Abstract
Transcriptome profiling enables discovery of gene networks that are altered in alcoholic brains. This technique has revealed involvement of the brain’s neuroimmune system in regulating alcohol abuse and dependence, and has provided potential therapeutic targets. In this review, we discuss Toll-like-receptor pathways, hypothesized to be key players in many stages of the alcohol addiction cycle. The growing appreciation of the neuroimmune system’s involvement in alcoholism has also led to consideration of crucial roles for glial cells, including astrocytes and microglia, in the brain’s response to alcohol abuse. We discuss current knowledge and hypotheses on the roles that specific neuroimmune cell types may play in addiction. Current strategies for repurposing US FDA-approved drugs for the treatment of alcohol use disorders are also discussed.
Keywords: : alcohol, drug repurposing, neuroimmune, transcriptome
Gene expression profiling is a powerful tool for defining key regulatory genes as well as molecular targets for potential drug development or repurposing. In particular, gene expression profiling has been instrumental in understanding how the neuroimmune system contributes to various aspects of alcohol dependence. Some of the first microarray analyses of cortical samples from human alcoholic brain identified differentially expressed genes associated with alcohol use disorder (AUD) [1–9]. Immune-related genes emerged as an unexpected category of abundantly changed genes in alcoholics [3]. This discovery catalyzed investigation into the role of immune signaling in alcohol dependence. Many of the immune-related genes were related to inflammatory responses such as production of cytokines, chemokines, glial activation and signaling components of the NF-κB inflammatory pathway [5]. Complementing the gene expression data, NF-κB and its p50 homodimer were shown to be upregulated in human alcoholic frontal cortex [10]. In addition, human alcoholics show positive correlations between alcohol craving and serum levels of lipopolysaccharides, peptidoglycans, cytokines and chemokines (e.g., IL-8, IL-1β), suggesting that activation of the innate immune system may regulate alcohol craving and consumption [11–13]. Knockout of some of the immune genes nominated by the gene expression studies (B2m, Cd14, Il1rn, Il6, Ctss and Ctsf) reduced ethanol consumption and preference in the 24-h two-bottle choice test in mice and provided behavioral validation for these candidate genes [14]. In contrast, the immune activator lipopolysaccharide (LPS) increased ethanol consumption in mice [15], indicating that enhancement of immune signaling is associated with increased drinking as suggested in humans. These preclinical and clinical evidences support the hypothesis that activation of inflammatory pathways is a key process in alcohol dependence and further implicate neuroimmune pathways as relevant targets to treat AUD.
In brain, alcohol induces neuroinflammatory and neurodegenerative changes partially mediated by innate immune activation [16,17]. The TLR pathway, which activates NF-κB and leads to the secretion of pro-inflammatory cytokines and chemokines, has been suggested to be a key regulator of alcohol-induced innate immune activation. It remains unclear which cell types are mediating this response in the CNS. Neurons, astrocytes and microglia release and respond to neuroimmune signaling components. To determine if cell-type specific modulation of neuroimmune signaling is important in alcohol dependence, it is necessary to understand the transcriptional changes that occur in each cell type after ethanol exposure. Transcriptomics has already been used to identify potential drugs for treating AUD [18], and transcriptome profiling in different cell types after ethanol exposure will further define discrete cellular effects in the brain.
The mechanisms by which alcohol triggers inflammation in the brain are only partially understood, complicated in part by the interplay between peripheral and central immune activation. Alcohol consumption increases intestinal permeability and levels of LPS, resulting in peripheral and central immune activation [11–12,15,19]. Moreover, the extent to which stress plays a role in central immune activation is another complicating factor. Literature has shown that stress increases brain cytokines which potentially act as neuromodulators of alcohol consumption (for review see [16]). An additional consideration is that withdrawal from alcohol is an extremely stressful event, causing stress-induced increases in CRF as well as cytokines [17]. Because of the potential effects of stress on central immune activation, the drinking studies cited in this review focus on ethanol administration (e.g., alcohol fed diet, two-bottle choice tests, operant self-administration) under non-stressful conditions, unless otherwise noted. Additionally, levels of cytokines and other neuroimmune mediators measured during alcohol withdrawal are not discussed here.
We will discuss the TLR neuroimmune pathway and its validated genomic targets as well as the potential for identifying new drug targets that are TLR- and cell-type specific. We also review alcohol-induced transcriptional changes in astrocytes, microglia and neurons and their role as immune regulators and cellular targets for future drug development. Finally, we discuss using cutting-edge technology, such as the Library of Integrated Network-based Cellular Signatures (LINCS) program and other genomics strategies, to predict drugs for the treatment of alcohol dependence. Although these techniques are still in their infancy, they provide promising examples of the future of drug repurposing as a strategy for advancing therapeutic options for AUD.
Toll-like receptors
TLRs & immune responses
Innate immunity is the body’s first defense against infection and is responsible for most peripheral and central inflammatory responses. Acute inflammatory stressors induce a large number of genomic/transcriptional responses, and unlike adaptive immunity, do not require genetic rearrangement to mount an inflammatory response [20]. Cells of the innate immune system rely on germ-line encoded receptors that recognize broadly defined molecular motifs known as pathogen-associated molecular patterns (PAMPs). Multiple receptor families are comprised of transmembrane pattern-recognition receptors (PRRs), which initiate inflammatory signaling in response to a wide variety of PAMPs. TLRs represent a class of PRRs that are critical for both innate immune reactions to pathogens and for initiating adaptive immunity [20]. Most TLRs rely on homodimerization to achieve specific agonist recognition [21]. In addition to detecting molecular motifs associated with microorganisms, TLRs recognize endogenous ligands referred to as damage- (or danger-) associated molecular patterns (DAMPs), including β-defensin, heat shock proteins 60 and 70, stathmin, HMGB1 and reactive oxygen species [22,23]. The recognition of specific PAMPs or DAMPs leads to receptor activation and initiation of multiple signaling cascades, eventually resulting in NF-κB pathway activation and immune-related gene expression changes.
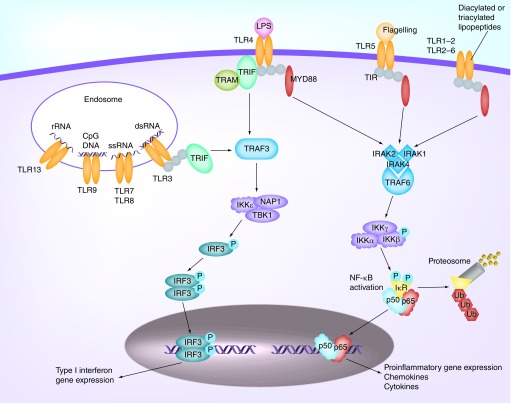
There are currently 11 known TLRs in humans and 13 in mice, and each receptor responds to specific classes of pathogens (Figure 1). TLRs 1–2, 4–6 and 11–13 are localized on the cell surface, whereas TLRs 3 and 7–9 are found on the endosome or lysosome compartments in the endoplasmic reticulum [24,25]. TLR2 recognizes lipoproteins, peptidoglycan (PGN) and dectin, and TLR2 dimerization with TLR1 or TLR6 allows for the discrimination between triacylated and diacylated lipoproteins, respectively. LPS primarily targets TLR4. TLR2 and TLR4 can also form oligomers, which recognize a broader range of microbial motifs [26]. TLR5 senses bacterial flagella and is commonly found in the intestine [27]. TLR11 responds to bacteria associated with urinary tract infections, such as uropathogenic Escherichia coli and a profilin-like protein from the parasite Toxoplasma gondii [28]. TLRs 3 and 7–9 recognize intracellular pathogen-derived nucleic acid motifs such as dsRNA, ssRNA and even mRNA from viruses, pathogens or infected cells [29]. TLR9 recognizes nonmethylated CpG islands of bacterial and viral mRNAs. PAMPs for TLRs 10, 12 and 13 are not yet well established. Recent evidence suggests that TLR10 may act as a modulatory receptor with mainly inhibitory effects by forming a heterodimer with TLR2 [30]. TLR12 was hypothesized to form a heterodimer with TLR11 to recognize a profilin-like protein from T. gondii [31].
Figure 1. . Toll-like receptors initiate inflammatory responses via two intracellular signaling transduction cascades.
TLRs initiate inflammatory responses via two intracellular signaling transduction cascades: MyD88-dependent pathway and MyD88-independent pathway. As shown in Figure 1, TLRs are activated by binding of PAMPs or DAMPs to initiate an innate immune response. After binding, all TLRs (except for TLR3) utilize the MyD88-dependent pathway, via recruitment of adaptor proteins in the cytoplasmic Toll/interleukin receptor (TIR) domain (MyD88, IRAK4 and IRAK) to activate NF-κB [32]. In contrast, TLR3 recruits TIR-domain-containing adaptor-inducing IFN-β (TRIF) and bifurcates to activate either type-I interferons or NF-κB [32]. Interestingly, TLR4 can also induce inflammatory responses via the MyD88-independent pathway by recruiting the TLR-adaptor proteins TRAM and TRIF [33].
TLRs & alcohol exposure
Differential expression of TLRs 1, 6 and 7, as well as genes downstream of NF-κB (e.g., Casp8, Fadd, Ikbkb, Kbkg, Tradd, Map3k1), were observed in mice predisposed to drink ethanol [34]. Microarray analysis of gene expression in Drosophila showed ethanol increases expression of genes in the Toll pathway [35,36], as seen in vertebrates. An increase in mRNA expression of Tlr 1, 2, 4 and 6–9 was detected in the liver of mice fed an ethanol-containing diet for 10 days [37]. Additionally, mice consuming ethanol for 5 weeks showed increased Tlr 2, 4 and 9 mRNA in the cerebellum [38]. Ten days of binge-like drinking increased brain mRNA and protein expression of TLRs 2–4 in rats [39]. Further support comes from study of human postmortem brain showing increased protein expression of TLRs 2–4, which correlated with lifetime alcohol consumption [39].
Despite the evidence that TLRs are key regulators of immune activity in response to alcohol, only a few studies have examined the role of TLRs in regulating behavioral responses to alcohol. Most research has focused on how ethanol changes TLR expression, in particular TLR4, due to its apparent ability to protect against ethanol-induced changes in immune gene expression. Knockout of TLR4 or its downstream signaling component, MYD88, as well as TLR4 antagonism by (+)-naloxone in mice decreased the duration of loss of righting reflex and motor impairment recovery time induced by ethanol [40]. A naturally TLR4-deficient mouse model C3H/HeJ showed a decrease in ethanol consumption compared with a control strain with a functional TLR4 receptor [41], although global knockout of TLR4 did not reduce ethanol consumption [42]. These studies highlight the potential role of TLR4 in ethanol's behavioral responses; however, it should be noted that these results cannot distinguish between peripheral and central TLR involvement. To determine if the immune targets in brain can directly modulate behavior, researchers injected siRNA into brain regions involved in alcohol abuse and monitored ethanol consumption. Injection of Tlr4 siRNA into the central amygdala of alcohol-preferring rats reduced operant administration of ethanol but not sucrose [43]. Furthermore, infusion of amplicons for Tlr4 or Mcp1 siRNA into the central amygdala or ventral tegmental area inhibited target gene expression and altered binge-like drinking behavior [44,45]. Amplicons for scrambled siRNA did not inhibit TLR4 or MCP-1 expression or reduce binge drinking, suggesting that a specific neuronal TLR4/MCP-1 signal may regulate initiation of voluntary alcohol self-administration [44].
Other types of TLRs may also regulate inflammatory responses to alcohol. In vitro studies showed that acute ethanol treatment enhances pro-inflammatory cytokine production in response to a variety of TLR agonists. For example, acute ethanol enhanced Poly I:C (TLR3 ligand), PGN (TLR2 ligand) and microbial methylated DNA CpG (TLR9 ligand) stimulated TNF-α, IL-6 and IL-12 synthesis in murine macrophages [46,47]. In mouse models of alcoholic liver disease, activation of TLR3 attenuated alcoholic liver injury by stimulating Kupffer and stellate cells to produce the anti-inflammatory cytokine IL-10 [48]. However, ethanol-induced TLR3 activation can also have neurodegenerative effects. For example, chronic ethanol exposure combined with TLR3 stimulation via Poly I:C led to an increase in pro-inflammatory cytokines in rat brain (e.g. TNF-α, IL-6, IL-1β and MCP-1) [49,50]. It should be noted that in Qin et al. [50], ethanol was administered intragastrically which could cause stress-induced increases in cytokine responses (for review see [16]). Similar to TLR4, TLR2 may have a brain region-specific effect on alcohol-induced immune responses. Mice chronically treated with ethanol for 5 months showed increased levels of cytokines and chemokines in the striatum; however, there was no change in TLR2 knockout mice, suggesting that TLR2 also regulates ethanol-induced pro-inflammatory gene expression [51]. Elimination of TLR2 in the striatum also appeared to protect against abstinence-induced anxiogenic behavior, evaluated by elevated plus-maze and dark and light box tests [51]. These studies suggest that TLR signaling is critical for alcohol-induced cytokine production and represent potential targets for regulating behavioral responses to alcohol.
TLR cell-type specificity & alcohol modulation
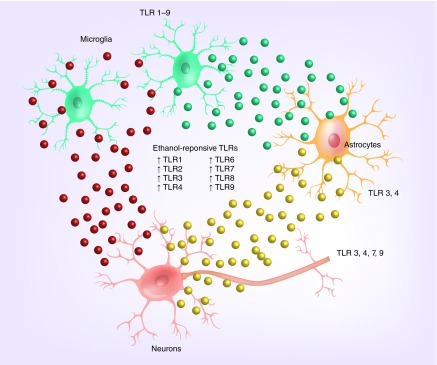
The cellular specificity of TLRs is an important consideration for developing drug targets that control alcohol-induced inflammatory gene expression. As shown in Figure 2, the expression of TLRs and related signaling proteins has been demonstrated in all cell types, with the strongest expression in glia (microglia, astrocytes and oligodendrocytes) and more limited expression in neurons [52–55]. Consequently, the majority of studies have examined how ethanol activates TLRs in glia. Ethanol directly activates microglia and astrocytes, resulting in stimulation of NF-κB, MAPKs, MyD88-independent pathways and inflammatory responses [42,55–58]. However, neurons likely play a regulatory role in TLR responses to alcohol-induced inflammation, most likely by stimulating microglia. For example, neuronal stimulation causes release of HMGB1, an endogenous ligand for TLR2, TLR4 and RAGE that has been shown to increase NF-κB-mediated transcription of pro-inflammatory cytokines [39,59].
Figure 2. . Alcohol-sensitive expression of Toll-like receptors and cell-type specificity.
Chronic alcohol abuse also activates microglia in the orbital frontal cortex of alcoholics and in the cortex of ethanol-treated mice [49]. In microglial cultures, ethanol upregulates TLR2 and TLR4 expression, causing increased production of inflammatory mediators [60]. Moreover, TLR4 expression and TLR2/4 association in cultured microglia appear to be necessary for alcohol-induced microglia activation and production of inflammatory mediators [38,57,60]. Facci et al. (2014) demonstrated that TLR 2-4 endogenous ligands prime microglia (but not astrocytes) across the central nervous system for ATP-dependent IL-1β release [61]. Microglia-specific priming may be linked to the increased IL-1β that is observed in ethanol-fed mice and human alcoholic frontal cortex [38], providing evidence that microglia are necessary mediators of alcohol-induced inflammatory responses.
Additionally, astrocytes may be important in TLR-mediated inflammatory signaling, although fewer studies to date have examined their specific role after chronic alcohol exposure. Ethanol treatment activated MAPK, NF-κB and AP-1 inflammatory pathways via a TLR4-dependent mechanism in cultured astrocytes [55]. Furthermore, ethanol-stimulated astrocytes produced IL-1R1 by activating the same inflammatory mediators and signaling cascades observed in the cerebral cortex of ethanol-fed animals [62]. Alfonso-Loeches et al. [63] linked astrocyte-mediated inflammatory responses to the TLR pathway by showing that TLR4 is critical for ethanol-induced inflammatory signaling. This group demonstrated that siRNA-mediated knockdown of TLR4 abolished alcohol activation of MAPK and NF-κB responses, which was also observed in TLR4-deficient cultures [63]. The studies above support the pivotal role of TLRs in astrocyte and microglia activation in ethanol-induced inflammation.
TLR-specific drugs alter alcohol consumption
Because alcohol activates astrocytes and microglia via TLR4 signaling, drug targets may require cell-type as well as TLR-specificity. One potential drug already being investigated is (+)-naloxone, which inhibits TLR4-mediated microglial activation and decreases proinflammatory cytokine expression [63]. Another microglia-related drug is minocycline, which reduces microglial and neuroimmune activation [64]. Minocycline also decreases ethanol consumption, albeit by an unknown mechanism [65]. It is hypothesized that the reduced drinking occurs via neuroimmune signaling because minocycline reduces mRNA expression of Il1β, Il6 and other cytokines linked to alcohol abuse in mouse cortex and hippocampus [66]. However, it remains unknown if minocycline could reduce cytokine expression via a TLR-dependent pathway. Phosphodiesterase-4 (PDE4) inhibition is also of interest because PDE4 inhibitors reduce alcohol consumption [67–69]. In addition, cAMP-specific PDE4 regulates LPS-TLR4 induced inflammatory cytokine expression in mouse models of liver fibrosis, a condition commonly attributed to alcohol abuse [70]. Finally, PDE4 inhibitors have been shown to reduce NF-κB binding, leading to decreased inflammatory cytokine production in macrophages [71]. A promising preliminary study by Avila et al. [72] showed that alcohol-induced activation of astrocytes and microglia in mouse brain was attenuated in PDE4b knockout mice and by pharmacological inhibition using a PDE4 inhibitor (rolipram, 5 mg/kg). A remaining question is if cell-specific TLR responses are critical to behaviors related to alcohol consumption. Further investigation of TLR- and cell-type specificity will be needed to determine if these are relevant molecular targets with therapeutic potential.
Cell-type specificity
Astrocytes
Alcohol-induced molecular changes in astrocytes
Astrocytes have diverse roles in the CNS, many of which are still being elucidated. As discussed earlier for TLR signaling, astrocytes play a substantial role in immune responses by producing and secreting inflammatory modulators. In addition, they are highly involved in neurotransmitter uptake and recycling. Astrocytes can also communicate with each other and influence neuronal functioning through Ca2+ signaling.
Chronic alcohol causes molecular changes in astrocytes that could affect their function. Some studies suggest alcohol dependence is related to a hyperexcitable state caused by excess extracellular glutamate [73]. Expression changes in astrocyte glutamate transporters are observed in alcoholics and alcohol-exposed rodents [74,75]. Upregulation of glutamate transporters in postmortem alcoholic brain could be a sign of compensation for increased levels of extracellular glutamate. In cultured astrocytes, ethanol exposure alters the distribution of the glutamate transporter GLAST, shifting GLAST expression from the cytoplasm to the plasma membrane [76]. Ethanol-treated rats display lower levels of the astrocyte glutamate transporter GLT-1 [77]. Modulating glutamate transporter levels or activity alters drinking behavior. Blocking astrocytic glutamate uptake via GLT-1 attenuates binge-drinking behavior in mice [78]. Adenosine-mediated glutamate signaling has also been implicated in alcohol abuse. Deletion or knockdown of ENT1, an adenosine transporter, leads to reduced expression of GLT-1 and an astrocyte-specific water channel, AQP4. Treatment with the β-lactam antibiotic ceftriaxone, a potent inducer of glutamate uptake in astrocytes, increases GLT-1 expression and decreases ethanol drinking in mice, an effect dependent on AQP4 expression [79]. Ceftriaxone blocks glutamate increases in the extracellular space and increases glutamine synthetase activity in the nucleus accumbens [77]. It is effective in reducing relapse drinking in alcohol-preferring rats after long-term ethanol dependence [80] and in attenuating withdrawal [81]. These studies all point to a central role for astrocyte glutamate uptake in drinking behavior. Ceftriaxone, or similar compounds that target GLT-1, could potentially be used to treat alcohol withdrawal and other AUD-related behaviors. Baclofen, which has anticraving effects in patients with alcohol dependence [82], also affects glutamate transporter expression, as it prevented changes in subcellular localization of GLAST in cultured alcohol-treated astrocytes. This indicates a possible link between astrocytic glutamate transport and baclofen’s anticraving effects [76]. However, other non-glutamatergic responses in astrocytes could also be modulated by long-term alcohol exposure and play a role in alcohol dependence. Astrocytes thus appear to be a viable cellular target for some alcohol-related drugs, supporting our earlier point of the relevance of examining cell-type specificity in alcohol-neuroimmune signaling.
Alcohol induces reactive astrocytes
When astrocytes respond to a pathological condition in the brain, they exhibit an evolutionarily conserved phenotype known as astrocyte reactivity. This response is characterized by upregulation of the intermediate filament and astrocyte-specific marker, GFAP, as well as cellular hypertrophy and elongated processes. Astrocyte reactivity has been observed in multiple brain regions, including the frontal cortex, hippocampus and cerebellum of ethanol-exposed mice and rats [49,83–88]. Differences in GFAP-positive astrocyte distributions in human brains have also been shown [89]. Alcohol-fed mice show increased Gfap mRNA [38]. Vimentin, another intermediate filament and indicator of astrocyte reactivity, was highly upregulated in a binge model of alcohol drinking in rats and was associated with alcohol-induced cell death [1]. Although the molecular, functional and behavioral consequences of alcohol-induced astrocyte reactivity are unknown, evidence suggests that this reactivity is commonly occurring in specific brain regions following alcohol exposure.
Astrocyte reactivity involves transcriptional induction triggered by signaling cascades that can result in long-lasting functional adaptations [90], which may be either beneficial or detrimental to neuronal functioning. For example, in the event of a traumatic brain injury, reactive astrocytes play both helpful and harmful roles. They can prevent infections and spread of cellular damage by the generation of a barrier across the injured area known as a glial scar. However, they can also block neuronal regeneration by secreting growth-inhibitory molecules to prevent axonal extensions. Reactive astrocytes can alter metabolism, disrupt the blood–brain barrier and alter ion and neurotransmitter uptake. They have the capacity to release various molecules that affect nearby cells in many ways. Many subtypes of reactive astrocytes exist, all precisely altering their function to adjust to their unique pathological environment. For example, acute brain injury induces a response distinct from that of astrocytes affected by slowly progressing neurodegenerative disorders; however, both types of conditions upregulate GFAP and exhibit hypertrophic cell bodies and processes [90]. Unique groups of genes are altered depending on the different models of astrocyte reactivity. In a study comparing gene expression changes in isolated reactive astrocytes induced by two distinct models (LPS injection and experimentally-induced stroke), 50% of the differential expression was specific to the injury model [91]. Many of the top upregulated genes in LPS-treated astrocytes also show differential expression in existing gene expression datasets from human alcoholics and alcohol-exposed mice from our laboratory (unpublished), indicating that alcohol-induced reactivity may overlap with functional changes in astrocytes induced by immune perturbation.
Alcohol-exposed astrocytes as immune regulators
Astrocyte reactivity and accompanying changes in expression of immune-related genes can cause release of pro- or anti-inflammatory mediators, depending on the injury or disease context [92]. Some studies have identified the specific inflammatory mediators released in response to ethanol. For example, acute ethanol treatment of cultured astrocytes causes upregulation of iNOS and COX-2 [56], and chronic ethanol activates the NLRP3 inflammasome in astrocytes from cerebral cortex and in primary cultures of astrocytes [93]. Ethanol also promotes TLR4 signaling in astrocytes and microglia, which leads to upregulation of inflammatory cytokines, including IL-1β, TNF-α, IL-6, iNOS and COX-2 [63]. Knockout of TLR4 partially prevents the upregulation of GFAP observed after chronic ethanol-stimulated inflammation, indicating that TLR4 plays a role but is not solely responsible for inducing astrocyte reactivity [63].
It is unclear how astrocyte-specific immune signaling regulates drinking and the development of alcohol dependence. Expression of Ccl2, an inflammatory chemokine, is increased in alcohol-exposed tissue [84]. Astrocytes are a prime source of CCL2, and immune stimulation can cause striking upregulation of Ccl2 in astrocytes [94]. Astrocyte-specific Ccl2 overexpression in the hippocampus reduces ethanol-induced depression of long-term potentiation [95]. Another study shows that Ccl2 and other inflammatory cytokines are increased in specific brain regions of adult, but not adolescent mice, after ethanol gavage treatment [84]. Immune-related changes that depend upon sex, including sexually dimorphic cytokine signaling, have been reported in astrocytes after alcohol exposure [96]. Thus, several factors may regulate distinct inflammatory mediators that influence the role of astrocytes in alcohol-neuroimmune responses.
Astrocyte Ca2+ signaling in drinking behavior
Secretion of neurotransmitters and other factors can activate astrocytic G-protein coupled receptors (GPCR), leading to release of intracellular Ca2+. Through Ca2+ waves, astrocytes can communicate with each other and influence neuronal activity. Astrocyte-neuronal Ca2+ signaling may be an important mechanism that is modulated by alcohol exposure. Studies have already shown that astrocyte activity can affect motivation for alcohol. The use of designer receptors activated by designer drugs (DREADDs) in nucleus accumbens core astrocytes led to the discovery that increased Ca2+ activity through DREADD activation reduces motivation for ethanol after a 3-week period of abstinence [88]. Blocking gap-junction hemichannels, through which astrocytes can propagate Ca2+ waves and release other molecules important for cell-to-cell communication [97], leads to increased motivation to self-administer ethanol after 3 weeks of abstinence. This indicates that modulating the activity of discrete populations of astrocytes could alter synaptic plasticity and influence drug-seeking behavior. Alcohol may also cause specific GPCR expression changes, altering Ca2+ signaling and astrocyte responses. Inflammatory mediators such as LPS, TGF-β1 and IFN-γ induce notable expression changes in several GPCRs, effector proteins and downstream signal transduction pathways in cultured astrocytes [98]. Considering that the effects of neuroimmune activation are still relatively unexplored and neuroimmune mediators affect many pathways and cellular processes, it may be useful to consider targeting more discrete immune signaling processes. For example, if alcohol-induced alterations in astrocyte Ca2+ signaling regulate alcohol consumption, then targeting astrocyte-specific GPCRs could be a useful therapeutic tool.
Alcohol-responsive transcriptome in astrocytes
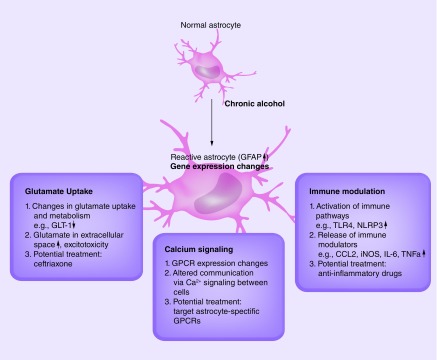
There are dynamic gene expression and transcriptional changes in reactive astrocytes in the face of immune modulation and alcohol exposure, and knowledge of the networks and pathways could reveal targeted cellular therapies for alcohol dependence. Microarray studies of postmortem frontal cortex from alcoholic patients showed altered expression of astrocyte-specific genes [9,99] and genes generally associated with glial function [3]. Astrocytes express many of the same genes as other cell types, and many innate immune genes are expressed in both microglia and astrocytes [100]. Most gene expression studies in brain have not determined which immune genes are altered in the different cell types. Astrocytes and microglia are often grouped and discussed together as the immune cells of the brain. However, considering the diverse nature and functions of reactive astrocytes, it is feasible that molecular alterations in astrocytes are very different from those in microglia. Figure 3 summarizes some of the possible alcohol-induced functional changes discussed in this review that could result from astrocyte transcriptional adaptations. Dissecting the alcohol-induced transcriptome changes in astrocytes will help define alcohol's discrete effects in brain and identify potential cellular pathways and targets.
Figure 3. . Possible alcohol-induced functional changes that potentially result from astrocyte transcriptional adaptations.
Microarray profiling of gene expression changes after acute ethanol exposure in cultured cortical astrocytes identified several immune and glial-specific genes, including Gfap [101]. Hsf1, a heat shock transcription factor, was activated in astrocytes, and expression profiling of heat-shock stressed astrocytes revealed overlapping changes between ethanol exposure and heat stress, indicating that the heat shock pathway is important in ethanol-induced gene expression changes. There was also a striking resemblance between expression changes in astrocytes and cultured hepatocytes after alcohol exposure, suggesting common signaling mechanisms between the two types of cells.
Studies utilizing longer ethanol exposures will provide more insight into the long-term molecular adaptations of astrocytes. There are also significant differences in gene expression between astrocytes in vitro and astrocytes isolated from mature brains [91]. Cultured astrocytes appear to exhibit signs of reactivity without any stimulation, while astrocytes co-cultured with neurons show resting morphology, suggesting that neuronal interactions are important for maintaining nonreactive states. Thus, in vivo transcriptome analysis in astrocytes isolated from chronic ethanol-treated animals is an important research direction.
Microglia
Alcohol activation of microglial genes
As previously noted, initial interest in alcohol-neuroimmune responses came from microarray studies that identified immune and inflammatory transcripts and pathways in alcohol-exposed tissue from mice, rats and human tissue [3–5,34,102–104]. Several immune genes found in these microarray studies were validated in rodent ethanol consumption paradigms, further corroborating a role for neuroimmune signaling in drinking behavior [14,105]. Microglia are the resident immune cells in the brain and many of the validated transcripts are expressed in or secreted from microglia (Cd14, Ctsf, Ctss, Il1rn, Il6), prompting interest in the role of this cell type in ethanol consumption [14,106]. Since then, many studies have found that in vitro ethanol treatment of microglia leads to activation and the release of pro-inflammatory cytokines, while chronic ethanol consumption results in microglial activation in rodents and human postmortem tissue [57,63,107–110].
Gene expression in isolated microglia
The alcohol-neuroimmune field is progressing rapidly, and there is increasing interest in cell-specific changes. Cell-type specific databases provide one tool to identify the cellular basis of the transcriptome changes by identifying the gene co-expression modules that are enriched in different cells. This strategy has also helped to identify discrete cells that are targeted depending on the ethanol drinking paradigms, time points and brain regions studied [111–114]. Because microglia make up only 5–15% of cells in the brain, it is likely that key microglial gene expression changes are missed when examining whole tissue [115], making it necessary to perform transcriptome studies in isolated microglia. It will also become important to identify subtypes of microglia in gene expression studies, as microglia can be both pro- and anti-inflammatory as well as play a role in neurogenesis and synaptic remodeling [116].
Drug identification & the transcriptome
Transcriptome studies have been helpful in identifying drugs that could potentially be used in the treatment of AUD. An insilico bioinformatics study examined gene expression data collected from adolescent and adult mice exposed to 20% ethanol using a 4 h/day, 4-day drinking-in-dark paradigm [18]. Several neuroimmune pathways related to microglial action were altered in the adult but not the adolescent mice. The study also showed that minocycline, a drug that modulates neuroimmune signaling and microglial activity, could be given intraperitoneally and cross the blood–brain barrier. Treatment with minocycline led to a significant reduction in ethanol consumption in the adult mice. This study highlights how transcriptome data can be successfully used to identify pathways that are altered by ethanol consumption and ultimately identify drugs that target these pathways.
Drug repurposing
Drug repurposing is the process of identifying a new therapeutic indication for an existing drug or drug candidate. The concept of US FDA-approved drug repurposing as a strategy for therapeutic development has received increased support because it can resurrect a failed drug or expand the number of clinical uses for a successful one [117–123]. The primary advantage is that the time to clinical trial can be dramatically reduced. There are numerous examples of successfully repurposed drugs that are beyond the scope of the current review and are only briefly highlighted here. A notable example of drug repurposing is buprenorphine which was used to treat moderate pain, then subsequently used for treating heroin and other opioid addictions [124]. Buspirone was initially marketed as an anxiolytic, but more recently has been identified as a potential candidate to treat drug addiction and compulsive behavior disorders based on its high affinity for D3 receptors (as well as 5-HT1A and D2 receptors) [125]. In addition, a study of nonhuman primates indicates that buspirone attenuates drug-taking relapse in abstinent animals [126]. Other general examples of repurposed drugs include sildenafil for erectile dysfunction (developed as an anti-hypertensive) [127], thalidomide (original indication: nonbarbiturate sedative-hypnotic), finasteride (original indication: prostatic hyperplasia) and chlorpromazine (original indication: antihistamine) that are also effective treatments for erectile dysfunction, leprosy, hair loss and psychosis, respectively [128–130]. There are a limited number of repositioned drugs that have been identified as potential AUD therapeutics. These include topiramate, which was originally used as an anticonvulsant drug used to treat epilepsy [131,132], naltrexone, an opiate receptor antagonist [133] and aripiprazole, an atypical antipsychotic, with potential for treating alcohol dependence [134]. And, relevant to the current review and the role of the neuroimmune system in AUD, there is evidence that ibudilast, a nonselective phosphodiesterase inhibitor, decreases alcohol consumption in animal models [135].
There is growing support for computational approaches to drug repurposing as outlined in recent reviews [136–139]. The NIH is currently funding multiple research institutions to refine and extend a database of human cellular responses. This funding supports the LINCS program, and includes funding to the Broad Institute which originally established the Connectivity Map (CMAP) database [140,141], cataloging the biological responses of approximately 1 million gene expression profiles using L1000 technology. This approach has been shown to identify existing drugs and elucidate modes of action for novel chemicals based on findings from expression studies [142]. L1000 technology is a high-throughput method to estimate genome-wide mRNA expression based on unique perturbations by drugs, ligands and other small molecules applied at different time points and doses in cell culture. Recently, the NeuroLINCS Center (University of California, CA, USA) was funded to include neuronal and glial signatures from normal and diseased-induced pluripotent stem cells, thus expanding the LINCS database to noncancerous cell lines.
In a successful application of this approach, data from the Connectivity Map (CMAP, [143]) were queried with the gene expression signatures of Alzheimer’s disease to identify compounds putatively beneficial for the disease [144]. A somewhat different approach used SNPs related to aversive memory combined with Ingenuity Pathway Analysis to predict drugs that might modulate memory, and the effectiveness of one of these drugs was verified in human studies [145]. The use of genomics to predict drugs for brain diseases is in its infancy, but these examples are quite promising for future therapeutic development. For example, these computational approaches, which integrate gene expression signatures of drugs and small molecules with disease, have been successful in identifying potential therapeutics for complex disorders such as obesity [146] and inflammatory bowel disease [147]. In addition, gabapentin, which has been shown to be effective in treating alcohol dependence and preventing relapse in clinical trials [148] was identified as a potential therapeutic target based on RNA-sequencing of human postmortem brain [149]. A key question is whether consistent changes in gene expression profiles and networks found in human alcoholics and animal models can be used to predict drugs that will normalize the targeted gene networks and provide new treatments for AUD. This research direction provides an important avenue for discovering novel compounds that target immune signaling in alcohol dependence.
Conclusion and future perspective
There is increasing evidence for neuroimmune system involvement in AUD. This work is also leading to a better appreciation for the roles of glial cells, including astrocytes and microglia, in the brain’s response to alcohol. Strategies for drug repurposing based upon transcriptional profiles for the treatment of AUD, particularly relevant to the neuroimmune system, represent an important area of emerging research.
Executive summary.
Toll-like receptors and alcohol
TLRs are key regulators of alcohol-induced neuroimmune activation
Alcohol exposure leads to changes in expression of many TLR pathway components
Genetic modulation of TLR signaling components can change alcohol behaviors
Evidence supports roles for neurons, astrocytes and microglia in TLR-mediated alcohol responses
Drugs targeting TLR pathways can modulate alcohol consumption
Cell-type specific changes
-
Astrocytes
– Alcohol exposure causes molecular changes in astrocytes that can affect crucial functions such as glutamate uptake
– Alcohol can induce astrocyte reactivity, a conserved astrocyte phenotype for responding to stress and disease
– Astrocytes may play key roles in immune modulation after alcohol exposure
– Astrocyte Ca2+ signaling could play a role in modulating alcohol-related behaviors
– Alcohol-induced transcriptional and functional changes in astrocytes are largely unknown
-
Microglia
– Alcohol induces expression of immune genes that are expressed in or secreted from microglia
– Microglial gene expression changes can be used to predict drugs that reduce alcohol consumption
– Alcohol-mediated changes in isolated microglia can elucidate their specific role in alcohol-immune responses
Drug repurposing
– Global transcription-based analyses identifying drug candidates for the treatment alcohol use disorders could lead to improved therapeutic strategies.
Acknowledgements
A special thanks to K Jameson for her help making figures and to J Mayfield for assistance editing the manuscript.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Kelso ML, Liput DJ, Eaves DW, Nixon K. Upregulated vimentin suggests new areas of neurodegeneration in a model of an alcohol use disorder. Neuroscience. 2011;197:381–393. doi: 10.1016/j.neuroscience.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J. Neurochem. 2004;90(5):1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Lewohl JM, Harris RA, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31(7):1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- 4.Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J. Neurochem. 2002;81(4):802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- 5.Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol. Clin. Exp. Res. 2000;24(12):1873–1882. [PubMed] [Google Scholar]
- 6.Flatscher-Bader T, Van Der Brug M, Hwang JW, et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J. Neurochem. 2005;93(2):359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- 7.Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J. Neurosci. Res. 2003;72(6):756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- 8.Farris SP, Harris RA, Ponomarev I. Epigenetic modulation of brain gene networks for cocaine and alcohol abuse. Front. Neurosci. 2015;9:176. doi: 10.3389/fnins.2015.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okvist A, Johansson S, Kuzmin A, et al. Neuroadaptations in human chronic alcoholics: dysregulation of the NF-kappaB system. PLoS ONE. 2007;2(9):e930. doi: 10.1371/journal.pone.0000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclercq S, Cani PD, Neyrinck AM, et al. Role of intestinal permeability and inflammation in the biological and behavioral control of alcohol-dependent subjects. Brain Behav. Immun. 2012;26(6):911–918. doi: 10.1016/j.bbi.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq S, De Saeger C, Delzenne N, De Timary P, Starkel P. Role of inflammatory pathways, blood mononuclear cells, and gut-derived bacterial products in alcohol dependence. Biol. Psychiatry. 2014;76(9):725–733. doi: 10.1016/j.biopsych.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Maes M, Lin A, Bosmans E, et al. Serotonin-immune interactions in detoxified chronic alcoholic patients without apparent liver disease: activation of the inflammatory response system and lower plasma total tryptophan. Psychiatry Res. 1998;78(3):151–161. doi: 10.1016/s0165-1781(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 14.Blednov YA, Ponomarev I, Geil C, Bergeson S, Koob GF, Harris RA. Neuroimmune regulation of alcohol consumption: behavioral validation of genes obtained from genomic studies. Addict. Biol. 2012;17(1):108–120. doi: 10.1111/j.1369-1600.2010.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA. Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain. Behav. Immun. 2011;25(Suppl. 1):S92–S105. doi: 10.1016/j.bbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breese GR, Sinha R, Heilig M. Chronic alcohol neuroadaptation and stress contribute to susceptibility for alcohol craving and relapse. Pharmacol. Ther. 2011;129(2):149–171. doi: 10.1016/j.pharmthera.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breese GR, Knapp DJ, Overstreet DH, Navarro M, Wills TA, Angel RA. Repeated lipopolysaccharide (LPS) or cytokine treatments sensitize ethanol withdrawal-induced anxiety-like behavior. Neuropsychopharmacology. 2008;33(4):867–876. doi: 10.1038/sj.npp.1301468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal RG, Owen JA, Levin PS, et al. Bioinformatics analyses reveal age-specific neuroimmune modulation as a target for treatment of high ethanol drinking. Alcohol. Clin. Exp. Res. 2014;38(2):428–437. doi: 10.1111/acer.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin L, Wu X, Block ML, et al. Systemic LPS causes chronic neuroinflammation and progressive neurodegeneration. Glia. 2007;55(5):453–462. doi: 10.1002/glia.20467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medzhitov R, Janeway C. Innate immune recognition: mechanisms and pathways. Immunol. Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 21.Takeda K, Akira S. TLR signaling pathways. Semin. Immunol. 2004;16(1):3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Bsibsi M, Bajramovic JJ, Vogt MH, et al. The microtubule regulator stathmin is an endogenous protein agonist for TLR3. J. Immunol. 2010;184(12):6929–6937. doi: 10.4049/jimmunol.0902419. [DOI] [PubMed] [Google Scholar]
- 23.Yu L, Wang L, Chen S. Endogenous Toll-like receptor ligands and their biological significance. J. Cell. Mol. Med. 2010;14(11):2592–2603. doi: 10.1111/j.1582-4934.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benias PC, Gopal K, Bodenheimer H, Theise ND. Hepatic expression of toll-like receptors 3, 4, and 9 in primary biliary cirrhosis and chronic hepatitis C. Clin. Res. Hepatol. Gastroenterol. 2012;36(5):448–454. doi: 10.1016/j.clinre.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki M, Sugimoto Y, Ohsaki Y, et al. Endosomal accumulation of Toll-like receptor 4 causes constitutive secretion of cytokines and activation of signal transducers and activators of transcription in Niemann-Pick disease type C (NPC) fibroblasts: a potential basis for glial cell activation in the NPC brain. J. Neurosci. 2007;27(8):1879–1891. doi: 10.1523/JNEUROSCI.5282-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith KD, Andersen-Nissen E, Hayashi F, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 2003;4(12):1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 28.Yarovinsky F, Zhang D, Andersen JF, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308(5728):1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- 29.Brencicova E, Diebold SS. Nucleic acids and endosomal pattern recognition: how to tell friend from foe? Front. Cell. Infect. Microbiol. 2013;3:37. doi: 10.3389/fcimb.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oosting M, Cheng SC, Bolscher JM, et al. Human TLR10 is an anti-inflammatory pattern-recognition receptor. Proc. Natl Acad. Sci. USA. 2014;111(42):E4478–E4484. doi: 10.1073/pnas.1410293111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raetz M, Kibardin A, Sturge CR, et al. Cooperation of TLR12 and TLR11 in the IRF8-dependent IL-12 response to Toxoplasma gondii profilin. J. Immunol. 2013;191(9):4818–4827. doi: 10.4049/jimmunol.1301301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Z, Mak TW, Sen G, Li X. Toll-like receptor 3-mediated activation of NF-kappaB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-beta. Proc. Natl Acad. Sci. USA. 2004;101(10):3533–3538. doi: 10.1073/pnas.0308496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald KA, Rowe DC, Barnes BJ, et al. LPS–TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003;198(7):1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulligan MK, Ponomarev I, Hitzemann RJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc. Natl Acad. Sci. USA. 2006;103(16):6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Study demonstrates cell-type specificity of neuroimmune signaling after ethanol exposure. Found that TLR4 and IL-1RI are targets of ethanol-induced inflammatory mediators in both brain and astrocyte cultures
- 35.Kong Y, Le Y. Toll-like receptors in inflammation of the central nervous system. Int. Immunopharmacol. 2011;11(10):1407–1414. doi: 10.1016/j.intimp.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 36.Troutwine BR, Ghezzi A, Pietrzykowski AZ, Atkinson NS. Alcohol resistance in Drosophila is modulated by the Toll innate immune pathway. Genes Brain Behav. 2016;15(4):382–389. doi: 10.1111/gbb.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustot T, Lemmers A, Moreno C, et al. Differential liver sensitization to toll-like receptor pathways in mice with alcoholic fatty liver. Hepatology. 2006;43(5):989–1000. doi: 10.1002/hep.21138. [DOI] [PubMed] [Google Scholar]
- 38.Lippai D, Bala S, Petrasek J, et al. Alcohol-induced IL-1β in the brain is mediated by NLRP3/ASC inflammasome activation that amplifies neuroinflammation. J. Leukoc. Biol. 2013;94(1):171–182. doi: 10.1189/jlb.1212659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crews FT, Qin L, Sheedy D, Vetreno RP, Zou J. High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biol. Psychiatry. 2013;73(7):602–612. doi: 10.1016/j.biopsych.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Lousberg EL, Moldenhauer LM, et al. Inhibiting the TLR4-MyD88 signalling cascade by genetic or pharmacological strategies reduces acute alcohol-induced sedation and motor impairment in mice. Br. J. Pharmacol. 2012;165(5):1319–1329. doi: 10.1111/j.1476-5381.2011.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris R, Blednov Y. Neuroimmune Genes and Alcohol Drinking Behavior. In: Cui C, Grandison L, Noronha A, editors. Neural-Immune Interactions in Brain Function and Alcohol Related Disorders. Springer; NY, USA: 2013. pp. 425–440. [Google Scholar]
- 42.Pascual M, Baliño P, Alfonso-Loeches S, Aragón CM, Guerri C. Impact of TLR4 on behavioral and cognitive dysfunctions associated with alcohol-induced neuroinflammatory damage. Brain. Behav. Immun. 2011;25(Suppl. 1):S80–S91. doi: 10.1016/j.bbi.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Yang AR, Kelly T, et al. Binge alcohol drinking is associated with GABAA alpha2-regulated Toll-like receptor 4 (TLR4) expression in the central amygdala. Proc. Natl Acad. Sci. USA. 2011;108(11):4465–4470. doi: 10.1073/pnas.1019020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.June HL, Liu J, Warnock KT, et al. CRF-amplified neuronal TLR4/MCP-1 signaling regulates alcohol self-administration. Neuropsychopharmacology. 2015;40(6):1549–1559. doi: 10.1038/npp.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valenta JP, Gonzales RA. Chronic intracerebroventricular infusion of monocyte chemoattractant protein-1 leads to a persistent increase in sweetened ethanol consumption during operant self-administration but does not influence sucrose consumption in Long-Evans rats. Alcohol. Clin. Exp. Res. 2016;40(1):187–195. doi: 10.1111/acer.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goral J, Choudhry MA, Kovacs EJ. Acute ethanol exposure inhibits macrophage IL-6 production: role of p38 and ERK1/2 MAPK. J. Leukoc. Biol. 2004;75(3):553–559. doi: 10.1189/jlb.0703350. [DOI] [PubMed] [Google Scholar]
- 47.Goral J, Kovacs EJ. In vivo ethanol exposure down-regulates TLR2-, TLR4-, and TLR9-mediated macrophage inflammatory response by limiting p38 and ERK1/2 activation. J. Immunol. 2005;174(1):456–463. doi: 10.4049/jimmunol.174.1.456. [DOI] [PubMed] [Google Scholar]
- 48.Byun JS, Suh YG, Yi HS, Lee YS, Jeong WI. Activation of toll-like receptor 3 attenuates alcoholic liver injury by stimulating Kupffer cells and stellate cells to produce interleukin-10 in mice. J. Hepatol. 2013;58(2):342–349. doi: 10.1016/j.jhep.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 49.Qin L, Crews FT. NADPH oxidase and reactive oxygen species contribute to alcohol-induced microglial activation and neurodegeneration. J. Neuroinflammation. 2012;9:5. doi: 10.1186/1742-2094-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin L, Crews FT. Chronic ethanol increases systemic TLR3 agonist-induced neuroinflammation and neurodegeneration. J. Neuroinflammation. 2012;9:130. doi: 10.1186/1742-2094-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pascual M, Baliño P, Aragón CM, Guerri C. Cytokines and chemokines as biomarkers of ethanol-induced neuroinflammation and anxiety-related behavior: role of TLR4 and TLR2. Neuropharmacology. 2015;89:352–359. doi: 10.1016/j.neuropharm.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 52.Bsibsi M, Ravid R, Gveric D, Van Noort JM. Broad expression of Toll-like receptors in the human central nervous system. J. Neuropathol. Exp. Neurol. 2002;61(11):1013–1021. doi: 10.1093/jnen/61.11.1013. [DOI] [PubMed] [Google Scholar]
- 53.Kielian T. Toll-like receptors in central nervous system glial inflammation and homeostasis. J. Neurosci. Res. 2006;83(5):711–730. doi: 10.1002/jnr.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lafon M, Megret F, Lafage M, Prehaud C. The innate immune facet of brain: human neurons express TLR-3 and sense viral dsRNA. J. Mol. Neurosci. 2006;29(3):185–194. doi: 10.1385/JMN:29:3:185. [DOI] [PubMed] [Google Scholar]
- 55.Blanco AM, Vallés SL, Pascual M, Guerri C. Involvement of TLR4/type I IL-1 receptor signaling in the induction of inflammatory mediators and cell death induced by ethanol in cultured astrocytes. J. Immunol. 2005;175(10):6893–6899. doi: 10.4049/jimmunol.175.10.6893. [DOI] [PubMed] [Google Scholar]; • Study demonstrates cell-type specificity of neuroimmune signaling after ethanol exposure. Found that TLR4 and IL-1RI are targets of ethanol-induced inflammatory mediators in both brain and astrocyte cultures.
- 56.Blanco AM, Pascual M, Valles SL, Guerri C. Ethanol-induced iNOS and COX-2 expression in cultured astrocytes via NF-kappa B. Neuroreport. 2004;15(4):681–685. doi: 10.1097/00001756-200403220-00021. [DOI] [PubMed] [Google Scholar]
- 57.Fernandez-Lizarbe S, Pascual M, Guerri C. Critical role of TLR4 response in the activation of microglia induced by ethanol. J. Immunol. 2009;183(7):4733–4744. doi: 10.4049/jimmunol.0803590. [DOI] [PubMed] [Google Scholar]
- 58.Scumpia PO, Kelly KM, Reeves WH, Stevens BR. Double-stranded RNA signals antiviral and inflammatory programs and dysfunctional glutamate transport in TLR3-expressing astrocytes. Glia. 2005;52(2):153–162. doi: 10.1002/glia.20234. [DOI] [PubMed] [Google Scholar]
- 59.Park C, Lee S, Cho IH, et al. TLR3-mediated signal induces proinflammatory cytokine and chemokine gene expression in astrocytes: differential signaling mechanisms of TLR3-induced IP-10 and IL-8 gene expression. Glia. 2006;53(3):248–256. doi: 10.1002/glia.20278. [DOI] [PubMed] [Google Scholar]
- 60.Fernandez-Lizarbe S, Montesinos J, Guerri C. Ethanol induces TLR4/TLR2 association, triggering an inflammatory response in microglial cells. J. Neurochem. 2013;126(2):261–273. doi: 10.1111/jnc.12276. [DOI] [PubMed] [Google Scholar]
- 61.Facci L, Barbierato M, Marinelli C, Argentini C, Skaper SD, Giusti P. Toll-like receptors 2, -3 and -4 prime microglia but not astrocytes across central nervous system regions for ATP-dependent interleukin-1β release. Sci. Rep. 2014;4:6824. doi: 10.1038/srep06824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vallés SL, Blanco AM, Pascual M, Guerri C. Chronic ethanol treatment enhances inflammatory mediators and cell death in the brain and in astrocytes. Brain Pathol. 2004;14(4):365–371. doi: 10.1111/j.1750-3639.2004.tb00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alfonso-Loeches S, Pascual-Lucas M, Blanco AM, Sanchez-Vera I, Guerri C. Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. J. Neurosci. 2010;30(24):8285–8295. doi: 10.1523/JNEUROSCI.0976-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan R, Xu F, Previti ML, et al. Minocycline reduces microglial activation and improves behavioral deficits in a transgenic model of cerebral microvascular amyloid. J. Neurosci. 2007;27(12):3057–3063. doi: 10.1523/JNEUROSCI.4371-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agrawal RG, Hewetson A, George CM, Syapin PJ, Bergeson SE. Minocycline reduces ethanol drinking. Brain. Behav. Immun. 2011;25(Suppl. 1):S165–S169. doi: 10.1016/j.bbi.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henry CJ, Huang Y, Wynne A, et al. Minocycline attenuates lipopolysaccharide (LPS)-induced neuroinflammation, sickness behavior, and anhedonia. J. Neuroinflammation. 2008;5:15. doi: 10.1186/1742-2094-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blednov YA, Benavidez JM, Black M, Harris RA. Inhibition of phosphodiesterase 4 reduces ethanol intake and preference in C57BL/6J mice. Front. Neurosci. 2014;8:129. doi: 10.3389/fnins.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu W, Lu T, Chen A, et al. Inhibition of phosphodiesterase-4 decreases ethanol intake in mice. Psychopharmacology (Berl.) 2011;218(2):331–339. doi: 10.1007/s00213-011-2290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wen RT, Zhang M, Qin WJ, et al. The phosphodiesterase-4 (PDE4) inhibitor rolipram decreases ethanol seeking and consumption in alcohol-preferring Fawn-Hooded rats. Alcohol. Clin. Exp. Res. 2012;36(12):2157–2167. doi: 10.1111/j.1530-0277.2012.01845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gobejishvili L, Barve S, Breitkopf-Heinlein K, et al. Rolipram attenuates bile duct ligation-induced liver injury in rats: a potential pathogenic role of PDE4. J. Pharmacol. Exp. Ther. 2013;347(1):80–90. doi: 10.1124/jpet.113.204933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hervé R, Schmitz T, Evain-Brion D, Cabrol D, Leroy MJ, Méhats C. The PDE4 inhibitor rolipram prevents NF-kappaB binding activity and proinflammatory cytokine release in human chorionic cells. J. Immunol. 2008;181(3):2196–2202. doi: 10.4049/jimmunol.181.3.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Avila DV, Gobejishvili L, Myers S, et al. Pathogenic role of phosphodiesterase 4B (PDE4B) in alcohol-induced neuroinflammation. FASEB J. 2015;29(1):771–718. [Google Scholar]
- 73.Crews FT. Immune function genes, genetics, and the neurobiology of addiction. Alcohol Res. 2012;34(3):355–361. [PMC free article] [PubMed] [Google Scholar]
- 74.Miguel-Hidalgo JJ, Waltzer R, Whittom AA, Austin MC, Rajkowska G, Stockmeier CA. Glial and glutamatergic markers in depression, alcoholism, and their comorbidity. J. Affect. Disord. 2010;127(1–3):230–240. doi: 10.1016/j.jad.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. FASEB J. 2002;16(1):27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- 76.Šerý O, Sultana N, Kashem MA, Pow DV, Balcar VJ. GLAST but not least – distribution, function, genetics and epigenetics of L-glutamate transport in brain-focus on GLAST/EAAT1. Neurochem. Res. 2015;40(12):2461–2472. doi: 10.1007/s11064-015-1605-2. [DOI] [PubMed] [Google Scholar]
- 77.Das SC, Yamamoto BK, Hristov AM, Sari Y. Ceftriaxone attenuates ethanol drinking and restores extracellular glutamate concentration through normalization of GLT-1 in nucleus accumbens of male alcohol-preferring rats. Neuropharmacology. 2015;97:67–74. doi: 10.1016/j.neuropharm.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith KL, John CS, Sypek EI, et al. Exploring the role of central astrocytic glutamate uptake in ethanol reward in mice. Alcohol. Clin. Exp. Res. 2014;38(5):1307–1314. doi: 10.1111/acer.12361. [DOI] [PubMed] [Google Scholar]
- 79.Lee MR, Ruby CL, Hinton DJ, et al. Striatal adenosine signaling regulates EAAT2 and astrocytic AQP4 expression and alcohol drinking in mice. Neuropsychopharmacology. 2013;38(3):437–445. doi: 10.1038/npp.2012.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rao P, Sari Y. Effectiveness of ceftriaxone treatment in preventing relapse-like drinking behavior following long-term ethanol dependence in P rats. J. Addict. Res. Ther. 2014;5 doi: 10.4172/2155-6105.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abulseoud OA, Camsari UM, Ruby CL, Kasasbeh A, Choi S, Choi DS. Attenuation of ethanol withdrawal by ceftriaxone-induced upregulation of glutamate transporter EAAT2. Neuropsychopharmacology. 2014;39(7):1674–1684. doi: 10.1038/npp.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simioni N, Preda C, Deken V, Bence C, Cottencin O, Rolland B. Characteristics of patients with alcohol dependence seeking baclofen treatment in France: a two-centre comparative cohort study. Alcohol Alcohol. 2016 doi: 10.1093/alcalc/agw011. Epub ahea dof print. [DOI] [PubMed] [Google Scholar]
- 83.Franke H, Kittner H, Berger P, Wirkner K, Schramek J. The reaction of astrocytes and neurons in the hippocampus of adult rats during chronic ethanol treatment and correlations to behavioral impairments. Alcohol. 1997;14(5):445–454. doi: 10.1016/s0741-8329(96)00209-1. [DOI] [PubMed] [Google Scholar]
- 84.Cynthia JM, Kane KDP, et al. Drew. Effects of ethanol on immune response in the brain: region specific changes in adolescent versus adult mice. Alcohol Clin. Exp. Res. 2014;38(2):384–391. doi: 10.1111/acer.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Satriotomo I, Miki T, Itoh M, Ameno K, Ijiri I, Takeuchi Y. Short-term ethanol exposure alters calbindin D28k and glial fibrillary acidic protein immunoreactivity in hippocampus of mice. Brain Res. 2000;879(1–2):55–64. doi: 10.1016/s0006-8993(00)02729-3. [DOI] [PubMed] [Google Scholar]
- 86.Evrard SG, Duhalde-Vega M, Tagliaferro P, Mirochnic S, Caltana LR, Brusco A. A low chronic ethanol exposure induces morphological changes in the adolescent rat brain that are not fully recovered even after a long abstinence: an immunohistochemical study. Exp. Neurol. 2006;200(2):438–459. doi: 10.1016/j.expneurol.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 87.Bull C, Syed WA, Minter SC, Bowers MS. Differential response of glial fibrillary acidic protein-positive astrocytes in the rat prefrontal cortex following ethanol self-administration. Alcohol. Clin. Exp. Res. 2015;39(4):650–658. doi: 10.1111/acer.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bull C, Freitas KC, Zou S, et al. Rat nucleus accumbens core astrocytes modulate reward and the motivation to self-administer ethanol after abstinence. Neuropsychopharmacology. 2014;39(12):2835–2845. doi: 10.1038/npp.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study shows that manipulating Ca2+ activity in specific astrocyte populations through the use of DREADDs can lead to changes in alcohol-seeking behavior in rats.
- 89.Cullen KM, Halliday GM. Chronic alcoholics have substantial glial pathology in the forebrain and diencephalon. Alcohol Alcohol. Suppl. 1994;2:253–257. [PubMed] [Google Scholar]
- 90.Ben Haim L, Carrillo-De Sauvage MA, Ceyzériat K, Escartin C. Elusive roles for reactive astrocytes in neurodegenerative diseases. Front. Cell. Neurosci. 2015;9:278. doi: 10.3389/fncel.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zamanian JL, Xu L, Foo LC, et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 2012;32(18):6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrates the diversity of transcriptional changes that reactive astrocytes can experience responding to various injuries and diseases.
- 92.Jensen CJ, Massie A, De Keyser J. Immune players in the CNS: the astrocyte. J. Neuroimmune Pharmacol. 2013;8(4):824–839. doi: 10.1007/s11481-013-9480-6. [DOI] [PubMed] [Google Scholar]
- 93.Alfonso-Loeches S, Ureña-Peralta JR, Morillo-Bargues MJ, Oliver-De La Cruz J, Guerri C. Role of mitochondria ROS generation in ethanol-induced NLRP3 inflammasome activation and cell death in astroglial cells. Front. Cell. Neurosci. 2014;8:216. doi: 10.3389/fncel.2014.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Madeddu S, Woods TA, Mukherjee P, Sturdevant D, Butchi NB, Peterson KE. Identification of glial activation markers by comparison of transcriptome changes between astrocytes and microglia following innate immune stimulation. PLoS ONE. 2015;10(7):e0127336. doi: 10.1371/journal.pone.0127336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bray JG, Reyes KC, Roberts AJ, Ransohoff RM, Gruol DL. Synaptic plasticity in the hippocampus shows resistance to acute ethanol exposure in transgenic mice with astrocyte-targeted enhanced CCL2 expression. Neuropharmacology. 2013;67:115–125. doi: 10.1016/j.neuropharm.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilhelm CJ, Hashimoto JG, Roberts ML, Bloom SH, Andrew MR, Wiren KM. Astrocyte dysfunction induced by alcohol in females but not males. Brain Pathol. 2015;26(4):433–451. doi: 10.1111/bpa.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bennett MV, Contreras JE, Bukauskas FF, Sáez JC. New roles for astrocytes: gap junction hemichannels have something to communicate. Trends Neurosci. 2003;26(11):610–617. doi: 10.1016/j.tins.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hamby ME, Coppola G, Ao Y, Geschwind DH, Khakh BS, Sofroniew MV. Inflammatory mediators alter the astrocyte transcriptome and calcium signaling elicited by multiple G-protein-coupled receptors. J. Neurosci. 2012;32(42):14489–14510. doi: 10.1523/JNEUROSCI.1256-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saba LM, Flink SC, Vanderlinden LA, et al. The sequenced rat brain transcriptome – its use in identifying networks predisposing alcohol consumption. FEBS J. 2015;282(18):3556–3578. doi: 10.1111/febs.13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pignataro L, Varodayan FP, Tannenholz LE, Protiva P, Harrison NL. Brief alcohol exposure alters transcription in astrocytes via the heat shock pathway. Brain Behav. 2013;3(2):114–133. doi: 10.1002/brb3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Flatscher-Bader T, Zuvela N, Landis N, Wilce PA. Smoking and alcoholism target genes associated with plasticity and glutamate transmission in the human ventral tegmental area. Hum. Mol. Genet. 2008;17(1):38–51. doi: 10.1093/hmg/ddm283. [DOI] [PubMed] [Google Scholar]
- 103.Kimpel MW, Strother WN, Mcclintick JN, et al. Functional gene expression differences between inbred alcohol-preferring and non-preferring rats in five brain regions. Alcohol. 2007;41(2):95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tabakoff B, Saba L, Kechris K, et al. The genomic determinants of alcohol preference in mice. Mamm. Genome. 2008;19(5):352–365. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blednov YA, Bergeson SE, Walker D, Ferreira VM, Kuziel WA, Harris RA. Perturbation of chemokine networks by gene deletion alters the reinforcing actions of ethanol. Behav. Brain Res. 2005;165(1):110–125. doi: 10.1016/j.bbr.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gehrmann J, Matsumoto Y, Kreutzberg GW. Microglia: intrinsic immuneffector cell of the brain. Brain Res. Brain Res. Rev. 1995;20(3):269–287. doi: 10.1016/0165-0173(94)00015-h. [DOI] [PubMed] [Google Scholar]
- 107.He J, Crews FT. Increased MCP-1 and microglia in various regions of the human alcoholic brain. Exp. Neurol. 2008;210(2):349–358. doi: 10.1016/j.expneurol.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crews F, Nixon K, Kim D, et al. BHT blocks NF-kappaB activation and ethanol-induced brain damage. Alcohol. Clin. Exp. Res. 2006;30(11):1938–1949. doi: 10.1111/j.1530-0277.2006.00239.x. [DOI] [PubMed] [Google Scholar]
- 109.Zou J, Crews F. Induction of innate immune gene expression cascades in brain slice cultures by ethanol: key role of NF-kappaB and proinflammatory cytokines. Alcohol. Clin. Exp. Res. 2010;34(5):777–789. doi: 10.1111/j.1530-0277.2010.01150.x. [DOI] [PubMed] [Google Scholar]
- 110.Lippai D, Bala S, Petrasek J, et al. Alcohol-induced IL-1beta in the brain is mediated by NLRP3/ASC inflammasome activation that amplifies neuroinflammation. J. Leukoc. Biol. 2013;94(1):171–182. doi: 10.1189/jlb.1212659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Osterndorff-Kahanek E, Ponomarev I, Blednov YA, Harris RA. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS ONE. 2013;8(3):e59870. doi: 10.1371/journal.pone.0059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Most D, Ferguson L, Blednov Y, Mayfield RD, Harris RA. The synaptoneurosome transcriptome: a model for profiling the emolecular effects of alcohol. Pharmacogenomics J. 2014;15(2):177–188. doi: 10.1038/tpj.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saba LM, Flink SC, Vanderlinden LA, et al. The sequenced rat brain transcriptome-its use in identifying networks predisposing alcohol consumption. FEBS J. 2015;282(18):3556–3578. doi: 10.1111/febs.13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Osterndorff-Kahanek EA, Becker HC, Lopez MF, et al. Chronic ethanol exposure produces time- and brain region-dependent changes in gene coexpression networks. PLoS ONE. 2015;10(3):e0121522. doi: 10.1371/journal.pone.0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carson MJ, Reilly CR, Sutcliffe JG, Lo D. Mature microglia resemble immature antigen-presenting cells. Glia. 1998;22(1):72–85. doi: 10.1002/(sici)1098-1136(199801)22:1<72::aid-glia7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 116.Salter MW, Beggs S. Sublime microglia: expanding roles for the guardians of the CNS. Cell. 2014;158(1):15–24. doi: 10.1016/j.cell.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 117.Aronson JK. Old drugs-new uses. Br. J. Clin. Pharmacol. 2007;64(5):563–565. doi: 10.1111/j.1365-2125.2007.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 119.Gelijns AC, Rosenberg N, Moskowitz AJ. Capturing the unexpected benefits of medical research. N. Engl. J. Med. 1998;339(10):693–698. doi: 10.1056/NEJM199809033391010. [DOI] [PubMed] [Google Scholar]
- 120.Okada Y. From the era of genome analysis to the era of genomic drug discovery: a pioneering example of rheumatoid arthritis. Clin. Genet. 2014;86(5):432–440. doi: 10.1111/cge.12465. [DOI] [PubMed] [Google Scholar]
- 121.Wang W, Yang S, Zhang X, Li J. Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics. 2014;30(20):2923–2930. doi: 10.1093/bioinformatics/btu403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J, Li Z, Fan X, Cheng Y. Drug–disease association and drug-repositioning predictions in complex diseases using causal inference-probabilistic matrix factorization. J. Chem. Inf. Model. 2014;54(9):2562–2569. doi: 10.1021/ci500340n. [DOI] [PubMed] [Google Scholar]
- 123.Ye H, Liu Q, Wei J. Construction of drug network based on side effects and its application for drug repositioning. PLoS ONE. 2014;9(2):e87864. doi: 10.1371/journal.pone.0087864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rodriguez-Arias M, Aguilar MA, Minarro J. Therapies in early development for the treatment of opiate addiction. Expert Opin. Invest. Drugs. 2015;24(11):1459–1472. doi: 10.1517/13543784.2015.1086746. [DOI] [PubMed] [Google Scholar]
- 125.Pich EM, Collo G. Pharmacological targeting of dopamine D3 receptors: possible clinical applications of selective drugs. Eur. Neuropsychopharmacol. 2015;25(9):1437–1447. doi: 10.1016/j.euroneuro.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 126.Newman AH, Blaylock BL, Nader MA, Bergman J, Sibley DR, Skolnick P. Medication discovery for addiction: translating the dopamine D3 receptor hypothesis. Biochem. Pharmacol. 2012;84(7):882–890. doi: 10.1016/j.bcp.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Galie N, Ghofrani HA, Torbicki A, et al. Sildenafil citrate therapy for pulmonary arterial hypertension. N. Engl. J. Med. 2005;353(20):2148–2157. doi: 10.1056/NEJMoa050010. [DOI] [PubMed] [Google Scholar]
- 128.Ahmed U, Jones H, Adams CE. Chlorpromazine for psychosis-induced aggression or agitation. Schizophr. Bull. 2011;37(5):890–891. doi: 10.1093/schbul/sbr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leyden J, Dunlap F, Miller B, et al. Finasteride in the treatment of men with frontal male pattern hair loss. J. Am. Acad. Dermatol. 1999;40(6 Pt 1):930–937. doi: 10.1016/s0190-9622(99)70081-2. [DOI] [PubMed] [Google Scholar]
- 130.Teo S, Resztak KE, Scheffler MA, et al. Thalidomide in the treatment of leprosy. Microbes Infect. 2002;4(11):1193–1202. doi: 10.1016/s1286-4579(02)01645-3. [DOI] [PubMed] [Google Scholar]
- 131.Johnson BA, Ait-Daoud N. Topiramate in the new generation of drugs: efficacy in the treatment of alcoholic patients. Curr. Pharm. Des. 2010;16(19):2103–2112. doi: 10.2174/138161210791516404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moore CF, Lycas MD, Bond CW, Johnson BA, Lynch WJ. Acute and chronic administration of a low-dose combination of topiramate and ondansetron reduces ethanol’s reinforcing effects in male alcohol preferring (P) rats. Exp. Clin. Psychopharmacol. 2014;22(1):35–42. doi: 10.1037/a0035215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ray LA, Chin PF, Miotto K. Naltrexone for the treatment of alcoholism: clinical findings, mechanisms of action, and pharmacogenetics. CNS Neurol. Disord. Drug Targets. 2010;9(1):13–22. doi: 10.2174/187152710790966704. [DOI] [PubMed] [Google Scholar]
- 134.Vergne DE, Anton RF. Aripiprazole: a drug with a novel mechanism of action and possible efficacy for alcohol dependence. CNS Neurol. Disord. Drug Targets. 2010;9(1):50–54. doi: 10.2174/187152710790966731. [DOI] [PubMed] [Google Scholar]
- 135.Bell RL, Lopez MF, Cui C, et al. Ibudilast reduces alcohol drinking in multiple animal models of alcohol dependence. Addict. Biol. 2015;20(1):38–42. doi: 10.1111/adb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen B, Butte AJ. Leveraging big data to transform target selection and drug discovery. Clin. Pharmacol. Ther. 2016;99(3):285–297. doi: 10.1002/cpt.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Frail DE, Brady M, Escott KJ, et al. Pioneering government-sponsored drug repositioning collaborations: progress and learning. Nat. Rev. Drug Discov. 2015;14(12):833–841. doi: 10.1038/nrd4707. [DOI] [PubMed] [Google Scholar]
- 138.Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Ther. 2013;93(4):335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 139.Leanza L, Manago A, Zoratti M, Gulbins E, Szabo I. Pharmacological targeting of ion channels for cancer therapy: In vivo evidences. Biochim. Biophys. Acta. 2015;1863(6 Pt B):1385–1397. doi: 10.1016/j.bbamcr.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 140.Lamb J. The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer. 2007;7(1):54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 141.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]; •• Seminal studies that provide a reference collection gene-expression profiles in response to various small molecule perturbations. Works provides a valuable tool for therapeutic development and drug repurposing.
- 142.Qu XA, Rajpal DK. Applications of Connectivity Map in drug discovery and development. Drug Discov. Today. 2012;17(23–24):1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 143.Broad Institute. www.broad.mit.edu/cmap/
- 144.Hajjo R, Setola V, Roth BL, Tropsha A. Chemocentric informatics approach to drug discovery: identification and experimental validation of selective estrogen receptor modulators as ligands of 5-hydroxytryptamine-6 receptors and as potential cognition enhancers. J. Med. Chem. 2012;55(12):5704–5719. doi: 10.1021/jm2011657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Papassotiropoulos A, Gerhards C, Heck A, et al. Human genome-guided identification of memory-modulating drugs. Proc. Natl Acad. Sci. USA. 2013;110(46):E4369–E4374. doi: 10.1073/pnas.1314478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu J, Lee J, Salazar Hernandez MA, Mazitschek R, Ozcan U. Treatment of obesity with celastrol. Cell. 2015;161(5):999–1011. doi: 10.1016/j.cell.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dudley JT, Sirota M, Shenoy M, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 2011;3(96):96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mason BJ, Quello S, Goodell V, Shadan F, Kyle M, Begovic A. Gabapentin treatment for alcohol dependence: a randomized clinical trial. JAMA Intern. Med. 2014;174(1):70–77. doi: 10.1001/jamainternmed.2013.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Mol. Psychiatry. 2015;20(11):1438–1447. doi: 10.1038/mp.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Work demonstrates novel convergent evidence for biological networks related to excessive alcohol consumption, which may prove fundamentally important in the development of pharmacotherapies for alcohol dependence.