Abstract
Algae are ubiquitous in the marine environment, and the ways in which they interact with bacteria are of particular interest in marine ecology field. The interactions between primary producers and bacteria impact the physiology of both partners, alter the chemistry of their environment, and shape microbial diversity. Although algal-bacterial interactions are well known and studied, information regarding the chemical-ecological role of this relationship remains limited, particularly with respect to quorum sensing (QS), which is a system of stimuli and response correlated to population density. In the microbial biosphere, QS is pivotal in driving community structure and regulating behavioral ecology, including biofilm formation, virulence, antibiotic resistance, swarming motility, and secondary metabolite production. Many marine habitats, such as the phycosphere, harbour diverse populations of microorganisms and various signal languages (such as QS-based autoinducers). QS-mediated interactions widely influence algal-bacterial symbiotic relationships, which in turn determine community organization, population structure, and ecosystem functioning. Understanding infochemicals-mediated ecological processes may shed light on the symbiotic interactions between algae host and associated microbes. In this review, we summarize current achievements about how QS modulates microbial behavior, affects symbiotic relationships, and regulates phytoplankton chemical ecological processes. Additionally, we present an overview of QS-modulated co-evolutionary relationships between algae and bacterioplankton, and consider the potential applications and future perspectives of QS.
Keywords: phytoplankton, bacterioplankton, signal language, algal-bacterial relationships, co-evolution, ecological behaviors
I. INTRODUCTION
Microbes comprise around 3.5–5.5×1017 g carbon or 4.0–6.0×1030 cells, which accounts for approximately half of the total biomass of living organisms on Earth (Whitman et al., 1998). Most microorganisms are found in the ocean, including bacteria, archaea, fungi, and some algae. These organisms drive oceanic energy fluxes process (Falkowski et al., 2008), which in turn influence their diversity and interactions (Strom, 2008). Marine microbes are critical for the biogeochemical cycling of elements due to their abundance, taxonomic diversity, and high potential metabolic activity (Azam and Malfatti, 2007). Marine algae, which are autotrophic and free-living aquatic plants, mediate primary production in the ocean, half of which are transformed into dissolved organic matter that is available for use by heterotrophic prokaryotes (Azam and Malfatti, 2007). Algae consumed by higher eukaryotes supply nourishment to microbial loop through algal exudates/decomposition, thus providing the base of the food web (Carrillo et al., 2006). In addition, algae have high photosynthetic efficiency, and diatoms alone account for approximately 20% of global photosynthesis (Rosenwasser et al., 2014). Consequently, algae play various ecological roles, including the cycling of organic matter (Armbrust, 2009), and help reduce the impact of global warming (Sayre, 2010).
Marine ecosystems are based on multiple interactions among organisms, which may be competitive, mutualistic, parasitic, or symbiotic. Due to the importance and pervasiveness of marine algae, there has been strong scientific interest in elucidating algal-bacterial interactions. Previous studies have focused on the biological, environmental, and physical aspects of these various types of interactions; however, few studies have investigated the role chemical signaling, largely because the field of marine chemical ecology is relatively new (Hay, 2009). Recent evidence shows that the regulation of behavior by chemical signals is not restricted in bacteria, but was also found between microorganisms and their hosts (Freestone, 2013). An example of this activity was proposed by Long et al. (2007), who suggested that under symbiotic conditions, small molecules might regulate the colony formation of the phytoplankton Phaeocystis globosa by altering energy flow, nutrient release, and carbon sequestration patterns. Hughes and Sperandio (2008) further noted that individual bacteria can alter their behaviour through chemical interactions between organisms in microbial communities, and these communications often occur at scales much bigger than traditionally recognized because of the organisms’ ability to respond to surrounding signals. Thus, in addition to affecting the behavior of both individuals and populations, chemical cues might also affect community organization and ecosystem functioning (Hay, 2009).
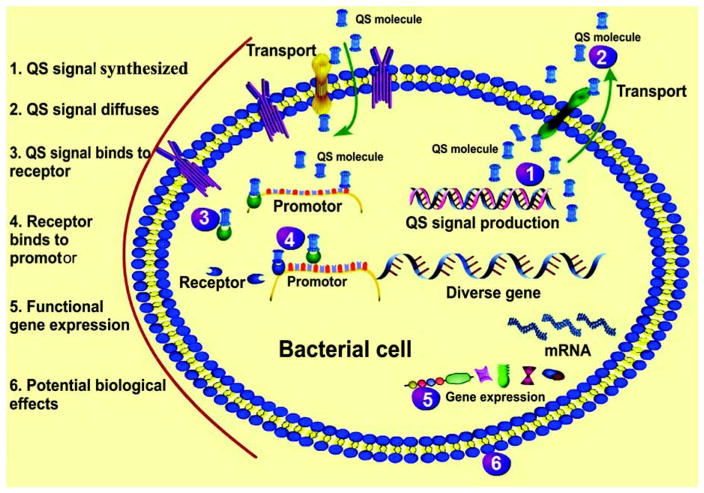
Quorum sensing (QS) is a common form of chemical signaling receiving increasing attention from marine ecologists. QS occurs within microbial populations in a density-dependent manner (Bassler, 2002), causes downstream changes in gene regulation, and modulates many biological functions such as bioluminescence, virulence factor expression, biofilm formation, sporulation, and bacterial conjugation (Waters and Bassler, 2005; Mangwani et al., 2012). To date, three main QS systems have been described (Table 1): (i) the LuxR/I-type system, (ii) the LuxS/AI-2 system, and (iii) the AI-3/epinephrine/norepinephrine system. The LuxR/I-type system uses acyl-L-homoserine lactone (AHL) as an autoinducer, and is predominantly used by gram-negative (G−) bacteria. The LuxS/AI-2 system uses peptide substances as the signal for interspecies communication, and is primarily used by gram-positive (G+) bacteria. The AI-3/epinephrine/norepinephrine system facilitates signaling among kingdoms. These systems allow bacteria to communicate across species boundaries or involved in interkingdom signaling. Taken G− bacteria as an example, Figure 1 shows the molecular formation mechanisms of QS and subsequent cascade response process.
TABLE 1.
QS signals and their characterization in marine microorganisms
| QS signal | Producing species | Chemical structure | Physical properties | Molecular synthase | Regulated protein | Ecological function | References | |
|---|---|---|---|---|---|---|---|---|
| AI-1 short-chain molecules |
C4-HSL | Vibrio harveyi | Homoserine lactone (HSL) ring with a variable acyl side chain.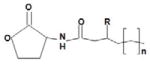
|
Molecular weight: from 171.9 Da for a C4- to 367.57 Da for an unsubstituted C 18-AHL (Decho et al., 2011). Solubility: low-molecular-weight AHLs tend to be relatively water soluble, and it is unlikely that communal responses based on their presence would ever be limited by their solubility. However, as molecular weights increase significantly above 520 g/mol, the saturation concentration of the AHL is expected to drop below 1μM AHL (Decho et al., 2011). Diffusivity: from 10 to 78 μm AHL (Decho et al., 2011). Dispersion: from 10 s micrometers to 100 s micrometers AHL (Decho et al., 2011). |
LuX-M | LuxR type protein | Bioluminescence | Waters and Bassler (2005) |
| Pseudomonas aeruginosa | RH II | Biofilm maturation | Waters and Bassler (2005) | |||||
| Aeromonas hydrophyla |
Ahy I
Asa I |
Biofilm formation, Enzyme production | Tebben et al. (2011) | |||||
| Serratia marcescens | SWrI | Swarming | Miller and Bassler (2001) | |||||
| C6-HSL | Erwinia stewartii G− | LuXI | LuxR type protein | Exopolysaccharide | Watson et al. (2002) | |||
| Vibrio fischeri G− | Light production | Nealson et al. (1970) | ||||||
| P. fischeri G− | Virulence | Eberhard et al. (1981) | ||||||
| A. salmonicida | Ahy I | LuxRtype protein | Biofilm formation | Swift et al. (1999) | ||||
| Chromobacterium Violaceum | CV I | Violacein, antibiotics, and enzyme production | Cha et al. (1998) | |||||
| Yersinia enterolcolitica, Y. pseudotuberculosis | Yen I, Yps I | Motility aggregation | Miller and Bassler (2001) | |||||
| Vibrio sp. (algae-associated) | LuxI | Settlement activity | Taylor et al. (2007) | |||||
| Vibrio alginolyticus | LuxI | / | Huang et al. (2007) | |||||
| C8-HSL | Agrobacterium tumefaciens | Tra I | Virulence factors | Fuqua et al. (2001) | ||||
| Y. pseudotuberculosis | Yps I | Potential regulate cells wimming | Miller and Bassler (2001) | |||||
| Roseobacter spp., Marinobacter sp. | / | / | Taylor et al. (2007) | |||||
|
|
|
|||||||
| AI-1 long-chain molecules |
C10-HSL | Vibrio anguillarum | Van I | LuxR | Virulence | Defoirdt et al. (2004) | ||
| C12-HSL | Vibrio alginolyticus | Las I | LuxR | Virulence formation | Pearson et al. (1994) | |||
| / | LuxR | / | Huang et al. (2007) | |||||
| C14-HSL | Roseobacter sp. | / | LuxR | / | Mohamed et al. (2008) | |||
|
| ||||||||
| AI-2 | Furanosyl borate diester | Vibrio harveyi | The proposed structure contains two fused five member rings containing one boron atom bridging the diester.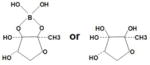
|
/ | Ai-2 | Bioluminescence | Bassler et al. (1993) | |
| V. cholerae | Ai-2 | Virulence | Chen et al. (2002) | |||||
| V. harveyi | Ai-2 | Virulence | Henke and Bassler (2004) | |||||
| 2-methy-2,3,3,4-tetrahydroxytetrahydrofuran | V. parahaemolyticus | Cqs A | Virulence | Miller et al. (2002) | ||||
| V. cholerae | Ai -2 | Virulence | Miller et al. (2004) | |||||
| Salmonella enterica | Virulence gene expression | Sperandio et al. (2001) | ||||||
| Escherichia coli | Type II secretion | Day and Maurelli (2001) | ||||||
| Shigella flexneri | LupX | Virulence factor virB expression | Mcnab et al. (2003) | |||||
| Streptococcus pordonii | Biofilm formation | Li et al. (2002) | ||||||
|
| ||||||||
| AI-3 | E. coli, Xanthomonas campestris | DSF:cis-11-methyl-2-dodecenoic acid. |
/ | LuxS | Lux P/Q type protein | Biofilm formation | Barber et al. (1997) | |
| Lux O type protein, mRNA-dependent regulation, such as 5RNAs | Reading and Sperandio (2006); Wang et al. (2004) | |||||||
|
| ||||||||
| Peptide autoinducers | Cyclic dipeptides |
Pseudomonas sp. Streptomyces griseus Staphylococcus aureus |
Diketopiperazines (DPKS)* cyclo (L-Ala-L-Val) and cyclo (L-Pro-L-Tyr).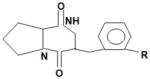
|
/ | LuxR | Activates AHL biosensors. *: the induction threshold for DKPS is higher than AI-1, which indicates that DKPS may not have a significant role in the marine environment |
Degrassi et al. (2002) | |
| Butyrolactonase | Waters and Bassler (2005) | |||||||
| Thiolactone | Lyon et al. (2000); Zhang and Dong (2004) | |||||||
|
| ||||||||
| Others | Bradyoxetin | Bradyrhizobium japonicum G−, Rhizobium sp., Alphaproteobacteria sp., Pseudomonas aeruginosa | 2-{4-[[4-(3-aminooxetan-2-yl)phenyl](amino)methyl]phenyl} oxetan-3-yl amine. |
/ | Nod gene | Nod gene | Nitrogen-fixing | Loh et al. (2002a,b) |
| Hydroxy palmitic acid methyl ester | Ralstonia solanacearum | PhcS-phcR | Iron concentration | Polysaccharide biosynthesis | Flavier et al. (1997) | |||
| Cys-11-methyl-2-dodecenoic acid | Xanthomonas campestris | / | Virulence | Zhang and Dong (2004) | ||||
| Non-identified compounds | Algae symbiotic bacterium | / | Competence, Morphogenesis | Decho et al. (2011) | ||||
Figure 1.
The molecular formation mechanisms of quorum sensing (QS) and the subsequent cascade response, using the example of gram-negative bacteria. The numbers represent different stages of the QS process. Arrows show the main direction of signal transport due to active transport or diffusion.
In marine microbiological field, previous studies have shown that many bacteria (Vibrio sp., Ochrobactrum sp., Shewanella sp., and Alteromonas sp.) possess autoinducer synthases and enable them to produce AHLs (Case et al., 2008; Cuadrado-Silva et al., 2013). These AHL molecules can regulate bacterial behavior and exhibit various ecological functions. Some bacteria belonging to the genera Vibrio, Shewanella, and Sulfitobacter synthesized or degraded AHLs and modulated the zoospore settlement of their host (Ulva sp.) (Tait et al., 2009). In the oligotrophic North Pacific Ocean, blue-green alga (cyanobacterium) Trichodesmium consortia regulate alkaline phosphatase activity and control phosphorus acquisition by secrete AI-1 and AI-2 signals (Van Mooy et al., 2012). Hmelo et al. (2011) reported that the addition of particulate organic carbon (POC) containing AHLs to bacterial cultures enhanced the functioning of bacterial hydrolytic enzymes involved in POC degradation, and suggested that there might be a link between hydrolytic enzymes and QS. Jatt et al. (2015) further identified six AHL signals (i.e. C4-HSL, 3OC6-HSL, C6-HSL, C10-HSL, C12-HSL and C14-HSL) from marine snow, and found that addition of exogenous AHLs enhanced the extracellular hydrolytic enzyme in Pantoea ananatis B9. They pointed out that AHL-based QS system could be involved in biosynthesis of extracellular alkaline phosphatase in P. ananatis B9. These studies indicated that QS was an important modulator to regulate the ecological functions of marine plankton by affecting related gene or protein expression. Besids these, in recents years, chemical mediators of QS and molecular crosstalk between bacteria and eukaryotes have been described in a wide range of symbiotic organisms (Pacheco and Sperandio, 2009), broadening our understanding about certain multi-species interactions.
In algal-bacterial microenvironments, chemical-mediated processes under natural conditions are not well known. Furthermore, information remains limited about the intercellular signaling that occurs during interactions between algae and associated microbes. In this review, we focus on the recent achievements in QS signals and discuss their ecological functions in symbiotic interactions between algae and bacteria. The aim is to expand current knowledge about algae-bacteria relationships from microsociological prespective.
II. THE PHYCOSPHERE
Bell and Mitchell (1972) coined the term “phycosphere” in microecology to describe the region that extends outward from an algal cell, chain, or colony of cells “in which bacterial growth is stimulated by algal exudates.” The phycosphere is analogous to the rhizosphere (where nutrients pass between the soil and roots), influencing the nutrient fluxes entering and exiting algal cells (Fig. 2). Within this microzone, planktonic bacteria are highly heterogeneous. Furthermore, ingestion, digestion, egestion, excretion, and exudation by other marine organisms create habitats characterized by the frequent and pervasive occurrence of microscale chemical gradients (Stocker and Seymour, 2012). Marine phytoplankton release substantial amounts of amino acids, simple sugars, complex polysaccharides, organic acids, and lipids into the phycosphere area (Jones and Cannon, 1986); similarly, bacteria generate organic and inorganic substrates that are used by host cells (Doucette, 1995). For example, cobalamin (or vitamin B12) cannot be synthesized by phytoplankton, but is an essential nutrient produced by bacterial communities associated with unicellular algae (Tang et al., 2010). Phytoplankton thus benefit by acquiring micronutrients from bacterial metabolism, which forms the basis of the symbiotic relationship between phytoplankton and bacteria (Geng and Belas, 2010). Positive consequences of this relationship include nutritional contributions, biological matter cycling, and creation of an ecological niche for symbiotic taxa. Conversely, harmful results include competition between taxa, the generation of harmful compounds, and deleterious metabolic impacts. Thus, the phycosphere serves as a hot spot for algal-bacterial communication, in which the diffusive surrounding microzone is a dynamic and fluid environment containing many different types of chemical fluxes (Fig. 2). Notably, some chemical signals, such as those involved in allelopathy and QS, are vital for constructing the heterogeneous and dynamic environment of the phycosphere that creates a harmonious microcommunity.
Figure 2.
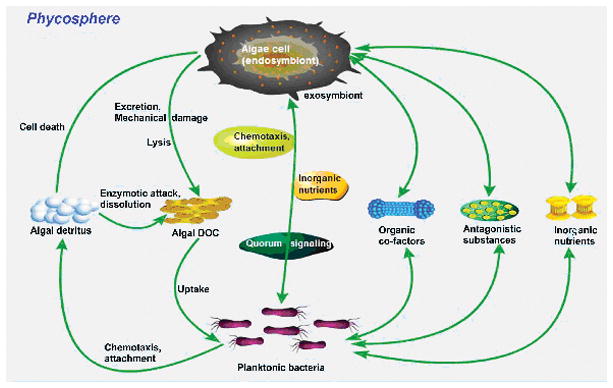
Flow chart of the biological processes in the phycosphere micro-environment. This schematic excludes a number of indirect interactions, such as bacterial metabolism and signaling substances (such as quorum sensing and allelochemicals). The arrows show the predominant direction of a process.
III. QS COMMUNICATION IN SYMBIOTIC ENVIRONMENTS
Various methods of communication exist within the symbiotic environments of bacteria and algae, including interbacterial and interalgal communication, and interkingdom signaling. Understanding how these two groups of organisms communicate may enhance our understanding about their behavioral ecology within phycospheres.
A. Bacterial intercellular communication
Many marine bacteria, such as proteobacteria, produce QS chemicals and participate in ecological processes. Bacteria from the Roseobacter clade (which often co-occur with algae) regulate primary metabolic processes through AHL molecules (Wagner-Dobler and Biebl, 2006). QS-based AHL contributes to the regulation of extracellular hydrolytic alkaline-phosphatase activity and is responsible for the degradation of sinking POC (Jatt et al., 2015); the authors speculated that variability in AHL-triggered POC hydrolysis might have a profound effect on marine food webs. Other chemical compounds (like five or six-membered ring structures with attached lipophilic carbon chains) also exhibit interkingdom signaling activity in prokaryotic and eukaryotic systems (Gerwick et al., 2013). Amin et al. (2015) unraveled a bacterial consortium associated with a globally distributed diatom, and showed that one sulfitobacter species promotes diatom cell division via the secretion of the hormone indole-3-acetic acid. This study provided evidence that these interactions are mediated through the production and exchange of infochemicals. Therefore, QS has multiple effects on bacterial behavior, and facilitates communication among bacterial species and with other organisms (Bassler, 2002).
B. Interalgal communication
Eukaryotes use a variety of signaling molecules for different physiological processes; however, compared to bacteria, information about algal cell signaling remains limited. Thus far, molecules identified to be involved in signaling include phosphorlipids, flavonoids, indole-3-acetic acid, and quorum substrates (Van Leeuwen et al., 2004; Sprague and Winans, 2006; Hassan and Mathesius, 2012; Amin et al., 2015). The most commonly used signaling processes by algae are allelochemicals and QS. Allelochemicals are primarily secondary metabolites that influence the survival, reproduction, and/or growth of algae (Graneli and Pavia, 2006). These allelopathic interactions have several critical roles in regulating the concentration and distribution of the target species, cell defense, and in nutrition procurement (Inderjit et al., 2011). Allelopathy also regulates the toxin production in some algal species, including Alexandrium spp. and Prymnesium spp. (Weissbach et al., 2011), and thus may help improve microalgal cultivation (Mendes and Vermelho, 2013).
Algae, particularly marine diatoms, also use the QS or structural analogues to regulate their own behavior. Among the QS analogues, pheromones were regarded as representative, since homoserine, fucoserratene, and ectocarpene were widely found in Gomphonema parvulum, Asterionella formosa, and Skeletonema costatum, respectively. These molecules mediate the sexuality and reproductive behavior of algae (Derenbach and Pesando, 1986; Pohnert and Boland, 1996; Hombeck and Boland, 1998). Other representative signaling molecules are hypothesized to function as infochemicals for green algae and diatoms. Examples of such substances include nitric oxide (NO) and 5,6-membered ring molecules (laurenciones, malyngamides, and honaucins). These signals can recruit cells to mount anti-oxidant or anti-inflammatory responses for protection against stress-induced cellular damage (Vardi et al., 2008; Gerwick et al., 2013). Though self-modulation of homeostasis and physiologyical responses via chemical cues in algae are evident, as discussed above, how the different signaling molecules secreted by algae, and their importance in intercellular interactions remain unknown. Futher efforts are of great urgent.
C. Interkingdom signaling
Interkingdom signaling in marine has only recently become an area of scientific interest. To date, research has primarily focused on bacteria, with limited focus on algae (Amin et al., 2012). Crosstalk between bacteria and algae has only been confirmed over the last decade (Bauer et al., 2005; Steinberg et al., 2011; Amin et al., 2015). At present, three types of interkingdom signaling molecules/mechanisms have been reported. First, lipid-based molecules commonly used in bacteria and algae, with poor solubility in aqueous conditions, which allows them to cross cell membranes freely in an energy-independent manner. Second, a certain structureally conserved molecules with related properties between the 3D structures and the functional domains of their regulators (Vannini et al., 2002). Third, bacterial QS molecules and algal pheromones (particularly in diatoms) with similar structures and functionality, which may facilitate their use as crosstalk signals between species. Hughes and Sperandio (2008) reviewed the third form of interkingdom signaling in terrestrial environments, and demonstrated both beneficial and detrimental interactions between eukaryotes and prokaryotes. It speculates that this form of signaling may have similar roles in aquatic environment (Decho et al., 2011).
1. Eukaryotic response to bacteria
Algae have developed the ability to sense bacterial QS signals, and have evolved multiple mechanisms for interpreting these QS signals to initiate physiological responses (Teplitski et al., 2004; Bauer et al., 2005; Joint et al., 2002, 2007). Amin et al. (2012) reported that two different bacterial species known to associate with diatoms could secrete different AHLs that enter and accumulate within host-body. It was proposed that once within the cells, these AHLs bind to their molecular targets, lead to different responses depending on whether the bacteria releasing the AHLs are symbiotic or algicidal. In addition to these molecule-dependent signaling regulatory mechanisms, algae also produce AHL mimics that can mislead or “confuse” bacteria (Hughes and Sperandio, 2008). QS signal exchange is required for co-existing hosts and bacteria to interact. The crosstalk of QS between bacteria and algae suggests that interkingdom sensing is widespread in marine environment.
2. Bacterial response to eukaryotes
Accumulating evidence supports the concept that signaling is not limited to intercellular bacterial communication, but is also used by microbes and their hosts. In terrestrial environments, the photosynthetic bacterium Rhodopseudomonas palustris uses a plant metabolite (p-coumarate) to produce QS signal molecules (such as p-coumaroyl-homoserine lactone) (Schaefer et al., 2008). This work further suggested that pC-HSL operates as both an intraspecies bacterial signal and as an interkingdom signal to host plants (Schaefer et al., 2008). AHLs that influence the behavior of various algae-associated bacteria have also been detected in marine ecosystem. For example, in the red alga Gracilaria chilensis, AHLs produced by the epiphyte Acrochaetium sp. control spore release (Weinberger et al., 2007). Subsequent research showed that 12% of Acrochaetium sp. strains found on the brown alga Coplomenia sinuosa inhibited QS production of the host alga (Kanagasabhapathy et al., 2009). These algal epibiotic bacteria may play an important role in the defensive mechanisms of their host by producing QSI or QSI-like compounds to suppress the settlement of other competitive bacteria. The foregoing examples suggest that bacteria and eukaryotes are able to sense one another’s signaling compounds under certain conditions. Identifying these substances and their molecular targets could improve our understanding of QS-based cross-talking in algae-bacteria symbionts (Zhou et al., 2014).
IV. ECOLOGICAL FUNCTIONS OF QS DURING ALGAL-BACTERIAL INTERACTIONS
Interactions between algae and bacteria do not occur in isolation. Specific organisms may be affected at certain times, based on the ratio between different ongoing stimulatory and inhibitory processes (Cole, 1982). Furthermore, the turbulence of marine environment makes life in the phycosphere substantially different to life in the terrestrial soil rhizosphere. Robust and efficient mechanisms are thus required to mediate the complex ecological relationship between algae and bacteria. As our understanding of QS signals improves, researchers are increasingly recognizing their role as intelligent cues. In the next section, we will discuss how QS affects certain algae and/or bacteria physiological features, such as nutrient acquisition, biofilm formation, ecological niche construction, and self-motility.
A. Nutrient acquisition
1. Carbon resources
Phytoplankton can provide a variety of carbon resources to heterotrophic bacteria, especially the dissolved organic carbon (DOC). There is a close link between DOC features (biomass and types) and bacterial diversity in the phycosphere. Amin et al. (2012) reported that different DOC compounds produced by diatom could regulate the microbial communities of associated symbionts. Among the DOCs, a typical example is glycolate (a type of water-soluble, low molecular weight compound). To some extent, glycolate can shape the structure of bacterial communities through the glycolate-producers in response to diatomic signaling molecules (Haynes et al., 2007). Bacteria may sense these signals, enhancing their ability to assimilate glycolate. In symbiotic environments, only bacteria that have the glycolate utilization gene glcD benefit from associations with glycolate-releasing phytoplankton (Lau and Armbrust, 2006). These bacteria use QS molecules to regulate their density and improve their ability to assimilate glycolate before it diffuses away or is consumed by competing glcD-containing bacteria (Leboulanger et al., 1997). For instance, during spring phytoplankton blooms, glcD transcripts increase when glycolate production peaks over the diel cycle (Lau et al., 2007). As a result, bacteria may respond to QS signals by modulating organic carbon metabolism based on glycolate availability.
2. Nitrogen resources
Nitrogen-fixing bacteria are critical in the nitrogen cycle because they convert dinitrogen gas into more available forms, such as ammoniacal-nitrogen. In leguminous plant, QS intercedes in the signal exchange process and may play a role in coordinating the nitrogen-fixing rhizobia during the establishment of the symbiosis (González and Marketon, 2003). In aquatic plants, some green algae such as Caulerpa taxifolia and Codium fragile capitalize on the nitrogen fixation capabilities of associated bacteria to support their invasion in oligotrophic environments (Chisholm et al., 1996). Wyss (2013) recently reported that bacteria use a quorum-sensing-like mechanism to sense algal culture density in Chlorella vulgaris, and repress nitrogen fixation gene expression at high algal culture density to deprive the algae of bioavailable nitrogen. Although the available examples are still limited, the knowledge obtained so far provides a foundation for understanding the role of QS in regulation of genes involved in nitrogen metabolism during host-bacterium interactions.
3. Sulfate resources
In addition to macronutrients, important microelements are also present in the phycosphere, such as dimethylsulfonio-propionate (DMSP), which is a major source of organic sulfur produced by dinoflagellates (Moran et al., 2003). DMSP is the favored source of reduced sulfur for bacteria from the Roseobacter clade, even though it is present at 107-fold lower concentrations than sulfate in seawater (Raina et al., 2009). Flagella have been shown to enable chemotaxis towards DMSP in some Roseobacter members, which prompted Belas et al. (2009) to suggest a link between this observed phenotype with QS substances. Geng and Belas (2010) further pointed out that DMSP exchange between Roseobacter and dinoflagellates involves a vir-gene-mediated Type 4 secretion system (T4SS) and/or QS by certain chemicals.
Another important micronutrient is tropoditheietic acid (TDA). TDA is a dual-sulfultropolone produced by oxidative ring expansion of phenylacetic acid via the shikimate-chorismate pathway (Geng et al., 2008; Geng and Belas, 2011). TDA prevents algicidal bacteria from damaging phytoplankton, and boosts symbiosis with Roseobacter by reducing competition with other species. In dinoflagellates, the associated bacterium Silicibacter sp. TM1040 produces TDA in a population density-dependent manner. Although TM1040 does not produce canonical AHL signals, molecular analysis revealed that other chemicals may replace AHLs in TM1040 (Geng and Belas, 2010, 2011), thus providing the link between QS and the observed phenotype (i.e. signal molecules participate in the regulation of TDA synthesis).
4. Iron resources
Marine microbial communities and structures are often regulated by the availability of iron, which is necessary for photosynthesis and respiration (Coale et al., 1996). Dissolved iron is present at an extremely low concentrations in seawater (10−10–10−9 moles kg−1) (Geider, 1999), and in some open ocean regions these low concentrations can limit primary productivity and bacterial growth (Tortell et al., 1999). In the last decade, iron acquisition has been a major focus of studies on algal-bacterial interactions because its scarcity in the marine environment has driven the evolution of numerous nutrient acquisition strategies in bacteria (Boyd and Ellwood, 2010). One strategy used by marine bacteria is the production and secretion of siderophores, which bind to iron, and thus increases its solubility (Vraspir and Butler, 2009). Bacteria then reacquire soluble Fe(III)-siderophore complexes by using specific outer-membrane transporters.
Unlike bacteria, eukaryotic phytoplankton that produce siderophores, or that directly take up bacterially derived Fe(III)–siderophore complexes, have not yet to be identified up to now. However, phytoplankton may obtain iron by using QS signaling to regulate the siderophore biosynthesis in siderophore-producing bacteria, and quickly mobilize iron assimilation mechanisms (Amin, 2010). There is also genomic evidence that some phytoplankton obtain iron from siderophores or other chelates using ferrireductase and cell-surface Fe (II)-transporters (Kustka et al., 2007). This dynamic process modulated by QS indicates that the regulation of QS signaling is ecologically driven. In addition, AHLs from QS may also be involved in iron uptake in algal populations. Different algal species use different forms of iron, often complexed with siderophores. In contrast, diatoms use iron complexed with porphyrins (Hutchins et al., 1999).
B. Biofilm formation and ecological niche construction
Algae are coated with specific communities of bacteria, fungi, protozoa, and other eukaryotes. In these epiphytic communities, “biological films” (or biofilms) are among the most common. The primary bacterial phyla participating in algal biofilm formation include proteobacteria (alpha-, beta-, and gamma-type), nitrospira, actinobacteria, acidobacteria, and firmicutes (Souza-Eqipsy et al., 2008). These “microepibionts” are multi-functional, and are involved in obtaining nutrients, acquiring new genetic traits, and providing some measure of chemical defense against pathogens (Wahl et al., 2012).
Biofilms are chemically mediated. The communication, construction, and breakdown of many algal biofilms are regulated by the symbiotic microbial QS system, which helps to control film formation, development, maturation, and dispersal (Parsek and Greenberg, 2005). Algae are phylogenetically and morphologically diverse organisms that often host multiple species in the stereoscopic biofilms on their surfaces. Diverse bacteria live within the biofilm; however, these bacteria are not randomly distributed among different algal species. Each algal species hosts and supports bacterial communities with different species compositions, and the same algal species tends to host the same bacterial community, even in different environments (Lachnit et al., 2009; Trias et al., 2012). This phenomenon may occur because the host species may have markedly different spatial environments, physiological states, and chemical factors (including QS metabolites), which determine the composition of the associated bacterial community. AHLs and their inhibitors influence the behavior of various biofilm-associated bacteria, confirming that marine epibiotic communities produce and use QS in signaling (Maximilien et al., 1998). Thus, it is important to determine the functional consequences and gene expression patterns associated with QS in biofilm communities, which will improve our understanding about the structure and function of “ecological biofilms”.
Biofilm also promote the construction of algal ecological niches. Newly generated bacteria or algae must locate suitable sites for colonization, which is a critical stage in the life cycle. Settlement takes place in three steps: contact, temporary adhesion, and irreversible adhesion (Fletcher and Callow, 1992). It is well-known that biofilms provide niches for new colonizers, and enables bacteria to coordinate and respond quickly to environmental changes. Over time, biofilm structure matures to reflect the cooperative division of labor in which multiple cells undergo specialization leading to complementary and synergistic behaviors (Chen, 2013). Meanwhile, mature biofilms modulate bacterial settlement by regulating the flow of energy and matter across the host surface, thus altering its chemical properties (Davey and O’toole, 2000). Ecologically, the concept of modulating bacterial settlement to occupy an appropriate niche represents a fascinating evolutionary strategy, as it provides a competitive advantage in addition to regulating the associated microbial community (Hentschel et al., 2001). Biofilms also regulate algal larval colonization by modifying the surfaces of potential settlement sites, as well as through the molecular signaling. In recent years, some studies have demonstrated that biofilms provide ideal sites for the metamorphosis of coralline algae (Tebben et al., 2011), release of diatoms (Zargiel and Swain, 2014), and swarming of dinoflagellates (Alagely et al., 2011).
The selection of colonization sites and associated behaviors are QS mediated (Joint et al., 2007). In some marine species, signal substances affect multiple life cycle functions, such as contributing towards the initiation of Hydroides elegans larval colonization with C6-, C12-, and 3-oxo-C8-HLS (Huang et al., 2007), promoting spores release with C4-HLS (Weinberger et al., 2007), and providing protection to algal surfaces with polybrominated 2-heptanone (Nylund et al., 2008). Certain examples of QS-mediated behaviors in microalgae are also implied. In the planktonic life-phase of two bacteria, Pseudomonas spp. and Rhizobium spp. (isolated from Botryococcus braunii), short chain AHLs (C4 or C6) were detected during biofilm formation (Rivas et al., 2010). Seven long-chain AHL producers (Psychrobacter cryohalolentis, Providencia sneebia, Pseudomonas stutzeri, Exiguobacterium sp. AT1b, Klebsiella oxytoca, Lysinibacillus sphaericus and Acinetobacter baumannii) isolated in our lab could promoter or inhibit Scrippsiella trochoidea growth (Lv et al., 2016).
In some cases, QS molecules could prevent certain bacteria from occupying certain niches. Algae reduce the harmful effects of bacterial overpopulation by interfering with bacterial QS systems controlling bacterial colonization through regulating biofilm formation (Steinberg et al., 1997; Dworjanyn et al., 1999). Some algae are able to stimulate, inhibit, and/or inactivate QS signals in bacteria by producing QS inhibitors or their analogues. The best-investigated example is that of an Australian red alga, Delisea pulchra, which produces metabolites known as halogenated furanones. These molecules interfere with AHLs through competitive inhibition at the LuxR-type receptor site, and selectively inhibits bacterial colonization and biofilm formation on algal surfaces (Mclean et al., 2004). Other bacteria and eukaryotes produce dipeptides that act as AHL mimics. For example, AHL mimics affect QS-regulated behaviors (like biofilm formation) in the genus Vibrio (Dickschat, 2010). These examples suggest that QS and QS-related molecules may help balance the niche of symbionts and the stability of their habitats.
C. Self-motility
Because bacteria live in diverse areas, they often need to migrate to favorable environments to acquire nutrients and information, and use a variety of strategies to find and exploit advantageous niches. Various molecular processes drive migration, including phototaxis (response to light), thermotaxis (response to temperature), aerotaxis (response to oxygen), and chemotaxis (response to chemical cues) (Gluch et al., 1995; Paerl, 1996). The most common and best studied of these migratory processes is chemotaxis. Chemotaxis allows bacteria to swim toward ideal conditions, thus providing bacteria with a competitive advantage in obtaining nutrition in natural environments (Wadhams and Armitage, 2004). Early work on chemotaxis in marine bacteria also suggested that this ability is a critical component of bacterial-phytoplanktonic interactions. To date, microfluidic visualized methods have been used to demonstrate that marine bacteria use chemotaxis to respond to chemicals released by certain phytoplankton, including Pfiesteria piscicida, Dunaliella tertiolecta, and Thallassiosira weissflogii (Miller and Belas, 2006; Stocker et al., 2008; Seymour et al., 2008, 2009). Marine bacteria may even use chemotaxis to track swimming phytoplankton cells (Barbara and Mitchell, 2003). For instance, Pseudoalteromonas haloplanktis and Shewanella putrefaciens bacteria respond to nutrient or oxygen gradients released by phytoplankton, attaining high swimming speeds and exhibiting quick directional changes to reach them (Stocker and Seymour, 2012). DMSP and amino acids present in dinoflagellate homogenates are essential attractant molecules for the marine bacterium Silicibacter sp. (strain TM1040), and the bacterial motility is an important factor in the symbiosis between dinoflagellates and bacteria (Miller et al., 2004; Miller and Belas, 2006). Genomic analyses further demonstrated that TM1040 contains genes that are necessary to sense and respond to chemical attractants (Moran et al., 2007; Geng and Belas, 2010). More than half of the sequenced genomes of Roseobacter sp., which is another free-living bacterium found in phycospheres, contain homologs to known genes for chemotaxis and chemoreceptors (Slightom and Buchan, 2009). In addition, Thar and Fenchel (2001) observed that free-swimming Thiovulum majus cells exhibit versatile chemotactic behavior, including both a phobic response and true chemotaxis in oxygen gradients.
Bacterial motility and chemotaxis are ecological behaviors that are partly regulated by social signals, such as QS, as well as other chemical signals. In terrestrial environments, the soil bacterium Serratia liquefaciens modulates swarming by using AHLs or their regulators (the gene surR or synthase swrI, which respond to C4- and C6 AHLs) (Lindum et al., 1998). Researchers have also found that certain marine bacteria, including Vibrio alginolyticus, S. putrefaciens, and P. haloplanktis, exhibit “run-reverse-flick” and “run-reverse” swimming strategies to trigger biofilm formation. These movements affect chemotactic drift speed and responses to surrounding signals (Taktikos et al., 2012, 2013). Daniels et al. (2004) suggested that QS mediates the swarming behavior of bacteria, with AHLs, diketopiperazines, and signal mimics causing the flagella-propelled movement of cells that are elongated, multinucleated, and hyper-flagellated.
In addition to modulating bacterial motility, QS also regulates the swimming ability of algal spores. Wheeler et al. (2006) demonstrated that QS-modulated biofilms use AHL-regulated chemokinesis to enhance algal motility, and that spore swimming speed was reduced by up to 27% and 47% within four minutes of adding 25 and 125 μmol L−1 of 3-oxo-C12-HSL, respectively. This effect was further enhanced by AHLs with long side chains (10–12 carbon atoms), which have low solubility in water (Yates et al., 2002). The cellular mechanism involved in this chemokinetic effect involves an influx of Ca2+ in the spores, which alters the flagella-driven pattern of movement in a Ca2+-dependent manner (Joint et al., 2007). The resultant decrease in swimming speed may effectively increase the chances of locating suitable sites for settlement by spores.
D. Others (virulence and reproduction)
Signaling molecules also guide other physiological processes in algal-bacterial symbiosis, including virulence factor production and reproductive behavior. For instance, dinoflagellate-associated Vibrio spp. sense and respond to small signaling molecules produced by host organisms. This interspecies communication influences pathogenic virulence, bacterial development and host infectious processes (Tsim et al., 1996). One example of this was provided by Natrah et al. (2011), who showed that microalgae Chlorella saccharophila and Chlamydomonas reinhardtii interfere with bacterial QS signals and reduce the virulence of the pathogen V. harveyi.
Reproductive parameters, such as diatom gametogenesis and cell division, are regulated by environmental chemical signals, with a similar effect to terrestrial pheromones. For instance, QS induces the switch from asexual to sexual reproduction in the centric diatom Thalassiosira weissflogii (Falciatore and Bowler, 2002). The timely formation of gametes is critical for the efficient operation of the phytoplanktonic sexual cycle. Using the Kolmogorov model, Peperzak (2006) speculated that QS regulates this process, whereby cells sense the number of conspecifics through the quantity of secreted secondary metabolites, and adjust the induction of gamete formation accordingly. Kouzuma and Watanabe (2015) suggested that bacteria secrete chemical signals (such as QS) to induce the morphogenesis and germination of algae. Hence, QS may regulate the switch to sexual reproduction by phytoplankton at high population densities, along with the synchronization of sexual activities, such as encystment.
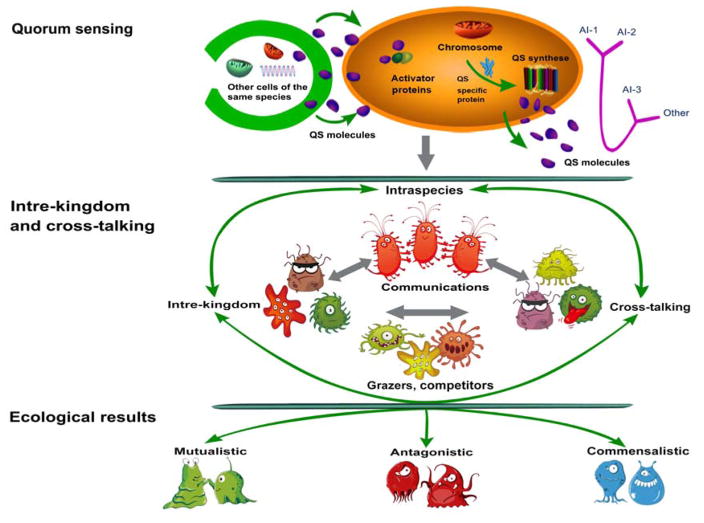
Therefore, QS signals are multi-functional modulators that regulate a variety of phenotypes in symbiotic relationships between phytoplankton and bacteria, primarily through three main cascading processes: i) QS molecule producion and transport in the bacterium cell; ii) microbial cross-talking (intraspecies and interspecies) by QS molecules; and iii) QS-mediated social behavior in algal-bacterial symbionts, including mutualistic, antagonistic, and commensalistic interactions (Fig. 3).
Figure 3.
Formation and multifunctional roles of QS in algal-bacterial symbiosis. Top: quorum sensing (QS) molecules are produced in the bacterium by QS synthesis and diffuse from the cell to enter neighboring bacteria. QS signaling molecules bind to the receptor polypeptide, leading to formation of active dimers. The receptor dimers bind to specific promoter sequences in the bacterial genome and activate the transcription of certain sets of genes. Middle: microbial communication by QS molecules, including intre-kingdom and intra-species communication, interspecies cross-talking, and other communication among different species. Bottom: QS-mediated social cooperation and conflict in algal-bacterial symbionts, such as mutualistic, antagonistic, and commensalistic symbionts.
V. ANTAGONISTIC INTERACTIONS
In some of the more complex and dynamic symbiotic relationships, algal holobionts occur in close proximity to various species, including fungi, bacteria, and protozoa. These relationships may be both beneficial and deleterious. Thus, competition is widespread among bacteria, as well as between algae and bacteria. Antagonistic interactions between different species may be equally important for the initiation and establishment of symbiotic relationships (Filzgerald, 1969; Duval and Margulis, 1995).
A. QS modulates antagonism between bacteria
Dynamic equilibrium exists among various species in bacterial community structures. To maintain the structural stability of a population and ensure population success, bacteria use antagonistic strategies, including the secretion of secondary metabolites, which may be harmful or lethal to the target cells (Hibbing et al., 2010). However, these metabolites may not be released at the concentrations necessary to produce toxic effects. Furthermore, the microbes secreting these molecules must deliver a toxic dose sufficiently high to be effective, while also minimizing subsequent self-exposure to potentially damaging toxin levels. QS may help realize this strategy, whereby antimicrobial release only occurs once a threshold number of antimicrobial-producing cells are present. Hence, it is not surprising that QS is used to modulate antimicrobial production and functioning in a large number of organisms (Hibbing et al., 2010).
Many competitive elements are also regulated by QS. For example, QS inhibitors are produced by some bacterial species to avoid succumbing to competition, thus allowing them to compete in the symbiotic environment without needing to produce antibiotics to any toxic metabolites that may be present. Recently, several QS inhibitors of this kind have been found. Honaucins, a structurally analogue of AHLs from a marine cyanobacterium, can inhibit bioluminescence in V. harveyi (Choi et al., 2012). Tumonoic acid produced by V. harveyi in non-toxic concentrations can modestly inhibit QS without affecting host growth (Clark et al., 2008). In addition, two QS inhibitors in the form of phenethylamide metabolites were identified in Halobacillus salinus. These two molecules prevented QS-regulated violacein biosynthesis by Chromobacterium violaceum at non-toxic concentrations by competing with AHLs for receptor binding (Teasdale et al., 2009). Among the QS inhibitors, PQ (2-n-pentyl-4-quinolinol) is impressive in that it can modify bacterial metabolism, shift the composition of bacterial populations, and further change their interactions in marine environment (Long et al., 2003). The effects of antimicrobials on interbacterial interactions may significantly modify the structure of the food web and biogeochemical dynamics, as bacteria use these enzymatic substances to interrupt QS signaling by other species that may compete for space and food. Furthermore, a link between enzymatic degradation and the ability to gain a competitive advantage has been demonstrated. Evidents showed that degradation enzymes such as lactonases, acylases and oxidoreductases affect the signaling processes of QS or degrade AHL molecules (Dong et al., 2001; Park et al., 2006), as bacterial strains that enzymatically inactivate AHLs and thus inhibit QS process were observed (Romero et al., 2011).
B. QS modulates antagonism between bacteria and algae
Competition between algae and bacteria is common because of food and space limitations. Bacteria use several strategies to obtain resources, including secreting toxins and producing compounds harmful to algae or other organisms that affect the algal lifecycle. Among these ecological strategies, the role of algicides has received much scientific focus, due to potential repurposing of these elements as biocontrol agents against harmful algal blooms (HABs) (Mayali and Azam, 2004; Kodama et al., 2006).
1. Bacterial effects on algae
Algicidal bacteria secrete chemical substances that inhibit or kill algae in phycospheres, and the biosynthesis and release of these algicides is regulated by AHLs. That is, QS is used by bacteria to control algicidal activity. For example, the flavobacterium Kordia algicidais releases a protease that acts against a subset of its symbiotic hosts, including Skeletonema, Thalassiosira, and Phaeodactylum spp. (Paul and Pohnert, 2011). This protease is only secreted when the bacterial population level reaches a certain threshold controlled by QS (Paul and Pohnert, 2011). Another clue is exemplified by the discovery of nine algicidal bacteria isolated from microalgae, which could inhibit several red-tide algae such as Prorocentrum donghaiense, P. globosa, Thalassiosira sp., and Heterosigma akashiwo, to different degress (Xu et al., 2012). Interestingly, these nine strains produce C6–C14 HSL, with their physiological behavior being controlled by HSL molecules (Xu et al., 2012). Furthermore, the inducer role of QS was also pointed out for bacteria producing and secreting algicide molecules (Demuez et al., 2015). Recently, data mining of the genome of a Rhodobacteraceae strain isolated from the microalga Prorocentrum donghaiense also provided hint of QS-regulated algicidal activity in bactera (Zheng et al., 2015).
Though many phenomenons of QS-regulated algicidal activity in bacteria are clearly observed, limited information on how the algal cell lysis process takes play hinders insights into the molecular mechanisms of QS-regulated algicidal activity. Up to now, limited studies arround AI-1 and AI-2 system have been preliminarily carried out. Two novel G+ bacterial strains (Zobellia sp. and Planomicrobium sp.) were shown to exhibit algicidal activity against the toxic dinoflagellate Gymnodinium catenatum, and the algicidal functions was modulated by AI-2 system (Skerratt et al., 2002). Nakashima et al. (2006) subsequently showed that PG-L-1 (a prodigiosin pigment produced by gamma-proteobacterium), exhibits algicidal activity against various red tide phytoplankton under AHL singals control. The authors suggested that AHL signals might also regulate the production of other algicidal molecules by marine bacteria. As a result, these bacteria may modulate blooms of harmful algae through a QS system (Nakashima et al., 2006). Recently, some authors investigated the potential role of AI-1/AI-2 systems in cultures of a Florida dinoflagellate Karenia sp. and associated algicidal bacteria; they concluded that algae-associated bacteria, including those that are algicidal, depend on autoinducer systems (Blair and Marshall, 2013). These mentioned above examples tell us, use of QS systems for this purpose would introduce a new paradigm for understanding algal-bacterial relationships and the biotic regulation of bloom dynamics.
2. Algal defense against bacteria
Many algae use defense strategies to protect themselves from antagonistic bacteria. These algae secrete metabolites that act as antibacterial substances and influence bacterial growth or biomass. Such metabolites include polyunsaturated fatty acid that inhibits algicidal bacteria (Lebeau and Robert, 2003), eicosapentaenoic acids that inhibit pathogens (Desbois et al., 2008), and polyunsaturated aldehydes that suppress unfavorable bacterial growth (Wichard et al., 2007). From a signaling regulation perspective, phytoplankton have an additional defense strategy in their arsenal. Like terrestrial plants, algae may be able to disrupt harmful bacterial behavior by interfering with QS signals. For instance, algae produce halogenated furanones, which are structural analogues of AHLs. These products protect algal surfaces by interfering with AHL-regulated processes, in addition to selectively inhibiting bacterial colonization and biofilm formation (Manefield et al., 2002). Another well-characterized natural compound is 5-4-5-bromomethylene-3-butyl-2-5H-furanone, which effectively inhibited AHL-regulated gene expression in several G− bacteria (Defoirdt et al., 2007). This compound was shown to inhibit AI-2 signaling by inactivating the LuxS enzyme, and interrupting the production of AI-2 through covalent modification (De Keersmaecker et al., 2006). Kjelleberg et al. (1996) further reported that eukaryotes produce cyclic dipeptides that act as AHL mimics, and affect QS-regulated behavior in symbiotic microbes. Other secondary metabolites (such as manoalide, brominated alkaloids, and kojil acid) are also antagonistic to QS through inhibiting QS production, degrading the LuxR activator, and blocking the LuxR-based reporter (Skindersoe et al., 2008; Dobretsov et al., 2011). Syrpas et al. (2014) demonstrated that the haloperoxidase mediated loss of β-keto-AHL activity in the benthic diatom (Nitzschia cf. pellucida) is caused by the final cleavage of the halogenated N-acyl chain of the signal molecules. Other algae, such as Asparagopsis taxiformis, produce QS inhibitors as a safeguard against biofilm formation (Jha et al., 2013). In these cases, algal hosts secrete compounds that mimic bacterial QS signals, allowing the hosts to manipulate (blocking or disrupting) bacterial QS-regulated gene expression. Rajamani et al. (2011) also suggested that the secretion of lumichrome by C. reinhardtii might serve as either a QS signal or interkingdom signal mimic capable of manipulating QS in bacteria possessing a LasR-like receptor.
Enzymes may also be used to modify QS signals to defend against unfavorable or harmful bacteria. A typical example is haloperoxidases, which regulate AHLs by modifying their acyl side chains, and prevent binding between QS factors and their related regulators. Borchardt et al. (2001) confirmed that one haloperoxidase family member, Vanadium haloperoxidases, interferes with QS by brominating AHLs. Some haloperoxidases deactivate AHLs on contact with the surface of bacteria (Butler and Sandy, 2009). Therefore, the study and characterization haloperoxidases is essential to understanding diatom defense systems against antagonistic bacteria (Amin et al., 2012).
The aforementioned examples show that algae and bacteria clearly use QS to regulate their own behavior, in addition to the behavior of other organisms. Prokaryotes and eukaryotes diffuse their signals and metabolites into the environment, which are then recognized as chemical cues by surrounding organisms to establish ecological niches, following the principle of “ecological regulation serves ecological function.”
3. Defense aganist eukaryotic grazers
Protozoans often graze on phytoplankton, which reduces phytoplankton biomass and the diversity of bacterioplankton communities. Marine bacteria have developed several mechanisms to protect themselves and their hosts against this type of predation (Jousset, 2012). Over the last 20 years, studies have shown that cyanobacteria from the genera Lyngbya and Microcoleus produce a wide range of metabolites (including the lipopeptide polyamide, malyngamides, and majusculamides) to prevent protozoa from consuming their mats (Nagle et al., 1996; Pennings et al., 1996; Capper et al., 2006; Berry et al., 2008). In addition, some species of marine heterotrophic bacteria exhibit anti-grazing-mediated behavior (Wietz et al., 2013). For instance, Pseudoalteromonas luteoviolacea produces an anti-grazing compound called purple pigment violaceins. At very low concentrations (nanomolar level), this compound triggers the autolysis of bacterivorous dinoflagellates, and may provide defense within biofilms (Matz et al., 2008). Vibrio cholera produces an extracellular protease that provides resistance against the flagellate Cafeteria roenbergensis and the ciliate Tetrahymena pyriformis (Vaitkevicius et al., 2008). In this form of self-defense, chemical signals serve as modulators and guides. V. cholerae also produces a QS-regulated antiprotozoal factor that prevents the flagellate Rhynchomonas nasuta from growing, which reduces grazing losses (Erken et al., 2011). Sun et al. (2013) further showed that polysaccharide production induced by the QS regulator HapR acts as inhibitor to suppress R. nasuta growth.
In addition to self-defense, symbiotic microbes protect their hosts through associated metabolites. The microalgae-associated bacterium Theonella swinboei and those of Paederus spp. protect their hosts by producing polyketides (Kellner and Dettner, 1996). Pseudomonas spp. use 2,4-diacetylphloroglucinol and pyoluteorin as antibiotics to prevent host infection by the pathogens Pseudoalteromonas elyakovii and Algicola bacteriolytica (Nagel et al., 2012). Marinobacter sp. uses a different host protection strategy, which involves secreting lipopolysaccharides to trigger early algal defense reactions by inducing oxidative bursts (Kupper et al., 2006). While it is well known that anti-molecules must reach a certain concentration to be effective, it remains unclear whether QS contributes to these processes. QS regulates the population density of bacteria based on the concentration of biological activators that they secrete. Thus, it is important to obtain direct proof of QS involvement in host-protection mechanisms to advance this area of research.
In summary, compounds that obstruct QS may have a positive or a negative effect on bacterial responses modulated by QS. Eukaryotic hosts or phytoplankton produce these QS-inhibiting compounds are listed in Table 2.
TABLE 2.
Natural QS inhibitors observed in algae and major symbiotic bacteria
| Species | Compound | Mode of action | Biological role | QS system affected | References |
|---|---|---|---|---|---|
| Red alga, Ahnfeltiopssis flabelliformis | α-D-galactopyranosyl-glycerol (Floridoside), betonicine, and isoethionic acid | Competes with AHL signals | Kim et al. (2007a, b) | ||
| Red alga, Galaxa uraceae, Laurencia sp. | Unidentified algal extract | AHL inhibitor | Skindersoe et al. (2008) | ||
| Green alga, Chlamydomonas reinhardtii | Unidentified AHL mimics, Lumichrome | Blocks AHL molecules | Rajamani et al. (2008); Teplitski et al. (2004) | ||
| Brown alga, Laminaria digitata | Oxidized halogen HOBr, Hypobromous acid | Deactivates AHL by interfering with QS genes | Borchardt et al. (2001) | ||
| Macroalga, Delisea pulchra | Furanone | MimicsAHLsignals, inhibits gene expression | Affects biofilm formation, swarming motility, toxin production, chemotaxis | Swr system of S. liquefaciens and other gram negative bacteria | Givskov et al. (1996); Gram et al. (1996); Manefield et al. (2000) |
| Macroalga, Chlorophyta, Caulerpa sp. | Unidentified extract | AHL inhibitor | Skindersoe et al. (2008) | ||
| Marine algal | Honaucin, coibacin, laurencione, tumononic acid, and malyngamide | QS inhibitor | Vibrio harveyi | Gerwick et al. (2013) | |
| Microalga, Chlamydomonas reinhardtii | Lumichrome | Mimics AHL signals | Rajamani et al.(2008) | ||
| Bacteria | |||||
| Penicillium spp. | Penicillic acid, patulin | Las and RhI system of P. aeruginosa | Rasmussen et al. (2005) | ||
| Bacillus spp., Agrobacterium tumefaciens, Arthrobacter sp. | Lactonase | Degradation of AHL signals | Romero et al.(2011) | ||
| Bacillus spp., Alteromanas sp., Tenacibaculum discolor strain; Nocardioides sp. Streptomycetes sp. | Acylase | Degradation of C4-HSL, 3-O-C12-HSL, and long chain AHLs | Kang et al. (2004); Nithya and Pandian (2010); Romero et al. (2011) | ||
| Bacillus megaterium | AHL-oxidase | Degradation of C4 HSL and 3-O-C12 HSL | Choudhary et al. (2007) | ||
| Burkholderia strain GC4 | AHL-oxidoreductase | Degradation of 3-O-C6 HSL | Chan et al. (2011) | ||
| Streptomycetes spp. | Lactones | Mimics AHL signals | Cho et al. (2001) | ||
| Xanthomonas campestris | Cys-11-methyl-2-dodecenoic acid | Mimicsfarnesoic acid signals of C. albicans | Zhang and Dong (2004) | ||
| Staphylococcus xylosus | RNA III inhibiting peptide | Competes with QS signals | Gov et al. (2004) | ||
| Halobacillus salinus | Phenethylamide | Antagonist of AHLs | Teasdale et al. (2009) | ||
| Cyanobacteria, Blennothrix cantharidosmum | Tumonoic acids | Competes with QS signals | Clark et al. (2008) | ||
VI. ENVIRONMENTAL FACTORS AFFECTING QUORUM BEHAVIOR
To understand quorum behavior, it is important to fully characterize microbial phenotypic plasticity in response to environmental factors. Natural environments are often heterogeneous, and feature strong spatial variation in both abiotic and biotic factors. For holobiont systems, all of the requirements for an organism (such as temperature, pH, salinity, light, and nutrients) are rarely optimal. Therefore, species have evolved to withstand intermittent, non-optimal conditions. In phycospheres (particularly in coastal environmnts), microorganisms are subject to various environmental threats, including ocean acidification, nutrient unavailability, temperature changes, water pollution, and eutrophication. These environmental threats damage holobiont health by decreasing the biodiversity of algae and surrounding aquatic microorganisms. Furthermore, these threats interfere with the symbiotic interactions between bacteria and algae, which impede cellular communication. These issues raise questions of how symbionts adapt to complex systems and how they modify their behavior in response to environmental changes.
To understand how environmental threats influence the symbiotic relationship between microbes and their hosts, a comprehensive characterization of how QS is used in symbiotic relationships is needed (Generous, 2014). In some extremophile ecosystems, there is considerable evidence about the function of QS. Averhoff and Muller (2010) reported that Halobacillus halophilus (a G+ bacterium) from a coastal salt marsh in Germany tolerates high-salt environments by forming biofilms and producing an extracellular polymeric substance (EPS), both of which are regulated by AHLs. Wenbin et al. (2011) confirmed that the A. ferrooxidans (5Z)-4-bromo-5-(bromomethylene)-2(5H)-furanone acts as a QS blocker of Cu2+ in other turbulent environments (such as polluted and high-pressure conditions). In addition, the deep-sea bacterium Photobacterium profundum SS9 contains a putative AI-2 signaling system; the comparative genomic studies have shown that this signal has approximately 35% sequence conservation with the LuxMN and AinSR systems in V. harveyi and Aliivibrio fischeri (Reen et al., 2006). This observation supports the concept that QS is critical in high-pressure environments. Other studies have shown that bacteria trigger QS and downstream reactions to adapt to various extreme environments, including thermophilic (Nichols et al., 2009), psychrophilic (Riley et al., 2008), and acidified (Montgomery et al., 2013) conditions.
Although interactions between algae and bacteria are well studied, knowledge about QS systems in extremophiles remains incomplete, the development of genomic sequencing techniques, bioinformatics analysis, and plasmid-based biosensors have been prompting the elucidation of the functional plasticity of QS in response to environmental cues.
VII. ROLE OF QS IN ALGAL AND BACTERIAL CO-EVOLUTION
Co-evolution is a form of evolutionary change that involves give-and-take between interacting species. Co-evolution dynamically shapes the intricate symbioses as well as the organization of interactions among free-living taxa, which ultimately affects populations, communities, and ecosystems (Palkovacs and Hendry, 2010; Thompson, 2012). The earliest evidence for ancient interactions between algae and microbes derives from the fossil record, confirming that phytoplankton and phytobacteria closely coexisted (Cernichiari et al., 1969). The first suggested incidence of co-evolution occurred between the red alga Prionitis and its gall-forming Roseobacter symbionts, based on comparison of the molecular phylogenies of the symbiont and its host (Ashen and Goff, 2000). Subsequently, genes from marine sponges and their associated bacteria, including a mitochondrial cytochrome oxidase subunit 1 gene, co1, and its bacterial homolog were identified (Dunn et al., 2002). Based on this gene, a phylogenetic tree was constructed for six putatively alpha-proteobacterial symbionts, which were found to correspond to a tree generated using sequences from associated host sponges. This finding implied that these two groups of organisms co-evolved (Dunn et al., 2002). Recent research has supplied further convincing evidence for co-evolution. Qiu et al. (2013) pointed out that red algae are one of the major players in eukaryotic genomic evolution because of their ability to act as “sinks” and “sources” for foreign genes through horizontal or vertical gene transfer and endosymbiosis, respectively. Hollants et al. (2013) provided evidence for non-random associations between Bryopsis and its flavobacteriaceae endosymbionts. Specifically, host species that are more closely related tend to contain genetically similar endosymbionts. Schaum and Collins (2014) further suggested that plasticity could be used to predict the magnitude of evolutionary responses by phytoplankton populations under global change.
Throughout the long evolutionary process, algal-bacterial symbionts lacked a central nervous system to aid in decision-making. Instead, these two groups rely on genetic regulatory networks to adapt and adjust their phenotypic states in response to the environment and selective pressures (Harrington and Sanchez, 2014). Among the tools available to these symbionts, QS might serve as a decision-making mechanism, because it provides an effective and complex strategy for regulating microbial social behaviors. In terrestrial soil-dwelling plants (such as opine), QS has been shown to indirectly impact the evolution between the host and their associated bacterium (Oger and Farrand, 2001). In this pairing, signaling affects certain clustered genotypes, such as the comQXPA locus, which has a large number of genetic polymorphisms and encodes the QS-transduction system that controls QS in Bacillus spp. (Tran et al., 2000; Stefanic et al., 2012).
In marine environment, co-evolution between algae and bacteria is regulated in part by “public goods” or “cheaters” (i.e., organisms that use but do not contribute resources), which arise from ecological and social interactions. These interactions generate selective pressure based on the frequency of certain traits within a population. These pressures help to maintain genes at a moderate frequency within a population or reduce the frequency of certain genes. For example, despite the benefits of siderophore production, only 40% of the population produces these molecules (Cordero and Polz, 2014). Over time, the siderophore operon has undergone genetic recombination from the population gene pool between the “cheaters” and the “producers” (Sandy and Butler, 2009; Amin, 2010). This phenomenon enhanced the co-evolution of these groups or the co-evolution of algae and their associated bacteria. This process is modulated by QS, because QS affects vibrioferrin production (Iqbal et al., 2012). Taylor et al. (2007) and Coelho et al. (2013) suggested that antagonistic co-evolution has occurred between algae and bacteria involved in QS-regulated biofilm formation, although the evidence for this phenomenon remains inconclusive.
In addition to co-evolution, algae and bacteria evolve independently, and this process is influenced by various eco-environmental factors, such as competition (Hibbing et al., 2010), cooperation (Zhang et al., 2009), genomic diversity (Cordero and Polz, 2014), and ocean acidification (Collins et al., 2014). Furthermore, the evolution of the QS system itselfis is influenced by social conflict (Eldar, 2011), different cues (Diggle et al., 2007), and dynamic environments. Therefore, it is necessary to investigate the combined evolutionary processes of algae, bacteria, and QS signals, because QS allows bacteria to adapt and respond to their social and physical environments (Cornforth et al., 2014). Eco-evolutionary feedback loops and cascades of genes, behaviors, communication, populations, and ecosystems should also be incorporated into studies on these phenomena.
VIII. POTENTIAL APPLICATION OF QS IN ALGAL ECOLOGICAL ISSUES
QS and QS-related mechanisms are of particular interest to the scientific community because of their possible biotechnological applications. At present, hundreds of patents involving QS and QS inhibitors exist globally (Jiang and Li, 2013). These applications are separated into three categories: (i) medical uses of AHLs and AHL analogs, (ii) agricultural uses of QS and QS-blockers, and (iii) QS compounds used to scale-up the production of microbial products. In marine ecology field, several diverse applications have been considered, including anti-biofouling (Dobrestov et al., 2009) and algal culturing (Mendes and Vermelho, 2013). According to the scope of this review, we focus on the ecological issue of harmful algal blooms (HABs), and discuss the potential use of QS to predict and/or control HABs.
Over the last 20 years, HABs caused by cyanobacteria and planktonic protists have increased in frequency worldwide, with considerable threats to human health and aquatic-based economies (Anderson et al., 2012). The severe socioeconomic impacts of HABs have prompted the development of technologies and approaches for their prediction and control. The detrimental effects of HAB species are primarily due to the highly toxic compounds produced and accumulated in the food web (Hallegraeff, 1993). Microoragnisms provide some of the necessary nutrients and resources for the toxin-producing species, during the process of cell growth and division in microbial populations. As is well-known that the density in microbial populations is modulated by QS, making it a target for anti-HABs treatment, developing QS inhibitors that block the biological function of QS may provide a way to control algae concentrations to limit the impacts of their toxins (Zhou et al., 2014). A QS-based method is an attractive alternative to antibiotics because it hinders the colonization of bacteria without removing native flora or increasing the risk of antibiotic resistance. Application of QS inhibitors for anti-HABs has been reported. For example, by using ethyl 2-methyl acetoacetate (EMA) as a signal inhibitor, Hong et al. (2008) successfully disrupted the equilibrium of cellular redox process to inhibit blue-green algae (Microcystis aeruginosa) growth. Polyunsaturated aldehydes (PUAs) acts as chemical signal to initiate cell death by affecting the QS-regulated bacteria symbiotic with diatoms, therefore, is a potential anti-HABs compound in bloom resistance and novel biosensors for predicting HABs (Ribalet et al., 2009; Vardi et al., 2008).
In preliminary experiments carried out by the authors, we screened hundreds of QS-producing bacteria and biofilm-forming bacteria isolated from dinoflagellates, Scrippsiella trochoidea and Gambierdiscus taxicus, using the reporter strains Chromobacterium violaceum CV026 and Agrobacterium tumefaciens A136. Bacterial behavioris known to be partly modulated by long- or short-chain AHL molecules and exhibits significant labor division (Chen, 2013). Some bacterial strains significantly inhibited algae or were algicidal; however, we have yet to determine whether these abilities are quorum-regulated (Wang, 2014; Tan et al., 2015). We are currently conducting genomic analyses and investigating the functional proteins of these strains. This information is expected to help clarify the underlying mechanisms about how HAB formation and development is influenced by bacterial behavior, which is, to some extent, regulated by AHL molecules (Wang and Zhou, 2015; Zhou et al., 2016). Amin et al. (2015) suggested that interactions based on signal substances demonstrate how the bacterial influence on phytoplankton physiology is linked to the global carbon cycle and algal bloom formation. Thus, future studies should focus on obtaining details about the efficacy and molecular mechanisms underlying QS in HABs.
IX. CONCLUSIONS AND FUTURE PERSPECTIVES
Signal-mediated behavior is omnipresent, and has a considerable impact on the structure and function of populations, communities, and ecosystems of symbionts (Zhang and Dong, 2004). Quorum and quorum-related signals are involved in regulating the physiological behavior of bacteria at multiple aspects, and facilitate indirect communication that extends throughout communities and modifies the organization of the food web, community structure, and ecosystem-wide events, such as nutrient and element cycling. In phycosphere environment, dynamic and complex interactions occur between different species of algae, bacteria, archaea, predators, and viruses, all of which co-exist through the mediation of signals. An overarching objective in the field of signaling biology is the comprehension of mechanisms that modulate marine biotic communication, involving the network of direct and indirect relationships that regulate community organization and ecosystem functioning. However, gaps remain in our knowledge about these mechanisms. For this field of research to realize its goals and fill these gaps, we should advance our knowledge in the following areas:
Understanding the specifics of cross-talking. One challenge faced by research on inter-kingdom relationships is differentiating between algal and bacterial metabolic responses to QS, particularly between their downstream interactions. Co-culture model systems could be developed to overcome this challenge, which would also facilitate the characterization of signaling molecules and the resulting responses.
Improved coupling between chemical and biomolecular techniques. In aquatic environments, chemical signals may only be present as complex mixtures at low concentrations, rather than as individual compounds, and may degrade quickly. These issues increase the challenge of separating them and characterizing their structure (Hay, 2009). Combining chemical and molecular methods would help to improve our understanding about the dynamics of these chemical languages. For instance, imaging mass spectrometry provides unprecedented opportunities to study signaling molecule-mediated phenomena visually (Shih et al., 2014). This visualization technology may help us identify more natural products. In addition, approaches based on metabolism and the use of combined metabolic models for algae and their associated bacteria are important to enhance our understanding of the complexity of holobiont systems (Dittami et al., 2014). This information could guide us to a thorough understanding of the biological effects induced by QS, and eventually allow us to assess QS-related co-evolution.
The fate of QS molecules in natural environment. Because QS relies on chemicals, a number of factors affect its rate, including abiotic hydrolysis, enzymatic degradation, and oxidation. These factors have a particular effect on odd-number chain AHLs (such as C7 AHLs) (Pedroza et al., 2014). Thus, knowledge about QS rates and range of functions under natural conditions could help us understand how the environment alters the rate of QS and associated sociobiological behaviors (Decho et al., 2011).
Screening, isolation, and identification of additional QS signal molecules, such as AI-3. Establishing whether AI-3 affects host cells and if so, whether AI-3 signaling is adrenergically mediated would generate both expected and unexpected results. This information would improve our understanding about the potential mechanisms that use these diverse QS signaling components.
Investigate QS systems in archaea. QS systems in the phycospheres of archaea receives comparatively less research focus than those in bacteria (Mackin, 2011). Archaea are important members in ocean and are critical for marine biogeochemistry process. The ecofunctions of archaea rely on population-level traits for survival and physiological activities. Therefore, archaeal QS requires further study. Social network involving QS in archaea may also provide new knowledge about the lifecycle and dynamic processes of algae. The lack of current knowledge about specific interactions between archaea and algae represents an exciting new area of research.
Application of QSIs and QQ signals to prevent HABs. The use of QSIs and QQs against HABs requires further investigation to characterize their efficacy, stability, and degradability in water bodies. Some QSIs and QQs may be toxic or may adversely affect the QS of favorable bacteria. Possible negative impacts on marine organisms or humans require investigation.
Investigate climate-related effects of QS processes. Marine environments are rapidly changing because of global climate change. Important questions include (i) how temperature increases will affect QS behavior, (ii) how ocean acidification will influence microbial communities and their interactions, and (iii) how extremophiles disrupt the social language of organisms in phycospheres. Identification of QS-related genes and hypotheses about the use of these genes by organisms to respond to their environments are now possible due to the development of genomic sequencing and bioinformatics databases.
Investigate practical applications of QS compounds. Compounds or chemicals, such as QSI and biofilm inhibitors, that may be used to resolve ecological issues (e.g., HABs) may be of great value and should be developed. For example, the unicellular alga, C. reinhardtii, inhibits bacterial QS and produces more than a dozen previously unidentified substances capable of activating LasR and cepR (but not luxR, AhyR, or CviR QS) reporter stains. Furthermore, C. reinhardtii extracts exhibit the highest biological activity in polar solvents (Teplitski et al., 2004). Thus, identification of these compounds could have a broad influence across scientific fields, as well as for management and exploitation of algal and bacterial populations globally, among many other uses.
In conclusion, research on QS between algae and bacteria continues to provide detailed knowledge about signal regulation mechanisms in algal-bacterial interactions. This field offers an exciting and open arena for dedicated research to improve our understanding of these intriguing and biologically relevant mechanisms, which underpin ecosystems and may provide economically beneficial remediation.
Acknowledgments
FUNDING
This study was supported by the NSFC (41476092), the Basic Research Program (JCYJ20150529164918736) and Key Project of Science & Technology (JSGG20140519113458237, CXZZ20150529165045063) of Shenzhen Science and Technology Innovation Committee. Funding was also provided by the Woods Hole Center for Oceans and Human Health through the National Science Foundation Grant OCE-1314642 and National Institute of Environmental Health Sciences Grant 1-P01-ES021923-01.
References
- Alagely A, Krediet CJ, Ritchie KB, Teplitski M. Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J. 2011;5:1609–1620. doi: 10.1038/ismej.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin SA. Doctor Scholar Thesis, California Sea Grant College Program. University of California; San Diego: San Diego State University; 2010. The role of siderophores in algal-bacterial interactions in the marine environment. [Google Scholar]
- Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, Morales RL, Berthiaume CT, Parker MS, Djunaedi B, Ingalls AE, Parsek MR, Moran MA, Armbrust EV. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;3:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Alpermann TJ, Cembella AD, Collos Y, Masseret E, Montresor M. The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae. 2012;14:10–35. doi: 10.1016/j.hal.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV. The life of diatoms in the world’s oceans. Nature. 2009;459:185–192. doi: 10.1038/nature08057. [DOI] [PubMed] [Google Scholar]
- Ashen JB, Goff LJ. Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Appl Environ Microbiol. 2000;66:3024–3030. doi: 10.1128/aem.66.7.3024-3030.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averhoff B, Muller V. Exploring research frontiers in microbiology-recent advances in halophilic and thermophilic extremophiles. Res Microbiol. 2010;161:506–514. doi: 10.1016/j.resmic.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- Barbara GM, Mitchell JG. Bacterial tracking of motile algae. FEMS Microbiol Lett. 2003;44:79–87. doi: 10.1111/j.1574-6941.2003.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, Dow JM, Williams P, Daniels MJ. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol. 1997;24:555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL. Small talk, cell-to-cell communication in bacteria. Cell. 2002;109:421–424. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signaling in Vibrio harveyi, sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bauer WD, Mathesius U, Teplitski M. Eukaryotes deal with bacterial quorum sensing. ASM News. 2005;71(3):129–135. [Google Scholar]
- Belas R, Horikawa E, Aizawa S, Suvanasuthi R. Genetic determinants of Silicibacter Sp TM1040 motility. J Bacteriol. 2009;191:4502–4512. doi: 10.1128/JB.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell W, Mitchell R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull. 1972;143:265–277. [Google Scholar]
- Berry JP, Gantar M, Perez MH, Berry G, Noriega FG. Cyanobacterial toxins as allelochemicals with potential applications as algaecides, herbicides and insecticides. Mar Drugs. 2008;6:117–146. doi: 10.3390/md20080007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair W, Marshall MS. PhD Thesis. College of Charleston; Charleston, SC: 2013. In vitro search for quorum sensing activity in batch cultures of harmful dinoflagellates and algicidal bacteria and improved bioassay methods for applications in marine matrices. [Google Scholar]
- Borchardt SA, Allain EJ, Michels JJ, Stearns GW, Kelly RF, McCoy WF. Reaction of acylated homoserine lactone bacterial signaling molecules with oxidized halogen antimicrobials. Appl Environ Microbiol. 2001;67:3174–3179. doi: 10.1128/AEM.67.7.3174-3179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd PW, Ellwood MJ. The biogeochemical cycle of iron in the ocean. Nat Geosci. 2010;3:675–682. [Google Scholar]
- Butler A, Sandy M. Mechanistic considerations of halogenating enzymes. Nature. 2009;460:848–854. doi: 10.1038/nature08303. [DOI] [PubMed] [Google Scholar]
- Capper A, Cruz-Rivera E, Paul VJ, Tibbetts IR. Chemical deterrence of a marine cyanobacterium against sympatric and non-sympatric consumers. Hydrobiologia. 2006;553:319–326. [Google Scholar]
- Carrillo P, Medina-Sanchez JM, Villar-Argaiz M, Delgado-Molina JA, Bullejos FJ. Complex interactions in microbial food webs stoichiometric and functional approaches. Limnetica. 2006;25(1–2):189–204. [Google Scholar]
- Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the proteobacteria. ISME J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- Cernichiari E, Muscatine L, Smith DC. Maltose excretion by symbiotic algae of Hydra viridis. Proc Biol Sci. 1969;173:557–576. [Google Scholar]
- Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- Chan KG, Atkinson S, Mathee K, Sam CK, Chhabra SR, Cámara M. Characterization of N-acylhomoserine lactone-degrading bacteria associated with Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011;11:51. doi: 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Master Scholar Thesis. Tsinghua University; Beijing, China: 2013. Cooperative division of labor between quorum-sensing bacteria within marine self-organized biofilm. [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Chisholm JRM, Dauga C, Ageron E, Grimont PAD. ‘Roots’ in mixotrophic algae. Nature. 1996;381:382. [Google Scholar]
- Cho KW, Lee HS, Rho JR, Kim TS, Mo SJ, Shin J. New lactone-containing metabolites from a marine derived bacterium of the genus Streptomyces. J Nat Prod. 2001;64:664–667. doi: 10.1021/np000599g. [DOI] [PubMed] [Google Scholar]
- Choi H, Mascuch SJ, Villa FA, Byrum T, Teasdale ME, Smith JE, Preskitt LB, Rowley DC, Gerwick L, Gerwick WH. Honaucins A-C, potent inhibitors of inflammation and bacterial quorum sensing: synthetic derivatives and structure-activity relationships. Chem Biol. 2012;19:589–598. doi: 10.1016/j.chembiol.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary PK, Keshvan N, Nguyen HQ, Peterson A, González JE, Haines DC. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry. 2007;46:14429–14437. doi: 10.1021/bi701945j. [DOI] [PubMed] [Google Scholar]
- Clark BR, Engene N, Teasdale ME, Rowley DC, Matainaho T, Valeriote FA, Gerwick WH. Natural product chemistry and taxonomy of the marine cyanobacterium Blennothrix cantharidosmum. J Nat Prod. 2008;71:1530–1537. doi: 10.1021/np800088a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coale KH, Johnson KS, Fitzwater SE, Gordon RM, Tanner S, Chavez FP, Ferioli L, Sakamoto C, Rogers P, Millero F, Steinberg P, Nightingale P, Cooper D, Cochlan WP, Landry MR, Constantinou J, Rollwagen G, Trasvina A, Kudela R. Amassive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature. 1996;383:495–501. doi: 10.1038/383495a0. [DOI] [PubMed] [Google Scholar]
- Coelho FJRC, Santos AL, Coimbra J, Almeida A, Cunha A, Cleary DFR, Calado R, Gomes NCM. Interactive effects of global climate change and pollution on marine microbes: the way ahead. Ecol Evol. 2013;3:1808–1818. doi: 10.1002/ece3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JJ. Interactions between bacteria and algae in aquatic ecosystems. Annu Rev Ecol Syst. 1982;13:291–314. [Google Scholar]
- Collins S, Rost B, Rynearson TA. Evolutionary potential of marine phytoplankton under ocean acidification. Evol Appl. 2014;7:140–155. doi: 10.1111/eva.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol. 2014;12:263–273. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- Cornforth DM, Popat R, McNally L, Gurney J, Scott PTC, Ivens A, Diggle SP, Brown SP. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci USA. 2014;111:4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado-Silva Carmen T, Leonardo C, Catalina AF, Oscar OE. Detection of quorum sensing systems of bacteria isolated from fouled marine organisms. Biochem Syst Ecol. 2013;46:101–107. [Google Scholar]
- Daniels R, Vanderleyden J, Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day WAJ, Maurelli AT. Shigella flexneri LuxS quorum sensing system modulates virB expression but is not essential for virulence. Infect Immun. 2001;69:15–23. doi: 10.1128/IAI.69.1.15-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker SC, Sonck K, Vanderleyden J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006;14(3):114–119. doi: 10.1016/j.tim.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Decho AW, Fery RL, Ferry JL. Chemical chellenges to bacterial AHL signaling in the environment. Chem Rev. 2011;111:86–99. doi: 10.1021/cr100311q. [DOI] [PubMed] [Google Scholar]
- Defoirdt T, Boon N, Bossier P, Verstraete W. Disruption of bacterial quorum sensing: an unexplored strategy to fight infections in aquaculture. Aquaculture. 2004;240:69–88. [Google Scholar]
- Defoirdt T, Miyamoto CM, Wood TK, Meighen EA, Sorgeloos P, Verstraete W, Bossier P. The natural furanone (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone disrupts quorum sensing-regulated gene expression in Vibrio harveyi by decreasing the DNA-binding activity of the transcriptional regulator protein luxR. Environ Microbiol. 2007;9(10):2486–2495. doi: 10.1111/j.1462-2920.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- Degrassi G, Aguilar C, Bosco M, Zahariev S, Pongor S, Venturi V. Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: cross-talk with quorum sensing bacterial sensors. Curr Microbiol. 2002;45:250–254. doi: 10.1007/s00284-002-3704-y. [DOI] [PubMed] [Google Scholar]
- Demuez M, Gonzalez-Fernandez C, Ballesteros M. Algicidal microorganisms and secreted algicides: new tools to induce microalgal cell disruption. Biotechnol Adv. 2015;33(8):1616–1625. doi: 10.1016/j.biotechadv.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Derenbach JB, Pesando D. Investigations into a small fraction of volatile hydrocarbons III. Two diatom cultures produce ectocarpene, a pheromone of brown algae. Mar Chem. 1986;19:337–341. [Google Scholar]
- Desbois A, Lebl T, Yan L, Smith V. Isolation and structural characterisation of two antibacterial free fatty acids from the marine diatom, Phaeodactylum tricornutum. Appl Microbiol Biotechnol. 2008;81:755–764. doi: 10.1007/s00253-008-1714-9. [DOI] [PubMed] [Google Scholar]
- Dickschat JS. Quorum sensing and bacterial biofilms. Nat Prod Rep. 2010;27:343–369. doi: 10.1039/b804469b. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philos Trans R Soc Lond B Biol Sci. 2007;1483:1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittami SM, Eveillard D, Tonon T. A metabolic approach to study algal-bacterial interactions in changing environments. Mol Ecol. 2014;23:1656–1660. doi: 10.1111/mec.12670. [DOI] [PubMed] [Google Scholar]
- Dobrestov S, Teplitski M, Paul V. Quorum sensing in the marine environment and its relationship to biofouling. Biofouling. 2009;25:413–427. doi: 10.1080/08927010902853516. [DOI] [PubMed] [Google Scholar]
- Dobretsov S, Teplitski M, Bayer M, Gunasekra S, Proksch P, Paul VJ. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling. 2011;27:893–905. doi: 10.1080/08927014.2011.609616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH. Quenching quorum-sensing dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- Doucette G. Interactions between bacteria and harmful algae: a review. J Nat Toxins. 1995;3:65–74. doi: 10.1002/nt.2620030202. [DOI] [PubMed] [Google Scholar]
- Dunn SR, Bythell JC, Le Tissier MDA, Burnett WJ, Thomason JC. Programmed cell death and cell necrosis activity during hyperthermic stress-induced bleaching of the symbiotic sea anemone Aiptasia sp. J Exp Mar Bio Ecol. 2002;272:29–53. [Google Scholar]
- Duval B, Margulis L. The microbial community of Ophrydium versatile colonies: endosymbionts, residents, and tenants. Symbiosis. 1995;18:181–210. [PubMed] [Google Scholar]
- Dworjanyn SA, de Nys R, Steinberg PD. Localization and surface quantification of secondary metabolites in the red alga Delisea pulchra. Mar Biol. 1999;133:727–736. [Google Scholar]
- Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Eldar A. Social conflict drives the evolutionary divergence of quorum sensing. Proc Natl Acad Sci USA. 2011;108:13635–13640. doi: 10.1073/pnas.1102923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erken M, Weitere M, Kjelleberg S, McDougald D. In situ grazing resistance of Vibrio cholerae in the marine environment. FEMS Microbiol Lett. 2011;76:504–512. doi: 10.1111/j.1574-6941.2011.01067.x. [DOI] [PubMed] [Google Scholar]
- Falciatore A, Bowler C. Revealing the molecular secrets of marine diatoms. Ann Rev Plant Biol. 2002;53:109–130. doi: 10.1146/annurev.arplant.53.091701.153921. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive earth’s biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- Filzgerald GP. Some factors in the competition or antagonism among bacteria, algae, and aquatic weeds. J Phycol. 1969;5(4):351–359. doi: 10.1111/j.1529-8817.1969.tb02625.x. [DOI] [PubMed] [Google Scholar]
- Flavier AB, Clough SJ, Schell MA, Denny TP. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol Microbiol. 1997;26:251–259. doi: 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- Fletcher RL, Callow ME. The settlement, attachment and establishment of marine algal spores. Br Phycol J. 1992;27:303–329. [Google Scholar]
- Freestone P. Communication between bacteria and their hosts. Scientifica (Cairo) 2013;2013:361073. doi: 10.1155/2013/361073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acylhomoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Geider RJ. Biological oceanography: complex lessons of iron uptake. Nature. 1999;400:815–816. [Google Scholar]
- Generous RA. Environmental threats to the symbiotic relationship of coral reefs and quorum sensing. J Sustain Dev. 2014;11:116–122. [Google Scholar]
- Geng HF, Belas R. Molecular mechanisms underlying rosebacter-phytoplankton symbioses. Curr Opin Biotechnol. 2010;21:332–338. doi: 10.1016/j.copbio.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Geng HF, Belas R. TdaA regulates tropodithietic acid synthesis by binding to the tdaC promoter region. J Bacteriol. 2011;193(15):4002–4005. doi: 10.1128/JB.00323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Bruhn JB, Nielsen KF, Gram L, Belas R. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl Environ Microbiol. 2008;74(5):1535–1545. doi: 10.1128/AEM.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwick L, Boudreau P, Choi H, Mascuch S, Villa FA, Balunas MJ, Malloy KL, Teasdale ME, Rowley DC, Gerwick WH. Interkingdom signaling by structurally related cyanobacterial and algal secondary metabolites. Phytochem Rev. 2013;12:459–465. [Google Scholar]
- Givskov M, de Nys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg PD, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluch MF, Typke D, Baumeister W. Motility and thermotactic responses of Thermotoga maritima. J Bacteriol. 1995;177:5473–5479. doi: 10.1128/jb.177.19.5473-5479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JE, Marketon MM. Quorum sensing in nitrogen-fixing rhizobia. Microbiol Mol Biol Rev. 2003;67(4):574–592. doi: 10.1128/MMBR.67.4.574-592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gov Y, Borovok I, Korem M, Singh VK, Jayaswal RK, Wilkinson BJ, Rich SM, Balaban N. Quorum sensing in Staphylococci is regulated via phosphorylation of three conserved histidine residues. J Biol Chem. 2004;279:14665–14672. doi: 10.1074/jbc.M311106200. [DOI] [PubMed] [Google Scholar]
- Gram L, de Nys R, Maximilien R, Givskov M, Steinberg P, Kjelleberg S. Inhibitory effects of secondary metabolites from the red alga Delisea pulchra on swarming motility of Proteus mirabilis. Appl Environ Microbiol. 1996;62:4284–4287. doi: 10.1128/aem.62.11.4284-4287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graneli E, Pavia H. Allelopathy in marine ecosystems (Chapter 18,ppabcxyzpp415–433) In: Reigosa MJ, Pedrol N, Gonzalez L, editors. Allelopathy: a physiological process with ecological implications. Springer Publisher; P.O Box 17 3300 A A Dordrecht, the Netherlands: 2006. [Google Scholar]
- Hallegraeff GM. A review of harmful algal blooms and their apparent global increase. Phycologia. 1993;32:79–99. [Google Scholar]
- Harrington KI, Sanchez A. Eco-evolutionary dynamics of complex social strategies in microbial communities. Integ Biol. 2014;7:e28230. doi: 10.4161/cib.28230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S, Mathesius U. The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot. 2012;63(9):3429–3444. doi: 10.1093/jxb/err430. [DOI] [PubMed] [Google Scholar]
- Hay ME. Marine chemical ecology: chemical signals and cues structure marine populations, communities, and ecosystems. Ann Rev Mar Sci. 2009;1:193–212. doi: 10.1146/annurev.marine.010908.163708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes K, Hofmann TA, Smith CJ, Ball AS, Underwood GJ, Osborn AM. Diatom-derived carbohydrates as factors affecting bacterial community composition in estuarine sediments. Appl Environ Microbiol. 2007;73(19):6112–6124. doi: 10.1128/AEM.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol. 2004;186:3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Schmid M, Wagner M, Fieseler L, Gernert C, Hacker J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol Lett. 2001;35:305–312. doi: 10.1111/j.1574-6941.2001.tb00816.x. [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmelo LR, Mincer TJ, Van Mooy BAS. Possible influence of bacterial quorum sensing on the hydrolysis of sinking particulate organic carbon in marine environments. Environ Microbiol Rep. 2011;3:682–688. doi: 10.1111/j.1758-2229.2011.00281.x. [DOI] [PubMed] [Google Scholar]
- Hollants J, Leliaert F, Verbruggen H, De Clerck O, Willems A. Host specificity and coevolution of Flavobacteriaceae endosymbionts within the siphonous green seaweed Bryopsis. Mol Phylogenet Evol. 2013;67:608–614. doi: 10.1016/j.ympev.2013.02.025. [DOI] [PubMed] [Google Scholar]
- Hombeck M, Boland W. Biosynthesis of the algal pheromone fucoserratene by the freshwater diatom Asterionella formosa (Bacillariophyceae) Tetrahedron. 1998;54:11033–11042. [Google Scholar]
- Hong Y, Hu HY, Xie X, Li FM. Responses of enzymatic antioxidants and non-enzymatic antioxidants in the cyanobacterium Microcystis aeruginosa to the allelochemical ethyl 2-methyl acetoacetate (EMA) isolated from reed (Phragmites communis) J Plant Physiol. 2008;165:1264–1273. doi: 10.1016/j.jplph.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Huang YL, Dobretsov S, Ki JS, Yang LH, Qian PY. Presence of acyl-homoserine lactone in subtidal biofilm and the implication in larval behavioral response in the polychaete Hydroides elegans. Microb Ecol. 2007;54:384–392. doi: 10.1007/s00248-007-9210-9. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins DA, Witter AE, Butler A, Luther GW., III Competition among marine phytoplankton for different chelated iron species. Nature. 1999;400:858–861. [Google Scholar]
- Inderjit Wardle DA, Karban R, Callaway RM. The ecosystem and evolutionary contexts of allelopathy. Trends Ecol Evol. 2011;26:655–662. doi: 10.1016/j.tree.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Iqbal HA, Feng ZY, Brady SF. Biocatalysts and their small molecule products from metagenomic studies. Curr Opin Chem Biol. 2012;16:109–116. doi: 10.1016/j.cbpa.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatt AN, Tang K, Liu J, Zhang Z, Zhang XH. Quorum sensing in marine snow and its possible influence on production of extracellular hydrolytic enzymes in marine snow bacterium Pantoea ananatis B9. FEMS Microbiol Ecol. 2015;91(2):1–13. doi: 10.1093/femsec/fiu030. [DOI] [PubMed] [Google Scholar]
- Jatt AN, Tang KH, Su Y, Zhang XH. Bacterial quorum sensing and its regulation of particulate organic carbon degradation. International Marine Microbiology Conference, Abstract; 22–25, May, 2015; Qsingdao, China. 2015. pp. 9–10. [Google Scholar]
- Jha B, Kavita K, Westphal J, Hartmann A, Schmitt-Kopplin P. Quorum sensing inhibition by Asparagopsis taxiformis, a marine macro alga: separation of the compound that interrupts bacterial communication. Mar Drugs. 2013;11(1):253–265. doi: 10.3390/md11010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Li M. Quorum sensing inhibitors: a patent review. Expert Opin Ther Pat. 2013;23(7):867–894. doi: 10.1517/13543776.2013.779674. [DOI] [PubMed] [Google Scholar]
- Joint I, Tait K, Wheeler G. Cross-kingdom signalling: exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Philos Trans R Soc Lond B Biol Sci. 2007;362:1223–1233. doi: 10.1098/rstb.2007.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint I, Tait K, Callow ME, Callow JA, Milton D, Williams P, Camara M. Cell-to-cell communication across the prokaryote-eukaryote boundary. Science. 2002;298:1207. doi: 10.1126/science.1077075. [DOI] [PubMed] [Google Scholar]
- Jones AK, Cannon RC. The release of micro-algal photosynthate and associated bacterial uptake and heterotrophic growth. Br Phycol J. 1986;21:341–358. [Google Scholar]
- Jousset A. Ecological and evolutive implications of bacterial defences against predators. Environ Microbiol. 2012;14:1830–1843. doi: 10.1111/j.1462-2920.2011.02627.x. [DOI] [PubMed] [Google Scholar]
- Kanagasabhapathy M, Yamazaki G, Ishida A, Sasaki H, Nagata S. Presence of quorum-sensing inhibitor like compounds from bacteria isolated from the brown alga Colpomenia sinuosa. Lett Appl Microbiol. 2009;49:573–579. doi: 10.1111/j.1472-765X.2009.02712.x. [DOI] [PubMed] [Google Scholar]
- Kang BR, Lee JH, Ko SJ, Lee YH, Cha JS, Cho BH, Kim YC. Degradation of acyl-homoserine lactone molecules by Acinetobacter sp strain C1010935-941. Can J Microbiol. 2004;50:935–941. doi: 10.1139/w04-083. [DOI] [PubMed] [Google Scholar]
- Kellner RL, Dettner K. Differential efficacy of toxic pederin in deterring potential arthropod predators of Paederus (Coleoptera: Staphylinidae) offspring. Oecologia. 1996;107:293–300. doi: 10.1007/BF00328445. [DOI] [PubMed] [Google Scholar]
- Kim C, Kim J, Park HY, McLean RJC, Kim CK, Jeon J, Yi SS, Kim YG, Lee YS, Yoon J. Molecular modeling, synthesis, and screening of new bacterial quorum-sensing antagonists. J Microbiol Biotechnol. 2007a;17:1598–1606. [PubMed] [Google Scholar]
- Kim JS, Kim YH, Seo YW, Park S. Quorum sensing inhibitors from the red alga, Ahnfeltiopsis labelliformis. Biotechnol Bioprocess Eng. 2007b;12:308–311. [Google Scholar]
- Kjelleberg S, Manefield M, Maximilien R, Givskov M, de Nys R, Gram L, Steinberg P. Eukaryotic interference with acyl homoserine lactone mediated gene expression in bacteria. Fifth European Marine Microbiology Symposium. Session: Nutritional and physiological state of single cells, populations and communities; 1996. Abstracts. [Google Scholar]
- Kodama M, Doucette G, Green D. Relationships between bacteria and harmful algae. In: Granéli E, Turner JT, editors. Ecology of harmful algae. Springer-Verlag; Heidelberg, Germany: 2006. pp. 243–255. [Google Scholar]
- Kouzuma A, Watanabe K. Exploring the potential of algae/bacteria interactions. Curr Opin Biotechnol. 2015;33:125–129. doi: 10.1016/j.copbio.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Küpper FC, Gaquerel E, Boneberg EM, Morath S, Salaün JP, Potin P. Early events in the perception of lipopolysaccharides in the brown alga Laminaria digitata include an oxidative burst and activation of fatty acid oxidation cascades. J Exp Bot. 2006;57:1991–1999. doi: 10.1093/jxb/erj146. [DOI] [PubMed] [Google Scholar]
- Kustka AB, Allen AE, Morel FMM. Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J Phycol. 2007;43:715–729. [Google Scholar]
- Lachnit T, Blümel M, Imhoff JF, Wahl M. Specific epibacterial communities on macroalgae: phylogeny matters more than habitat. Aqua Biol. 2009;5:181–186. [Google Scholar]
- Lau WW, Armbrust EV. Detection of glycolate oxidase gene glcD diversity among cultured and environmental marine bacteria. Environ Microbiol. 2006;8:1688–1702. doi: 10.1111/j.1462-2920.2006.01092.x. [DOI] [PubMed] [Google Scholar]
- Lau WW, Keil RG, Armbrust EV. Succession and diel transcriptional response of the glycolate-utilizing component of the bacterial community during a spring phytoplankton bloom. Appl Environ Microbiol. 2007;73:2440–2450. doi: 10.1128/AEM.01965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau TL, Robert JMR. Diatom cultivation and biotechnologically relevant products. II Current and putative products. Appl Microbiol Biotechnol. 2003;60:624–632. doi: 10.1007/s00253-002-1177-3. [DOI] [PubMed] [Google Scholar]
- Leboulanger C, Oriol L, Jupin H, Desolasgros C. Diel variability of glycolate in the eastern tropical Atlantic Ocean. Deep Sea Res A. 1997;44:2131–2139. [Google Scholar]
- Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002;184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindum PW, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-L-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Carlson RW, York WS, Stacey G. Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc Natl Acad Sci USA. 2002a;99:14446–14451. doi: 10.1073/pnas.222336799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Pierson EA, Pierson LS, 3rd, Stacey G, Chatterjee A. Quorum sensing in plant-associated bacteria. Curr Opin Plant Biol. 2002b;5:285–290. doi: 10.1016/s1369-5266(02)00274-1. [DOI] [PubMed] [Google Scholar]
- Long JD, Smalley GW, Barsby T, Anderson JT, Hay ME. Chemical cues induce consumer-specific defenses in a bloom-forming marine phytoplankton. Proc Natl Acad Sci USA. 2007;104:10512–10517. doi: 10.1073/pnas.0611600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RA, Qureshi A, Faulkner DJ, Azam F. 2-n-Pentyl-4-quinolinol produced by a marine Alteromonas sp and its potential ecological and biogeochemical roles. Appl Environ Microbiol. 2003;69:568–576. doi: 10.1128/AEM.69.1.568-576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H, Zhou J, Cai ZH. The dynamic variation process of quorum sensing bacterium in a Scrippsiella Trochoidea. Bloom Ecol Sci. 2016 (in Chinese, abstract in English). In press. [Google Scholar]
- Lyon GJ, Mayville P, Muir TW, Novick RP. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc Natl Acad Sci USA. 2000;97:13330–13335. doi: 10.1073/pnas.97.24.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackin C. Honors Scholar Thesis. University of Conneticut-Storrs; CT: 2011. Quorum sensing in archaea. [Google Scholar]
- Manefield M, Harris L, Rice SA, de Nys R, Kjelleberg S. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl Environ Microbiol. 2000;66:2079–2084. doi: 10.1128/aem.66.5.2079-2084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, Rasmussen TB, Henzter M, Andersen JB, Steinberg P, Kjelleberg S, Givskov M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology (NY) 2002;148:1119–1127. doi: 10.1099/00221287-148-4-1119. [DOI] [PubMed] [Google Scholar]
- Mangwani N, Dash HR, Chauhan A, Das S. Bacterial quorum sensing: functional features and potential applications in biotechnology. J Mol Microbiol Biotechnol. 2012;22(4):215–227. doi: 10.1159/000341847. [DOI] [PubMed] [Google Scholar]
- Matz C, Webb JS, Schupp PJ, Phang SY, Penesyan A, Egan S, Steinberg P, Kjelleberg S. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS ONE. 2008;3:e2744. doi: 10.1371/journal.pone.0002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximilien R, deNys R, Holmström C, Gram L, Givskov M. Chemical mediation of bacterial surface colonisation by secondary metabolites from the red alga Deliseapulchra. Aqua Microb Ecol. 1998;15:233–246. [Google Scholar]
- Mayali X, Azam F. Algicidal bacteria in the sea and their impact on algal blooms. J Eukaryot Microbiol. 2004;51:139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x. [DOI] [PubMed] [Google Scholar]
- Mclean RJ, Pierson LS, 3rd, Fuqua C. A simple screening protocol for the identification of quorum signal antagonists. J Microbiol Methods. 2004;58:351–360. doi: 10.1016/j.mimet.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Mcnab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes LBB, Vermelho AB. Allelopathy as a potential strategy to improve microalgae cultivation. Biotechnol Biofuels. 2013;6:152. doi: 10.1186/1754-6834-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Miller TR, Belas R. Motility is involved in Silicibacter sp TM1040 interaction with dinoflagellates. Environ Microbiol. 2006;8:1648–1659. doi: 10.1111/j.1462-2920.2006.01071.x. [DOI] [PubMed] [Google Scholar]
- Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R. Chemotaxis of Silicibacter sp TM1040 towards dinoflagellate products. Appl Environ Microbiol. 2004;70:4692–4701. doi: 10.1128/AEM.70.8.4692-4701.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NM, Cicirelli EM, Kan J, Chen F, Fuqua C, Hill RT. Diversity and quorum-sensing signal production of Proteobacteria associated with marine ponges. Environ Microbiol. 2008;10:75–86. doi: 10.1111/j.1462-2920.2007.01431.x. [DOI] [PubMed] [Google Scholar]
- Montgomery K, Charlesworth JC, Lebard R, Visscher PT, Burns BP. Quorum sensing in extreme environments. Life. 2013;3:131–148. doi: 10.3390/life3010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 2007;73(14):4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Gonzalez JM, Kiene RP. Linking a bacterial taxon to sulfur cycling in the sea, studies of the marine Roseobacter group. Geomicrobiol J. 2003;20:375–388. [Google Scholar]
- Nagel K, Schneemann I, Kajahn I, Labes A, Wiese J, Imhoff JF. Beneficial effects of 2,4-diacetylphloroglucinol-producing pseudomonads on the marine alga Saccharina latissima. Aqua Microb Ecol. 2012;67:239–249. [Google Scholar]
- Nagle DG, Paul VJ, Roberts MA. Ypaoamide, a new broadly acting feeding deterrent from the marine cyanobacterium Lyngbya majuscula. Tetrahedron Lett. 1996;37:6263–6266. [Google Scholar]
- Nakashima T, Miyazaki Y, Matsuyama Y, Muraoka W, Yamaguchi K, Oda T. Producing mechanism of an algicidal compound against red tide phytoplankton in a marine bacterium gamma-proteobacterium. Appl Microbiol Biotechnol. 2006;73:684–690. doi: 10.1007/s00253-006-0507-2. [DOI] [PubMed] [Google Scholar]
- Natrah F, Kenmegne M, Wiyoto W, Sorgeloos P, Bossier P, Defoirdt T. Effects of microalgae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture. 2011;317:53–57. [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Johnson M, Chou C, Kelly R. Temperature, not LuxS, mediates AI-2 formation in hydrothermal habitats. FEMS Microbiol Lett. 2009;68:173–181. doi: 10.1111/j.1574-6941.2009.00662.x. [DOI] [PubMed] [Google Scholar]
- Nithya C, Pandian SK. The in vitro antibiofilm activity of selected marine bacterial culture supernatants against Vibrio spp. Arch Microbiol. 2010;192:843–854. doi: 10.1007/s00203-010-0612-6. [DOI] [PubMed] [Google Scholar]
- Nylund GM, Cervin G, Persson F, Hermansson M, Steinberg PD, Pavia H. Seaweed defense against bacteria: a poly-brominated 2-heptanone from the red alga Bonnemaisonia hamifera inhibits bacterial colonization at natural surface concentrations. Mar Ecol Prog Ser. 2008;369:39–50. [Google Scholar]
- Oger P, Farrand SK. Co-evolution of the agrocinopine opines and the agrocinopine-mediated control of TraR, the quorum-sensing activator of the Ti plasmid conjugation system. Mol Microbiol. 2001;41(5):1173–1185. doi: 10.1046/j.1365-2958.2001.02584.x. [DOI] [PubMed] [Google Scholar]
- Pacheco AR, Sperandio V. Inter-kingdom signaling: chemical language between bacteria and host. Curr Opin Microbiol. 2009;12:192–198. doi: 10.1016/j.mib.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl HW. Microscale physiological and ecological studies of aquatic cyanobacteria: macroscale implications. Microsci Res Tech. 1996;33:47–72. doi: 10.1002/(SICI)1097-0029(199601)33:1<47::AID-JEMT6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Palkovacs EP, Hendry AP. Eco-evolutionary dynamics: intertwining ecological and evolutionary processes in contemporary time. F1000 Biol Rep. 2010;18(2):1–5. doi: 10.3410/B2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Hwang BJ, Shin MH, Kim JA, Kim HK, Lee JK. N-acylhomoserine lactonase producing Rhodococcus spp with different AHL-degrading activities. FEMS Microbiol Lett. 2006;261:102–108. doi: 10.1111/j.1574-6968.2006.00336.x. [DOI] [PubMed] [Google Scholar]
- Parsek MR, Greenberg EP. SocioMicrobiol: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Paul C, Pohnert G. Interactions of the algicidal bacterium Kordia algicida with diatoms: regulated protease excretion for specific algal lysis. PLoS ONE. 2011;6:e21032. doi: 10.1371/journal.pone.0021032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroza CJ, Flórez AM, Ruiz OS, Orduz S. Enzymatic hydrolysis of molecules associated with bacterial quorum sensing using an acyl homoserine lactonase from a novel Bacillus thuringiensis strain. Antonie Van Leeuwenhoek. 2014;105(1):253–264. doi: 10.1007/s10482-013-0072-5. [DOI] [PubMed] [Google Scholar]
- Pennings SC, Weiss AM, Paul VJ. Secondary metabolites of the cyanobacterium Microcoleus lyngbyaceus and the sea hare Stylocheilus longicauda: palatability and toxicity. Mar Biol. 1996;126:735–743. [Google Scholar]
- Peperzak L. Below the Kolmogorov scale: the non-turbulent sex life of phytoplankton. African J Mar Sci. 2006;28:261–264. [Google Scholar]
- Pohnert G, Boland W. Biosynthesis of the algal pheromone hormosirene by the freshwater diatom Gomphonema parvulum (Bacillariophyceae) Tetrahedron. 1996;52:10073–10082. [Google Scholar]
- Qiu H, Yoon HS, Bhattacharya D. Algal endosymbionts as vectors of horizontal gene transfer in photosynthetic eukaryotes. Front Plant Sci. 2013;4:366. doi: 10.3389/fpls.2013.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina JB, Tapiolas D, Willis BL, Bourne DG. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol. 2009;75:3492–3501. doi: 10.1128/AEM.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani S, Bauer WD, Robinson JB, Farrow JM., III The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum-sensing receptor. Mol Plant Microb Interact. 2008;21:1184–1192. doi: 10.1094/MPMI-21-9-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani S, Teplitski M, Kumar A, Krediet CJ, Sayre RT, Bauer WD. N-acyl homoserine lactone lactonase, AiiA, inactivation of quorum-sensing agonists produced by Chlamydomonas reinhardtii (Chlorophyta) and characterization of aiiA transgenic algae. J Phycol. 2011;47:1219–1227. doi: 10.1111/j.1529-8817.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen TB, Skindersoe ME, Bjarnsholt T, Phipps RK, Christensen KB, Jensen PO, Andersen JB, Koch B, Larsen TO, Hentzer M, Eberl L, Hoiby N, Givskov M. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology. 2005;151:1325–1340. doi: 10.1099/mic.0.27715-0. [DOI] [PubMed] [Google Scholar]
- Reading NC, Sperandio V. Quorum sensing: the many languages of bacteria. FEMS Microbiol Lett. 2006;254:1–11. doi: 10.1111/j.1574-6968.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- Reen F, Almagro-Moreno S, Ussery D, Boyd E. The genomic code: inferring Vibrionaceae niche specialization. Nat Rev Microbiol. 2006;4:1–8. doi: 10.1038/nrmicro1476. [DOI] [PubMed] [Google Scholar]
- Ribalet F, Vidoudez C, Cassin D, Pohnert G, Ianora A, Miralto A, Casotti R. High plasticity in the production of diatom-derived polyunsaturated aldehydes under nutrient limitation: physiological and ecological implications. Protist. 2009;160:444–451. doi: 10.1016/j.protis.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Riley M, Staley J, Danchin A, Wang TZ, Brettin TS, Hauser LJ, Land ML, Thompson LS. Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genomics. 2008;9:210. doi: 10.1186/1471-2164-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MO, Vargas P, Riquelme CE. Interactions of Botryococcus braunii cultures with bacterial biofilms. Microb Ecol. 2010;60:628–635. doi: 10.1007/s00248-010-9686-6. [DOI] [PubMed] [Google Scholar]
- Romero M, Martin-Cuadrado A, Roca-Rivada A, Cabello AM, Otero A. Quorum quecnching in cultivable bacteria from dense marine cosatal mircobial communitites. FEMS Microbiol Lett. 2011;75:205–217. doi: 10.1111/j.1574-6941.2010.01011.x. [DOI] [PubMed] [Google Scholar]
- Rosenwasser S, Graff van Creveld S, Schatz D, Malitsky S, Tzfadia O, Aharoni A, Levin Y, Gabashvili A, Feldmesser E, Vardi A. Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc Natl Acad Sci USA. 2014;111(7):2740–2745. doi: 10.1073/pnas.1319773111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy M, Butler A. Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre R. Microalgae: The potential for carbon capture. BioScience. 2010;60(9):722–727. [Google Scholar]
- Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- Schaum CE, Collins S. Plasticity predicts evolution in a marine alga. Proc Biol Sci. 2014;281(1793) doi: 10.1098/rspb.2014.1486. pii: 20141486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour JR, Ahmed T, Marcos Stocker R. A microfluidic chemotactic assay for assessing the behavior of microbes within diffusing nutrient patches. Limnol Oceanogr Methods. 2008;6:477–488. [Google Scholar]
- Seymour JR, Ahmed T, Stocker R. Bacterial chemotaxis towards the extracellular products of the toxic phytoplankton Heterosigma akashiwo. J Plankton Res. 2009;31:1557–1561. [Google Scholar]
- Shih CJ, Chen PY, Liaw CC, Lai YM, Yang YL. Bringing microbial interactions to light using imaging mass spectrometry. Nat Prod Rep. 2014;31:739–755. doi: 10.1039/c3np70091g. [DOI] [PubMed] [Google Scholar]
- Skerratt JH, Bowman JP, Hallegraeff G, James S, Nichols PD. Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar Ecol Prog Ser. 2002;244:1–15. [Google Scholar]
- Skindersoe ME, Ettinger-Epstein P, Rasmussen TB, Bjarnsholt T, de Nys R, Givskov M. Quorum sensing antagonism from marine organisms. Mar Biotechnol. 2008;10:56–63. doi: 10.1007/s10126-007-9036-y. [DOI] [PubMed] [Google Scholar]
- Slightom RN, Buchan A. Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl Environ Microbiol. 2009;75:6027–6037. doi: 10.1128/AEM.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Egipsy V, González-Toril E, Zettler E, Amaral-Zettler L, Aguilera A, Amils R. Prokaryotic community structure in algal photosynthetic biofilms from extreme acidic streams in Río Tinto (Huelva, Spain) Int Microbiol. 2008;11(4):251–260. doi: 10.2436/20.1501.01.69. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Giron JA, Kaper JB. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague GF, Winans SC. Eukaryotes learn how to count: quorum sensing by yeast. Genes & Dev. 2006;20:1045–1049. doi: 10.1101/gad.1432906. [DOI] [PubMed] [Google Scholar]
- Stefanic P, Decorosi F, Viti C, Petito J, Cohan FM, Mandic-Mulec I. The quorum sensing diversity within and between ecotypes of Bacillus subtilis. Environ Microbiol. 2012;14:1378–1389. doi: 10.1111/j.1462-2920.2012.02717.x. [DOI] [PubMed] [Google Scholar]
- Steinberg PD, Rice SA, Campbell AH, McDougald D, Harder T. Interfaces between bacterial and eukaryotic “neuroecology”. Integr Comp Biol. 2011;51(5):794–806. doi: 10.1093/icb/icr115. [DOI] [PubMed] [Google Scholar]
- Steinberg PD, Schneider R, Kjelleberg S. Chemical defenses of seaweeds against microbial colonization. Biodegradation. 1997;8:211–220. [Google Scholar]
- Stocker R, Seymour JR. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev. 2012;76:792–812. doi: 10.1128/MMBR.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R, Seymour JR, Samadani A, Hunt DH, Polz MF. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA. 2008;105:4209–4214. doi: 10.1073/pnas.0709765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom SL. Microbial Ecology of ocean biogeochemistry: a community perspective. Science. 2008;320:1043–1045. doi: 10.1126/science.1153527. [DOI] [PubMed] [Google Scholar]
- Sun S, Kjelleberg S, McDougald D. Relative contributions of Vibrio polysaccharide and quorum sensing to the resistance of Vibrio cholerae to predation by heterotrophic protists. PLoS ONE. 2013;8:e56338. doi: 10.1371/journal.pone.0056338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, Lynch MJ, Fish L, Kirke DF, Tomas JM, Stewart GSAB, Williams P. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect Immun. 1999;67:5192–5199. doi: 10.1128/iai.67.10.5192-5199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrpas M, Ruysbergh E, Blommaert L, Vanelslander B, Sabbe K, Vyverman W, De Kimpe N, Mangelinckx S. Haloperoxidase mediated quorum quenching by Nitzschia cfpellucida: study of the metabolization of N-acyl homoserine lactones by a benthic diatom. Mar Drugs. 2014;12:352–367. doi: 10.3390/md12010352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait K, Williamson H, Atkinson S, Williams P, Camara M, Joint I. Turnover of quorum sensing signal molecules modulate cross-kingdom signalling. Environ Microbiol. 2009;11:1792–1802. doi: 10.1111/j.1462-2920.2009.01904.x. [DOI] [PubMed] [Google Scholar]
- Taktikos J, Stark H, Zaburdaev V. How the motility pattern of bacteria affects their dispersal and chemotaxis. PLoS ONE. 2013;8(12):e81936. doi: 10.1371/journal.pone.0081936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taktikos J, Zaburdaev V, Stark H. Collective dynamics of model microorganisms with chemotactic signaling. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;85:051901. doi: 10.1103/PhysRevE.85.051901. [DOI] [PubMed] [Google Scholar]
- Tan SJ, Zhou J, Zhu XS, Yu SC, Zhan WG, Wang B, Cai ZH. An association network analysis among microeukaryotes and bacterioplankton reveals algal bloom dynamics. J Phycol. 2015;51(1):120–132. doi: 10.1111/jpy.12259. [DOI] [PubMed] [Google Scholar]
- Tang YZ, Koch F, Gobler CJ. Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc Natl Acad Sci USA. 2010;107:20756–20761. doi: 10.1073/pnas.1009566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71(2):295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale ME, Liu J, Wallace J, Akhlaghi F, Rowley DC. Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl Environ Microbiol. 2009;75:567–572. doi: 10.1128/AEM.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebben J, Tapiolas DM, Motti CA, Abrego D, Negri AP, Blackall LL, Steinberg PD, Harder T. Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS ONE. 2011;6(4):e19082. doi: 10.1371/journal.pone.0019082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Chen H, Rajamani S, Gao M, Merighi M, Sayre RT, Robinson JB, Rolfe BG, Bauer WD. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 2004;134:137–146. doi: 10.1104/pp.103.029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thar R, Fenchel T. True chemotaxis in oxygen gradients of the sulfur-oxidizing bacterium Thiovulum majus. Appl Environ Microbiol. 2001;67(7):3299–3303. doi: 10.1128/AEM.67.7.3299-3303.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. The role of coevolution. Science. 2012;335(6067):410–411. doi: 10.1126/science.1217807. [DOI] [PubMed] [Google Scholar]
- Tortell PD, Maldonado MT, Granger J, Price NM. Marine bacteria and biogeochemical cycling of iron in the oceans. FEMS Microbiol Lett. 1999;29:1–11. [Google Scholar]
- Tran LSP, Nagai T, Itoh Y. Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol Microbiol. 2000;37:1159–1171. doi: 10.1046/j.1365-2958.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- Trias R, García-Lledó A, Sánchez N, López-Jurado JL, Hallin S, Bañeras L. Abundance and composition of epiphytic bacterial and archae alammonia oxidizers of marine red and brown macroalgae. Appl Environ Microbiol. 2012;78:318–325. doi: 10.1128/AEM.05904-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsim ST, Wong JT, Wong YH. Effects of dibutyryl cAMP and bacterial toxins on indoleamine-induced encystment of dinoflagellates. Neurosignals. 1996;5:22–29. doi: 10.1159/000109170. [DOI] [PubMed] [Google Scholar]
- Vaitkevicius K, Rompikuntal PK, Lindmark B, Vaitkevicius R, Song T, Wai SN. The metalloprotease PrtV from Vibrio cholerae. FEBS J. 2008;275:3167–3177. doi: 10.1111/j.1742-4658.2008.06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen W, Okrész L, Bögre L, Munnik T. Learning the lipid language of plant signalling. Trends Plant Sci. 2004;9:378–384. doi: 10.1016/j.tplants.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Van Mooy BAS, Hmelo LR, Sofen LE, Campagna SR, May AL, Dyhrman ST, Heithoff A, Webb EA, Momper L, Mincer TJ. Quorum sensing control of phosphorus acquisition in Trichodesmium consortia. ISME J. 2012;6:422–429. doi: 10.1038/ismej.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi A, Bidle KD, Kwityn C, Hirsh DJ, Thompson SM, Callow JA, Falkowski P, Bowler C. A diatom gene regulating nitric-oxide signaling and susceptibility to diatom-derived aldehydes. Curr Biol. 2008;18:895–899. doi: 10.1016/j.cub.2008.05.037. [DOI] [PubMed] [Google Scholar]
- Vraspir JM, Butler A. Chemistry of marine ligands and siderophores. Ann Rev Mar Sci. 2009;1:43–63. doi: 10.1146/annurev.marine.010908.163712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Ann Rev Microbiol. 2006;60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- Wahl M, Goecke F, Labes A, Dobretsov S, Weinberger F. The second skin: ecological role of epibiotic biofilms on marine organisms. Front Microbiol. 2012;3:1–21. doi: 10.3389/fmicb.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. Master Scholar Thesis. Tsinghua University; Beijing, China: 2014. Effects of microbes on growth and toxicity of Gambierdiscus spp. [Google Scholar]
- Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu JL, Tay L, Fang RX, Zhang LH. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol. 2004;51:903–912. doi: 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhou J. Draft genome sequence of Citrobacter freundiistrain ST2, a γ-proteobacterium that produces N-acylhomoserine lactones. Genomics Data. 2015;6:234–236. doi: 10.1016/j.gdata.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Watson WT, Minogue TD, Val DL, von Bodman SB, Churchill ME. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol Cell. 2002;9:685–694. doi: 10.1016/s1097-2765(02)00480-x. [DOI] [PubMed] [Google Scholar]
- Weinberger F, Beltran J, Correa JA, Lion U, Pohnert G. Spore release in acrochaetium sp (Rhodophyta) is bacterially controlled. J Phycol. 2007;43:235–241. [Google Scholar]
- Weissbach A, Rudström M, Olofsson M, Béchemin C, Icely J, Newton A, Tillmann U, Legrand C. Phytoplankton allelochemical interactions change microbial food web dynamics. Limnol Oceanogr. 2011;56:899–909. [Google Scholar]
- Wenbin N, Dejuan Z, Feifan L, Lei Y, Peng C, Xiao XY, Hongyu L. Quorum-sensing system in Acidithiobacillusferrooxidans involved in its resistance to Cu2+ J Appl Microbiol. 2011;53:84–91. doi: 10.1111/j.1472-765X.2011.03066.x. [DOI] [PubMed] [Google Scholar]
- Whalan S, Webster NS. Sponge larval settlement cues: the role of microbial biofilms in a warming ocean. Sci Rep. 2014;4:4072. doi: 10.1038/srep04072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Tait K, Taylor A, Brownlee C, Joint I. Acyl-homoserine lactones modulate the settlement rate of zoospores of the marine alga Ulva intestinalis via a novel chemokinetic mechanism. Plant Cell Environ. 2006;29(4):608–618. doi: 10.1111/j.1365-3040.2005.01440.x. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes, the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichard T, Gerecht A, Boersma M, Poulet SA, Wiltshire K, Pohnert G. Lipid and fatty acid composition of diatoms revisited: rapid wound activated change of food quality parameters influences herbivorous copepod reproductive success. Chem Bio Chem. 2007;8:1146–1153. doi: 10.1002/cbic.200700053. [DOI] [PubMed] [Google Scholar]
- Wietz M, Duncan K, Patin NV, Jensen PR. Antagonistic interactions mediated by marine bacteria: the role of small molecules. J Chem Ecol. 2013;39(7):879–891. doi: 10.1007/s10886-013-0316-x. [DOI] [PubMed] [Google Scholar]
- Wyss Sarah C. A thesis presented to the Honors Tutorial College. Ohio University; 2013. Design of a cross-domain quorum sensing pathway for algae biofuel applications; p. 3. [Google Scholar]
- Xu LY, Li Z, Han XT, Cui ZS, Guo XC, Li XZ. Screening of microalgae associated bacteria with quorum sensing system and their algicidal activity. Oceanol Limnol Sinica. 2012;43(6):1149–1155. (In Chinese, English abstract) [Google Scholar]
- Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Cámara M, Smith H, Williams P. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun. 2002;70(10):5635–546. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zargiel KA, Swain GW. Static vs dynamic settlement and adhesion of diatoms to ship hull coatings. Biofouling. 2014;30:115–129. doi: 10.1080/08927014.2013.847927. [DOI] [PubMed] [Google Scholar]
- Zhang LH, Dong YH. Quorum sensing and signal interference: diverse implications. Mol Microbiol. 2004;53(6):1563–1571. doi: 10.1111/j.1365-2958.2004.04234.x. [DOI] [PubMed] [Google Scholar]
- Zhang QG, Buckling A, Ellis RJ, Godfray HC. Coevolution between cooperators and cheats in a microbial system. Evolution. 2009;63:2248–2256. doi: 10.1111/j.1558-5646.2009.00708.x. [DOI] [PubMed] [Google Scholar]
- Zheng L, Cui Z, Xu L, Sun C, Powell RJ, Hill RT. Draft genome sequence of Rhodobacteraceae strain PD-2, an algicidal bacterium with a quorum-sensing system, isolated from the marine microalga Prorocentrum donghaiense. Genome Announc. 2015;3(1):e01549–14. doi: 10.1128/genomeA.01549-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chen GF, Zhu XS, Chen L, Cai ZH. A review of the relationship between algae and bacteria in harmful algal blooms. Acta Ecol Sinica. 2014;34:269–281. (In Chinese, English abstract) [Google Scholar]
- Zhou J, Lao YM, Cai ZH. Draft genome sequence of Providencia sneebia strain ST1, a quorum sensing bacterium associated with marine microalgae. J Genomics. 2016 doi: 10.7150/jgen.14051. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]