Abstract
Eicosanoids are inflammatory signaling lipids that are biosynthesized in response to cellular injury or threat. They were originally thought to be pro-inflammatory molecules, but members of at least one subclass, the lipoxins, are able to resolve inflammation. One step in lipoxin synthesis is the oxygenation of arachidonic acid by 15-lipoxygenase (15-LOX). 15-LOX contains two domains: a Ca2+ binding PLAT domain and a catalytic domain. 15-LOX is a soluble cytosolic protein until binding of Ca2+ to the PLAT domain promotes translocation to the membrane surface. The role of 15-LOX structural dynamics in this translocation has remained unclear. We investigated the dynamics of 15-LOX isoform B (15-LOX-2) upon binding of Ca2+ and ligands, as well as upon membrane association using hydrogen–deuterium exchange mass spectrometry (HDX-MS). We used HDX-MS to probe the solvent accessibility and backbone flexibility of 15-LOX-2, revealing significant differences in deuterium incorporation between the PLAT and catalytic domains, with the PLAT domain demonstrating higher flexibility. Comparison of HDX for 15-LOX-2 in the presence and absence of Ca2+ indicates there are few differences in structural dynamics. Furthermore, our HDX results involving nanodisc-associated 15-LOX-2 suggest that significant structural and dynamic changes in 15-LOX-2 are not required for membrane association. Our results also show that a substrate lipid binding to the active site in the catalytic domain does induce changes in incorporation of deuterium into the PLAT domain. Overall, our results challenge the previous hypothesis that Ca2+ binding induces major structural changes in the PLAT domain and support the hypothesis that is interdomain communication in 15-LOX-2.
In response to cellular injury or threat, chemical mediators are released from the cell to activate immune and inflammatory responses. One of these responses is the biosynthesis of eicosanoids, inflammatory signaling lipids.1 Originally, eicosanoids were thought to possess only pro-inflammatory properties. However, members of one subclass identified as lipoxins are able to resolve inflammation.2 The first step in lipoxin synthesis is the oxygenation of arachidonic acid (AA) by 15-lipoxygenase (15-LOX).3 15-LOX is a non-heme iron-containing dioxygenase that oxygenates polyunsaturated fatty acids (PUFA) containing cis,cis-1,4-pentadiene moieties.4 The primary substrate for lipoxygenases is arachidonic acid (AA) in mammals and linoleic acid (LA) in plants. The primary 15-LOX reaction product, 15-hydroperoxyeicosatetraenoic acid (15-HpETE), is further deoxygenated by 5-LOX into (15S)-epoxytetraene. This epoxy intermediate is converted into lipoxin A4 or B4 by hydrolases.5 The lipoxins have been shown to resolve inflammation by limiting polymorphonuclear neutrophil migration through the endothelium.6 However, 15-LOX has also been involved with an increased level of inflammation by propagating the formation of atherosclerotic plaques.7 In addition, levels of 15-LOX and its enzymatic products are elevated in carotid lesions.8 In mouse models of hyperlipidemia, the levels of 15-LOX expression were positively correlated with that of low-density lipoprotein oxidation.8 Thus, the increase in the level of lipid oxidation due to 15-LOX catalysis results in an escalation of inflammatory responses and promotes the transformation of macrophages to foam cells, an important step in the formation of atherosclerotic plaques.9
Humans express two isoforms of 15-LOX, A (15-LOX-1) and B (15-LOX-2).10 They share 38% sequence identity, but their substrate specificity and cellular distribution differ significantly.11 15-LOX-2 primarily produces 15-HpETE when oxygenating AA, whereas 15-LOX-1 produces 10–20% of 12-HpETE (12-hydroperoxyeicosatetraenoic acid) in addition to 15-HpETE.12 Members of the LOX superfamily utilize the same chemical mechanism for oxygenation. Briefly, the reaction is initiated by oxidation of the catalytic iron, followed by a hydrogen abstraction on the central carbon of the pentadiene moiety, forming a lipid radical.13 One molecule of dioxygen is then incorporated into the lipid radical forming a peroxyl radial that is reduced by the catalytic iron center and later released as the product of hydroperoxide. The oxidation of the free radical occurs two carbons removed from the site of hydrogen abstraction. The different forms of LOX in mammals are named for the positional specificity of oxygenation of their primary products. Thus, 15-LOX primarily generates 15-HpETEs, while 5-LOX primarily generates 5-HpETE. In addition to employing a common reaction mechanism, mammalian LOXs share several structural features. Currently, four mammalian LOX crystal structures have been determined: rabbit 15-LOX-1 in complex with an inhibitor [Protein Data Bank (PDB) entry 1LOX],14,15 human 15-LOX-2 in complex with the substrate mimic, C8E4 (PDB entry 4NRE),16 human stable 5-LOX (which is 5-LOX mutated for improved enzymatic half-life) (PDB entry 3O8Y),17 and the catalytic domain of porcine 12-LOX with a specific inhibitor bound (PDB entry 3D3L).18 Mammalian LOXs contain approximately 650 amino acids comprising two domains. The first is a C2-like polycystin-1, lipoxygenase α-toxin (PLAT) domain, and the second is the catalytic domain. The PLAT domain is a β-barrel-like structure that binds Ca2+, while the catalytic domain is mainly comprised of α-helices with a hydrophobic active site containing the catalytic iron.16−18 In the crystal structure of human 15-LOX-2 with C8E4 bound, C8E4 is positioned in a U-shaped conformation in the active site. When AA is superimposed onto the C8E4 location, the C-13 atom of AA is positioned adjacent to the catalytic iron, in an appropriate position for hydrogen abstraction, and subsequent oxygenation at C-15.16
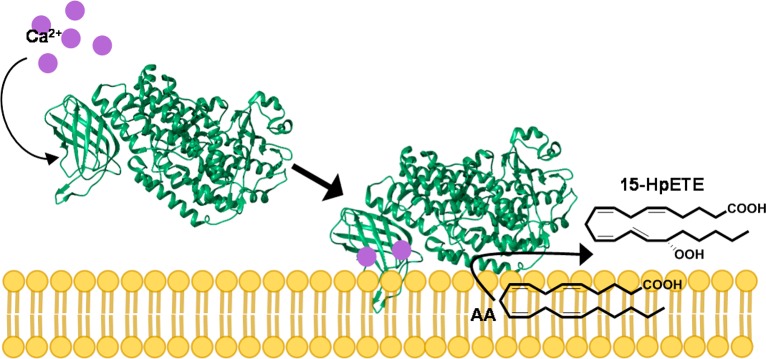
In intact cells, binding f Ca2+ to the PLAT domain results in translocation of 15-LOX from the cytosol to the cellular membrane (Figure 1).19 Studies have suggested that upon binding of Ca2+ to LOXs, the PLAT domain undergoes local conformational changes that result in an increased level of exposure of hydrophobic amino acids to the protein surface. However, these changes have yet to be fully elucidated.20 The structure of 15-LOX-2 contains a loop in the PLAT domain consisting mainly of prolines and leucines. It is proposed that this hydrophobic loop may become stabilized in a solvent-exposed position upon Ca2+ binding, allowing it to act as a membrane anchor.16
Figure 1.
15-LOX-2 reaction in intact cells. Ca2+ concentrations in the cell increase in response to stress. The Ca2+ binds to the PLAT domain of 15-LOX-2, resulting in a translocation from the cytosol to the membrane. 15-LOX-2 then binds its substrate, AA, and oxygenates it to form 15-HpETE. 15-HpETE is further metabolized by downstream enzymes to form lipoxins, which initiate the resolution of inflammation.
Small angle X-ray scattering indicates a high degree of flexibility between the PLAT and catalytic domains.21,22 However, the crystal structure and scattering data reveal little about the role that structural dynamics play in the translocation of 15-LOX-2 to the membrane, regulatory mechanisms, or allosteric interactions. In this report, we investigated the structural changes in 15-LOX-2 upon binding of Ca2+ and ligands, as well as upon membrane association using hydrogen–deuterium exchange mass spectrometry (HDX-MS). Despite the resolution limit of this technique, dictated by the length of peptides generated in protein digests, the insight such data provide into structural dynamics is highly informative, especially when combined with the crystal structure.23 HDX-MS is a solution-based technique that is not limited by the size or flexibility of the protein and can therefore provide essential information for understanding the role of structural dynamics in 15-LOX-2 regulatory and enzymatic mechanisms. In this study, we used HDX-MS to map changes in the structural dynamics of 15-LOX-2 onto the crystal structure as a consequence of substrate binding and membrane association.
Materials and Methods
Materials
Buffer salts and common chemicals of the highest quality were purchased from commercial resources. All polyunsaturated fatty acids were obtained from Cayman Chemical. Phospholipids were purchased from Avanti Polar Lipids. Competent cells used in transformations were purchased from Novagen. 15-LOX-2 and MSP1e3d1 plasmids were provided by M. Newcomer (Department of Biological Sciences, Louisiana State University, Baton Rouge, LA).
Protein Expression and Purification
15-LOX-2 with a hexa-His tag was co-expressed with Escherichia coli YjgD protein in a pET-Duet-1 plasmid. The plasmid was transformed into Rossetta 2 (DE3) competent cells and grown in LB cultures overnight. Small overnight cultures were diluted (1:200) into a 1 L volume of Terrific Broth in 2 L Erlenmeyer flasks. Cultures were grown at 37 °C and 200 rpm until an OD600 of 1.0 was reached. The temperature was decreased to 18 °C, and cells were grown overnight for leaky expression of 15-LOX-2. Cells were harvested by centrifugation and stored at −80 °C.
Protein purification of 15-LOX-2 was adapted from a previously published protocol.16 Cells were lysed with 2 mL of B-PER Bacterial Protein Extraction buffer (Thermo Fisher Scientific) per gram of cell pellet supplemented with 10 μL of Problock 2D protease inhibitor cocktail (Gold Biotechnology) per milliliter of total lysate and 2 mg of DNase-1 (Sigma-Aldrich). Lysis was performed for approximately 30 min at 4 °C, and the sample was further lysed by probe sonication (1 s pulse, 2 min on, and 3 min off) on ice until the sample was no longer viscous. Cellular debris was removed by centrifugation (approximately 200000 rcf) for 30 min. Imidazole was added to the supernatant to a final concentration of 10 mM. The supernatant was incubated for 30 min at 4 °C with approximately 10 mL of Co2+-NTA resin (Thermo Fisher Scientific) equilibrated with column buffer [50 mM Tris, 500 mM KCl, and 10 mM imidazole (pH 8.0)] and applied to a gravity column. Resin was washed with column buffer containing 20 mM imidazole (10 × resin volume). Protein was eluted from resin utilizing column buffer containing 150 mM imidazole (3 × resin volume). Protein was concentrated to 1.5 mL using Amicon centrifugation filters with a molecular weight cutoff (MWCO) of 30 kDa and then loaded onto a HiLoad 16/600 Superdex-200 pg column (GE Healthcare) equilibrated with 20 mM Tris and 150 mM KCl (pH 8.0). The maximum peak fractions were collected at approximately 80 mL and concentrated. 15-LOX-2 purification was verified utilizing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and Western blots probing for hexa-His-tagged protein.
15-LOX-2 Enzymatic Activity
15-LOX-2 oxygenates AA, resulting in the formation of conjugated double bonds on the product; therefore, the change in ultraviolet (UV) absorbance from AA to the HpETE product was monitored by a PerkinElmer Lambda 45 UV/vis spectrometer at 25 °C. The assays were completed in 20 mM Tris-HCl, 150 mM KCl, and 0.5 mM CaCl2 (pH 7.5). 15-LOX-2 was added to activity assay buffer to a final concentration of 0.5 μM. AA to a final concentration of 80 μM (from 28 mM AA in a 100% ethanol stock solution) was then added to the buffer to initiate the reaction. An increase at 238 nm was monitored for 3 min. The products from the assay were extracted by acidifying the reaction buffer to pH 4.0 and adding 1 volume of dichloromethane. The phases were separated, and the organic phase was extracted with water two additional times. The dichloromethane was then evaporated with a stream of nitrogen, and the remaining product was dissolved in a small volume of a 1:1 methanol/water mixture. Reverse-phase high-performance liquid chromatography (HPLC) was performed using a Varian ProStar HPLC system with a kinetex 5 μ C18 100A column (Phenomenex) using a mobile phase of acetonitrile, water, and formic acid [70:30:0.4 (v/v/v)]. Elution times were compared against that of an HpETE standard mixture.
15-LOX-2 Phospholipid Specificity
PIP Strips and SphingoStrips were used to identify the phospholipid 15-LOX-2 prefers for membrane association. Strips were used according to the manufacturer’s instructions (Thermo Fisher Scientific). Briefly, strips were blocked with 3% BSA in TBS-T buffer [20 mM Tris-HCl, 150 mM NaCl, and 0.1% Tween 20 (pH 8.0)] and gently rocked for 1 h at approximately 25 °C. Then strips were incubated with 1 μg/mL 15-LOX-2 in TBS-T buffer with 3% BSA and 0.5 mM CaCl2 for 2 h at approximately 25 °C. Strips were rinsed thoroughly with TBS-T buffer, and protein binding was visualized with a mouse anti-penta-His monoclonal antibody (1:3000 dilution) and an AP-conjugated anti-mouse secondary antibody (1:10000 dilution).
Nanodisc Assembly
Nanodiscs were assembled using membrane scaffolding protein (MSP1e3d1), and the phospholipids phosphatidylserine and phosphatidylcholine. pMSP1E3D1 in a pET28 vector was transformed into Rosetta 2 (DE3) competent cells and grown in 5 mL LB cultures overnight. The overnight cultures were added to 1 L of Terrific broth with 30 μg/mL kanamycin. Expression was induced with 1 μM isopropyl 1-thio-β-d-galactopyranoside at an OD600 of 1.0, and cells were grown for an additional 4 h at 37 °C. Cells were harvested by centrifugation and frozen at −80 °C. Cell pellets were resuspended in lysis buffer [50 mM Tris, 300 mM NaCl, and 1% Triton X-100 (pH 8.0)] supplemented with two Complete, mini, EDTA free Protease Inhibitor cocktail tablets and DNase I. Lysozyme (0.2 mg/mL) was added to the lysate and the mixture stirred at 4 °C for 1.5 h. The lysate was further lysed by probe sonication (1 s pulses, 2 min on, and 3 min off). Cellular debris was cleared by centrifugation, and the supernant was applied to a Ni-NTA gravity column equilibrated with 50 mM Tris and 300 mM NaCl (pH 8.0). Resin was washed with 10 column volumes of equilibration buffer and 10 column volumes of equilibration buffer containing 50 mM imidazole. MSP1e3d1 was eluted from the resin with 3 column volumes of equilibration buffer with 200 mM imidazole. Protein was concentrated using Amicon centrifugation filters with a MWCO of 10 kDa.
Nanodiscs were prepared by dialyzing MSP1e3d1 into nanodisc buffer [20 mM Tris and 150 mM KCl (pH 8.0)] and adding cholate to a final concentration of 100 mM. A mixture of phospholipids (3:1 phosphatidylcholine/phosphatidylserine) were added to the solution in a 160:1 molar ratio with MSP1e3d1. The solution was shaken for 1 h at 4 °C. Nanodisc buffer was added until the cholate concentration decreased to 25 mM; then 0.1 g/mL biobeads were added, and the solution was shaken for 1 h. The mixture was shaken at 4 °C with subsequent additions of biobeads (0.2 g/mL after 2 h, 0.2 g/mL after 4 h). The mixture was filtered to remove biobeads 4 h after the last addition of biobeads. Nanodisc formation was monitored by size exclusion chromatography with a Superose-12 column connected to the UV detector set for 280 nm. Fractions collected from the column were analyzed by SDS–PAGE and subsequent Western blotting to confirm the presence of MSP1E3D1 and 15-LOX-2.
Identification of Peptic Fragments of 15-LOX-2
The protein digest of 15-LOX-2 was optimized for greatest peptide sequence coverage and the highest signal-to-noise ratio for peptide ion envelopes. Protein digestion was performed under the quenching conditions of the hydrogen–deuterium (H/D) exchange experiment (7 min, pH 2.3, 0 °C). A 4.5:1 pepsin:15-LOX-2 molar ratio provided optimal sequence identification. The peptides generated were injected onto an Aeris PEPTIDE 3.6 μ XB-C18 column (50 mm × 2.6 mm, Phenomenex) connected to a ThermoFinnigan LTQ Linear Ion Trap with an electrospray ionization source for separation and tandem mass spectrometry. The peptides were separated using a 5 to 50% acetonitrile/H2O gradient containing 0.4% formic acid over 25 min (at 0.1 mL/min). The peptides were sequenced in positive ion mode by tandem MS/MS collision-induced dissociation. The resulting fragmentation patterns were analyzed using the computational program, PEAKS (Bioinformatics Solutions Inc.). Peptide identities determined by PEAKS were confirmed by comparing experimental fragmentation to theoretical patterns. Theoretical peptide identities were determined by the parent mass from experimental data and the 15-LOX-2 FASTA sequence using ExPASy-PeptideMass software. Theoretical fragmentation patterns were created with ProteinProspector program MS-Product.
H/D Exchange Mass Spectrometry
Deuterium exchange was initiated by the addition of 45 μL of D2O to 5 μL of 15-LOX-2 (50 μM). The deuterium/protein solution was incubated for 12 periods of time ranging from 15 s to 8 h at approximately 25 °C. The exchange was quenched with the addition of 50 μL of quench buffer [100 mM K2PO4 and 10 mM TCEP (pH 2.3)] and transferred to ice. Pepsin (2 μL of a 20 mg/mL solution) was added immediately after quenching and incubated on ice for 7 min. All samples were prepared individually and analyzed by mass spectrometry the same day. Samples with 0.5 mM Ca2+ had CaCl2 added (as 1 μL of a 12.5 mM CaCl2 solution in H2O) to protein solution 2 min before exchange was initiated. Samples containing AA had 1 μL of AA (in 100% ethanol) added 30 s before the addition of D2O. The concentration of the AA stock solution was such that only 1 μL of the stock solution was added to reach the desired AA sample concentration to ensure the ethanol effect was not observed. Deuterium incorporated during on-exchange can back-exchange for hydrogen upon addition of aqueous quench buffer. Controls for deuterium back-exchange were conducted by digesting 15-LOX-2 using the same conditions created for peptide identification and H/D exchange. The resulting peptides were lyophilized and then incubated in D2O. Lyophilization and incubation in D2O were repeated, and these 100% deuterated peptides were injected onto the mass spectrometer using the same method for time point samples. By this method, the back-exchange was calculated to be between 15 and 40% depending on the peptide’s innate exchange rates, elution time, and length.
HPLC and ESI-MS were used to determine the amount of deuterium incorporated into each 15-LOX-2-derived peptide at tested time points. The HPLC injection loop, column, and all chromatographic buffers were bathed in an ice bath to prevent a high level of back-exchange. The peptides were separated over 13 min with a 5 to 50% acetonitrile/H2O gradient with a constant level of 0.4% formic acid. The peptides were separated using a ThermoFinnigan Surveyor HPLC system with a C-18 reverse-phase column. Buffer salts were diverted to waste, and an additional wash step of 10 min with a 2-propanol-containing solution (50% acetonitrile/40% 2-propanol/10% H2O/0.4% formic acid) was added to the chromatographic profile for samples containing lipids. Mass spectra were recorded on a ThermoFinnigan LTQ linear ion trap using a positive ion electrospray ionization source. The detector was set to scan between m/z 300 and 2000 and recorded at an enhanced scan rate to improve the signal:noise ratio. The resulting peptide ion envelopes were analyzed by the computational program HDExaminer. Each sample was tested two or three times, and deuterium incorporation for that peptide was averaged. The total number of deuterium incorporated (D) is plotted versus time (t) and fit to the sum of first- or second-order rate terms. This allows amplitudes (An) and exchange rates (k) to be calculated (eq 1). The rates will be classified into rapid, intermediate, and slow exchange categories. The change in deuterium incorporation rate between the different sample states will indicate changes in structural dynamics and protein interactions.
| 1 |
The exchange amplitude at time zero (Afast) represents the number of deuteriums incorporated in the first 15 s. The value of Afast is calculated by subtracting the sum of the fitted amplitudes in the slow kinetic phases (>15 s) from the total number of exchangeable sites. Inasmuch as Afast is not a fitted parameter, no errors are reported. Given that the errors in the fitted amplitudes, A1–A3, are typically <10%, the errors in Afast are estimated to be <20–30%. Peptides were considered to have significant changes between protein states when they exhibited a change in amplitude of 10% of the exchangeable backbone protons or experienced a 10-fold change in the rate of exchange. Calculated amplitudes and rate constants for peptides directly discussed throughout this work are presented in Figures S1–S3.
Results
15-LOX-2 Peptide Map
The 15-LOX-2 digestion procedure was optimized to create a peptide map that yielded the greatest sequence coverage. Optimization of the digestion procedure included testing different proteases at varying molar ratios with 15-LOX-2, the addition of denaturants to the quenching buffer, and varying protein–protease incubation times. A pepsin digest of 15-LOX-2 with tandem MS created a peptide map providing 72% sequence coverage and 68 measurable peptides ranging from 5 to 20 residues for the 15-LOX-2 enzyme (Figure 2). The peptide coverage decreased slightly upon addition of AA and nanodiscs because of charge competition in electrospray ionization and overlap of 15-LOX-2 peptides with peptides derived from the membrane scaffolding protein component of the nanodiscs. All of the peptides identified in Figure 2 were analyzed for H/D exchange MS experiments. Results for the peptides in the PLAT domain and active site that illuminate the roles of structural dynamics in 15-LOX-2 regulation and function are the focus of this discussion. Additional peptide exchange profiles that provide an overall view of the structural dynamics of 15-LOX-2 are presented in Figures S4–S7.
Figure 2.
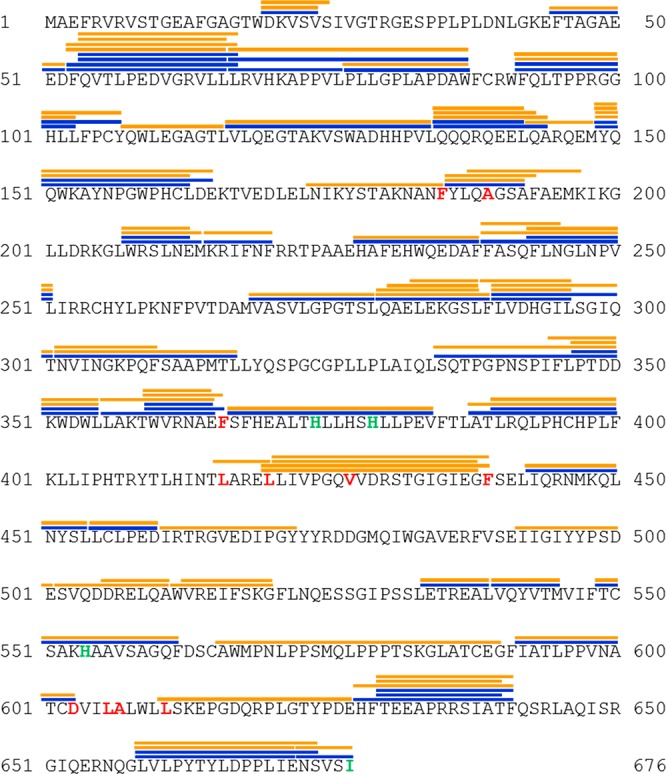
Peptide map of 15-LOX-2. Peptides were generated by pepsin digest and identified through tandem mass spectrometry. Peptides with measurable ion envelopes are shown as orange lines (in chelated and Ca2+-bound states) and blue lines (in a nanodisc-associated state) above the peptide sequence. Residues that are conserved in mammalian LOXs and thought to be part of substrate binding are colored red, while residues that coordinate the catalytic iron are colored green.
Spatial Resolution of the Structural Dynamics of 15-LOX-2
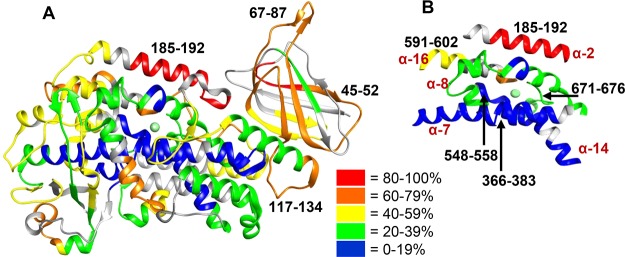
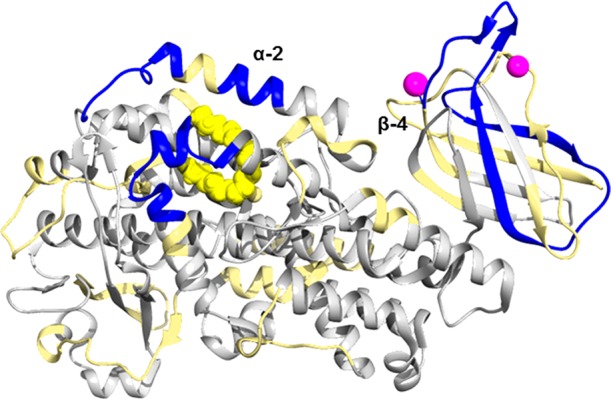
Investigating the rate of deuterium incorporation in peptides informs the stability of partially water-exposed secondary structural elements that undergo local unfolding. The level of deuteration of 15-LOX-2 with no bound Ca2+ or AA was mapped onto a crystal structure of 15-LOX-2 (PDB entry 4NRE, which has Ca2+ bound to the PLAT domain and the substrate mimic, C8E4, bound to the active site of the catalytic domain) (Figure 3 and Figure S8).16 A wide variety (5–90%) of levels of deuterium incorporation were observed for 15-LOX-2 peptides. These data indicate the two domains of 15-LOX-2 exhibit significant differences in deuterium incorporation. In general, peptides derived from the PLAT domain, such as peptide 45–52 and 67–87, exhibit an overall high level of incorporation of deuterium, with most exhibiting 35–50% exchange within 15 s and 40–80% exchange within 10 min of D2O incubation, indicating a relatively high level of solvent exposure and flexibility, whereas many peptides in the catalytic domain exhibit low incorporation rates of only 0–20% even after D2O incubation for 8 h, indicating limited solvent exposure and less flexibility (Figure 3A). Nevertheless, there are peptides in the catalytic domain that exhibit a high level of exchange. One of the highest levels of deuterium incorporation involves α-helix 2 (peptide 185–192) in which 77% of the available amide protons exchange within 10 min of D2O incubation. This helix is thought to be flexible so that the active site can accommodate different substrates (Figure 3B).24
Figure 3.
Structural dynamics of ligand and substrate free 15-LOX-2. (A) Backbone amide deuteration levels of 15-LOX-2 peptides following incubation for 1 h in D2O are mapped onto the crystal structure (PDB entry 4NRE)16 to provide information about the general flexibility and solvent exposure. Gray indicates regions that could not be identified. Peptides mentioned in this paper are designated by their sequence range. (B) Deuterium levels of the five helices that form the active site and coordinate the catalytic iron are shown. Helices are labeled by their numerical order in LOXs.
As anticipated, peptides that are involved in coordination of the catalytic iron, such as peptides 366–383 and 548–558, exhibit a low level of deuterium incorporation of <20% throughout the 8 h deuterium incubation, indicating that the area is highly protected from solvent exposure. The only iron-coordinating peptide that exhibited >18% deuterium incorporation contains the C-terminal end of 15-LOX-2 (peptide 671–676), which exchanges >22% of the backbone amide hydrogens at later time points (>30 min) (Figure 3B). It is also notable that results for peptide 117–134 indicate that the linker between the PLAT and catalytic domain is both flexible and solvent-exposed as a steady increase in the level of exchange occurs throughout time points ending with >80% of the hydrogens being exchanged by 8 h.
Major Structural Changes Are Not Required for Membrane Association
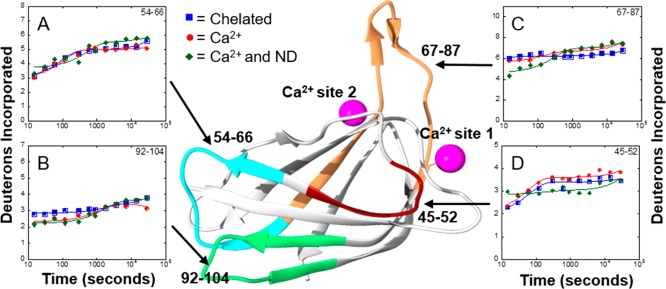
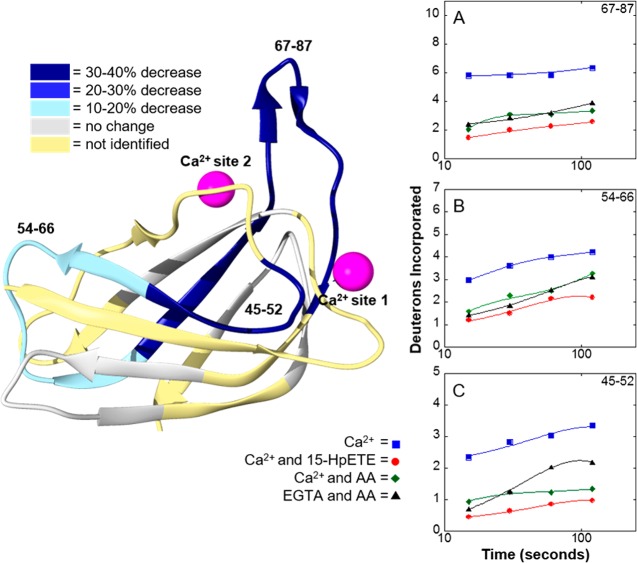
The crystal structure of 15-LOX-2 shows two Ca2+ binding sites located in the PLAT domain.16 Ca2+ binding promotes the protein’s translocation from the cytosol to the membrane.25 To investigate the structural dynamics of Ca2+ binding that might promote membrane association, full backbone H/D exchange kinetics of 15-LOX-2 was performed in the presence and absence of Ca2+. Unfortunately, peptides that contain residues of Ca2+ binding site 1 could not be analyzed because of the low signal:noise ratio. However, peptide 67–87, which had a stronger signal and could be analyzed, is important in structuring Ca2+ binding site 2 (Figure 4C). Surprisingly, there are no significant changes in deuterium incorporation in this peptide upon Ca2+ binding. In fact, the only peptide to show a change in deuterium incorporation due to Ca2+ binding was peptide 92–104, which exhibited a small decrease in the level of deuterium incorporation during the first 5 min of H/D exchange (Figure 4B). Comparison of H/D exchange for 15-LOX-2 and 15-LOX-2 with bound Ca2+ indicates there are no significant changes (>10% change) in structural dynamics upon binding Ca2+.
Figure 4.
Impact of Ca2+ binding and membrane association on H/D exchange for 15-LOX-2. The number of deuteriums incorporated on peptides vs time was plotted to obtain H/D exchange kinetics for 15-LOX-2 peptides in a Ca2+ free state (blue squares), a Ca2+-bound state (red circles), and a nanodisc-associated state (green triangles). Peptides are labeled by their primary sequence and mapped onto the PLAT domain of 15-LOX-2 by color: (A) peptide 54–66, (B) peptide 92–104, (C) peptide 67–87, and (D) peptide 45–52.
15-LOX-2 has previously been shown to associate with nanodiscs in a Ca2+-dependent manner.16 In addition to investigating how Ca2+ binding may affect the structural dynamics of 15-LOX-2 in solution, H/D exchange kinetics of 15-LOX-2 associated with nanodiscs was recorded to identify regions involved in membrane association or that undergo conformational changes upon membrane association. Preliminary experiments with PIP and Sphingo strips suggested that 15-LOX-2 preferably associates with phosphatidylserine over other phospholipids (Figure S9). Therefore, phosphatidylserine was added to the nanodiscs to promote 15-LOX-2 association.
Prior to H/D exchange, association of 15-LOX-2 with nanodiscs was verified using size exclusion chromatography as monitored using a 280 nm UV detector. A peak was observed at approximately 8.8 mL when purified nanodiscs were run on a Superose-12 10/300 GL column. Purified 15-LOX-2 eluted at 13.7 mL, while the peak for 15-LOX-2 associated with nanodiscs in the presence of Ca2+ eluted at approximately 8.5 mL in the same column. The fractions from each peak were analyzed via SDS–PAGE (Figure S10). A band indicative of 15-LOX-2 (76 kDa) and a band indicative of the nanodisc scaffold protein (MSP1e3d1, 37 kDa) were observed from the peak at 8.5 mL (Figure S11), further confirming the association of 15-LOX-2 with nanodiscs in the presence of Ca2+.
H/D exchange of Ca2+-bound 15-LOX-2 in solution and associated with nanodiscs was compared, revealing only slight changes in H/D exchange kinetics when 15-LOX-2 is membrane-associated (Figure 4). Peptide 67–87 in nanodisc-associated 15-LOX-2 showed a 12% decrease in the level of deuterium incorporation during the first 2 min of exchange compared to 15-LOX-2 in solution (Figure 4C). The kinetic profile of peptide 45–52 changed slightly upon nanodisc association, with the extent of fast exchange in nanodisc-associated 15-LOX-2 only one deuteron higher than that of 15-LOX-2 with Ca2+. However, over long exchange times, the level of deuteration of nanodisc-associated 15-LOX-2 remained constant, ending with a slightly lower level of deuterium incorporation (Figure 4D) relative to that of free protein. No other changes in 15-LOX-2 peptide H/D kinetics were observed upon comparison of cytosolic 15-LOX-2 and nanodisc-associated 15-LOX-2. These results indicate that membrane association has an only minimal effect on the structural dynamics of 15-LOX-2. Substituting DDM for nanodiscs as a membrane mimic and varying the Ca2+ concentration yielded similar results (Figure S12).
Impact of Substrate Binding on Structural Dynamics
To potentially determine lipid binding sites and map conformational changes upon substrate binding, H/D exchange experiments were performed with Ca2+-bound 15-LOX-2 in the presence of AA. Because 15-LOX-2 is active in this experiment, generation of the product (15-HpETE) interfering with the data interpretation was a concern. Therefore, activity assays were conducted under conditions similar to H/D exchange sample conditions. These activity assays indicated that only 10% of AA is converted to 15-HpETE by 15-LOX-2 after approximately 200 s. Therefore, we focused only on data collected following incubation of 15-LOX-2 in deuterium for <120 s in the presence of AA. As a control, H/D exchange was performed for 15-LOX-2 in the presence of 15-HpETE.
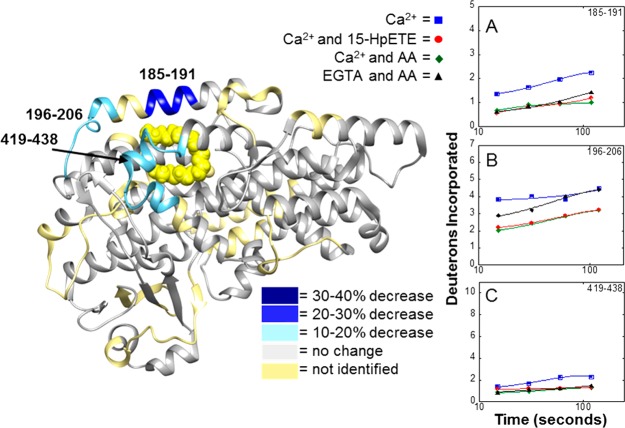
When Ca2+-bound 15-LOX-2 is incubated with AA or 15-HpETE, six peptides (peptides 45–52, 54–67, 67–87, 185–191, 196–206, and 418–438) demonstrate a similar and significant reduction in the level of deuterium incorporation compared to that of Ca2+-bound 15-LOX-2 alone (Figure 5). Two of the peptides that experienced a reduction in their level of deuterium incorporation are part of the active site. Peptide 185–191, which is part of α-helix 2 (the exterior of the active site), displayed a reduction in its level of deuterium incorporation of 21% after H/D exchange for 120 s (Figure 5A). Peptide 419–438, part of the active site entrance, experienced a 12% reduction after H/D exchange for the same period of time (Figure 5C). This is likely due to the lipids binding to the active site and shielding the peptide backbone from H/D exchange, which indicates that both the lipid substrate and the lipid product are binding in a similar manner to 15-LOX-2. As an additional control, the level of deuterium incorporation at 15 s was measured at increasing concentrations of AA to test if decreases in the level of deuterium incorporation were due to AA micelle formation. The smooth dose-dependent trend over the critical micelle concentration suggests that the decreases in the level of deuterium incorporation are not caused by 15-LOX-2 interacting with an AA micelle (Figure S13).
Figure 5.
Deuterium exchange in the catalytic domain in 15-LOX-2 upon lipid binding. H/D exchange was performed for 15-LOX-2 in four different states: (1) 15-LOX-2 with Ca2+ present (blue squares), (2) 15-LOX-2 with Ca2+ and AA present (green diamonds), (3) 15-LOX-2 with Ca2+ and 15-HpETE present (red circles), and (4) Ca2+ free 15-LOX-2 with AA present (black triangles). The exchange profiles for peptides exhibiting changes of >10% after exchange for 2 min are shown: (A) peptide 185–191, (B) peptide 196–206, and (C) peptide 419–438.
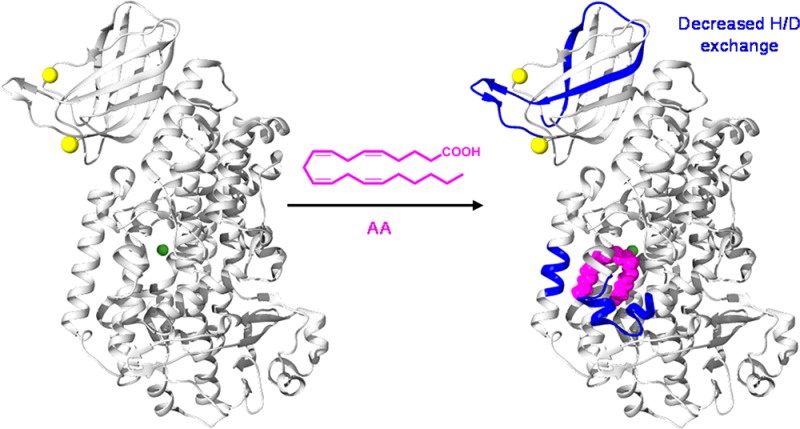
One of the most interesting results is that binding of either AA or 15-HpETE to the active site in the catalytic domain induces changes in deuterium incorporation in peptides 45–52, 54–76, and 67–87, which are located in the PLAT domain (Figure 6). Peptide 67–87 showed a 32% reduction in its level of deuterium incorporation after H/D exchange for 120 s in Ca2+-bound 15-LOX-2 incubated with AA relative to Ca2+-bound 15-LOX-2 alone. This suggests that binding of AA to the catalytic domain stabilizes or induces conformational changes in the PLAT domain.
Figure 6.
Deuterium exchange in the PLAT domain in 15-LOX-2 upon lipid binding. Addition of the substrate and product to 15-LOX-2 resulted in unexpected decreases in the level of H/D exchange in the PLAT domain. H/D exchange was performed for 15-LOX-2 in four different states: (1) 15-LOX-2 with Ca2+ present (blue squares), (2) 15-LOX-2 with Ca2+ and AA present (green diamonds), (3) 15-LOX-2 with Ca2+ and 15-HpETE present (red circles), and (4) chelating agent treated 15-LOX-2 with AA present (black triangles). The exchange profiles for peptides exhibiting changes of >10% after exchange for 2 min are shown: (A) peptide 67–87, (B) peptide 54–66, and (C) peptide 45–52.
To test if the decreases in the level of deuterium incorporation in peptides derived from the PLAT domain in the presence of AA are Ca2+-dependent, 15-LOX-2 was chelated with EGTA before the addition of AA followed by H/D exchange kinetics. Two of the three peptides in the PLAT domain (peptides 54–66 and 67–87) demonstrated no change in deuterium incorporation, indicating that neither peptide is affected by removal of Ca2+. The third peptide (peptide 45–52) demonstrated an increased level of deuterium incorporation in the absence of Ca2+ (Figure 6C), suggesting that this peptide is destabilized upon Ca2+ removal in the presence of AA.
Discussion
Conformational Dynamics of Native 15-LOX-2
Information provided by HDX-MS is particularly useful when combined with crystal structure data. A crystal structure provides an indispensable “snapshot” of a protein conformation in a static state. Experimental data from HDX-MS can be used to characterize local dynamics and motions of the protein of interest in solution at ambient temperature. In the 15-LOX-2 crystal structure with C8E4 bound, the PLAT and catalytic domains are dominated by stable secondary structures; the PLAT domain is composed of sandwiched β-sheets and the catalytic domain of α-helical bundles (see Figure 3).4 For native 15-LOX-2 in solution, the deuteration levels of the two domains after H/D exchange for 1 h indicate significantly different degrees of flexibility. The peptides from the PLAT domain exhibited relatively higher levels of deuterium incorporation at all the time points for D2O incubation, indicating that the PLAT domain is globally dynamic to the extent that the β-strands of the barrel become transiently free from their normal β-sheet hydrogen bonding patterns enabling facile H/D exchange (Figure 3A).
Another region of 15-LOX-2 that exhibited a high level of deuteration upon longer D2O incubations (>30 min) was the long loop that connects the PLAT domain and catalytic domain (Figure 3A). In many mammalian LOX crystal structures, the domains adopt a similar overall arrangement with the PLAT domain resting atop the catalytic domain, giving the LOXs a 100 Å long cylindrical topology.16,17,26 This suggests the possibility of an extensive interaction interface between the two domains. However, in this work, the peptide backbone on the loop (peptide 117–134) experiences high exchange rates and peptides involved in the domain interface (peptide 92–104) exhibit a moderate deuterium incorporation level. This implies that peptide 117–134 is dynamic and lacks stabilizing interactions, while the domain interface experiences a degree of solvent exposure, contrary to what might be supposed from the crystal structures, suggesting few or weak interactions between these domains in solution.
The H/D exchange observed in the 15-LOX-2 active site reveals a low level of exchange even after D2O incubation for 8 h on the active site interior (α-7 and α-14), indicating well-ordered structures with few local structural fluctuations (Figure 3B). The higher levels of deuterium incorporation within the first 2 min of D2O incubation at the active site entrance and exterior (α-2, peptide 185–191; α-8, peptide 419–438; and α-16, peptide 591–602) indicate a moderate degree of flexibility. This flexibility is necessary for 15-LOX-2 to coordinate the binding of lipids with different chain lengths and degrees of unsaturation, such as AA or dihomo-γ-linolenic acid.
H/D Exchange of 15-LOX-2 Reveals Significant Structural Changes Are Not Required for Membrane Association
15-LOX-2 is a cytosolic protein until binding of Ca2+ to the PLAT domain results in translocation to the membrane, regulating cellular compartmentalization.19 The nature of the structural changes resulting in LOX membrane association has yet to be determined. Previous H/D exchange studies that involved the C2 domain, which is structurally and functionally similar to the PLAT domain, in cytosolic phospholipase A2 (cPLA2) suggested that binding of Ca2+ to the C2 domain of cPLA2 resulted in an overall decrease in domain flexibility.27 However, in this work, we found that the addition of Ca2+ to the 15-LOX-2 PLAT domain did not result in significant changes in the rate of incorporation of deuterium into any region of the protein that we were able to analyze (Figure 4).
In addition, nanodisc-associated 15-LOX-2 did not exhibit significant differences in H/D exchange kinetics compared to cytosolic 15-LOX-2 (Figure 4). In fact, the only peptide to show a slight decrease in its level of deuterium incorporation in this experiment was peptide 67–87 (Figure 4B). This peptide spans a unique hydrophobic loop (P73–A86) that protrudes from the PLAT domain and is proposed to act as a membrane anchor.16 The H/D exchange kinetics obtained in this study for nanodisc-associated 15-LOX-2 eliminates the possibility of the hydrophobic PLAT loop inserting deeply and stably into the membrane but leaves open the possibility of the hydrophobic loop transiently sampling the membrane in a way that still allows water accessibility and H/D exchange. Enzymes that access the substrate by moving along the membrane surface can do so through either a scooting mode, in which the enzyme “scoots” through a single membrane surface, or a hopping mode, in which an enzyme–membrane collision is required for each turnover event. Previous work with nanodisc-associated 15-LOX-2 suggests that human 15-LOX-2 “hops” along the membrane surface, which explains the minimal change in H/D exchange upon nanodisc association.28 As a reflection of the preference of 15-LOX-2 for phosphatidylserine in membrane association (Figure S9), it is plausible that binding of Ca2+ to the PLAT domain adds positive charge for associating with phosphatidylserine, without significantly changing the structure and dynamics of the protein. 15-LOX-2 phospholipid specificity was assessed with PIP and Sphingo strips, which is a nonquantitative approach, particularly when protein–membrane interactions are of only modest affinity. Future experiments for quantifying 15-LOX-2 membrane associations will be required to assess this membrane association model. However, this proposed membrane-associating method is similar to the mechanism proposed for certain other peripheral proteins with C2 domains such as protein kinase C. In protein kinase C, small structural changes have been observed upon Ca2+ binding. Instead, phosphatidylserine fills an incomplete coordination sphere of Ca2+, making Ca2+ a bridge between the C2 domain and the phospholipid.29−31 It is plausible that Ca2+ binding in the PLAT domain of 15-LOX-2 plays a similar role in coordinating association to the plasma membrane.
Conformational Changes in the 15-LOX-2 PLAT Domain Resulting from Binding of Lipid Substrates to the Active Site
Traditionally, PLAT domains were primarily thought to function only as membrane-associating domains.32 However, recent work suggests a plausible communication site between the PLAT and catalytic domains.33 There is a conserved π-cation interaction observed in all mammalian LOXs. In 15-LOX-2, this interaction is between W109 on β-strand 7 and K175 on α-helix 2. A crystal structure of truncated 12S-LOX (lacking its PLAT domain) revealed significant positional shifts in α-helix 2 upon removal of the PLAT domain, implying the physical restraint imposed by the PLAT domain may play a role in positioning α-helix 2.18 In addition, mutations involved with interdomain interactions in rabbit 12/15-LOX resulted in a decrease in catalytic activity and protein stability.34 Site mutations in the domain interface in 5-LOX increased enzymatic activity, implying an important role for PLAT and catalytic domain communication in the LOX catalytic mechanism.35
One of the more interesting results of our studies was the changes in H/D exchange in the PLAT domain upon binding of the lipid substrate to the catalytic domain. We found decreases in the level of deuterium incorporation in peptides involved in the active site [peptides 185–191, 196–206, and 419–438 (Figures 5 and 7)] and the PLAT domain [peptides 45–52, 54–66, and 67–86 (Figures 6 and 7)] in the presence of AA and 15-HpETE. The extents of H/D exchange in 15-LOX-2 peptides in the PLAT domain and active site are highly dependent on AA concentration (Figure S13). While there are observable differences in H/D exchange at low AA concentrations (10 μM), the most significant results were observed when the concentration of AA was ≥250 μM. Previous work established the Km value for 15-LOX-2 to be 1.9 μM,16 which means the active site in 15-LOX-2 for the H/D exchange studies is saturated. Exactly how occupation of the active site alters the structure of the PLAT domain is not established by our data. The level of deuteration in peptides 92–104 and 117–134 suggests that there is a degree of solvent accessibility in the domain interface (Figure 3). Previous work demonstrated that optimal activity in rabbit 12/15-LOX required tight association between the PLAT and catalytic domains.34 It is possible that binding of AA to the active site changes the structural dynamics of α-2, which increases the number of interactions between the domains, therefore inducing a change in the PLAT domain structural dynamics. While further work is required to clarify the relationship between AA and the PLAT domain, our results support the hypothesis of important interdomain communications, as it appears that binding of the substrate lipid to the active site results in changes in H/D exchange profiles in the PLAT domain.
Figure 7.
Changes in deuterium incorporation resulting from the presence of AA and 15-HpETE. The crystal structure of 15-LOX with bound Ca2+ and C8E4 is used to depict connections between the PLAT and catalytic domains.16 Peptides that exhibited a decrease in the level of deuterium incorporation of >10% upon AA addition following incubation in D2O for 2 min are colored blue. Peptides with no changes are colored gray, and unidentified peptides are colored tan. C8E4 is colored yellow to demonstrate the putative orientation of AA in the 15-LOX-2 active site, and Ca2+ is colored purple.
An alternative explanation for decreases in the level of deuterium incorporation in the PLAT domain is binding of AA to a secondary allosteric site proposed to be between the PLAT and catalytic domains.36 LOXs have been shown be allosterically regulated by their enzymatic products.37 However, the allosteric site in 15-LOX-2 is highly specific for 13-HODE, the oxygenated product of linoleic acid, binding 13-HODE but not 15-HpETE or 12-HpETE.36 This suggests it is unlikely the decreases in the level of deuterium incorporation in the PLAT domain are due to direct binding of AA to the secondary allosteric site. In addition, a nonspecific hydrophobic interaction between an AA micelle and the PLAT domain seems unlikely as the shielding of H/D exchange that results from the presence of AA was not observed because of the presence of nanodiscs or detergent micelles (Figure 4 and Figure S12).
Conclusions
HDX-MS has allowed us to observe the structural dynamics of 15-LOX-2 in various liganded and membrane-associated states. For Ca2+-bound 15-LOX-2 in both cytosolic and membrane-associated states, the PLAT domain is more flexible and solvent-exposed than the catalytic domain. Most intriguingly, there is strong structural coupling between the membrane binding PLAT and catalytic domains in 15-LOX-2, suggesting a coordination of membrane binding and catalytic activities. The general lack of changes in deuterium incorporation patterns upon binding of Ca2+ and subsequent membrane association indicates that major changes in the structure of 15-LOX-2 are not required for membrane association as was previously suggested.16 This leads to questions about the nature of the domain–membrane interactions. One possible explanation is that membrane association is promoted by binding of Ca2+ to the PLAT domain by increasing its overall positive charge, activating interaction with the negatively charged headgroup of membrane-associated phosphatidylserine.
Acknowledgments
We thank the staff at the Vanderbilt University Mass Spectrometry Core Lab and Dr. Laura Busenlehner (deceased December 2014) for technical support. In addition, we thank Dr. Marcia Newcomer, who provided plasmids to initiate the project and much useful advice.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00559.
Tables of amplitudes and rate constants for peptides (Figures S1–S3), additional deuterium incorporation profiles (Figures S4–S7), additional heat maps of the 15-LOX-2 structure (Figure S8), images from PIP and SphingoStrips (Figure S9), SDS–PAGE of 15-LOX-2 associated with nanodiscs (Figure S10), SEC profile of 15-LOX-2 associated with nanodiscs (Figure S11), dependence of deuterium incorporation on DDM concentration (Figure S12), and dependence of deuterium incorporation on AA concentration (Figure S13) (PDF)
Raw data used to calculate the percent of deuteration (XLSX)
Author Present Address
⊥ M.K.T.: Department of Chemistry, The University of Alabama, Tuscaloosa, AL 35487.
This work was funded by National Institutes of Health Grant RO1 GM030910 to R.N.A. and C.R.S.
The authors declare no competing financial interest.
Author Status
∥ Deceased June 18, 2015.
Supplementary Material
References
- Dennis E. A.; Norris P. C. (2015) Eicosanoid Storm in Infection and Inflammation. Nat. Rev. Immunol. 15, 511–523. 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliberti J.; Hieny S.; Reis e Sousa C.; Serhan C. N.; Sher A. (2002) Lipoxin-mediated inhibition of IL-12 production by DCs: a mechanism for regulation of microbial immunity. Nat. Immunol. 3, 76–82. 10.1038/ni745. [DOI] [PubMed] [Google Scholar]
- Serhan C. N.; Hamberg M.; Samuelsson B. (1984) Lipoxins: Novel series of biologically active compounds formed from arachidonic acid in human leukocytes (lipoxygenase interaction products/conjugated tetraenes/human neutrophils). Proc. Natl. Acad. Sci. U. S. A. 81, 5335–5339. 10.1073/pnas.81.17.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer M. E.; Brash A. R. (2015) The structural basis for specificity in lipoxygenase catalysis. Protein Sci. 24, 298–309. 10.1002/pro.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda N.; Yamamoto S.; Fitzsimmons B. J.; Rokach J. (1987) Lipoxin synthesis by arachidonate 5-lipoxygenase purified from porcine leukocytes. Biochem. Biophys. Res. Commun. 144, 996–1002. 10.1016/S0006-291X(87)80062-1. [DOI] [PubMed] [Google Scholar]
- Takano T.; Clish C. B.; Gronert K.; Petasis N.; Serhan C. N. (1998) Lipoxin Analogues Inhibit Neutrophil-mediated Leakage Permeability Neutrophil-mediated Changes in Vascular Permeability Are Inhibited by Topical Application of Aspirin-triggered 15-epi-lipoxin A 4 and Novel Lipoxin B 4 Stable Analogues. J. Clin. Invest. 101, 819–826. 10.1172/JCI1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendobry S. M.; et al. (1997) Attenuation of diet-induced atherosclerosis in rabbits with a highly selective 15-lipoxygenase inhibitor lacking significant antioxidant properties. Br. J. Pharmacol. 120, 1199–1206. 10.1038/sj.bjp.0701007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harats D.; Shaish A.; George J.; Mulkins M.; Kurihara H.; Levkovitz H.; Sigal E. (2000) Overexpression of 15-Lipoxygenase in Vascular Endothelium Accelerates Early Atherosclerosis in LDL Receptor-Deficient Mice. Arterioscler., Thromb., Vasc. Biol. 20, 2100–2105. 10.1161/01.ATV.20.9.2100. [DOI] [PubMed] [Google Scholar]
- Rydberg E. K.; et al. (2004) Hypoxia Increases LDL Oxidation and Expression of 15-Lipoxygenase-2 in Human Macrophages. Arterioscler., Thromb., Vasc. Biol. 24, 2040–2045. 10.1161/01.ATV.0000144951.08072.0b. [DOI] [PubMed] [Google Scholar]
- Brash A. R.; Boeglin W. E.; Chang M. S. (1997) Discovery of a second 15S-lipoxygenase in humans. Proc. Natl. Acad. Sci. U. S. A. 94, 6148–6152. 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C. D.; Chen X.-S.; Johnson E. N.; Zhao L. (2002) Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediators 68–69, 303–312. 10.1016/S0090-6980(02)00036-9. [DOI] [PubMed] [Google Scholar]
- Bryant R. W.; Bailey J. M.; Schewes T.; Rapoports S. M. (1982) Positional Specificity of a Reticulocyte Lipoxygenase. J. Biol. Chem. 257, 6050–6055. [PubMed] [Google Scholar]
- Zoia L.; Perazzini R.; Crestini C.; Argyropoulos D. S. (2011) Understanding the radical mechanism of lipoxygenases using 31P NMR spin trapping. Bioorg. Med. Chem. 19, 3022–3028. 10.1016/j.bmc.2011.02.046. [DOI] [PubMed] [Google Scholar]
- Gillmor S. A.; Villaseñor A.; Fletterick R.; Sigal E.; Browner M. F. (1997) The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat. Struct. Biol. 4, 1003–1009. 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- Choi J.; Chon J. K.; Kim S.; Shin W. (2008) Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins: Struct., Funct., Genet. 70, 1023–1032. 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- Kobe M. J.; Neau D. B.; Mitchell C. E.; Bartlett S. G.; Newcomer M. E. (2014) The structure of human 15-lipoxygenase-2 with a substrate mimic. J. Biol. Chem. 289, 8562–9. 10.1074/jbc.M113.543777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N. C.; et al. (2011) The structure of human 5-lipoxygenase. Science (Washington, DC, U. S.) 331, 217–219. 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.; Mueser T. C.; Marnett L. J.; Funk M. O. (2012) Crystal Structure of 12-Lipoxygenase Catalytic Domain-Inhibitor Complex Identifies a Substrate Binding Channel for Catalysis. Structure 20, 1490–1497. 10.1016/j.str.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther M.; Wiesner R.; Kuhn H. (2004) Investigations into Calcium-dependent Membrane Association of 15-Lipoxygenase-1. J. Biol. Chem. 279, 3717–3725. 10.1074/jbc.M309564200. [DOI] [PubMed] [Google Scholar]
- Kulkarni S.; Das S.; Funk C. D.; Murray D.; Cho W. (2002) Molecular Basis of the Specific Subcellular Localization of the C2-like Domain of 5-Lipoxygenase. J. Biol. Chem. 277, 13167–13174. 10.1074/jbc.M112393200. [DOI] [PubMed] [Google Scholar]
- Hammel M.; Walther M.; Prassl R.; Kuhn H. (2004) Structural Flexibility of the N-terminal b-Barrel Domain of 15-Lipoxygenase-1 Probed by Small Angle X-ray Scattering. Functional Consequences for Activity Regulation and Membrane Binding. J. Mol. Biol. 343, 917–929. 10.1016/j.jmb.2004.08.076. [DOI] [PubMed] [Google Scholar]
- Mittal M.; Hasan M.; Balagunaseelan N.; Fauland A.; Wheelock C.; Rådmark O.; Haeggström J. Z.; Rinaldo-Matthis A. (2017) Investigation of calcium-dependent activity and conformational dynamics of zebra fish 12-lipoxygenase. Biochim. Biophys. Acta, Gen. Subj. 1861, 2099–2111. 10.1016/j.bbagen.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Pirrone G. F.; Iacob R. E.; Engen J. R. (2015) Applications of Hydrogen/Deuterium Exchange MS from 2012 to 2014. Anal. Chem. 87, 99–118. 10.1021/ac5040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins P. M.; Murphy R. C. (2012) Cholesteryl ester acyl oxidation and remodeling in murine macrophages: formation of oxidized phosphatidylcholine. J. Lipid Res. 53, 1588–97. 10.1194/jlr.M026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilty I.; Logan A.; Vickers P. J. (1999) Differential characteristics of human 15-lipoxygenase isozymes and a novel splice variant of 15S-lipoxygenase. Eur. J. Biochem. 266, 83–93. 10.1046/j.1432-1327.1999.00818.x. [DOI] [PubMed] [Google Scholar]
- Gilbert N. C.; et al. (2012) Conversion of human 5-lipoxygenase to a 15-lipoxygenase by a point mutation to mimic phosphorylation at Serine-663. FASEB J. 26, 3222–3229. 10.1096/fj.12-205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-H.; et al. (2008) Calcium Binding Rigidifies the C2 Domain and the Intradomain Interaction of GIVA Phospholipase A 2 as Revealed by Hydrogen/Deuterium Exchange Mass Spectrometry. J. Biol. Chem. 283, 9820–9827. 10.1074/jbc.M708143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender G.; Schexnaydre E. E.; Murphy R. C.; Uhlson C.; Newcomer M. E. (2016) Membrane-dependent Activities of Human 15-LOX-2 and Its Murine Counterpart. J. Biol. Chem. 291, 19413–19424. 10.1074/jbc.M116.741454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111. 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Corbalan-Garcia S.; Gómez-Fernández J. C. (2014) Signaling through C2 domains: More than one lipid target. Biochim. Biophys. Acta, Biomembr. 1838, 1536–1547. 10.1016/j.bbamem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Verdaguer N.; Corbalan-Garcia S.; Ochoa W. F.; Fita I.; Gó Mez-Ferná Ndez J. C. (1999) Ca 2+ bridges the C2 membrane-binding domain of protein kinase Cá directly to phosphatidylserine. EMBO J. 18, 6329–6338. 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer M.; Gilbert N. (2010) Location, Location, Location: Compartmentalization of Early Events in Leukotriene Biosynthesis. J. Biol. Chem. 285, 25109–25114. 10.1074/jbc.R110.125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eek P.; et al. (2012) Structure of a Calcium-dependent 11R-Lipoxygenase Suggests a Mechanism for Ca2+ Regulation. J. Biol. Chem. 287, 22377–22386. 10.1074/jbc.M112.343285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.; et al. (2011) Tight association of N-terminal and catalytic subunits of rabbit 12/15-lipoxygenase is important for protein stability and catalytic activity. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1811, 1001–1010. 10.1016/j.bbalip.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Rakonjac Ryge M.; et al. (2014) A mutation interfering with 5-lipoxygenase domain interaction leads to increased enzyme activity. Arch. Biochem. Biophys. 545, 179–185. 10.1016/j.abb.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Joshi N.; et al. (2013) Kinetic and structural investigations into the allosteric and pH effect on the substrate specificity of human epithelial 15-lipoxygenase-2. Biochemistry 52, 8026–35. 10.1021/bi4010649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecksler A. T.; Kenyon V.; Deschamps J. D.; Holman T. R. (2008) Substrate Specificity Changes for Human Reticulocyte and Epithelial 15-Lipoxygenases Reveal Allosteric Product Regulation. Biochemistry 47, 7364–7375. 10.1021/bi800550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.