Abstract
The characterization of polyubiquitin chains has been an analytical challenge for several decades. It has been shown that anchored and unanchored polyubiquitin chains with different isopeptide linkages and lengths exhibit a wide range of profoundly different cellular functions. However, structure function studies have been hindered by the difficulty of characterizing these complex chain structures. This report presents a broadly applicable workflow to characterize ubiquitin tetramers without the need for genetic mutations or reiterative immunoprecipitations. We use a top-down proteomic strategy that exploits ETciD activation on an orbitrap Fusion Lumos, and manual interpretation aided by graphical interpretation of mass shifts to facilitate characterization of chain topography and lysine linkage sites. Our workflow differentiates all topological features of the numerous isomers of tetraubiquitin, which have molecular masses in excess of 34,000 Da, and identifies linkage sites in these branched proteins.
INTRODUCTION
Polyubiquitination is a post-translational modification with many functions within eukaryotic cells. Both anchored (attached to a substrate protein) and unanchored chains have been shown to play major roles in DNA repair, [1] protein degradation, [2,3,4] cancer morphology, [5] and immune response. [6,7] It is now well accepted that the size of polymers and the sites of lysine linkages between their subunits determine their physiologic effects. Structures of ubiquitin (Ub) chains and the relationship of structures to functions have been difficult to characterize because of their nontraditional linkages, redundant sequences and heavy masses. [8] In a previous study by this group, a novel strategy was developed to characterize isomeric unbranched and branched ubiquitin trimers by top-down mass spectrometry. [9] The present report extends our successful strategy to a broad group of unbranched and branched tetrameric chains.
Tetrameric ubiquitins have been of particular interest following the demonstration [10] that a chain of four ubiquitins connected by linkages through lysines at position 48 is the minimum signal for efficient targeting of proteins to the proteasome for degradation. Other functions have been assigned to tetramers comprising other linkages. [11] Recent studies have proposed several techniques for the elucidation of tetrameric ubiquitin chains. [6,7,12,13,14] One method demonstrated for elucidation of polyubiquitin chains uses linkage site-specific deubiquitinating enzymes (DUBs) to differentiate lysine isopeptide attachment and topology. [14] This method is reported to provide good results, however many different DUBs must be used and each must be used in a separate experiment. Here we report a sensitive proteomic workflow that can be applied post-analysis to spectra of components of cell lysates. It provides exact molecular mass determination and complete sequence characterization for most ubiquitin tetramers.
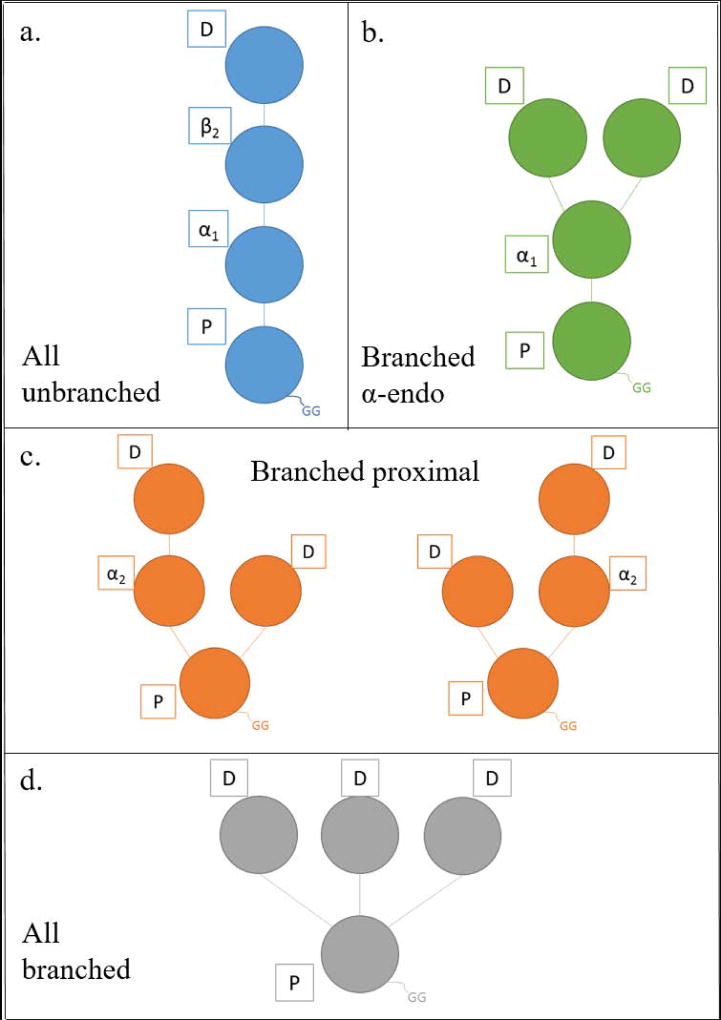
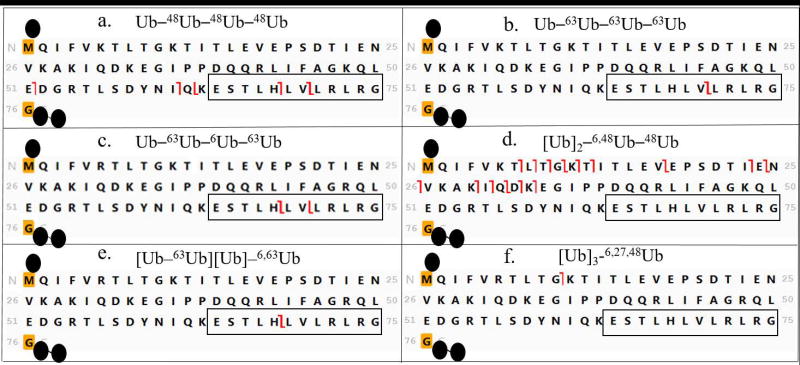
Tetraubiquitin (Ub4) chains make up a large family of isomers. Each isopeptide linkage can be formed with one of seven different lysines in ubiquitin (see examples in Figure 1) and the chain can comprise homogeneous or heterogeneous linkages. Considering possible combinations of linkages among the seven lysines, 819 isomeric tetramers are predicted. (Linkage at M1 does not produce a branched polypeptide and is not considered here.) Without specifying linkage sites, there are four general topologies that a tetrameric chain can take (Figure 2). These topologies are a critical component of the relationship between tandem mass spectra and the sequence of these branched proteins.
Figure 1.
Figure 2.
The objective of the current study is to develop a structured workflow for interpreting top-down mass spectra of unanchored tetraubiquitins to ascertain topology and linkage sites, and to test and demonstrate this approach across all tetramer topologies. The strategy is tested on six synthetic standards whose chemical structures are shown in Figure 1. The strategy requires extensive fragmentation across the branched polypeptides, provided here by electron transfer dissociation on an orbitrap Fusion Lumos mass spectrometer.
METHODS
Synthesis of ubiquitin tetramers
All ubiquitin tetramers were assembled from the respective recombinant Ub monomers (Figure 1) using linkage-specific enzymes as described [13,15,16,17] or, in case of [Ub]3–6,27,48Ub, by combining this methodology with a nonenzymatic chain assembly approach. [18] Molecular masses differ (See Supplemental Table 1 and Supplemental Table 2) because residue replacements (K → R) were used in several cases as chain-terminating mutations to control the synthesis.
LC-MS/MS
Intact tetramers were diluted to 0.03 mg/mL in Solvent A (97.5% water, 2.5% ACN and 0.1% formic acid). The chromatography was performed using an Ultimate 3000 ultra-high performance liquid chromatograph (ThermoFisher Scientific, San Jose, CA) interfaced to a orbitrap Fusion Lumos Tribrid mass spectrometer (ThermoFisher Scientific). Five µL (~4.4 pmol) was injected, concentrated and desalted on a PepSwift Monolith trap (200 µm × 5 mm) for 5 min at 99% Solvent A before separation on a ProSwift RP-4H column (100 µm × 25 cm) (ThermoFisher Scientific) with a gradient of 30% to 50% solvent B (75% ACN, 25% water, 0.1% formic acid) over 15 min. Precursor and fragment ion masses were acquired with a resolution of 120,000 at m/z 200 using “intact protein mode” with 1mtorr ion routing multipole pressure. Data dependent MS/MS was carried in top-N mode (N=2) with a precursor list of m/z values calculated for each tetramer. Isolated parents ions were fragmented using electron transfer dissociation supplemented with collisionally induced dissociation (ETciD) with a 3 msec ETD reaction time and supplemental activation at 10% normalized CID and averaging 20 microscans.
Processing the spectra
Precursor and fragment ions were deconvoluted using Xtract 3.0 (Thermo Scientific). Fragment ions were matched against modified sequences of monoubiquitin using ProSight Lite [19] with a mass tolerance equal to or less than 9 ppm. ProSight Lite provides graphic interpretation of tandem mass spectra, classifies fragment ions as a, b, c, x, y and z and provides a probability for each modified structure based on fragmentation. Because our polymers contain structural redundancy, an analysis of unique fragments was required and these were assigned manually with support from fragment ions illustrated on the sequence of ubiquitin by the software.
Nomenclature
To facilitate characterization by mass spectrometry, the different moieties in the polymer must be further distinguished. Traditionally the Ub with a free C-terminus is called the proximal Ub, designated as Pin Figure 2. The Ub attached most distant from the proximal Ub is known as the distal (D) moiety. [16] Intermediate ubiquitins are termed endo. [20] In this mass-based analysis of tetramers there are two different types of endo Ubs. The endo Ub attached by its C-terminus directly to a lysine on the proximal Ub is defined as an α-endo Ub. In unbranched tetramers the α-endo Ub has only the proximal mass attached to its C-terminus and is designated with a subscript 1 (i.e. α1-endo Ub). In the general case (αn-endo Ub) the subscript n designates the number of Ubs attached directly and indirectly to the C-terminus of the endo moiety in question. The β-endo Ub is attached by its C-terminus directly to a lysine on an α-endo Ub. The masses of two Ub moieties are attached to the β-endo Ub’s C-terminus in a tetramer. Thus it is designated with a subscript 2 (i.e. β2-endo Ub). This nomenclature can be extended to higher polyubiquitins. For anchored chains, we propose the addition of an S (for substrate) within the subscript (e.g. β2S-endo Ub).
RESULTS AND DISCUSSION
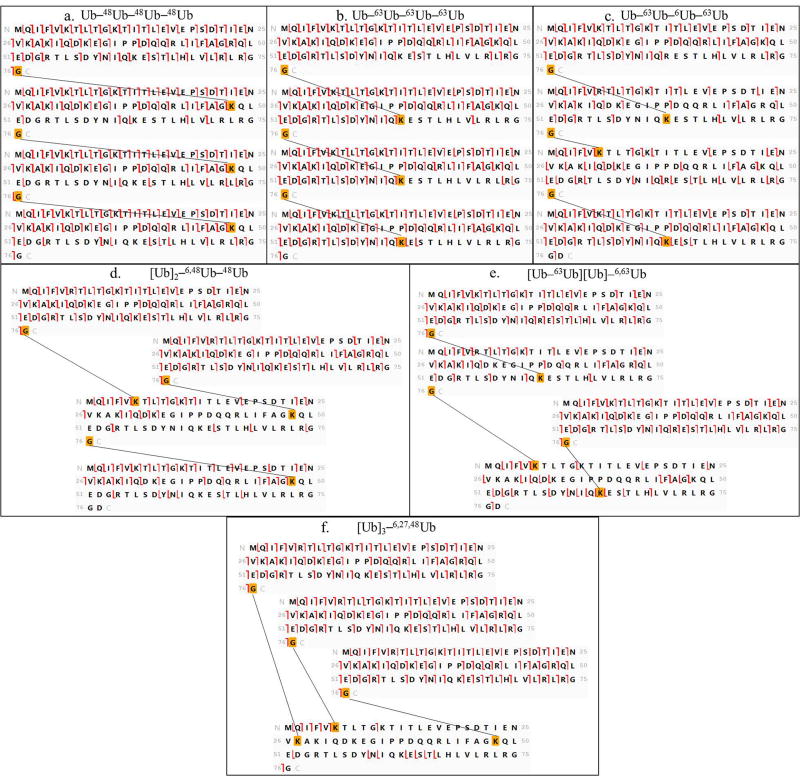
Six tetramers were synthesized for this study, representing the different topologies seen in Figure 2. Three unbranched tetramers were studied (Figure 2a), Ub–48Ub–48Ub–48Ub, Ub–63Ub–63Ub–63Ub and Ub–63Ub–6Ub–63Ub (chain nomenclature as in [15]). A branched topology in which an α1-endo Ub carries two distal ubiquitins, [Ub]2–6,48Ub–48Ub, is represented in Figure 2b. Tetramer [Ub–63Ub][Ub]–6,63Ub with two isopeptide linkages on the proximal Ub is shown in Figure 2c. In this case one branch comprises a distal moiety and the other branch contains an α2-endo Ub linked to a distal unit. Finally, a tetramer [Ub]3–6,27,48Ub with three distal Ubs linked to the proximal Ub was synthesized, shown in Figure 2d.
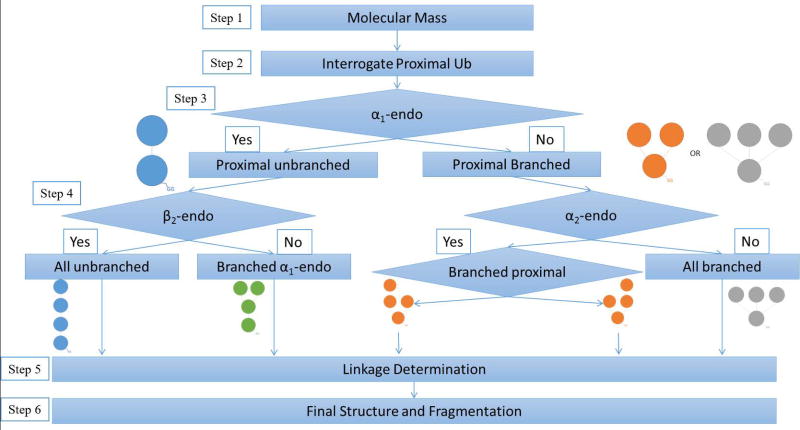
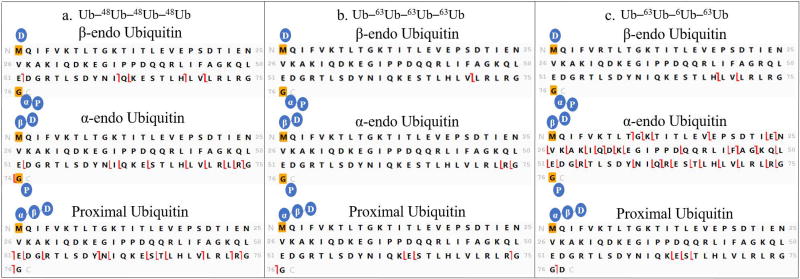
The workflow presented in Figure 3 extends the strategy of our previously published work on trimers of Ub [9], in which the monoubiquitin sequence is used as a template to assess the fragmentation patterns of each of the Ub moieties present in the tetramer. ProSight Lite software [19] assigns fragment ions from the tandem spectrum to a template sequence, in our case the sequence of monoubiquitin. The software allows custom mass additions to be made to any amino acid in the template sequence, and masses equivalent to one, two or three Ub moieties are added to simulate various units in the topology as labeled in Figure 2. The fragmentation patterns provided by the software are used to assign manually the topology of each tetramer. Once the topology is deciphered, specific linkage sites can be assigned by inspection of fragmentation patterns assigned to the template sequence of each subunit. Only c and z ions are considered on the templates, because b and y ions were found to introduce uninformative complexity, for example, assignments corresponding to internal fragment ions. [21,22] A tandem mass spectrum of Ub–63Ub–6Ub–63Ub is shown in Supplementary Figure 1 to show the quality of the spectra obtained. Fragment ions are recorded all across the mass range between m/z 200 and 34,500 Da. Assignments shown were made using ProSight Lite. The molecular ion isotope window and the chromatogram are also presented in the Figure.
Figure 3.
This workflow is specific to tetrameric Ub chain lengths, and is slightly different from the top-down workflows we have reported for ubiquitin dimers [23] and trimers. [9] Therefore Step 1, molecular mass determination (Figure 3), is necessary before moving on to any subsequent step. The intact masses of our standard compounds were all confirmed as tetramers within 2 ppm mass error (Supplemental Table 1). As an overview, in Step 2 the spectrum is mapped onto the proximal moiety (distinguished by a free carboxyl terminus) to confirm it as the spectrum of an unanchored polyUb. The proximal Ub can also be interrogated in this step to recognize the attachment site closest to the distinctive C-terminus. Steps 3 and 4 then test for α1-endo, α2-endo and β2-endo subunits based on diagnostic ions. Once the presence/absence and nature of any endo Ubs are known, a general topological shape can be assigned. After the full topology is determined, linkage sites are identified by inspection of fragmentation patterns assigned to the templates that present each subunit with its modifications (Step 5). In Step 6 the deduced structure is confirmed as that with the highest number of unique fragments.
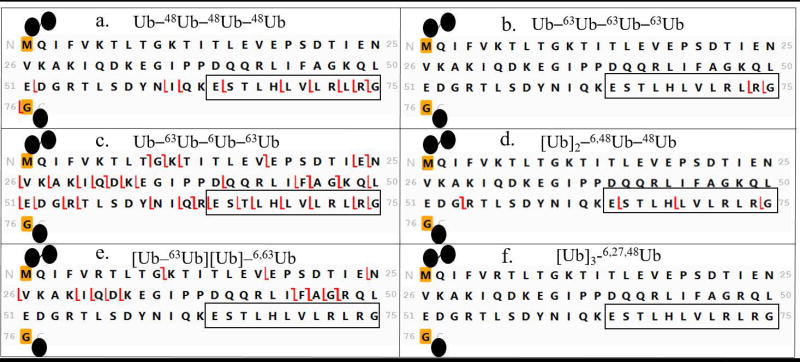
In Step 2 (Figure 3) the proximal moiety is used as the template for fragment assignment by ProSight Lite. The proximal moiety (see Figure 2) is unique in the ubiquitin tetramer as the only Ub with a free carboxyl terminus. As illustrated in Figure 4 fragmentation was mapped with high density on the proximal unit templates for all our synthetic tetramers. This confirms that each sample is a polyubiquitin. The fragmentation pattern examined in Step 2 also provides information about the lysine residue closest to the C-terminus that is modified as a linkage site. All the z ions mapped on the template (the unmodified sequence of proximal Ub) can be formed only by cleavage in the unmodified portion of the sequence. Thus formation of z ions can be traced back from the carboxyl terminus through the sequence until it ceases in the vicinity of a lysine residue that is modified (linked to another subunit). Modified lysines are boxed in black in Figure 4, based on this analysis. It should be pointed out that c ions mapped in this initial match with the proximal Ub sequence can also be formed by fragmentation in the distal moiety and in some other subunits. Thus c ions are potentially redundant and cannot be used to provide information in Step 2. In subsequent steps the other mass additions on the proximal Ub and the overall topology are determined.
Figure 4.
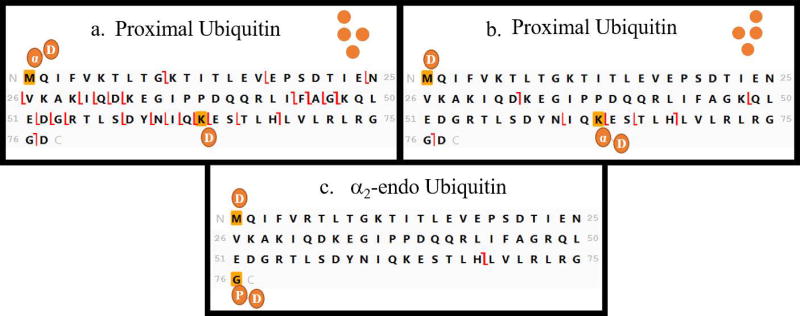
Step 3 is the first step in determining the topology. In this step, diagnostic ions are sought that will reveal any α1-endo subunit present. A Ub in the α1-endo position is attached to a proximal Ub through the glycine residue at its carboxyl terminus (see Figure 2). This connectivity is represented in the template (Figure 5) by a circle and the mass of a Ub moiety is attached in silico to the carboxyl terminus. In a tetramer two other Ub moieties will be attached to another lysine(s) in any α1-endo subunit, and these are represented by placing the mass of two Ub on the N-terminal methionine. (In reality these Ub would be attached at any of the lysines 63, 48, 33, 29, 27, 11 or 6.) In characterizing the topology of a native polyubiquitin the masses of all Ubs added as trial modifications would be the same in the analyses illustrated in Figures 5 and 6. Because synthetic tetramers with residue substitutions (Figure 1) were used here, appropriate masses were selected (Supplementary Table 2) for trial modifications in Figures 5 and 6. Fragment ions from the tandem spectrum are mapped to the trial template, and any c or z ions that are observed to be formed by fragmentation between K63 and the modified carboxyl terminus are diagnostic for the presence of an α1-endo subunit. The regions that should be examined for diagnostic ions are boxed in Figure 5. Ions formed by cleavage between K63 and G76 will be mapped only when the C-terminus is bonded to another Ub moiety. If there is branching on the proximal Ub, no fragment ions will be mapped in this region, indicating that no subunit defined as α1-endo Ub is present (Figure 5e and f). In this study the structures that do not have an α1-endo Ub are the branched proximal [Ub–63Ub][Ub]–6,63Ub (Figure 5e) and the all branched [Ub]3–6,27,48Ub (Figure 5f).
Figure 5.
Figure 6.
Step 4 (Figure 3) is the final step needed to determine the topology of an unknown tetramer, and it is applied to tetramers with or without an α-endo subunit as defined in Step 3. In Step 4 we use the graphic display of fragmentation to test for another Ub moiety in the chain that also carries two branching sites. In this step the substituted moiety will be characterized as an α2-endo or a β2-endo Ub. (See Figure 2.) As in Step 3, the Ub sequence is modified by addition of Ub masses at the amino and carboxyl termini to provide a template that can allow interrogation for isopeptide modification of the intermediate lysines. In this case the mass of two Ub is added to the carboxyl terminus and the mass of one Ub is added to the amino terminus methionine (Figure 6). This simulates α2-endo and β2-endo moieties, which carry the mass of two Ub at the C-terminus. Fragmentation detected between K63 and the C-terminus confirms the attachment of two Ub at the carboxyl terminus. The tandem mass spectra of the four tetramers synthesized with an α2-endo or β2-endo Ub contain such diagnostic ions (Figure 6a, b, c, and e), and diagnostic ions are absent in the spectra of standards that do not contain either of these subunits (Figure 6d and f).
Diagnostic ions are detected in Steps 3 and 4 in spectra of all six synthetic standards, which correctly define the topological classification of each tetramer. Knowledge of the topological shape is necessary before the linkage sites can be determined, since each topology requires trial modifications at different positions to map fragment ions in its template (Step 5).
After the topology of the tetramer is characterized, information can be sought about the linkage sites. One approach is to reiteratively add Ub masses at each lysine in the template and accept the isomer that maps the most unique or non-redundant fragment ions. Unique fragmentation is determined manually in this study, as discussed in Steps 2–5. In a more general approach, the appropriate Ub masses are added to the N- and C-termini of each subunit (Figures 7–9) and fragmentation is mapped on the correct template. Templates of each subunit (carrying trial modifications dictated by topology) in each of the three unbranched tetramers (Figure 2a) are shown in Figure 7. Ubiquitin moieties added as trial modifications are labelled as P, D, α or β in the circles. Masses for these specific Ub vary because synthetic standards have been used. Supplementary Table 2 provides the exact masses for each case. In these tetramers sites of attachment must be defined in the proximal, α-endo and β-endo moieties. The panels show fragment maps for each of the three moieties in each of the three unbranched tetramers. In each of the panels, fragmentation can be seen to proceed from the carboxyl terminus back toward, but not beyond, the first lysine that is modified by an isopeptide linkage. The same successful strategy has been used to decipher structures of ubiquitin trimers. [9] As seen in Figure 7c this is particularly informative for the K6 linkage site in the α1-endo moiety of Ub–63Ub–6Ub–63Ub.
Figure 7.
Figure 9.
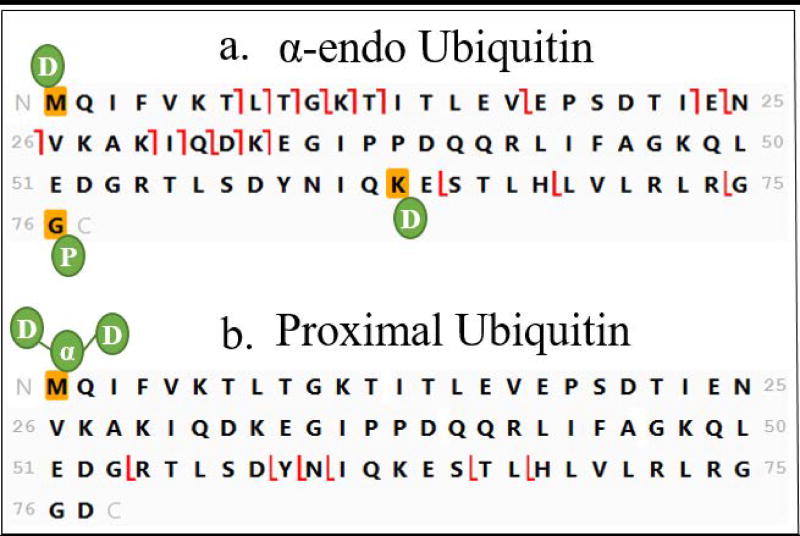
In tetramers with branched α1-endo topology (Figure 2b), linkage sites must be determined in only two subunits, the proximal and α-endo moieties. Determination of the linkage site on the proximal Ub proceeds analogously to that of the proximal moieties in the all unbranched tetramers illustrated in Figure 7; the mass of three Ub moieties is added to the N-terminus and the template is inspected for c and z ion formation from the carboxyl terminus back into the chain. As seen in Figure 8b this fragmentation proceeds past K63 and terminates before K48. Thus the α-endo Ub is proposed to be attached to K48 in the proximal Ub. The α-endo Ub contains two linkage sites in this topology and must also be interrogated. Here the template is constructed (Figure 8a) by adding the mass of one distal Ub on the N-terminus, the mass of the proximal Ub on the C-terminus, and one Ub on K63 (the lysine closest to the C-terminus). Observation of a series of c and z ions formed by fragmentation beyond K6 but not in residues 1–5 locates one linkage site at K6. The combination of c ions and z ions localizes the second attachment to K48 or K63. In Figure 5d we can see that fragmentation occurs between K63 and K48 (c ion formed at G53 and z ion at R54) on the α-endo Ub, which eliminates K63 as a possible linkage assignment, definitively allocating the attachment to K48.
Figure 8.
In the tetramer topology group called branched proximal (Figure 2c) the proximal Ub carries two linkage sites. One of these comprises a mono Ub moiety and the other a diUb chain. Graphic mapping of fragment ions in the tandem spectrum of synthetic [Ub–63Ub][Ub]–6,63Ub onto the template of the proximal moiety in Step 2 (Figure 3c) clearly assigned one branch at K63. To discern whether the dimer or the monomer was linked at K63, dimer and monomer masses were added alternately to M1 or K63 and the template that allowed assignment of more unique fragment ions was chosen as the correct orientation. This comparison is illustrated in Figures 9a and 9b, where it can be seen that localization of the dimer mass at the N-terminus and the monomer at K63 (Figure 9a) allows more fragmentation to be mapped than the configuration represented in figure 9b. This template also allows K6 to be assigned as the attachment site for diUb, distinguished by c fragmentation at G10.
To initiate template-based analysis of the linkage site on the α2-endo Ub in the branched proximal tetramer [Ub–63Ub][Ub]–6,63Ub, the mass of two Ubs is added to the C-terminus and the mass of the distal Ub is added to the N-terminus (Figure 9c). The fragmentation pattern supports linkage at K63. Thus analysis by graphic fragmentation indicates that this tetramer is [Ub–63Ub][Ub]–6,63Ub with two isopeptide linkages on the proximal Ub. This assignment is further confirmed in Step 6.
The last topological category to consider in interpreting the spectrum of an unknown ubiquitin tetramer is that in which the proximal Ub carries three monoubiquitin moieties attached at three different sites (called here the all branched tetramer). Examination of the template for the proximal moiety in Step 2 (Figure 4f) indicates that formation of z ions proceeds from G76 back past K63 to terminate before K48. This indicates that K48 is a linkage site and K63 is not. Addition of the masses of a single Ub and two Ubs to M1 and K63, respectively, in the proximal sequence (Figure 10a–b) produces a fragmentation map that supports K6 as a linkage site. Reversing the modifications in the template to two Ubs on M1 and one on K63 of the proximal sequence accommodates no informative fragmentation. Thus MS/MS evidence supports linkages at K6 and K48. No information is available on the third linkage site of the synthetic standard (K27). The absence of fragmentation suitable for this last assignment is also evident when the spectrum is mapped onto the correct structure in Figure 11f. Confirming fragmentation is seen for linkages at K48 (detailed in Step 2 and Step 6) and K6 (detailed in Step 5). The last moiety is limited to linkage at K11, K27, K29, or K33, with insufficient fragmentation to distinguish further. This workflow is compatible with any future developments in activation that will provide the missing fragmentation.
Figure 10.
Figure 11.
After the structure of each of the synthetic standards is characterized using the strategy outlined above, a final evaluation is made in Step 6. In Figure 11 the fragment ions recorded in the MS/MS spectra are assigned to the structures deduced in Steps 1–5. These can be compared with patterns assigned to incorrect structures. The correct structure should have a high fragment density and the highest number of unique or non-redundant fragment ions. All structures shown in Figure 11 yielded more extensive unique fragmentation patterns than those of incorrect alternatives.
In a preliminary study, successful interrogation of the tandem mass spectrum of pentameric polyubiquitin Ub–48Ub–48Ub–48Ub–48Ub [22] indicates that the strategy developed and tested here and in reference 9 can be further extended to pentamers.
CONCLUSIONS
Here we report a sensitive proteomic workflow that can be applied post-analysis to spectra of components of cell lysates. The top-down mass spectrometric strategy presented in this study correctly classifies Ub tetramer chains into one of four possible topologies. Assignment of the topology permits fragmentation to be visualized on an appropriate template and allows linkage sites to be identified on each ubiquitin moiety within the chain. The success of the approach depends on achieving extensive fragmentation. Sufficient fragmentation was recorded using ETciD on a tribrid orbitrap Fusion Lumos to assign topology to all the standards analyzed and to assign all linkage sites in three of the four topologic groups of unanchored ubiquitin tetramers. The most highly branched isomer fragmented most poorly. We have every confidence that our strategy for assigning topology and linkage sites to polyubiquitins will be even more convenient and effective as activation techniques are developed to provide more nearly complete fragmentation of heavy proteins. This is a workflow that can also be readily modified for ubiquitin-like polymers (e.g. SUMO) and for mixed polymers. We are currently developing an analogous strategy to characterize anchored polyubiquitins.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants GM021248, GM065334, and OD019938. We thank Dulith Abeykoon for providing a sample of the pentameric ubiquitin.
References
- 1.Chen J, Chen ZJ. Regulation of NF-κB by ubiquitination. Curr. Opin. Immunology. 2013;25:4. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amerik AY, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao R, Nanduri P, Rao Y, Panichelli RS, Ito A, Yoshida M, Yao TP. Proteasomes Activate Aggresome Disassembly and Clearance by Producing Unanchored Ubiquitin Chains. Molecular Cell. 2013;51:819. doi: 10.1016/j.molcel.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varshavsky A. The Ubiquitin System, an Immense Realm. Annu. Rev. Biochem. 2012;81:167. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 5.Vogel RI, Coughlin K, Scotti A, Iizuka Y, Anchoori R, Roden RBS, Marastoni M, Bazzaro M. Simultaneous inhibition of deubiquitinating enzymes ( DUBs ) and autophagy synergistically kills breast cancer cells. Oncotarget. 2015;6:4159. doi: 10.18632/oncotarget.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto E, Yamanaka Y, Ishikawa A, Aoki-Kawasumi M, Mito-Yoshida M, Ohmura-Hoshino M, Matsuki Y, Kajikawa M, Hirano H, Ishido S. Contribution of lysine 11-linked ubiquitination to MIR2-mediated major histocompatibility complex class I internalization. J. Biol. Chem. 2010;285:35311. doi: 10.1074/jbc.M110.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strachan J, Roach L, Sokratous K, Tooth D, Long J, Garner TP, Searle MS, Oldham NJ, Layfield R. Insights into the molecular composition of endogenous unanchored polyubiquitin chains. J. Proteome Res. 2012;11:1969. doi: 10.1021/pr201167n. [DOI] [PubMed] [Google Scholar]
- 9.Lee AE, Geis-Asteggiante L, Dixon EK, Kim Y, Kashyap TR, Wang Y, Fushman D, Fenselau C. Preparing to read the ubiquitin code: characterization of ubiquitin trimers by top-down mass spectrometry. J. of Mass Spec. 2016;51:315. doi: 10.1002/jms.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannon J, Nakasone M, Fushman D, Fenselau C. Proteomic Identification and Analysis of K63-Linked Ubiquitin Conjugates. Anal. Chem. 2012;84:10121. doi: 10.1021/ac302675y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannon JR, Martinez-fonts KL, Robotham SA, Matouschek AT, Brodbelt JS. Top down 193 nm ultraviolet photodissociation mass spectrometry for simultaneous determination of polyubiquitin chain length and topology. Anal. Chem. 2015;87:1812. doi: 10.1021/ac5038363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hospenthal MK, Mevissen TET, Komander D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest) Nat. Protoc. 2015;10:349. doi: 10.1038/nprot.2015.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Mixed-linkage ubiquitin chains send mixed messages. Structure. 2013;21:727. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 2002;324:637. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 17.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004;279:7055. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 18.Castañeda CA, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. Nonenzymatic Assembly of Natural Polyubiquitin Chains of Any Linkage Composition and Isotopic Labeling Scheme. J. Am. Chem. Soc. 2011;133:17855. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fellers RT, Greer JB, Early BP, Yu X, LeDuc RD, Kelleher NL, Thomas PM. ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics. 2015;15:1235. doi: 10.1002/pmic.201570050. http://prosightlite.northwestern.edu/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansour W, Nakasone MA, von Delbrück M, Yu Z, Krutauz D, Reis N, Kleifeld O, Sommer T, Fushman D, Glickman MH. Disassembly of Lys11- and mixed-linkage polyubiquitin conjugates provide insights into function of proteasomal deubiquitinases Rpn11 and Ubp6. J. Biol. Chem. 2015;290:4688. doi: 10.1074/jbc.M114.568295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frese CK, Altelaar AFM, Van Den Toorn H, Nolting D, Griep-Raming J, Heck AJR, Mohammed S. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 2012;84:9668. doi: 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- 22.Sobott F, Watt SJ, Smith J, Edelmann MJ, Kramer HB, Kessler BM. Comparison of CID versus ETD based MS/MS fragmentation for the analysis of protein ubiquitination. J. Am. Soc. Mass Spectrom. 2009;20:1652. doi: 10.1016/j.jasms.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Lee AE. Characterization of Polyubiquitin Chains by Mass Spectrometry Ph.D. Thesis. University of Maryland; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.