SUMMARY
Hematopoietic development requires the transcription factor GATA-2, and GATA-2 mutations cause diverse pathologies including leukemia. GATA-2-regulated enhancers regulate Gata2 expression in hematopoietic stem/progenitor cells and control hematopoiesis. The +9.5 kb enhancer activates transcription in endothelium and hematopoietic stem cells (HSCs), and its deletion abrogates HSC generation. The −77 kb enhancer activates transcription in myeloid progenitors, and its deletion impairs differentiation. Since +9.5−/− embryos are HSC-deficient, it was unclear whether the +9.5 functions in progenitors or if GATA-2 expression in progenitors solely requires −77. We further dissected the mechanisms using −77;+9.5 compound heterozygous (CH) mice. The embryonic lethal CH mutation depleted megakaryocyte-erythrocyte progenitors (MEPs). While the +9.5 suffices for HSC generation, the −77 and +9.5 must reside on one allele to induce MEPs. The −77 generated Burst Forming Unit-Erythroid through induction of GATA-1 and other GATA-2 targets. The enhancer circuits controlled signaling pathways that orchestrate a GATA factor-dependent blood development program.
Keywords: GATA-2, Enhancer, Hematopoiesis, Erythroid, Myeloid, Progenitor, Mouse model
INTRODUCTION
Genetic networks governing complex developmental processes are commonly established and maintained by master regulatory transcription factors (Orkin and Zon, 2008). When stem and progenitor cells differentiate into blood cells, GATA-2 serves this function (Tsai et al., 1994). GATA-2 also controls non-hematopoietic processes, including pregnancy (Rubel et al., 2016), neurogenesis (Zhou et al., 2000), renal function (Yu et al., 2014) and prostate cancer (Vidal et al., 2015). Gata2 nullizygous mouse embryos are severely anemic and die at ~E10.5 (Tsai et al., 1994). Studies with Gata2+/− mice revealed that GATA-2 ensures steady-state HSC levels in adults (Ling et al., 2004; Rodrigues et al., 2005). Targeted deletion of an intronic Gata2 enhancer 9.5 kb downstream of the transcription start site (+9.5) abrogated HSC genesis in the aorta-gonad-mesonephros (AGM) region, yielding ~E14 lethality (Gao et al., 2013; Johnson et al., 2012). Analysis of conditional Gata2 knockout mice yielded similar results (de Pater et al., 2013; Lim et al., 2012). In addition to regulating HSC genesis and function, GATA-2 confers myeloid progenitor differentiation (Johnson et al., 2015) and Granulocyte Macrophage Progenitor (GMP) function (Rodrigues et al., 2008). Myeloid progenitors lacking the Gata2 enhancer 77 kb upstream of the start site (−77) generate macrophages, but not erythroid or granulocytic cells, ex vivo (Johnson et al., 2015).
Human heterozygous GATA2 mutations cause primary immunodeficiency, myelodysplastic syndrome (MDS), acute myeloid leukemia (AML) and vascular/lymphatic dysfunction (Dickinson et al., 2011; Hahn et al., 2011; Hsu et al., 2011; Ostergaard et al., 2011), reinforcing the importance of GATA-2 mechanisms derived from mouse models (Gao et al., 2013; Johnson et al., 2012; Johnson et al., 2015; Ling et al., 2004; Rodrigues et al., 2005; Tsai et al., 1994). Mutations in the DNA binding C-terminal zinc finger inhibit binding (Hahn et al., 2011), while +9.5 enhancer mutations reduce GATA2 expression (Hsu et al., 2013; Johnson et al., 2012). Mutations also occur in the N-terminal zinc finger that is not required for DNA binding (Ping et al., 2017). GATA-2 promotes AML cell proliferation (Katsumura et al., 2016) and mediates leukemogenic activity in a mouse model involving Tet2 deficiency and mutant Flt3(ITD) expression (Shih et al., 2015). Leukemogenic cohesin mutants increase GATA-2 chromatin occupancy in human HSPCs, and GATA-2 is implicated in the cohesin mutant-induced differentiation block (Mazumdar et al., 2015).
The results described above indicate that GATA-2 levels/activity must be stringently regulated to ensure normal hematopoiesis. Considering the low frequency of GATA motif (WGATAR) occupancy in cells (<0.1%), based on genome-wide chromatin occupancy analysis (Fujiwara et al., 2009; Kang et al., 2012), and differential GATA-2 occupancy in distinct systems (Beck et al., 2013; DeVilbiss et al., 2014; Dore et al., 2012; Fujiwara et al., 2009; Li et al., 2011; Wilson et al., 2010; Wu et al., 2014b), it is instructive to compare mechanisms governing GATA-2 expression/function in different cell types. A major mechanism determining GATA-2 levels involves the +9.5 and −77 enhancers (Gao et al., 2013; Grass et al., 2006; Johnson et al., 2012; Johnson et al., 2015). Although GATA-2 occupies both enhancers, only the +9.5 triggers HSC genesis in the mouse embryo. Transcriptomes from Gata2 enhancer knockout mice (Gao et al., 2013; Johnson et al., 2012; Johnson et al., 2015), Gata2 conditional knockout mice (de Pater et al., 2013) and GATA-2-knockdown endothelial cells (Linnemann et al., 2011) suggest GATA-2-regulated genetic networks differ considerably.
To compare GATA-2 expression and function in different contexts, we developed a compound heterozygous (CH) model bearing +9.5 and −77 enhancer mutations on different Gata2 alleles. Dissecting mechanisms in this system and in −77−/− erythroid precursors revealed that while the +9.5 suffices to trigger HSC genesis, both enhancers must reside on a single allele to induce megakaryocyte erythrocyte progenitors (MEPs). GATA-2-dependent transcriptomes in myelo-erythroid progenitors and erythroid precursors differed greatly. A critical −77 enhancer function involved establishing circuits required for BFU-E activity, the vital erythroid precursor that ensures steady-state erythropoiesis and red blood cell regeneration in anemic stress (Koury, 2014).
RESULTS
Genetic Interactions Between Gata2 Enhancers Reveal a Mechanism Underlying a Hierarchical Blood Development Program
Whereas the +9.5 enhancer confers Gata2 expression in hemogenic endothelium and HSPCs (Gao et al., 2013; Johnson et al., 2012), the −77 enhancer increases Gata2 expression in myeloid progenitors, but not HSCs (Johnson et al., 2015). However, both enhancers contain conserved GATA motifs, which can be GATA-2-occupied. To test whether the distinct mechanisms establish unique GATA-2-regulated enhancer activities in vivo, we compared enhancer functions in distinct sectors of the hematopoietic hierarchy. As the +9.5 enhancer deletion abrogates HSC genesis in the mouse embryo and is lethal at ~E14 (Gao et al., 2013; Johnson et al., 2012), it was unclear whether it functions in myeloid progenitors (Figure 1A). The −77 enhancer deletion is lethal after E15.5 (Johnson et al., 2015). +9.5+/− and −77+/− mouse strains are viable, and adult steady-state hematopoiesis is not impaired (Johnson et al., 2012; Johnson et al., 2015). We tested whether downregulating GATA-2 in endothelium and HSCs (+9.5+/−) and myeloid progenitors (−77+/−) concomitantly impacts hematopoiesis in a way predictable by phenotypes of individual enhancer homozygous mutations or uniquely.
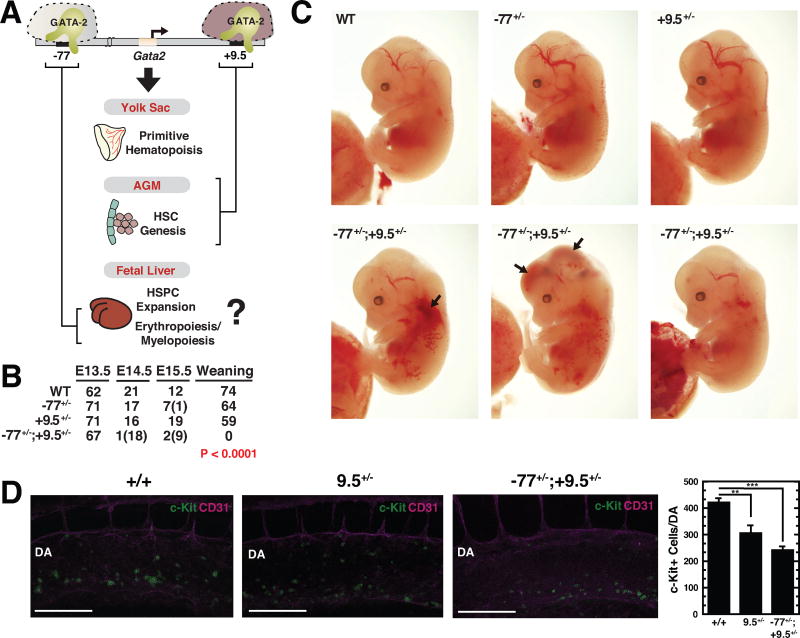
Figure 1. Unique Constellation of Hematopoietic Deficits in CH Embryos.
(A) Model depicting stage-specific requirement for Gata2 −77 and +9.5 enhancers during developmental hematopoiesis. As the +9.5 enhancer deletion abrogates HSC genesis in the AGM of +9.5−/− embryos, whether it exerts essential activities in fetal liver hematopoietic cells was unclear.
(B) Genotypes of WT, +9.5+/−, −77+/− and CH embryos at timed developmental stages and at time of weaning. The numbers of dead embryos at each developmental stage are shown in parentheses. Significance was analyzed using the Chi-squared test.
(C) Representative E13.5 CH embryos exhibiting reduced liver size and hemorrhage (arrows). (D) Representative whole-mount staining of E10.5 WT and CH embryos showing CD31+ cells (magenta) and c-Kit+ cells (green) within the dorsal aorta (DA). Scale bar, 100 µm. Quantitation of c-Kit+ cells within the DA (WT [n = 5], +9.5+/− [n = 4] and CH [n = 4]). Graphs depict mean ± SEM; **p < 0.01, ***p < 0.001. Significance was analyzed using the 2-tailed unpaired Student’s t test.
We considered the following models: (i) +9.5 might also function in myeloid progenitors, and compound heterozygous (CH) mice would phenocopy −77−/− mice. (ii) +9.5 and −77 might function in mutually exclusive contexts, and CH mice would resemble individual enhancer heterozygous mice. (iii) CH mutations might compromise hematopoiesis in a way not predictable from individual enhancer mutations. To test these models, CH mice, bearing a mutant +9.5 enhancer on one allele and a mutant −77 on the other allele, were produced by mating +9.5+/− and −77+/− mice to yield WT, +9.5+/−, −77+/− and CH progeny. Unlike +9.5+/− and −77+/− genotypes, no CH mice were alive at weaning (Figure 1B). The CH embryos were alive at E13.5, but most died by E14.5. Gata2 is expressed in endothelial cells (Linnemann et al., 2011; Wilson et al., 1990) and confers vascular integrity (Johnson et al., 2012; Lim et al., 2012). +9.5−/− but not −77−/−, embryos, exhibit hemorrhage (Johnson et al., 2012; Johnson et al., 2015). CH fetal liver size was reduced, and hemorrhaging was apparent (Figure 1C). HSC genesis in the +9.5−/− AGM is defective, as HSC-containing clusters within the AGM are absent, the mutant AGM lacks long-term repopulating HSCs in transplant assays (Gao et al., 2013) and the fetal liver does not acquire long-term repopulating HSCs (Johnson et al., 2012). −77−/− embryos produce normal HSC levels in the AGM (Johnson et al., 2015). A homozygous +9.5 mutation ablates HSC-containing c-Kit+ cells residing in clusters that emerge in the AGM (Gao et al., 2013). By contrast, c-Kit+ cell quantitation with E10.5 +9.5+/− and CH embryos revealed 29% (p = 0.010) and 43% (p < 0.0001) decreases vs. WT embryos respectively (Figure 1D). As the +9.5+/− and CH results did not differ significantly, the CH mutation did not exacerbate the partial inhibitory effect of +9.5 allele loss on c-Kit+ cell emergence in the AGM.
+9.5−/− and −77−/− embryos are anemic with reduced fetal liver size and a deficiency of Ter119+ cells (Johnson et al., 2012; Johnson et al., 2015). At E13.5, CH liver cellularity was 4.4-fold lower than WT littermates (Figure 2A). +9.5+/− and −77+/− fetal livers were comparable to WT. Unlike the dearth of HSCs in +9.5−/− fetal livers (Johnson et al., 2012), HSCs in −77−/− fetal liver are higher than WT (Johnson et al., 2015). As the −77 homozygous mutation did not reduce HSCs or impact Gata2 expression in Lineage− Sca1+c-Kit+ (LSK) cells, it appears that the −77 enhancer does not function in HSCs. However, Gata2 expression was not quantified in −77−/− HSCs (LSK CD150+CD48−), and if the −77 enhancer contributed modestly to Gata2 expression in HSCs, deleting the −77 enhancer from one allele and the +9.5 enhancer from the other, might impact HSC levels. Flow cytometric analysis of fetal liver HSCs and MPPs (Figure S1) revealed a 2.9-fold reduction in +9.5+/− HSCs (p = 0.003) and a 2.5-fold increase in MPPs (p = 2 × 10−6) versus WT littermates (Figure 2B). CH and +9.5+/− embryos contained indistinguishable HSC and MPP numbers, despite the 4.1-fold difference in CH liver cellularity (p < 2 × 10−17). The failure of the −77 enhancer deletion to exacerbate HSC loss caused by +9.5 deletion from the other allele (Figs. 1D and 2B) provides further evidence that the −77 is dispensable in HSCs.
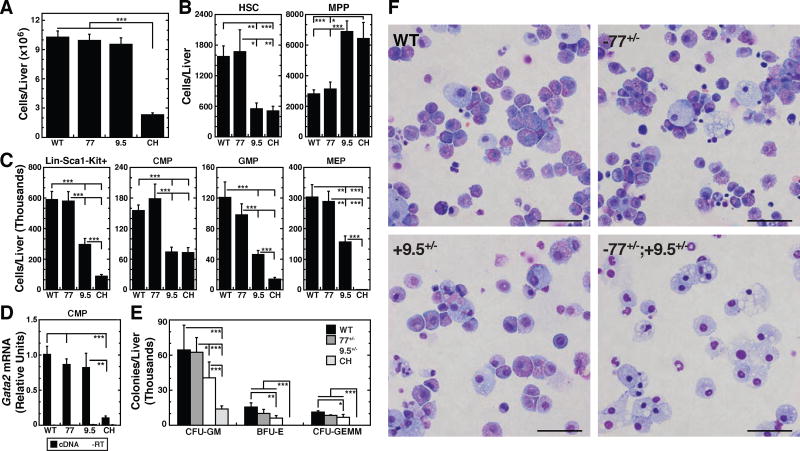
Figure 2. Myelo-erythroid Progenitor Functional Defect in CH Fetal Liver.
(A) Total fetal liver cells in E13.5 fetal livers (WT [n = 23]; −77+/− [n = 19]; +9.5+/− [n = 32]; CH [n = 27]).
(B) Quantitation of HSCs (Lin−Mac1+CD41−CD48−CD150+Sca1+Kit+) and MPPs (Lin− Mac1+CD41−CD48−CD150−Sca1+Kit+) from flow cytometric analysis of E13.5 fetal livers (WT [n = 12]; −77+/− [n = 6]; +9.5+/− [n = 6]; CH [n = 9]).
(C) Quantitation of Lin−Sca1−Kit+ cells, CMPs (Lin−CD34+FcRlowKit+Sca1−), GMPs (Lin− CD34+FcRhighKit+Sca1−), and MEPs (Lin−CD34−FcRlowKit+Sca1−) from E13.5 fetal livers (WT [n = 20]; −77+/− [n = 8]; +9.5+/− [n = 20]; CH [n = 20]).
(D) Gata2 mRNA quantitation in E13.5 fetal liver CMPs (WT [n = 19]; −77+/− [n = 8]; +9.5+/− [n = 8]; CH [n = 6]).
(E) CFU activity of E13.5 fetal liver hematopoietic progenitors. Graphs are adjusted for fetal liver cellularity. Graphs show mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001. Significance was analyzed using the 2-tailed unpaired Student’s t test.
(F) Representative images of Wright-Giemsa stained cells isolated from all colonies at time of counting. Scale bars, 50 µm.
Reduced GATA-2 expression in −77−/− myeloid progenitors caused a several-fold decrease in MEPs and biased CMP and GMP activities to differentiate predominantly into macrophages, but did not affect HSCs (Johnson et al., 2015). As CH embryos have modestly reduced HSC numbers, based on flow cytometry of fetal liver (2.9 fold) and AGM confocal imaging (41%), we considered how this might impact myeloid progenitors. Flow cytometric analysis of Lin−Sca1−Kit+ myeloid progenitors (Figure S2) revealed +9.5+/− fetal livers had 2-fold fewer progenitors, which involved similar reductions of CMP (2.1-fold; p = 1 × 10−6), GMP (2.6-fold; p = 0.0007) and MEP (1.9-fold; p = 0.002) (Figure 2C). Myeloid progenitors were 6.6-fold lower in CH vs. WT embryos (p = 1.8 × 10−11), with the difference involving disproportionate losses of GMP (8.6-fold; p = 3.7 × 10−6) and MEP (379-fold; p = 3.2 × 10−9). The 2.2-fold CMP decrease in CH (p = 1.2 × 10−6), resembled +9.5+/− embryos. Despite the similar CMP numbers, Gata2 mRNA levels in +9.5+/− CMPs did not differ significantly from that of WT or 77+/− CMPs. Gata2 mRNA was 9.6-fold lower (p = 0.008) in CH CMPs (Figure 2D) and 5.3 fold lower in 77−/− CMPs than WT (Johnson Sci. Adv 2015).
Consistent with myeloid progenitor reductions and MEP depletion, CH fetal livers lacked BFU-E and CFU-GEMM (Figure 2E). Colonies containing granulocytes and/or macrophages (enumerated as CFU-GM) were reduced significantly. Giemsa staining of cells within CFU-GM colonies after 8 days of culture revealed abundant macrophages and neutrophils in CH colonies, with a deficiency of the myeloid precursors common in WT, 77+/− and 9.5+/− colonies (Figure 2F). The genetic interaction between +9.5 and −77 enhancer alleles in myeloid progenitors, but not HSCs, revealed a novel mechanism in which one Gata2 allele must harbor both enhancers to induce MEPs, the critical red blood cell and platelet precursor (Akashi et al., 2000). Both enhancers function in progenitors to induce MEPs. These results highlight the uniqueness of the CH model, relative to other Gata2 enhancer mutants, Gata2 conditional knockout and Gata2 whole body knockout models, vis-à-vis phenotypes and utility for dissecting mechanisms underlying GATA-2 function and hematopoiesis.
Gata2 −77 Enhancer-dependent Erythroid Maturation In Vivo and Ex Vivo
GATA-2 is expressed in erythroid precursors (Leonard et al., 1993; Weiss et al., 1994), and as GATA-1 expression rises, it represses Gata2 transcription through a GATA switch mechanism (Bresnick et al., 2010; Grass et al., 2003). However, the consequences of premature GATA-2 loss on erythroid precursor function are unclear. Unanswered questions include whether mechanisms mediating GATA-1 upregulation are sustained, whether GATA-2-deficient erythroid precursors precociously differentiate or die and if GATA-2-deficient erythroid precursors acquire an ectopic fate change?
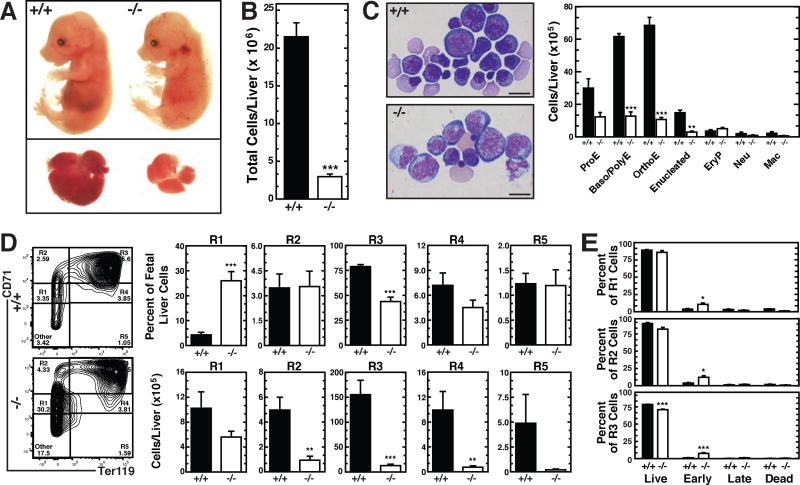
As CH embryos lacked MEPs, the CH model could not be used to ascertain the impact of reduced GATA-2 expression on MEP-derived erythroid precursors. We used the −77−/− model characterized by reduced, but detectable, MEPs to address this issue. −77−/− E13.5 fetal livers have reduced percentages of R1 (CD71medTer119−) and R2 (CD71highTer119−) erythroid precursors, suggesting defective erythroid precursor genesis, proliferation and/or survival (Johnson et al., 2015). The prior study did not establish whether reduced erythroid precursors reflected impaired upstream progenitors or an erythroid cell-instrinsic −77 enhancer requirement. −77−/− E14.5 fetal livers were pale and smaller than WT (Figure 3A). −77−/− fetal liver cellularity was reduced 7.2-fold (Figure 3B), with major decreases in basophilic and orthochromatic erythroblasts, as well as reticulocytes, despite normal numbers of nucleated primitive red cells (EryP) (Figure 3C). Flow cytometric analysis of erythroid maturation revealed an increased percentage of R1 cells in −77−/− fetal livers (from 4.2 to 26%, p = 0.0001) (Figure 3D). Though the percentage of −77−/− R1 cells decreased at E13.5 (Johnson et al., 2015), at E14.5, the proportion of −77−/− R1 cells increased concomitant with decreased R3 cells (Figure 3D). When adjusted for liver size, there was little to no difference in the number of E14.5 R1 cells per −77−/− liver vs. WT littermates (p = 0.31). By contrast, −77−/− R2-R5 cells declined sharply (R2, 8.2-fold, p = 0.004; R3, 14-fold, p < 10−5; R4, 9.7-fold, p = 0.002; R5, 14-fold, p = 0.087), indicating a maturation defect (Figure 3D). Annexin VDRAQ7 flow cytometric analysis (Figure S3) revealed −77−/− R1, R2 and R3 fetal liver cell populations contained early, but not late, apoptotic cells (Figure 3E). Most of the cells (~90%) were live and morphologically normal. The profound maturation defect provided rationale for dissecting how the −77 enhancer controls the erythroid maturation program.
Figure 3. −77 Gata2 Enhancer Promotes Eythroid Maturation In Vivo.
(A) Representative E14.5 −77+/+ and −77−/− embryos and fetal livers.
(B) Quantitation of cell numbers in E14.5 fetal livers (−77+/+ [n = 7], −77−/− [n = 7]).
(C) Representative Wright-Giemsa images and quantitation of −77+/+ and −77−/− E14.5 total fetal liver populations. Scale bar, 20 µm. ProE, proerythroblast; Baso/Poly, basophilic/polychromatophilic erythroblast; OrthoE, orthochromatic erythroblast; EryP, primitive erythroid; Neu, neutrophil; Mac, macrophage. (−77+/+ [n = 3], −77−/− [n = 3]).
(D) Representative flow cytometric analysis and quantitation of R1-R5 populations from E14.5 fetal livers (−77+/+ [n = 7], −77−/− [n = 7]).
(E) Quantitation of early (AnnexinV+DRAQ7−) and late (AnnexinV+DRAQ7+) apoptotic cells and dead cells (Annexin V−DRAQ7+) from R1–R3 populations by flow cytometry (−77+/+ [n = 4]; −77−/− [n = 5]). Graphs show mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001. Significance was analyzed using the 2-tailed unpaired Student’s t test.
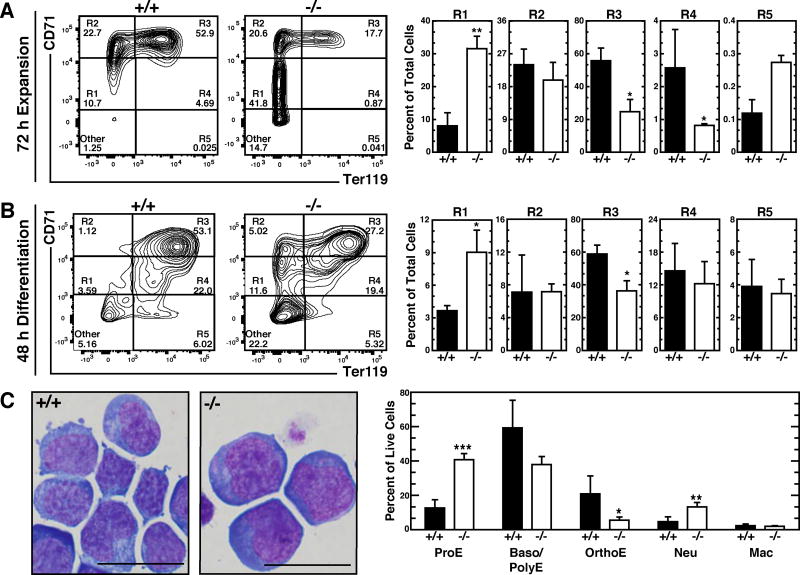
To test for intrinsic maturation and/or survival defects, lineage-depleted fetal liver progenitors were cultured for 72 hours in expansion medium containing Stem Cell Factor (SCF) and Erythropoietin (Epo). Flow cytometric analysis of CD71 and Ter119 revealed a maturation blockade for the mutants. After 72 hours ex vivo culture, WT cells distributed largely in R2 (24%) and R3 (55%) populations (proerythroblasts and basophilic/polychromatic erythroblasts, respectively), with small percentages in R1 (8%) and R4 (2.6%) (Figure 4A). By contrast, 32% of −77−/− cells persisted in the R1 population, with reduced R3 (24%) and R4 (0.8%) cells, recapitulating the maturation blockade detected with uncultured livers (Figure 4A). The expanded cells were cultured in high Epo-containing medium (6 U/ml) to promote erythroid maturation. Resembling the expansion culture, −77−/− cells had reduced R3 versus WT (−77−/−, 36%; −77+/+, 59%, p < 0.05), and R1 cells accumulated in the 77−/− population vs. WT (9 and 3.6%, respectively, p = 0.043) (Figure 4B). CD71−Ter119− (non-erythroid) cells persisted in the −77−/− population (22 and 3.4%, respectively, p < 10−5). Giemsa staining and morphological analysis indicated that −77−/− cultured cells contained a higher percentage of proerythroblasts (WT, 13%; −77−/−, 41%, p = 0.003) concomitant with reduced more mature basophilic/polychromatophilic (59 versus 38%) and orthochromatic (14 versus 4.6%, p = 0.047) erythroblasts (Figure 4C). Thus, the erythroid maturation blockade involves an intrinsic defect in −77−/− precursors that cannot be rescued with high Epo.
Figure 4. −77 Gata2 Enhancer Promotes Erythroid Maturation Ex Vivo.
Representative flow cytometric analysis and quantitation of R1–R5 populations from E14.5 fetal livers cultured in (A) expansion medium for 72 h (−77+/+ [n = 5]; −77−/− [n = 4]) or (B) expansion media for 72 h then differentiation medium for 48 h (−77+/+ [n=4]; −77−/− [n = 4]).
(C) Representative Wright-Giemsa images and quantitation of −77+/+ and −77−/− E14.5 lineage negative cells cultured in expansion system for 72 h. Scale bar, 20 µm. ProE, proerythroblast; Baso/Poly, basophilic/polychromatophilic erythroblast; OrthoE, orthochromatic erythroblast; Neu, neutrophil; Mac, macrophage (−77+/+ [n = 6]; −77−/− [n = 6]). Graphs show means ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001. Significance was analyzed using the 2-tailed unpaired Student’s t test.
Generating the Vital Erythroid Precursor Burst Forming Unit-Erythroid
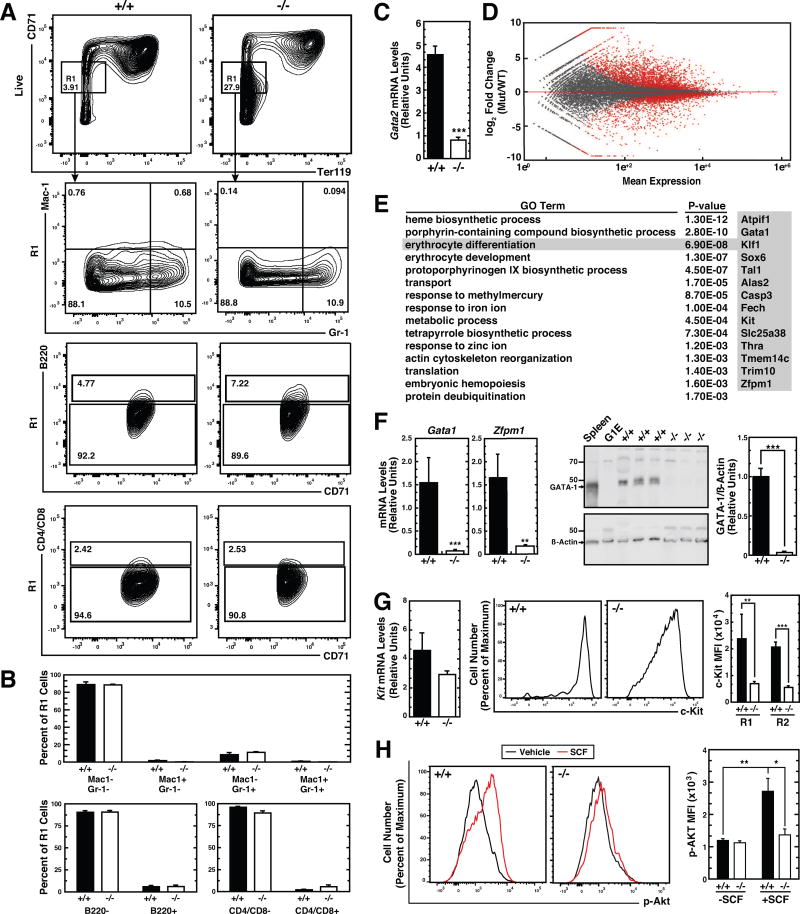
Considering the large decline of R2–R5 cells in −77−/− fetal livers, we analyzed mechanisms underlying the maturation blockade by comparing transcriptomes of FACS-isolated 77−/− and WT R1 cells from E14.5 livers (Figure 5A). Monocytes (Mac1+Gr1−), granulocytes (Mac1+Gr1+), B cells (B220+) and T cells (CD4/CD8+) were very low in both populations. Eythroid precursors comprised 80–85% of R1 population cells (Figure 5B). Gata2 mRNA levels were reduced 5.7-fold (p < 10−5) in −77−/− R1 cells (Figure 5C). RNA-seq was conducted with three −77−/− or WT R1 samples, and transcriptomes were compared with DESeq2 (Love et al., 2014). This analysis revealed 2805 and 2519 upregulated and downregulated (p < 0.05) genes, respectively, in −77−/− R1 cells (Figure 5D). Gene ontology analysis of the downregulated (≥2 fold) genes yielded categories relevant to erythropoiesis, including heme biosynthesis and erythrocyte differentiation (Figure 5E). The erythrocyte differentiation category included genes encoding proteins with critical erythroid functions, such as GATA-1, KLF1, c-Kit (Kit), and FOG-1 (Zfpm1). qRT-PCR confirmed strongly reduced Gata1 and Zfpm1 expression in −77−/− R1 cells. −77−/− R1 cells lacked GATA-1 protein (Figure 5F).
Figure 5. −77 Gata2 Enhancer-dependent Transcriptome Establishes Developmental Signaling.
(A) Representative flow cytometric analysis of erythroid cells from E14.5 fetal liver using cell surface markers CD71 and Ter119. The R1 (CD71lowTer119−) population was also evaluated for presence of monocytes (Mac1+Gr1−), granulocytes (Mac1+Gr1+), B cells (B220+) and T cells (CD4/CD8+).
(B) Quantitation of Mac1-Gr1, B220, and CD4/CD8 cells in R1 populations (−77+/+ [n = 4]; −77−/− [n = 4]).
(C) Quantitation of Gata2 mRNA levels in E14.5 fetal liver R1 cells (−77+/+ [n = 4]; −77−/− [n = 6]).
(D) MA plot of RNA-seq–based comparison of R1 transcriptomes from −77+/+ and −77−/− E14.5 fetal livers (−77+/+ [n = 3]; −77−/− [n = 3]). Red points indicate down- or up-regulated genes (false discovery rate (FDR) <0.05).
(E) Gene Ontology analysis of genes downregulated in −77−/− R1 cells. The most enriched biological processes are shown with corresponding p values. Genes comprising the erythrocyte differentiation category are shown.
(F) Quantitation of Gata1 and Zfpm1 mRNA levels (−77+/+ [n = 4]; −77−/− [n = 6]) and GATA-1 protein levels (−77+/+ [n = 6]; −77−/− [n = 5]) in E14.5 fetal liver R1 cells.
(G) Quantitation of Kit mRNA levels (−77+/+ [n = 4]; −77−/− [n = 6]) and cell surface c-Kit mean fluorescence intesity (MFI) (−77+/+ [n = 8]; −77−/− [n = 5]) in E14.5 fetal liver R1 cells.
(H) Quantitation of p-Akt MFI after stimulation with 10 ng/ml SCF in −77+/+ and −77−/− Ter119− cells (WT [n = 7]; −77−/− [n = 6]). Graphs show mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001. Significance was analyzed using the 2-tailed unpaired Student’s t test.
The GATA-2 target gene Kit (Jing et al., 2008; Munugalavadla et al., 2005) encodes the c-Kit receptor tyrosine kinase that controls HSPC and erythroid precursor proliferation, differentiation and survival (Chabot et al., 1988; Hewitt et al., 2015; Munugalavadla and Kapur, 2005; Wojchowski et al., 2010). c-Kit is activated by Stem Cell Factor (SCF) (Huang et al., 1990), a critical determinant of hematopoiesis (Ding and Morrison, 2013). Kit mRNA was modestly downregulated 1.5-fold, and cell surface c-Kit was 3.4-fold (p = 0.009) reduced in E14.5 −77−/− R1 cells (Figure 5G). We tested whether −77-dependent Kit expression in erythroid precursors establishes developmental signaling that is limiting in −77−/− cells using a phospho-flow cytometry assay (Hewitt et al., 2015; McIver et al., 2016). −77−/− and WT fetal liver Ter119− cells were serum-starved, treated with SCF or vehicle and p-Akt was quantified. While SCF increased p-Akt 2.3-fold (p = 0.003) in WT cells, −77−/− cells were non-responsive (Figure 5H). Basal p-Akt levels were equivalent in unstimulated WT and −77−/− cells. These results support a model in which loss of a single enhancer (−77) creates severe defects in erythroid precursors involving defective signaling and transcriptional circuitry required for erythroid maturation.
Though prior studies suggested that GATA-2 transcriptionally induces GATA-1 expression (Bresnick et al., 2010), the GATA-2 contribution to GATA-1 expression in vivo is unclear. Since −77 increases GATA-1 expression in erythroid precursors, we assumed it would control a large GATA-1 target gene ensemble. While GATA-1 represses certain GATA-2-activated genes, e.g. Kit, GATA-1 might sustain expression of others. Thus, −77−/− transcriptomic defects might reflect defects in genes regulated exclusively by GATA-2, by GATA-1 and/or jointly by GATA-1 and GATA-2.
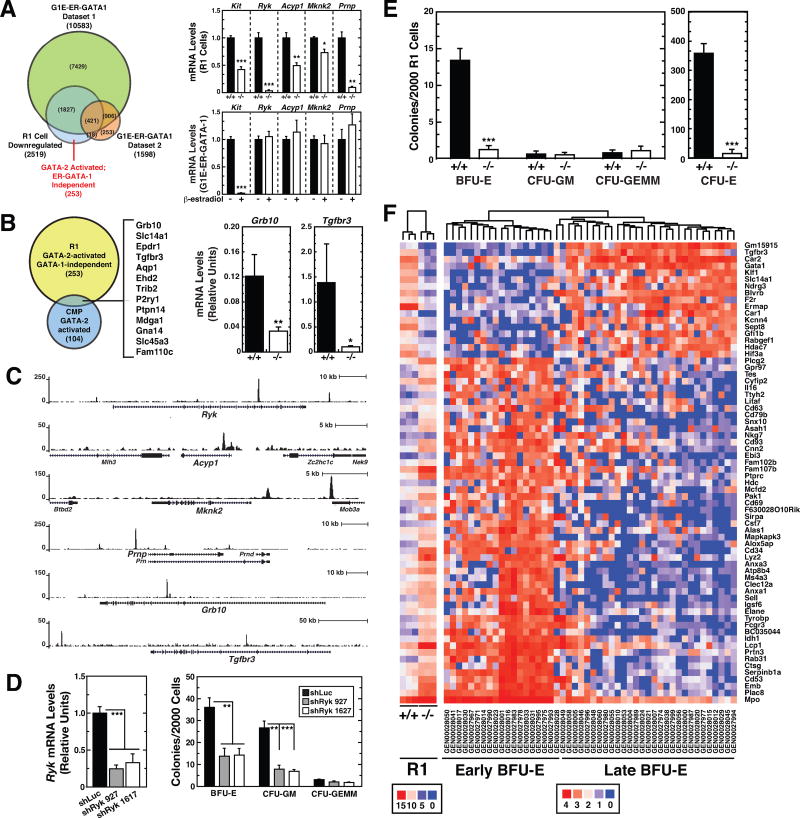
To assess the contribution of distinct regulatory modes to the −77−/− erythroid precursor transcriptomic defects, we compared the R1 cell RNA-seq data with GATA-1-regulated genes in G1E-ER-GATA-1 cells. The G1E-ER-GATA-1 data was derived from RNA-seq analysis 48 h post-ER-GATA-1 activation (Tanimura et al., 2016) and microarray analysis 24 h after ER-GATA-1 activation (DeVilbiss et al., 2013; Pope and Bresnick, 2013) (Datasets 1 and 2, respectively) (Figure 6A). In R1 cells, −77 loss decreased Gata2 expression, which reduced Gata1 expression. In G1E-ER-GATA-1 cells, genetic complementation with β-estradiol-activated ER-GATA-1 (Gregory et al., 1999) represses Gata2 transcription (Grass et al., 2003). Of the 2519 genes downregulated in −77−/− R1 cells, representing GATA-2 and/or GATA-1-activated genes, 731 were GATA-1-activated in G1E-ER-GATA-1 cells. 1376 genes downregulated in R1 cells were GATA-1-repressed, and therefore are GATA-2-activated/GATA-1-repressed. In combined GATA-2 and GATA-1 deficiency, e.g. in −77−/− R1 cells, these genes are not highly expressed. Of the remaining 412 genes, 253 are expressed in G1E-ER-GATA-1 cells but not ER-GATA-1-regulated. These 253 genes are candidate direct GATA-2-activated genes, without a GATA-2 requirement for maintenance, since their expression is sustained in G1E-ER-GATA-1 cells in which GATA-2 transcription is repressed by GATA-1. After GATA-1 represses Gata2 transcription in G1E-ER-GATA-1 cells, GATA-1 might sustain their expression. We quantified mRNAs in G1E-ER-GATA-1 cells before and after ER-GATA-1 induction. While ER-GATA-1 repressed the established GATA-2 target gene Kit 47-fold (p < 0.0001), Ryk, Acyp1, Mknk2 and Prnp expression were not significantly affected (Figure 6A), demonstrating the existence of GATA-2 target genes not regulated by GATA-1 in the complementation assay.
Figure 6. −77 Gata2 Enhancer Requirement for Burst Forming Unit-Erythroid.
(A) Venn diagram depicting overlap between genes regulated by GATA-1 in G1E-ER-GATA-1 cells and by the −77 enhancer in R1 cells. Changes in R1 cell RNA-seq read counts are shown for representative GATA-2-activated genes. Also shown are the relative mRNA levels of these genes in G1E-ER-GATA-1 cells with and without β-estradiol treatment. G1E-ER-GATA-1 Dataset 1; Gene expression changes following 48 h induction of GATA-1 activity by β-estradiol quantified by RNA-seq. G1E-ER-GATA-1 Dataset 2; Gene expression changes following 24 h induction of GATA-1 activity by β-estradiol. Transcripts were quantified by Agilent-028005 SurePrint G3 Mouse GE 8×60K Microarray (Pope and Bresnick, 2013).
(B) Venn diagram depicting overlap between GATA-2-activated genes in R1 cells and CMPs (GSE69786). Genes common to both categories are listed. Reductions in Grb10 and Tgfbr3 mRNA expression in −77−/− R1 cells were confirmed by qRT-PCR (−77+/+ [n = 4]; −77−/− [n = 6]).
(C) GATA-2 ChIP-seq profiles in GATA-1-null G1E erythroid precursor cells mined from dataset GSM722387.
(D) CFU activity of lineage-depleted E14.5 fetal liver cells following shRNA-mediated knockdown of Ryk. Ryk mRNA was quantified by qRT-PCR 3 days post-infection with control (shLuc) or Ryk shRNA-exressing retrovirus. 2000 cells were plated in methylcellulose within 16 h post-infection. Colonies were enumerated on day 8 (n = 6). (E) CFU activity of E14.5 fetal liver R1 cells quantified after 2 (CFU-E) or 8 (BFU-E, CFU-GM and CFU-GEMM) days in methylcellulose. Graphs show mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001. Significance was analyzed using the 2-tailed unpaired Student’s t test.
(F) Cluster analysis of −77+/+ and −77−/− R1 gene expression and comparison with “early” and “late” BFU-E gene expression signature (Gao et al., 2016).
We described deregulated expression of 133 genes in E13.5 CMPs from −77−/− fetal livers (Johnson et al., 2015); 104 were downregulated. Comparison of the 253 R1 cell and 104 CMP genes yielded an overlap of 13 (Figure 6B). qRT-PCR analysis confirmed Grb10 and Tgfbr3 expression were reduced in −77−/− R1 cells (Figure 6B). The signaling adapter GRB10 is implicated in promoting HSC self-renewal and regeneration (Yan et al., 2016). Tgfbr3, which encodes TGFβ receptor type III (Wang et al., 1991), is a marker of late BFU-Es (Gao et al., 2016). GATA-2 occupied GATA-2-activated genes Ryk, Acyp1, Mknk2, Prnp, Grb10 and Tgfbr3 in hematopoietic or erythroid progenitors (Figure 6C and Figure S5).
Ryk encodes an atypical Wnt ligand receptor (Green et al., 2014). Wnt5a induction of HSC quiescence (Nemeth et al., 2007) and protection against myeloablative injury (Povinelli et al., 2015) is blocked by Ryk inhibition (Povinelli and Nemeth, 2014). HSC self-renewal is reduced in Ryk−/− mice (Famili et al., 2016). However, Ryk function in erythroid precursors has not been reported. Colony assays were conducted with lineage-depleted WT fetal liver cells using two distinct shRyk-expressing retroviruses to downregulated Ryk. The shRNAs reduced Ryk mRNA by 75% (shRyk 927) and 67% (shRyk 1617) and decreased BFU-E and CFU-GM by 60–62% and 70–75%, respectively (Figure 6D). Thus, Ryk is one component of the GATA-2-regulated network that generates BFU-E and CFU-GM.
In E13.5 −77−/− fetal livers, BFU-Es were 11-fold lower, and R1 cells were 4-fold lower than WT (Johnson et al., 2015). However, BFU-Es were readily detectable in −77−/− fetal livers. It was not clear if deficient −77−/− BFU-E activity reflects defective R1 cells or impaired function of progenitors to generate BFU-E. We quantified the differentiation potential of purified 77−/− R1 cells. Two days after plating identical cell numbers, CFU-Es decreased 20-fold (p < 0.0001) with −77−/− cells (Figure 6E). By Day 8, −77−/− BFU-Es were 10-fold lower (p < 0.0001), which was comparable to our prior result with total fetal liver cells. These results provide evidence for an intrinsic defect in R1 cells that impairs their BFU-E-generating capacity. The R1 cells generated little to no CFU-GM and CFU-GEMM colonies (Figure 6E). The −77 enhancer is therefore a crucial determinant of BFU-E and its progeny, CFU-E (Figure 7), and −77−/− erythroid precursors did not undergo a fate switch to generate myeloid progeny.
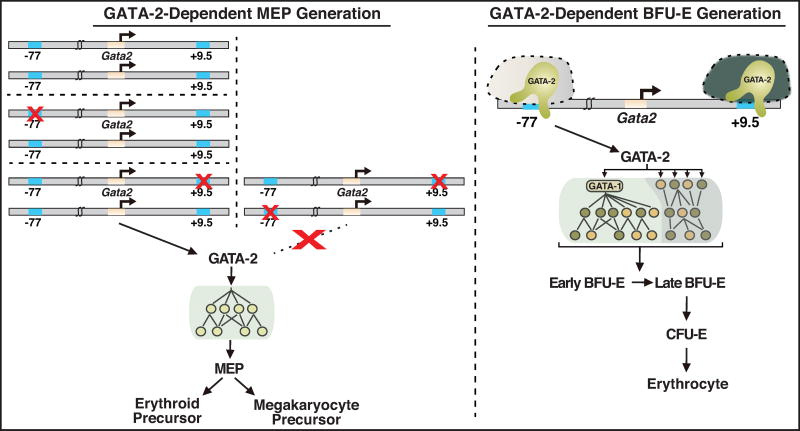
Figure 7. Model for Gata2 Enhancer-dependent Megakarycytic Erythrocyte Progenitor and Burst Forming Unit-Erythroid Generation.
Left, GATA-2-dependent MEP generation. Both −77 and +9.5 enhancers must reside on a single allele to activate Gata2 transcription to establish a genetic network that supports the genesis and/or survival of MEPs, the precursor to erythrocytes and megakaryocytes. Right, GATA-2-dependent BFU-E generation. The −77 enhancer activates Gata2 transcription in erythroid precursors, resulting in a GATA-2-dependent genetic network that supports the genesis and/or survival of BFU-E, the vital erythroid precursor required for erythrocyte development and regeneration. This network parses into two components: (i) GATA-2 activates Gata1 transcription, and GATA-1 regulates a large ensemble of genes that establish the erythroid cell transcriptome. (ii) GATA-2 activates a smaller target gene cohort without a requirement to upregulate GATA-1. In aggregate, the genes establish signaling pathways that support proliferation, differentiation and survival and the vast constituents that comprise the unique phenotype of erythroblasts and their reticulocyte and erythrocyte progeny.
The gene signatures of early and late BFU-Es have been described, and comparison of the two transcriptomes revealed high Tgfbr3 as a hallmark of late BFU-Es (Gao et al., 2016). As BFU-Es and Tgfbr3 expression are reduced in −77−/− R1 cells, we compared WT and −77−/− R1 cell transcriptomes with those of early and late BFU-Es. This analysis revealed −77−/− R1 cells exhibit an early BFU-E signature (Figure 6F). The WT R1 cell signature significantly correlated with the late BFU-E signature (r = 0.62, p < 0.0001) and negatively correlated with the early BFU-E signature (r = −0.34, p = 0.005). The −77−/− R1 signature positively correlated with the early BFU-E signature (r = 0.73, p < 0.0001) and negatively correlated with the late BFU-E signature (r = −0.42, p = 0.0004). The transcriptome alterations arising from −77 enhancer deletion indicate the −77 enhancer and GATA-2 are vital determinants of early erythroid precursor maturation and the genesis of BFU-E activity.
DISCUSSION
Recurring GATA2 mutations in cancer, immunological and vascular/lymphatic disorders highlight critical GATA-2 functions in human biology and pathology (Dickinson et al., 2014; Katsumura et al., 2017; Spinner et al., 2014). Since GATA2 mutations impair DNA binding (Hahn et al., 2011; Katsumura et al., 2016; Katsumura et al., 2014) or reduce GATA-2 expression (Hsu et al., 2013; Johnson et al., 2012), and elevated GATA2 expression can correlate with poor prognosis of AML (Vicente et al., 2012), strict control of GATA-2 levels/activity ensures normal hematopoiesis. Many GATA2 pathology phenotypes are predictable from concepts derived from mouse models (Gao et al., 2013; Johnson et al., 2012; Johnson et al., 2015; Ling et al., 2004; Rodrigues et al., 2005; Tsai et al., 1994). Gata2 enhancer mutants represent unique systems to unravel how GATA-2 expression is maintained within a critical physiological window and how altered GATA-2 expression impacts HSPC genesis/function. The +9.5 enhancer mutant revealed GATA-2 activity to trigger HSC generation in the AGM and to confer vascular integrity (Gao et al., 2013; Johnson et al., 2012). The −77 enhancer mutant revealed GATA-2 activity to confer myeloid progenitor differentiation into diverse progeny (Johnson et al., 2015). The −1.8 enhancer mutant revealed its requirement to maintain, but not establish, Gata2 repression during erythroid maturation (Snow et al., 2010). The −2.8 (Snow et al., 2011) and −3.9 (Sanalkumar et al., 2014) mutants revealed how cis-elements predicted to be important, based on conserved factor binding motifs, factor occupancy, and enhancer activity in artificial assays, may not be critical in vivo.
Herein, we described a Gata2 enhancer CH mutant model with one allele lacking the +9.5 enhancer and the other lacking the −77, which is embryonic lethal at ~E14.5. This unique model and the −77−/− model led to the discovery of mechanisms responsible for establishing two vital hematopoietic progenitor populations - MEP and BFU-E (Figure 7). The mechanisms were not predictable from those dictating HSC emergence (Gao et al., 2013). The Gata2 enhancers had both functional similarities and differences in vivo. As mutation or disruption via chromosomal rearrangement of both enhancers are implicated in human MDS and AML, and other pathologies (Groschel et al., 2014; Hsu et al., 2013; Johnson et al., 2012; Katsumura et al., 2017; Yamazaki et al., 2014), insights from this model will inform human enhanceropathies.
While the +9.5, but not the −77, triggers HSC genesis in the AGM, MEP production requires both enhancers on a single allele to increase Gata2 transcription. The simultaneous mutation of a single copy of each enhancer in trans abrogates Gata2 expression and MEP production. MEPs are implicated as myeloid leukemia-initiating cells (Dang et al., 2017; Zhang et al., 2017). Considering GATA-2 function in distinct sectors of the hematopoietic hierarchy, and its requirement for MEPs, it is attractive to consider how alterations in GATA-2-regulated circuits influence MEP genesis and function. Such alterations may involve mutations that reduce GATA-2 activity or expression or those impacting expression/activity of constituents of the GATA-2-regulated genetic network.
In addition to the enhancer-dependent MEP genesis mechanism, −77 deletion depleted BFU-Es. Major progress has been made in identifying BFU-E self-renewal regulators, including glucocorticoid receptor, peroxisome proliferation-activated receptor-α and the RNA binding protein ZFP36L2 (Flygare et al., 2011; Gao et al., 2016; Lee et al., 2015). Despite little to no alterations in expression of mRNAs encoding these factors in −77−/− R1 cells (≤ 1.5 fold), these mutant cells lacked BFU-E activity.
The −77 enhancer induced expression of GATA-1, a large ensemble of erythroid genes and previously undescribed GATA-2 targets. The latter genes included Ryk, Tgfbr3 and Grb10, encoding hematopoietic signaling components (Famili et al., 2016; Gao et al., 2016; Povinelli and Nemeth, 2014; Povinelli et al., 2015; Randrianarison-Huetz et al., 2010; Yan et al., 2016), and other factors not implicated in red cell biology. Based on the large reduction in Ryk expression in −77−/− R1 cells and reduced CFU activity upon Ryk knockdown, Ryk signaling may constitute an important determinant of erythroid precursor cell maturation and/or function. The −77 targets reveal a new dimension of GATA-2 function to control erythroid precursor genesis/function that could not be inferred from GATA-2 mechanisms in other systems.
A prior transcriptomic analysis identified early and late BFU-E signatures (Gao et al., 2016). Although it is assumed that early BFU-Es progress to a late BFU-E state, no mutants that ablate or enhance this process have been described. As defective BFU-E maturation/function may underlie pathologies, such as Epo-resistant anemias (Gao et al., 2016; Lee et al., 2015), the mechanisms are of great interest. The −77−/− R1 cell transcriptome resembled early BFU-E, suggesting the −77 controls BFU-E maturation, and the mutant cells lacked BFU-E activity. The diverse constituents of the −77-regulated genetic network include new targets for manipulation of erythroid precursors, e.g. to stimulate red blood cell regeneration.
In summary, CH and −77−/− models revealed mechanisms governing the production of vital hematopoietic progenitors. Why do the −77 and +9.5 enhancers need to reside on the same allele to generate myeloid progenitors, while the +9.5 has a unique function in HSC genesis? The +9.5 enhancer may exist in an active state in hemogenic endothelium and in HSCs, while the −77 enhancer complex has not yet been commissioned. The +9.5 active state would persist as myeloid progenitors develop, and the −77 active state would be acquired. In a myeloid progenitor with diminished developmental potential in comparison to HSCs, presumably, the distinct epigenomic environments create new obstacles and opportunities for gene control. The requirement for +9.5 and −77 enhancers on the same allele points to a model in which Gata2 transcription has a dual enhancer requirement to establish and/or maintain the active state. Why is the developmental stage- and cell type-specific dual enhancer mechanism required for generating a progenitor, but not a stem cell? It will be instructive to identify the shared and the developmental stage- and cell type-specific, components mediating the unique enhancer activities.
EXPERIMENTAL PROCEDURES
Mouse Crosses and Timed Matings
−77+/−;+9.5+/− CH embryos were generated by mating +9.5+/− and −77+/− mice. Pregnant females were euthanized with CO2, and the uterus was removed into IMDM (Gibco) containing 10% FBS (Gemini). Embryo viability was scored by the presence of a beating heart and were photographed with an Olympus Szx16 stereomicroscope. Animal experiments were performed with the ethical approval of the Association for the Assessment and Accreditation of Laboratory Animal Care at UW-Madison.
Flow Cytometry
Fetal liver cells from E13.5 or E14.5 embryos were dissociated and resuspended in PBS with 2% FBS and passed through 25 µm cell strainers to obtain single-cell suspensions. Detailed protocols can be found in the Supplemental Experimental Procedures.
Colony Forming Unit Assays
Dissociated cells from E13.5 or E14.5 fetal livers were plated in duplicate in Methocult M3434 complete media (StemCell Technologies) at 2 × 104 cells per 35 mm plate or 2000 sorted R1 cells per 35 mm plate. Plates were incubated for 7–8 days, and colonies were enumerated. For Giemsa staining, cells were isolated from the plates using PBS containing 2% FBS, centrifuged for 10 min, resuspended in PBS containing 25% FBS and centrifuged (CytoSpin4, ThermoFisher Scientific) for 10 min at 500 rpm onto slides.
Gene Expression Analysis
RNA was purified from fetal livers or sorted cells using TRIzol (Invitrogen). cDNA was synthesized by Moloney murine leukemia virus reverse transcription (RT). Real-time PCR was conducted with SYBR green master mix. Detailed protocols can be found in the Supplemental Experimental Procedures.
Primary Cell Isolation and Culture
Erythroid precursors were enriched from E14.5 fetal livers with EasySep negative selection Mouse Hematopoietic Progenitor Enrichment Kit (StemCell Technologies). Detailed protocols can be found in the Supplemental Experimental Procedures.
Statistics
For quantification of cells, colonies, or mRNA, the results are presented as mean ± SEM. Determination of significance was conducted using the 2-tailed unpaired Student’s t test. A p value <0.05 was considered significant. Single cell RNA-Seq data from a prior report (Gao et al., 2016) were averaged for each of the 68 signature genes for each of the Gata1hi and Gata1lo cells. RNA-Seq data from our R1 cell expression profiles were averaged for the −77+/+ and −77−/− samples. Pairwise correlation was computed and plotted using cor.test() and the pairs() function in R (http://www.r-project.org) statistical computing environment (Wu et al., 2014a).
Supplementary Material
Acknowledgments
The work was supported by NIH DK68634 and DK50107 (to E.H.B.), U01 HG007019 (to C.N.D. and E.H.B.), and Carbone Cancer Center Support Grant P30 CA014520. We thank Caroline Jiayun Liang for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
C.M., K.D.J. and E.H.B. conceived all studies and designed experiments. Experiments were conducted by C.M., K.D.J., X.G., K.R.K. and S.M. E.A.R. conducted Giemsa staining. Computational/statistical analysis was conducted by IMO. The manuscript was drafted by C.M., K.D.J. and E.H.B. and revised and approved by all authors.
ACCESSION NUMBERS
The accession number for the RNA-seq data from CD71medTer119− R1 cells reported in this paper is GEO: GSE96059
The authors declare no competing financial interests.
References
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Beck D, Thoms JA, Perera D, Schutte J, Unnikrishnan A, Knezevic K, Kinston SJ, Wilson NK, O'Brien TA, Gottgens B, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335:88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- Dang J, Nance S, Ma J, Cheng J, Walsh MP, Vogel P, Easton J, Song G, Rusch M, Gedman AL, et al. AMKL chimeric transcription factors are potent inducers of leukemia. Leukemia. 2017 doi: 10.1038/leu.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater E, Kaimakis P, Vink CS, Yokomizo T, Yamada-Inagawa T, van der Linden R, Kartalaei PS, Camper SA, Speck N, Dzierzak E. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210:2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVilbiss AW, Boyer ME, Bresnick EH. Establishing a hematopoietic genetic network through locus-specific integration of chromatin regulators. Proc Natl Acad Sci U S A. 2013;110:E3398–3407. doi: 10.1073/pnas.1302771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVilbiss AW, Sanalkumar R, Johnson KD, Keles S, Bresnick EH. Hematopoietic transcriptional mechanisms: from locus-specific to genome-wide vantage points. Exp Hematol. 2014;42:618–629. doi: 10.1016/j.exphem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Griffin H, Bigley V, Reynard LN, Hussain R, Haniffa M, Lakey JH, Rahman T, Wang XN, McGovern N, et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 2011;118:2656–2658. doi: 10.1182/blood-2011-06-360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Milne P, Jardine L, Zandi S, Swierczek SI, McGovern N, Cookson S, Ferozepurwalla Z, Langridge A, Pagan S, et al. The evolution of cellular deficiency in GATA2 mutation. Blood. 2014;123:863–874. doi: 10.1182/blood-2013-07-517151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famili F, Perez LG, Naber BA, Noordermeer JN, Fradkin LG, Staal FJ. The non-canonical Wnt receptor Ryk regulates hematopoietic stem cell repopulation in part by controlling proliferation and apoptosis. Cell Death Dis. 2016;7:e2479. doi: 10.1038/cddis.2016.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare J, Rayon Estrada V, Shin C, Gupta S, Lodish HF. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood. 2011;117:3435–3444. doi: 10.1182/blood-2010-07-295550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Johnson KD, Chang YI, Boyer ME, Dewey CN, Zhang J, Bresnick EH. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210:2833–2842. doi: 10.1084/jem.20130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lee HY, da Rocha EL, Zhang C, Lu YF, Li D, Feng Y, Ezike J, Elmes RR, Barrasa MI, et al. TGF-beta inhibitors stimulate red blood cell production by enhancing self-renewal of BFU-E erythroid progenitors. Blood. 2016;128:2637–2641. doi: 10.1182/blood-2016-05-718320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Jing H, Kim S-I, Martowicz ML, Pal S, Blobel GA, Bresnick EH. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Nusse R, van Amerongen R. The role of Ryk and Ror receptor tyrosine kinases in Wnt signal transduction. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promoter erythroid cell survival by regulating bcl-xl expression. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC, Babic M, Lin M, Carmagnac A, Lee YK, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt KJ, Kim DH, Devadas P, Prathibha R, Zuo C, Sanalkumar R, Johnson KD, Kang YA, Kim JS, Dewey CN, et al. Hematopoietic Signaling Mechanism Revealed from a Stem/Progenitor Cell Cistrome. Mol Cell. 2015;59:62–74. doi: 10.1016/j.molcel.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AP, Johnson KD, Falcone EL, Sanalkumar R, Sanchez L, Hickstein DD, Cuellar-Rodriguez J, Lemieux JE, Zerbe CS, Bresnick EH, et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 2013;121:3830–3837. S3831–3837. doi: 10.1182/blood-2012-08-452763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, Frucht DM, Vinh DC, Auth RD, Freeman AF, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E, Nocka K, Beier DR, Chu TY, Buck J, Lahm HW, Wellner D, Leder P, Besmer P. The hematopoietic growth factor KL is encoded by the Sl locus and is the ligand of the c-kit receptor, the gene product of the W locus. Cell. 1990;63:225–233. doi: 10.1016/0092-8674(90)90303-v. [DOI] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Hsu AP, Ryu MJ, Wang J, Gao X, Boyer ME, Liu Y, Lee Y, Calvo KR, Keles S, et al. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest. 2012;122:3692–3704. doi: 10.1172/JCI61623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Kong G, Gao X, Chang Y-I, Hewitt KJ, Sanalkumar R, Prathibha R, Ranheim EA, Dewey CN, Zhang J, et al. Cis-regulatory mechanisms governing stem and progenitor cell transitions. Science Advances. 2015 doi: 10.1126/sciadv.1500503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YA, Sanalkumar R, O'Geen H, Linnemann AK, Chang CJ, Bouhassira EE, Farnham PJ, Keles S, Bresnick EH. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012;32:226–239. doi: 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura KR, Bresnick EH, Group GFM. The GATA factor revolution in hematology. Blood. 2017;129:2092–2102. doi: 10.1182/blood-2016-09-687871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura KR, Ong IM, DeVilbiss AW, Sanalkumar R, Bresnick EH. GATA Factor-Dependent Positive-Feedback Circuit in Acute Myeloid Leukemia Cells. Cell Rep. 2016;16:2428–2441. doi: 10.1016/j.celrep.2016.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura KR, Yang C, Boyer ME, Li L, Bresnick EH. Molecular basis of crosstalk between oncogenic Ras and the master regulator of hematopoiesis GATA-2. EMBO Rep. 2014;15:938–947. doi: 10.15252/embr.201438808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury MJ. Abnormal erythropoiesis and the pathophysiology of chronic anemia. Blood Rev. 2014;28:49–66. doi: 10.1016/j.blre.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Lee HY, Gao X, Barrasa MI, Li H, Elmes RR, Peters LL, Lodish HF. PPAR-alpha and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature. 2015;522:474–477. doi: 10.1038/nature14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M, Brice M, Engel JD, Papayannopoulou T. Dynamics of GATA-1 transcription factor expression during erythroid differentiation. Blood. 1993;82:1–9. [PubMed] [Google Scholar]
- Li L, Jothi R, Cui K, Lee JY, Cohen T, Gorivodsky M, Tzchori I, Zhao Y, Hayes SM, Bresnick EH, et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12:129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KC, Hosoya T, Brandt W, Ku CJ, Hosoya-Ohmura S, Camper SA, Yamamoto M, Engel JD. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest. 2012;122:3705–3717. doi: 10.1172/JCI61619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann AK, O'Geen H, Keles S, Farnham PJ, Bresnick EH. Genetic framework for GATA factor function in vascular biology. Proc Natl Acad Sci U S A. 2011;108:13641–13646. doi: 10.1073/pnas.1108440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, Corces MR, Flynn RA, Buenrostro JD, Chan SM, et al. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 2015;17:675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver SC, Katsumura KR, Davids E, Liu P, Kang YA, Yang D, Bresnick EH. Exosome complex orchestrates developmental signaling to balance proliferation and differentiation during erythropoiesis. Elife. 2016;5 doi: 10.7554/eLife.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munugalavadla V, Kapur R. Role of c-Kit and erythropoietin receptor in erythropoiesis. Crit Rev Oncol Hematol. 2005;54:63–75. doi: 10.1016/j.critrevonc.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Nemeth MJ, Topol L, Anderson SM, Yang Y, Bodine DM. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proc Natl Acad Sci U S A. 2007;104:15436–15441. doi: 10.1073/pnas.0704747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ, Dafou D, Kilo T, Smithson S, Lunt P, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat Genet. 2011;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- Ping N, Sun A, Song Y, Wang Q, Yin J, Cheng W, Xu Y, Wen L, Yao H, Ma L, et al. Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia. 2017;31:195–202. doi: 10.1038/leu.2016.162. [DOI] [PubMed] [Google Scholar]
- Pope NJ, Bresnick EH. Establishment of a cell-type-specific genetic network by the mediator complex component Med1. Mol Cell Biol. 2013;33:1938–1955. doi: 10.1128/MCB.00141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povinelli BJ, Nemeth MJ. Wnt5a regulates hematopoietic stem cell proliferation and repopulation through the Ryk receptor. Stem Cells. 2014;32:105–115. doi: 10.1002/stem.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povinelli BJ, Srivastava P, Nemeth MJ. Related-to-receptor tyrosine kinase receptor regulates hematopoietic stem and progenitor sensitivity to myelosuppressive injury in mice. Exp Hematol. 2015;43:243–252 e241. doi: 10.1016/j.exphem.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randrianarison-Huetz V, Laurent B, Bardet V, Blobe GC, Huetz F, Dumenil D. Gfi-1B controls human erythroid and megakaryocytic differentiation by regulating TGF-beta signaling at the bipotent erythro-megakaryocytic progenitor stage. Blood. 2010;115:2784–2795. doi: 10.1182/blood-2009-09-241752. [DOI] [PubMed] [Google Scholar]
- Rodrigues NP, Boyd AS, Fugazza C, May GE, Guo Y, Tipping AJ, Scadden DT, Vyas P, Enver T. GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood. 2008;112:4862–4873. doi: 10.1182/blood-2008-01-136564. [DOI] [PubMed] [Google Scholar]
- Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, Scadden DT. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- Rubel CA, Wu SP, Lin L, Wang T, Lanz RB, Li X, Kommagani R, Franco HL, Camper SA, Tong Q, et al. A Gata2-Dependent Transcription Network Regulates Uterine Progesterone Responsiveness and Endometrial Function. Cell Rep. 2016;17:1414–1425. doi: 10.1016/j.celrep.2016.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanalkumar R, Johnson KD, Gao X, Boyer ME, Chang YI, Hewitt KJ, Zhang J, Bresnick EH. Mechanism governing a stem cell-generating cis-regulatory element. Proc Natl Acad Sci U S A. 2014;111:E1091–1100. doi: 10.1073/pnas.1400065111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27:502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JW, Trowbridge JJ, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. A single cis element maintains repression of the key developmental regulator Gata2. PLoS Genet. 2010;6:e1001103. doi: 10.1371/journal.pgen.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JW, Trowbridge JJ, Johnson KD, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. Context-dependent function of "GATA switch" sites in vivo. Blood. 2011;117:4769–4772. doi: 10.1182/blood-2010-10-313031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner MA, Sanchez LA, Hsu AP, Shaw PA, Zerbe CS, Calvo KR, Arthur DC, Gu W, Gould CM, Brewer CC, et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood. 2014;123:809–821. doi: 10.1182/blood-2013-07-515528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura N, Miller E, Igarashi K, Yang D, Burstyn JN, Dewey CN, Bresnick EH. Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO Rep. 2016;17:249–265. doi: 10.15252/embr.201541465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Vicente C, Vazquez I, Conchillo A, Garcia-Sanchez MA, Marcotegui N, Fuster O, Gonzalez M, Calasanz MJ, Lahortiga I, Odero MD. Overexpression of GATA2 predicts an adverse prognosis for patients with acute myeloid leukemia and it is associated with distinct molecular abnormalities. Leukemia. 2012;26:550–554. doi: 10.1038/leu.2011.235. [DOI] [PubMed] [Google Scholar]
- Vidal SJ, Rodriguez-Bravo V, Quinn SA, Rodriguez-Barrueco R, Lujambio A, Williams E, Sun X, de la Iglesia-Vicente J, Lee A, Readhead B, et al. A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell. 2015;27:223–239. doi: 10.1016/j.ccell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Lin HY, Ng-Eaton E, Downward J, Lodish HF, Weinberg RA. Expression cloning and characterization of the TGF-beta type III receptor. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- Wilson DB, Dorfman DM, Orkin SH. A nonerythroid GATA-binding protein is required for function of the human preproendothelin-1 promoter in endothelial cells. Mol Cell Biol. 1990;10:4854–4862. doi: 10.1128/mcb.10.9.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wojchowski DM, Sathyanarayana P, Dev A. Erythropoietin receptor response circuits. Curr Opin Hematol. 2010;17:169–176. doi: 10.1097/MOH.0b013e328338008b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014a;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Morrissey CS, Keller CA, Mishra T, Pimkin M, Blobel GA, Weiss MJ, Hardison RC. Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis. Genome Res. 2014b;24:1945–1962. doi: 10.1101/gr.164830.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD, Yamamoto M. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25:415–427. doi: 10.1016/j.ccr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Himburg HA, Pohl K, Quarmyne M, Tran E, Zhang Y, Fang T, Kan J, Chao NJ, Zhao L, et al. Deletion of the Imprinted Gene Grb10 Promotes Hematopoietic Stem Cell Self-Renewal and Regeneration. Cell Rep. 2016;17:1584–1594. doi: 10.1016/j.celrep.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Moriguchi T, Souma T, Takai J, Satoh H, Morito N, Engel JD, Yamamoto M. GATA2 regulates body water homeostasis through maintaining aquaporin 2 expression in renal collecting ducts. Mol Cell Biol. 2014;34:1929–1941. doi: 10.1128/MCB.01659-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kong G, Rajagopalan A, Lu L, Song J, Hussaini M, Zhang X, Ranheim EA, Liu Y, Wang J, et al. p53−/− synergizes with enhanced NrasG12D signaling to transform megakaryocyte-erythroid progenitors in acute myeloid leukemia. Blood. 2017;129:358–370. doi: 10.1182/blood-2016-06-719237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yamamoto M, Engel JD. GATA2 is required for the generation of V2 interneurons. Development. 2000;127:3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.