Abstract
Around 10-15% of patients diagnosed with frontotemporal dementia (FTD) have a positive family history for FTD with an autosomal dominant pattern of inheritance. Since the identification of mutations in MAPT (microtubule-associated protein tau gene) in 1998, over 10 other genes have been associated with FTD spectrum disorders, discussed in this review. Along with MAPT, mutations in GRN (progranulin) and C9orf72 (chromosome 9 open reading frame 72) are the most commonly identified in FTD cohorts. The association of FTD and motor neuron disease (MND) can be caused by mutations in C9orf72 and other genes, such as TARDBP (TAR DNA-binding protein), FUS (fused in sarcoma), UBQLN2 (ubiquilin 2). Multisystem proteinopathy is a complex phenotype that includes FTD, Paget disease of the bone, inclusion body myopathy and MND, and can be due to mutations in VCP (valosing containing protein) and other recently identified genes.
Keywords: frontotemporal lobar degeneration, frontotemporal dementia, genetics, amyotrophic lateral sclerosis
Abstract
Cerca de 10-15% dos pacientes diagnosticados com demência frontotemporal (FDFT) têm uma história familiar positiva para DFT com um padrão de herança autossômico dominante. Desde a identificação de mutações em MAPT (proteína tau associada a microtúbulos), em 1998, mais de 10 outros genes já foram associados a doenças do espectro da DFT, que são discutidas nesta revisão. Junto com MAPT, mutações em GRN (progranulina) e C9orf72 (chromosome 9 open reading frame 72) são as mais comumente identificadas em casuísticas de DFT. A associação de DFT com doença do neurónio motor (DNM) pode ser causada por mutações em C9orf72 e outros genes, tais como TARDBP (TAR DNA-binding protein), FUS (fused in sarcoma), UBQLN2 (ubiquilina 2) e outros genes. Proteinopatia multissistêmica é um fenótipo complexo que inclui DFT, doença de Paget óssea, miopatia com corpúsculos de inclusão e DNM, e pode ser devida a mutações em VCP (valosin containing protein) e outros genes recentemente identificados.
INTRODUCTION
The term frontotemporal dementia (FTD) refers to a group of diseases characterzed by focal degeneration of temporal and/or frontal lobes of the brain. Clinically, FTD is subclassified into a behavioral variant (bvFTD) and into two language variants, named agrammatic or nonfluent variant of Primary Progressive Aphasia (nfvPPA) and semantic variant of PPA (svPPA).1,2 Progressive supranuclear palsy (PSP) and corticobasal syndrome (CBS) often overlap clinically with FTD spectrum disorders, as behavioral symptoms and/or language impairment similar to those seen in FTD can appear in patients with PSP or CBS, leading to the inclusion of both (by some authors) within the clinical spectrum of FTD.3 There is also significant overlap between FTD and motor neuron disease (MND), as around 15% of patients with bvFTD are also diagnosed with amyotrophic lateral sclerosis (ALS).4
Whereas FTD is used to characterize clinical syndromes, the term Frontotemporal Lobar Degeneration (FTLD) is used in the neuropathological classification of this group of disorders. FTLD is subclassified according to the main aggregated protein found in neuronal and/or glial inclusions into: FTLD-tau (tau protein), FTLD-TDP (TAR DNA binding protein with 43kDA, or TDP-43) and FTLD-FUS (fused in sarcoma protein, or FUS).5 FTLD-TDP represents around 50% of FTLD cases, FTLD-tau around 45%, while FTLD-FUS is observed in 5-10% of FTLD cases. The main protein has not been identified in a small subset of cases, and these are diagnosed as FTLD-U, since the inclusions are immunoreactive to antibodies against proteins from the ubiquitin-proteasome system, but not to tau, TDP-43 or FUS.5 Again, overlap with PSP, CBS and MND is seen, as FTLD-tau might underlie PSP and CBS, and inclusions with TDP-43 and FUS can be found in MND.
FTD has an important genetic component, as around 40% of patients have at least one first-degree relative with an FTD spectrum disease, and about 10-15% have a family history of FTD suggesting an autosomal dominant pattern of inheritance.6
Since identification of mutations in the gene of microtubule-associated protein tau (MAPT) as a cause of monogenic FTD in 1998,7 mutations in over ten other genes have been identified, such as in the progranulin gene (GRN) in 2006, and in C9orf72 (Chromosome 9 open reading frame 72) in 2011.8-11 Mutations in MAPT, GRN and C9orf72 are the most commonly reported in FTD cohorts worldwide.
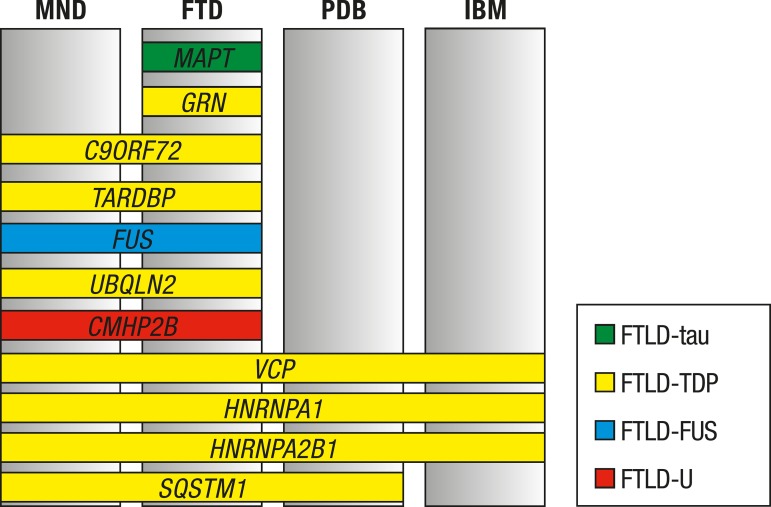
Mutations in MAPT and GRN typically present as FTD phenotypes, whereas hexanucleotide repeat expansions in C9orf72 are associated with FTD and MND (Figure 1). Mutations in other genes, such as in those encoding TDP-43 (TARDBP) or FUS (FUS) also present clinically as FTD and/or MND.12
Figure 1.
Genes associated with monogenic FTD with clinical and neuropathological correlations
MND: motor neuron disease; FTD: frontotemporal dementia; PDB: Paget disease ofthe bone; IBM: inclusion body myositis. Each color refers to a FTLD subtype. Please see text for more information regarding neuropathology. Even though TDP-43 pathology is present in UBQLN2 mutations, inclusions immunostained by antibodies against ubiquilin 2 have also been reported, and ubiquilin 2 might be the major abnormal protein in this mutation. Mutations in HNRNP1 and HNRNPA2B1 are still lacking neuropathological studies confirming TDP-43 pathology in the central nervous system.
FTD can also be part of a complex phenotype called multisystem proteinopathy (MSP) that can be caused by single mutations, such as in VCP. MSP is a rare syndrome characterized by a combination of the following phenotypes: bvFTD, MND, Paget disease of the bone and/or inclusion body myopathy.13
GENES ASSOCIATED WITH MONOGENIC FORMS OF FTD
MAPT GENE. The MAPT gene is located on chromosome 17q21.1. It encodes the microtubule-associated protein tau, which stabilizes and promotes the assembly of microtubules.14
There are six isoforms of tau in the normal brain, generated from the alternative splicing of exons 2, 3 and 10.14 Exons 9-12 encode the microtubule-binding domains and, depending on the inclusion or exclusion of exon 10 from the transcripts, the isoforms may have three (3R) or four repetitions (4R) of these domains. In normal brain, 3R and 4R isoforms coexist in equal proportions (1:1), and changes in this ratio are believed to lead to neurodegeneration.7,14
MAPT mutations can be divided into two groups, with different pathogenic mechanisms.7,14 The first group, composed of missense mutations and deletions, modifies tau protein and its function in order to increase or decrease its interaction with microtubules. Some of these mutations may lead to a greater tendency to form tau filaments that form toxic aggregates. The second group of mutations (located in exon 10 or flanking regions) interfere with the alternative splicing of exon 10, changing the 3R:4R tau ratio (increasing 4R tau), which leads to an increase in filamentous inclusions and is associated with neurodegeneration.14
More than 44 pathogenic mutations (missense, deletions, silent or intronic mutations) have been reported in the literature.15 The pathogenic gene mutations are clustered in exons 9-13, with the exception of two mutations in exon 1.15
In FTD cohorts with more than 100 patients, MAPT mutations were found in 3-11% of cases (9-38% among familial FTD cases, and 0-3% among sporadic FTD cases).9,12,16-21 MAPT mutations cause disease with autosomal dominant inheritance and penetrance greater than 95%.22
The mean age of onset of symptoms varies between 46 and 57 years, and the age at onset ranges between 17 and 75 years.12,16 Onset after 70 years of age is rare.12 The duration of the disease until death is also variable (on average 8 to 10 years), but cases with duration of disease ranging from one to 25 years have been described.12,16,17
BvFTD, with or without Parkinsonism, is the most commonly reported phenotype.12,16 PSP12 and svPPA17 are also relatively frequent, whereas CBS, nfvPPA12 and early-onset dementia of the Alzheimer type23 are less frequently observed. The Alzheimer's disease (AD) phenotype has been described in MAPT mutations, but the neuropathological diagnosis was not AD in any of the cases reported.23
Parkinsonism is reported in 25-56% of cases, and oculomotor abnormalities in 13-29%.12,16,24 Oculomotor changes appear to differentiate MAPT mutations from GRN or C9orf72 mutations, as they are infrequent in the latter.12,16
The presence of DNM is very rare in MAPT mutations. Recently, a new mutation (p.D348G) was found, a kindred with lower MND and respiratory failure, but without signs of cognitive impairment.25 Progressive hypoventilation has also been reported in the only family described to date with a homozygous MAPT mutation (p.S352L) although in the homozygous mutation, symptom onset occurred much earlier and both siblings died before five years of age.26
Structural neuroimaging studies have shown that the atrophy is more significant in anteromedial temporal regions in patients with symptomatic MAPT mutations.27 Atrophy is also observed in the frontal lobes, particularly in the orbitofrontal regions, and is typically symmetric (contrasting with the asymmetry observed in GRN mutations).27
The neuropathological changes described in MAPT mutations are heterogeneous, but typically neuronal or glial, and neuronal inclusions with hyperphosphorylated tau are observed.28 Mutations located in exons 9, 11, 12 and 13 are generally associated with inclusions in neurons that contain predominantly 3R tau, whereas those located in exons 1 and 10 or in introns flanking exons 9 and 10 tend to generate inclusions in neurons and glial cells in which 4R is predominant.28
GRN GENE. The progranulin gene is located in the 17q21.32 region and is composed of 13 exons. The gene encodes progranulin, a protein with 593 amino acids which can be cleaved into granulin peptides (A, B, C, D, E, F, and P).29 Progranulin is mainly expressed in epithelial and hematopoietic cells; and is expressed in neurons and microglia in the nervous system. The protein has properties as a neurotrophic growth factor and in neuronal survival (protecting neurons from toxic insults in laboratory models). Furthermore, it participates in inflammatory processes (such as wound healing, microglia activation and inhibition of TNF-α), tumorigenesis, and is increased in diabetes mellitus.29
Pathogenic mutations in GRN lead to loss of function, and cause disease through haploinsufficiency.8 Null mutations in GRN are most frequently nonsense, or insertions or deletions that generate a frameshift mutation with premature stop codon formation and subsequent nonsense-mediated mRNA decay. Mutations located in splicing or translation initiation sites, and (partial and total) gene deletions have also been reported.15,29 Missense mutations have also been described, but there is evidence of pathogenicity for only a few of these - affecting the production, secretion or function of progranulin - demonstrated by functional studies, including: p.A9D, p.C139R, p.R432C and p.C521Y.30
Experiments done in animal and cell culture models have shown that progranulin deficiency causes reduction in synaptic density, decreased survival of neurons, increased susceptibility of neurons to stressors, and abnormally increased inflammatory responses, which might be the mechanisms underlying neurodegeneration.29
Over 70 mutations have been reported in the literature.15 In FTD cohorts with more than 100 patients, mutations in GRN were found in 4-12% of cases (7-28% among familial and 1-4% among sporadic FTD cases).12,16-20,31 Mutations have an autosomal dominant pattern of inheritance, with an estimated penetrance in 50-60% at 60 years and greater than 90% at age 70.20,31,32
The mean age of onset of symptoms ranges from 57 to 62 years, and age at onset ranges from 35 to 87 years,12,16-20,31 Besides the phenotypic variability among families, there is great variability in the clinical presentation among individuals of the same family, with differences in age of onset of symptoms varying by up to 20 years.32 The duration of symptoms is on average 5-9 years (range 1-23 years).12,16,17,19,31
The most common clinical presentation is bvFTD, followed in order of frequency by nfvPPA and CBS.12,16,19,31 Mutations in GRN were the most frequent cause of familial CBS in some series.12,31 Other PPA variants, such as semantic variant and mixed variant, have been reported in a few cases.19 Alzheimer-type dementia, Parkinson's disease and dementia with Lewy bodies are infrequent phenotypes; and PSP is rare.31,33,34 The bvFTD with MND presentation is also very rare and has been reported only in a few isolated cases.20,31
Parkinsonism is observed in 20-50% of patients.12,16,32 One group reported an increased frequency of visual hallucinations (around 25%) in individuals with GRN mutations,12 but other groups failed to confirm the same finding, and found frequencies of hallucinations and/or psychosis of between 5 and 12%.19,32 In addition to executive dysfunction signs (typical of bvFTD), neuropsychological assessment may show signs of parietal lobe dysfunction, such as dysgraphia, dyscalculia, apraxia and/or visuospatial deficits.19
A homozygous GRN mutation was reported in two siblings with adult-onset neuronal ceroid lipofuscinosis, showing that homozygous mutations in GRN lead to a completely different phenotype of heterozygous mutations. Neuronal ceroid lipofuscinosis is a lysosomal storage disease, and this finding suggests that progranulin participates in cellular processes related to lysossomes.35
Structural neuroimaging studies indicate that the asymmetry is more significant in GRN than in MAPT or C9orf72 mutations.19,36 Atrophy is more prominent in posterior temporal and parietal regions, but also extends to the frontal lobes.27 Parietal regions are significantly more atrophied in GRN than in MAPT mutations.
Periventricular subcortical hyperintensities indicating involvement of the white matter on T2-weighted and FLAIR MRI images were observed in 20-40% of patients, and are found mainly in regions of maximal atrophy such as the frontal and parietal lobes.37,38 The hyperintensities can be confluent, and affect U-fibers as well as subcortical white matter.37 The neuropathological substrates of these white matter changes remain unknown, but are possibly related to demyelination and/or microglia activation.38
GRN null mutations lead to reductions in the levels of progranulin in plasma and cerebrospinal fluid, both in symptomatic and pre-symptomatic individuals.39 Missense mutations may also be associated with reduced levels of progranulin, similar to those of null mutations, with intermediate values between those found in null mutations and controls, or even with normal levels, which would be a limiting factor for the use of plasma progranulin measurement as a screening method for GRN mutations.39
Mutations in GRN lead to accumulation of intracellular inclusions with hyperphosphorylated TDP-43 and, in the neuropathological study of brains of individuals with GRN mutations, the pattern of TDP-43 inclusions is consistent with FTLD-TDP type A.40 In FTLD-TDP type A, neuronal cytoplasmic inclusions immunoreactive to antibodies against TDP-43 are frequent, as are short dystrophic neurites where abnormalities are mainly found in layer 2 of the cortex.40 The mechanism by which a deficiency in progranulin leads to neurodegeneration and abnormal TDP-43 accumulation in the brain is not yet known. It is possible that the abnormal accumulation of TDP-43 is related to increased susceptibility to external stressors than nerve cells present in progranulin deficiency, since the TDP-43 protein is involved in cellular processes activated during stress, such as the formation of stress granules, while the solubility and aggregation TDP-43 change during cellular stress.29
C9ORF72 AND OTHER GENES ASSOCIATED WITH FTD-MND
C9ORF72 gene. GGGGCC hexanucleotide repeat expansions in an intronic region of C9orf72 were discovered in 2011 by two independent groups as a cause of FTD, FTD-MND and ALS.9,10
The C9orf72 gene is located on chromosome 9p21.1. The function of the C9ORF72 protein is still not well understood but there is evidence to suggest it regulates processes related to the endosomal system and autophagy, one of the protein degradation pathways.41
The mechanisms by which GGGGCC expansions lead to neurodegeneration remain unclear, but important discoveries have been made. One of the most important findings was that the hexanucleotide repeats form proteins with repeats of dipeptides (polyGA [glycine-alanine], polyGP [glycine-proline], polyGR [arginine-glycine], polyPR [proline-arginine] and polyPA [proline-alanine]) through repeat-associated non-ATG-initiated translation (RAN translation).42 Based on the currently available evidence, it is believed that the hexanucleotide repeat expansion in C9orf72 leads to neurodegeneration either by a dual mechanism, i.e. through RNA- and polypeptide-mediated toxicity, or by toxicity mediated by polypeptides only.43
The exact number of repetitions required to cause disease has not been established due to the fact that the currently available technologies for sequencing are not able to sequence thousands of base pairs per read. In general, it is accepted that 30 or more hexanucleotide repeats are pathogenic, although it is estimated that the number of repeats in most patients is around 700-4400.9,10,44,45
The inheritance pattern is typically autosomal dominant, but the expansion has also been found in homozygosity, with a similar phenotype to that seen in heterozygous expansions.46 The penetrance is high, but is probably incomplete, as the mutation can be found in cognitively normal elderly and it is estimated that 0.1-0.2% of the normal population carries the pathogenic expansion.44,45 A multicenter study estimated penetrance of 58% at age 58 years and close to 100% at age 80.45
Diseases caused by repeat expansions can be associated with the anticipation phenomenon. In the case of expansions in C9orf72, the study of anticipation is limited by the lack of techniques to accurately measure the number of expansions, as previously mentioned. Another difficulty for studies of anticipation (and penetrance) is that the expansion is unstable, and even monozygotic twins, and different tissue samples from the same individual, might have expansions with different numbers of repeats.44,47 In addition, there is evidence that mosaicism is age dependent, which further complicates the interpretation of the number of repeats.47 Nonetheless, a few studies have suggested there is anticipation, with the age of onset of symptoms in individuals of a generation occurring on average 7-11 years earlier than in the previous generation.48,49
In Europe, the mutation was found in 10% of FTD cases (18% of familial FTD and 6% of sporadic FTD) and in 9% of cases of ALS (37% of familial ALS and 6% of sporadic ALS).45,50 In familial FTD-MND, 56% had the mutation, and in sporadic FTD-MND, 14% had the mutation.50
The most commonly observed phenotypes are bvFTD, FTD-MND and ALS.9,10 The three phenotypes can appear in individuals of the same family, but within a family only one or two of these phenotypes might also appear. Phenotypic heterogeneity is also observed, with age of onset of symptoms varying by more than 20 years, and duration of disease varying by more than 10 years, within a single kindred.49
The mean age at symptom onset in bvFTD varies between 49 and 67 years, and age at onset ranges from 18 to 76 years.12,21,48,50,51 Clinically, bvFTD caused by this mutation is generally similar to that of sporadic cases, but its onset can occur slightly earlier.48 Additionally, some studies suggest that psychotic symptoms are more common in patients with the mutation than in sporadic cases, particularly in the early stages of disease. The neuropsychological profile is typical of bvFTD, characterized by executive dysfunction, but decline in episodic memory and visuospatial skills were reported in some patients.21 Parkinsonism occurs in 12-35% of patients and is less frequent than in MAPT or GRN mutations.12,16,18
PPA is not a common presentation, and represented 0-20% of FTD cases due to C9orf72 repeat expansions in different cohorts.12,45,51 However, both svPPA and nfvPPA have been reported with this mutation, even associated with MND. MND is found in about 30% of cases of bvFTD and usually presents as ALS.50 The mean age of onset of ALS is 56.8 years, ranging from 27 to 89, and is similar to sporadic ALS cases.45
Aside from FTD, FTD-MND, or MND, other phenotypes are infrequent or rare in this mutation. There have been reports of AD, including cases with neuropathological confirmation in individuals with C9orf72 repeat expansions, but in most cohorts, the mutation was found in <1% of AD cases.33,44 Other phenotypes have been described in a few cases, or as case reports, such as Parkinson's disease (including one with neuropathological confirmation), CBS with or without ALS, PSP with or without ALS, dementia with Lewy bodies, cerebellar ataxia, and ALS with multiple sclerosis.52-55 Repeat expansions in C9orf72 were also found in about 2% of patients diagnosed with Huntington-like syndrome, and this may be the most common mutation in this diagnostic group.44
The duration of symptoms is variable, with mean duration reported in different studies of between 3 and 15 years (disease duration ranging from 1 to 24 years).12,16,18,51 Onset of symptoms is sometimes difficult to identify accurately, as in some cases, behavioral changes occur slowly years or decades before a dementia diagnosis can be established.49 The rate of progression of the disease also varies significantly, and both slowly and rapidly progressive forms of bvFTD caused by mutation in C9orf72 have been described.56,57
The atrophy in cases with the C9orf72 mutation predominates in the frontal region, but also affects the temporal and parietal lobes.21,36,51 This atrophy is typically symmetrical, and in some cases there can be minimal atrophy, particularly early in the disease course.18 The temporal lobes are generally less affected than in GRN or MAPT mutations. Some studies have also found atrophy in the thalamus and cerebellum.36,51
In terms of neuropathology, the mutation is associated with the presence of TDP-43 positive inclusions similar to those found in FTLD-TDP type B and type A (either type or in combination).18,58 FTLD-TDP type A is the one also observed in GRN mutations, as previously mentioned, and FTLD-TDP type B is the pathology typically seen in FTD-MND cases, and is characterized by a moderate amount of neuronal cytoplasmic inclusions and sparse dystrophic neurites.40 There are also inclusions immunoreactive to antibodies against p62 but negative for TDP-43, particularly in the pyramidal layer of the hippocampus and cerebellum granular cell layer.58 These inclusions are immunoreactive to antibodies against dipeptide repeat proteins, such as polyGA, polyGP, and polyGR.42 The interactions between the repeat expansion and TDP-43 protein are still unknown, but a study with cell cultures suggested that dipeptide repeat proteins interfere with the ubiquitin-proteasome system, which can affect the homeostasis of the TDP-43 protein.59
Recent studies have identified the presence of C9orf72 hexanucleotide repeat expansions in patients who also had pathogenic mutations in other genes, such as GRN, MAPT, FUS and TARDBP.60 This existence of coexisting mutations is infrequent (less than 2% of cases in a FTD study), but can alter the clinical presentation of the mutations, such as age at onset of symptoms.60
TARDBP gene. The TARDBP gene is located on chromosome 1p36.22 and encodes TDP-43. TDP-43 is a protein that binds to DNA and RNA, and has multiple functions in RNA metabolism, including regulation of transcription and translation, splicing, nuclear-cytoplasmic transport, and formation of stress granules.61
To date, 34 pathogenic mutations have been reported, mainly in cases of familial and sporadic ALS.15 Most TARDBP mutations are missense, though insertions, deletions and nonsense mutations have been found in some cases.62 Cells with cytoplasmic inclusions immunostained with antibodies against TDP-43 typically do not exhibit TDP-43 in the nucleus (where TDP-43 is found in normal cells). This suggests that loss of function in the nucleus and/or toxic effects of the protein in the cytoplasm might underlie neurodegeneration.61
The pattern of inheritance is typically autosomal dominant with incomplete penetrance, but individuals with homozygous mutations have been reported.62
Mutations in TARDBP cause around 3% of familial and 1.5% of sporadic forms of ALS, but are infrequent causes of FTD (with or without MND). Less than 20 probands have been reported in the literature with FTD or dementia.62,63 Within the FTD spectrum, FTD-MND, svPPA-MND, bvFTD, CBS and svPPA (including patients with right temporal variant bvFTD and a "bitemporal" bvFTD variant) have been reported.63,64 There is great variability in age of onset of symptoms, with reports of onset ranging from between 29 and 77 years.63
Mutations were also found in patients with clinical diagnosis of Parkinson's disease, ALS with parkinsonism and ALS with dementia of the Alzheimer type.65,66 Atypical presentations were reported, such as bvFTD with chorea and supranuclear gaze palsy, and progressive anarthria.67,68
In a patient with svPPA with a TARDBP mutation, Gelpi et al. found small intracytoplasmic neuronal inclusions and short neurites immunostained with antibodies against ubiquitin, and TDP-43 (in a pattern similar to FTLD-TDP type A).69
FUS gene. The FUS gene encodes the fused in sarcoma protein (FUS). This protein belongs to a family of DNA/RNA binding proteins called FET. FUS is involved in various roles related to RNA metabolism, including regulation of transcription, splicing, nuclear-cytoplasmic transport and stress granule formation, besides DNA repair and cellular proliferation.61,70
Twenty-three FUS mutations have been reported.15 Most mutations are missense but insertions and deletions (some causing frameshift mutations) have also been found.70 Mutations in this gene seem to cause neurodegeneration both through gain of toxic function, since the FUS mutant protein and overexpression of the normal FUS protein appear to be toxic in animal models, and loss of function mechanisms, because the mutant protein can bind and suppress the functions of normal FUS and of other proteins.61,70
Mutations in FUS most often present as MND, and are found in about 5% of the familial cases and <1% of sporadic cases of ALS, including juvenile ALS.62 Dementia is considered rare in FUS mutations and has been reported in only a few families. FTD-MND and ALS-dementia (with or without parkinsonism) have been reported in five families to date,70,71 and bvFTD without MND was reported in only two probands (though functional evidence of pathogenicity is still lacking for the mutations in these latter cases).64,70
The neuropathological evaluation of ALS cases with FUS mutations show cytoplasmic inclusions immunoreactive to antibodies against FUS in neurons and glial cells.70
UBQLN2 gene. The UBQLN2 (ubiquilin 2) is located on chromosome Xp11.21.72 Ubiquilin 2 participates in the ubiquitin-proteasome system, comprising part of cellular protein degradation pathways, and mutations in UBQLN2 appear to affect the function of the ubiquitin-proteasome system.72
The inheritance pattern is X-linked dominant, which means that both men and women can be affected.72 The penetrance is high, but might be incomplete.163
The age of onset is variable, but appears to be significantly lower among men (median of 33 years in men and 44.5 years in women).72 Most carriers develop ALS, but some cases with the FTD-MND phenotype have been reported.72 Mutations in UBQLN2 appear to be rare in FTD cohorts, and there is only one FTD patient without DNM reported in the literature.73 A kindred with spastic tetraparesis and dementia with behavioral symptoms and a p.P497L mutation was also reported, with symptom onset occurring before 10 years of age in males and between 20-30 years in females.74 Only one patient from this family developed ALS.
Studies have shown inclusions immunoreactive to antibodies against ubiquilin 2 in spinal motor neurons and in the hippocampus,72 which appear to be co-localized with TDP-43 and FUS.75 Ubiquilin 2 positive inclusions are not specific to the cases with mutations in this gene, also being found in cases of ALS and FTD-MND without mutations in UBQLN2.72
CHMP2B gene. The CHMP2B gene (charged multivesicular body protein 2B) is located on chromosome 3p11.2. The CHMP2B protein is part of the ESCRT III complex (endosomal sorting complex required for transport-III), which participates in protein degradation pathways associated with endosomes and lysossomes.76
Nonsense mutations in CHMP2B are rare genetic causes of FTD, and have been reported in Danish and Belgian families with autosomal dominant patterns of inheritance.15,76,77 The most commonly reported phenotype is bvFTD, with a mean age at onset of 68 years and mean disease duration of 10 years.77
Missense mutations in the gene have been described in patients with MND (particularly with primary muscle atrophy), FTD-MND, and isolated cases of svPPA and CBS.76,78 However, it should be noted that the pathogenicity of these mutations have not been proven unequivocally and most missense mutations have been reported in sporadic cases.
The neuropathological finding in CHMP2B mutations is FTLD-U, indicating that this mutation is associated with an as yet unknown proteinopathy.5
Other genes associated with the FTD-MND spectrum. Mutations in other genes that cause MND were also associated with the FTD-MND phenotype in a few cases or reports of isolated cases.
Mutations in the optineurin gene (OPTN) are known to cause open angle glaucoma, and more recently, mutations were identified in cases of ALS and FTD-MND.79 Variants in CHCHD10 (Coiled-Coil-Helix-Coiled-Coil-Helix Domain Containing 10), a gene that encodes a mitochondrial protein, were recently identified in a few families with MND and FTD-MND.80
The FTD-MND phenotype was also reported in SOD1 (superoxide dismutase),81 DCTN1 (dynactin 1),82 ANG (angiogenin),83 SIGMAR1 (Sigma Non-Opioid Intracellular Receptor 1),84 and DJ-1 (PARK 7) mutations.85 Mutations in SOD1 are the most common genetic cause of ALS, but are rarely associated with cognitive or behavioral symptoms.81 Mutations in DCTN1 are typically associated with Perry syndrome, an autosomal dominant disease characterized by parkinsonism, hypoventilation, weight loss and depression.82 Variants in DCTN1 and in ANG are considered risk factors for ALS.82,83 Mutations in DJ-1 cause early-onset Parkinson's disease, with an autosomal recessive pattern of inheritance (PARK 7).85 Mutations in SIGMA1R were initially described in cases of juvenile ALS.84
GENES ASSOCIATED WITH MULTISYSTEM PROTEINOPATHIES
Mutations in VCP were discovered in 2004 were found to cause a complex phenotype, which initially included inclusion body myopathy, Paget disease of the bone and bvFTD and consequently it was commonly referred to by the acronym IBMPFD.86 In 2010 however, a study employing exome sequencing found that mutations in VCP could also cause familial ALS, thus expanding the phenotypes associated with VCP mutations.87 Other reports subsequently suggested that these families could occasionally present systemic manifestations such as heart, liver, peripheral nervous system involvement, among others, and therefore the acronym originally used was no longer deemed adequate. Hence, Benatar et al. proposed that this group of phenotypes be renamed "Multisystem proteinopathy" (MSP).13
Mutations in VCP are the most frequent cause of MSP, but mutations in HNRNP1 and HNRNPA2B1 were also recently identified.88 Mutations in the SQSTM1 gene have not been associated with inclusion body myopathy, but have since been found in other phenotypes of MSP.89 Other MSP-causing genes will likely be identified in the future, since mutations in these four genes cannot explain all cases of familial MSP.
VCP gene. The VCP gene (valosin containing protein) is located on chromosome 9p13.3. The valosin-containing protein is an AAA + protein (ATPase associated with a variety of cellular activities), which participates in multiple cellular processes, such as degradation of ubiquitinated proteins by the ubiquitin-proteasome system and autophagy.86
There are currently 18 known missense mutations of the gene.15 Mutations in VCP seem to affect the protein degradation pathways, which can lead to neurodegeneration.86
Transmission occurs in autosomal dominant pattern of inheritance, with variable penetrance depending on the phenotype (around 30% of patients develop bvFTD, 85-90% develop inclusion body myopathy, and 45-50% have Paget's disease).90
VCP mutations are found in less than 2% of FTD cases and are responsible for 50-75% of cases of familial MSP.21,89,91 The onset of symptoms of myopathy (mean age 43 years) and Paget disease (mean age 42 years) usually occur earlier than the onset of FTD symptoms (mean age 55 years).90 Mutation carriers can develop all three phenotypes (12%), a combination of two phenotypes (50%) or only one of the phenotypes (38%).90 Only 3% develop FTD alone.90
The neuropsychiatric manifestations are compatible with bvFTD in most cases, but there are reports of svPPA and dementia of the Alzheimer type.92 Disease duration is, on average, 19 years after the onset of myopathy or Paget's disease, but only 6 years, on average, after dementia onset.92
Mutations in VCP were found in about 2% of familial cases of ALS, and around 9% of individuals with mutations in this gene develop ALS.87,92
The neuropathological findings in VCP mutations are consistent with FTLD-TDP type D, characterized by frequent short dystrophic neurites, frequent neuronal intranuclear inclusions and sparse neuronal cytoplasmic inclusions.40
HNRNPA2B1 and HNRNP1 genes. The HNRNPA2B1 and HNRNP1 genes (heterogeneous nuclear ribonucleoprotein A2/B1 and A1, respectively) encode ribonucleoproteins that participate in the RNA metabolism.13,88
The mutations described to date are located in the proteins' prion-like domain and are thought to facilitate the proteins' fibrilization and incorporation into stress granules, as well as induce the formation of pathologic cytoplasmic inclusions.88
Mutations in these genes are rare causes of familial MSP (0-12%), as well as familial and sporadic ALS.13,88,91
There are no reports with neuropathological findings in these mutations, but one study found cytoplasmic inclusions immunoreactive to antibodies against TDP-43, hnRNPA2/B1, and hnRNPA1 in muscle biopsies of patients with a mutation.88
SQSTM1 gene. The SQSTM1 gene (sequestosome 1) encodes the p62 protein, which participates in degradation of ubiquitinated proteins by autophagy, and is also involved in cell differentiation, apoptosis and immune responses.93
Mutations in SQSTM1 are missense (predominantly), nonsense or deletions. It is believed that the mutations affect the binding of p62 protein, and therefore, the degradation of proteins.93
The most frequently observed phenotype in SQSTM1 mutations is Paget disease where mutations are found in 25-50% of cases of familial and 5-10% of sporadic cases of Paget disease.94 Cases of sporadic and familial ALS, familial bvFTD and familial FTD-MND with SQSTM1 mutations, have also been identified.95 Mutations have been described in families with the bvFTD, Paget disease, and/or ALS phenotypes, but bvFTD and ALS have also been observed in families with no history of Paget disease.93,95
Mutations in SQSTM1 were found in 0.9 to 3% of cases of ALS and FTD (familial and sporadic), and in less than 5% of the familial MSP.89,93,95 The onset of symptoms in FTD ranged from between 48 and 73 years.89,93 SQSTM1 mutations have been identified in rare patients diagnosed with nfvPPA, svPPA, PSP and CBS, but the pathogenicity of these mutations has yet to be confirmed.96
Inclusions immunoreactive to antibodies against p62, TDP-43 and, in some cases phosphorylated TDP-43 in neurons and glial cells, have been described in patients with mutations in SQSTM1.96
FINAL CONSIDERATIONS
It should be noted that despite the important discoveries made to date on the genetics of FTD, there is still much to be discovered, as a significant percentage of familial FTD (around 50-60%) cannot be explained by mutations in the currently known genes.12,48 Some of these familial FTD cases without an identified gene might be explained by mutations in genes not typically related to FTD, but that have been reported with the bvFTD phenotype. In fact, mutations in PSEN1 (presenilin 1) and PSEN2 (Presenilin 2) that cause autosomal dominant AD, have been found in families with the bvFTD phenotype.97 Dementia with frontotemporal features has also been reported in mutations of other genes, such as in PRNP (prion protein), that cause genetic prion disease.98 The use of next generation sequencing technologies to sequence gene panels, or even whole genomes or exomes, might be useful for identifying pathogenic mutations in familial FTD.
With the identification of key genes associated with FTD, researchers have turned their attention to characterizing the pre-symptomatic stage of the disease in an effort to identify biomarkers that can be used to evaluate response to (future) therapies. Several studies have been conducted with different mutations and have identified mild cognitive symptoms, changes in brain glucose metabolism or structural neuroimaging, and/or changes in functional connectivity in pre-symptomatic mutation carriers.99 A recent study suggested structural changes might appear at least ten years (and up to 25 years) before the onset of symptoms.100
The ultimate goal of research in pre-symptomatic and symptomatic individuals with monogenic forms of FTD is to develop treatments that modify the course of the disease or even prevent its onset. Among the mutations associated with FTD, the discovery of treatments for GRN mutations are the most promising. As GRN mutations cause disease through haploinsufficiency, compounds are being investigated to increase gene expression (such as with the use of histone deacetylase inhibitors) or protein secretion. A multicenter phase-two clinical trial with a histone deacetylase inhibitor (FRM-0334) in patients with GRN mutations is ongoing (Clinicaltrials.gov NCT02149160).
Footnotes
This study was conducted at the Cognitive and Behavioral Neurology Unit, Department of Neurology, Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo, SP, Brazil.
Disclosure: The author reports no conflicts of interest.
REFERENCES
- 1.Gorno-Tempini ML, Hillis AE, Weintraub S, et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rascovsky K, Hodges JR, Knopman D, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Josephs KA. Frontotemporal dementia and related disorders: deciphering the enigma. Ann Neurol. 2008;64:4–14. doi: 10.1002/ana.21426. [DOI] [PubMed] [Google Scholar]
- 4.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 5.Mackenzie IR, Neumann M, Bigio EH, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman JS, Farmer JM, Wood EM, et al. Comparison of family histories in FTLD subtypes and related tauopathies. Neurology. 2005;65:1817–1819. doi: 10.1212/01.wnl.0000187068.92184.63. [DOI] [PubMed] [Google Scholar]
- 7.Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 8.Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 9.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruts M, Kumar-Singh S, Van Broeckhoven C. Progranulin mutations in ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Curr Alzheimer Res. 2006;3:485–491. doi: 10.2174/156720506779025251. [DOI] [PubMed] [Google Scholar]
- 12.Le Ber I, Camuzat A, Guillot-Noel L, et al. C9ORF72 repeat expansions in the frontotemporal dementias spectrum of diseases: a flow-chart for genetic testing. J Alzheimers Dis. 2013;34:485–499. doi: 10.3233/JAD-121456. [DOI] [PubMed] [Google Scholar]
- 13.Benatar M, Wuu J, Fernandez C, et al. Motor neuron involvement in multisystem proteinopathy: implications for ALS. Neurology. 2013;80:1874–1880. doi: 10.1212/WNL.0b013e3182929fc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 15.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Langenhove T, van der Zee J, Gijselinck I, et al. Distinct clinical characteristics of C9orf72 expansion carriers compared with GRN, MAPT, and nonmutation carriers in a Flanders-Belgian FTLD cohort. JAMA Neurol. 2013;70:365–373. doi: 10.1001/2013.jamaneurol.181. [DOI] [PubMed] [Google Scholar]
- 17.Pickering-Brown SM, Rollinson S, Du Plessis D, et al. Frequency and clinical characteristics of progranulin mutation carriers in the Manchester frontotemporal lobar degeneration cohort: comparison with patients with MAPT and no known mutations. Brain. 2008;131:721–731. doi: 10.1093/brain/awm331. [DOI] [PubMed] [Google Scholar]
- 18.Boeve BF, Boylan KB, Graff-Radford NR, et al. Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain. 2012;135:765–783. doi: 10.1093/brain/aws004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck J, Rohrer JD, Campbell T, et al. A distinct clinical, neuropsychological and radiological phenotype is associated with progranulin gene mutations in a large UK series. Brain. 2008;131:706–720. doi: 10.1093/brain/awm320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gass J, Cannon A, Mackenzie IR, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 21.Mahoney CJ, Beck J, Rohrer JD, et al. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain. 2012;135:736–750. doi: 10.1093/brain/awr361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boeve BF. Progress on progranulin. Arch Neurol. 2010;67:145–147. doi: 10.1001/archneurol.2009.320. [DOI] [PubMed] [Google Scholar]
- 23.Carney RM, Kohli MA, Kunkle BW, et al. Parkinsonism and distinct dementia patterns in a family with the MAPT R406W mutation. Alzheimers Dement. 2014;10:360–365. doi: 10.1016/j.jalz.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitwell JL, Jack CR Jr., Boeve BF, et al. Voxel-based morphometry patterns of atrophy in FTLD with mutations in MAPT or PGRN. Neurology. 2009;72:813–820. doi: 10.1212/01.wnl.0000343851.46573.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Fonzo A, Ronchi D, Gallia F, et al. Lower motor neuron disease with respiratory failure caused by a novel MAPT mutation. Neurology. 2014;82:1990–1998. doi: 10.1212/WNL.0000000000000476. [DOI] [PubMed] [Google Scholar]
- 26.Nicholl DJ, Greenstone MA, Clarke CE, et al. An English kindred with a novel recessive tauopathy and respiratory failure. Ann Neurol. 2003;54:682–686. doi: 10.1002/ana.10747. [DOI] [PubMed] [Google Scholar]
- 27.Rohrer JD, Ridgway GR, Modat M, et al. Distinct profiles of brain atrophy in frontotemporal lobar degeneration caused by progranulin and tau mutations. Neuroimage. 2010;53:1070–1076. doi: 10.1016/j.neuroimage.2009.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson BC, Goedert M. Frontotemporal Dementia Caused by MAPT mutations: A chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol. 2015;41:24–46. doi: 10.1111/nan.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinberger G, Capell A, Haass C, Van Broeckhoven C. Mechanisms of granulin deficiency: lessons from cellular and animal models. Mol Neurobiol. 2013;47:337–360. doi: 10.1007/s12035-012-8380-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson AM, Gass J, Petrucelli L, Rademakers R. Progranulin axis and recent developments in frontotemporal lobar degeneration. Alzheimers Res Ther. 2012;4:4–4. doi: 10.1186/alzrt102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benussi L, Ghidoni R, Pegoiani E, Moretti DV, Zanetti O, Binetti G. Progranulin Leu271LeufsX10 is one of the most common FTLD and CBS associated mutations worldwide. Neurobiol Dis. 2009;33:379–385. doi: 10.1016/j.nbd.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Rademakers R, Baker M, Gass J, et al. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C-->T (Arg493X) mutation: an international initiative. Lancet Neurol. 2007;6:857–868. doi: 10.1016/S1474-4422(07)70221-1. [DOI] [PubMed] [Google Scholar]
- 33.Wojtas A, Heggeli KA, Finch N, et al. C9ORF72 repeat expansions and other FTD gene mutations in a clinical AD patient series from Mayo Clinic. Am J Neurodegener Dis. 2012;1:107–118. [PMC free article] [PubMed] [Google Scholar]
- 34.Tremolizzo L, Bertola F, Casati G, Piperno A, Ferrarese C, Appollonio I. Progressive supranuclear palsy-like phenotype caused by progranulin p.Thr272fs mutation. Mov Disord. 2011;26:1964–1966. doi: 10.1002/mds.23749. [DOI] [PubMed] [Google Scholar]
- 35.Smith KR, Damiano J, Franceschetti S, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitwell JL, Weigand SD, Boeve BF, et al. Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain. 2012;135:794–806. doi: 10.1093/brain/aws001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caroppo P, Le Ber I, Camuzat A, et al. Extensive white matter involvement in patients with frontotemporal lobar degeneration: think progranulin. JAMA Neurol. 2014;71:1562–1566. doi: 10.1001/jamaneurol.2014.1316. [DOI] [PubMed] [Google Scholar]
- 38.Kelley BJ, Haidar W, Boeve BF, et al. Prominent phenotypic variability associated with mutations in Progranulin. Neurobiol Aging. 2009;30:739–751. doi: 10.1016/j.neurobiolaging.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghidoni R, Paterlini A, Benussi L. Circulating progranulin as a biomarker for neurodegenerative diseases. Am J Neurodegener Dis. 2012;1:180–190. [PMC free article] [PubMed] [Google Scholar]
- 40.Mackenzie IR, Neumann M, Baborie A, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farg MA, Sundaramoorthy V, Sultana JM, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ash PE, Bieniek KF, Gendron TF, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizielinska S, Isaacs AM. C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function? Curr Opin Neurol. 2014;27:515–523. doi: 10.1097/WCO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beck J, Poulter M, Hensman D, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet. 2013;92:345–353. doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooper-Knock J, Higginbottom A, Connor-Robson N, et al. C9ORF72 transcription in a frontotemporal dementia case with two expanded alleles. Neurology. 2013;81:1719–1721. doi: 10.1212/01.wnl.0000435295.41974.2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, et al. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 2013;12:978–988. doi: 10.1016/S1474-4422(13)70210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gijselinck I, Van Langenhove T, van der Zee J, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 49.Hsiung GY, DeJesus-Hernandez M, Feldman HH, et al. Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p. Brain. 2012;135:709–722. doi: 10.1093/brain/awr354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Zee J, Gijselinck I, Dillen L, et al. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat. 2013;34:363–373. doi: 10.1002/humu.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sha SJ, Takada LT, Rankin KP, et al. Frontotemporal dementia due to C9ORF72 mutations: clinical and imaging features. Neurology. 2012;79:1002–1011. doi: 10.1212/WNL.0b013e318268452e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lesage S, Le Ber I, Condroyer C, et al. C9orf72 repeat expansions are a rare genetic cause of parkinsonism. Brain. 2013;136:385–391. doi: 10.1093/brain/aws357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindquist SG, Duno M, Batbayli M, et al. Corticobasal and ataxia syndromes widen the spectrum of C9ORF72 hexanucleotide expansion disease. Clin Genet. 2013;83:279–283. doi: 10.1111/j.1399-0004.2012.01903.x. [DOI] [PubMed] [Google Scholar]
- 54.Murray ME, DeJesus-Hernandez M, Rutherford NJ, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–690. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ismail A, Cooper-Knock J, Highley JR, et al. Concurrence of multiple sclerosis and amyotrophic lateral sclerosis in patients with hexanucleotide repeat expansions of C9ORF72. J Neurol Neurosurg Psychiatry. 2013;84:79–87. doi: 10.1136/jnnp-2012-303326. [DOI] [PubMed] [Google Scholar]
- 56.Khan BK, Yokoyama JS, Takada LT, et al. Atypical, slowly progressive behavioural variant frontotemporal dementia associated with C9ORF72 hexanucleotide expansion. J Neurol Neurosurg Psychiatry. 2012;83:358–364. doi: 10.1136/jnnp-2011-301883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kandiah N, Sengdy P, Mackenzie IR, Hsiung GY, de Jesus-Hernandez M, Rademakers R. Rapidly progressive dementia in a Chinese patient due to C90RF72 mutation. Can J Neurol Sci. 2012;39:676–677. doi: 10.1017/s0317167100015481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mackenzie IR, Frick P, Neumann M. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta Neuropathol. 2014;127:347–357. doi: 10.1007/s00401-013-1232-4. [DOI] [PubMed] [Google Scholar]
- 59.Yamakawa M, Ito D, Honda T, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet. 2015;24:1630–1645. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- 60.van Blitterswijk M, Baker MC, DeJesus-Hernandez M, et al. C9ORF72 repeat expansions in cases with previously identified pathogenic mutations. Neurology. 2013;81:1332–1341. doi: 10.1212/WNL.0b013e3182a8250c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34:812–826. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- 63.Floris G, Borghero G, Cannas A, et al. Clinical phenotypes and radiological findings in frontotemporal dementia related to TARDBP mutations. J Neurol. 2015;262:375–384. doi: 10.1007/s00415-014-7575-5. [DOI] [PubMed] [Google Scholar]
- 64.Huey ED, Ferrari R, Moreno JH, et al. FUS and TDP43 genetic variability in FTD and CBS. Neurobiol Aging. 2012;33:1016–1016. doi: 10.1016/j.neurobiolaging.2011.08.004. e9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cannas A, Borghero G, Floris GL, et al. The p.A382T TARDBP gene mutation in Sardinian patients affected by Parkinson's disease and other degenerative parkinsonisms. Neurogenetics. 2013;14:161–166. doi: 10.1007/s10048-013-0360-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujita Y, Ikeda M, Yanagisawa T, Senoo Y, Okamoto K. Different clinical and neuropathologic phenotypes of familial ALS with A315E TARDBP mutation. Neurology. 2011;77:1427–1431. doi: 10.1212/WNL.0b013e318232ab87. [DOI] [PubMed] [Google Scholar]
- 67.Kovacs GG, Murrell JR, Horvath S, et al. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov Disord. 2009;24:1843–1847. doi: 10.1002/mds.22697. [DOI] [PubMed] [Google Scholar]
- 68.Praline J, Vourc'h P, Guennoc AM, Veyrat-Durebex C, Corcia P. Co-occurrence of progressive anarthria with an S393L TARDBP mutation and ALS within a family. Amyotroph Lateral Scler. 2012;13:155–157. doi: 10.3109/17482968.2011.598168. [DOI] [PubMed] [Google Scholar]
- 69.Gelpi E, van der Zee J, Estrada AT, van Broeckhoven C, Sanchez-Valle R. TARDBP mutation p.Ile383Val associated with semantic dementia and complex proteinopathy. Neuropathol Appl Neurobiol. 2014;40:225–230. doi: 10.1111/nan.12063. [DOI] [PubMed] [Google Scholar]
- 70.Deng H, Gao K, Jankovic J. The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol. 2014;10:337–348. doi: 10.1038/nrneurol.2014.78. [DOI] [PubMed] [Google Scholar]
- 71.Yan J, Deng HX, Siddique N, et al. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology. 2010;75:807–814. doi: 10.1212/WNL.0b013e3181f07e0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Synofzik M, Maetzler W, Grehl T, et al. Screening in ALS and FTD patients reveals 3 novel UBQLN2 mutations outside the PXX domain and a pure FTD phenotype. Neurobiol Aging. 2012;33:2949–2949. doi: 10.1016/j.neurobiolaging.2012.07.002. e13-7. [DOI] [PubMed] [Google Scholar]
- 74.Fahed AC, McDonough B, Gouvion CM, et al. UBQLN2 mutation causing heterogeneous X-linked dominant neurodegeneration. Ann Neurol. 2014;75:793–798. doi: 10.1002/ana.24164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williams KL, Warraich ST, Yang S, et al. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527–2527. doi: 10.1016/j.neurobiolaging.2012.05.008. e3-10. [DOI] [PubMed] [Google Scholar]
- 76.Isaacs AM, Johannsen P, Holm I, Nielsen JE. Frontotemporal dementia caused by CHMP2B mutations. Curr Alzheimer Res. 2011;8:246–251. doi: 10.2174/156720511795563764. consortium FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 78.van Blitterswijk M, Vlam L, van Es MA, et al. Genetic overlap between apparently sporadic motor neuron diseases. PLoS One. 2012;7:e48983. doi: 10.1371/journal.pone.0048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kamada M, Izumi Y, Ayaki T, et al. Clinicopathologic features of autosomal recessive amyotrophic lateral sclerosis associated with optineurin mutation. Neuropathology. 2014;34:64–70. doi: 10.1111/neup.12051. [DOI] [PubMed] [Google Scholar]
- 80.Bannwarth S, Ait-El-Mkadem S, Chaussenot A, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–2345. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Battistini S, Giannini F, Greco G, et al. SOD1 mutations in amyotrophic lateral sclerosis. Results from a multicenter Italian study. J Neurol. 2005;252:782–788. doi: 10.1007/s00415-005-0742-y. [DOI] [PubMed] [Google Scholar]
- 82.Munch C, Rosenbohm A, Sperfeld AD, et al. Heterozygous R1101K mutation of the DCTN1 gene in a family with ALS and FTD. Ann Neurol. 2005;58:777–780. doi: 10.1002/ana.20631. [DOI] [PubMed] [Google Scholar]
- 83.van Es MA, Diekstra FP, Veldink JH, et al. A case of ALS-FTD in a large FALS pedigree with a K17I ANG mutation. Neurology. 2009;72:287–288. doi: 10.1212/01.wnl.0000339487.84908.00. [DOI] [PubMed] [Google Scholar]
- 84.Luty AA, Kwok JB, Dobson-Stone C, et al. Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration-motor neuron disease. Ann Neurol. 2010;68:639–649. doi: 10.1002/ana.22274. [DOI] [PubMed] [Google Scholar]
- 85.Annesi G, Savettieri G, Pugliese P, et al. DJ-1 mutations and parkinsonism-dementia-amyotrophic lateral sclerosis complex. Ann Neurol. 2005;58:803–807. doi: 10.1002/ana.20666. [DOI] [PubMed] [Google Scholar]
- 86.Weihl CC. Valosin containing protein associated fronto-temporal lobar degeneration: clinical presentation, pathologic features and pathogenesis. Curr Alzheimer Res. 2011;8:252–260. doi: 10.2174/156720511795563773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim HJ, Kim NC, Wang YD, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rubino E, Rainero I, Chio A, Rogaeva E, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kimonis VE, Fulchiero E, Vesa J, Watts G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta. 2008;1782:744–748. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 91.Le Ber I, Van Bortel I, Nicolas G, et al. hnRNPA2B1 and hnRNPA1 mutations are rare in patients with "multisystem proteinopathy" and frontotemporal lobar degeneration phenotypes. Neurobiol Aging. 2014;35:934–934. doi: 10.1016/j.neurobiolaging.2013.09.016. e5-6. [DOI] [PubMed] [Google Scholar]
- 92.Mehta SG, Khare M, Ramani R, et al. Genotype-phenotype studies of VCP-associated inclusion body myopathy with Paget disease of bone and/or frontotemporal dementia. Clin Genet. 2013;83:422–431. doi: 10.1111/cge.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Le Ber I, Camuzat A, Guerreiro R, et al. SQSTM1 Mutations in French Patients With Frontotemporal Dementia or Frontotemporal Dementia With Amyotrophic Lateral Sclerosis. JAMA Neurol. 2013;70:1403–1410. doi: 10.1001/jamaneurol.2013.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bolland MJ, Cundy T. Paget's disease of boné: clinical review and update. J Clin Pathol. 2013;66:924–927. doi: 10.1136/jclinpath-2013-201688. [DOI] [PubMed] [Google Scholar]
- 95.Fecto F, Yan J, Vemula SP, Liu E, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 96.van der Zee J, Van Langenhove T, Kovacs GG, et al. Rare mutations in SQSTM1 modify susceptibility to frontotemporal lobar degeneration. Acta Neuropathol. 2014;128:397–410. doi: 10.1007/s00401-014-1298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Larner AJ, Doran M. Genotype-phenotype relationships of presenilin-1 mutations in Alzheimer's disease: an update. J Alzheimers Dis. 2009;17:259–265. doi: 10.3233/JAD-2009-1042. [DOI] [PubMed] [Google Scholar]
- 98.Nitrini R, Rosemberg S, Passos-Bueno MR, et al. Familial spongiform encephalopathy associated with a novel prion protein gene mutation. Ann Neurol. 1997;42:138–146. doi: 10.1002/ana.410420203. [DOI] [PubMed] [Google Scholar]
- 99.Dopper EG, Rombouts SA, Jiskoot LC, et al. Structural and functional brain connectivity in presymptomatic familial frontotemporal dementia. Neurology. 2013;80:814–823. doi: 10.1212/WNL.0b013e31828407bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rohrer JD, Nicholas JM, Cash DM, et al. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol. 2015;14:253–262. doi: 10.1016/S1474-4422(14)70324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]