregation of these potentially pathogenic mutations, Mizuno et al. [25] studied the relatives of probands with the p.R75P mutation. Those with clinical CADASIL and typical MRI findings had the same p.R75P mutation, and the asymptomatic relative did not carry the mutation. For p.D80G, two sisters and one brother with clinical CADASIL typical syndrome and extensive WMH shared the same mutation, and the other asymptomatic brother did not share it. In addition, the symptomatic brother had GOM in his skin biopsy, unlike the asymptomatic one. Segregation analysis was not performed for p.R61W or p.R213K.
Of the ten probands that carried one of these four mutations, 80% were Asian, 60% had the p.R75P mutation and just one of the probands (p.D80G) had temporal pole involvement.
Polyphen-2 analysis indicated that p.R61W, p.D80G, and p.R75P could be possible damaging mutations (Table 4).
Table 4.
Polyphen-2 results and domain localization of the selected mutations.
| Mutation | Score | Confidence | Domain | Prediction |
|---|---|---|---|---|
| R61W | 0.773 | Sensitivity: 0.76 | EGF-like 1 | Possibly damaging |
| Especificity: 0.86 | ||||
| R75P | 0.884 | Sensitivity: 0.71 | EGF-like 1 | Possibly damaging |
| Especificity: 0.89 | ||||
| D80G | 0.694 | Sensitivity: 0.78 | EGF-like 2 | Possibly damaging |
| Especificity: 0.85 | ||||
| R213K | 0.171 | Sensitivity: 0.89 | EGF-like 5 | Benign |
| Especificity: 0.72 |
3. Discussion
In this systematic review, we conducted a thorough search for cysteine-sparing NOTCH3 missense mutations in patients with typical clinical CADASIL syndrome in order to determine whether these mutations could be considered pathogenic and to describe the patients that carry this type of mutation.
We excluded nonsense and insertion/deletion mutations because they lead to a numerical cysteine alteration, as Rutten et al. described in a previous article [8].
The literature and gene databases describe 34 patients with 25 different cysteine-sparing NOTCH3 missense mutations. These 25 mutations included two in exons that encode for the ICD. The patients presented typical clinical CADASIL syndrome with migraine, psychiatric disturbances, early onset of stroke and/or dementia, and diffuse WMH in the MRI.
We believe there are four possible pathogenic cysteine-sparing NOTCH3 missense mutations: p.R61W [23], p.R75P [25], p.D80G [27], and p.R213K [33]. These mutations were located in EGF-like repeats. We think they are possible pathogenic mutations because patients presented typical clinical CADASIL syndrome and MRI profiles, no additional mutations were found in the gene, they do not represent low-frequency polymorphisms or subpolymorphic variants, and GOM deposits were found in their skin biopsy. A familial co-segregation was observed for p.R61W [23], p.D80G [27] and p.R75P [8,25], but not for p.R213K, because symptomatic relatives were not evaluated. In addition, Polyphen-2 analysis predicted that p.R61W, p.D80G, and p.R75P, but not p.R213K could be possibly damaging mutations. Taking into account the information published in the literature these mutations can be considered possible pathogenic mutations.
Of the 10 probands with these four mutations and CADASIL patients with cysteine mutations, non-significant clinical or demographic differences were observed in migraine, stroke, or dementia. However, WMH in the anterior part of the temporal pole were practically absent in these cysteine-sparing NOTCH3 missense mutations. As mentioned previously, WMH in the temporal pole has a reported 89–95% sensitivity and 80–86% specificity for CADASIL in Caucasian patients [14] and a prevalence of 44.8–71% [16] in Asian populations. Therefore, considering that 80% of patients in the review were Asians, at least four to five patients with temporal pole involvement should be expected in CADASIL due to cysteine mutations. Therefore, we believe that absent temporal pole involvement could be a characteristic of cysteine-sparing NOTCH3 missense mutations.
Rutten et al. performed an extensive search of all types of NOTCH3 mutations in order to provide information on their interpretation [8]. Regarding the cysteine-sparing NOTCH3 missense mutations, they studied 10 articles with 11 different mutations corresponding to p.R61W, p.R75P, p.Q151E, p.H170R, p.A202V, p.R213K, p.V237M, p.T577A, p.S978R, p.A1020P, and p.Y1098S. They concluded that these mutations were not associated with CADASIL, but p.R75P fulfilled all their criteria: (1) a complete evaluation of the NOTCH3 gene to rule out a typical cysteine-altering mutation in another exon; (2) the mutations were not polymorphisms; (3) a familial co-segregation was proven; and (4) the clinical diagnosis was confirmed through the observation of GOM deposits in the skin biopsy.
This systematic review updates the interesting work of Rutten et al. [8], taking into consideration three of the four criteria that they applied, excluding the lack of familiar segregation criterion [8]. We were able to add new information and conclusions stemming from the publication of new articles with a total of 25 different mutations, in contrast to the 11 found previously by Rutten et al. Of the four mutations considered potentially pathogenic in our systematic review, p.R75P was already mentioned by Rutten et al. in their reference to Mizuno et al. In this review, we added information from two new articles with a total of four new patients. Regarding p.R213K, Rutten et al. did not consider this mutation as potentially pathogenic because familial co-segregation was not proven. However, we did not define familial co-segregation as an exclusion criterion because it is not always possible to analyze relatives or there may be no live relatives.
The genetic analysis technique was not specified for p.R61W, so Rutten et al. suggested the possibility of using a technique that would not detect other mutations.
Finally, p.D80G was not studied previously by Rutten et al., but applying their criteria and ours, it is a pathogenic mutation.
In several cases, the conclusions of these articles are supported by bioinformatics and functional studies that have revealed the possible pathogenic effect of the mutations [37,39]. Mizuno et al. suggested that changes in NOTCH3 structure could be related to the replacement of an amino acid by proline (without cysteine involvement), because three-dimensional structure studies have shown that it helps to stabilize the β-sheet [25]. This may lead to a conformational change in the protein. Another study observed that several of these atypical mutations were associated with structural changes in the NOTCH3 receptor [39], similar to the mechanism observed for typical mutations involving cysteines. Thus, in addition to changes in the number of cysteines, mutations involving other amino acids could also lead to misfolding of the receptor. In the same way, in vitro studies with p.R75P, p.D80G, and delta88–91 have shown significantly enhanced aggregation similar to that of cysteine mutations [27].
With regard to limitations, we should consider the low number of cases reported so far, as well as the high number of Asian reports that could skew the frequencies of different features in this population. In order to avoid this last limitation, we compared some important characteristics (such as the temporal pole affectation) to their frequency in the Asian population.
Another limitation is that the analyses of p.R213K [33] to rule out other mutations in the NOTCH3 gene were performed using single-strand conformational polymorphism (SSCP) analysis, which does not detect other mutations, unlike the standard and reliable Sanger sequencing. In the case ofp.R61W, the genetic analysis technique was not specified.
Furthermore, we did not consider a small deletion (delta88–91) that does not directly involve a cysteine residue in the ECD of NOTCH3 [39]. This would have enriched our review, but could have compromised the reliability of our conclusions. This deletion was reported in an Italian family with typical clinical CADASIL syndrome and diffuse WMH, without anterior temporal pole involvement. In addition, the proband’s skin biopsy showed GOM deposits. This mutation was also observed in the only symptomatic relative. The asymptomatic relatives did not carry any NOTCH3 mutations.
Another limitation was combining ECD and ICD mutations in our review. The aim here was to review typical clinical CADASIL syndrome with cysteine-sparing NOTCH3 missense mutations to determine whether they were associated with CADASIL. In any case, ICD mutations were not considered pathogenic because GOM deposits were absent.
4. Materials and Methods
An extensive literature search was performed up to November 2016 on PubMed, Google Scholar, EMBASE, LILACS, Trip Database, and the Cochrane Library, as well as in “The Human Gene Mutation Database” (http://www.hgmd.cf.ac.uk/ac/index.php), with the key words: “CADASIL cysteine”, “NOTCH3 cysteine”, and “NOTCH3 polymorphism”, in order to collect data about cysteine-sparing NOTCH3 missense mutations that were associated with typical clinical CADASIL syndrome.
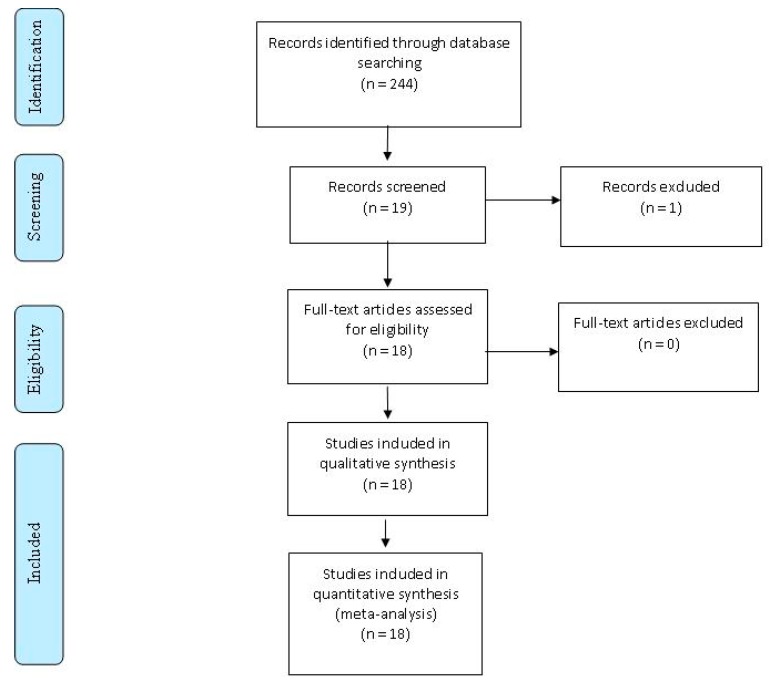
A total of 244 studies were found. Probands had typical clinical CADASIL syndrome and a cysteine-sparing NOTCH3 missense mutation (Figure 1) in 18 articles. Studies that analyzed other diseases or endophenotypes, such as patients with diffuse WMH instead of a typical clinical CADASIL syndrome, were excluded.
Figure 1.
Flow diagram of the literature review. This detailed flow diagram depicts the search and selection processes.
Only those articles that described missense mutations confirming an absence of cysteine involvement were selected. Therefore, nonsense, intronic, insertion, and deletion mutations were excluded because they can lead to cysteine number alterations, through the shortening of the protein (in the case of nonsense mutations) [40,41] and or through changes in the open reading frame (in the case of insertion-deletions) as described previously [8,42].
In addition, we excluded several studies describing mutations that were already known polymorphisms, suggesting a negative pathogenic role, as well as an article by Ueda et al. [16], as it was difficult to identify who was the proband and who was the relative of the patient with the typical clinical CADASIL syndrome.
We collected data on epidemiological, clinical, genetic, and neuroimaging characteristics, such as temporal pole involvement, as well as existence of skin biopsies. We used the IBM SPSS statistics software package, version 22, for the frequencies analysis.
Mutations were considered potentially pathogenic when they fulfilled the following criteria: (1) the patients had typical clinical CADASIL syndrome; (2) the patients had diffuse WMH in MRI; (3) the study analyzed the 33 exons of the NOTCH3 gene to rule out other pathogenic mutations; (4) the mutation had a MAF < 0.1%, since CADASIL is a low prevalence disease and it is therefore necessary to rule out low-frequency polymorphisms (MAF < 5%) and subpolymorphic variants (frequency 0.1–1.0%) that could represent rare variants and not disease-causing mutations. To establish the MAF, we used the ExAC database, containing genetic information on 60,706 unrelated individuals (http://exac.broadinstitute.org), and the 1000 Genomes Project database, containing the genetic information of 1000 individuals from different ethnicities (http://www.internationalgenome.org/1000-genomes-browsers/); and lastly (5) the patient had GOM deposits in the skin biopsy because this represents a specificity of 100% for CADASIL diagnosis. Finally we analyzed with Polyphen-2 tool the pathogenic prediction of the selected mutations that we considered potentially pathogenic (http://genetics.bwh.harvard.edu/pph2/).
5. Conclusions
We believe that the cysteine-sparing NOTCH3 missense mutations p.R61W, p.R75P, p.D80G, and p.R213K could be potentially pathogenic, and we found familial co-segregation forp.R61W, p.R75P, and p.D80G. Different studies confirmed an altered NOTCH3 structure or proaggregatory properties due to these changes. Thus, cysteine-sparing NOTCH3 missense mutations are associated with typical clinical CADASIL syndrome and a typical MRI profile, mostly without anterior temporal pole involvement.
Further studies are necessary to clarify the role of these cysteine-sparing NOTCH3 missense mutations in CADASIL to be able to interpret them properly to reach a correct diagnosis. Likewise, an improved understanding of this kind of mutation would be important to clarify whether other mechanisms, apart from disulfide bond-mediated misfolding due to an odd number of cysteines, play a major role in the development of CADASIL, such as a receptor misfolding produced by other amino acids.
Acknowledgments
The Stroke Genetics and Pharmacogenomics Laboratory is part of the International Stroke Genetics Consortium (ISGC, www.strokegenetics.com) and coordinates the Spanish Stroke Genetics Consortium (Genestroke, www.genestroke.com). Israel Fernández-Cadenas is supported by the Miguel Servet programme (CP12/03298), Instituto de Salud Carlos III. This study was funded by the Miguel Servet grant (Pharmastroke project: CP12/03298). The Neurovascular Research Laboratory is supported by the INVICTUS Network of the Spanish Ministry of Health (Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III). The IMIM-Hospital del Mar Neurology Service is supported in part by the Spanish Ministry of Health (Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III FEDER, RD12/0042/0020).
Abbreviations
| CADASIL | Cerebral autosomal dominant arteriopathy with subcortical infarcts and Leukoencephalopathy |
| ECD | Extracellular domain |
| EGF | Epidermal growth factor |
| ICD | Intracellular domain |
| MRI | Magnetic resonance imaging |
| WMH | White matter hyperintensities |
| GOM | Granular osmiophilic material |
| MAF | Minor allele frequency |
| ExAC | Exome aggregation consortium |
Author Contributions
Elena Muiño and Israel Fernández-Cadenas conceived and wrote the paper; Cristina Gallego-Fabrega, Natalia Cullell, Caty Carrera, Nuria Torres, Jurek Krupinski, Jaume Roquer, and Joan Montaner contributed by correcting the paper in their respective fields of biotechnology, biology, genetics and neurology.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tournier-Lasserve E., Joutel A., Melki J., Weissenbach J., Lathrop G.M., Chabriat H., Mas J.L., Cabanis E.A., Baudrimont M., Maciazek J., et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy maps to chromosome 19q12. Nat. Genet. 1993;3:256–259. doi: 10.1038/ng0393-256. [DOI] [PubMed] [Google Scholar]
- 2.Joutel A., Corpechot C., Ducros A., Vahedi K., Chabriat H., Mouton P., Alamowitch S., Domenga V., Cecillion M., Marechal E., et al. NOTCH3 mutations in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), a mendelian condition causing stroke and vascular dementia. Ann. N. Y. Acad. Sci. 1997;826:213–217. doi: 10.1111/j.1749-6632.1997.tb48472.x. [DOI] [PubMed] [Google Scholar]
- 3.Dziewulska D., Lewandowska E. Pericytes as a new target for pathological processes in CADASIL. Neuropathology. 2012;32:515–521. doi: 10.1111/j.1440-1789.2011.01290.x. [DOI] [PubMed] [Google Scholar]
- 4.Duering M., Karpinska A., Rosner S., Hopfner F., Zechmeister M., Peters N., Kremmer E., Haffner C., Giese A., Dichgans M., et al. Co-aggregate formation of CADASIL-mutant NOTCH3: A single-particle analysis. Hum. Mol. Genet. 2011;20:3256–3265. doi: 10.1093/hmg/ddr237. [DOI] [PubMed] [Google Scholar]
- 5.Meng H., Zhang X., Yu G., Lee S.J., Chen Y.E., Prudovsky I., Wang M.M. Biochemical characterization and cellular effects of CADASIL mutants of NOTCH3. PLoS ONE. 2012;7:e44964. doi: 10.1371/journal.pone.0044964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opherk C., Duering M., Peters N., Karpinska A., Rosner S., Schneider E., Bader B., Giese A., Dichgans M. CADASIL mutations enhance spontaneous multimerization of NOTCH3. Hum. Mol. Genet. 2009;18:2761–2767. doi: 10.1093/hmg/ddp211. [DOI] [PubMed] [Google Scholar]
- 7.Cognat E., Baron-Menguy C., Domenga-Denier V., Cleophax S., Fouillade C., Monet-Leprêtre M., Dewerchin M., Joutel A. Archetypal Arg169Cys mutation in NOTCH3 does not drive the pathogenesis in cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy via a loss-of-function mechanism. Stroke. 2014;45:842–849. doi: 10.1161/STROKEAHA.113.003339. [DOI] [PubMed] [Google Scholar]
- 8.Rutten J.W., Haan J., Terwindt G.M., van Duinen S.G. Interpretation of NOTCH3 mutations in the diagnosis of CADASIL. Expert Rev. Mol. Diagn. 2014;14:593–603. doi: 10.1586/14737159.2014.922880. [DOI] [PubMed] [Google Scholar]
- 9.Joutel A. Transgenic and knock-out mice to probe function and dysfunction of the mutated gene, NOTCH3, in the cerebrovasculature. Bioessays. 2011;33:73–80. doi: 10.1002/bies.201000093. [DOI] [PubMed] [Google Scholar]
- 10.Chabriat H., Vahedi K., Iba-Zizen M.T., Joutel A., Nibbio A., Nagy T.G., Krebs M.O., Julien J., Dubois B., Ducrocq X., et al. Clinical spectrum of CADASIL: A study of 7 families. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Lancet. 1995;346:934–939. doi: 10.1016/S0140-6736(95)91557-5. [DOI] [PubMed] [Google Scholar]
- 11.Moreton F.C., Razvi S.S.M., Davidson R., Muir K.W. Changing clinical patterns and increasing prevalence in CADASIL. Acta Neurol. Scand. 2014;130:197–203. doi: 10.1111/ane.12266. [DOI] [PubMed] [Google Scholar]
- 12.Adib-Samii P., Brice G., Martin R.J., Markus H.S. Clinical spectrum of CADASIL and the effect of cardiovascular risk factors on phenotype: Study in 200 consecutively recruited individuals. Stroke. 2010;41:630–634. doi: 10.1161/STROKEAHA.109.568402. [DOI] [PubMed] [Google Scholar]
- 13.Cumurciuc R., Guichard J.-P., Reizine D., Gray F., Bousser M.G., Chabriat H. Dilation of Virchow-Robin spaces in CADASIL. Eur. J. Neurol. 2006;13:187–190. doi: 10.1111/j.1468-1331.2006.01113.x. [DOI] [PubMed] [Google Scholar]
- 14.O’Sullivan M., Jarosz J.M., Martin R.J., Deasy N., Powell J.F., Markus H.S. MRI hyperintensities of the temporal lobe and external capsule in patients with CADASIL. Neurology. 2001;56:628–634. doi: 10.1212/WNL.56.5.628. [DOI] [PubMed] [Google Scholar]
- 15.Markus H.S., Martin R.J., Simpson M.A., Dong Y.B., Ali N., Crosby A.H., Powell J.F. Diagnostic strategies in CADASIL. Neurology. 2002;59:1134–1138. doi: 10.1212/WNL.59.8.1134. [DOI] [PubMed] [Google Scholar]
- 16.Ueda A., Ueda M., Nagatoshi A., Hirano T., Ito T., Arai N., Uyama E., Mori K., Nakamura M., Shinriki S., et al. Genotypic and phenotypic spectrum of CADASIL in Japan: The experience at a referral center in Kumamoto University from 1997 to 2014. J. Neurol. 2015;262:1828–1836. doi: 10.1007/s00415-015-7782-8. [DOI] [PubMed] [Google Scholar]
- 17.Ishiko A., Shimizu A., Nagata E., Takahashi K., Tabira T., Suzuki N. NOTCH3 ectodomain is a major component of granular osmiophilic material (GOM) in CADASIL. Acta Neuropathol. 2006;112:333–339. doi: 10.1007/s00401-006-0116-2. [DOI] [PubMed] [Google Scholar]
- 18.Chabriat H., Joutel A., Dichgans M., Tournier-Lasserve E., Bousser M.G. Cadasil. Lancet Neurol. 2009;8:643–653. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- 19.Joutel A., Favrole P., Labauge P., Chabriat H., Lescoat C., Andreux F., Domenga V., Cécillon M., Vahedi K., Ducros A., et al. Skin biopsy immunostaining with a NOTCH3 monoclonal antibody for CADASIL diagnosis. Lancet. 2001;358:2049–2051. doi: 10.1016/S0140-6736(01)07142-2. [DOI] [PubMed] [Google Scholar]
- 20.Shan S., He X., He L., Wang M., Liu C. Coexistence of congenital left ventricular aneurysm and prominent left ventricular trabeculation in a patient with LDB3 mutation: A case report. J. Med. Case Rep. 2017;11:229. doi: 10.1186/s13256-017-1405-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dastsooz H., Nemati H., Fard M.A.F., Fardaei M., Faghihi M.A. Novel mutations in PANK2 and PLA2G6genes in patients with neurodegenerative disorders: Two case reports. BMC Med. Genet. 2017;18:87. doi: 10.1186/s12881-017-0439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendioroz M., Fernández-Cadenas I., del Río-Espinola A., Rovira A., Solé E., Fernández-Figueras M.T., Garcia-Patos V., Sastre-Garriga J., Domingues-Montanari S., Alvarez-Sabín J., et al. A missense HTRA1 mutation expands CARASIL syndrome to the Caucasian population. Neurology. 2010;75:2033–2035. doi: 10.1212/WNL.0b013e3181ff96ac. [DOI] [PubMed] [Google Scholar]
- 23.Frosch M.P. A 46-year-old man with migraine, aphasia, and hemiparesis and similarly affected family members. N. Engl. J. Med. 2009;360:1656–1665. doi: 10.1056/NEJMcpc0810839. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y., Choi E.J., Choi C.G., Kim G., Choi J.H., Yoo H.W., Kim J.S. Characteristics of CADASIL in Korea: A novel cysteine-sparing NOTCH3 mutation. Neurology. 2006;66:1511–1516. doi: 10.1212/01.wnl.0000216259.99811.50. [DOI] [PubMed] [Google Scholar]
- 25.Mizuno T., Muranishi M., Torugun T., Tango H., Nagakane Y., Kudeken T., Kawabe K., Oshima F., Yaoi T., Itoh K., et al. Two Japanese CADASIL families exhibiting NOTCH3 mutation R75P not involving cysteine residue. Intern. Med. 2008;47:2067–2072. doi: 10.2169/internalmedicine.47.1391. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z., Yuan Y., Zhang W., Lv H., Hong D., Chen B., Liu Y., Luan X., Xie S., Wu S. NOTCH3 mutations and clinical features in 33 mainland Chinese families with CADASIL. J. Neurol. Neurosurg. Psychiatry. 2011;82:534–539. doi: 10.1136/jnnp.2010.209247. [DOI] [PubMed] [Google Scholar]
- 27.Wollenweber F.A., Hanecker P., Bayer-Karpinska A., Malik R., Bäzner H., Moreton F., Muir K.W., Müller S., Giese A., Opherk C., et al. Cysteine-sparing CADASIL mutations in NOTCH3 show proaggregatory properties in vitro. Stroke. 2015;46:786–792. doi: 10.1161/STROKEAHA.114.007472. [DOI] [PubMed] [Google Scholar]
- 28.Ungaro C., Mazzei R., Conforti F.L., Sprovieri T., Servillo P., Liguori M., Citrigno L., Gabriele A.L., Magariello A., Patitucci A., et al. Cadasil: Extended polymorphisms and mutational analysis of the NOTCH3 gene. J. Neurosci. Res. 2009;87:1162–1167. doi: 10.1002/jnr.21935. [DOI] [PubMed] [Google Scholar]
- 29.Ge W., Kuang H., Wei B., Bo L., Xu Z., Xu X., Geng D., Sun M. A novel cysteine-sparing NOTCH3 mutation in a Chinese family with CADASIL. PLoS ONE. 2014;9:e104533. doi: 10.1371/journal.pone.0104533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ampuero I., Alegre-Abarrategui J., Rodal I., España A., Ros R., Sendón J.L.L., Galloway E.G., Cervelló A., Caminero A.B., Zabala A., et al. On the diagnosis of CADASIL. J. Alzheimers Dis. 2009;17:787–794. doi: 10.3233/JAD-2009-1112. [DOI] [PubMed] [Google Scholar]
- 31.Roy B., Maksemous N., Smith R.A., Menon S., Davies G., Griffiths L.R. Two novel mutations and a previously unreported intronic polymorphism in the NOTCH3 gene. Mutat. Res. 2012;732:3–8. doi: 10.1016/j.mrfmmm.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Uchino M., Hirano T., Uyama Y.H.E. Cerebral Autosomal Dominant Arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) and CADASIL-like disorders in Japan. Ann. N. Y. Acad. Sci. 2002;977:273–278. doi: 10.1111/j.1749-6632.2002.tb04826.x. [DOI] [PubMed] [Google Scholar]
- 33.Santa Y., Uyama E., Chui D.H., Arima M., Kotorii S., Takahashi K., Tabira T. Genetic, clinical and pathological studies of CADASIL in Japan: A partial contribution of NOTCH3 mutations and implications of smooth muscle cell degeneration for the pathogenesis. J. Neurol. Sci. 2003;212:79–84. doi: 10.1016/S0022-510X(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 34.Abramycheva N., Stepanova M., Kalashnikova L., Zakharova M., Maximova M., Tanashyan M., Lagoda O., Fedotova E., Klyushnikov S., Konovalov R., et al. New mutations in the NOTCH3 gene in patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL) J. Neurol. Sci. 2015;349:196–201. doi: 10.1016/j.jns.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Homer V., George P.M., Toit S., Davidson J.S., Wilson C.J. Novel human pathological mutations. Hum. Genet. 2007;121:645–652. [PubMed] [Google Scholar]
- 36.Scheid R. Cysteine-sparing NOTCH3 mutations: CADASIL or CADASIL variants? Neurology. 2008;71:774–777. doi: 10.1212/01.wnl.0000324928.44694.f7. [DOI] [PubMed] [Google Scholar]
- 37.Fouillade C., Chabriat H., Riant F., Mine M., Arnoud M., Magy L., Bousser M.G., Tournier-Lasserve E., Joutel A. Activating NOTCH3 mutation in a patient with small-vessel-disease of the brain. Hum. Mutat. 2008;29:452. doi: 10.1002/humu.9527. [DOI] [PubMed] [Google Scholar]
- 38.Bersano A., Dotti M.T., Candelise L. Considerations on a mutation in the NOTCH3 gene sparing a cysteine residue: A rare polymorphism rather than a CADASIL variant. Funct. Neurol. 2012;27:247–252. [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachakis D., Tsaniras S.C., Ioannidou K., Baumann M., Kossida S. A series of NOTCH3 mutations in CADASIL, insights from 3D molecular modelling and evolutionary analyses. J. Mol. Biochem. 2014;3:97–105. [PMC free article] [PubMed] [Google Scholar]
- 40.Mazzei R., Conforti F.L., Lanza P.L., Sprovieri T., Lupo M.R., Gallo O., Patitucci A., Magariello A., Caracciolo M., Gabriele A.L., et al. A novel NOTCH3 gene mutation not involving a cysteine residue in an Italian family with CADASIL. Neurology. 2004;63:561–564. doi: 10.1212/01.WNL.0000133399.37716.84. [DOI] [PubMed] [Google Scholar]
- 41.Arboleda-Velasquez J.F., Manent J., Lee J.H., Tikka S., Ospina C., Vanderburg C.R., Frosch M.P., Rodríguez-Falcón M., Villen J., Gygi S., et al. Hypomorphic Notch 3 alleles link Notch signaling to ischemic cerebral small-vessel disease. Proc. Natl. Acad. Sci. USA. 2011;108:128–135. doi: 10.1073/pnas.1101964108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez-Cadenas I., Andreu A.L., Gamez J., Gonzalo R., Martín M.A., Rubio J.C., Arenas J. Splicing mosaic of the myophosphorylase gene due to a silent mutation in McArdle disease. Neurology. 2003;61:1432–1434. doi: 10.1212/WNL.61.10.1432. [DOI] [PubMed] [Google Scholar]